Abstract
Background
Qnr genes are known to confer a low-level resistance to fluoroquinolone in Enterobacteriaceae. They are often found on the same resistance plasmids as extended spectrum β-lactamase (ESBL) and constitute the most common antibiotic resistance mechanism. This study aimed to detect the presence of qnr genes in ESBL-producing E. coli and Klebsiella spp.
Methods
From May 2013 to July 2015, 91 E. coli and 64 Klebsiella spp. strains with phenotypic resistance to quinolone were collected from several specimens and analyzed for the detection of qnrA, qnrB, qnrS genes and the β-lactamase resistance genes (blaCTX-M, blaTEM, blaSHV) using simplex and multiplex PCR.
Results
In the present study, 107 (69%; 61 E. coli and 46 Klebsiella spp.) of 155 bacterial strains tested were found harboring at least one qnr gene consisting of 74 (47.74%) qnrB, 73 (47.10%) qnrS and 4 (2.58%) qnrA. Of the 107 strains encoding qnr genes, 102, 96 and 52 carried CTX-M1, TEM and SHV type ESBL respectively.
Conclusion
This study identified quinolone resistance (qnr) gene in ESBL-producing E. coli and Klebsiella spp. in Togo. These finding which suggest a possible resistance to quinolone are of high interest for better management of patients and control of antimicrobial resistance in Togo.
Electronic supplementary material
The online version of this article (10.1186/s13756-019-0552-0) contains supplementary material, which is available to authorized users.
Keywords: E. coli, Klebsiella spp., ESBL, Qnr gene, Togo
Background
Quinolones and β-lactams are classes of extensively used molecules worldwide in the treatment of many infectious diseases [1]. Quinolones are synthetic antibiotics used for infections involving Gram-negative bacteria such as Enterobacteriaceae. Fluoroquinolones have broad-spectrum intrinsic activity greater than quinolones [2].
Three main mechanisms of quinolone resistance have been described: i) the accumulation of mutations in the genes encoding quinolone target DNA gyrase and topoisomerase IV; ii) a decrease of intracellular concentration of fluoroquinolones by porins down-regulation or modification of the efflux pumps activity, iii) the acquisition of plasmid resistance genes [2].
The acquisition of plasmid-mediated quinolone resistance genes (PMQR) leads to the protection of quinolone’s targets by qnr proteins belonging to the pentapeptide repeat (PRP) family and the hydrolysis of quinolones by the aac (6′)-Ib-cr protein [2, 3]. Mechanism of plasmid-mediated quinolone resistance leads to a low level of fluoroquinolone resistance and facilitates the selection of mutant strains with a high level of fluoroquinolone resistance [3, 4].
Since the discovery of plasmid quinolone resistance genes, a large number of qnr alleles have been found on plasmids or bacterial chromosome. About 100 qnr genes variant have been described mainly from Enterobacteriaceae, and grouped into 5 distinct families: qnrA, qnrB, qnrC, qnrD and qnrS [3, 5].
Several surveys, based on molecular approaches, have found a strong association between qnr-positive and ESBL-positive isolates [5–8]. The presence of qnr genes in ESBL-producing Enterobacteriaceae has been reported in Europe, United States, Asia and Africa [9–13]. In Niger, qnr genes (9.5% of qnrA, 26.2% of qnrB and 64.3% of qnrS) were found in ESBL-producing Enterobacteriaceae among fecal commensal of children with severe malnutrition [14].
In Togo, results from previous studies revealed the presence of beta-lactamase gene CTX-M1 (95.73%), TEM (82.31%) and SHV (45.12%) in ESBL-producing E. coli and Klebsiella spp. The production of ESBL was associated with high co-resistance to fluoroquinolone (93% for ciprofloxacin), aminoglycosides (76.36% for gentamicin) and trimethoprim/sulfamethoxazole (95.65%) [15, 16]. This high level of multidrug resistance suggests acquisition of plasmid-mediated antibiotic resistance factors in these strains. PMQR determinants as qnr genes was also usually found in multidrug resistance plasmid among Enterobacteriaceae producing-ESBL especially E. coli and species of Klebsiella [3, 6, 17, 18]. In this study we are interested in fluoroquinolone resistance. Here, we report the frequency of qnrA, qnrB and qnrS genes in ESBL-producing E. coli and Klebsiella spp.
Methods
Samples collection and identification
Well characterized Escherichia coli and Klebsiella spp. strains were collected during a prospective study from May 2013 to July 2015 in the bacteriology laboratory of the National Institute of Hygiene (INH) in Lomé, Togo. This public health institute is specialized in biomedical analysis, epidemiological surveillance, immunization, water, and food quality control. Strains were isolated from various pathological specimens including urine, vaginal swabs, pus, and sperm samples. Standard microbiological methods were used to isolate and purify bacterial strains on Mac-Conkey or Eosin Methylene Blue (EMB) media. Strains were identified using the API 20E identification system (API 20 E, Identification System for Enterobacteriaceae and others non-fastidious Gram-negative rod; BioMérieux, Marcy-Etoile, France). The API 20 E system is a standardized technique allowing only the biochemical identification of an Enterobacterial strain using an isolated colony.
Susceptibility test and ESBL phenotype detection
Antibiotic susceptibility test was performed and interpreted according to the 2014 recommendations of Antibiogram Committee of the French Society of Microbiology [19].
Antibiotics were purchased from BioRad (Marnes-la-Coquette, France) and included amoxicillin + clavulanate (AMC, 20/10 μg), piperacillin-tazobactam (TZP, 75/10 μg), cefoxitin (FOX, 30 μg), ceftriaxon (CRO, 30 μg), ceftazidim (CAZ, 30 μg), cefotaxim (CTX, 30 μg), cefepim (FEP, 30 μg), aztreonam (ATM, 30 μg), imipenem (IPM, 10 μg), amikacin (AKN, 30 μg), gentamicin (G, 15 μg), nalidixic acid (NA, 30 μg) ciprofloxacin (CIP, 5 μg), trimethoprim-sulfamethoxazole (SXT 1.25/23,75 μg), fosfomycin (FOS, 50 μg), doxycycline (DOX, 30 μg).
All isolates were subjected to the double disc synergy test for ESBL detection [20]. The presence of ESBL is detected by a synergy between ceftazidim and cefotaxim or ceftriaxon discs and amoxicillin + clavulanic acid disc.
E. coli ATCC 25922 strain was used as a control for antibiotic susceptibility testing.
Escherichia coli and Klebsiella spp. strains resistant to at least one third generation cephalosporin (cephalosporin, ceftazidim, ceftriaxon or cefotaxim) were collected in a storage medium (trypticase soy broth TCS) and stored at − 80 °C. Samples were then sent under strict transportation conditions (in triple packaging boxes with ice packs), to Molecular Biology Laboratory of CERBA/LABIOGENE in Ouagadougou, Burkina Faso for qnr and ESBL genes detection on in January 2018.
Extraction of bacterial DNA
Rapid DNA extraction was performed using a boiling technique. Shortly, strains from TCS broth were reactivated on TCS agar for 18–24 h and two or three isolated colonies were inoculated in Luria Bertani (LB, 2 mL). After 18–24 h of overnight culture, LB broth were centrifuged at 10000 rpm/min for 10 min and the pellet suspended in 500 μL of phosphate buffer (100 mM, pH 7) to cell-wall weakening. The mix was heated at 100 °C for 15 min in a water bath to release bacterial nucleic acid.
DNA was then precipitated in 250 μL of absolute ethanol, washed twice in 1000 μL of ethanol 75%, dried and re-suspended in 100 μL of sterile water.
PCR amplification
DNA samples (5 μL) were subjected to multiplex PCR in a 25 μL reaction mixture as previously described by Robicsek [21] for Qnr genes and Dallenne [22] for ESBL genes blaTEM and blaSHV using GeneAmp® PCR System 9700 (Applied Biosystems, California USA).
Qnr genes (qnrA, qnrB and qnrS) amplification was performed using the following thermal cycling profile: 32 cycles consisting of 45 s at 95 °C for denaturation, 45 s at 53 °C for annealing and 60 s at 72 °C for extension.
For the blaTEM and blaSHV ESBL genes, multiplex PCR amplification conditions were as follows: initial denaturation step at 94 °C for 10 min; 30 cycles of denaturation at 94 °C for 40s, annealing at 60 °C for 40s, extension at 72 °C for 1 min, followed by a final extension step at 72 °C for 7 min. However, the amplification of BlaCTX-M-G1 was carried out as previously described by Pagani [23] in 25 μL reaction mixture according to the following PCR program: initial denaturation at 96 °C for 10 min. 35 cycles of denaturation at 94 °C for 1 min, annealing at 50 °C for 1 min and extension at 72 °C for 1 min. Final extension at 72 °C for 10 min. Negative (DNA from E. coli ATCC 25922) and positive controls (DNA from qnr B and S genes positive strains) were used to check potential unspecific amplification. Specific sequences primers provided by Applied Biosystems (California, USA) are shown in Table 1. DNA fragments were analyzed by electrophoresis in a 2% agarose gel at 100 V for 1 h in TBE 1X containing ethidium bromide using 100-bp DNA ladder (Promega, USA) as a size marker.
Table 1.
Primers used for PCR amplification of qnr and bla genes identification
| Bla Genes | Sequence (5′ – 3′) | Size (pb) | References |
|---|---|---|---|
| qnrA |
For: ATTTCTCACGCCAGGATTTG Rev.: GATCGGCAAAGGTTAGGTCA |
516 | [21] |
| qnrB |
For: GATCGTGAAAGCCAGAAAGG Rev.: ACGATGCCTGGTAGTTGTCC |
469 | [21] |
| qnrS |
For: ACGACATTCGTCAACTGCAA Rev.: TAAATTGGCACCCTGTAGGC |
417 | [21] |
| TEM |
For: CATTTCCGTGTCGCCCTTATTC Rev.: CGTTCATCCATAGTTGCCTGAC |
800 | [22] |
| SHV |
For: AGCCGCTTGAGCAAATTAAAC Rev.: ATCCCGCAGATAAATCACCAC |
713 | [22] |
| CTX-M-G1 |
For: GTTACAATGTGTGAGAAGCAG Rev.: CCGTTTCCGCTATTACAAAC |
1000 | [23] |
Statistical analysis
Statistical analysis was performed using Epi Info Version 7.1.1.14 software. Fisher’s exact test was used for comparison and the difference was statistically significant when p < 0.05.
Results
Bacterial strains
A sample of 155 strains, 91 E. coli and 64 Klebsiella spp. (55 Klebsiella pneumoniae and 9 Klebsiella oxytoca) resistant to at least one third generation cephalosporin (ceftazidim, cefotaxim or ceftriaxon) were collected during the study period. Bacteria were isolated from urine 91/155 (58.71%), vaginal samples 38/155 (24.52%), wound swabs 15/155 (9.69%), semen samples 6/155 (3.87%), urethral curettage 2/155 (1.29%), sputum 1/155 (0.65%), stool 1/155 (0.65%) and joint fluid 1/155 (0.65%).
Antibiotic susceptibility profile
All E. coli strains were resistant to ceftriaxon and cefotaxim and 97.80% to ceftazidim. The resistance rates to other β-lactam antibiotics were 2.20% for imipenem (very low levels), 17.59% for piperacillin-tazobactam and 25.35% for cefoxitin. Quinolones, nalidixic acid and ciprofloxacin were very inactive with rates of 96.67% and 94.51% respectively. Among aminoglycosides, the strains were more resistant to gentamicin (75.82%) in contrast to amikacin which showed very low levels of resistance (3.30%).
All Klebsiella spp. strains were resistant to cefepim. Resistance to ceftazidime and ceftriaxone was 98.44% (63/64) each one, and to cefotaxime 96.88% (62/64). Nalidixic acid, ciprofloxacin, doxycycline and trimethoprim-sulfamethoxazole were also inactive antibiotics with at least a resistance rate of 90%. Only imipenem, amikacin and fosfomycin were very active on Klebsiella spp. strains with a low resistance rate (< 5%). The resistance profile to other beta-lactams and other antibiotics for all isolates is presented in Table 2.
Table 2.
Antibiotic susceptibility profile
| ATB | E. coli | Klebsiella spp | Total | |||
|---|---|---|---|---|---|---|
| R (n/N) | R (%) | R (n/N) | R (%) | R (n/N) | R (%) | |
| TZP | 16/91 | 17.58 | 19/24 | 29.69 | 35/155 | 22.58 |
| FOX | 18/71 | 25.35 | 12/52 | 23.08 | 30/123 | 24.39 |
| CAZ | 89/91 | 97.80 | 63/64 | 98.44 | 152/155 | 98.06 |
| CRO | 91/91 | 100 | 63/64 | 98.44 | 154/155 | 99.35 |
| CTX | 91/91 | 100 | 62/64 | 96.88 | 153/155 | 98.71 |
| FEP | 89/91 | 97.80 | 64/64 | 100 | 153/155 | 98.71 |
| ATM | 90/91 | 98.90 | 63/64 | 98.44 | 153/155 | 98.71 |
| IMP | 2/91 | 2.20 | 1/64 | 1.56 | 3/155 | 1.94 |
| G | 69/91 | 75.82 | 51/64 | 79.69 | 120/155 | 77.42 |
| AKN | 3/91 | 3.30 | 1/64 | 1.56 | 4/155 | 2.58 |
| NA | 87/90 | 96.67 | 51/63 | 80.95 | 138/155 | 90.20 |
| CIP | 86/91 | 94.51 | 58/64 | 90.63 | 144/155 | 92.90 |
| FOS | 4/90 | 4.44 | 3/64 | 4.69 | 7/154 | 4.55 |
| SXT | 85/90 | 94.44 | 56/61 | 96.72 | 144/151 | 95.36 |
| DOX | 86/89 | 96.63 | 58/63 | 92.06 | 144/152 | 94.74 |
ATB antibiotic, R resistant, TZP piperacillin-tazobactam, FOX cefoxitin, CAZ ceftazidim, CRO ceftriaxon, CTX cefotaxim, FEP cefepim, ATM aztreonam, IPM imipenem, G gentamicin, AKN amikacin, NA nalidixic acid, CIP ciprofloxacin, FOS fosfomycin, SXT trimethoprim / sulfamethoxazole
The ESBL were phenotypically detected in 87/91 (95.60%) E. coli and 62/64 (96.88%) Klebsiella spp. strains.
Distribution of qnr genes
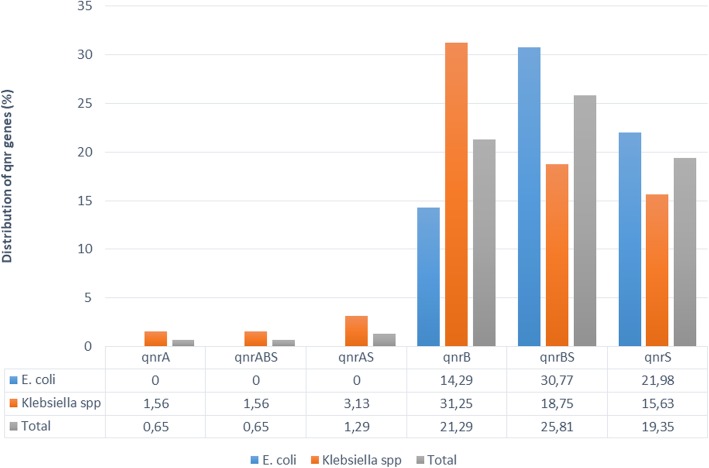
Electrophoresis analysis revealed 107 strains (61 E. coli and 46 Klebsiella spp.) harboring at least one qnr gene: 74 (47.74%) qnrB (41 E. coli and 33 Klebsiella spp.), 73 (47.10%) qnrS (48 E. coli and 25 Klebsiella spp.) and 4 (2.58%) qnrA (Klebsiella spp. only). However, any qnr genes were not detected in 48 strains. The concomitant presence of two or three qnr genes was detected. An additional figure showed agarose gel electrophoresis in more detail (see Additional file 1). Proportions of qnrBS combinations were 30.77% and 18.75% respectively in E. coli and Klebsiella spp.; qnrAS was observed in 3.13% of Klebsiella spp. while the triple association qnrABS was found in one Klebsiella spp. strains. QnrA was not found in E. coli strains. The prevalence of qnr genes was higher in Klebsiella spp. 71.88% (46/64) compared to E. coli 67.03% (61/91) strains. Distribution of qnr genes in bacterial species is shown in Fig. 1.
Fig. 1.
Distribution of qnr genes in bacterial species
Antibiotic susceptibility profile of qnr-positive strains
Out of the 107 (61 E. coli + 46 Klebsiella spp.) strains encoding qnr genes, 95/105 (90.48%) were resistant to nalidixic acid (58 E. coli + 37 Klebsiella spp.) and 100/107 (93.46%) were resistant to ciprofloxacin (57 E. coli + 43 Klebsiella spp.); 105/107 (98.13%) resistant to ceftazidim (59 E. coli + 46 Klebsiella spp.) and 106/107 (99.07%) to ceftriaxon (61 E. coli + 45 Klebsiella spp.). The resistance rates to gentamicin were 82.24% (88/107).
These strains remained however susceptible to imipenem (97.20%), amikacin (97.20%), and fosfomycin (95.33%). The resistance profile is presented in Table 3.
Table 3.
Antibiotic susceptibility profile of qnr positive strains
| ATB | E. coli | Klebsiella spp. | Total | ||
|---|---|---|---|---|---|
| n/N | % | ||||
| NA | R | 58/60 | 37/45 | 95/105 | 90.48 |
| S | 2/60 | 8/45 | 10/105 | 9.52 | |
| CIP | R | 57/61 | 43/46 | 100/107 | 93.46 |
| S | 4/61 | 3/46 | 7/107 | 6.54 | |
| CAZ | R | 59/61 | 46/46 | 105/107 | 98.13 |
| S | 2/61 | 0/46 | 2/107 | 1.87 | |
| CRO | R | 61/61 | 45/46 | 106/107 | 99.07 |
| S | 0/61 | 1/46 | 1/107 | 0.93 | |
| IMP | R | 2/61 | 1/46 | 3/107 | 2.80 |
| S | 59/61 | 45/46 | 104/107 | 97.20 | |
| AKN | R | 2/61 | 1/46 | 3/107 | 2.80 |
| S | 59/61 | 45/46 | 104/107 | 97.20 | |
| G | R | 48/61 | 40/46 | 88/107 | 82.24 |
| S | 13/61 | 6/46 | 19/107 | 17.76 | |
| FOS | R | 4/61 | 1/46 | 5/107 | 4.67 |
| S | 57/61 | 45/46 | 102/107 | 95.33 | |
R resistant, S sensible, CAZ ceftazidim, CRO ceftriaxon, IPM imipenem, G gentamicin, AKN amikacin, NA nalidixic acid, CIP ciprofloxacin, FOS fosfomycin
Most isolates that were resistant to ciprofloxacin and nalidixic acid encoded qnrB and qnrS alone or in association but no qnr genes was detected in 29 E. coli and 15 Klebsiella spp. strains resistant to ciprofloxacin and nalidixic acid. Nevertheless, isolates encoding qnrB or qnrS were also identified among nalidixic acid-susceptible (10/105) and ciprofloxacin-susceptible strains (7/107) (Table 4).
Table 4.
Co-existence of qnr gene and bla gene in E. coli and Klebsiella spp.
| Species | E. coli (N = 61) | Klebsiella spp (N = 46) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Qnr gene | qnrB (n = 13) | qnrBS (n = 28) | qnrS (n = 20) | Total 1 | qnrA (n = 1) | qnrABS (n = 1) | qnrAS (n = 2) | qnrB (n = 20) |
qnrBS
(n = 12) |
qnrS
(n = 10) |
Total 2 | |
| NA | R | 12 | 28 | 18 | 58 | 1 | 1 | 2 | 17 | 10 | 6 | 37 |
| S | 1 | 0 | 1 | 2 | 0 | 0 | 0 | 2 | 2 | 4 | 8 | |
| CIP | R | 11 | 28 | 18 | 57 | 1 | 1 | 2 | 20 | 11 | 8 | 43 |
| S | 2 | 0 | 2 | 4 | 0 | 0 | 0 | 0 | 1 | 2 | 3 | |
| ESBL genes | CTX-M1 | 2 | 4 | 1 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| SHV CTX-M1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 1 | 4 | |
| TEM | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| TEM CTX-M1 | 6 | 16 | 14 | 36 | 0 | 1 | 0 | 5 | 2 | 3 | 11 | |
| TEM SHV | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 2 | 0 | 3 | |
| TEM SHV CTX-M1 | 5 | 8 | 3 | 16 | 1 | 0 | 2 | 14 | 5 | 6 | 28 | |
R resistant, S sensible, NA nalidixic acid, CIP ciprofloxacin, ESBL extended spectrum β-lactamase, qnr quinolone resistance
Distribution of qnr genes among ESBL-producing isolates
PCR was performed to determine the presence of ESBL genes and all strains, positive or not to the double disc synergy test, carried at least one genes blaTEM, blaSHV and/or blaCTX-M1. All E. coli and Klebsiella spp. strains qnr positive were ESBL-producing. Within the 107 strains encoding qnr genes (61 E. coli + 46 Klebsiella spp.), 102 carried CTX-M1 (59 E. coli + 43 Klebsiella spp.), 96 carried TEM (54 E. coli + 42 Klebsiella spp.) and 52 carried SHV (17 E. coli + 35 Klebsiella spp.).
Results revealed that qnr subtypes (A, B, S) could co-exist alone or in association with blaCTX-M1, blaTEM and blaSHV. Among E. coli strains, qnrBS combinations was most frequently associated with TEM/CTX-M1 combinations and among Klebsiella spp., the most frequent association was qnrB plus TEM/SHV/CTX-M1 (Table 4).
Discussion
Plasmid-mediated quinolone resistance may facilitate the spread and increase frequency of quinolone-resistant strains. Until now qnr genes have been widely detected in different parts of the world. Such data are not available in Togo. This is the first study which reports the frequency and diversity of qnr genes among ESBL-producing Enterobacteriaceae in Togo.
The highest rate was found among Klebsiella spp. (71.88%) and E. coli (67.03%). Three qnr groups were detected and are described in this report. Among all the isolates detected, qnrB (47.74%) and qnrS (47.10%) were the most predominant, followed by qnrA (2.58%).
These frequencies found in this study are higher than those reported in Côte d’Ivoire where qnr genes were found at 31.2% in E. coli and 20.5% in Klebsiella spp. with 14.6% for qnrB, 9.9% for qnrA and 2.7% for qnrAS [10]. Always in Côte d’Ivoire, others authors found 50.54% of qnr genes in Klebsiella pneumoniae (71.73% qnrB, 26.08% qnrS and 2.17% qnrA) [24].
In Niger, qnr genes were also reported at 93.3% in Klebsiella spp. and 44.4% in E. coli with 64.3% for qnrS, 26.2% for qnrB and 9.5% for qnrA [14].
In Morocco, qnrB was found at 23%, qnr A at 10% and qnrS at 3% in 50% Klebsiella spp. and 18.7% E. coli [9]. In 2014 in Moroccan community enterobacteria, the prevalence of qnr gene was 2.6% (1.7% qnrS1 and 0.9% qnrB) [25]. These genes are usually plasmid mediated and can easily spread among the members of Enterobacteriaceae, through gene transfer mechanisms [3, 6, 17, 18]. Results of plasmid isolation test and conjugation experiments in different studies indicated that these qnr gene were carried by conjugative plasmid of high molecular weight. These determinants can be transferred between bacteria, thus realizing the epidemic spread of quinolone resistance through horizontal gene transfer [11, 13, 25–28]. However, due to the lack of financial resources, conjugation experiments or hybridization confirming the presence of target genes on plasmids were not performed in the present study.
Among strains encoding the qnr gene in our study, 90.48% were resistant to nalidixic acid, 93.46% to ciprofloxacin, 98.13% to ceftazidim and 99.07% to ceftriaxon. These rates are higher than those observed in Morocco (57% for nalidixic acid and 78% ciprofloxacin, 100% for ceftazidim and 71% for cefotaxim) [9]. In Mexico, the resistance rate among qnr positive pediatric strains was 41.1% for nalidixic acid, 29.4% for ciprofloxacin, 82.3% for ceftazidim and 100% for cefotaxim [13]. In this study, higher resistance rate of the qnr positive strains against nalidixic acid and ciprofloxacin could be explained by the concomitant presence of two or three qnr gene groups (43/107, 40.20%), also found in Algeria and in Vietnam, thus inducing an additive effect on the minimal inhibiting concentration (MICs) of these different molecules. In addition, qnr positive isolates showed more resistance to gentamicin (82.24%). This may be explained by the fact that plasmid-mediated quinolone resistance is associated with integrons bearing resistance determinants to several other antibiotics such as beta-lactams and aminoglycosides [3, 4, 27, 29]. The qnr genes were identified among isolates which were susceptible to nalidixic acid and ciprofloxacin.
This result has clinical implications since the acquisition of the qnr genes by quinolone susceptible ESBL-producing strains could lead to selection of ciprofloxacin and cephalosporin resistant strains an increasing the mutant prevention concentration (MPC) [6, 17]. However no qnr genes was detected in 29 E. coli and 15 Klebsiella spp. strains resistant to ciprofloxacin and nalidixic acid, can suggest the presence of another mechanism of resistance to quinolones such as mutations in the gyrase and topoisomerase IV genes [2, 6, 30].
The presence of ESBL and some of the quinolone-resistant genes in the same mobile genetic elements could explain the co-resistance to beta-lactams and fluoroquinolones. Our results showed that all E. coli and Klebsiella spp. strains qnr positive were ESBL-producing. Among qnr-positive strains, 102 produce CTX-M1, 52 SHV and 96 produce TEM.
Previous studies showed that qnr-positive strains frequently expressed ESBL [9–11, 28, 31, 32]. The strong association between PMQR gene and blaCTX-M-15 and blaTEM-116 was detected in clinical Enterobacterial isolates from Iran [31]. In Mexico, characterization of adult qnr-positive isolates indicated that the SHV ESBL-type (SHV-12, − 5, and 2a) was the most prevalent (81.6%) followed by CTX-M-15 (44.9%) [28]; but in pediatric isolates, CTX-M-15 was the most predominant (70.5%) [13]. However, in both bacterial population, combination of ESBL and qnr genes may be pointing to a co-selection of cephalosporin and quinolone resistance. QnrA1 and qnrS1 have previously been found to be associated with blaCTX-M-9, blaSHV-12 and blaSHV-92 among Enterobacterial isolates in Spain [11]. QnrB was observed to be co-produced with CTX-M-15 in Algeria strains of E. coli [33]. A similar result was recently found in Klebsiella pneumoniae isolates from Côte d’Ivoire, but the type of ESBL was not determined [24]. Qnr genes are usually found in multi-resistance plasmids linked to other resistance determinants, beta-lactamase genes have been conspicuously common [5, 6, 25, 26].
Our results also revealed that qnr subtypes could co-exist alone or in association with beta-lactamase genes.
Among E. coli strains, qnrBS combinations was most frequently associated with blaTEM/CTX-M1 combinations and among Klebsiella spp., the most frequent association was qnrB plus blaTEM/SHV/CTX-M1. The blaCTX-M1, plasmid-mediated class A ESBL [34–36] expression was observed to be currently more frequent in a double or triple combination with blaTEM and blaSHV. This genes combination suggests a progressive consolidation of resistance genes on a single mobile genetic element (plasmids, integrons, etc.).
These findings raise the hypothesis that the qnr genes detected in these strains could also have the same plasmid location. Plasmid isolation and conjugation experiments should be then investigated to confirm the presence of target genes on plasmids. Our results also suggested the community emergence of PMQR determinants (qnr gene) that contributed to the development and spread of fluoroquinolone resistance in E. coli and Klebsiella spp. isolates in Togo. The presence of these determinants in the outpatient is worrisome, due of the potential spread of plasmids in a scenario of uncontrolled oral quinolone usage, which can compromise therapeutic options and therefore concern for public health.
The potential limitations of this study were the absence of data on minimal inhibiting concentration (MIC) for nalidixic acid and ciprofloxacin to determine the level of bacterial resistance to these antibiotics and also the bias of including only ESBL strains and the absence of molecular typing of strains and sequence analysis of the different genes.
Altogether, the results of the present study underline the frequency of qnr determinants associated to fluoroquinolones resistance among E. coli and Klebsiella spp ESBL-producing strains in Togo and identifies the presence of qnr genes in quinolone-susceptible strains which could lead to in vivo selection of ciprofloxacin-resistant strains.
Conclusion
This first report of qnrA, qnrB and qnrS gene among ESBL-producing E. coli and Klebsiella spp. from Togo, extends upon similar finding in many countries supporting the wide distribution of qnr genes.
The results revealed a high rate of qnrB and qnrS alone or in combination and a higher association with blaTEM/CTX-M1 and blaTEM/SHV/CTX-M1 combinations. These qnr genes positive strains were highly resistant to nalidixic acid, ciprofloxacin, ceftazidim, ceftriaxon and gentamycin. However, they remain susceptible to imipenem, amikacin and fosfomycin.
The plasmid-mediated quinolone resistance genes and their association with cephalosporin resistance mediated by ESBL contribute to the spread of multidrug resistance due to their easy transfer between bacteria. Their wide dissemination impairs treatment outcome of common infections in community and hospitals settings. These finding suggest the strengthening of the public health policies in Togo in order to prevent, monitor and control antimicrobial resistance through the implementation of an antibiotic resistance surveillance system. Further studies on sequence analysis of the ESBL gene amplicons are also needed to determine different resistance profile of ESBL-producing bacteria in Togo.
Additional file
Agarose gel electrophoresis (2%) used for the separation of multiplex PCR products. M: molecular size marker (100 bp ladder, Promega, USA); line 1, 8, 9, 12: negative; line 2, 4, 5, 7, 10, 11, 14, 15, 17, 18, 19, 20: qnr B + qnr S genes: line 3: qnr A + qnrB + qnr S genes; line 6: qnrS genes; line 13: qnrB genes; line 21: negative control and line 22: positive control qnrB genes. Qnr A (517 bp), qnrB (469 bp), qnr S (417 bp). (PDF 325 kb)
Acknowledgements
FDS is grateful to the 2018 Mwalimu Nyerere African Union Scholarship scheme for an enrolment in PhD in molecular biology and genetics at the University of Ouaga I Prof Joseph Ki-Zerbo, Burkina Faso. We also thank the WAEMU Commission, through the PACER II program granted the molecular biology laboratory CERBA/LABIOGENE University Ouaga I Prof. Joseph Ki-Zerbo for the realization of molecular biology analysis.
We thank Kossi Komlan and Maman Issaka for their advice and suggestions.
Abbreviations
- AKN
Amikacin
- AMC
Amoxicillin + clavulanate
- ATCC
American Type Culture Collection
- ATM
Aztreonam
- CASFM/EUCAST
Comité d’Antibiogramme de la Socièté Française de Microbiologie/EUropean Committee on Antimicrobial Susceptibility Testing
- CAZ
Ceftazidim
- CERBA
Centre de Recherche Biomoléculaire Pietro Annigoni
- CIP
Ciprofloxacin
- CRO
Ceftriaxon
- CTX
Cefotaxim
- CTX-M
Cefotaximase-Munich
- DNA
Desoxyribonucleic Acid
- DOX
Doxycyclin
- EMB
Eosin Methylene Blue
- ESBL
Extended Spectrum Beta-Lactamase
- ESTBA
Ecole Supérieure des Techniques Biologique et Alimentaire
- FEP
Cefepim
- FOS
Fosfomycin
- FOX
Cefoxitin
- G
Gentamycin
- IMP
Imipenem
- INH
Institut National d’Hygiène
- LABIOGENE
Laboratoire de Biologie Moléculaire et de Génétique Moléculaire
- NA
Nalidixic acid
- PMQR
Plasmid Mediated Quinolone Resistance
- PRP
Pentapeptid Repeat Protein
- qnr
quinolone resistance
- SHV
Sulfhydryl Variable
- SXT
trimethoprim/sulfamethoxazole
- TCS
Trypticase Soy
- TEM
Temoniera
- TZP
Piperacillin-tazobactam
Authors’ contributions
FDS, AYS and JS designed the study; FDS, STS and AKO performed the experiments, analyzed the data and drafting the manuscript. FDS, DOY, AMD, ABK, SK and JS were involved in critically reviewing the manuscript. All authors have read and approved the final version.
Funding
Not applicable.
Availability of data and materials
The database analyzed during the study is available on reasonable request from the corresponding author.
Ethics approval and consent to participate
This study received the INH approval for the transfer of strains, to the molecular biology laboratory of CERBA/ LABIOGENE, University Ouaga I Professor Joseph Ki-Zerbo, Burkina Faso. The institutional ethic committee of CERBA/LABIOGENE reviewed and approved the study protocol.
Consent for publication
Not applicable. This study does not contain any individual or personal data.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Fortune Djimabi Salah, Phone: 00226 62 52 39 86, Email: fortunedavi@yahoo.fr.
Serge Théophile Soubeiga, Email: s.soubeiga@labiogene.fr.
Abdoul Karim Ouattara, Email: ak_ouattara@labiogene.fr.
Adodo Yao Sadji, Email: adodosadji@yahoo.fr.
Amana Metuor-Dabire, Email: ametuordabire@yahoo.fr.
Dorcas Obiri-Yeboah, Email: castella.oy@gmail.com.
Abiba Banla-Kere, Email: kerebanla@yahoo.fr.
Simplice Karou, Email: simplicekarou@hotmail.com.
Jacques Simpore, Email: jacques.simpore@labiogene.fr.
References
- 1.Adriaenssens N, Coenen S, Versporten A, Muller A, Minalu G, Faes C, et al. European surveillance of antimicrobial consumption (ESAC): outpatient antibiotic use in Europe (1997-2009) J Antimicrob Chemother. 2011;66(Suppl 6):vi3–v12. doi: 10.1093/jac/dkr453. [DOI] [PubMed] [Google Scholar]
- 2.Jacoby GA. Mechanisms of resistance to quinolones. Clin Infect Dis. 2005;41(Suppl 2):S120–S126. doi: 10.1086/428052. [DOI] [PubMed] [Google Scholar]
- 3.Strahilevitz J, Jacoby GA, Hooper DC, Robicsek A. Plasmid-mediated quinolone resistance: a multifaceted threat. Clin Microbiol Rev. 2009;22(4):664–689. doi: 10.1128/CMR.00016-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muylaert A, Mainil J, editors. Résistances aux fluoroquinolones: la situation actuelle. Annales de Médecine Vétérinaire; 2013: Université de Liège.
- 5.Nordmann P, Poirel L. Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J Antimicrob Chemother. 2005;56(3):463–469. doi: 10.1093/jac/dki245. [DOI] [PubMed] [Google Scholar]
- 6.Jacoby GA, Strahilevitz J, Hooper DC. Plasmid-mediated quinolone resistance. Microbiol Spectr. 2014;2(5). [DOI] [PMC free article] [PubMed]
- 7.Jacoby GA, Walsh KE, Mills DM, Walker VJ, Oh H, Robicsek A, et al. qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob Agents Chemother. 2006;50(4):1178–1182. doi: 10.1128/AAC.50.4.1178-1182.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robicsek A, Jacoby GA, Hooper DC. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis. 2006;6(10):629–640. doi: 10.1016/S1473-3099(06)70599-0. [DOI] [PubMed] [Google Scholar]
- 9.Bouchakour M, Zerouali K, Gros Claude JD, Amarouch H, El Mdaghri N, Courvalin P, et al. Plasmid-mediated quinolone resistance in expanded spectrum beta lactamase producing enterobacteriaceae in Morocco. J Infect Dev Ctries. 2010;4(12):779–803. doi: 10.3855/jidc.796. [DOI] [PubMed] [Google Scholar]
- 10.Guessennd N, Bremont S, Gbonon V, Kacou-Ndouba A, Ekaza E, Lambert T, et al. Qnr-type quinolone resistance in extended-spectrum beta-lactamase producing enterobacteria in Abidjan, Ivory Coast. Pathologie-biologie. 2008;56(7–8):439–446. doi: 10.1016/j.patbio.2008.07.025. [DOI] [PubMed] [Google Scholar]
- 11.Lavilla S, Gonzalez-Lopez JJ, Sabate M, Garcia-Fernandez A, Larrosa MN, Bartolome RM, et al. Prevalence of qnr genes among extended-spectrum beta-lactamase-producing enterobacterial isolates in Barcelona, Spain. J Antimicrob Chemother. 2008;61(2):291–295. doi: 10.1093/jac/dkm448. [DOI] [PubMed] [Google Scholar]
- 12.Poirel L, Leviandier C, Nordmann P. Prevalence and genetic analysis of plasmid-mediated quinolone resistance determinants QnrA and QnrS in Enterobacteriaceae isolates from a French university hospital. Antimicrob Agents Chemother. 2006;50(12):3992–3997. doi: 10.1128/AAC.00597-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silva-Sanchez J, Cruz-Trujillo E, Barrios H, Reyna-Flores F, Sanchez-Perez A, Bacterial Resistance C, et al. Characterization of plasmid-mediated quinolone resistance (PMQR) genes in extended-spectrum beta-lactamase-producing Enterobacteriaceae pediatric clinical isolates in Mexico. PLoS One. 2013;8(10):e77968. doi: 10.1371/journal.pone.0077968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moumouni A, Diagbouga S, Nadembèga C, Metuor Dabire A, Ouattara K, Zohoncon T, et al. Quinolone Resistance (qnr) genes in fecal carriage of extended Spectrum beta-lactamases producing Enterobacteria isolated from children in Niger. Curr Res Microbiol Biotechnol. 2017;5(1):953–957. [Google Scholar]
- 15.Diagbouga S, Salah FD, Sadji AY, Metuor Dabire A, Nadembega C, Banla Kere A, et al. Detection of high prevalence of TEM/SHV/CTX-M genes in ESBL producing and multidrug resistant Klebsiella Pneumoniae and Klebsiella Oxytoca. J Clin Diagn Res. 2016;4(1:130):p7. [Google Scholar]
- 16.Salah FD, Diagbouga S, Metuor Dabire A, Sadji AY, Nadembega C, Moumouni A, et al. First detection of Resistance genes encoding extended Spectrum β-lactamase producing Escherichia coli at Lomé, Togo. Arch Clin Microbiol. 2017;7(6):p7. [Google Scholar]
- 17.Martinez-Martinez L, Pascual A, Jacoby GA. Quinolone resistance from a transferable plasmid. Lancet. 1998;351(9105):797–799. doi: 10.1016/S0140-6736(97)07322-4. [DOI] [PubMed] [Google Scholar]
- 18.Tran JH, Jacoby GA. Mechanism of plasmid-mediated quinolone resistance. Proc Natl Acad Sci. 2002;99(8):5638–5642. doi: 10.1073/pnas.082092899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.CASFM/EUCAST . Recommendations 2014 V1.0 Mai 2014. Paris, FRANCE: Socièté Française de Microbiologie; 2014. p. 114. [Google Scholar]
- 20.Jarlier V, Nicolas MH, Fournier G, Philippon A. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis. 1988;10(4):867–878. doi: 10.1093/clinids/10.4.867. [DOI] [PubMed] [Google Scholar]
- 21.Robicsek A, Strahilevitz J, Sahm DF, Jacoby GA, Hooper DC. Qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob Agents Chemother. 2006;50(8):2872–2874. doi: 10.1128/AAC.01647-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother. 2010;65(3):490–495. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- 23.Pagani L, Dell'Amico E, Migliavacca R, D'Andrea MM, Giacobone E, Amicosante G, et al. Multiple CTX-M-type extended-spectrum beta-lactamases in nosocomial isolates of Enterobacteriaceae from a hospital in northern Italy. J Clin Microbiol. 2003;41(9):4264–4269. doi: 10.1128/JCM.41.9.4264-4269.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tahou JE, Guessennd N, Sokouri PD, Gbonon V, Konan F, Kouadio J, et al. Antimicrobial Resistance of Klebsiella pneumoniae -ESBL producing strains isolated from clinical specimens in Abidjan (cote de Ivoire) Microbiol Res J Int. 2017;20(2):1–7. doi: 10.9734/MRJI/2017/34085. [DOI] [Google Scholar]
- 25.Jamali L, Haouzane F, Bouchakour M, Oufrid S, Ghazlane Z, El Mdaghri N, et al. Prévalence des gènes de résistance plasmidique aux quinolones chez des entérobactéries communautaires isolées au Maroc [Prevalence of plasmid mediated quinolone resistance genes among enterobacteria isolates in Moroccan community] Int J Innov Sci Res. 2014;11(2):387–399. [Google Scholar]
- 26.Barguigua A, El Otmani F, Talmi M, Reguig A, Jamali L, Zerouali K, et al. Prevalence and genotypic analysis of plasmid-mediated β-lactamases among urinary Klebsiella pneumoniae isolates in Moroccan community. J Antibiotics. 2013;66(1):11. doi: 10.1038/ja.2012.91. [DOI] [PubMed] [Google Scholar]
- 27.Minh Vien LT, Baker S, Phuong Thao LT, Phuong Tu LT, Thu Thuy C, Thu Nga TT, et al. High prevalence of plasmid-mediated quinolone resistance determinants in commensal members of the Enterobacteriaceae in Ho Chi Minh City. Vietnam J Med Microbiol. 2009;58(12):1585–1592. doi: 10.1099/jmm.0.010033-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silva-Sanchez J, Barrios H, Reyna-Flores F, Bello-Diaz M, Sanchez-Perez A, Rojas T, et al. Prevalence and characterization of plasmid-mediated quinolone Resistance genes in extended-Spectrum β-lactamase–producing Enterobacteriaceae isolates in Mexico. Microb Drug Resist. 2011;17(4):497–505. doi: 10.1089/mdr.2011.0086. [DOI] [PubMed] [Google Scholar]
- 29.Iabadene H, Messai Y, Ammari H, Ramdani-Bouguessa N, Lounes S, Bakour R, et al. Dissemination of ESBL and Qnr determinants in Enterobacter cloacae in Algeria. J Antimicrob Chemother. 2008;62(1):133–136. doi: 10.1093/jac/dkn145. [DOI] [PubMed] [Google Scholar]
- 30.Redgrave LS, Sutton SB, Webber MA, Piddock LJV. Fluoroquinolone resistance: mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol. 2014;22(8):438–445. doi: 10.1016/j.tim.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 31.Azargun R, Sadeghi MR, Soroush Barhaghi MH, Samadi Kafil H, Yeganeh F, Ahangar Oskouee M, et al. The prevalence of plasmid-mediated quinolone resistance and ESBL-production in Enterobacteriaceae isolated from urinary tract infections. Infect Drug Resist. 2018;11:1007–1014. doi: 10.2147/IDR.S160720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sedighi I, Arabestani MR, Rahimbakhsh A, Karimitabar Z, Alikhani MY. Dissemination of extended-spectrum β-lactamases and quinolone resistance genes among clinical isolates of uropathogenic Escherichia coli in children. Jundishapur J Microbiol. 2015;8(7):e19184. doi: 10.5812/jjm.19184v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Betitra Y, Teresa V, Miguel V, Abdelaziz T. Determinants of quinolone resistance in Escherichia coli causmg community-acquired urinary tract infection in Bejaia, Algeria. Asian Pac J Trop Med. 2014;7:462–467. doi: 10.1016/S1995-7645(14)60075-4. [DOI] [PubMed] [Google Scholar]
- 34.Bonnet R. Growing group of extended-spectrum beta-lactamases: the CTX-M enzymes. Antimicrob Agents Chemother. 2004;48(1):1–14. doi: 10.1128/AAC.48.1.1-14.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poirel L, Decousser J-W, Nordmann P. Insertion sequence ISEcp1B is involved in expression and mobilization of a blaCTX-M β-lactamase gene. Antimicrob Agents Chemother. 2003;47(9):2938–2945. doi: 10.1128/AAC.47.9.2938-2945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poirel L, Naas T, Nordmann P. Genetic support of extended-spectrum beta-lactamases. Clin Microbiol Infect. 2008;14(Suppl 1):75–81. doi: 10.1111/j.1469-0691.2007.01865.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Agarose gel electrophoresis (2%) used for the separation of multiplex PCR products. M: molecular size marker (100 bp ladder, Promega, USA); line 1, 8, 9, 12: negative; line 2, 4, 5, 7, 10, 11, 14, 15, 17, 18, 19, 20: qnr B + qnr S genes: line 3: qnr A + qnrB + qnr S genes; line 6: qnrS genes; line 13: qnrB genes; line 21: negative control and line 22: positive control qnrB genes. Qnr A (517 bp), qnrB (469 bp), qnr S (417 bp). (PDF 325 kb)
Data Availability Statement
The database analyzed during the study is available on reasonable request from the corresponding author.