Abstract
Argonaute (Ago) proteins are key players in RNA interference in eukaryotes, where they function as RNA-guided RNA endonucleases. Prokaryotic Argonautes (pAgos) are much more diverse than their eukaryotic counterparts but their cellular functions and mechanisms of action remain largely unknown. Some pAgos were shown to use small DNA guides for endonucleolytic cleavage of complementary DNA in vitro. However, previously studied pAgos from thermophilic prokaryotes function at elevated temperatures, which limits their potential use as a tool in genomic applications. Here, we describe two pAgos from mesophilic bacteria, Clostridium butyricum (CbAgo) and Limnothrix rosea (LrAgo), that act as DNA-guided DNA nucleases at physiological temperatures. In comparison with previously studied pAgos, CbAgo and LrAgo do not show strong preferences for the 5′-nucleotide in guide DNA and can use not only 5′-phosphorylated but also 5′-hydroxyl DNA guides. Both CbAgo and LrAgo can tolerate guide/target mismatches in the seed region, but are sensitive to mismatches in the 3′-guide region. Both pAgos can perform programmable endonucleolytic cleavage of double-stranded DNA substrates, showing enhanced activity at AT-rich regions and at elevated temperatures. The biochemical characterization of mesophilic pAgo proteins paves the way for their use for DNA manipulations both in vitro and in vivo.
INTRODUCTION
Argonaute proteins are an integral part of the eukaryotic RNA interference machinery. They bind small noncoding RNAs and utilize them for guided cleavage of complementary RNA targets or indirect gene silencing by recruiting additional factors (1–3). Argonaute proteins are also found in bacterial and archaeal genomes (4–6). Structural and biochemical studies of a few prokaryotic Ago (pAgo) proteins—predominantly from thermophilic bacterial and archaeal species—showed that they can function as endonucleases in vitro (7–11) and may provide cell defense against foreign genetic elements in vivo (8,12,13). pAgos are thus proposed to act as a bacterial system of innate immunity acting against invasive genetic elements (5,6,12–15).
Prokaryotic Argonaute proteins can be classified into several clades, including long pAgos (further subdivided into two clades, long-A and long-B), short pAgos and PIWI-RE proteins (Supplementary Figure S1A) (4,6,16). All pAgos characterized so far belong to the long pAgo clade and include the N, PAZ, MID and PIWI domains (except for AfAgo that has lost its N and PAZ domains), involved in guide binding, target recognition and catalysis; the same domains are also present in all eukaryotic Argonautes (eAgos). The MID and PAZ domains are responsible for binding of the 5′-end and 3′-end, respectively, of a guide nucleic acid molecule (7,17–21). In contrast to eAgos that use exclusively small RNA guides, the majority of characterized pAgos bind single-stranded DNA (ssDNA) guides, including AaAgo (from Aquifex aeolicus) (10), AfAgo (Archaeoglobus fulgidus) (22), TtAgo (Thermus thermophilus) (13), PfAgo (Pyrococcus furiosus) (8) and MjAgo (Methanocaldococcus jannaschii) (21). RsAgo (Rhodobacter sphaeroides) (12) and MpAgo (Marinitoga piezophila) (7) bind small RNA guides. The specificity towards RNA or DNA guides cannot be inferred from the pAgo sequence and has to be determined experimentally. Binding of the guide to pAgo changes its conformation to expose nucleotides in the seed region (2–8 nt from the guide 5′-end) in solution in an A-form helix to facilitate target recognition (7,20,21). While eAgos universally recognize complementary RNA as a target, studies of a few long pAgos suggested that they employ their guides to bind DNA targets (7,8,11,12,19,22,23). Complementary interactions between the guide and double-stranded DNA (dsDNA) target require unwinding of the DNA helix; however, pAgos lack intrinsic helicase activity, so the molecular mechanism of this step remains to be understood.
The PIWI domain of most long-A pAgos has an RNase H-like fold with the active site containing the DEDX (X = N, D or H) catalytic tetrad, which endows these proteins with endonuclease activity (4,6). Dissociation of the processed target strand after cleavage was shown to be the rate-limiting step in catalysis in eAgos, which could be overcome at increased temperatures in the case of pAgos from thermophilic prokaryotes (19,24–26). Some pAgos such as RsAgo have substitutions in the catalytic tetrad making them deficient in endonuclease activity (12). The active site of the PIWI domain slices the complementary target at a single site, between the tenth (10′) and eleventh (11′) nucleotides starting from the guide 5′-end (7–10,13). In contrast to the Cas9 protein that has two distinct endonuclease domains allowing it to cut both strands of dsDNA upon target recognition, pAgos have only one active site, so only one DNA strand can be cleaved by a single complex. Mismatches between the guide and target molecules in the seed region decrease the target binding rate by previously studied Agos and perturb the slicer activity, while mismatches adjacent to the cleavage site abolish it (9,17,24–27). In addition to guide-dependent processing of ssDNA targets, several thermophilic pAgos were shown to perform slow guide-independent cleavage of dsDNA termed chopping (8,11,28). Short DNAs generated by the chopping activity of pAgos might then be used as guides for further specific target cleavage. Chopping was proposed to facilitate the onset of immunity against foreign DNA (11,28).
pAgos are programmable endonucleases that may potentially be used as a tool for DNA manipulations in vitro and in vivo, including molecular cloning and genome editing applications (14,16,29). However, catalytically active pAgos characterized to date were isolated from thermophilic species and function at elevated temperatures. Furthermore, it is not clear if pAgos are able to process DNA substrates at moderate temperature without additional partners due to their inability to unwind dsDNA. These concerns have cast doubt on the practicality of pAgos as a tool for DNA manipulation (30,31). Here, we describe two Argonaute proteins from mesophilic prokaryotes, bacillus Clostridium butyricum (CbAgo) and cyanobacterium Limnothrix rosea (LrAgo). We show that both pAgos are DNA-guided DNA nucleases that function at much lower temperatures than other pAgos studied to date. We characterize activities of CbAgo and LrAgo under a wide range of conditions and reveal functional differences in the efficiency and fidelity of DNA processing by the two proteins. Finally, we demonstrate that CbAgo and LrAgo can perform precise guide-dependent cleavage of dsDNA when supplied with two guides targeting both strands of the dsDNA target. Discovery of programmable pAgo endonucleases that are able to process DNA targets at moderate temperatures opens the way for their use as a tool in DNA technology.
MATERIALS AND METHODS
Protein expression and purification
Nucleotide sequences of CbAgo (WP_058142162.1; C. butyricum strain TK520) and LrAgo (WP_075892274.1; L. rosea strain IAM M-220) were codon-optimized using IDT Codon Optimization Tool for expression in E. coli, the genes were synthesized by the IDT core facility and cloned into p-ET28b expression vectors in frame with the N-terminal His6 tag. Catalytically dead mutants were obtained by site-directed mutagenesis using QuikChange Lightning Multi mutagenesis kit (Agilent). Expression was carried out as described in ref. (32) with minor modifications. Briefly, E. coli BL21(DE3) cells carrying the expression plasmid were adapted to high ionic strength conditions by overnight cultivation in the LBN medium at 37°C. The cells were transferred into fresh LBN (1:500 inoculation) supplemented with 1 mM betaine and aerated at 37°C until OD600 0.6. The cultures were cooled down to 18°C, induced with 0.25 mM IPTG and aerated for 12 h at 18°C. The cells were collected by centrifugation and stored at –80°C for further protein purification.
Cell pellet was resuspended in Ni-NTA chromatography buffer A (50 mM Tris–HCl, 0.5 M NaCl, 20 mM imidazole, 5% glycerol, 1 mM TCEP pH 7.5) supplemented with EDTA-free protease inhibitor cocktail (Roche) and disrupted by a high-pressure homogenizer at 18000 psi. The lysate was clarified by centrifugation at 32000 g for 30 min and the supernatant was loaded onto a HisTrap HP column (GE Healthcare). The column was washed extensively with buffer A, then with buffer A containing 60 mM imidazole, and the proteins were eluted with buffer A containing 300 mM imidazole. Fractions containing pAgos were concentrated by ultrafiltration using an Amicon 50K filter unit (Millipore) and purified on a Superose 6 10/300 GL column (GE Healthcare) equilibrated with buffer GF (10 mM HEPES–NaOH pH 7.0, 0.5 M NaCl, 5% glycerol, 1 mM DTT). Fractions containing pAgos were pooled and loaded onto a Heparin FF column (GE Healthcare) equilibrated with buffer GF, washed with at least 10 column volumes of the same buffer and eluted with a linear NaCl gradient (0.5–1 M). Samples containing CbAgo and LrAgo (eluted at 650–700 mM NaCl) were aliquoted and flash-frozen in liquid nitrogen. The purity of the final protein samples was assessed by denaturing PAGE followed by silver staining. Control experiments demonstrated that the purified pAgo proteins are essentially free of associated nucleic acids. The protein concentration was determined by the Qubit protein assay kit (Thermo Fischer Scientific).
Nucleic acid cleavage assays
Most cleavage assays were performed at the 5:2:1 pAgo:guide:target molar ratio at 37°C, unless otherwise indicated. All guide and target oligonucleotide sequences are shown in Supplementary Table S1. 500 nM CbAgo or LrAgo was mixed with 200 nM guide DNA in reaction buffer containing 10 mM HEPES–NaOH pH 7.0, 100 mM NaCl, 5% glycerol and 5 mM MnCl2, and incubated at 37°C for 10 min for guide loading. Target DNA was added to 100 nM. The reaction was stopped after indicated time intervals by mixing the samples with equal volumes of stop solution (8 M urea, 20 mM EDTA, 0.005% Bromophenol Blue, 0.005% Xylene Cyanol). The cleavage products were resolved by 19% denaturing PAGE, stained with SYBR Gold (Invitrogen), visualized with Typhoon FLA 9500 or ChemiDoc XP, and analyzed by the ImageQuant (GE Healthcare) and Prism 8 (GraphPad) software. For analysis of temperature dependence of ssDNA cleavage, all samples were incubated at indicated temperatures simultaneously using a PCR thermocycler (T100, Bio-Rad).
Kinetic analysis of ssDNA cleavage was performed under single-turnover or multiple-turnover conditions using ssDNA substrates containing a fluorescent dye in the 5′-end (Cy5, for analysis of fully complementary 18 nt gDNA) or 3′-end (Cy3, for analysis of 18 nt gDNA with mismatches or 14 nt gDNA). In single-turnover reactions, preformed binary pAgo:guide complex was added in an excess to the ssDNA target (final concentrations: 1 μM pAgo, 250 nM gDNA, 50 nM target DNA for CbAgo; 500 nM pAgo, 500 nM gDNA, 100 nM target DNA for LrAgo), and the kinetics of cleavage was monitored over time. The samples were separated by PAGE and labeled DNA target and products were visualized with Typhoon FLA 9500 in corresponding fluorescence channels. The data were fitted to the single-exponential equation: C = Cmax × (1 – exp(–kobs × t)), where C is the efficiency of cleavage at a given time point, Cmax is the maximum cleavage, and kobs is the observed rate constant. In multiple-turnover reactions, the ssDNA target was added in an excess to the binary pAgo:guide complex (60 nM pAgo preloaded with 120 nM gDNA, plus 300 nM target DNA, final concentrations), and the kinetics of cleavage was fitted according to the equation: C = B × (1 – exp(–kburst × t)) + V × t, where B is the burst fraction, kburst is the observed cleavage rate in the burst fraction, and V is the steady-state velocity of ssDNA cleavage. Calculation of the confidence intervals for all kinetic constants was performed by the Delta method in the Prism 8 (GraphPad) software.
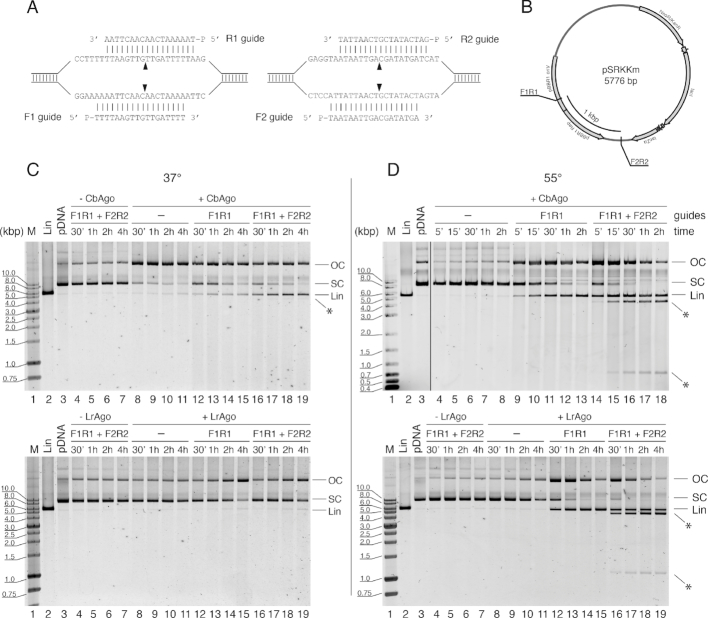
In plasmid cleavage assays with several guide molecules (Figure 6, Supplementary Figure S7), pAgo samples were independently loaded with each of the guides at equimolar ratios and combined together to the final concentration of 200 nM. For analysis of the chopping activity (Figure 7), the final pAgo concentration was 500 nM (plus 500 nM gDNA when indicated). Plasmid pSRKKm (33) was added to the reaction mixtures at 2 nM final concentration, followed by incubation for indicated time intervals at either 37°C or 55°C. The reactions were stopped by treatment with Proteinase K at 4°C, the samples were mixed with 6xPurple Loading Dye (New England Biolabs) and the cleavage products were resolved by native 1% agarose gel electrophoresis.
Figure 6.
CbAgo and LrAgo act as programmable DNA-nucleases for double-stranded DNA cleavage in vitro. (A) Target regions of the pSRKKm plasmid with two pairs of DNA guides (F1R1 and F2R2). Black triangles indicate the predicted cleavage sites. (B) Scheme of the pSRKKm plasmid with the positions of the two target regions. (C) Plasmid cleavage by CbAgo (top) and LrAgo (bottom) at 37°C. (D) Plasmid cleavage at 55°C. The reactions were performed with no guides, one pair of guides (F1R1) or two pairs of guides for indicated time intervals. Control reactions contained no pAgo proteins. Note that under the conditions of our assay, the supercoiled form of this plasmid migrated slower on the agarose gel than the linear plasmid, which identity was confirmed by comparison with the plasmid treated with a restriction endonuclease (KpnI, lane 2 in all panels). FR, forward and reverse guide DNAs; M, molecular weight marker; Lin, linearized plasmid; OC, open circular plasmid; SC, supercoiled plasmid; positions of specific cleavage products are indicated with asterisks.
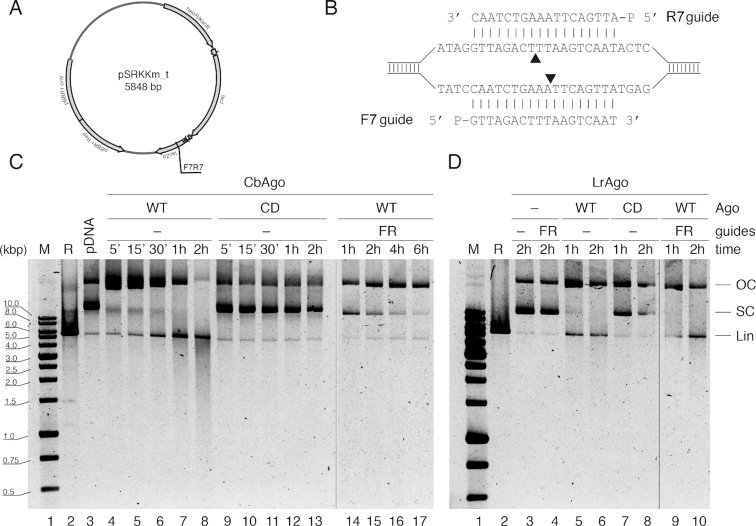
Figure 7.
Guide-independent cleavage of plasmid DNA by CbAgo and LrAgo at 37°C. (A) Scheme of the pSRKKm_t plasmid (in comparison with pSRKKm, it contains an insert of 227 nt between the NdeI and KpnI sites, see Supplementary Table S1). (B) The plasmid site corresponding to the forward (F) and reverse (R) gDNAs. Black triangles indicate the predicted cleavage sites. (C and D) Kinetics of pSRKKm_t processing by CbAgo (C) or LrAgo (D) in the presence or absence of DNA guides. WT, wild-type; CD, catalytically dead pAgos; R, linear plasmid obtained after KpnI treatment; all other designations are the same as in Figure 6.
EMSA assays
Reaction mixtures containing the required components (500 nM pAgo preloaded with 500 nM guides, plus 100 nM target DNA) were incubated in the reaction buffer at 37°C for 10 min, then mixed with 5× loading buffer (5× TBE, 50% glycerol) and resolved by 10% native PAGE buffered with TBE at 4°C. Nucleic acids were stained with SYBR Gold and visualized using Typhoon FLA 9500.
K d measurements
Apparent dissociation constants (Kd,app) for guide DNA binding were determined using double-filter assay as described (34). Briefly, 10 pM 5′-[P32]-labeled oligonucleotide was mixed with increasing amounts of CbAgo or LrAgo in 50 μl of binding buffer (10 mM HEPES–NaOH pH 7.0, 100 mM NaCl, 5% glycerol, 100 μg/ml BSA, 5 mM MnCl2) and incubated at 37°C for 40 min. The samples were filtered through nitrocellulose membrane (Merck–Millipore) layered on top of nylon Hybond N+ (GE Healthcare) equilibrated with the ice-cold binding buffer, using the Bio-Dot Microfiltration apparatus (Bio-Rad). After three washing steps with 200 μl of the same buffer the membranes were removed, air-dried and analyzed by phosphorimaging, using the ImageQuant (GE Healthcare) and Prism 8 (GraphPad) software. The data were better fitted with the Hill equation with a Hill coefficient of 2–2.5 rather than with a standard binding isotherm (P < 0.0001, the extra sum-of-squares F test), possibly reflecting partial inactivation of pAgo in the lower range of concentrations. For the competition binding assay, increasing amounts of unlabeled 5′-OH or 5′-P guide DNA were added to the 50 μl reaction mixture containing 100 pM of radiolabeled 5′-P DNA guide, followed by the addition of 100 pM LrAgo protein. After 1 h incubation at 37°C, the samples were processed as described above. The data were fitted using the one-site competitive binding model (35).
RESULTS
CbAgo and LrAgo use small DNA guides for endonucleolytic cleavage of ssDNA targets at ambient temperature
Many members of the Ago family in both pro- and eukaryotes rely on the RNaseH-like active site in their PIWI domain for endonucleolytic cleavage of nucleic acid targets, while others have substitutions in the catalytic tetrad which likely make them inactive as nucleases (see Introduction). Based on phylogenetic analysis of prokaryotic Ago proteins (4), we selected two pAgos from mesophilic bacteria, cyanobacterium L. rosea (LrAgo) and anaerobic bacillus C. butyricum (CbAgo), belonging to different subclades of long pAgos (Supplementary Figure S1A). Sequence alignments showed that both LrAgo and CbAgo have four conserved catalytic resides (DEDD) in the PIWI domain suggesting that they potentially have catalytic endonuclease activity (Supplementary Figure S1B).
To study biochemical properties of CbAgo and LrAgo we expressed and purified each protein. Codon-optimized genes encoding both proteins were chemically synthesized and cloned. In addition to the wild-type proteins, we obtained their catalytically inactive variants with substitutions of one or two out of four catalytic tetrad residues (D541A/D611A in CbAgo and D516A in LrAgo, Supplementary Figure S1B). The proteins were expressed in E. coli cells and purified using sequential Ni-NTA-affinity, size-exclusion and cation-exchange chromatography (see Supplementary Figure S2 and Materials and Methods for details). Examination of purified pAgos showed high purity of the samples with a single band of the expected molecular weight (Supplementary Figure S2).
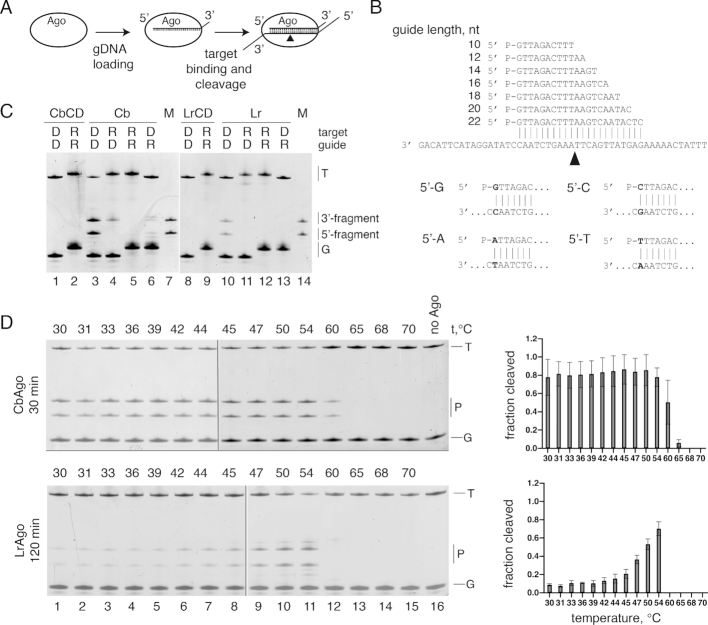
We first studied the nucleic acid specificity of CbAgo and LrAgo in an in vitro cleavage assay using synthetic oligonucleotides (Figure 1A). CbAgo and LrAgo were loaded with 18 nt 5′-phosphorylated DNA or RNA guides followed by the addition of complementary 50 nt long single-stranded DNA or RNA targets (Figure 1B, Supplementary Table S1). After incubation at 37°C in reaction buffer containing divalent metal ions (Mn2+ in most experiments) the products were resolved on denaturing gel and visualized by SYBR Gold staining. In reactions containing guide DNA (gDNA) and target DNA (tDNA), we observed DNA cleavage by both pAgos at a single site between target positions 10′ and 11′, identical to all previously studied Ago proteins, resulting in the appearance of the 27 nt long 3′-fragment and 23 nt long 5′-fragment of the target DNA (Figure 1C). No cleavage products were observed in the absence of pAgo proteins (Figure 1D and Supplementary Figure S3). In this and subsequent experiments, the efficiency of DNA cleavage was lower for LrAgo than for CbAgo; we therefore used longer incubation times for LrAgo in most experiments. Cleavage required the intact catalytic tetrad in the PIWI domain, and point mutations in the tetrad eliminated the activity of both CbAgo and LrAgo (Figure 1C). No substrate cleavage was observed with other combinations of guide and target molecules (gRNA-tDNA, gDNA-tRNA and tRNA-gRNA) for LrAgo, and only very weak target cleavage was observed in the gRNA-tDNA and gDNA-tRNA reactions for CbAgo (Figure 1C).
Figure 1.
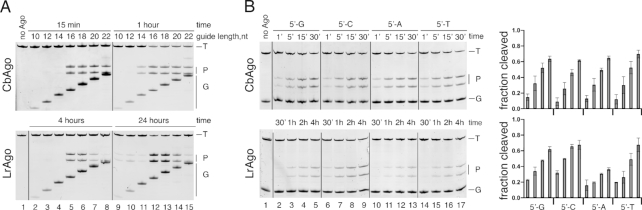
CbAgo and LrAgo are DNA-dependent DNA-endonucleases. (A) Scheme of the in vitro assay. (B) Guide and target oligonucleotides. DNA guides and targets were used in most experiments; 18 nt guide and 50 nt target RNAs were additionally used for analysis of pAgo specificity (see panel C). Black triangle indicates the predicted cleavage site between target positions 10′ and 11′ (corresponding to the guide positions 10 and 11 starting from the 5′-end). Four guide-target pairs with different nucleotides in the guide 5′-end are shown at the bottom. (C) Analysis of the cleavage specificity of pAgos. Wild-type (WT) or catalytically dead (CD) CbAgo and LrAgo were loaded with 18 nt DNA (D) or RNA (R) guides and incubated with 50 nt ssDNA or RNA targets for 2 hours at 37°C. Positions of the guides (G), targets (T) and cleavage products (P) are indicated on the right of the gels. The marker (M) lane contains synthetic oligonucleotides corresponding to the expected cleavage products (23 nt 5′-end and 27 nt 3′-end target fragments). (D) Temperature dependence of ssDNA cleavage by CbAgo and LrAgo. The assay was performed for 30 min (CbAgo) or 2 h (LrAgo) at indicated temperatures. Quantification of the cleavage efficiencies (the percentage of target cleavage) is shown on the right.
Analysis of the temperature dependence of the reaction revealed that CbAgo had comparable levels of endonuclease cleavage between 30 and 54°C and still retained some activity at 60°C (Figure 1D). LrAgo was significantly less active than CbAgo at most temperatures, but its activity was stimulated at 50–54°C, followed by complete inactivation at 60°C (Figure 1D). Thus, CbAgo and LrAgo are active DNA nucleases at moderate temperatures and are also sufficiently stable to perform DNA cleavage at elevated temperature.
We next analyzed the effects of divalent cations and other reaction conditions on guide-dependent DNA cleavage. LrAgo had the highest endonuclease activity in the presence of Mn2+, was much less active with Mg2+ and was inactive in the presence of Co2+, Cu2+ or Zn2+. CbAgo was active with both Mn2+ and Mg2+, and could also catalyze target cleavage in the presence of Co2+ (Supplementary Figure S3B). Titration of Mn2+ ions showed that CbAgo was equally active between 0.03 and 5 mM Mn2+, while LrAgo required increased Mn2+ concentrations (>1 mM; Supplementary Figure S3C), significantly higher than needed for CbAgo or PfAgo (100 μM, (8)). Mn2+ is an abundant cation in cyanobacterial cells (36), possibly explaining this unusual property of LrAgo.
Analysis of ssDNA cleavage at various ionic strength conditions revealed that LrAgo was active at NaCl concentrations up to 750 mM, while CbAgo retained high level of activity even at 1 M NaCl (Supplementary Figure S3D). Finally, pH did not noticeably influence the efficiency of DNA cleavage within the tested range (6.8–8.0), analyzed for LrAgo (Supplementary Figure S3E).
In summary, in contrast to previously characterized pAgos from thermophilic species that are active only at high temperatures, CbAgo and LrAgo are mesophilic DNA-guided DNA nucleases that can perform DNA cleavage under a wide range of conditions and can potentially be active in eukaryotic cells at 37°C.
Kinetic analysis of ssDNA cleavage by CbAgo and LrAgo
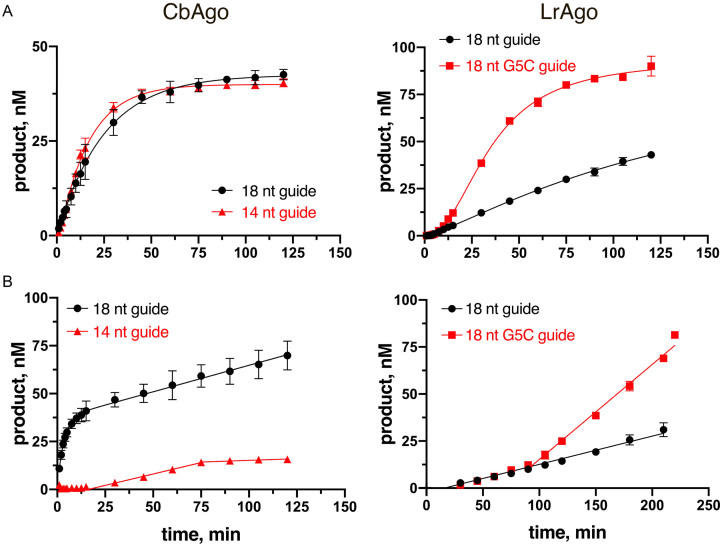
To further explore the catalytic properties of CbAgo and LrAgo, we measured the kinetics of ssDNA cleavage at 37°C. The reactions were performed with 18 nt gDNA and complementary 50 nt tDNA (Figure 2) under either single-turnover conditions (when the binary complex of pAgo with gDNA was present in excess over tDNA) or multiple-turnover conditions (when tDNA was present in excess over the binary complex). We found that during single round of catalysis, CbAgo can cleave the ssDNA target much faster than LrAgo, with kobs values of 0.0398 min−1 (95% confidence interval, CI: 0.03588–0.04398 min−1) and 0.00463 min−1 (95% CI: 0.00451–0.00530 min−1), respectively, corresponding to the reaction half-times of 17.4 and 150 min (Figure 2A, compare left and right panels). Interestingly, LrAgo also had a lower maximum cleavage efficiency in comparison with CbAgo (43 ± 1.7% of ssDNA cleavage in comparison with 85 ± 2.8% for CbAgo), suggesting that a fraction of complexes formed by LrAgo was catalytically inactive (Figure 2A, right panel).
Figure 2.
Kinetic analysis of ssDNA cleavage by CbAgo and LrAgo at 37°C. (A) Comparison of the observed rates of target DNA cleavage by CbAgo and LrAgo. The reactions were performed under single-turnover conditions (1 μM CbAgo, 250 nM gDNA, 50 nM tDNA; 500 nM LrAgo, 500 nM gDNA, 100 nM tDNA) with fully complementary 18 nt gDNAs for both pAgos, or with 14 nt gDNA for CbAgo (left), or with 18 nt gDNA containing the G5C mismatch for LrAgo (right). The kobs values were determined from the single-exponential fits of the data (only the exponential phase of the cleavage kinetics was used for fitting in the case of the mismatched gDNA for LrAgo). (B) Analysis of the steady-state kinetics of target DNA cleavage with the same combinations of guide and target molecules. The reactions were performed under multiple-turnover conditions at the 1:2:5 pAgo:guide:target molar ratio (60 nM pAgo, 120 nM gDNA, 300 nM tDNA). The linear parts of the reaction kinetics were used for calculation of the steady-state velocities of DNA cleavage. Means and standard deviations from 3 independent experiments are shown.
The kinetics of target cleavage by CbAgo and LrAgo was very different under multiple-turnover conditions. For CbAgo, we observed a two-phase kinetics with initial burst followed by a slower linear increase in product formation (Figure 2B, left panel). As demonstrated previously for other Ago proteins (24–26), the initial burst phase likely corresponds to the first round of catalysis, following rapid formation of the ternary pAgo/gDNA/tDNA complex, while the linear phase corresponds to the multiple-round steady-state kinetics of ssDNA cleavage.
From the burst amplitude, we estimated that the concentration of active binary CbAgo/gDNA complexes was 37 nM (out of 60 nM CbAgo taken in the reaction), corresponding to ∼60% of active CbAgo in our preparations. The steady-state velocity of ssDNA cleavage by CbAgo under these conditions was 0.277 nM × min−1 (95% CI: 0.232–0.319 nM × min−1), corresponding to the kobs value of 0.0075 min−1 (based on the 37 nM effective concentration of the binary complex), 5-times slower than the rate of ssDNA cleavage under single-turnover conditions (0.0398 min−1; see above). This type of reaction kinetics is indicative of slow dissociation of the product complex, which becomes rate-limiting under multiple-round conditions, as previously observed for eAgos (24–26). The kobs value determined in this assay (0.0075 min−1) likely corresponds to the rate-limiting step of complex dissociation and thus gives an estimate of the half-life time of the product complex (∼90 min).
For LrAgo, no burst phase was detected under multiple-turnover conditions, with a linear steady-state kinetics of target cleavage observed after a lag phase (Figure 2B, right). The absence of burst might reflect a slower rate of the ternary complex formation and/or catalysis by LrAgo in comparison with CbAgo, so that product dissociation was no more rate-limiting in the steady-state reaction. Similarly, a lag phase without burst was previously observed during multiple-turnover DNA cleavage by thermophilic TtAgo, for which product dissociation was not rate-limiting likely because the high temperature promoted complex disassembly (37).
The steady-state velocity of target DNA cleavage by LrAgo was 0.181 nM × min−1 (95% CI: 0.1466–0.2156 nM × min−1) at 60 nM LrAgo concentration, only slightly lower than for CbAgo (0.277 nM × min−1). While we could not determine the fraction of active complexes of LrAgo in this assay due to the absence of burst, it is likely comparable to CbAgo, thus giving a similar steady-state rate of catalysis (kobs of 0.0075 min−1 for CbAgo, see above). Notably, this is comparable to the single-turnover rate of ssDNA cleavage by LrAgo (0.00463 min−1) further suggesting that the catalytic step may be rate-limiting for LrAgo. In summary, it can be concluded that CbAgo and LrAgo significantly differ in their properties at the steps of target DNA binding and cleavage,and may have different rate-limiting steps in catalysis; however, the resulting steady-state rates of ssDNA cleavage are similar for both pAgos.
Optimal parameters of guide DNAs utilized by CbAgo and LrAgo
All eAgos and pAgos studied to date bind nucleic acid guides of specific lengths (7,11–13,38). Furthermore, many eAgos and pAgos were also reported to have a bias for a particular nucleotide residue at the first position of the guide molecule, which is accommodated in a protein pocket formed by the MID domain (12,18,21,39,40). To determine the optimal parameters of guide DNA structure for pAgo-mediated target cleavage, we performed the cleavage reaction with guides of different structure (Figure 1B and Supplementary Table S1).
We first explored the role of the guide length by testing a series of DNA guides from 10 to 22 nt long that shared identical sequences at their 5′-ends, so that the predicted cleavage site was the same for all guides (Figure 1B). 16–18 nt guides led to efficient target cleavage by both pAgos (Figure 3A). The cleavage efficiency for LrAgo was much lower with longer guides suggesting that extended duplex formation may prevent target cleavage (Figure 3A). The use of shorter guides greatly diminished the reaction efficiency. However, even the shortest 10 and 12 nt guides were able to direct proper target cleavage by LrAgo (but not CbAgo), which could be detected after prolonged incubation (24 h, right panels in Figure 3A).
Figure 3.
Effects of the guide length and 5′-nucleotide identity on DNA cleavage by CbAgo and LrAgo. (A) Cleavage assay with guide DNAs of varied length. (B) Preferences for the 5′-guide nucleotide. Quantification of the cleavage efficiencies is shown on the right. All experiments were performed at the 5:2:1 pAgo:guide:target molar ratio in reaction buffer containing Mn2+ ions at 37°C. Means and standard deviations from two to three independent measurements are shown.
Changes in the guide length may decrease the efficiency of target DNA cleavage in several ways, including changes in the rates of target DNA search and binding, DNA cleavage in the ternary complex, or product complex dissociation. To test which of these steps may be affected in the case of short guide DNAs, we analyzed the kinetics of ssDNA cleavage by CbAgo with 14 nt gDNA, which had a markedly lower efficiency of cleavage in comparison with 18 nt gDNA (see Figure 3A, upper panel). Under single-turnover conditions, the observed rate of tDNA cleavage with this gDNA was almost identical to the 18 nt gDNA (kobs of 0.0522 min−1; 95% CI: 0.0476–0.0571 min−1) (Figure 2A, left). In contrast, the kinetics of product accumulation with 14 nt gDNA was dramatically slower under multiple-round conditions (Figure 2B, left). In particular, a prolonged lag phase (>15 min) was observed instead of burst, followed by steady-state product formation. These changes may probably result from a decreased rate of the formation of catalytically active complex containing suboptimal gDNA. At the same time, the steady-state velocity of tDNA cleavage with 14 nt gDNA (0.238 nM × min−1; 95% CI: 0.172–0.303 nM × min−1) was comparable with the velocity measured for 18 nt gDNA (0.277 nM × min−1) suggesting that CbAgo turnover may not be significantly changed with the shorter gDNA. The steady-state phase was followed by a slowdown and plateau in the product accumulation, possibly as a result of irreversible complex inactivation with this gDNA, the reasons for which remain unknown (for example, short gDNA might gradually dissociate from the binary complex with pAgo and anneal to the free tDNA strand, thus forming the gDNA–tDNA duplex inaccessible for CbAgo).
To determine if CbAgo and LrAgo have a preference for the first nucleotide of the guide DNA, we also tested four guide variants with different 5′-terminal nucleotides but otherwise identical sequences (Figure 1B). All four guides were able to direct cleavage of complementary targets by both CbAgo and LrAgo, with only slightly lower efficiency of target cleavage with the 5′-A guide observed for LrAgo (Figure 3B). Thus, the optimal guide length for CbAgo and LrAgo is comparable to other pAgo and eAgo proteins, and the two pAgo proteins can likely direct cleavage of any desired sequence making them a flexible tool for DNA manipulation.
CbAgo and LrAgo can use both 5′-P and 5′-OH guides for target cleavage, but with different precision
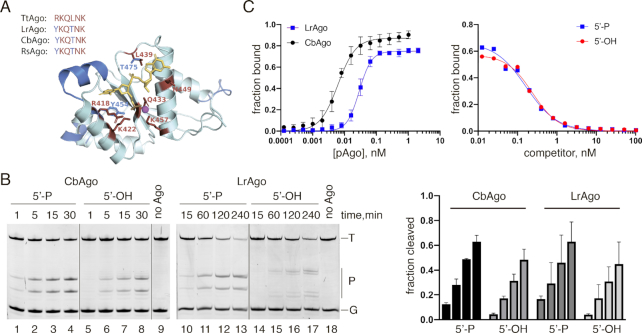
All eukaryotic and the majority of prokaryotic Ago proteins – including the best studied TtAgo and RsAgo—strongly prefer 5′-phosphorylated nucleic acid guides due to multiple contacts formed between the phosphate group and amino acid residues in the 5′-end binding pocket of the Ago MID domain (Figure 4A) (17,18,20,21,39–41). However, pAgo from M. piezophila (MpAgo) has a unique 5′-end binding pocket that confers it the ability to bind 5′-OH guides and exclude 5′-phosphorylated molecules (7). Analysis of the 5′-binding pocket in the MID domain of LrAgo and CbAgo revealed substitutions of two out of six amino acid residues in the specific motif involved in interactions with the Me2+ ion and the guide 5′-phosphate relative to TtAgo (Figure 4A).
Figure 4.
CbAgo and LrAgo can utilize both 5′-phosphorylated and 5′-hydroxyl DNA guides at 37°C. (A) A 3D model of the LrAgo MID domain aligned to the structure of TtAgo in complex with gDNA (with bound Mg2+ ion; PDB: 3HO1). The model was built using the SWISS-MODEL portal. Amino acid residues of the conserved MID-domain motif (red; shown for various pAgos above the structure (4)) and Mg2+ ion (magenta) involved in interactions with the first two guide nucleotides (yellow) are highlighted. Elements of the secondary structure and amino residues specific to LrAgo are shown in blue. (B) Programmable ssDNA cleavage by CbAgo and LrAgo in the presence of either 5′-P or 5′-OH guides. The reactions were performed at the 5:2:1 pAgo:guide:target molar ratio for indicated time intervals. (C) (Left) Binding of 18 nt phosphorylated DNA guide by CbAgo and LrAgo. The fraction of bound DNA was plotted against protein concentration and fitted using the model of specific binding with the Hill slope. (Right) Analysis of gDNA binding by a competition assay. Radiolabeled gDNA (100 pM) was combined with increasing amounts of unlabeled 5′-P (blue) or 5′-OH (red) competitor, and incubated with LrAgo (100 pM). The fraction of bound DNA was plotted against competitor concentration and fitted using the one-site competitive binding model. Means and standard deviations from 3 independent experiments are shown.
We studied the ability of CbAgo and LrAgo to use 5′-P and 5′-OH guides in the cleavage reaction. Surprisingly, both CbAgo and LrAgo could use 5′-OH DNA guides to cleave the target DNA, with only slightly lower efficiency in comparison to 5′-P guides of identical sequence (Figure 4B). Interestingly, when LrAgo was loaded with the 5′-OH DNA guide, target cleavage was observed 1–2 nucleotides downstream of the canonical site, between target positions 11′-12′ and 12′-13′ relative to the guide 5′-end (Figure 4B). The ability of pAgos to use 5′-OH guides to cleave target ssDNA was lost at 55°C (Supplementary Figure S4), suggesting that interactions with the 5′-phosphate are required to stabilize the binary pAgo-guide complex at elevated temperature.
We further measured the apparent equilibrium dissociation constants (Kd,app) for guide binding by LrAgo and CbAgo using a filter-based titration assay (Figure 4С). LrAgo associated with 5′-phosphorylated guides with an Kd,app value of 34.8 pM (95% CI: 29.3–41.2 pM) demonstrating a much higher affinity to nucleic acid guides in comparison with other studied pAgos (see Discussion). CbAgo also revealed exceptionally high affinity to 5′-P guides, even exceeding that of LrAgo (Kd,app of 6.2 pM; 95% CI: 5.3–7.3 pM). The real Kd values for guide binding by these pAgos are likely even lower, given that the effective concentration of pAgo samples is <100% and because of the detection limits of measurements.
Since LrAgo revealed changes in the cleavage position with the 5′-OH guide, we compared the ability of LrAgo to bind 5′-P and 5′-OH guides by a competition assay, in which LrAgo was incubated with a mixture of 5′-P32-labeled guide and increasing amounts of unlabeled guide of the same sequence either containing or lacking the 5′-phosphate. Surprisingly, LrAgo revealed essentially the same affinities towards both variants of the guide DNA in this assay, with the Kd,app values of 33.3 pM (95% CI 25.1–43.9 pM) and 45.6 pM (95% CI 33.7–61.7 pM) for the 5′-P and 5′-OH guides, respectively (Figure 4C). Electrophoretic mobility shift assay (EMSA) also showed that LrAgo can form binary complexes with the 5′-OH and 5′-P guides with comparable efficiencies (Supplementary Figure S5).
Taken together, the results demonstrate that CbAgo and LrAgo are able to bind both 5′-P and 5′-OH DNA guides and use them for target DNA cleavage. Unexpectedly, the nature of the 5′-guide group is also critical for defining the exact position of the cleavage site by LrAgo.
CbAgo and LrAgo tolerate mismatches in the seed region, but are sensitive to mismatches in the 3′-portion of guide DNA
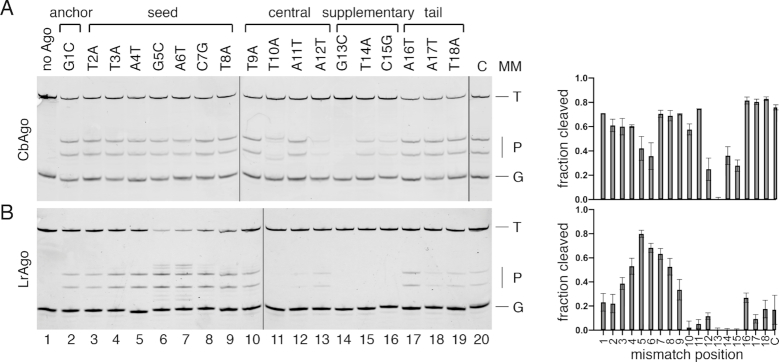
Previous studies of eAgos and several pAgos, including AfAgo, TtAgo, RsAgo and MpAgo, showed that mismatches between the guide and target strands may have large effects on the target recognition (9,17,22,24,27,42–45). In particular, even a single mismatch in the seed region within 2–8 nts of the guide can impair target binding and silencing by many eAgos as well as pAgos (e.g. (9,17,18)). To study the effect of mismatches on target cleavage we designed a set of DNA guides, each containing a single mismatched nucleotide at a certain position (Supplementary Table S1), and tested them in the cleavage reaction with CbAgo and LrAgo (Figure 5).
Figure 5.
Effects of mismatches in the guide-target duplex on the slicing activity of CbAgo (A) and LrAgo (B). The assay was performed at the 5:5:1 Ago:guide:target molar ratio. Mismatch positions in different guide regions are designated above the gels; nucleotide substitutions relative to the standard guide sequence are indicated (see Supplementary Table S1 for complete guide sequences). The reactions were performed at 37°C for 30 min for CbAgo and for 2 h for LrAgo. Quantification of the data (means and standard deviations from three independent experiments) is shown on the right. T, target; P, cleavage products; G, guide; C, control reactions with guide variants containing no substitutions.
Surprisingly, mismatches in the seed region had little or no effect on the efficiency of target cleavage by CbAgo (Figure 5A) and substantially stimulated the activity of LrAgo (Figure 5B). For LrAgo, mismatches at positions 5 and 6 of the seed region also induced target cleavage at several additional sites located closer to the guide 3′-end (between target positions 11′–12′, 12′–13′ and 13′–14′) (Figure 5B). Mismatches downstream of the cleavage site, in the 3′-supplementary region of gDNA (guide positions 12–15), strongly decreased the efficiency of target cleavage by both CbAgo and LrAgo (Figure 5A and B). Mismatches at the site of cleavage (guide positions 10–11) also led to a strong decrease in target cleavage by LrAgo (Figure 5B). However, in the case of CbAgo, mismatch at position 11 had no significant effect on cleavage, while mismatch at position 10 shifted the cleavage site one nucleotide closer to the guide 3′-end (between target positions 11′ and 12′) (Figure 5A). Thus, in contrast to the majority of Ago proteins studied to date, target cleavage by CbAgo and LrAgo is not inhibited and can be even stimulated by mismatches in the seed region but is decreased by mismatches in the 3′-supplementary guide region.
The effects of mismatches on target DNA cleavage might be explained by changes in the rates of ternary complex formation, catalysis, or product dissociation (pAgo recycling). To discriminate between these possibilities, we analyzed target DNA binding by LrAgo, for which various mismatches had either stimulatory or inhibitory effect on catalysis, using EMSA (Supplementary Figure S6). In addition to the fully complementary guide-target pair, we tested mismatches that increased or decreased the rate of cleavage. In all cases, almost all target DNA was bound by guide-loaded LrAgo, regardless of the presence and position of the mismatched nucleotide (Supplementary Figure S6). Efficient binding of target DNA by LrAgo indicates that mismatches do not prevent ternary complex formation and suggests that ternary complexes may possibly undergo slow isomerization required for catalysis after initial target recognition (see Discussion). Indeed, the time sufficient for target binding was significantly shorter than the time required for target cleavage by LrAgo (see Figures 2A and 5B).
We further measured single- and multiple-turnover kinetics of target DNA cleavage by LrAgo with gDNA containing a mismatch at the 5th position (G5C). The mismatch significantly increased the rate of catalysis under single-turnover conditions (kobs of 0.0343 min−1 (95% CI: 0.0256–0.0444 min−1) in comparison with 0.00463 min−1 for the fully complementary 18 nt gDNA) (Figure 2A, LrAgo), demonstrating that it affects the catalytic step of the reaction. In addition, a lag phase in product formation was observed for the mismatched gDNA, suggesting that the mismatch also affects initial steps of enzyme-substrate complex formation/isomerization prior to slicing. Furthermore, the maximum efficiency of target cleavage with the mismatched gDNA was significantly higher than with the fully complementary gDNA (90.1 ± 5.3% and 43.0 ± 1.6% of tDNA cleavage, respectively), suggesting that the mismatch promotes formation of the productive enzyme-substrate complex.
Under multiple turnover conditions, the mismatch increased the steady-state velocity of target DNA cleavage (V = 0.521 nM × min−1 (95% CI: 0.474–0.568 nM × min−1), in comparison with 0.181 nM × min−1 for the fully complementary gDNA; measured at 60 nM LrAgo concentration), after a lag phase possibly reflecting slow formation of the catalytically competent complex (Figure 2B). This effect could be explained by stimulation of catalysis (detected in the single-turnover experiments) and/or stimulation of product dissociation and LrAgo turnover. Further experiments are required to distinguish between these possibilities.
Overall, our data suggest that mismatches may have complex effects on ssDNA processing by pAgos, by affecting ternary complex formation and/or isomerization, and also by changing the rates of catalysis and dissociation of the cleavage products.
Guide-directed cleavage of double-stranded DNA by CbAgo and LrAgo
While CbAgo and LrAgo can efficiently cut ssDNA substrates, the cleavage of dsDNA likely presents a bigger challenge for pAgos because the DNA duplex has to be unwound to form the ternary complex with guide-loaded pAgo. However, pAgos do not have helicase domains and, unlike the Cas9 protein, cannot perform DNA melting. Indeed, previous studies of TtAgo and PfAgo from thermophilic prokaryotes observed guide-dependent dsDNA cleavage only at elevated temperatures, which likely facilitated DNA melting (8,13). In the next set of experiments we analyzed processing of double-stranded plasmid substrates by CbAgo and LrAgo under various conditions.
We first tested the ability of CbAgo and LrAgo to cut supercoiled plasmid DNA at 37°C. The plasmid pSRKKm was incubated with empty (unloaded) pAgos, or with pAgos loaded with DNA guides designed to target both DNA strands, either at one or at two plasmid sites separated by ∼1000 base pairs (Figure 6A and B). No plasmid relaxation or linearization was observed in the absence of pAgos (Figure 6C, upper panel, lanes 4–7). In the absence of gDNAs, CbAgo relaxed the plasmid resulting in accumulation of the open circular form of DNA, likely containing single-stranded nicks in one or two strands, with very low level of plasmid linearization (Figure 6C, left panel, lanes 8–11). In the presence of one or two pairs of gDNAs, the efficiency of plasmid linearization was increased but a significant fraction of the plasmid remained in the open circular form (lanes 12–19). Only faint amounts of linear fragments resulting from specific plasmid cleavage at the two target sites were visible when the reaction was performed with two guide pairs (lanes 16–19). LrAgo revealed almost no activity with the plasmid substrate under these conditions, with some plasmid relaxation observed at long time points in the presence of guide molecules (Figure 6C, lower panel, lanes 12–19).
We therefore searched for conditions that would enhance the ability of pAgos to use DNA guides for specific cleavage of dsDNA, in particular, by increasing the temperature of the reaction. Importantly, we observed no plasmid processing by either LrAgo or CbAgo in the absence of guide molecules at 55°C (Figure 6D, lanes 4–8 in the upper panel for CbAgo, lanes 8–11 in the lower panel for LrAgo). Thus, guide-independent relaxation activity, observed for CbAgo at 37°C, is suppressed at elevated temperature. Furthermore, the linear plasmid product was formed with high efficiency when the plasmid was incubated with CbAgo or LrAgo pre-loaded with one pair of guide molecules targeting one site of the plasmid (Figure 6D, lanes 9–13 for CbAgo, lanes 12–15 for LrAgo). When the reaction was performed with CbAgo or LrAgo loaded with two pairs of guide molecules corresponding to two sites of the plasmid, both pAgos cut plasmid DNA into two linear molecules with the size of fragments corresponding to the sites targeted by guide DNAs (Figure 6D, lanes 14–18 for CbAgo, lanes 16–19 for LrAgo).
We further analyzed the dependence of plasmid cleavage on the GC-content of the target sites. In addition to the first set of guides, which targeted plasmid regions with ∼20% GC-content (see Figure 6), we designed two guide sets corresponding to different target regions of the pSRKKm plasmid with ∼35% and ∼50% GC-content at the sites of guide binding (Supplementary Figure S7A and B). The most efficient cleavage was observed for the first set of guides with the lowest GC-content (Supplementary Figure S7C, lanes 4 and 7). The cleavage efficiency was somewhat lower for the 35% GC target regions (in particular, a fraction of plasmid remained in the open circular form for LrAgo) (lanes 5 and 8). The cleavage efficiency was significantly decreased at 50% GC but CbAgo was still able to cut the plasmid into the linear fragments of the expected lengths (lanes 6 and 9).
Therefore, both CbAgo and LrAgo can perform dsDNA cleavage—likely by cutting each DNA strand independently of the other—with the precision similar to restriction endonucleases or the Cas9 nuclease, but without strict sequence requirements (although the cleavage efficiency depends on the GC-content of DNA at the cleavage site).
Guide-free CbAgo and LrAgo can process plasmid DNA
Thermophilic pAgos, TtAgo and MjAgo, were shown to process double-stranded plasmid DNA in a guide-independent manner (‘chopping’ activity), generating small DNA fragments that could be further loaded into pAgos and used as guides for subsequent target cleavage (11,28). In the previous experiments, which were performed at relatively low pAgo concentrations (200 nM, in comparison with micromolar concentrations used for TtAgo and MjAgo), we observed guide-independent plasmid relaxation—but not chopping—by free CbAgo and no plasmid processing by free LrAgo at 37°C. Therefore, to detect possible chopping activity of CbAgo and LrAgo, we analyzed plasmid DNA cleavage at higher pAgo concentrations (500 nM).
Under these conditions, incubation of the plasmid with empty CbAgo at 37°C resulted in rapid plasmid relaxation (5 min time point; Figure 7C, lane 4). The open circular form gradually disappeared after prolonged incubation, accompanied by appearance of the linear form and a smear of shorter DNA products, indicative of the chopping activity (Figure 7C, lanes 5–8). This activity was dependent on the active site of CbAgo since no plasmid processing was observed with catalytically dead (CD) CbAgo mutant (lanes 9–13). The chopping activity of wild-type CbAgo was suppressed when it was loaded with a pair of guides corresponding to the target plasmid (Figure 7B and С, lanes 14–17). No guide-dependent plasmid linearization was observed under these conditions with this guide pair (comparable to the inefficient plasmid linearization observed with the F1R1 pair of gDNAs in Figure 6C, upper panel, lanes 12–15).
Similarly to CbAgo, wild-type, but not catalytically dead, LrAgo could relax supercoiled plasmid in the absence of guide molecules (Figure 7D, compare lanes 5, 6 and 7, 8). The linear form and shorter DNA fragments that migrated on the gel as a light smear also appeared in the reaction with wild-type LrAgo. The nicking and linearization activities of LrAgo were little affected by the addition of DNA guides (lanes 9 and 10), indicating that under these conditions LrAgo likely cuts the plasmid at random positions in a guide-independent manner.
Therefore, at high concentrations both CbAgo and LrAgo can process double-stranded plasmid DNA in a guide-independent manner but for CbAgo this activity can be suppressed when it is loaded with guide molecules, which makes it a better candidate for targeted DNA cleavage.
DISCUSSION
The majority of previously characterized pAgo proteins were derived from thermophilic bacterial and archaeal species and therefore are optimally active at high temperatures. In this study we present detailed characterization of pAgo proteins from mesophilic bacteria C. butyricum and L. rosea. We show that CbAgo and LrAgo are active endonucleases that can be programmed with small DNA guides to process target DNA substrates with high precision at moderate temperatures. Below, we compare the properties of CbAgo and LrAgo and suggest that they can be used for development of tools for manipulation of DNA in vitro and in vivo.
Both CbAgo and LrAgo are long catalytically active pAgos containing the complete DEDD catalytic tetrad in their PIWI domains. They perform precise DNA-guided slicing of ssDNA substrates in vitro at a wide range of temperatures (from 30 to 60°C for CbAgo), and prefer Mn2+ as the catalytic ion. CbAgo is significantly faster than LrAgo at 37°C and reveals higher specific activity when acting on double-stranded DNA substrates. Based on these and other properties discussed below, CbAgo appears a promising candidate for various genomic applications.
Guide DNA binding and ssDNA target cleavage by CbAgo and LrAgo
Both CbAgo and LrAgo bind small DNA guides with exceptionally high affinities, with Kds in picomolar range. This markedly exceeds affinities previously reported for other pAgos (e.g. Kds of ∼3 nM for MjAgo, ∼1 nM for RsAgo) (17,46). CbAgo and LrAgo show no obvious preference toward 5′-guide nucleotide during target cleavage indicating that both proteins can be programmed with DNA guides of any sequence permitting flexible choice of the target site. The efficiency of DNA cleavage by CbAgo and LrAgo is significantly reduced if guide length is below 16 nucleotides. However, slow target slicing can still be observed with shorter guides, down to 10 nt for LrAgo, when the scissile bond is not flanked by base-paired nucleotides. In comparison, the shortest length of the guide-target duplex required for target cleavage by fly RISC was 12 bp, although the efficiency of RNA cleavage was greatly diminished with this guide length (26). Similarly to LrAgo, TtAgo was shown to use 9–10 nt DNA guides for target DNA cleavage, but this activity disappeared at increased temperatures (9). Thus, base-pairing around the cleavage position likely helps to stabilize target binding in the active site of Ago proteins.
In contrast to other studied Agos, both CbAgo and LrAgo are able to utilize 5′-OH guides for target cleavage with almost the same efficiency as 5′-P guides. The majority of eAgos and pAgos were reported to use 5′-P guides, and multiple contacts between the 5′-P group and the 5′-binding pocket in the MID domain are observed in the structures of several Ago-guide binary complexes (Figure 4A) (17,18,20,21,39–41). A notable exception is the RNA-guided MpAgo that binds exclusively to 5′-OH guides (7), and several other pAgos predicted to have a similar 5′-binding pocket (4,7). Interestingly, although eAgos universally associate with 5′-P guides in vivo, human Ago2 was shown to cut mRNA targets when bound to non-phosphorylated small RNA guides in vitro (47), suggesting that the ability of various Ago proteins to use non-phosphorylated guides may be underexplored.
Our recent bioinformatic analysis revealed several subtypes of the MID domain with substitutions of key residues involved in interactions with the 5′-group of guide molecule (4); most of them have not been characterized experimentally. Relative to TtAgo, CbAgo and LrAgo contain substitutions of two conserved residues in the 5′-end binding pocket (Figure 4A). However, these substitutions are also present in RsAgo that was shown to recognize 5′-phosphorylated guide molecules, similarly to TtAgo (12). Furthermore, homology-based structural modeling suggests that interactions of the guide 5′-end with the MID pocket are overall very similar for TtAgo and LrAgo (Figure 4A). Thus, contacts with other parts of the guide molecule may compensate for the loss of interactions with the 5′-phosphate in LrAgo and CbAgo. Yet, interactions with the 5′-phosphate can stabilize the complexes under suboptimal conditions, such as increased temperature, as demonstrated for CbAgo that is not able to use 5′-OH guides to cut targets at 55°C.
In the case of LrAgo, the cleavage site is shifted 1–2 nucleotides downstream from the guide 5′-end in the absence of the 5′-phosphate group in the guide molecule. Changes in the slicing position were also observed for hAgo2 with nonphosphorylated guides (47). Such changes might be caused by sliding of the guide-target duplex in the active site in the absence of the 5′-P-MID interactions. Interestingly, MjAgo also demonstrated noncanonical target DNA cleavage at several positions around the canonical cleavage site, suggesting that the guide-target duplex can be bound in several registers in the case of this pAgo (11,21,23). The role of the 5′-P-MID interactions in guide positioning in MjAgo remains to be established. Our results indicate that the guide 5′-phosphate can help to determine the correct register of the guide/target duplex relative to the active site of pAgo.
The rates of single-turnover ssDNA cleavage by ternary complexes of CbAgo and LrAgo are slower than the rates of mRNA cleavage measured for the fly or mouse RISC complex (kobs of 0.0071 s−1, or 0.426 min−1) (26). At the same time, similarly to the previously studied eAgos (24–26), the rate of multiple-turnover catalysis by CbAgo is likely limited at the step of product dissociation after target DNA cleavage. LrAgo distinguishes from CbAgo and eAgos by the absence of burst during multiple-turnover DNA cleavage, suggesting that catalytic complex formation and/or target cleavage, but not product dissociation, may be rate-limiting for this pAgo.
Previous studies of eAgos and pAgos demonstrated the importance of complementarity between the guide and the target for efficient repression: mismatches in the seed region usually reduce repression, while mismatches in the 3′-part (downstream of the cleavage site) are tolerated without significant loss of efficiency (9,17,22,24,27,42–45). In comparison with these studies, mismatches between the guide and target molecules have unusual effects on target cleavage by CbAgo and LrAgo. Mismatches in the seed region do not significantly affect (for CbAgo) or even stimulate (for LrAgo) target DNA cleavage. Interestingly, recent analysis of zAgo2 from zebrafish demonstrated that it was inactive with perfectly complementary targets but a mismatch in the seed region stimulated target RNA cleavage (48). Mismatches in the seed region may possibly affect target positioning and/or conformational changes in the active site of zAgo2 and LrAgo during catalysis. Indeed, single-turnover kinetic analysis demonstrated that mismatches in the seed can increase the rate of catalysis in the ternary complex of LrAgo. Structural studies of pAgos revealed that one of the active site residues, the catalytic glutamate, is unplugged in free pAgos and must be plugged in for catalytic metal binding and scissile bond cleavage following target binding (14,19,37). It can be speculated that this or other isomerization step(s) may be affected by mismatches between the guide and target strands in the complex.
On the whole, changes in the guide-target complementarity can have diverse effects on catalysis by pAgos. Mismatches in the seed region may promote target DNA cleavage, by stimulating the catalytic step of the reaction and, possibly, increasing the rate of product dissociation, but may also decrease the rate of catalytic complex formation. In contrast to most other Ago proteins studied to date, mismatches in the 3′-supplementary guide region strongly inhibit target DNA cleavage by CbAgo and LrAgo; similar effects were recently reported for TtAgo (31). Therefore, in the case of these pAgos propagation of the guide-target duplex, rather than initial interactions in the seed region, may control the fidelity of target recognition. More thorough kinetic analysis is needed to decipher the complete catalytic mechanism for both pAgos, and to fully understand the observed effects of changes in the guide structure on the target DNA cleavage.
Double-stranded DNA cleavage by CbAgo and LrAgo
Both CbAgo and LrAgo can relax supercoiled plasmid DNA in a guide-independent manner, likely by accommodating both DNA strands within the catalytic cleft for the single-strand nicking. Plasmid relaxation is followed by slow processing of plasmid DNA, resulting in its linearization and further degradation, corresponding to the ‘chopping’ activity previously described for TtAgo and MjAgo (11,28). The chopping activity may complicate the use of pAgos as specific genome editing tools, as recently discussed for TtAgo and NgAgo (pAgo from Natronobacterium gregoryi) (31). However, the chopping activity of CbAgo is markedly suppressed if it is preloaded with guide molecules. Furthermore, the guide-independent plasmid processing is suppressed at low concentrations of CbAgo and LrAgo, possibly because of their inefficient binding to dsDNA under these conditions.
Both CbAgo and LrAgo can precisely cut dsDNA at one or more sites when programmed with corresponding guides. This opens the way for development of novel pAgo-based tools for DNA manipulations in vitro and in vivo. Previous attempts to use thermophilic pAgos, such as PfAgo or TtAgo, as programmable nucleases were limited by their low activity at ambient temperature, which required heating the samples with concomitant DNA denaturation (13,29). We showed that mesophilic pAgos can target specific sites in plasmid DNA at lower temperatures, with low efficiency at 37°C (CbAgo only) and with high efficiency at 55°C (both CbAgo and LrAgo)—the temperature compatible with many in vitro applications. Most efficient plasmid cleavage was observed for AT-rich regions but it was also detectable for target regions with higher GC content (up to 50% GC for CbAgo).
One obvious application is the use of mesophilic pAgos in recombinant DNA technology, by analogy with restriction endonucleases but with potential ability to target any site of interest. In contrast to restriction endonucleases and Cas9, guide-directed pAgo can cut only one DNA strand in the dsDNA duplex. Therefore, two strands of DNA can be targeted by pAgo loaded with two guides independently, so ‘sticky’ ends of any desired configuration can be produced. In contrast to Cas nucleases, CbAgo or LrAgo do not require the presence of any specific motifs (such as PAM, protospacer adjacent motif) in the guide or target DNAs which may enable DNA targeting with a single-nucleotide resolution. Furthermore, short DNA oligonucleotides utilized by CbAgo and LrAgo as guide molecules are much easier to synthesize compared to longer RNA guides required for Cas nucleases.
Important problems that need to be solved to allow the use of pAgos in genetic technologies such as genome editing include finding of conditions that would allow specific guide loading and efficient targeting of dsDNA of any sequence, preferably independent of its AT-content, in vivo in eukaryotic cells. In contrast to Cascade complexes and Cas nucleases of the CRISPR systems, pAgos do not unwind dsDNA so they might be assisted by auxiliary cellular factors that could perform DNA melting or recruit pAgos to ssDNA regions. Indeed, DNA cleavage by TtAgo was recently shown to be stimulated by SSB or UvrD helicase (49). Furthermore, negative DNA supercoiling might facilitate the formation of ternary pAgo complexes at locally unwound plasmid or genomic regions (8,13). Analysis of in vivo DNA processing by mesophilic pAgos and its dependence on various cellular activities will be an important goal of future studies.
Supplementary Material
ACKNOWLEDGEMENTS
We thank A. Oguienko for assistance with figure preparation, D. Esyunina and M. Petrova for experimental support and helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Higher Education of the Russian Federation [14.W03.31.0007]. Funding for open access charge: Ministry of Science and Higher Education of the Russian Federation [14.W03.31.0007].
Conflict of interest statement. None declared.
REFERENCES
- 1. Joshua-Tor L. The Argonautes. Cold Spring Harb. Symp. Quant. Biol. 2006; 71:67–72. [DOI] [PubMed] [Google Scholar]
- 2. Peters L., Meister G.. Argonaute proteins: mediators of RNA silencing. Mol. Cell. 2007; 26:611–623. [DOI] [PubMed] [Google Scholar]
- 3. Pratt A.J., MacRae I.J.. The RNA-induced silencing complex: a versatile gene-silencing machine. J. Biol. Chem. 2009; 284:17897–17901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ryazansky S., Kulbachinskiy A., Aravin A.A.. The expanded universe of prokaryotic Argonaute proteins. mBio. 2018; 9:e01935-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Makarova K.S., Wolf Y.I., van der Oost J., Koonin E.V.. Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol. Direct. 2009; 4:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Swarts D.C., Makarova K., Wang Y., Nakanishi K., Ketting R.F., Koonin E.V., Patel D.J., van der Oost J.. The evolutionary journey of Argonaute proteins. Nat. Struct. Mol. Biol. 2014; 21:743–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kaya E., Doxzen K.W., Knoll K.R., Wilson R.C., Strutt S.C., Kranzusch P.J., Doudna J.A.. A bacterial Argonaute with noncanonical guide RNA specificity. PNAS. 2016; 113:4057–4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Swarts D.C., Hegge J.W., Hinojo I., Shiimori M., Ellis M.A., Dumrongkulraksa J., Terns R.M., Terns M.P., van der Oost J.. Argonaute of the archaeon Pyrococcus furiosus is a DNA-guided nuclease that targets cognate DNA. Nucleic Acids Res. 2015; 43:5120–5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang Y., Juranek S., Li H., Sheng G., Tuschl T., Patel D.J.. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008; 456:921–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yuan Y.R., Pei Y., Ma J.B., Kuryavyi V., Zhadina M., Meister G., Chen H.Y., Dauter Z., Tuschl T., Patel D.J.. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. Cell. 2005; 19:405–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zander A., Willkomm S., Ofer S., van Wolferen M., Egert L., Buchmeier S., Stockl S., Tinnefeld P., Schneider S., Klingl A. et al.. Guide-independent DNA cleavage by archaeal Argonaute from Methanocaldococcus jannaschii. Nat. Microbiol. 2017; 2:17034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Olovnikov I., Chan K., Sachidanandam R., Newman D.K., Aravin A.A.. Bacterial argonaute samples the transcriptome to identify foreign DNA. Mol. Cell. 2013; 51:594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Swarts D.C., Jore M.M., Westra E.R., Zhu Y., Janssen J.H., Snijders A.P., Wang Y., Patel D.J., Berenguer J., Brouns S.J.J. et al.. DNA-guided DNA interference by a prokaryotic Argonaute. Nature. 2014; 507:258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lisitskaya L., Aravin A.A., Kulbachinskiy A.. RNA interference and beyond: structure and functions of prokaryotic Argonaute proteins. Nature Communications. 2018; 9:5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Koonin E.V. Evolution of RNA- and DNA-guided antivirus defense systems in prokaryotes and eukaryotes: common ancestry vs convergence. Biol. Direct. 2017; 12:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hegge J.W., Swarts D.C., van der Oost J.. Prokaryotic Argonaute proteins: novel genome-editing tools. Nat. Rev. Microbiol. 2017; 16:5–11. [DOI] [PubMed] [Google Scholar]
- 17. Liu Y., Esyunina D., Olovnikov I., Teplova M., Kulbachinskiy A., Aravin A.A., Patel D.J.. Accommodation of helical imperfections in Rhodobacter sphaeroides Argonaute ternary complexes with guide RNA and target DNA. Cell Rep. 2018; 24:453–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Miyoshi T., Ito K., Murakami R., Uchiumi T.. Structural basis for the recognition of guide RNA and target DNA heteroduplex by Argonaute. Nat. Commun. 2016; 7:11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sheng G., Zhao H., Wang J., Rao Y., Tian W., Swarts D.C., van der Oost J., Patel D.J., Wang Y.. Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage. PNAS. 2014; 111:652–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Y., Sheng G., Juranek S., Tuschl T., Patel D.J.. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008; 456:209–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Willkomm S., Oellig C.A., Zander A., Restle T., Keegan R., Grohmann D., Schneider S.. Structural and mechanistic insights into an archaeal DNA-guided Argonaute protein. Nat. Microbiol. 2017; 2:17035. [DOI] [PubMed] [Google Scholar]
- 22. Parker J.S., Parizotto E.A., Wang M., Roe S.M., Barford D.. Enhancement of the seed-target recognition step in RNA silencing by a PIWI/MID domain protein. Mol. Cell. 2009; 33:204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zander A., Holzmeister P., Klose D., Tinnefeld P., Grohmann D.. Single-molecule FRET supports the two-state model of Argonaute action. RNA Biol. 2014; 11:45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Salomon W.E., Jolly S.M., Moore M.J., Zamore P.D., Serebrov V.. Single-Molecule imaging reveals that argonaute reshapes the binding properties of its nucleic acid guides. Cell. 2015; 162:84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wee L.M., Flores-Jasso C.F., Salomon W.E., Zamore P.D.. Argonaute divides its RNA guide into domains with distinct functions and RNA-binding properties. Cell. 2012; 151:1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haley B., Zamore P.D.. Kinetic analysis of the RNAi enzyme complex. Nat. Struct. Mol. Biol. 2004; 11:599–606. [DOI] [PubMed] [Google Scholar]
- 27. Doxzen K.W., Doudna J.A.. DNA recognition by an RNA-guided bacterial Argonaute. PLoS One. 2017; 12:e0177097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Swarts D.C., Szczepaniak M., Sheng G., Chandradoss S.D., Zhu Y., Timmers E.M., Zhang Y., Zhao H., Lou J., Wang Y. et al.. Autonomous generation and loading of DNA guides by bacterial Argonaute. Mol. Cell. 2017; 65:985–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Enghiad B., Zhao H.. Programmable DNA-guided artificial restriction enzymes. ACS Synth. Biol. 2017; 6:752–757. [DOI] [PubMed] [Google Scholar]
- 30. Lee S.H., Turchiano G., Ata H., Nowsheen S., Romito M., Lou Z., Ryu S.M., Ekker S.C., Cathomen T., Kim J.S.. Failure to detect DNA-guided genome editing using Natronobacterium gregoryi Argonaute. Nat. Biotechnol. 2017; 35:17–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. O’Geen H., Ren C., Coggins N.B., Bates S.L., Segal D.J.. Unexpected binding behaviors of bacterial Argonautes in human cells cast doubts on their use as targetable gene regulators. PLoS One. 2018; 13:e0193818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oganesyan N., Ankoudinova I., Kim S.H., Kim R.. Effect of osmotic stress and heat shock in recombinant protein overexpression and crystallization. Protein Expr. Purif. 2007; 52:280–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Khan S.R., Gaines J., Roop R.M. 2nd, Farrand S.K.. Broad-host-range expression vectors with tightly regulated promoters and their use to examine the influence of TraR and TraM expression on Ti plasmid quorum sensing. Appl. Environ. Microbiol. 2008; 74:5053–5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wong I., Lohman T.M.. A double-filter method for nitrocellulose-filter binding: application to protein-nucleic acid interactions. PNAS. 1993; 90:5428–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Connolly B.A., Liu H.H., Parry D., Engler L.E., Kurpiewski M.R., Jen-Jacobson L.. Assay of restriction endonucleases using oligonucleotides. Methods Mol. Biol. 2001; 148:465–490. [DOI] [PubMed] [Google Scholar]
- 36. Keren N., Kidd M.J., Penner-Hahn J.E., Pakrasi H.B.. A light-dependent mechanism for massive accumulation of manganese in the photosynthetic bacterium Synechocystis sp. PCC 6803. Biochemistry. 2002; 41:15085–15092. [DOI] [PubMed] [Google Scholar]
- 37. Wang Y., Juranek S., Li H., Sheng G., Wardle G.S., Tuschl T., Patel D.J.. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009; 461:754–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ghildiyal M., Zamore P.D.. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 2009; 10:94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Elkayam E., Kuhn C.D., Tocilj A., Haase A.D., Greene E.M., Hannon G.J., Joshua-Tor L.. The structure of human argonaute-2 in complex with miR-20a. Cell. 2012; 150:100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Matsumoto N., Nishimasu H., Sakakibara K., Nishida K.M., Hirano T., Ishitani R., Siomi H., Siomi M.C., Nureki O.. Crystal Structure of Silkworm PIWI-Clade Argonaute Siwi Bound to piRNA. Cell. 2016; 167:484–497. [DOI] [PubMed] [Google Scholar]
- 41. Schirle N.T., MacRae I.J.. The crystal structure of human Argonaute2. Science. 2012; 336:1037–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dahlgren C., Zhang H.Y., Du Q., Grahn M., Norstedt G., Wahlestedt C., Liang Z.. Analysis of siRNA specificity on targets with double-nucleotide mismatches. Nucleic Acids Res. 2008; 36:e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Doench J.G., Sharp P.A.. Specificity of microRNA target selection in translational repression. Genes Dev. 2004; 18:504–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sun G., Wang J., Huang Y., Yuan C.W., Zhang K., Hu S., Chen L., Lin R.J., Yen Y., Riggs A.D.. Differences in silencing of mismatched targets by sliced versus diced siRNAs. Nucleic Acids Res. 2018; 46:6806–6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sheng G., Gogakos T., Wang J., Zhao H., Serganov A., Juranek S., Tuschl T., Patel D.J., Wang Y.. Structure/cleavage-based insights into helical perturbations at bulge sites within T. thermophilus Argonaute silencing complexes. Nucleic Acids Res. 2017; 45:9149–9163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Willkomm S., Zander A., Grohmann D., Restle T.. Mechanistic insights into archaeal and human argonaute substrate binding and cleavage properties. PLoS One. 2016; 11:e0164695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rivas F.V., Tolia N.H., Song J.J., Aragon J.P., Liu J., Hannon G.J., Joshua-Tor L.. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005; 12:340–349. [DOI] [PubMed] [Google Scholar]
- 48. Chen G.R., Sive H., Bartel D.P.. A seed mismatch enhances Argonaute2-Catalyzed cleavage and partially rescues severely impaired cleavage found in fish. Mol. Cell. 2017; 68:1095–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hunt E.A., Evans T.C. Jr, Tanner N.A.. Single-stranded binding proteins and helicase enhance the activity of prokaryotic argonautes in vitro. PLoS One. 2018; 13:e0203073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.