Abstract
Objective
To evaluate association of patients’ clinicopathological data with expression of nicotinamide nucleotide transhydrogenase (NNT) and naturally occurring antisense RNA of the same gene locus (NNT-AS1) in breast cancer samples.
Materials and Methods
In the current case-control study, mean expressions of NNT and NNT-AS1 were assessed in 108 breast tissue samples including 54 invasive ductal carcinoma samples and 54 adjacent non-cancerous tissues (ANCTs) by quantitative reverse transcription-polymerase chain reaction (qRT-PCR).
Results
NNT expression was not significantly different between tumor tissues and ANCTs. However, NNT-AS1 expression was significantly down-regulated in tumor tissues compared to ANCTs (expression ratio=0.51, P=0.01). NNT-AS1 expression was significantly higher in estrogen receptor (ER) negative samples, in comparison with ER positives (P=0.01). No considerable difference was found in the gene expressions between other subcategories of patients. Considerable correlations were detected between expression levels of these two genetic loci in both tumor tissues and ANCTs.
Conclusion
In the current study, for the first time we simultaneously assessed expression of NNT and NNT-AS1 in breast cancer tissues. This study highlights association of ER status with dysregulation of NNT-AS1 in breast cancer tissues. Future researches are necessary to explore the function of this long non-coding RNA (lncRNA) in the pathogenesis of breast cancer.
Keywords: Breast Cancer, Long Non-Coding RNA, NNT
Introduction
Breast cancer, as the most frequent type of female cancer, has prompted several investigators to find diagnostic or prognostic biomarkers among long noncoding RNAs (lncRNAs) (1). This significant fraction of human transcriptome contributes to several aspects of cell physiology; so that dysregulation of them leads to development of cancer (2). An important subgroup of lncRNAs includes natural antisense transcripts (NATs) which are transcribed from the opposite DNA strand in relation to the sense transcripts, so their nucleotide sequences are complementary to the protein coding mRNA and the latter molecule is regulated by them. NATs can either inhibit or activate expression of the sense transcript (3).
Recent studies have highlighted contribution of Nicotinamide nucleotide transhydrogenase (NNT) and the related NAT in the pathogenesis of some human cancer types. NNT has an indispensible role in the homeostasis of NADH and NADPH (4). The significant contribution of NNT in mitochondrial antioxidant pathways shields cells from oxidative stress (5). NNT silencing decreases ability of cancer cells to preserve NAD+ and NADPH levels and suppresses their proliferation and aggressive behavior possibly via alteration of HIF-1α and HDAC1-dependent pathways (4). The related antisense transcript (NNT-AS1) participates in proliferation, migration, invasion and metastasis of colorectal cancer (CRC) cells (6). In hepatocellular carcinoma (HCC), NNT-AS1 expression has enhanced cell proliferation and inhibited cycle arrest as well as apoptosis through modulation of miR- 363/CDK6 axis (7).
NNT loss has led to accumulation of acetylated catabolic substances of polyamines and a subsequent diminution of spermine and spermidine. Polyamine catabolism would mutually be elicited by oxidative stress and over-produce hydrogen peroxide, resulting in a malicious cycle that accelerates reactive oxygen species (ROS) production (8, 9). In spite of the appreciated role of these processes in pathogenesis of breast cancer (8), no study has yet assessed the simultaneous significance of NNT and NNT-AS1 dysregulation in breast cancer. Consequently, we conducted current study to assess expression of NNT and NNT-AS1 in breast cancer tissues compared to ANCTs in association with patients’ clinicopathological data, to find whether their transcript levels are altered in breast cancer in parallel, or they can be used as biomarkers of breast cancer.
Materials and Methods
Patients
For the current case-control study, a total of 108 breast tissue samples -including 54 invasive ductal carcinoma samples and 54 ANCTs- were excised during surgery from patients hospitalized in Farmanieh and Sina Hospitals (Tehran, Iran) during January 2017-January 2018. Patients with definite diagnosis of invasive ductal carcinoma were included in the study. Those with other types of breast cancer and those received chemo-/radio-therapy before surgery were excluded from the study. All patients signed the written informed consent forms. The study protocol was permitted by the Ethical Committee of Shahid Beheshti University of Medical Sciences (IR.REC. SBMU.1397.764), Iran. Clinical and demographical data of patients were collected through evaluation of medical reports and interviews with patients.
Expression analysis
Total RNA was extracted from tumor tissues and ANCTs using the TRIzol™ reagent (Invitrogen, USA) based on the company guidelines. In brief, we homogenized 75 mg of tissues in 1 ml TRIzol™ reagent, followed by RNA precipitation using isopropanol and washing it in 75% ethanol (10). After assessment of quality and quantity of the isolated total RNAs, a proportion of each RNA sample was converted to cDNA, using the RevertAid First Strand cDNA Synthesis Kit (TaKaRa, Japan). Transcript levels of NNT and NNT-AS1 genes were compared between tumor tissues and ANCTs using rotor gene 6000 Real-Time PCR System (Corbett Research, Australia). TaqMan Fast Universal PCR Master Mix (Applied Biosystems, USA) was used for the expression study. The Hypoxanthineguanine phosphoribosyl transferase (HPRT) gene was used for normalization of the gene and lncRNA expressions. The nucleotide sequences of primers are as follow:
-
HPRT1-
F: 5ˊ-AGCCTAAGATGAGAGTTC-3ˊ,
R: 5ˊ-CACAGAACTAGAACATTGATA-3ˊ,
FAM-CATCTGGAGTCCTATTGACATCGC-TAMRA;
-
NNT1-
F: 5ˊ-AGCCACCTTCTGTGTTACTTGC-3ˊ,
R: 5ˊ-TAGCCCAGAGCTGCCATGAC-3ˊ,
FAM-TCAACCGTCAGGCTGCCACTGCTG-TAMRA
-
NNT-AS1-
F: 5ˊ-CTTCCACTCTCGGGGACAGG-3ˊ,
R: 5ˊ-GCACCAGGTTTGATTGACAAGG-3ˊ,
FAM-TTGTCTCTGCCTCGGCCTGCGG-TAMRA.
PCR efficiency and threshold cycle (Ct) values were obtained to quantify relative expression of each genetic locus in tumor tissues and ANCTs.
Statistical analysis
SPSS software version 18.0 (SPSS Inc., USA) was used for statistical analysis. Ct values obtained from qRT-PCR experiments were adjusted based on the PCR efficiency values. Association of patients’ data with relative expression of the gene or lncRNA (down-/ up-regulation in the tumor samples vs. ANCTs) was evaluated using Chi-square test. Relative expression of each genetic loci in each tumor sample was calculated using EfficiencyCt reference/ EfficiencyCt target formula. Data is presented as mean ± SD. The difference in these values between individual groups of patients was evaluated using Tukey’s honest significance test. The pairwise correlation between relative transcription levels of the genetic loci in each set of samples (tumor tissues and ANCTs) was calculated using the regression model. For all statistical analyses, P<0.05 was regarded as significant. The receiver operating characteristic (ROC) curve was plotted to assess the power of genetic loci expression levels for diagnosis of disease status in breast samples. The Youden index (j) was applied to get the highest difference between sensitivity (true-positive rate) and 1-specificity (falsepositive rate).
Results
Overall demographic and clinical information of patients
Demographic and clinical information of the study participants are reported in Table 1.
Transcript levels of NNT and NNT-AS1 in tumor tissues and ANCTs
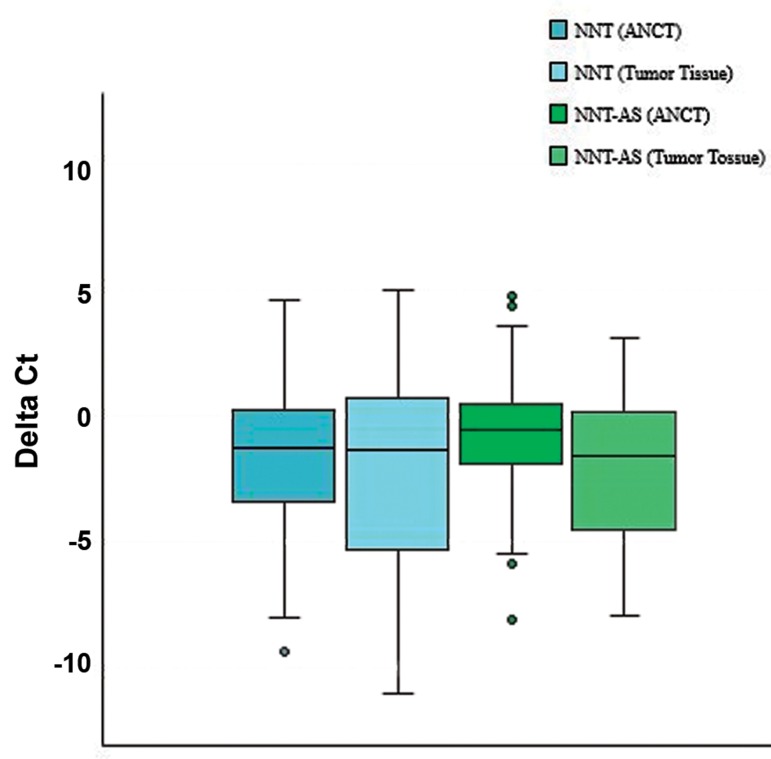
NNT expression level was not significantly different between tumor tissues and ANCTs. However, NNT-AS1 expression was significantly down-regulated in tumor tissues compared to ANCTs (expression ratio=0.51, P=0.01). Figure 1 shows relative expression of NNT and NNT-AS1 in each set of samples, as described by -ΔCt values (Ct reference-Ct target).
Table 1.
General demographic and clinical data of patients
| Variables | Values | |
|---|---|---|
| Age (Y) | 51.79 ± 13.54 (29-81) | |
| Menarche age (Y) | 13 ± 1.65 (10-18) | |
| Menopause age (Y) | 44.91 ± 14.91 (38-60) | |
| First pregnancy age (Y) | 18.04 ± 8.36 (14-32) | |
| Breast feeding duration (months) | 41.62 ± 34.1 (3-120) | |
| Positive family history for other cancers | 17 | |
| Cancer stage | ||
| I | 30.8 | |
| II | 28.8 | |
| III | 30.8 | |
| IV | 9.6 | |
| Overall grade | ||
| I | 17 | |
| II | 49 | |
| III | 34 | |
| Mitotic rate | ||
| I | 45.2 | |
| II | 42.9 | |
| III | 11.9 | |
| Tumor size | ||
| <2 cm | 32 | |
| ≥2 cm, <5cm | 66 | |
| ≥5 cm | 2 | |
| Estrogen receptor | ||
| Positive | 87.8 | |
| Negative | 12.2 | |
| Progesterone receptor | ||
| Positive | 77.1 | |
| Negative | 22.9 | |
| HER2/Neu expression | ||
| Positive | 25 | |
| Negative | 75 | |
Data are presented as mean ± SD (range) or %.
Fig.1.
Relative transcription of NNT and NNT-AS1 in tumor tissues (n=54) and ANCTs (n=54).
NNT; Nicotinamide nucleotide transhydrogenase, NNT-AS1; Nicotinamide nucleotide transhydrogenaseantisense 1, ANCTs; Adjacent non-cancerous tissues, and ct; Threshold cycle.
Association between relative expression of genetic loci and patients’ clinicopathological information
Based on the transcriptions in each tumor sample compared to related ANCT (<1 or >1), we categorized patients to up-/down-regulated groups. Then, we evaluated associations between such values and patients’ clinicopathological data. No association was found between relative expression of genetic loci in tumor tissue vs. ANCT and any of tumor characteristics (Table 2).
Table 2.
Association of relative transcriptions in tumor tissues compared to ANCTs, with patients’ clinicopathological data
| Parameters | NNTUp-regulation | NNTDown-regulation | P value | NNT-AS1Up-regulation | NNT-AS1Down-regulation | P value | |
|---|---|---|---|---|---|---|---|
| Age | 0.4 | 0.79 | |||||
| <55 Y | 15 (45.5) | 18 (54.5) | 12 (35.3) | 22 (64.7) | |||
| ≥55 Y | 6 (33.3) | 12 (66.7) | 7 (38.9) | 11 (61.1) | |||
| Stage | 0.9 | 0.76 | |||||
| I | 6 (37.5) | 10 (64.5) | 5 (33.3) | 10 (66.7) | |||
| II | 5 (35.7) | 9 (64.3) | 7 (46.7) | 8 (53.3) | |||
| III | 7 (46.7) | 8 (53.3) | 5 (56.3) | 11 (43.7) | |||
| IV | 2 (50) | 2 (50) | 1 (25) | 3 (75) | |||
| Histological grade | 0.38 | 0.97 | |||||
| I | 5 (62.5) | 3 (37.5) | 3 (37.5) | 5 (62.5) | |||
| II | 8 (34.8) | 15 (65.2) | 8 (36.4) | 14 (63.6) | |||
| III | 5 (37.5) | 8 (62.5) | 5 (33.3) | 10 (66.7) | |||
| Mitotic rate | 0.57 | 0.26 | |||||
| I | 8 (42.1) | 11 (57.9) | 9 (50) | 9 (50) | |||
| II | 5 (33.3) | 10 (66.7) | 4 (23.5) | 13 (76.5) | |||
| III | 3 (60) | 2 (40) | 2 (40) | 3 (60) | |||
| Tumor size | 0.67 | 0.31 | |||||
| <2 | 7 (43.8) | 9 (56.2) | 7 (43.8) | 9 (56.2) | |||
| 2-5 | 12 (38.7) | 19 (61.3) | 10 (32.3) | 21 (67.7) | |||
| >5 | 0 (0) | 1 (100) | 1 (100) | 0 (0) | |||
| ER status | 0.84 | 0.33 | |||||
| Positive | 17 (40.5) | 21 (58.3) | 15 (36.6) | 26 (63.4) | |||
| Negative | 2 (40) | 3 (60) | 1 (16.7) | 5 (83.3) | |||
| PR status | 0.92 | 0.54 | |||||
| Positive | 15 (41.7) | 21 (58.3) | 13 (37.1) | 22 (62.9) | |||
| Negative | 4 (40) | 6 (60) | 3 (27.3) | 8 (72.7) | |||
| HER2 status | 0.51 | 0.54 | |||||
| Positive | 4 (33.3) | 8 (66.7) | 3 (27.3) | 8 (72.7) | |||
| Negative | 15 (44.1) | 19 (54.3) | 13 (37.1) | 22 (62.9) | |||
NNT; Nicotinamide nucleotide transhydrogenase, NNT-AS1;Nicotinamide nucleotide transhydrogenase-antisense 1, ANCTs; Adjacent noncancerous tissues, ER; Estrogen receptor, PR; Progesterone receptor, and HER2; Human epidermal growth factor receptor 2.
We also calculated relative expression of each genetic loci in tumor samples using EfficiencyCt reference/ EfficiencyCt target formula and compared these values between tumor subgroups. We detected significantly higher expression of NNT-AS1 in ER negative samples compared to ER positive cases (P=0.01). No remarkable difference was found between transcription levels of the other patient subcategories (Table 3).
Table 3.
Association of transcription levels in tumor tissues with tumors characteristics (mean ± SD values of EfficiencyCt reference- EfficiencyCt target arepresented)
| Parameters | NNT | P value | NNT-AS1 | P value | |
|---|---|---|---|---|---|
| Age | |||||
| <55 vs. ≥55 Y | 71.97 (221.19) vs. 2.6 (4.63) | 0.18 | 33.96 (127.22) vs. 2.79 (4.94) | 0.29 | |
| ER status | |||||
| ER(+) vs. ER(-) | 54.76 (198.15) vs. 20.84 (45.58) | 0.68 | 10.58 (39.1) vs. 119.63 (289.12) | 0.01 | |
| PR status | |||||
| PR(+) vs. PR(-) | 63.53 (212.69) vs. 11.72 (33.89) | 0.42 | 12.23 (42) vs. 57.81 (213.71) | 0.15 | |
| HER2 status | |||||
| HER2 (+) vs. HER2 (-) | 1.17 (2.78) vs. 68.49 (215.27) | 0.28 | 1.53 (4.68) vs. 32.06 (123.7) | 0.4 | |
| Tumor grade | |||||
| Grade I vs. II | 84.71 (222.77) vs. 28.58 (131.89) | 0.76 | 5.61 (7.02) vs. 13.89 (52.76) | 0.98 | |
| Grade I vs. III | 84.71 (222.77) vs. 75.51 (253.41) | 0.99 | 5.61 (7.02) vs. 53.38 (182.01) | 0.58 | |
| Grade II vs. III | 28.58 (131.89) vs. 75.51 (253.41) | 0.74 | 13.89 (52.76) vs. 53.38 (182.01) | 0.53 | |
NNT; Nicotinamide nucleotide transhydrogenase, NNT-AS1; Nicotinamide nucleotide transhydrogenase-antisense 1, ct; Threshold cycle, ER; Estrogen receptor, PR; Progesterone receptor, and HER2; Human epidermal growth factor receptor 2. Data are presented as mean (SD) values.
Correlations between transcript levels of NNT and NNT-AS1 in tumor tissues and ANCTs
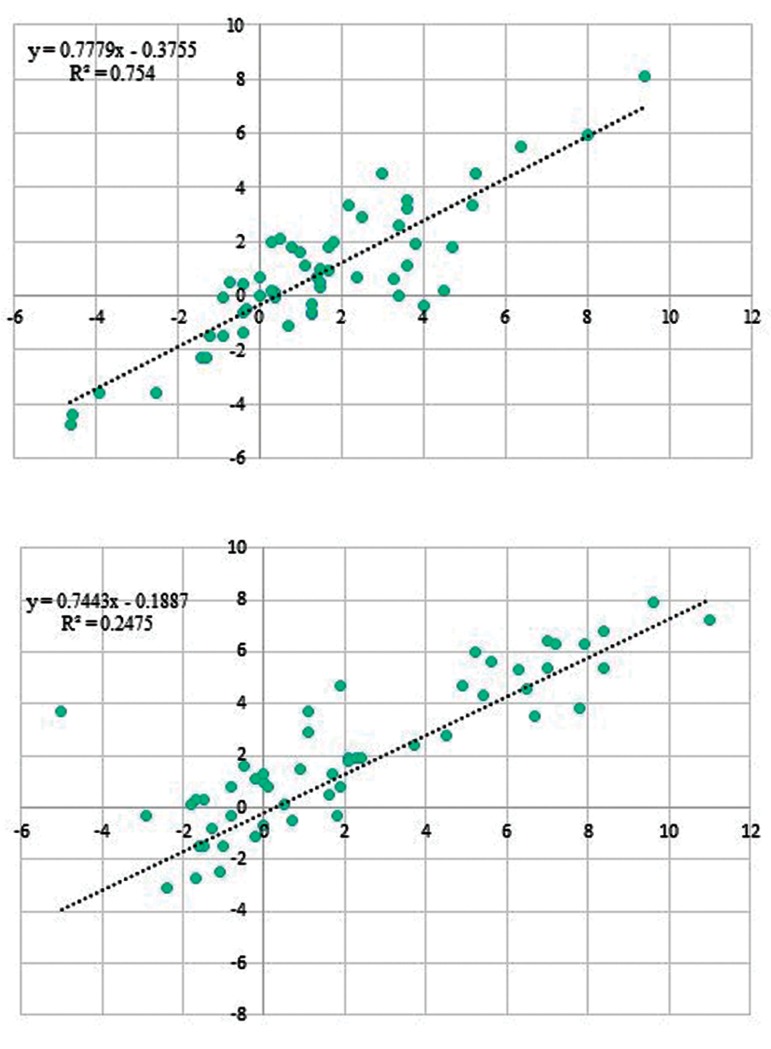
Correlations between transcript level of NNT and its naturally occurring antisense was assessed in both tumor and ANCT samples. Transcription levels of NNT were correlated with the expression of NNTAS1 in both ANCT and tumor samples (Fig .2A, B, respectively). Considering R2 values, the correlation was stronger in ANCTs compared to tumor tissues.
Fig.2.
Correlation of NNT and NNT-AS1 transcription levels. A. Shows the correlation in ANCTs and B. Shows the correlations in tumor tissues.
NNT; Nicotinamide nucleotide transhydrogenase, NNT-AS1; Nicotinamide nucleotide transhydrogenase- antisense 1, and ANCTs; Adjacent noncancerous tissues.
Receiver operating characteristic curve analysis
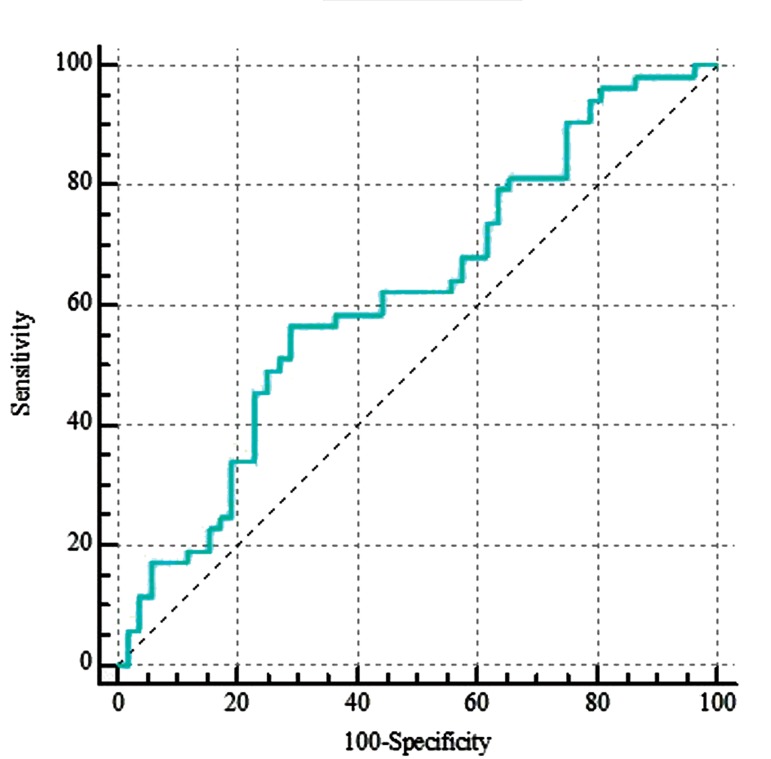
The power of NNT-AS1 expression in prediction of disease status in breast samples was evaluated using ROC curve (Fig .3). Assessment of this lncRNA transcription level shows 71.2% specificity and 56.6% sensitivity for breast cancer diagnosis.
Fig.3.
Results of receiver operating characteristic (ROC) curve assessment analysis for NNT-AS1 performance in breast cancer diagnosis. The values were analysed based on EfficiencyCt reference - EfficiencyCt target .
Discussion
In the current study, for the first time we simultaneously assessed expression of NNT and NNT-AS1 in breast cancer tissues in comparison with ANCTs and found downregulation of NNT-AS1 in tumor tissues, in spite of detecting similar level of NNT expression in tumor tissues and ANCTs. NNT-AS1 has previously been shown to exert oncogenic effects in CRC, HCC, osteosarcoma and cervical cancer (6, 7, 11, 12). However, expression of this lncRNA was significantly decreased in patients with ovarian cancer as well as the human ovarian cancer cell lines. Moreover, in vitro studies has shown that NNT-AS1 silencing enhances cell migration and invasion, while it suppresses apoptosis (13). So, the observed down-regulation of NNT-AS1 in the current study is consistent with the previously reported dysregulation of this molecule in ovarian cancer. This lncRNA is transcribed in the opposite direction of NNT gene and it has no intersection with latter gene nucleotide sequence (14). Most recently, Li et al. (14) demonstrated overexpression of NNT-AS1 in breast tumor tissues compared to ANCTs in correlation with patients’ survival and HER2, but not ER, status. In vitro experiments showed that NNT-AS1 contributes to breast cancer pathogenesis via altering miR-142-3p/ZEB1 axis. The inconsistency between our results and their results might be due to a possible difference in the mean age of the study participants or an ethnic-based modulator of transcription. They have shown that ZEB1 is positively regulated by NNT-AS1. In our previous study, on the same cohort of patients, we failed to demonstrate any significant change in ZEB1 expression between tumor tissues and ANCTs (15). So, we hypothesized that NNT-AS1 might participate in the pathogenesis of breast cancer through other mechanisms, including regulation of NNT expression. Significant correlation between NNT and NNT-AS1 expressions, especially in non-tumor tissues, implies the presence of a feed-forward loop between these two genetic loci which should be assessed in future studies.
In spite of totally down-regulation of NNT-AS1 in tumor tissues compared to ANCTs, we demonstrated higher expression of it in ER negative tumor samples, compared to ER positive samples, likely suggesting the importance of this lncRNA in pathogenesis of ER negative breast cancers. Future studies are needed to evaluate expression of NNT-AS1 in larger cohorts of patients with regards to hormone receptor status.
Finally, we evaluated the power of NNT-AS1 expression in prediction of the disease status in breast samples. Although transcription level of this genetic locus is not individually a sensitive marker for prediction of breast cancer, it might increase specificity of other putative panels of gene expression.
Conclusion
The current study shows down-regulation of NNT-AS1 in breast cancer tissues compared to ANCTs in association with ER status. Future studies are necessary to explore function of this lncRNA in the pathogenesis of breast cancer
Acknowledgments
The current study was financially supported by Shahid Beheshti University of Medical Sciences (grant number: 15033), Iran. There is no conflict of interest in this study.
Author’s Contributions
S.S.G., V.K.O., M.D.O, S.G.-F; Contributed to conception and design. M.T.; Contributed to all experimental work, data and statistical analysis, as well as interpretation of data. M.D.O., S.G.-F.; Supervised the experiments. S.G.-F.; Drafted the manuscript, which was revised by M.D.O. and S.S.G. All authors read and approved the final manuscript.
References
- 1.Tasharrofi B, Soudyab M, Nikpayam E, Iranpour M, Mirfakhraie R, Sarrafzadeh S, et al. Comparative expression analysis of hypoxiainducible factor-alpha and its natural occurring antisense in breast cancer tissues and adjacent noncancerous tissues. Cell Biochem Funct. 2016;34(8):572–578. doi: 10.1002/cbf.3230. [DOI] [PubMed] [Google Scholar]
- 2.Nikpayam E, Tasharrofi B, Sarrafzadeh S, Ghafouri-Fard S. The role of long non-coding RNAs in ovarian cancer. Iran Biomed J. 2017;21(1):3–15. doi: 10.6091/.21.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fang Y, Fullwood MJ. Roles, Functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics. 2016;14(1):42–54. doi: 10.1016/j.gpb.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ho HY, Lin YT, Lin G, Wu PR, Cheng ML. Nicotinamide nucleotide transhydrogenase (NNT) deficiency dysregulates mitochondrial retrograde signaling and impedes proliferation. Redox Biol. 2017;12:916–928. doi: 10.1016/j.redox.2017.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chortis V, Taylor AE, Doig CL, Walsh MD, Meimaridou E, Jenkinson C, et al. Nicotinamide Nucleotide Transhydrogenase as a novel treatment target in adrenocortical carcinoma. Endocrinology. 2018;159(8):2836–2849. doi: 10.1210/en.2018-00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Q, Yang L, Hu X, Jiang Y, Hu Y, Liu Z, et al. Upregulated NNT-AS1, a long noncoding RNA, contributes to proliferation and migration of colorectal cancer cells in vitro and in vivo. Oncotarget. 2017;8(2):3441–3453. doi: 10.18632/oncotarget.13840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu YB, Jiang Q, Yang MY, Zhou JX, Zhang Q. Long noncoding RNA NNT-AS1 promotes hepatocellular carcinoma progression and metastasis through miR-363/CDK6 axis. Oncotarget. 2017;8(51):88804–88814. doi: 10.18632/oncotarget.21321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pirinen E, Kuulasmaa T, Pietilä M, Heikkinen S, Tusa M, Itkonen P, et al. Enhanced polyamine catabolism alters homeostatic control of white adipose tissue mass, energy expenditure, and glucose metabolism. Mol Cell Biol. 2007;27(13):4953–4967. doi: 10.1128/MCB.02034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chopra S, Wallace HM. Hydrogen peroxide induces the catabolism of polyamines in human breast cancer cells. Biochem Soc Trans. 1996;24(2):230S–230S. doi: 10.1042/bst024230s. [DOI] [PubMed] [Google Scholar]
- 10.Rio DC, Ares M Jr, Hannon GJ, Nilsen TW. Purification of RNA using TRIzol (TRI reagent) Cold Spring Harb Protoc. 2010;2010(6):pdb–pdb. doi: 10.1101/pdb.prot5439. prot5439. [DOI] [PubMed] [Google Scholar]
- 11.Hua F, Liu S, Zhu L, Ma N, Jiang S, Yang J. Highly expressed long non-coding RNA NNT-AS1 promotes cell proliferation and invasion through Wnt/beta-catenin signaling pathway in cervical cancer. Biomed Pharmacother. 2017;92:1128–1134. doi: 10.1016/j.biopha.2017.03.057. [DOI] [PubMed] [Google Scholar]
- 12.Ye H, Lin J, Yao X, Li Y, Lin X, Lu H. Overexpression of Long Non- Coding RNA NNT-AS1 Correlates with Tumor Progression and Poor Prognosis in Osteosarcoma. Cell Physiol Biochem. 2018;45(5):1904–1914. doi: 10.1159/000487966. [DOI] [PubMed] [Google Scholar]
- 13.Huang Y, Shi J, Xu Y. Long non-coding RNA NNT-AS1 contributes to cell proliferation, metastasis and apoptosis in human ovarian cancer. Oncol Lett. 2018;15(6):9264–9270. doi: 10.3892/ol.2018.8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Lv M, Song Z, Lou Z, Wang R, Zhuang M. Long non-coding RNA NNT-AS1 affects progression of breast cancer through miR- 142-3p/ZEB1 axis. Biomed Pharmacother. 2018;103:939–946. doi: 10.1016/j.biopha.2018.04.087. [DOI] [PubMed] [Google Scholar]
- 15.Nikpayam E, Soudyab M, Tasharrofi B, Sarrafzadeh S, Iranpour M, Geranpayeh L, et al. Expression analysis of long non-coding ATB and its putative target in breast cancer. Breast Dis. 2017;37(1):11–20. doi: 10.3233/BD-160264. [DOI] [PubMed] [Google Scholar]