Short abstract
A defining feature of neurodegenerative diseases is the abnormal and excessive loss of neurons. One molecule that is particularly important in promoting neuronal death in a variety of cell culture and in vivo models of neurodegeneration is histone deacetylase-3 (HDAC3), a member of the histone deacetylase family of proteins. As a step towards understanding how HDAC3 promotes neuronal death, we conducted a proteomic screen aimed at identifying proteins that were regulated by HDAC3. HDAC3 was overexpressed in cultured rat cerebellar granule neurons (CGNs) and protein lysates were analyzed by mass spectrometry. Of over 3000 proteins identified in the screen, only 21 proteins displayed a significant alteration in expression. Of these, 12 proteins were downregulated whereas 9 proteins were upregulated. The altered expression of five of these proteins, TEX10, NPTX1, TFG, TSC1, and NFL, along with another protein that was downregulated in the proteomic screen, HIP1R, was confirmed using Western blots and commercially available antibodies. Because antibodies were not available for some of the proteins and since HDAC3 is a transcriptional regulator of gene expression, we conducted RT-PCR analysis to confirm expression changes. In separate analyses, we also included other proteins that are known to regulate neurodegeneration, including HDAC9, HSF1, huntingtin, GAPDH, FUS, and p65/RELA. Based on our proteomic screen and candidate protein approach, we identify three genes, Nptx1, Hip1r, and Hdac9, all known to regulate neurodegeneration that are robustly regulated by HDAC3. Given their suggested roles in regulating neuronal death, these genes are likely to be involved in regulating HDAC3-mediated neurotoxicity.
Impact statement
Neurodegenerative diseases are a major medical, social, and economic problem. Recent studies by several laboratories have indicated that histone deacetylase-3 (HDAC3) plays a key role in promoting neuronal death. But the downstream mediators of HDAC3 neurotoxicity have yet to be identified. We conducted a proteomic screen to identify HDAC3 targets the results of which have been described in this report. Briefly, we identify Nptx1, Hip1r, and Hdac9 as genes whose expression is altered by HDAC3. Investigating how these genes are involved in HDAC3 neurotoxicity could shed valuable insight into neurodegenerative disease and identify molecules that can be targeted to treat these devastating disorders.
Keywords: Histone deacetylase, neurons, neuronal survival, histone deacetylase-3, neurodegeneration, Huntington’s disease
Introduction
A defining feature of neurodegenerative diseases is the abnormal and excessive loss of neurons. Research conducted over the past decade has shown that treatment with pharmacological inhibitors of histone deacetylases (HDAC) protect against neuronal loss in a wide variety of neurodegenerative diseases.1–3 A limitation of the inhibitors used in most of the studies demonstrative neuroprotective action is that they are not selective with regard to which member of the HDAC family of proteins they inhibit. As a result, it has been unclear which HDAC(s) are abnormally activated in neurodegenerative condition and contribute to neuronal loss. A growing body of evidence indicates that HDAC3, a member of the Class I family of HDACs, is particularly important in the promotion of neurodegeneration.1–3 First demonstrated in cultured cerebellar granule neurons (CGNs) and in a cell culture model of Huntington’s disease (HD),4,5 a growing number of studies have implicated or demonstrated HDAC3 involvement in neurodegeneration. Pharmacological inhibition of HDAC3 using more recently developed and highly selective inhibitors protects against neuronal loss and behavioral deficits in mouse models shown in literature.6–10 Similar protection against disease pathogenesis has been described in other inherited polyglutamine-expansion disorders, such as spinocerebellar ataxias (SCA).11 HDAC3 inhibition was also found to protect CA1 hippocampal neurons against oligomeric beta-amyloid-induced impairment of synaptic plasticity and against neuronal loss in cell culture and mouse model of ischemic stroke.12,13 A substantial increase in HDAC3 expression along with nuclear localization has been observed soon after the induction of ischemic stroke in mice which if prevented by HDAC3 knockdown protected cortical neurons from OGD-induced death.12,14
HDAC3 is highly expressed in the developing and mature brain.15,16 Indeed, conditional ablation of HDAC3 in the mouse brain leads to serious neurodevelopmental problems and death within a day to six weeks depending on the extent of ablation.17 While being necessary for normal brain development, HDAC3 can be converted into a neurotoxic protein through its phosphorylation by GSK3β, its disassociation from normal huntingtin protein, and its interaction with HDAC1.4,5,18 Exactly how the HDAC3–HDAC1 association promotes neuronal death is not known. Interestingly, the toxic effect of HDAC3 is selective for neurons.4 We have previously described that while primary neurons and neuroblastoma cell lines are sensitive to HDAC3 toxicity, other primary cell types and non-neuronal cell lines are not.4
As a step towards understanding the mechanism by which HDAC3 promotes neuronal death, we conducted a proteomic analysis to identify genes that were regulated in CGNs by overexpressed HDAC3. As we describe below, this analysis resulted in the identification of several proteins whose expression is altered in neurons by elevated HDAC3. After conducting validation experiments, we analyzed the expression of these genes in HEK293T cells overexpressing HDAC3. We previously described that HEK293T cells are not sensitive to HDAC3 toxicity.4
Materials and methods
Materials
Unless stated otherwise, all cell culture media were purchased from Thermo Fisher Scientific (Waltham, MA), and chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MO). Poly-L-Lysine for primary neuronal cultures was purchased from R&D Systems (Minneapolis, MN).
Generation of HDAC3 adenovirus
GFP-encoding adenovirus (Ad-GFP) was described previously.19 HDAC3-FLAG adenovirus (Ad-HDAC3) was generated using ViraPower adenoviral expression system (Thermo Fisher Scientific) following the manufacturer’s protocol. Briefly, an HDAC3-FLAG encoding fragment of DNA was amplified using a CMV-HDAC3-FLAG plasmid (Addgene, Cambridge, MA; plasmid number 13819). The primers used are as follows.
Forward: 5-GGGGACAAGTTTGTACAAAAAAGCAG GCTTCACCATGGCCAAGACCGTGGCCTAT-3;
Reverse: 5-GGGGACCACTTTGTACAAGAAAGCTGG GTGTTATTACTTATCGTCGTCATCCTTGTAATC-3.
The fragment containing the HDAC3 coding sequence and the FLAG sequence was cloned into the pDONR221 shuttle vector and then transferred into the adenoviral vector pAd/CMV/V5-DEST. After sequencing, the construct was linearized by PacI and transfected to HEK293A cells (ATCC, Manassas, VA) for virus amplification. The virus was purified using CsCl density gradient centrifugation. CsCl was removed by dialysis against phosphate-buffered saline (PBS) at 4°C for 24 h. After purification, the viral stocks were adjusted to 3 × 1010 pfu/mL. The multiplicity of infection for CGNs was 10.
Cell culture and viability assay
CGNs were prepared as previously described using 7- to 8-day-old Wistar rats.20 The neuronal cultures were plated in Basal Eagle Medium (BME) in 24-well dishes (1 × 106 cells/well for viability assay) or 60 mm dishes (12 × 106 cells/dish for RT-PCR or Western blotting). Five days later, the CGNs were infected with Ad-HDAC3 or Ad-GFP for 2 h in serum-free BME medium containing 25 mM KCl (HK). The virus was then removed and the CGNs were further cultured for 28 or 32 h. The infection rate for HDAC3 and GFP viruses in CGNs was approximately 30% and 40%, respectively. To compare viability, the cells were fixed using 4% paraformaldehyde in PBS and immunocytochemistry was performed. GFP antibody (Santa Cruz Biotechnology, Dallas, TX; catalog # sc-9996) and FLAG antibody (Sigma, catalog # F1804) were utilized to detect GFP and HDAC3, respectively. The secondary antibody Dylight 594 (catalog # 115–585-146) was purchased from Jackson ImmunoResearch Laboratories (West Grove, PA). Staining with 4′6-diamidino-2-phenylindole hydrochloride (DAPI) was used to quantify cell viability. Cells with condensed or fragmented nuclei were scored as dead. For each condition, more than 200 infected cells were counted.
HEK293T cells (ATCC) were maintained in DMEM containing 10% FBS, 50 μg/mL streptomycin, and 50 U/mL penicillin. The cells were transfected with GFP plasmid or HDAC3-FLAG plasmid 24 h after plating using EndoFectin (GeneCopoeia, Rockville, MD) following manufacturer’s instructions. Thirty hours after transfection, the cells were collected for RT-PCR or Western blotting.
RNA preparation and RT-PCR
RNA was extracted from cultured CGNs or HEK293T cells using TRIzol RNA isolation reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. cDNA was prepared from 3 μg of RNA using the Verso cDNA Synthesis Kit (Thermo Scientific). PCR was performed with GoTaq Green Master Mix (Promega, Madison, WI) and repeated at least three times using cells from separate cultures. The primers used for PCR amplification with CGNs samples were as follows: M4k3 Forward: TGTGTGACCTCTGGGCTGTG; M4k3 Reverse: ATGCGG CTCTGTCTCCTTCC; Ppp2r2d Forward: AATAAAGGCC GCGCTCACTC; Ppp2r2d Reverse: ACAGCCTGATGGT CCCCTTG; Tsc1 Forward: AACATGGGCCTGACA CACCA; Tsc1 Reverse: TCGGAGGGTCCGGATCTCAT; Tex10 Forward: GTCTCGCTGGCAAATGCCTC; Tex10 Reverse: ACAAAAGCTGCACCAGACGC; Smarca2 Forward: AAAGGAGGTGCCAAGACGCT; Smarca2 Reverse: TGGGCCTGCAGATCCTGATG;
Tfg Forward: TAATGGCCAGCCAAGACCCC; Tfg Reverse: AGCCTGCTGCTGGTACTGTT; Nptx1 Forward: CTACACCAGCGGATCAGCGA; Nptx1 Reverse: CTCCG CCTAGAGTGTCCTGC; Rab23 Forward: GGTGGTGGT AGGGAATGGGG; Rab23 Reverse: GCTCGAATGTGTC TGCTCCG; Ptprn Forward: CTGGTCTGCCTGCTGTTG TTGAG; Ptprn Reverse: CCATCTCGGGAACCAAA CTGC; Plxnb1 Forward: ACTGCTCCAGCCACCCTTTT; Plxnb1 Reverse: ACCAGCACACACTCACTGCT; Gpd1 Forward: GGAAGACATCGGGGGCAGAA; Gpd1 Reverse: TGCGGAAGTTGGGTGTTTGC;
Sec62 Forward: GCTGAACAAAGACACCAGGCT; Sec62 Reverse: TCCCTGGCTGGCGTGTAAAT; Exd2 Forward: GGAAGCACTGGAATCGCAGC;
Exd2 Reverse: GAGATGGCATGGCAGGAGGT; Psme2 Forward: GCCTTGTGGGGTTCGCTTG; Psme2 Reverse: TCCTTGGAGGCCTTGGCTAC;
Nf l Forward: TGCGTACTCCAGCTACTCCG; Nf l reverse: CAGGTTGCGCAGAGTCTCCT; Hip1r Forward: CAGTT CGACAAGACGCAGGC; Hip1r Reverse: GATGGCTG TGTTGAGCTGCC; HDAC3 Forward: GAATTCACCA TGGAGGCCATTAGTGAGGAGCTTCC; HDAC3 Reverse: GAATTCAATCTCCACATCGCTTTCCTTG; Hdac9 Forward: AAATCTATTGAACAACTGAAGCAACCAG GC; Hdac9 Reverse: AGCTCATTCCAAATGGTGTCAC TGTCCACC; c-Jun Forward: GATGGAAACGACCTT CTACG; c-Jun Reverse: GTTGAAGTTGCTGAGGTTGG; Actin Forward: GAGAGGGAAATCGTGCGTGAC; Actin Reverse: CATCTGCTGGAAGGTGGACA. The primers used for PCR amplification using HEK293T samples were as follows: M4k3 Forward: TCATGAACTTGATCTGC AACTGGA; M4k3 Reverse: TGAGCCATCTCGTTCAC CATTT; Ppp2r2d Forward: GCGAGTCCACGGCGAA TTTT; Ppp2r2d Reverse: GTCCCACACCTTCACCGACA; Tsc1 Forward: ACTGGACCCACTTTGGAGGC; Tsc1 Reverse: TATCCGCAGCTCCGCAATCA; Tex10 Forward: GGCTGCCATGCTTATCGGGA; Tex10 Reverse: CTCTG CAGCATCAACCTCTTTCT; Smarca2 Forward: GATCCA GGCGGGCATGTTTG; Smarca2 Reverse: GGGCGGCCTC TTCTCTTCTT;
Tfg Forward: ATCAGGTTTCAGGGCCACCC; Tfg Reverse: ATTGCTCGCCTGGTACTGCT; Nptx1 Forward: AGCCTGCCAGAGATGTACGC; Nptx1 Reverse: ACATT GCCGGACAGAGCCTT; Rab23 Forward: ATGGGACA CTGCAGGTCAGG; Rab23 Reverse: ACCGGAGTGACTT CCACCAG; Ptprn Forward: ATCGGCTTCCACAACC ACCA; Ptprn Reverse: GCCGCATCTGGCTGCAC; Plxnb1 Forward: ACGTATCCGTGAGCGTGGAG; Plxnb1 Reverse: CTCATGGAGAGGCGACCCTG; Gpd1 Forward: CCCCC AAATGTGTTCATCGGC; Gpd1 Reverse: CACTGCAGAA GAGCTTGGCG;
Sec62 Forward: AGCGGATCCAGGAAGTTGGT; Sec62 Reverse: ACATACACCTCATTTCCATCCAGAA; Exd2 Forward: GTGCCTGGACCTCCGATACC;
Exd2 Reverse: GGGCTGTCCATCAGGAGCAT; Psme2 Forward: TCTTTTCCAGGAGGCTGAGGAAT; Psme2 Reverse: CACCAAGGCCCGGTAATCCA;
Nf l Forward: GTTCAAGAGCCGCTTCACCG; Nf l reverse: ATCGGCCAAAGACCTGGGAG; Hip1r Forward: ATTC TGGGCACACACCACGA; Hip1r Reverse: GCACTG GCCTGAGGACATCT; Hdac9 Forward: GTGCCATCCC AGCTCAATGC; Hdac9 Reverse: AGGTGACTGCCT GGTTGCTT; c-Jun Forward: CAGTCCAGCAACGGG CACAT; c-Jun Reverse: CTTTTTCGGCACTTGGAGGCA;
Actin Forward: CTGGGACGACATGGAGAAAA; Actin Reverse: AAGGAAGGCTGGAAGAGTGC.
Annealing temperature for each gene was optimized to reduce bias caused by primer mismatch. Amplifying cycles for each mRNA analyzed was adjusted to make sure the reaction is within the exponential amplification phage based on quantification of the signals using a Gel Logic 200 Imaging System (Kodak, Rochester, NY).
Western blot analysis
CGNs or HEK293T cells were lysed in cell lysis buffer and Western blot was performed using standard procedures as we have previously detailed.21–23 After blotting on to PVDF membrane, the membrane was first blocked with 5% nonfat milk in TBST buffer (0.05% Tween-20 in 50 mM tris-buffered saline, pH 7.4) at room temperature for 1 h after which it was incubated with primary antibody (1:1000 dilution) at 4°C overnight, followed by secondary antibody (1:10,000 dilution) for 1 h at room temperature. The following primary antibodies were obtained from Santa Cruz Biotechnology: TSC1 (catalog # SC-377386), TEX10 (catalog # SC-398384), TFG (catalog # SC-515054), NPTX1 (catalog # SC-374199), PTPRN (catalog # SC-390101), PLXNB1 (catalog # SC-28372), NFL (catalog # SC-20012), PSME2 (catalog # SC-390563), p65 (catalog # SC-372), HTT (catalog # SC-47757), FUS (catalog # SC-47711), and GAPDH (catalog # SC-47724). HDAC3 antibody (catalog # OAAB09604) was from Aviva Systems Biology (San Diego, CA). c-JUN antibody (catalog # 9165) and HSF1 antibody (catalog # 4356) were from Cell Signaling Technology (Danvers, MA). HDAC9 antibody (catalog # AP1109A) was purchased from Abgent (San Diego, CA) Goat anti-Rabbit IgG secondary antibody (catalog # 31460) and Goat anti-Mouse IgG, IgM secondary antibody (catalog # 31444) were purchased from Thermo Fisher Scientific. HIP1R antibody (catalog # 16814–1-AP) was obtained from Proteintech (Rosemont, IL). Immuno-reactive bands were detected with Clarity Western ECL substrate (BioRad, Hercules, CA). The Western blot analysis for every protein was repeated at least three times using lysates from separate cultures of neurons or HEK293T cells.
Proteomic analysis
For proteomic analysis, 40 μg proteins from each sample were run on an SDS-PAGE gel for a distance of approximately 10 mm. The gel was stained using Coomassie Blue (R-250) for 1 h. After destaining with 30% methanol, the stained bands representing the full range of protein were cut as a gel slab and transferred to a 1.5 ml tube. The short run and utilization of a single gel slab permitted the digestion and analyses of all cellular proteins together. The single gel slab from different lanes was digested overnight with trypsin (Thermo Fisher Scientific) following reduction and alkylation with DTT and iodoacetamide. The samples then underwent solid-phase extraction cleanup with an Oasis HLB plate (Waters, Milford, MA) and the resulting samples were analyzed by Liquid chromatography-tandem mass spectrometry (LC-MS/MS), using an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific) coupled to an Ultimate, 3000 RSLC-Nano liquid chromatography system (Thermo Fisher Scientific). Samples were injected into a 75 μm i.d., 50 cm long EasySpray column (Thermo Fisher Scientific), and eluted with a gradient of 1–28% buffer B over 60 min. Buffer A contained 2% (v/v) acetonitrile (ACN) and 0.1% formic acid in water, and buffer B contained 80% (v/v) ACN, 10% (v/v) trifluoroethanol, and 0.1% formic acid in water. The mass spectrometer operated in positive ion mode with a source voltage of 2.3 kV and an ion transfer tube temperature of 275°C. Mass spectrometry (MS) scans were acquired at 120,000 resolution in the Orbitrap and up to 10 MS/MS spectra were obtained in the ion trap for each full spectrum acquired using higher energy collisional dissociation (HCD) for ions with charges 2–7. Dynamic exclusion was set for 25 s after an ion was selected for fragmentation.
Raw MS data files were converted to a peak list format and analyzed using the central proteomics facilities pipeline (CPFP), version 2.0.3.24,25 Peptide identification was performed using the X!Tandem26 and Open MS Search Algorithm (OMSSA)27 search engines against the rat protein database from Uniprot, with common contaminants and reversed decoy sequences appended.28 Fragment and precursor tolerances of 20 ppm and 0.6 Da were specified, and three missed cleavages were allowed. Carbamidomethylation of Cys was set as a fixed modification and oxidation of Met was set as a variable modification. Label-free quantitation of proteins across samples was performed using SINQ normalized spectral index software.29
Bioinformatics
Differentially expressed proteins identified by proteomic analysis were subjected to functional annotation by Ingenuity Pathway Analysis (IPA; http://www.ingenuity.com). IPA is a powerful analysis tool that builds on Ingenuity Knowledge Base and identifies significant pathways and potential regulatory networks associated with differentially expressed proteins.
Statistical analysis
GraphPad Prism 5 software was used for data analyses and the generation of graphs. Unless otherwise mentioned in the figure legends, statistical analysis was done using two-tailed t test (Student’s t test) and the results were shown as mean ± SD. P < 0.1 was considered as statistically significant for Western blotting and RT-PCR analyses.
Results
Proteomic analysis of CGNs overexpressing HDAC3
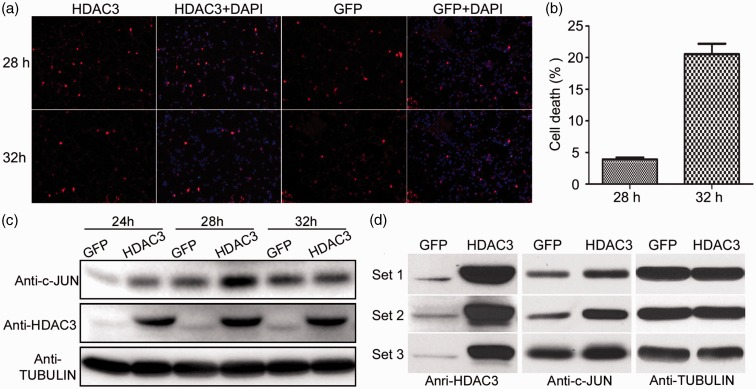
Initially, we conducted a time course analysis to identify a time point at which cell death was not significant, but which was followed by significant cell death. The rationale was that at this time point, molecular alterations responsible for inducing cell death would be apparent. For the time course, CGNs were infected with equal amounts of Ad-HDAC3 or Ad-GFP for 2 h in HK after which the virus was removed and cell viability assay was performed at 28, 32, 36, and 40 h. Based on these experiments, we found that there was a sharp increase of cell death in Ad-HDAC3-infected cultures between 28 and 32 h. Repetition of these experiments confirmed that while only 3.9% of the HDAC3-overexpressing neurons were apoptotic at 28 h, and at 32 h, the number was 20.6% (Figure 1(a) and (b)). Western blot analysis further confirmed that expression of c-JUN, a transcription factor that is often used as a marker of neuronal apoptosis, was elevated in HDAC3-overexpressing CGNs compared to GFP control at 28 h, but not at 24 and 32 h (Figure 1(c)). Based on these results, we chose to prepare protein lysates from neuronal cultures 28 h after removal of Ad-HDAC3. Lysates from three separate CGN cultures transduced with Ad-GFP or Ad-HDAC3 were prepared. To ensure that the cultures were responding appropriately to ectopic HDAC3 expression, we analyzed an aliquot of the protein lysates for the expression of c-JUN. We confirmed that c-JUN expression was stimulated by Ad-HDAC3 in all three sets of lysates (Figure 1(d)).
Figure 1.
Time-course analyses of HDAC3 neurotoxicity. (a) CGNs were infected with Ad-HDAC3 or Ad-GFP for 2 h in HK, and immunocytochemistry was performed using FLAG or GFP antibody at 28, 32, 36, and 40 h after infection. Immunofluorescence images showed that the number of HDAC3-overexpressing CGNs dramatically reduced at 32 h compared with 28 h. (b) Cell death was quantified by counting the number of infected cells displaying condensed or fragmented nuclei and expressed as % (apoptotic cells vs. total cells). Results are normalized to control cultures infected with Ad-GFP which showed little or no cell death. 3.9% of HDAC3-overexpressing CGNs underwent apoptosis at 28 h, while this number increased to 20.6% at 32 h. P < 0.05. (c) Lysates from CGN cultures over expressing human HDAC3 for 24, 28, and 32 h were subjected to Western blot analysis using a c-JUN antibody. Robust elevation of c-JUN level was seen at 28 h. (d) For proteomic analysis, CGNs infected by Ad-HDAC3 or Ad-GFP were lysed 28 h after removal of the virus. Protein samples were prepared separately from three biological replicates. Western blot using both antibodies to HDAC3 and c-JUN confirmed that HDAC3 was robustly overexpressed and c-JUN was upregulated by HDAC3. (A color version of this figure is available in the online journal.)
We electrophoresed the lysates briefly on a denaturing gel and a single slab containing all proteins was cut out and sent for proteomic analyses using label-free MS-based approach. A total of 3292 proteins were identified in GFP expressing samples, whereas in HDAC3-overexpressing samples, 3212 were identified. Of these, only 21 proteins showed significant level of changes in HDAC3-overexpressed CGNs compared with GFP-expressing neurons as defined by a change of at least 2-fold and P < 0.05 (Table 1).
Table 1.
Differentially expressed proteins identified by proteomic analysis in HDAC3-overexpressing CGNs. As described in the text, the proteomic analysis was conducted using three sets of lysates prepared from independent neuronal cultures infected either with Ad-HDAC3 or Ad-GFP. Ratio of expression between HDAC3-expressing and GFP-expressing lysates is shown along with p-value, the molecular weight of the protein in daltons (MW [Da]). PSMs (Peptide Spectrum Matches) indicate the number of spectra assigned to peptides that contributed to the inference of the protein. Peptide seqs (peptide sequences) shows the number of different unique peptide sequences that were identified for the protein. Coverage (%) represents the percentage of the protein sequence that was covered by the peptides identified for the protein. ‘HDAC3-1 only’ indicates that the protein was only identified from HDAC3-1 sample but not from GFP-1. Similarly, ‘GFP-2 only’ indicates that the protein was only identified from GFP-2 sample but not from HDAC3-2.
| Protein | Description | HDAC3-1/GFP-1 | HDAC3-2/GFP-2 | HDAC3-3/GFP-3 | HDAC3/GFP | P | MW (Da) | PSMs | Peptide Seqs | Coverage (%) | Validation by WB | Validation by PCR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Q924I2 | M4K3_RAT Mitogen-activated protein kinase 3 GN=Map4k3 | 2.41 | 4.21 | 6.60 | 4.04 | 0.048 | 98910.3 | 14 | 6 | 12.7 | — | Does not match(no change) |
| P56932 | 2ABD_RAT Serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B delta isoform GN=Ppp2r2d | HDAC3-1 only | 2.00 | 5.86 | 3.61 | 0.039 | 52075.6 | 9 | 8 | 23.2 | — | Match |
| D3ZLD5 | D3ZLD5_RAT Protein Golga3 GN=Golga3 | 1.3 | 4.09 | 4.50 | 2.73 | 0.023 | 162878 | 22 | 16 | 16.2 | — | — |
| Q9Z136 | TSC1_RAT Hamartin GN=Tsc1 | 2.42 | 3.02 | 2.70 | 0.025 | 129284 | 7 | 4 | 6.3 | Match | Does not match(no change) | |
| D4A401 | D4A401_RAT Protein Tex10 GN=Tex10 | 7.75 | GFP-2 only | 1.34 | 2.65 | 0.045 | 105482 | 7 | 4 | 5.6 | Match | Does not match(no change) |
| E9PTG1 | E9PTG1_RAT Protein Smarca2 GN=Smarca2 | 3.05 | HDAC3-2 only | 2.28 | 2.49 | 0.004 | 182708 | 7 | 7 | 5.5 | — | Match |
| Q5U2U5 | Q5U2U5_RAT Perilipin GN=Plin2 | 1.71 | GFP-2 only | 2.28 | 2.34 | 0.014 | 46360.7 | 10 | 3 | 10.9 | — | — |
| Q6AYR1 | Q6AYR1_RAT Protein Tfg GN=Tfg | 3.08 | 4.19 | 1.19 | 2.33 | 0.038 | 43177.1 | 28 | 8 | 39.9 | Match | Does not match(no change) |
| P47971 | NPTX1_RAT Neuronal pentraxin-1 GN=Nptx1 | 1.65 | 1.78 | 3.26 | 2.11 | 0.015 | 47312.9 | 29 | 11 | 21.5 | Match | Match |
| D3ZRM5 | D3ZRM5_RAT Protein Rab23 GN=Rab23 | 0.70 | 0.34 | 0.43 | 0.5 | 0.046 | 26670.6 | 14 | 7 | 43.5 | — | Does not match(no change) |
| Q63259 | PTPRN_RAT Receptor-type tyrosine-protein phosphatase-like N GN=Ptprn | 0.39 | 0.59 | 0.49 | 0.49 | 0.01 | 106449 | 11 | 5 | 7.7 | Does not match(opposite) | Does not match(no change) |
| D3ZDX5 | D3ZDX5_RAT Protein Plxnb1 GN=Plxnb1 | 0.48 | 0.47 | HDAC3-3 only | 0.47 | 0.022 | 232294 | 7 | 5 | 3.6 | Does not match(no change) | Match |
| O35077 | GPDA_RAT Glycerol-3-phosphate dehydrogenase [NAD(+)], cytoplasmic GN=Gpd1 | 0.55 | HDAC3-2 only | 0.47 | 0.41 | 0.016 | 37518.3 | 12 | 7 | 21.8 | — | Match |
| Q7TP42 | Q7TP42_RAT Ab2-292 GN=Sec62 | 0.56 | 0.20 | 0.40 | 0.38 | 0.011 | 68068.1 | 20 | 7 | 10.4 | — | Match |
| P19527 | NFL_RAT Neurofilament light polypeptide GN=Nefl | 0.34 | 0.49 | 0.28 | 0.37 | 0.017 | 61460.1 | 38 | 17 | 35.6 | Match | Does not match(no change) |
| B2GUW4 | B2GUW4_RAT Exdl2 protein GN=Exd2 | 0.25 | GFP-2 only | 0.53 | 0.35 | 0.015 | 74193.1 | 10 | 6 | 11.9 | — | Does not match(no change) |
| P11348 | DHPR_RAT Dihydropteridine reductase GN=Qdpr | 0.35 | 0.14 | 0.38 | 0.31 | 0.017 | 25607.9 | 31 | 7 | 41.1 | — | — |
| F1LY09 | F1LY09_RAT Protein Actr3b (Fragment) GN=Actr3b | 0.87 | 0.16 | 0.13 | 0.30 | 0.040 | 35900.4 | 13 | 9 | 27.9 | — | — |
| Q63798 | PSME2_RAT Proteasome activator complex subunit 2 GN=Psme2 | 0.14 | 0.36 | 0.41 | 0.29 | 0.001 | 26894.1 | 21 | 8 | 36.6 | Does not match(no change) | Does not match(no change) |
| Q63610 | TPM3_RAT Tropomyosin alpha-3 chain GN=Tpm3 | 0.42 | 0.03 | 0.57 | 0.29 | 0.038 | 29060.7 | 13 | 25 | 68.1 | — | — |
| P39069 | KAD1_RAT Adenylate kinase isoenzyme 1 GN=Ak1 | 0.34 | 0.02 | 0.51 | 0.27 | 0.018 | 21624.2 | 26 | 11 | 54.1 | — | — |
Result validation using Western blot
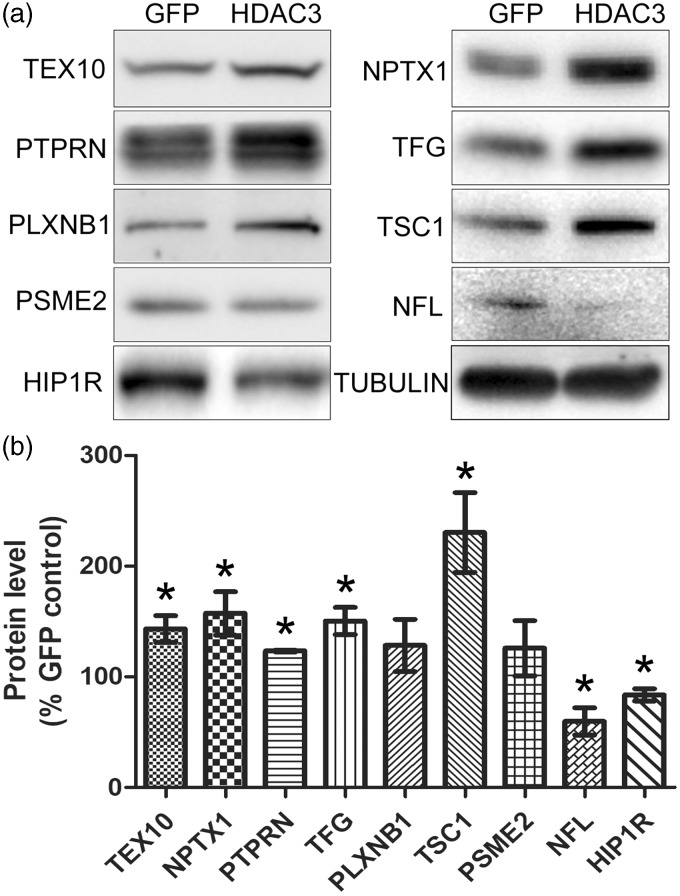
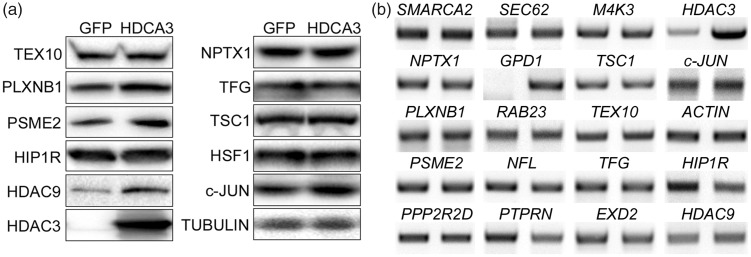
Based on antibody availability, eight of the differentially expressed proteins were chosen for validation. While five of these proteins, TEX10, NPTX1, TFG, TSC1, and NFL showed a pattern that was consistent with the results from proteomic analysis, expression of PSME2 and PLXNB1 displayed no significant changes. The expression of one protein, PTPRN, showed an increase in expression with HDAC3 overexpression, rather than a downregulation as revealed by proteomic analyses (Figure 2). We extended our analyses to include HIP1R. Besides being implicated in the regulation of neurodegeneration previously, our proteomic analyses showed that it was robustly downregulated with a ratio of HDAC3/GFP at 0.33 albeit with a P value of 0.052 which was marginally over the cut-off for the rest of the 21 differentially expressed proteins. Immunoblot analysis confirmed that HIP1R was downregulated by HDAC3 (Figure 2).
Figure 2.
Result validation by Western blotting. (a) Eight of the 21 differentially expressed proteins identified by the proteomic analysis were selected for validation by Western blotting. (b) The reactive bands on the Western blots were normalized to the loading control, α-TUBULIN, and then expressed as % of GFP control. TEX10, NPTX1, TFG, TSC1, and NFL showed consistent results with proteomics. However, level of PSME2 and PLXNB1 was not significantly altered and PTPRN was elevated by HDAC3, which are different from the proteomic data. Expression level of HIP1R, a protein identified by the proteomic analysis but falls off our criteria (HDAC3/GFP ratio = 0.33, P = 0.052), was suppressed by HDAC3, which is in line with the proteomic result.
Result validation using RT-PCR
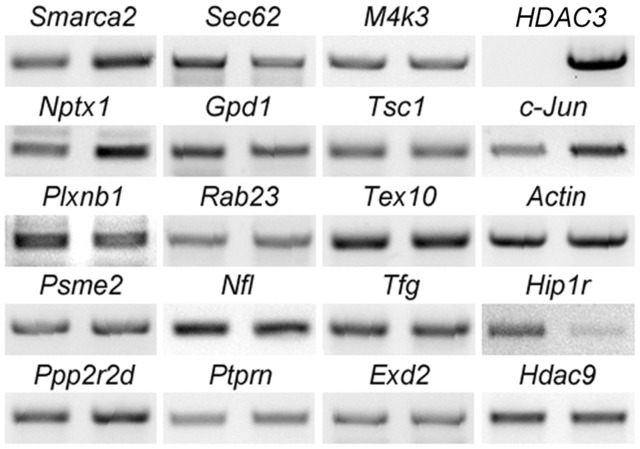
Since HDAC3 is widely regarded as a transcriptional repressor, we also examined mRNA expression of 15 of the 21 differentially expressed proteins in CGN cultures transduced with GFP or HDAC3, including the 8 proteins that we had validated by Western blots (Figure 3). The choice of the 15 proteins was based on a literature search which indicated their involvement in neuronal function or cellular signaling events. The pattern of change for six of the genes (Ppp2r2d, Smarca2, Nptx1, Plxnb1, Gpd1, and Sec62) was clearly consistent with the pattern observed by proteomic analysis. The mRNA expression of nine other proteins did not display significant changes. RT-PCR analysis also indicated that mRNA levels of Hdac9 and Hip1r were downregulated by HDAC3 (Figure 3).
Figure 3.
Result validation using RT-PCR. RNA was isolated from CGN cultures infected with Ad-GFP or Ad-HDAC3. Expression of 15 of the 21 differentially expressed proteins was examined by RT-PCR analyses. mRNA level changes for six genes, including Ppp2r2d, Smarca2, Gpd1, Nptx1, Plxnb1, and Sec62, displayed consistent patterns with proteomic findings. The nine other proteins along with HDAC9 showed no significant changes at mRNA levels. Hip1r showed a dramatic downregulation by HDAC3. HDAC3, c-Jun, and Actin were run as controls for sample quality. For each of the genes, left panel: GFP control sample; right panel: HDAC3-overexpressing sample.
Analysis of expression of other proteins of interest
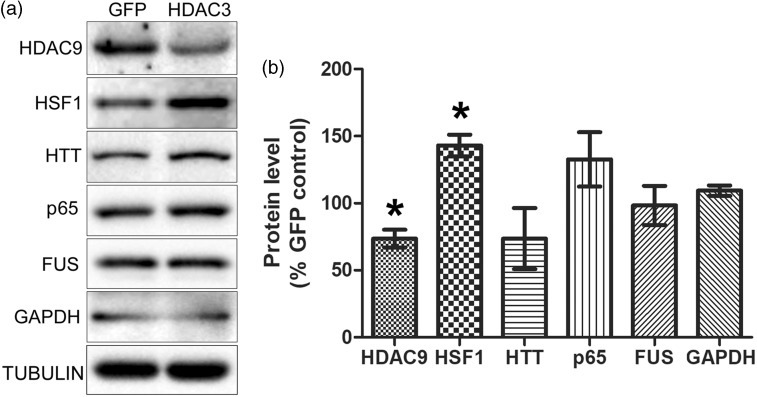
In addition to proteins identified in the MS screen, we examined other protein targets that of interest (Figure 4). These proteins were chosen because we or other labs have found them to regulate or be associated with neurodegeneration. These included HDAC9 (a Class II HDAC protein with neuroprotectiveactivity1,30), HSF1 (a neuroprotective transcription factor21,31,32), huntingtin (HTT, mutations of this neuroprotective protein cause HD5,33,34), p65/RELA (an NF-κB subunit that can promote both neuronal survival and death35–37), FUS (mutations and increased expression cause neurodegeneration38,39), and GAPDH (a glycolytic protein implicated in promoting neuronal death40,41). We found that expression of HDAC9 was downregulated by HDAC3 expression (Figure 4). Surprisingly, given their neuroprotective actions, the expression of HSF1 was increased in HDAC3-overexpressing cultures (Figure 4). The levels of HTT, p65, FUS, and GAPDH did not change appreciably (Figure 4).
Figure 4.
Regulation of expression of other proteins of interest by HDAC3. (a) CGN cultures were infected with Ad-GFP or Ad-HDAC3 and lysates were prepared. Expression levels of six other proteins that are involved in regulating in neurodegeneration were analyzed by Western blot analyses. (b) Quantitative analysis with the Western blots showed that compared to GFP-overexpressing control samples, HDAC9 was downregulated whereas HSF1 was upregulated in HDAC3-overexpressing CGNs. No significant changes were observed for HTT, p65, FUS, and GAPDH.
Analyses of neuronal HDAC3-regulated genes in HEK293T cells
Overexpression of HDAC3 is selectively toxic to neurons and cell lines of neuronal origin.4 As an attempt to identify changes in gene expression that are related to neurotoxicity, we extended our analyses of mRNA and proteins that were deregulated in neurons to HEK293T cells, which we previously described, are not sensitive to the toxic effect of HDAC3 overexpression.4 Similar to the proteomic and Western analysis of CGNs, c-JUN was upregulated by HDAC3 in HEK293T cells also (Figure 5(a)). Interestingly however, PLXNB1, PSME2, and HDAC9 which are downregulated by HDAC3 in CGNs were significantly elevated in HEK293T cells (Figure 5(a)). The other six proteins (TEX10, NPTX1, TFG, TSC1, HIP1R, and HSF1), which were found to be differentially expressed in CGNs-overexpressing HDAC3, showed no significant changes in expression in HEK293T cells (Figure 5(a)). We were not able to detect PTPRN and NFL in HEK293T cells using the same antibodies.
Figure 5.
Protein/gene regulation by HDAC3 in HEK293T cells. HEK293T cells were transfected with HDAC3-FLAG or GFP plasmid and then 30 h later harvested for mRNA or protein lysates. (a) Protein lysates were analyzed by Western blotting using antibodies to proteins shown in the figure. Western blot analysis showed that protein levels of c-JUN, PSME2, PLXNB1, and HDAC9 significantly increased in HDAC3-overexpressing HEK293T cells compared to GFP control. No significant changes were observed from other proteins. (b) RNA was subjected to RT-PCR analysis using primers against the mRNA shown in the figure. Expression of GPD1 mRNA was dramatically upregulated by HDAC3 in HEK293T cells, whereas levels of HIP1R and PTPRN were downregulated. No significant changes were found in other genes. For each of the genes, left panel: GFP control sample; right panel: HDAC3-overexpressing sample.
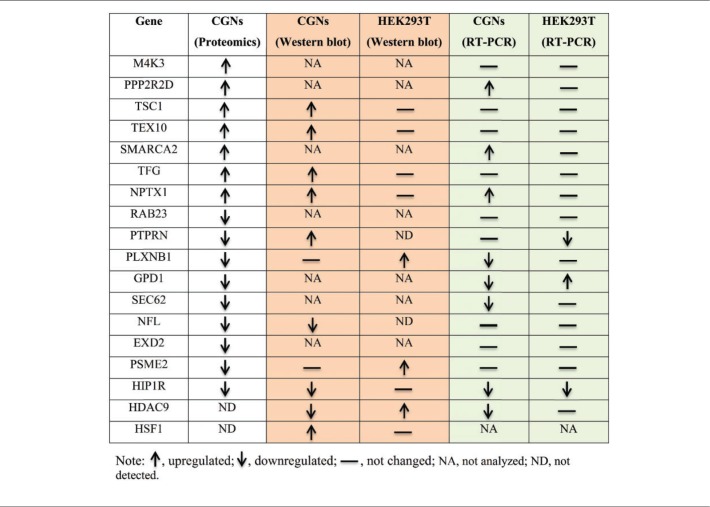
We also conducted RT-PCR analysis of RNA isolated from HDAC3-overexpressing HEK293T cells to examine expression of mRNAs that showed differential expression in CGNs (Figure 5(b)). As observed in CGNs, the level of HIP1R mRNA was reduced by HDAC3 in HEK293T cells (Figure 5(b)). On the other hand, expression of GPD1 mRNA, which was downregulated in CGNs, was robustly increased by HDAC3 in HEK293T cells. Similarly, PTPRN, also showed a different expression pattern in HEK293 cells—its level was not changed in CGNs, but decreased in HEK293T cells. No significant changes were observed in the expression of mRNA for the six other genes (PPP2R2D, SMARCA2, NPTX1, PLXNB1, SEC62, and HDAC9) that were regulated by HDAC3 in CGNs. Finally, in both CGNs and HEK293T cells, gene expression levels of M4K3, TSC1, TEX10, TFG, RAB23, NFL, EXD2, and PSME2 were not altered by HDAC3. These data show that HDAC3 regulates gene expression differently in neurons versus HEK293T cells. Further work will be necessary to determine the extent to which genes that are regulated differently in neurons compared with HEK293T cells contribute to the selective toxicity of HDAC3 to neurons.
Pathways and functional analysis
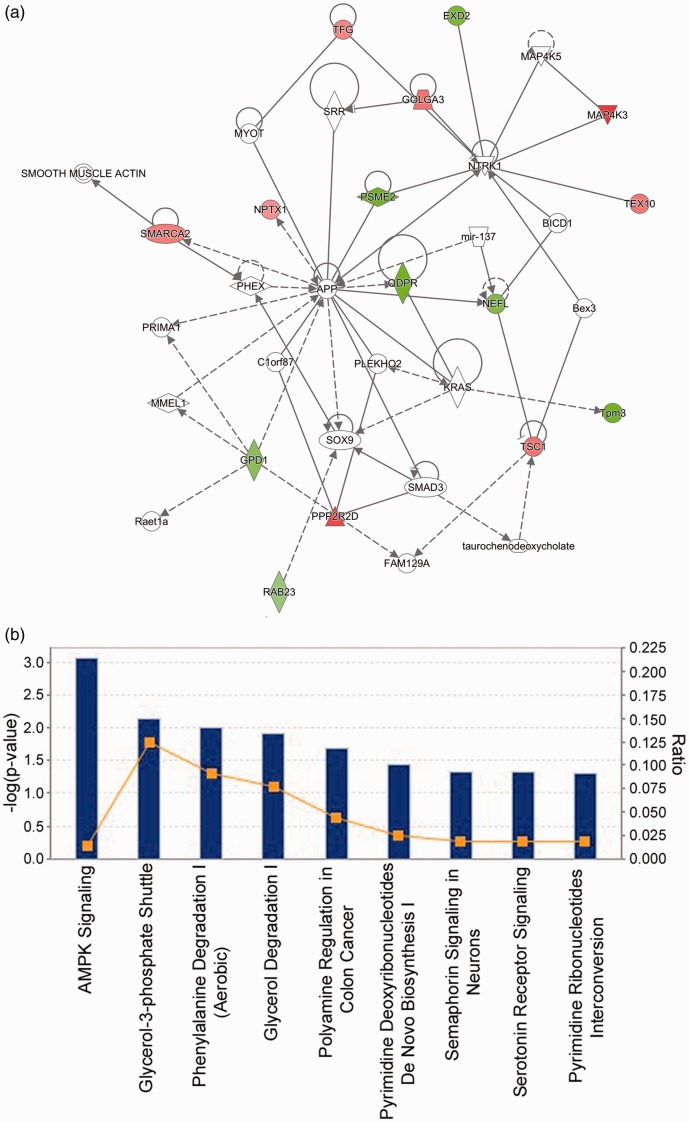
To discover significant pathways and potential regulatory networks associated with the differentially expressed proteins, we conducted IPA analysis. Functional analysis by IPA shows that the differentially expressed proteins identified from HDAC3-overexpressing neurons are involved in various molecular and cellular functions, including cellular assembly and organization, cell cycle, cell death and survival, cell morphology, and cell-to-cell signaling and interaction. Fifteen of the 21 differentially expressed proteins are linked in a protein interaction network (Figure 6(a)). IPA also shows that 8 of the 21 proteins (NFL, TSC1, NPTX1, PTPRN, PLXNB1, SMARCA2, AK1, and QDPR) play critical roles in neuronal development and function. Top canonical pathways the 21 proteins participate in include AMPK signaling, semaphorin signaling in neurons, and serotonin receptor signaling as shown by IPA (Figure 6(b)).
Table 2.
Qualitative comparison of the pattern of gene expression changes obtained from proteomic, Western blot, and RT-PCR analyses. (A color version of this table is available in the online journal.)
Figure 6.
Pathway and functional annotation. (a) IPA analysis showed that the differentially expressed proteins in HDAC3-overexpressing CGNs are associated with cell-to-cell signaling and interaction, cellular assembly and organization, and cellular function and maintenance. Fifteen out of the 21 identified proteins are involved in this network. Green color shows downregulation by HDAC3 while red color represents upregulation. The color intensity indicates the degree of protein level change. Solid lines in the network indicate direct interactions between proteins and dashed lines imply indirect interactions. Geometric shapes represent various general functional protein groups (diamond for enzyme, oval for transcription regulator, trapezoid for transporter, inverted triangle for kinase, double circle for complex/group, and circle for others). Proteins in white shapes are not part of our data set but have relationships with our proteins in the network. (b) Top IPA canonical pathways targeted by HDAC3 in CGNs are presented. These pathways were ranked according to their −log (P value) (blue bars). A ratio (orange square) indicates the number of identified proteins found in each pathway over the total number of proteins in that pathway. (A color version of this figure is available in the online journal.)
Discussion
Findings from several cell culture and in vivo models indicate that HDAC3 plays a critical role in promoting neuronal death.4,5,7,9,16 However, exactly how this neurotoxicity is mediated is not known. As a step towards understanding the underlying mechanisms, we conducted a proteomic analysis of CGNs overexpressing HDAC3. We have previously described that overexpression of HDAC3 promotes death of otherwise healthy CGN cultures and knockdown of HDAC3 protects against low potassium (LK)-induced death of these neurons.4 Although HDAC3 is a transcriptional repressor, its direct targets would be best identified by methods such as RNA-Seq which identifies changes in RNA expression. Such RNA-Seq analyses for transcriptional targets of HDAC3 have been conducted by other labs using other non-neuronal systems. One potential problem though is the large numbers of mRNA alterations that are detected (generally in the thousands), making it difficult to identify targets that are causally involved in key biological or cellular actions. In this study, we utilized an MS-based proteomic screen, which typically yields a manageable number of changes in protein expression. Moreover, it is the changes in protein expression rather than mRNA alteration that ultimately mediate neurotoxicity or other effects of HDAC3. In our proteomic analysis, we failed to detect c-JUN, a protein that we have found to be regulated by HDAC3 and whose increased expression by HDAC3 was used to confirm that the protein lysates that we were analyzing were from cultures that were responding appropriately. The reason for this is unclear but could be due to the utilization of a single gel slab and in-gel digestion which may have reduced sensitivity of our mass spectrometric analysis. It is also known that signals of low-abundant peptides are often not picked for sequencing by mass spectrometers when many highly abundant peptides exist in the sample and this may have also contributed.
Although HDAC3 is generally believed to be a transcriptional repressor, similar numbers of proteins were identified after GFP and HDAC3 overexpression in CGNs. Of the approximately 3000 proteins identified by an MS analysis, only 21 displayed significantly altered expression with HDAC3 overexpression. Of these proteins, nine were upregulated. It is possible that these changes are secondary to the repression of direct targets of HDAC3. It is noteworthy, however, that recent studies suggest that HDAC3 may also participate in stimulating gene expression.42 Both upregulation and downregulation of gene expression in comparable numbers has also been described in cells treated with Class I HDAC inhibitors.43 One of the upregulated proteins of particular interest is NPTX1, a protein that is exclusively localized to the central nervous system.44 NPTX1 expression is increased in CGNs induced to die by LK-treatment.45,46 Expression of NPTX1 is also increased in the brain following hypoxia injury and this increase plays a critical role in hypoxic-ischemic neuronal injury.47–49 Indeed, genetic deletion of NPTX1 expression protects mice against neuronal loss resulting from cerebral hypoxia-ischemia and delays neurodegeneration and extends life span in a mouse model of Sandhoff disease, a lysosomal storage disorder characterized by cognitive impairment and ataxia.50–52 NPTX1 has also been found to be necessary for amyloid-beta-induced neuronal loss both in cultured cortical neurons and in the mouse brain.53 Interestingly, both in LK-treated CGNs and following hypoxia, the increase in NPTX1 expression requires GSK3β activity,45,54 which we have previously described, is also required for HDAC3 neurotoxicity.4 Although further work is needed, these observations suggest that HDAC3 neurotoxicity could be mediated through the stimulation of NPTX1. Another protein that has also been connected to the regulation of neuronal death is HIP1R, which through its association with the clathrin complex is thought to link endocytic machinery to the actin cytoskeleton.55,56 A more recent study has found that HIP1R plays a critical role in dendritic development and formation of excitatory synapses.57 Relevant to this study is the finding that HIP1R promotes cell death through interaction with the pro-apoptotic BCL2 protein, BAK.58 Given this, it is surprising that rather being induced by HDAC3, our MS, immunoblot, and RT-PCR analyses reveal that suppression of HIP1R by HDAC3. Mutations of HIP1R are associated with Parkinson’s disease although whether these mutations increase or reduce HIP1R function is not known.59
Because of the limited sensitivity of an MS-based protein identification, we complemented our unbiased proteomic analyses with the analyses of several other proteins that we and others have previously described as regulators of neuronal death, including HDAC9, HSF1, p65/RELA, HTT, FUS, and GAPDH.21,30,31,35,36,38–41 Among these, only HDAC9 showed reduced expression in HDAC3-overexpressing neurons. We have previously shown that HDRP, a smaller form of HDAC9 generated by alternative splicing, plays a key role in promoting neuronal survival and protecting them from apoptotic stimuli.30,60 Moreover, overexpression of HDRP suppresses HDAC3 neurotoxicity.18 Others have found that HDAC9 expression is downregulated after motor neuron denervation and promotes dendritic grown in cortical neurons.61,62 It is possible that suppression of HDAC9 expression is involved in HDAC3-mediated neurotoxicity.
We previously described that primary neurons and cell lines of neuronal origin are selectively sensitive to elevated HDAC3.4 To examine whether the gene expression changes were observed in cells that are not sensitive to HDAC3 toxicity, we used the HEK293T cell line. As described in the results section, most of the mRNA and protein changes that we identified were not observed in HEK293T cells (see Table 2 for qualitative summary of changes in CGNs versus HEK293T cells). This suggests that HDAC3 regulates gene expression differently in different cell types and that at least some of the changes observed in neurons are related to their sensitivity to elevated HDAC3. Consistent with our proposal that altered expression of NPTX1 and HIP1R are targets of HDAC3-mediated neurodegeneration, the expression of both these genes was different in neurons at the protein level (Figure 5(a) and Table 2). Additionally, TSC1 also showed strikingly different patterns of expression in the two cell types after HDAC3 overexpression (Table 2). TSC1 is a negative regulator of mTOR signaling, and reduction of mTOR signaling has been implicated in neurodegenerative diseases.63,64
In sum, we have identified three proteins, previously implicated in the regulation of neurodegeneration, that are regulated by HDAC3 in neurons—NPTX1, HIP1R, and HDAC9. Further analyses, such as ChIP assays and promoter activity assays, will be necessary to determine if the regulation by HDAC3 is direct or through genes that are directly regulated by HDAC3. Given the growing evidence suggesting that HDAC3 is involved in multiple neurodegenerative conditions, investigating the extent to which NPTX1, HIP1R, and HDAC9 are involved in its neurotoxic effect would be clinically significant.
Acknowledgement
We thank Dr. Andrew Lemoff from UTSW Proteomics Core for the assistance with proteomics analyses.
Author contributions
ZQ conducted all the experiments and helped in the preparation of the manuscript. SRD wrote the manuscript and helped in the design of experiments.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
This research was supported by NIH grant R01 NS 40408 to SRD.
References
- 1.D'Mello SR. Histone deacetylases as targets for the treatment of human neurodegenerative diseases. Drug News Perspect 2009; 22:513–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sleiman SF, Basso M, Mahishi L, Kozikowski AP, Donohoe ME, Langley B, Ratan RR. Putting the 'HAT' back on survival signalling: the promises and challenges of HDAC inhibition in the treatment of neurological conditions. Expert Opin Investig Drugs 2009; 18:573–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kazantsev AG, Thompson LM. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat Rev Drug Discov 2008; 7:854–68. [DOI] [PubMed] [Google Scholar]
- 4.Bardai FH, D'Mello SR. Selective toxicity by HDAC3 in neurons: regulation by Akt and GSK3beta. J Neurosci 2011; 31:1746–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bardai FH, Verma P, Smith C, Rawat V, Wang L, D'Mello SR. Disassociation of histone deacetylase-3 from normal huntingtin underlies mutant huntingtin neurotoxicity. J Neurosci 2013; 33:11833–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas EA. Involvement of HDAC1 and HDAC3 in the pathology of polyglutamine disorders: therapeutic implications for selective HDAC1/HDAC3 inhibitors. Pharmaceuticals 2014; 7:634–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jia H, Pallos J, Jacques V, Lau A, Tang B, Cooper A, Syed A, Purcell J, Chen Y, Sharma S, Sangrey GR, Darnell SB, Plasterer H, Sadri-Vakili G, Gottesfeld JM, Thompson LM, Rusche JR, Marsh JL, Thomas EA. Histone deacetylase (HDAC) inhibitors targeting HDAC3 and HDAC1 ameliorate polyglutamine-elicited phenotypes in model systems of Huntington's disease. Neurobiol Dis 2012; 46:351–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas EA, Coppola G, Desplats PA, Tang B, Soragni E, Burnett R, et al. The HDAC inhibitor 4b ameliorates the disease phenotype and transcriptional abnormalities in Huntington's disease transgenic mice. Proc Natl Acad Sci U S A 2008; 105:15564–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jia H, Wang Y, Morris CD, Jacques V, Gottesfeld JM, Rusche JR, et al. The effects of pharmacological inhibition of histone deacetylase 3 (HDAC3) in Huntington's disease mice. PLoS One 2016; 11:e0152498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suelves N, Kirkham-McCarthy L, Lahue RS, Gines S. A selective inhibitor of histone deacetylase 3 prevents cognitive deficits and suppresses striatal CAG repeat expansions in Huntington's disease mice. Sci Rep 2017; 7:6082–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chou AH, Chen SY, Yeh TH, Weng YH, Wang HL. HDAC inhibitor sodium butyrate reverses transcriptional downregulation and ameliorates ataxic symptoms in a transgenic mouse model of SCA3. Neurobiol Dis 2011; 41:481–8. [DOI] [PubMed] [Google Scholar]
- 12.Chen YT, Zang XF, Pan J, Zhu XL, Chen F, Chen ZB, et al. Expression patterns of histone deacetylases in experimental stroke and potential targets for neuroprotection. Clin Exp Pharmacol Physiol 2012; 39:751–8. [DOI] [PubMed] [Google Scholar]
- 13.Krishna K, Behnisch T, Sajikumar S. Inhibition of histone deacetylase 3 restores amyloid-beta oligomer-induced plasticity deficit in hippocampal CA1 pyramidal neurons. J Alzheimers Dis 2016; 51:783–91. [DOI] [PubMed] [Google Scholar]
- 14.Yang X, Wu Q, Zhang L, Feng L. Inhibition of histone deacetylase 3 (HDAC3) mediates ischemic preconditioning and protects cortical neurons against ischemia in rats. Front Mol Neurosci 2016; 9:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Broide RS, Redwine JM, Aftahi N, Young W, Bloom FE, Winrow CJ. Distribution of histone deacetylases 1-11 in the rat brain. J Mol Neurosci 2007; 31:47–58. [DOI] [PubMed] [Google Scholar]
- 16.Thomas EA. Focal nature of neurological disorders necessitates isotype-selective histone deacetylase (HDAC) inhibitors. Mol Neurobiol 2009; 40:33–45. [DOI] [PubMed] [Google Scholar]
- 17.Norwood J, Franklin JM, Sharma D, D'Mello SR. Histone deacetylase 3 is necessary for proper brain development. J Biol Chem 2014; 289:34569–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bardai FH, Price V, Zaayman M, Wang L, D'Mello SR. Histone deacetylase-1 (HDAC1) is a molecular switch between neuronal survival and death. J Biol Chem 2012; 287:35444–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Louis Sam Titus ASC, Yusuff T, Cassar M, Thomas E, Kretzschmar D, D'Mello SR. Reduced expression of Foxp1 as a contributing factor in Huntington's disease. J Neurosci 2017; 37:6575–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calissano P, Matrone C, Amadoro G. Apoptosis and in vitro Alzheimer disease neuronal models. Commun Integr Biol 2009; 2:163–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verma P, Pfister JA, Mallick S, D'Mello SR. HSF1 protects neurons through a novel trimerization- and HSP-independent mechanism. J Neurosci 2014; 34:1599–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dastidar SG, Narayanan S, Stifani S, D'Mello SR. Transducin-like enhancer of Split-1 (TLE1) combines with Forkhead box protein G1 (FoxG1) to promote neuronal survival. J Biol Chem 2012; 287:14749–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dastidar SG, Landrieu PM, D'Mello SR. FoxG1 promotes the survival of postmitotic neurons. J Neurosci 2011; 31:402–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trudgian DC, Mirzaei H. Cloud CPFP: a shotgun proteomics data analysis pipeline using cloud and high performance computing. J Proteome Res 2012; 11:6282–90. [DOI] [PubMed] [Google Scholar]
- 25.Trudgian DC, Thomas B, McGowan SJ, Kessler BM, Salek M, Acuto O. CPFP: a central proteomics facilities pipeline. Bioinformatics 2010; 26:1131–2. [DOI] [PubMed] [Google Scholar]
- 26.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics 2004; 20:1466–7. [DOI] [PubMed] [Google Scholar]
- 27.Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, et al. Open mass spectrometry search algorithm. J Proteome Res 2004; 3:958–64. [DOI] [PubMed] [Google Scholar]
- 28.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 2007; 4:207–14. [DOI] [PubMed] [Google Scholar]
- 29.Trudgian DC, Ridlova G, Fischer R, Mackeen MM, Ternette N, Acuto O, et al. Comparative evaluation of label-free SINQ normalized spectral index quantitation in the central proteomics facilities pipeline. Proteomics 2011; 11:2790–7. [DOI] [PubMed] [Google Scholar]
- 30.Morrison BE, Majdzadeh N, Zhang X, Lyles A, Bassel-Duby R, Olson EN, et al. Neuroprotection by histone deacetylase-related protein. Mol Cell Biol 2006; 26:3550–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagai Y, Fujikake N, Popiel HA, Wada K. Induction of molecular chaperones as a therapeutic strategy for the polyglutamine diseases. Curr Pharm Biotechnol 2010; 11:188–97. [DOI] [PubMed] [Google Scholar]
- 32.Kondo N, Katsuno M, Adachi H, Minamiyama M, Doi H., Matsumoto S, et al. Heat shock factor-1 influences pathological lesion distribution of polyglutamine-induced neurodegeneration. Nat Commun 2013; 4:1405. [DOI] [PubMed] [Google Scholar]
- 33.Liu JP, Zeitlin SO. Is Huntingtin dispensable in the adult brain? J Huntingtons Dis 2017; 6:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dragatsis I, Levine MS, Zeitlin S. Inactivation of Hdh in the brain and testis results in progressive neurodegeneration and sterility in mice. Nat Genet 2000; 26:300–6. [DOI] [PubMed] [Google Scholar]
- 35.Pizzi M, Spano P. Distinct roles of diverse nuclear factor-kappaB complexes in neuropathological mechanisms. Eur J Pharmacol 2006; 545:22–8. [DOI] [PubMed] [Google Scholar]
- 36.Sarnico I, Lanzillotta A, Benarese M, Alghisi M, Baiguera C, Battistin L, et al. NF-kappaB dimers in the regulation of neuronal survival. Int Rev Neurobiol 2009; 85:351–62. [DOI] [PubMed] [Google Scholar]
- 37.Koulich E, Nguyen T, Johnson K, Giardina C, D'mello S. NF-kappaB is involved in the survival of cerebellar granule neurons: association of IkappaBbeta [correction of Ikappabeta] phosphorylation with cell survival. J Neurochem 2001; 76:1188–98. [DOI] [PubMed] [Google Scholar]
- 38.Sama RR, Ward CL, Bosco DA. Functions of FUS/TLS from DNA repair to stress response: implications for ALS. ASN Neuro 2014; 6:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guerrero EN, Wang H, Mitra J, Hegde PM, Stowell SE, Liachko NF, et al. TDP-43/FUS in motor neuron disease: complexity and challenges. Prog Neurobiol 2016; 145–146:78–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tatton WG, Chalmers-Redman RM, Elstner M, Leesch W, Jagodzinski FB, Stupak DP, et al. Glyceraldehyde-3-phosphate dehydrogenase in neurodegeneration and apoptosis signaling. J Neural Transm Suppl 2000;( 60):77–100. [DOI] [PubMed] [Google Scholar]
- 41.Berry MD, Boulton AA. Glyceraldehyde-3-phosphate dehydrogenase and apoptosis. J Neurosci Res 2000; 60:150–4. [DOI] [PubMed] [Google Scholar]
- 42.Sun Z, Feng D, Fang B, Mullican SE, You SH, Lim HW, et al. Deacetylase-independent function of HDAC3 in transcription and metabolism requires nuclear receptor corepressor. Mol Cell 2013; 52:769–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Glaser KB, Staver MJ, Waring JF, Stender J, Ulrich RG, Davidsen SK. Gene expression profiling of multiple histone deacetylase (HDAC) inhibitors: defining a common gene set produced by HDAC inhibition in T24 and MDA carcinoma cell lines. Mol Cancer Ther 2003; 2:151–63. [PubMed] [Google Scholar]
- 44.Schlimgen AK, Helms JA, Vogel H, Perin MS. Neuronal pentraxin, a secreted protein with homology to acute phase proteins of the immune system. Neuron 1995; 14:519–26. [DOI] [PubMed] [Google Scholar]
- 45.Enguita M, DeGregorio-Rocasolano N, Abad A, Trullas R. Glycogen synthase kinase 3 activity mediates neuronal pentraxin 1 expression and cell death induced by potassium deprivation in cerebellar granule cells. Mol Pharmacol 2005; 67:1237–46. [DOI] [PubMed] [Google Scholar]
- 46.DeGregorio-Rocasolano N, Gasull T, Trullas R. Overexpression of neuronal pentraxin 1 is involved in neuronal death evoked by low K(+) in cerebellar granule cells. J Biol Chem 2001; 276:796–803 [DOI] [PubMed] [Google Scholar]
- 47.Al Rahim M, Thatipamula S, Hossain MA. Critical role of neuronal pentraxin 1 in mitochondria-mediated hypoxic-ischemic neuronal injury. Neurobiol Dis 2013; 50:59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hossain MA. Hypoxic-ischemic injury in neonatal brain: involvement of a novel neuronal molecule in neuronal cell death and potential target for neuroprotection. Int J Dev Neurosci 2008; 26:93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hossain MA, Russell JC, O'Brien R, Laterra J. Neuronal pentraxin 1: a novel mediator of hypoxic-ischemic injury in neonatal brain. J Neurosci 2004; 24:4187–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thatipamula S, Al Rahim M, Zhang J, Hossain MA. Genetic deletion of neuronal pentraxin 1 expression prevents brain injury in a neonatal mouse model of cerebral hypoxia-ischemia. Neurobiol Dis 2015; 75:15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Al Rahim M, Hossain MA. Genetic deletion of NP1 prevents hypoxic-ischemic neuronal death via reducing AMPA receptor synaptic localization in hippocampal neurons. J Am Heart Assoc 2013; 2:e006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hooper AWM, Alamilla JF, Venier RE, Gillespie DC, Igdoura SA. Neuronal pentraxin 1 depletion delays neurodegeneration and extends life in Sandhoff disease mice. Hum Mol Genet 2017; 26:661–73. [DOI] [PubMed] [Google Scholar]
- 53.Abad MA, Enguita M, DeGregorio-Rocasolano N, Ferrer I, Trullas R. Neuronal pentraxin 1 contributes to the neuronal damage evoked by amyloid-beta and is overexpressed in dystrophic neurites in Alzheimer's brain. J Neurosci 2006; 26:12735–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Russell JC, Kishimoto K, O'Driscoll C, Hossain MA. Neuronal pentraxin 1 induction in hypoxic-ischemic neuronal death is regulated via a glycogen synthase kinase-3alpha/beta dependent mechanism. Cell Signal 2011; 23:673–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Metzler M, Legendre-Guillemin V, Gan L, Chopra V, Kwok A, McPherson PS, et al. HIP1 functions in clathrin-mediated endocytosis through binding to clathrin and adaptor protein 2. J Biol Chem 2001; 276:39271–6. [DOI] [PubMed] [Google Scholar]
- 56.Legendre-Guillemin V, Metzler M, Lemaire JF, Philie J, Gan L, Hayden MR, et al. Huntingtin interacting protein 1 (HIP1) regulates clathrin assembly through direct binding to the regulatory region of the clathrin light chain. J Biol Chem 2005; 280:6101–8. [DOI] [PubMed] [Google Scholar]
- 57.Peng L, Yang Q, Xu X, Du Y, Wu Y, Shi X, et al. Huntingtin-interacting protein 1-related protein plays a critical role in dendritic development and excitatory synapse formation in hippocampal neurons. Front Mol Neurosci 2017; 10:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim JH, Yoon S, Won M, Sim SH, Ko JJ, Han S, et al. HIP1R interacts with a member of Bcl-2 family, BCL2L10, and induces BAK-dependent cell death. Cell Physiol Biochem 2009; 23:43–52. [DOI] [PubMed] [Google Scholar]
- 59.Li NN, Tan EK, Chang XL, Mao XY, Zhang JH, Zhao DM, et al. Genetic association study between STK39 and CCDC62/HIP1R and Parkinson's disease. PLoS One 2013; 8:e79211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang X, Chen HM, Jaramillo E, Wang L, D'Mello SR. Histone deacetylase-related protein inhibits AES-mediated neuronal cell death by direct interaction. J Neurosci Res 2008; 86:2423–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mejat A, Ramond F, Bassel-Duby R, Khochbin S, Olson EN, Schaeffer L. Histone deacetylase 9 couples neuronal activity to muscle chromatin acetylation and gene expression. Nat Neurosci 2005; 8:313–21. [DOI] [PubMed] [Google Scholar]
- 62.Sugo N, Oshiro H, Takemura M, Kobayashi T, Kohno Y, Uesaka N, et al. Nucleocytoplasmic translocation of HDAC9 regulates gene expression and dendritic growth in developing cortical neurons. Eur J Neurosci 2010; 31:1521–32. [DOI] [PubMed] [Google Scholar]
- 63.Chong ZZ, Shang YC, Wang S, Maiese K. Shedding new light on neurodegenerative diseases through the mammalian target of rapamycin. Prog Neurobiol 2012; 99:128–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maiese K, Chong ZZ, Shang YC, Wang S. mTOR: on target for novel therapeutic strategies in the nervous system. Trends Mol Med 2013; 19:51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]