Abstract
Ebola virus (EBOV) is a non-segmented, negative-sense RNA virus (NNSV) in the family Filoviridae, and is recognized as one of the most lethal pathogens in the planet. For RNA viruses, cellular or virus-encoded RNA helicases play pivotal roles in viral life cycles by remodelling viral RNA structures and/or unwinding viral dsRNA produced during replication. However, no helicase or helicase-like activity has ever been found to associate with any NNSV-encoded proteins, and it is unknown whether the replication of NNSVs requires the participation of any viral or cellular helicase. Here, we show that despite of containing no conserved NTPase/helicase motifs, EBOV VP35 possesses the NTPase and helicase-like activities that can hydrolyse all types of NTPs and unwind RNA helices in an NTP-dependent manner, respectively. Moreover, guanidine hydrochloride, an FDA-approved compound and inhibitor of certain viral helicases, inhibited the NTPase and helicase-like activities of VP35 as well as the replication/transcription of an EBOV minigenome replicon in cells, highlighting the importance of VP35 helicase-like activity during EBOV life cycle. Together, our findings provide the first demonstration of the NTPase/helicase-like activity encoded by EBOV, and would foster our understanding of EBOV and NNSVs.
INTRODUCTION
Ebola virus (EBOV) is a filamentous, enveloped, non-segmented, negative-sense RNA virus, belonging to the genus Ebolavirus in the family Filoviridae of the order Mononegavirales. EBOV is highly pathogenic, whose infection causes severe Ebola virus disease (EVD), also known as Ebola haemorrhagic fever (EHF), in humans with high mortality rates between 25 and 90% (1–3). From 2014 to 2016, West Africa has experienced the largest Ebola outbreak in the history that has resulted in more than 28 000 cases and over 11 000 deaths (4) (https://www.cdc.gov/vhf/ebola/outbreaks/2014-west-africa/index.html). Moreover, the most recent Ebola outbreak is still on-going and deteriorating in DR Congo, which has caused 1290 EVD cases and 833 deaths until 16 April 2019 (http://www.who.int/ebola/situation-reports/drc-2018/en/). Together with another filovirus, Marburg virus (MARV), EBOV has been classified as a Category A priority pathogen, which imposes a significant threat to global public health (https://www.niaid.nih.gov/research/emerging-infectious-diseases-pathogens). Thus far, no approved drug or vaccine is commercially available against EBOV.
Besides filoviruses, non-segmented, negative-sense RNA viruses (NNSVs) in the order Mononegavirales include numerous important human and zoonotic pathogens, such as rabies virus, Nipah virus, respiratory syncytial virus, human parainfluenza viruses, measles virus, mumps virus, etc. (5,6). NNSVs share common organizations of viral genomic RNAs and similar strategies of transcription and replication (5). For filoviruses, the (–)-sense viral genomic RNA is approximately 19 kb in length and encodes seven open reading frames (ORFs) for all the viral proteins, i.e. nucleoprotein (NP), viral protein 35 (VP35), VP40, glycoprotein (GP), VP30, VP24, and the large polymerase protein (L) (7–9). Upon viral entry into host cells, the released viral genomic (–)-RNAs are used as the templates for the transcription of individual viral mRNAs that are translated into filoviral proteins. The viral genomic (–)-RNAs are then switched to replicate the antigenomic positive-sense (+)-RNAs, which are used as the templates for the production of progeny viral genomic (–)-RNAs (7). The transcription and replication of EBOV RNAs are carried out by the viral ribonucleoprotein (RNP) complex that contains the RNA-dependent RNA polymerase (RdRP) L, the polymerase cofactor VP35 (10,11), as well as NP and VP30 (12), which represent the minimal elements required for EBOV transcription and replication (13).
For most RNA viruses including NNSVs, viral genomic, antigenomic and messenger RNAs contain multiple cis-acting elements that play critical roles in viral RNA replication, transcription, translation, and packaging during viral life cycles (14,15). Similar with cellular RNAs, these highly structured RNA elements need to be properly folded to be functional. However, the correct folding of RNA molecules is challenging, since RNAs could be kinetically trapped in misfolded states that are relatively stable in thermodynamics (16,17). To facilitate the correct folding of RNAs, a variety of RNA remodelling proteins, such as RNA helicases and RNA chaperones, are encoded by cells and viruses. These proteins function to destabilize RNA-RNA or RNA–DNA base-pairings to aid RNAs’ proper folding or refolding (18–20). In addition, they are believed to participate in the unwinding of viral double-stranded RNA (dsRNA) intermediates produced during replication and/or transcription, thereby facilitating the recycling of viral RNA templates for more efficient RNA synthesis. Thus far, a wide range of positive-sense RNA viruses and dsRNA viruses, including picornavirus (21), norovirus (22), flavivirus (23,24), alphavirus (25), coronavirus (26) and reovirus (27), have been found to encode their own RNA helicases and/or RNA chaperones (28,29). However, it is unknown whether NNSVs including filoviruses encode any protein with RNA remodelling activity or the replication/transcription of NNSVs requires the participation of any viral RNA remodelling activity. This apparent discrepancy hampers our understanding of this large group of important pathogenic viruses.
Filoviral VP35 is an essential polymerase L cofactor that is analogous to the P proteins of other NNSVs (5). In addition, VP35 is a modular multifunctional protein that contains dsRNA-binding and oligomerization domains, and plays pivotal roles in filoviral replication and transcription, nucleocapsid assembly, and evasion of host antiviral defenses (7,30–34). Here, we show that EBOV VP35 has an unexpected nucleoside triphosphatase (NTPase) activity, which can hydrolyze all kinds of ribonucleotide triphosphates (NTPs). Strikingly, EBOV VP35 can function like a helicase to unwind RNA helices from 5′ to 3′ in an NTP-dependent manner. Moreover, we have found that guanidine hydrochloride (GuHCl), a well-known helicase inhibitor (22,34,35) and a U.S. FDA-approved small compound to treat the symptoms of muscle weakness and fatigability associated with Eaton-Lambert syndrome (https://www.accessdata.fda.gov/scripts/cder/daf/index.cfm?event=overview.process&ApplNo=001546) (36,37) can inhibit the NTPase and helicase-like activities of VP35 in a dose-dependent manner. More importantly, GuHCl has also been found to exhibit inhibitory effect on the replication/transcription of a stable EBOV minigenome replicon in cultured human cells, highlighting the functional significance of VP35′s NTPase and helicase-like activities in EBOV life cycle, and implying that targeting the helicase-like activity of VP35 can be a novel strategy to develop antivirals against EBOV.
MATERIALS AND METHODS
Plasmid construction
The construction of pFastBac HTB-MBP and pFastBac HTB-MBP-EBOV VP35, pFastBac HTB-MBP-Enterovirus type 71 (EV71) 2C, pFastBac HTB-MBP-Zika virus (ZIKV) NS3 have been described previously (38). The cDNA fragment of EBOV VP35 (GenBank accession no.: AF086833.2) was amplified by polymerase chain reaction (PCR) from the plasmid containing full-length EBOV cDNA. The cDNA fragments of EBOV VP35, EV71 2C and ZIKV NS3 were cloned into the vector pFastBac HTA or pFastBac HTB-MBP that was originated from pFastBac HTB (Invitrogen, Carlsbad, CA, USA), respectively. The mutations were generated as previously described (39). The resulting plasmids were subjected to Bac-to-Bac baculovirus system to express the recombinant proteins with an MBP or His6 fused at the N-terminal. The primers used in this study are shown in Supplementary Table S1.
Expression and purification of recombinant proteins
The expression and purification of proteins from baculovirus system were performed as previously described (38,40). Briefly, Sf9 cells were infected with the recombinant baculoviruses and harvested at 3 days post-infection. Cell pellets were re-suspended, lysed by sonication and subject to centrifugation for 30 min at 11 000 g to remove debris. To get rid of the possible contaminant co-purified from MBP–VP35 via binding to RNA, the supernatant was treated with RNase A (Omega) at the final concentration of 0.1 μg/μl for 4 h. Then, the protein in the supernatant was purified using amylase affinity chromatography (New England BioLabs, Ipswich, MA, USA) according to the manufacturer's protocol. For His6-fusion protein, the protein in the supernatant was purified by Ni-NTA agarose column (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer's protocol. All the purified proteins were concentrated using Amicon Ultra-30 filters (Millipore, Schwalbach, Germany). After that, the store buffer was exchanged to 50 mM 2-[4-(2-hydroxyethyl)-1-piperazinyl] ethanesulfonic acid (HEPES)–KOH (pH8.0). All proteins were quantified by the Bradford method and stored at –80°C in aliquots. Proteins were separated on 10% SDS-PAGE and visualized by Coomassie blue.
Size Exclusion Chromatography
The affinity-purified protein sample was concentrated by tangential flow filtration using Amicon Ultra centrifugal filters (Merck) to 1 mg/ml for further analysis. For size exclusion chromatography, concentrated protein sample was mixed with BSA control and loaded onto a Superdex 200 increase 10/300 GL column (GE Healthcare) after pre-equilibration with buffer containing 50 mM HEPES-KOH (pH 8.0). Chromatography was taken with BioLogic DuoFlow system (Bio-Rad) at a flow rate of 1 ml/min. Peak analysis was performed using the ASTRA software package (BioLogic Chromatography Systems).
NTP binding and NTPase assays
The recombinant baculovirus-infected Sf9 cells were re-suspended, lysed by sonication and subject to centrifugation for 30 min at 11 000 g to remove debris. The protein in the supernatant was pulled down by using 5′-ATP agarose (Sigma-Aldrich) according to the manufacture's protocol. The purified ATP-bound protein was analyzed by western blotting with anti-MBP antibody. NTPase activities were determined via measuring the released inorganic phosphate during NTP hydrolysis using a direct colorimetric assay as previously described (21).
Gel mobility shift assay
Gel mobility shift assay was performed in 50 mM HEPES–KOH (pH 8.0), 100 mM NaCl, 2 mM MgCl2, 1 mM tRNA, 2 mM DTT, 20 U RNasin, in a total volume of 10 μl reaction with the indicated amount of proteins and 0.1 pmol dsRNA or ssRNA. The dsRNA and ssRNA were labeled with DIG-UTP (Roche) by in vitro transcription and derived from 200-nt EGFP. Reactions were incubated for 30 min at 25°C. The reactions were terminated by the addition of 2.5 μl 5× sample buffer [20 mM Tris–HCl (pH 8.0), 30% glycerol and 0.1% bromophenol blue]. The nucleic acid–protein complexes were separated by electrophoresis on 1.5% agarose gels and transferred to Hybond-A nylon membrane (GE Healthcare). After that, the membrane was subjected to cross-linking with 120°C and was incubated with anti-DIG-alkaline phosphatase antibody (Roche), followed by incubating with CDP-STAR (Roche) for 15 min at 37°C. The signals are then detected by X-ray film (Fujifilm, Tokyo, Japan).
Preparation of oligonucleotide helix substrates
RNA helix, DNA helix and RNA–DNA hybrids were prepared by annealing two complementary nucleic acid strands. One was labeled at the 5′ end with hexachloro-fluorescein (HEX)-labeled, and the other strand was unlabeled. HEX-labeled oligonucleotide strands were purchased from TaKaRa (Dalian, China). Unlabeled DNA strands were synthesized by Invitrogen, and unlabeled RNA strands were in vitro transcribed using T7 RNA polymerase (Promega, Madison, WI). The in vitro transcribed RNA strands were purified by Poly-Gel RNA Extraction Kit (Omega bio-tek, Guangzhou, China) according to the manufacturer's instructions. The two strands were mixed in a proper ratio, and annealed through heating and gradually cooling as previously described (21,27). The resulting duplexes were examined by 15% native-PAGE gels to make sure that all the single-stranded RNA or DNA was annealed in a ratio of 1:1. The standard (R*/R) by annealing a 42-nt HEX-labeled single -stranded RNA1 and a 54-nt non-labeled single-stranded RNA2. The 3’-tailed and 5′-tailed RNA helixes by annealing a 42-nt HEX-labeled single-stranded RNA1 and a 48-nt non-labeled single-stranded RNA3 (3′-tailed) or RNA4 (5′-tailed). (D*/D) by annealing a 28-nt HEX-labeled single-stranded DNA1 and a 49-nt non-labeled single stranded DNA2, (D/R*) by annealing a 42-nt HEX-labeled single-stranded RNA1 and a 30-nt non-labeled single-stranded DNA3, (R*/D) by annealing a 42-nt HEX-labeled single-stranded RNA1 and a 54-nt non-labeled DNA4. All oligonucleotides used in this study are listed in Supplementary Table S2.
Nucleic acid helix unwinding assay
The standard helix unwinding assay was performed as previously described (27) with minor modifications. Briefly, 20 pmol of recombinant protein and 0.1 pmol of HEX-labeled helix substrate were added to a mixture containing 50 mM HEPES–KOH (pH 8.0), 2 mM MgCl2, 100 mM NaCl2 and 20 U RNasin (Promega). After incubation at 37°C for 60 min, the reaction was terminated by adding 5× loading buffer [100 mM Tris–HCl, 1% SDS, 50% glycerol, and bromophenol blue (pH 7.5)]. The mixtures were then electrophoresed on 15% native-PAGE gels, followed by scanning with a Typhoon 9500 imager (GE Healthcare, Piscataway, NJ, USA).
GuHCl treatment
GuHCl (Sigma-Aldrich) was serially diluted to the indicated concentrations and added to EBOV minigenome replicon seeded in a 24-well plate and a 96-well plate. The culture supernatants and cell lysates from the 24-well plate were harvested at 96 h post-treatment for measuring GLuc activity and RNA levels, and cells in the 96-well plate were used for the cell viability assay (41).
Cell viability assay
A CellTiter-Glo luminescent cell viability assay kit (Promega, Madison, WI, USA) was used to measure the cell viability after drug treatment. 100 μl of CellTiter-Glo assay solution was added directly into the culture medium of the cells grown in a 96-well plate. Incubate the plate on a shaker for 30 min in dark and then transfer 100 μl of the supernatant into a 96-well white plate. Luminescence was then measured by the use of a VarioSkan Flash reader (Thermo Fisher Scientific).
Luciferase assay
A BioLux Gaussia luciferase assay kit (New England BioLabs, Ipswich, MA, USA) was used to measure the GLuc activity. Culture supernatants were harvested and centrifuged at 10 000 g for 3 min. A 20-μl volume of supernatant was added into a 96-well white plate, and 50 μl of GLuc assay solution was added and mixed with culture supernatant immediately before measurement of the luminescence by the use of a VarioSkan Flash reader (Thermo Fisher Scientific).
RNA isolation and qRT-PCR
Total cellular RNA was isolated using TRIzol reagent (Tiangen, Beijing, China). RT was performed using a ReverTra Ace qPCR RT kit following the protocol of the manufacturer (Toyobo, Kyoto, Japan). Quantitative PCR was carried out utilizing SYBR green real-time PCR master mix (Toyobo). The RT and qPCR primers were described previously (41).
Western blotting
Cells were washed twice in cold PBS and lysed in lysis buffer [50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1% NP40, 0.25% deoxycholate and a protease inhibitor cocktail (Roche)], and the lysates were subjected to 10% SDS-PAGE and Western blotting according to our standard procedures (42) with the relevant antibodies. The anti-MBP and anti-β-actin antibodies were purchased from MBL Co. Ltd. The anti-NP, anti-VP30 and anti-VP35 antibodies were raised in rabbits as previously described (41).
Northern blotting and RNA-immunoprecipitation (RNA-IP)
Northern blot was performed as previously described (42) with minor modification. Five μg of total RNAs were electrophoresed on denaturing 1.2% agarose gels with 2.2 M formaldehyde, and then capillary transferred to Hybond-A nylon membrane (GE Healthcare). The membranes were hybridized with the DIG-labeled RNA probe at 65°C for 12 h, and then incubated with anti-DIG-alkaline phosphatase antibody (Roche), followed by incubation with CDP-STAR (Roche) at 37°C for 10 min. The signals were detected by radiography on X-ray film (FujiFilm, Tokyo, Japan). The DIG-RNA probe targeting the negative-strand of EBOV 3′UTR 1–200 nt were produced via in vitro transcription using DIG RNA labeling mix (Roche).
RNA-IP was performed as previously described (42) with minor modification. Briefly, cells were lysed in a lysis buffer [20 mM Tris–HCl (pH 7.4), 200 mM NaCl, 2.5 mM MgCl2, 0.5% Triton X-100, 0.5 U/ml RNase inhibitor (Promega) and a protease inhibitor cocktail (Roche)] at 4°C for 30 min. Lysates were clarified at 12 000 g for 10 min at 4°C and the post-nuclear lysates were pre-cleared by incubation with protein-A/G agarose beads (Roche) at 4°C for 2 h. Then the pre-cleared lysates were incubated with antibodies (anti-Flag or anti-IgG) together with protein-A/G agarose beads (Roche) at 4°C for 12 h. The antibody-bound complexes were washed for five times with the same lysis buffer. Finally, proteins or RNAs were extracted from the complexes and analyzed by northern or western blotting as describe above.
RESULTS
EBOV VP35 has NTP-binding and NTPase activities
Previous studies have revealed that EBOV VP35 is a multifunctional protein with dsRNA-binding activity that is important for its ability to suppress immune responses (31,43,44). To further characterize EBOV VP35, it was fused with maltose-binding protein at its N-terminal (MBP–VP35) and then purified (Supplementary Figure S1). Previous studies have uncovered that EBOV VP35 can be oligomerized (33,45). To examine if the purified MBP–VP35 is oligomeric (33,46), MBP–VP35 was purified via MBP column and ion-exchange (Supplementary Figure S2A), and then subjected to the size-exclusion chromatography analysis using a Superdex 200 column. The purified MBP–VP35 proteins were eluted in the major peak that should correspond to a molecular mass of ∼600 kDa, while monomeric BSA was used as a control that was eluted in its correct size of ∼67 kDa (Supplementary Figure S2B). Although the effect of shape on elution on size exclusion column makes it hard to calculate the exact molecular mass of the MBP–VP35 complex, these results indicate that the purified recombinant MBP–VP35 should be in an oligomeric form.
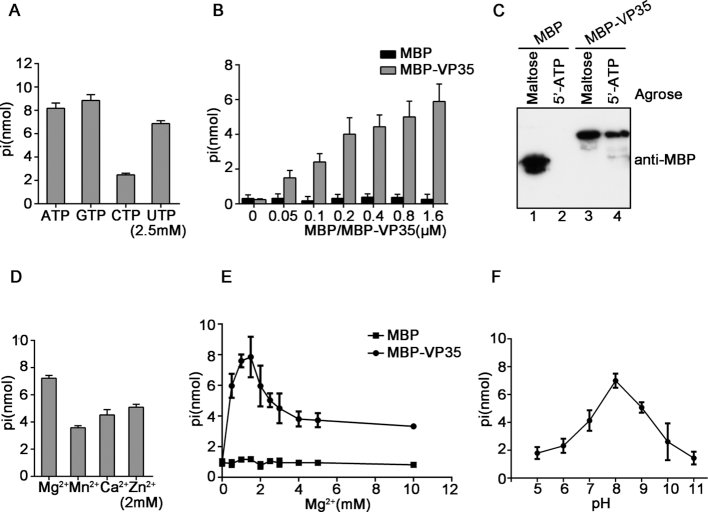
Because a number of viral RNA remodelling proteins also have NTPase activity (22,34,47), we next examined if EBOV VP35 has NTPase activity that can hydrolyze NTPs (ATP, GTP, CTP and UTP) by using a sensitive colorimetric assay that measures the total amount of free orthophosphate released after ATP hydrolysis. Our data showed that MBP–VP35 hydrolyzed all four types of NTPs, with a preference on ATP, GTP and UTP over CTP (Figure 1A). Moreover, when increasing concentrations of MBP–VP35 were incubated with ATP, MBP–VP35 can efficiently hydrolyze ATP in a dose-dependent manner (Figure 1B; Supplementary Figure S3). Furthermore, because VP35 has NTPase activity, it is intriguing to examine if this protein has NTP-binding activity. Our data show that 5′-ATP agarose can readily pull-down MBP–VP35 but not MBP alone (Figure 1C), showing that VP35 does bind to ATP. Of note, MBP protein was expressed and purified exactly same as MBP–VP35, and showed negligible NTP hydrolysis and binding.
Figure 1.
EBOV VP35 has NTP-binding and NTPase activities. (A) 20 pmol MBP–VP35 was reacted with the indicated NTPs (2.5 mM). The NTPase activity of MBP–VP35 was measured as nanomoles of released inorganic phosphate by using a sensitive colorimetric assay. (B) 2.5 mM ATP was incubated with MBP–VP35 or MBP alone at the increasing concentrations, and the ATPase activity was determined. (C) MBP or MBP–VP35 was pulled down by using maltose- or 5′-ATP agarose, followed by SDS-PAGE and immunoblotting using anti-MBP antibody. (D–F) 20 pmol MBP–VP35 was reacted with 2.5 mM ATP at 2 mM indicated divalent metal ions (D), the indicated concentrations of MgCl2 (E), or the indicated pH (F). MBP alone was used as the negative control. Error bars represent standard deviation (SD) values from three separate experiments.
After determining that EBOV VP35 has NTPase activity, we then examined its optimal biochemical reaction conditions, including different salt content, Mg2+ concentrations, and pH, by using ATP as the substrate. Our data showed that 2 mM Mg2+, Mn2+, Ca2+ or Zn2+ could support the ATPase activity of VP35, and their efficiencies were as follow: Mg2+>Zn2+>Ca2+>Mn2+ (Figure 1D). MBP–VP35 showed the optimal ATPase activity in the presence of 1.5 mM Mg2+, while higher concentrations of Mg2+ had some inhibitory effect on its ATPase activity; and more importantly, the optimal ATPase activity of VP35 requires the presence of Mg2+, while VP35 had no ATPase activity in absence of divalent metallic ion (Figure 1E), similarly with other NTPases (22,48). Additionally, we observed that MBP–VP35 achieved its optimal ATPase activity at pH 8.0 (Figure 1F).
Altogether, our data show that EBOV VP35 has both NTP-binding and NTPase activities.
EBOV VP35 shows RNA helix unwinding activity in an NTP-dependent manner
To further confirm the dsRNA-binding activity of MBP–VP35, we incubated the protein with an artificial dsRNA that was constructed by annealing two complementary digoxin (DIG)-labeled RNA strands that are derived from 5′ 200-nt egfp ORF, followed by the gel mobility shift assay. Our data show that while MBP alone showed no dsRNA binding activity, MBP–VP35 did bind to the RNA helix substrate in a dose-dependent manner (Supplementary Figure S4A and B). Moreover, we generated three EBOV-specific RNA helix substrates that contain 1–200 nts of EBOV 5′-UTR, 1–200 nts of 3′-UTR, and 1–200 nts of NP ORF of EBOV genomic RNA. Our data show that VP35 can bind to these EBOV-specific RNA helices (Supplementary Figure S4C-E).
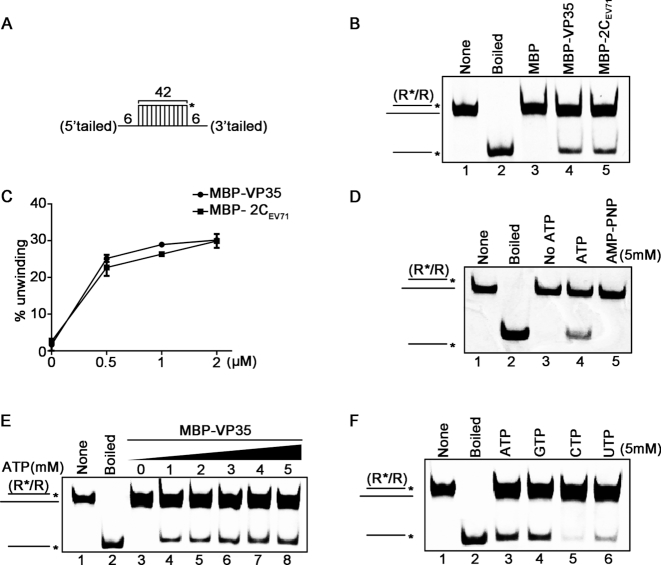
Since EBOV VP35 has dsRNA-binding, NTP-binding and NTPase activities, we speculated that VP35 may serve as a putative viral RNA remodelling protein. To examine if VP35 has RNA helix unwinding activity, we constructed the standard HEX-labeled RNA helix substrate with both 3′ and 5′ single-stranded tails by annealing a short 42-nt HEX-labeled RNA1 and a long non-labeled 54-nt RNA2 (as illustrated in Figure 2A), which was commonly used to determine the helix-unwinding activity previously (34). The helix-unwinding assay was performed by incubating the RNA helix substrate with MBP–VP35 in a standard unwinding reaction mixture, followed by the separation of the substrate strands via gel electrophoresis. Our data showed that the HEX-labeled RNA strand was efficiently released from the RNA helix substrate in the presence of MBP–VP35 (Figure 2B, lane 4), whereas the same helix substrate was not separated when the negative control (MBP alone) was present (Figure 2B, lane 3). Of note, the unwinding reactions were performed in the reaction mixture containing ATP and MgCl2; and the boiled helix substrate or the addition of MBP-fusion EV71 2C, a well-characterized viral RNA helicase, was used as positive control for helix unwinding. And the helix-unwinding activity of VP35 is almost as robust as that of EV71 2C (Figure 2C).
Figure 2.
EBOV VP35 has the NTP-dependent RNA helix unwinding activity. (A) Schematic illustration of the standard RNA helix substrate (R*/R). Asterisks indicate the HEX-labeled strands. (B) The standard RNA helix substrate (0.1 pmol) was reacted with each indicated protein (20 pmol). And the unwinding activity was assessed via gel electrophoresis and scanning on a Typhoon 9500 imager. Non-boiled reaction mixture (lane 1) and reaction mixture with MBP alone (lane 3) were used as negative controls, and boiled reaction mixture (lane 2) and reaction mixture with MBP-fusion EV71 2C (lane 5) were used as positive controls. (C) The unwinding activities at different MBP–VP35 or MBP-2C concentrations were plotted as the percentage of the released RNA from the total RNA helix substrate (Y-axis) at each protein concentration (X-axis). (D) MBP–VP35 (20 pmol) was reacted with the standard RNA helix substrate (0.1 pmol) in reaction mixture in the absence (lane 3) or presence (lane 4) of 5 mM ATP, or in the presence of 5 mM AMP–PNP (lane 5), followed by unwinding assay. (E) The RNA helix unwinding assay as described in (A) was performed in the presence of increasing concentrations of ATP. (F) The RNA helix unwinding assay was performed in the presence of each indicated NTP (5mM). For (C), error bars represent SD values from three separate experiments.
We then examined whether the RNA helix unwinding activity of VP35 is ATP-dependent. Our data showed that MBP–VP35 could only unwind the RNA helix in the presence of ATP (Figure 2D, lane 4), but not in the absence of ATP (lane 3) or the presence of AMP–PNP, a non-hydrolysable ATP analog (lane 5). Moreover, our results show that the presence of ATP can promote the helix-unwinding activity of VP35 in a dose-dependent manner (Figure 2E). Additionally, all four types of NTPs could enable VP35 to unwind the RNA helix substrate, and the preference of the helix unwinding activity of VP35 on different NTPs is consistent with that of the VP35 NTPase, as CTP is the least favorable for both activities (Figure 2F and Figure 1A).
To further verify the activities of VP35, we also produced His6-tagged VP35 (His-VP35). Our results clearly showed that His-VP35 have the NTPase and helix-unwinding activities (Supplementary Figure S5). Moreover, because VP35 is an RNA-binding protein, to exclude the possibility that some contaminant can be co-purified with MBP–VP35 via cellular RNA, we used RNase A to treat MBP–VP35 proteins during the process of protein purification to remove any possible RNAs, followed by protein purification, and the RNA-free MBP–VP35 showed NTPase and helix-unwinding activities (Supplementary Figure S6).
Taken together, our findings show that EBOV VP35 has an NTP-dependent RNA helix-unwinding activity.
EBOV VP35 directs RNA helix unwinding in the 5′ to 3′ directionality
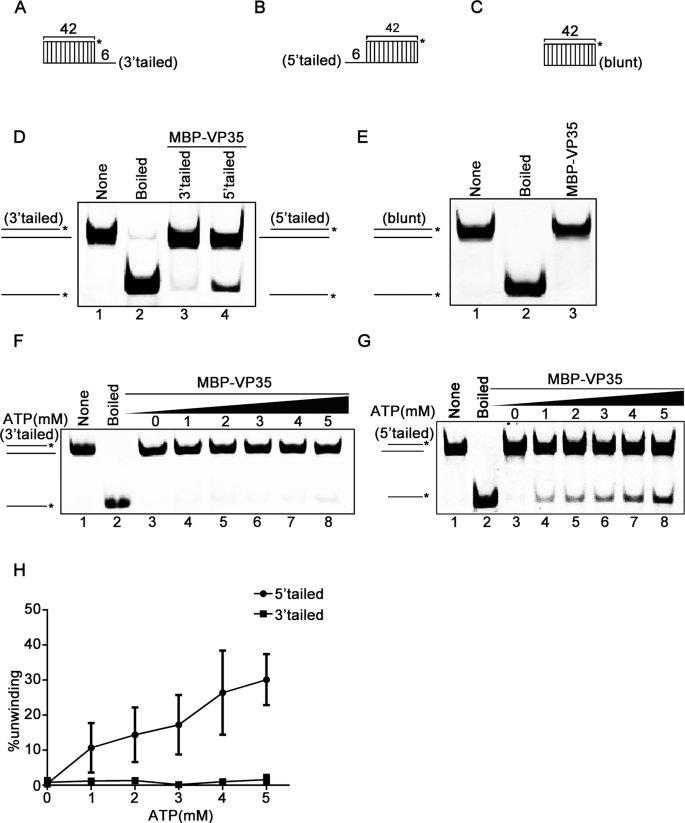
The directionality of helix unwinding is a basic property of helicases. After finding that EBOV VP35 has the RNA helix-unwinding activity, we sought to assess its helix unwinding directionality. For this purpose, we used a classic assay by constructing three different RNA helix substrates that are 3′-tailed (Figure 3A), 5′-tailed (Figure 3B), and blunt ended (Figure 3C), respectively. The three helix substrates were then subjected to the RNA helix-unwinding assay by being reacted with purified MBP–VP35. Our results showed that MBP–VP35 efficiently unwound 5′-tailed RNA helix, but barely unwound 3′-tailed one (Figure 3D). Moreover, it could not unwind the blunt-ended RNA helix (Figure 3E), like other viral helicases such as EV71 2C (34). To further determine the unwinding directionality of VP35, we evaluated its helix-unwinding activity in each direction in the presence of increasing concentrations of ATP. Our data showed that the increasing concentrations of ATP failed to promote the unwinding of 3′-tailed helix substrate (Figure 3F), but efficiently promoted the unwinding of 5′-tailed helix substrate in a dose-responsive manner (Figure 3G and 3H). In conclusion, our data demonstrated that EBOV VP35 is able to unwind RNA helix in the directionality of 5′ to 3′.
Figure 3.
EBOV VP35 unwinds RNA helix in the 5′ to 3′ directionality. (A–C) Schematic illustrations of the RNA helix substrates with 3′-tailed (A), 5′-tailed (B), and blunt ends (C). Asterisks indicate the HEX-labeled strand. (D and E) MBP–VP35 (20 pmol) was reacted with 0.1 pmol 3′-tailed (lane 3) or 0.1 pmol 5′-tailed (lane 4) RNA helix substrate (D), or 0.1 pmol helix substrate with blunt ends (E). (F and G) MBP–VP35 (20 pmol) was reacted with 0.1 pmol 3′-tailed (F) or 5′-tailed (G) RNA helix substrate in the presence of increasing concentrations of ATP. (H) The unwinding activities were plotted as the percentage of the released RNA from the total 3′-tailed (F) or 5′-tailed (G) RNA helix substrate (Y-axis) at each ATP concentration (X-axis). The error bars represent SD values from three separate experiments.
Characterization of the RNA helix-unwinding activity of VP35
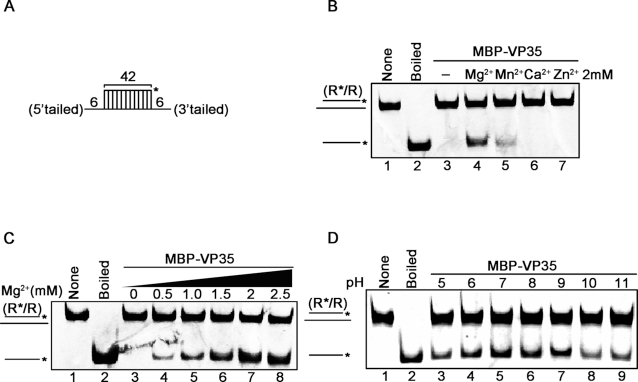
To further characterize the optimal biochemical reaction conditions of EBOV VP35, the purified MBP–VP35 was reacted with the standard RNA helix substrate (Figure 4A) under different conditions. We observed that the RNA helix-unwinding activity of MBP–VP35 required the presence of Mg2+ (Figure 4B, lane 4), while the presence of Mn2+ could also support the helix-unwinding by VP35 in a much lesser extent (Figure 4B, lane 5). On the other hand, although Ca2+ and Zn2+ have been found to support the NTPase activity of VP35 (Figure 1D), these two ions could not support the helix-unwinding activity of VP35 (Figure 4B, lanes 6 and 7). Moreover, increasing concentrations of Mg2+ could promote the helix-unwinding activity of VP35 in a dose-responsive manner (Figure 4C). Besides, the optimal helix-unwinding activity of MBP–VP35 was determined to be at pH values of 7–8 (Figure 4D, lanes 5 and 6), consistent with the optimal pH for the NTPase activity of VP35 (Figure 1F).
Figure 4.
Optimal biochemical reaction conditions for the RNA helix unwinding activity of EBOV VP35. (A) Schematic illustration of the standard RNA helix substrate (R*/R). Asterisks indicate the HEX-labeled strands. (B–D) MBP–VP35 (20 pmol) was reacted with 0.1 pmol standard RNA helix substrate in the presence of each indicated divalent metal ion (2 mM for each) (B), increasing concentrations of Mg2+ (C), or indicated pH values (D).
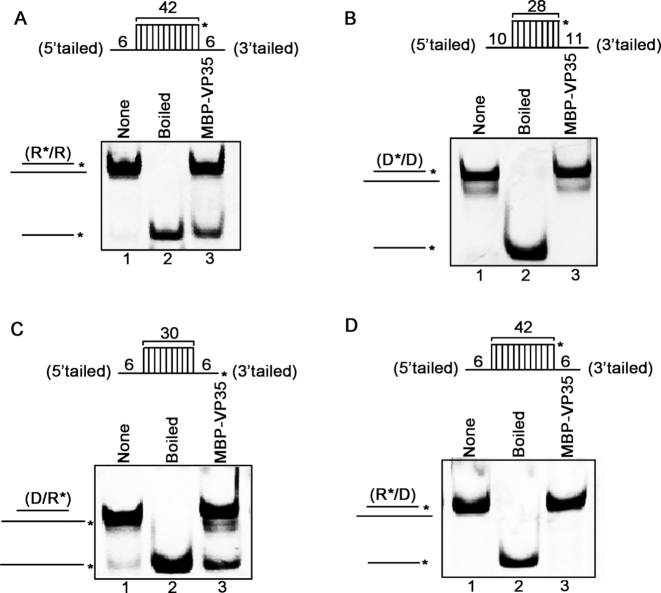
Because some virus-encoded RNA helix-unwinding proteins can also unwind DNA helix and/or RNA–DNA hybrids (22), we sought to examine the possibility for EBOV VP35. To this end, we generated four different nucleic acid helix substrates, which are RNA helix R*/R, DNA helix D*/D, RNA–DNA hybrids with longer RNA or DNA strand (D/R* or R*/D, as illustrated in Figure 5A–D, upper panels), respectively. Each of the four different helix substrates was incubated with MBP–VP35 and subjected to the standard helix-unwinding assay. Our results showed that VP35 could unwind RNA helix R*/R (Figure 5A) and RNA–DNA hybrid D/R* (Figure 5C), both of which have longer RNA strands. On the other hand, VP35 could not unwind DNA helix D*/D (Figure 5B) or RNA–DNA hybrid R*/D (Figure 5D) that have longer DNA strand. This finding indicates that the helix-unwinding activity of EBOV VP35 requires the presence of protruded single-stranded RNA in the helix substrates, which is consistent with its potential roles in the RNA replication and/or transcription of an RNA virus.
Figure 5.
EBOV VP35 unwinds RNA-protruded nucleic acid helices. (A–D) MBP–VP35 (20 pmol) was reacted with 0.1 pmol of standard RNA helix (R*/R) (A), DNA helix (D*/D) (B), or DNA/RNA hybrid with longer RNA strand (D/R*) (C) or longer DNA strand (R*/D) (D). For (A–D), schematic illustrations of the helix substrates are illustrated in the upper panels. Asterisks indicate the HEX-labeled strand.
The NTPase activity is required for the helix-unwinding but not dsRNA-binding activity of VP35
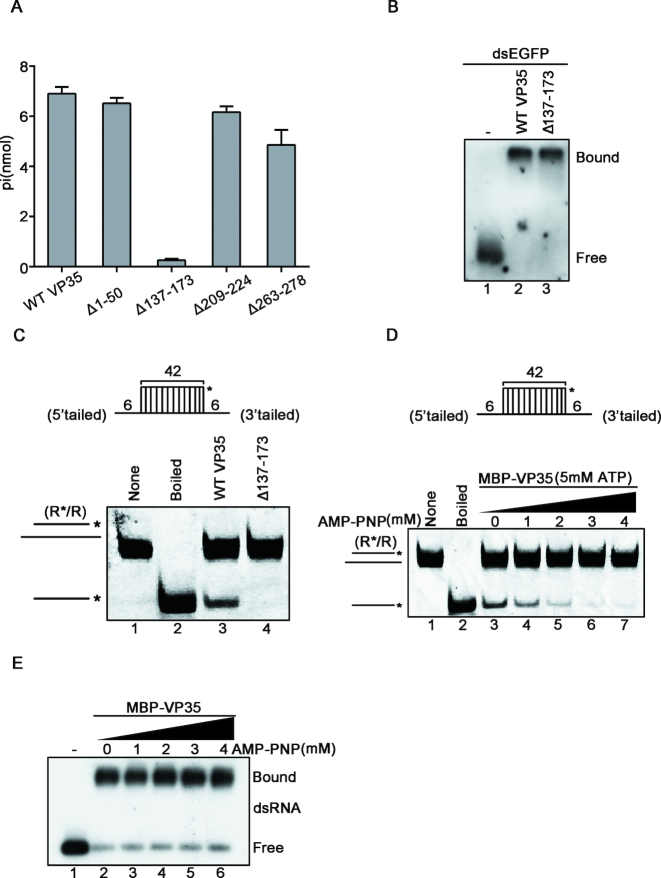
Because the RNA helix-unwinding activity of EBOV VP35 is NTP-dependent, we sought to determine the region responsible for the NTPase activity within VP35. Our data showed that VP35 mutant with the deletion of a.a. 137–173 (Δ137–173) failed to hydrolyze ATP (Figure 6A), indicating that this region, which is highly conserved within multiple filoviruses (Supplementary Figure S7), is crucial for the ATPase of VP35. Furthermore, we examined the helix-unwinding and dsRNA-binding activities of the VP35 deletion mutant, and found that while MBP–VP35Δ137–173 retained its dsRNA binding activity (Figure 6B), it failed to unwind RNA helix even in the presence of ATP (Figure 6C), ruling out the possibility that the observed NTPase and helicase-like activities are caused by unknown contaminant via dsRNA.
Figure 6.
The NTPase activity of EBOV VP35 is required for its helix unwinding. (A) 20 pmol MBP-fusion WT, Δ1–50, Δ137–173, Δ209–224 and Δ263–278 with 2.5 mM ATP, respectively. The NTPase activity of each protein was measured as nanomoles of released inorganic phosphate by using a sensitive colorimetric assay. (B) 0.1 pmol DIG-labeled dsRNA substrate was incubated with 20 pmol of each indicated protein, and the complex was analyzed by gel electrophoresis, transferred to membranes and then incubated with anti-DIG antibody conjugated with alkaline phosphatase. Protein-bound and free RNA strands are indicated. (C) Upper panel: Schematic illustration of the standard RNA helix substrate (R*/R); asterisks indicate the HEX-labeled strand. Lower panel: The RNA helix unwinding assays were performed by incubating 0.1 pmol standard helix substrate with 20 pmol of each indicated protein. (D) MBP–VP35 (20 pmol) was reacted with 0.1 pmol of standard RNA helix (R*/R) in the presence of 5 mM ATP with increasing concentrations of AMP–PNP. (E) 0.1 pmol DIG-labeled dsRNA substrate was incubated with 20 pmol of MBP–VP35 in the presence of increasing concentrations of AMP–PNP, and the complex was analyzed by gel electrophoresis, transferred to membranes and then incubated with anti-DIG antibody conjugated with alkaline phosphatase.
In addition, we further assessed the relation of the NTPase activity with the dsRNA-binding and helix-unwinding activities of VP35 by specifically blocking the NTPase activity of VP35 using AMP–PNP, the non-hydrolysable ATP analog. Consistent with our data by using MBP–VP35Δ137–173, blocking the NTPase via AMP–PNP treatment efficiently blocked the helix-unwinding activity of MBP–VP35 in a dose-dependent manner (Figure 6D), but did not inhibit the dsRNA-binding activity (Figure 6E). Together, our data show that the NTPase activity is required for the helix-unwinding activity but not the dsRNA-binding activity of EBOV VP35.
The RNA-binding activity of VP35 is required for its helix-unwinding activity
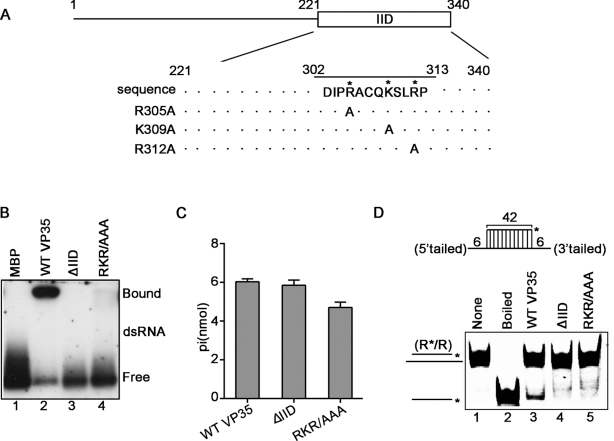
The RNA-binding activity is important for the RNA helix-unwinding activity of helicase. We sought to assess whether the dsRNA-binding activity of VP35 can affect its RNA helix-unwinding activity. To this end, two VP35 mutants were constructed, including a deletion mutant that deletes the dsRNA-binding domain (named as interferon inhibitory domain, IID) of VP35 (ΔIID) (49–51) and the point mutant (RKR/AAA) that has its three critical dsRNA-binding sites (R305, K309 and R312) being mutated to alanine (Figure 7A; Supplementary Figure S8)(50). These mutant proteins were then expressed and purified as recombinant MBP-fusion proteins (Supplementary Figure S9). Our results showed that both ΔIID and RKR/AAA MBP–VP35 mutants completely lost their dsRNA-binding activities (Figure 7B). Moreover, to examine if VP35 can bind to EBOV-specific dsRNA, we ectopically expressed WT or RKR/AAA mutant Flag-VP35 in 293T cells together with EBOV 3′-UTR1–200 dsRNA, followed by RNA-IP using anti-Flag antibody. Our data show that WT but not mutant Flag-VP35 could bind to EBOV 3′-UTR1–200 dsRNA (Supplementary Figure S10).
Figure 7.
The dsRNA-binding sites of EBOV VP35 are required for its helix unwinding. (A) Schematic illustration of the IID and critical dsRNA-binding residues in VP35. Asterisks indicate the sites that are replaced with alanine. (B) 0.1 pmol DIG-labeled dsRNA substrate was incubated with 20 pmol of each indicated protein, and the complex was analyzed by gel electrophoresis, transferred to membranes and then incubated with anti-DIG antibody conjugated with alkaline phosphatase. Protein-bound and free RNA strands are indicated. (C) 20 pmol MBP-fusion WT, ΔIID or RKR/AAA VP35 was reacted with 2.5 mM ATP, respectively. The NTPase activity of each protein was measured as nanomoles of released inorganic phosphate by using a sensitive colorimetric assay. (D) Upper panel: Schematic illustration of the standard RNA helix substrate (R*/R); asterisks indicate the HEX-labeled strand. Lower panel: The RNA helix unwinding assays were performed by incubating 0.1 pmol standard helix substrate with 20 pmol of each indicated protein.
In addition, we examined the RNA helix-unwinding and NTPase activities of the dsRNA-binding-defective mutant (ΔIID or RKR/AAA) VP35. Our results show that VP35 mutants retained their NTPase activities (Figure 7C). On the other hand, MBP–VP35WT can unwinds RNA helix substrate, while the MBP–VP35 mutants failed to do so (Figure 7D).
Together with the data presented previously (Figure 6), our results show that the dsRNA-binding and NTPase activities of VP35 are independent with each other, while the helix-unwinding activity requires both dsRNA-binding and ATPase activities, further confirming that the observed helicase-like and ATPase activities are VP35-specific.
GuHCl inhibits the ATPase and RNA helix-unwinding activities of VP35
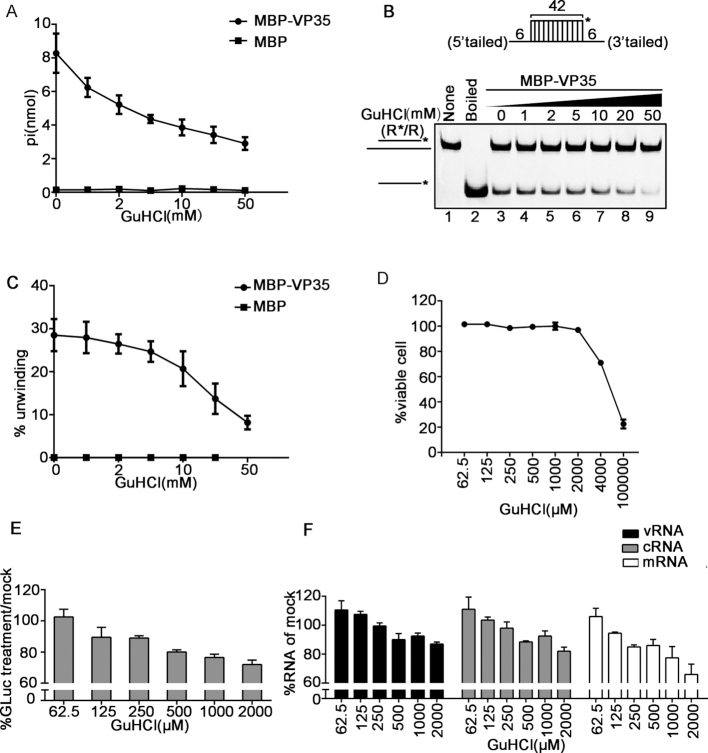
Previous studies reported that GuHCl can inhibit the NTPase and helix-unwinding activities of some helicases, such as EV71 2C and human norovirus NS3, as well as the RNA replication of enterovirus and norovirus. Thus, we sought to assess whether the NTPase and helicase-like activities of EBOV VP35 can also be inhibited by this compound. To this end, the ATPase and helix-unwinding assays were conducted in the presence of MBP–VP35 and increasing concentrations of GuHCl. Our data showed that GuHCl could inhibit the ATP hydrolysis (Figure 8A) and the RNA helix-unwinding by VP35 in a dose-dependent manner (Figure 8B and C). It is worth to note that the inhibitory effects of GuHCl on the ATPase and helicase-like activities of EBOV VP35 are relatively moderate, similarly with the observed effects on EV71 2C and norovirus NS3 (22,34). Altogether, GuHCl is an inhibitor of the NTPase and helicase-like activities of EBOV VP35 in vitro.
Figure 8.
GuHCl inhibits the ATPase and RNA helix unwinding activities of VP35, and the replication/transcription of EBOV minigenome. (A) The ATPase activity of MBP–VP35 was performed in the presence of increasing concentrations of GuHCl. MBP protein alone was used as the negative-control. (B) MBP–VP35 (20 pmol) was reacted with 0.1 pmol standard RNA helix (as illustrated in the upper panel) in the presence of increasing concentrations of GuHCl. (C) The unwinding activities in (B) were plotted as percentages of the released HEX-labeled RNA from the total RNA helix (Y-axis) at indicated GuHCl concentrations (X-axis). MBP protein alone was used as the negative-control. (D–F) The cells harbouring EBOV minigenome replicon were treated with indicated concentrations of GuHCl (X-axis) for 96 h. The number of viable cells was determined and the cell viability (Y-axis) was calculated as a percentage of that of the mock-treated cells (D). GLuc activities (E) and RNA levels (F) were measured at 96 h post-treatment of GuHCl with the indicated concentrations. Minigenomic viral RNA (vRNA), complementary vRNA (cRNA, replication intermediate), and viral mRNA (mRNA). Values (Y-axis) were expressed as percentages of those of mock-treated cells.
GuHCl inhibits the replication/transcription of EBOV minigenome replicon
Helicase activities are important for the replication of diverse viruses (52), thus it would be intriguing to determine whether GuHCl can also inhibit the replication of EBOV. Because the live virus experiments of EBOV can only be conducted in the biosafety level 4 (BSL-4) facilities, EBOV minigenome (MG) replicon systems are usually used to study EBOV replication under common BSL-2 conditions (53,54). To this end, we utilized a stable EBOV minigenome replicon system (EBOV replicon cells) that can stably replicate and transcribe the EBOV minigenomic RNA [the Gaussia luciferase (GLuc)-encoding ORF was flanked by the minimal cis-elements located at the two ends of EBOV genome] in Huh7 cells stably expressing NP, VP35, VP30, and L (41). The EBOV replicon cells were treated with GuHCl at the indicated concentrations for 96 h. After that, we examined the cell viability at the doses we applied, and found no obvious cytotoxicity until the concentration of GuHCl up to 2 mM (Figure 8D). And 1 mM GuHCl did not affect the expression levels of NP, VP30, VP35 and β-actin (Supplementary Figure S11A). Then, the replication and transcription of the EBOV minigenome replicon were examined by measuring the levels of GLuc activity, minigenomic viral RNA (vRNA), cRNA (complementary to vRNA; replication intermediate), and viral mRNA. Our data show that both the replication and transcription of the EBOV minigenome were inhibited by GuHCl treatment in a dose-dependent manner (Figure 8E and F).
We further examined whether the effect of GuHCl is specific to the NTPase and helicase-like activities of VP35 or due to some general denaturing effect. EBOV VP35 has been previous reported to antagonize interferon (IFN) response and be a viral suppressor of RNA interference (VSR) (30,43,44). Our data show that GuHCl had little effect on IFN response in Huh7 cells with or without EBOV minigenome (Supplementary Figure S11B) and did not affect the VSR activity of VP35 (Supplementary Figure S11C). Moreover, the effect of GuHCl to Zika virus (ZIKV) NS3, which is a well-known monomeric RNA helicase (23), and found that GuHCl affected neither the helicase activity of ZIKV NS3 in vitro nor the replication of ZIKV in 293T cells (Supplementary Figure S12). These data confirm that the effects of GuHCl to EBOV VP35 and EBOV replicon are specific.
Of note, the minigenome replication/transcription seemed be more sensitive to GuHCl treatment than the biochemical activities of VP35. It is possible that because EBOV minigenomic RNA replication is a multi-round process, a slight inhibition in each round can result in an accumulated effect. Besides, when comparing with transiently transfected minigenome system, this stable EBOV minigenome replicon has been reported to show high level of active minigenome replication, and be relatively stable and even insensitive to IFN treatment or RNA interference, probably due to the persistent replication and the strong viral RNP stability of this minigenome (41). Therefore, the inhibitory effect of GuHCl on the replication and transcription of EBOV minigenome is quite remarkable. In conclusion, our results show that GuHCl is able to inhibit the replication and transcription of the EBOV minigenome replicon in cultured human cells at the concentrations without apparent cytotoxicity.
DISCUSSION
RNA remodelling proteins, including RNA helicases and RNA chaperones, are generally believed to play critical roles in every cellular process involving RNAs by facilitating the correct folding and refolding of RNA molecules (18,55). Herein we report for the first time that the EBOV-encoded multifunctional protein VP35 has NTPase and helicase-like activities that can bind to NTPs, hydrolyze all types of NTPs and unwind RNA helices in an NTP-dependent manner. Moreover, our work shows that GuHCl, an FDA-approved small molecule drug, is able to inhibit the NTPase/helicase-like activities of VP35 in vitro, and more importantly, the replication and transcription of a stable EBOV minigenome replicon in cultured human cells.
VP35 has been reported to play pivotal roles in multiple processes of filoviral replication and pathogenesis. And it is intriguing to ask how the NTPase/helicase-like activity of VP35 functions in the life cycle of EBOV. VP35 is a cofactor of polymerase L and an essential component of filoviral RNP complex, which carries out the EBOV RNA replication and transcription. Thus, it is plausible that VP35 can work together with NP and L to mediate the unwinding of dsRNA intermediates produced during viral RNA replication/transcription, and/or can aid the rearrangement of the cis-acting elements within EBOV mRNA and genomic/antigenomic RNAs, thereby facilitating the transcription, translation and encapsidation of EBOV RNAs or allowing viral RNAs to switch among distinct processes. However, owing to technical limitations, virus-encoded RNA remodeling activities are usually studied in vitro, and it is mostly infeasible to measure the exact roles of RNA helicases on the structures and functionalities of viral RNAs in infected cells (56). Indeed, GuHCl, an FDA-approved small compound and well-known inhibitor of certain viral helicases, including enterovirus 2C and norovirus NS3, is able to inhibit the NTPase/helicase-like activity of EBOV VP35 as well as EBOV minigenome replication/transcription in cells, implying the functional importance of the VP35-associated RNA remodeling activity in EBOV replication. Future advances in techniques may overcome the technical barriers and provide mechanistic and dynamic views of how VP35 remodels viral RNA molecules at different processes of EBOV life cycle.
According to the conventional view, helicases contain NTPase activity, utilize the energy of ATP binding and hydrolysis to melt nucleic acid base-pairings, and are thought to participate in most ATP-dependent rearrangements of structured RNAs. Helicases are generally classified into six superfamilies (SFs), designated SF1 to SF6, on the basis of conserved helicase motifs. Interestingly, although EBOV VP35 does not contain any conventional helicase motifs (57,58), the NTPase/helicase-like activities of VP35 have most basic biochemical characteristics of canonical RNA helicases, including the NTP-dependency, the requirement of divalent metallic ion, and the directionality of helix unwinding, leading to the question whether the canonical conserved motifs are not necessary for a protein to possess the NTPase and RNA remodeling activities. Factually, Ectropis obliqua picorna-like virus (EoV) 2C protein contains the conserved motif A, which is commonly recognized as the core NTP binding and NTPase active site of SF3 helicases (21), but the NTPase activity of EoV 2C is not dependent on this motif. In addition, the capsid protein VP5 of Helicoverpa armigera cypovirus-5 (HaCPV-5; genus Cypovirus, family Reoviridae) has the NTPase activity, which is also Mg2+-dependent and can hydrolyze all kinds of NTPs and dNTPs, but contains no conserved NTPase/helicase motifs (27,47). Thus, our current study, together with the previous studies, imply that some of the NTPase and RNA remodeling activities may not strictly rely on conserved motifs in linear sequences of amino acids, but probably determined by more sophisticated active sites formed in tertiary protein structures. In the current study, we have determined that the middle aa. 137–173 region of EBOV VP35 is critical for its NTPase activity. This middle region does not belong to the N-terminal oligomerization domain and the C-terminal IID, both of whose structures have been resolved (45,50). Interestingly, although the structure of the VP35 middle region has not been determined yet, the sequence alignment of VP35 proteins from multiple filoviruses, including Zaire ebolavirus (EBOV), Sudan ebolavirus (SUDV), Reston ebolavirus (RESTV), MARV, and Lloviu virus (LLOV) shows that this region is highly conserved within Filoviridae (Supplementary Figure S7A). And our predicted secondary structure from PSI-blast based secondary structure prediction (PSIPRED) revealed that this region contains a β-strand and an α-helix (Supplementary Figure S7B) (59,60). Besides, the motif scan by using the PROSITE profiles via the MyHits (https://myhits.isb-sib.ch/cgi-bin/motif_scan) suggested that the amino acid T149 of EBOV VP35 is a possible protein kinase C (PKC) phosphorylation site (61). These analyses highlight the importance of this region in the structure and function of filoviral VP35s. Future studies should reveal whether the β-strand and α-helix as well as the putative PKC phosphorylation site are critical for the NTPase/helicase-like activity of VP35.
An interesting question raised by the current study is that: if RNA remodelling activities are so important to viruses, why helicase or helicase-like activity has never been found in NNSVs before? Indeed, numerous, if not all, DNA viruses and positive-sense RNA viruses have been found to encode their own helicases, which share the conserved NTPase/helicase signature motifs with various host helicases of diverse organisms ranging from prokaryotes to eukaryotes (62,63). Based on the conserved motifs, putative helicases can be predicted by bioinformatic algorithms before being biochemically confirmed and characterized. And the lack of conserved helicase signature motifs within EBOV VP35 should be the reason why the NTPase/helicase-like activities were not predicted and identified previously. Moreover, the high conservation of helicase signature motifs suggests that various viral and host helicases share similar origins. On the other hand, the NTPase/helicase-like activities of EBOV VP35 should be evolved independently, suggesting that filoviruses (and probably other NNSVs) have an earlier evolutionary divergence with positive-sense RNA viruses before the latter ‘acquire’ canonical helicases. More importantly, in the absence of a canonical helicase, EBOV manages to obtain a helicase-like activity instead, highlighting its functional necessity to EBOV. Given that all the members of Filoviridae encode VP35 proteins, which are functionally analogous to the P proteins of other NNSVs (5), future studies by us and others should investigate whether the NTPase/helicase activities are general to other filoviral VP35 as well as NNSV P proteins.
Virus-encoded helicases have long been considered as potential targets for antivirals due to their importance in viral replication (35). Previous studies have reported that GuHCl can effectively inhibit the helicase activities of enterovirus 2C and human norovirus NS3, as well as the replication of enterovirus and human norovirus (22,64–66). Similarly, using the stable EBOV minigenome as the model, we also found that the helicase-like activity of VP35 and the replication of EBOV minigenome in cells can be significantly inhibited with negligible cell toxicity by GuHCl. Guanidines are ubiquitously present in nature, and can function as mediators of specific non-covalent binding in various catalytic processes (64,67). Due to the physicochemical characteristics, guanidine derivatives have been used for drug development against diverse diseases, some of which are potential antivirals against hepatitis C virus (HCV), human immunodeficiency virus (HIV), and flaviviruses (64,67–69), and GuHCl is a U.S. FDA-approved small compound drug for the treatment of Eaton-Lambert syndrome (36,37). Considering the presence of multiple guanidine derivatives, the modification and screening of various guanidine-containing compounds can probably identify antiviral compounds with better inhibitory effects on EBOV VP35 and EBOV replication.
In conclusion, this study provides the first demonstration of the NTPase/helicase-like activity associated with an NNSV-encoded protein, EBOV VP35, and finds that GuHCl is an inhibitor of not only the in vitro NTPase and helicase-like activities of EBOV VP35, but also the replication/transcription of EBOV minigenome in human cells. These findings uncover an unexpected novel function of EBOV VP35, extend the view of RNA remodelling proteins, provide the evidence that guanidine-containing compounds may serve as potential candidates for anti-EBOV/filovirus drug development by targeting VP35, and shed light on the understanding of EBOV and filovirus replication and pathogenesis.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to thank Dr Cheng-Feng Qin (Beijing, China) for providing reagents and Dr Jie Cui (Wuhan, China) for helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Strategic Priority Research Program of Chinese Academy of Sciences [XDPB0301 to X.Z.]; National Science and Technology Major Project [2018ZX10101004 to X.Z.]; National Natural Science Foundation of China [31761130075, 31670161 to X.Z.]; Newton Advanced Fellowship from the Academy of Medical Sciences, UK [NAF005\1002 to X.Z.]; Science and Technology Bureau of Wuhan [2018060401011309 to X.Z.]; Advanced Customer Cultivation Project of Wuhan National Biosafety Laboratory [2018ACCP-MS11 to Y.Q.]. Funding for open access charge: National Natural Science Foundation of China [31761130075, 31670161 to X.Z.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Feldmann H., Geisbert T.W.. Ebola haemorrhagic fever. Lancet. 2011; 377:849–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gatherer D. The 2014 Ebola virus disease outbreak in West Africa. J. Gen. Virol. 2014; 95:1619–1624. [DOI] [PubMed] [Google Scholar]
- 3. Baize S., Pannetier D., Oestereich L., Rieger T., Koivogui L., Magassouba N., Soropogui B., Sow M.S., Keita S., De Clerck H. et al.. Emergence of Zaire Ebola virus disease in Guinea. N. Engl. J. Med. 2014; 371:1418–1425. [DOI] [PubMed] [Google Scholar]
- 4. Liu Y., Sun Y., Wu W., Li A., Yang X., Zhang S., Li C., Su Q., Cai S., Sun D. et al.. Serological Investigation of Laboratory-Confirmed and Suspected Ebola Virus Disease Patients During the Late Phase of the Ebola Outbreak in Sierra Leone. Virol. Sin. 2018; 33:323–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jamin M., Yabukarski F.. Nonsegmented negative-sense RNA viruses-structural data bring new insights into nucleocapsid assembly. Adv. Virus Res. 2017; 97:143–185. [DOI] [PubMed] [Google Scholar]
- 6. Ortin J., Martin-Benito J.. The RNA synthesis machinery of negative-stranded RNA viruses. Virology. 2015; 479–480:532–544. [DOI] [PubMed] [Google Scholar]
- 7. Muhlberger E. Filovirus replication and transcription. Future Virol. 2007; 2:205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ning Y.J., Deng F., Hu Z., Wang H.. The roles of ebolavirus glycoproteins in viral pathogenesis. Virol. Sin. 2017; 32:3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rougeron V., Feldmann H., Grard G., Becker S., Leroy E.M.. Ebola and Marburg haemorrhagic fever. J. Clin. Virol. 2015; 64:111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Luthra P., Jordan D.S., Leung D.W., Amarasinghe G.K., Basler C.F.. Ebola virus VP35 interaction with dynein LC8 regulates viral RNA synthesis. J .Virol. 2015; 89:5148–5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Prins K.C., Binning J.M., Shabman R.S., Leung D.W., Amarasinghe G.K., Basler C.F.. Basic residues within the ebolavirus VP35 protein are required for its viral polymerase cofactor function. J. Virol. 2010; 84:10581–10591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kirchdoerfer R.N., Moyer C.L., Abelson D.M., Saphire E.O.. The Ebola virus VP30-NP interaction is a regulator of viral RNA synthesis. PLoS Pathog. 2016; 12:e1005937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kirchdoerfer R.N., Abelson D.M., Li S., Wood M.R., Saphire E.O.. Assembly of the Ebola virus nucleoprotein from a chaperoned VP35 complex. Cell Rep. 2015; 12:140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mehedi M., Hoenen T., Robertson S., Ricklefs S., Dolan M.A., Taylor T., Falzarano D., Ebihara H., Porcella S.F., Feldmann H.. Ebola virus RNA editing depends on the primary editing site sequence and an upstream secondary structure. PLoS Pathog. 2013; 9:e1003677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sugai A., Sato H., Yoneda M., Kai C.. Gene end-like sequences within the 3′ non-coding region of the Nipah virus genome attenuate viral gene transcription. Virology. 2017; 508:36–44. [DOI] [PubMed] [Google Scholar]
- 16. Lorsch J.R. RNA chaperones exist and DEAD box proteins get a life. Cell. 2002; 109:797–800. [DOI] [PubMed] [Google Scholar]
- 17. Woodson S.A. Taming free energy landscapes with RNA chaperones. RNA Biol. 2010; 7:677–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Musier-Forsyth K. RNA remodeling by chaperones and helicases. RNA Biol. 2010; 7:632–633. [DOI] [PubMed] [Google Scholar]
- 19. Rajkowitsch L., Chen D., Stampfl S., Semrad K., Waldsich C., Mayer O., Jantsch M.F., Konrat R., Blasi U., Schroeder R.. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 2007; 4:118–130. [DOI] [PubMed] [Google Scholar]
- 20. Bleichert F., Baserga S.J.. The long unwinding road of RNA helicases. Mol. Cell. 2007; 27:339–352. [DOI] [PubMed] [Google Scholar]
- 21. Cheng Z., Yang J., Xia H., Qiu Y., Wang Z., Han Y., Xia X., Qin C.F., Hu Y., Zhou X.. The nonstructural protein 2C of a Picorna-like virus displays nucleic acid helix destabilizing activity that can be functionally separated from its ATPase activity. J. Virol. 2013; 87:5205–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li T.F., Hosmillo M., Schwanke H., Shu T., Wang Z., Yin L., Curry S., Goodfellow I.G., Zhou X.. Human Norovirus NS3 Has RNA Helicase and Chaperoning Activities. J. Virol. 2018; 92:e01606-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jain R., Coloma J., Garcia-Sastre A., Aggarwal A.K.. Structure of the NS3 helicase from Zika virus. Nat. Struct. Mol. Biol. 2016; 23:752–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lam A.M., Keeney D., Eckert P.Q., Frick D.N.. Hepatitis C virus NS3 ATPases/helicases from different genotypes exhibit variations in enzymatic properties. J. Virol. 2003; 77:3950–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Karpe Y.A., Aher P.P., Lole K.S.. NTPase and 5′-RNA triphosphatase activities of Chikungunya virus nsP2 protein. PLoS One. 2011; 6:e22336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee N.R., Kwon H.M., Park K., Oh S., Jeong Y.J., Kim D.E.. Cooperative translocation enhances the unwinding of duplex DNA by SARS coronavirus helicase nsP13. Nucleic Acids Res. 2010; 38:7626–7636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang J., Cheng Z., Zhang S., Xiong W., Xia H., Qiu Y., Wang Z., Wu F., Qin C.F., Yin L. et al.. A cypovirus VP5 displays the RNA chaperone-like activity that destabilizes RNA helices and accelerates strand annealing. Nucleic Acids Res. 2014; 42:2538–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kadare G., Haenni A.L.. Virus-encoded RNA helicases. J. Virol. 1997; 71:2583–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang J., Xia H., Qian Q., Zhou X.. RNA chaperones encoded by RNA viruses. Virol. Sin. 2015; 30:401–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cardenas W.B., Loo Y.M., Gale M. Jr., Hartman A.L., Kimberlin C.R., Martinez-Sobrido L., Saphire E.O., Basler C.F.. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J. Virol. 2006; 80:5168–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kimberlin C.R., Bornholdt Z.A., Li S., Woods V.L. Jr., MacRae I.J., Saphire E.O.. Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:314–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Leung D.W., Prins K.C., Basler C.F., Amarasinghe G.K.. Ebolavirus VP35 is a multifunctional virulence factor. Virulence. 2010; 1:526–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reid S.P., Cardenas W.B., Basler C.F.. Homo-oligomerization facilitates the interferon-antagonist activity of the ebolavirus VP35 protein. Virology. 2005; 341:179–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xia H., Wang P., Wang G.C., Yang J., Sun X., Wu W., Qiu Y., Shu T., Zhao X., Yin L. et al.. Human enterovirus nonstructural protein 2CATPase functions as both an RNA helicase and ATP-independent RNA chaperone. PLoS Pathog. 2015; 11:e1005067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pfister T., Wimmer E.. Characterization of the nucleoside triphosphatase activity of poliovirus protein 2C reveals a mechanism by which guanidine inhibits poliovirus replication. J. Biol. Chem. 1999; 274:6992–7001. [DOI] [PubMed] [Google Scholar]
- 36. Oh S.J., Lee Y.W., Rutsky E.. Eaton-Lambert syndrome: reflex improvement with guanidine. Arch. Phys. Med. Rehabil. 1977; 58:457–459. [PubMed] [Google Scholar]
- 37. Oh S.J., Kim K.W.. Guanidine hydrochloride in the Eaton-Lambert syndrome. Electrophysiologic improvement. Neurology. 1973; 23:1084–1090. [DOI] [PubMed] [Google Scholar]
- 38. Wang Q., Han Y., Qiu Y., Zhang S., Tang F., Wang Y., Zhang J., Hu Y., Zhou X.. Identification and characterization of RNA duplex unwinding and ATPase activities of an alphatetravirus superfamily 1 helicase. Virology. 2012; 433:440–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Qi N., Cai D., Qiu Y., Xie J., Wang Z., Si J., Zhang J., Zhou X., Hu Y.. RNA binding by a novel helical fold of B2 protein from wuhan nodavirus mediates the suppression of RNA interference and promotes B2 dimerization. J. Virol. 2011; 85:9543–9554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dai S., Zhang T., Zhang Y., Wang H., Deng F.. Zika virus baculovirus-expressed virus-like particles induce neutralizing antibodies in mice. Virol. Sin. 2018; 33:213–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tao W., Gan T., Guo M., Xu Y., Zhong J.. Novel stable Ebola virus minigenome replicon reveals remarkable stability of the viral genome. J. Virol. 2017; 91:e01316-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Qiu Y., Xu Y., Zhang Y., Zhou H., Deng Y.Q., Li X.F., Miao M., Zhang Q., Zhong B., Hu Y. et al.. Human virus-derived small RNAs can confer antiviral immunity in mammals. Immunity. 2017; 46:992–1004. [DOI] [PubMed] [Google Scholar]
- 43. Basler C.F., Wang X., Muhlberger E., Volchkov V., Paragas J., Klenk H.D., Garcia-Sastre A., Palese P.. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:12289–12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Haasnoot J., de Vries W., Geutjes E.J., Prins M., de Haan P., Berkhout B.. The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog. 2007; 3:e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ramaswamy V.K., Di Palma F., Vargiu A.V., Corona A., Piano D., Ruggerone P., Zinzula L., Tramontano E.. Insights into the homo-oligomerization properties of N-terminal coiled-coil domain of Ebola virus VP35 protein. Virus Res. 2018; 247:61–70. [DOI] [PubMed] [Google Scholar]
- 46. Chanthamontri C.K., Jordan D.S., Wang W., Wu C., Lin Y., Brett T.J., Gross M.L., Leung D.W.. The Ebola viral protein 35 N-terminus is a parallel tetramer. Biochemistry. 2019; 58:657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yang J., Qian Q., Li T.F., Yang X., Won S.J., Zhou X.. Cypovirus capsid protein VP5 has nucleoside triphosphatase activity. Virol. Sin. 2017; 32:328–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Frick D.N., Banik S., Rypma R.S.. Role of divalent metal cations in ATP hydrolysis catalyzed by the hepatitis C virus NS3 helicase: magnesium provides a bridge for ATP to fuel unwinding. J. Mol. Biol. 2007; 365:1017–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Leung D.W., Ginder N.D., Fulton D.B., Nix J., Basler C.F., Honzatko R.B., Amarasinghe G.K.. Structure of the Ebola VP35 interferon inhibitory domain. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:411–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Leung D.W., Prins K.C., Borek D.M., Farahbakhsh M., Tufariello J.M., Ramanan P., Nix J.C., Helgeson L.A., Otwinowski Z., Honzatko R.B. et al.. Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35. Nat. Struct. Mol. Biol. 2010; 17:165–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zinzula L., Esposito F., Pala D., Tramontano E.. dsRNA binding characterization of full length recombinant wild type and mutants Zaire ebolavirus VP35. Antiviral Res. 2012; 93:354–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lam A.M., Frick D.N.. Hepatitis C virus subgenomic replicon requires an active NS3 RNA helicase. J. Virol. 2006; 80:404–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hoenen T., Feldmann H.. Reverse genetics systems as tools for the development of novel therapies against filoviruses. Expert Rev. Anti Infect. Ther. 2014; 12:1253–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hoenen T., Groseth A., de Kok-Mercado F., Kuhn J.H., Wahl-Jensen V.. Minigenomes, transcription and replication competent virus-like particles and beyond: reverse genetics systems for filoviruses and other negative stranded hemorrhagic fever viruses. Antiviral Res. 2011; 91:195–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pyle A.M. RNA helicases and remodeling proteins. Curr. Opin. Chem Biol. 2011; 15:636–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jankowsky E. RNA helicases at work: binding and rearranging. Trends Biochem. Sci. 2011; 36:19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Frick D.N. Helicases as antiviral drug targets. Drug News Perspect. 2003; 16:355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Singleton M.R., Dillingham M.S., Wigley D.B.. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007; 76:23–50. [DOI] [PubMed] [Google Scholar]
- 59. Buchan D.W., Minneci F., Nugent T.C., Bryson K., Jones D.T.. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 2013; 41:W349–W357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jones D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999; 292:195–202. [DOI] [PubMed] [Google Scholar]
- 61. Woodgett J.R., Gould K.L., Hunter T.. Substrate specificity of protein kinase C. Use of synthetic peptides corresponding to physiological sites as probes for substrate recognition requirements. Eur. J. Biochem. 1986; 161:177–184. [DOI] [PubMed] [Google Scholar]
- 62. Fairman-Williams M.E., Guenther U.P., Jankowsky E.. SF1 and SF2 helicases: family matters. Curr. Opin. Struct. Biol. 2010; 20:313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gorbalenya A.E., Koonin E.V., Donchenko A.P., Blinov V.M.. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989; 17:4713–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Saczewski F., Balewski L.. Biological activities of guanidine compounds, 2008 - 2012 update. Expert Opin. Ther. Pat. 2013; 23:965–995. [DOI] [PubMed] [Google Scholar]
- 65. Lyons T., Murray K.E., Roberts A.W., Barton D.J.. Poliovirus 5′-terminal cloverleaf RNA is required in cis for VPg uridylylation and the initiation of negative-strand RNA synthesis. J. Virol. 2001; 75:10696–10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rocha-Pereira J., Jochmans D., Debing Y., Verbeken E., Nascimento M.S., Neyts J.. The viral polymerase inhibitor 2′-C-methylcytidine inhibits Norwalk virus replication and protects against norovirus-induced diarrhea and mortality in a mouse model. J. Virol. 2013; 87:11798–11805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Saczewski F., Balewski L.. Biological activities of guanidine compounds. Expert Opin. Ther. Pat. 2009; 19:1417–1448. [DOI] [PubMed] [Google Scholar]
- 68. Wang J., Wang Y., Li Z., Zhan P., Bai R., Pannecouque C., Balzarini J., De Clercq E., Liu X.. Design, synthesis and biological evaluation of substituted guanidine indole derivatives as potential inhibitors of HIV-1 Tat-TAR interaction. Med. Chem. 2014; 10:738–746. [DOI] [PubMed] [Google Scholar]
- 69. Frick D.N. The hepatitis C virus NS3 protein: a model RNA helicase and potential drug target. Curr. Issues Mol. Biol. 2007; 9:1–20. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.