Abstract
Background
Campylobacter jejuni is the leading cause of bacterial gastroenteritis (campylobacteriosis) in humans worldwide, and the most frequent pathogen associated with Guillain-Barré syndrome (GBS) and Miller-Fisher syndrome (MFS). The study was designed in order to assess similarities between genomes of Campylobacter jejuni strains, isolated from children suffering from acute diarrhea in northeastern Poland, in comparison to C. jejuni genomes stored in public databases. The analysis involved phylogeny, resistome and virulome. In addition, the Campylobacter PubMLST database was used to estimate the prevalence of the analyzed C. jejuni sequence type (STs) in other countries.
Results
Campylobacter jejuni ST50, ST257 and ST51 represented 5.3%, 4.5% and 2.2% of the PubMLST records, respectively. Overall, strains representing the STs showed common resistance to tetracyclines (51.3%) and fluoroquinolones (31.8%), mediated through the tetO gene (98.2%) and point mutation (T86I) in the gyrA gene (100%). However, the latter was present in all our isolates. The major differences in virulence patterns concerned serotypes, lipooligosaccharide (LOS) classes and certain clinically relevant genes.
Conclusions
Campylobacter jejuni ST50, ST51 and ST257 are among the top ten of STs isolated in Europe. WGS revealed diversity of serotypes and LOS classes in ST50 strains, that deserves further clinical and epidemiological investigations as it might be related to a risk of post-infectious neurological sequels such as Guillain-Barré syndrome. Additionally, the results implicate lower pathogenic potential and distinct transmission chains or reservoirs for C. jejuni ST51 isolates responsible for campylobacteriosis in northeastern Poland.
Electronic supplementary material
The online version of this article (10.1186/s13099-019-0313-x) contains supplementary material, which is available to authorized users.
Keyword: Campylobacteriosis, WGS, MLST, Virulence, Antibiotic resistance
Background
Campylobacter jejuni is the leading cause of bacterial gastroenteritis (campylobacteriosis) in humans worldwide, with more than 200,000 annual cases in European Union (EU) [1, 2]. The natural reservoirs of C. jejuni are intestinal tracts of many wild and agriculture-associated birds and mammals. Therefore, faecal contamination from carrier animals is considered to be a primary source of this pathogen in the environment and food products. Although the transmission routes of C. jejuni are not fully recognized, chickens are considered to be the major source for transmission to humans. In addition, outbreaks of campylobacteriosis are often linked to consumption of unpasteurized milk and contaminated water [3].
In general, campylobacteriosis is a self-limiting illness, however, rare but life-threatening neurological sequels such as Guillain-Barré syndrome (GBS) and Miller-Fisher syndrome (MFS) may occur in patients [4]. For instance, it was estimated that 31.0% of the GBS cases may be ascribed to a previous campylobacteriosis [5]. Yet in contrast to other bacterial enteropathogens, C. jejuni does not possess numerous classical virulence factors. Cytolethal distending enterotoxin (Cdt) is the only virulence determinant located on C. jejuni chromosome, however, its role in the pathogenesis is still not clear [6]. Nevertheless (i) flagella based motility and chemotaxis as well as secretion of invasive antigens (Cia) (ii) polysaccharide capsule (CPS) (iii) lipooligosaccharide (LOS), and (iv) various proteins associated with adhesion, colonization and infection of host cells, were recognized as important factors for C. jejuni pathogenicity [7, 8]. Additionally, CPS and LOS are also implicated with post-gastroenteritis GBS and MFS [4, 9]. Therefore, the identification and profiling of C. jejuni virulence determinants are crucial for risk assessment of infections caused by this pathogen.
Although antimicrobial therapy is not routinely recommended to treat campylobacteriosis, in severe and prolonged or immunocompromised cases, fluoroquinolones and macrolides are agents of choice [10]. Also, tetracyclines and aminoglycosides can be used as alternatives. However, an increasing resistance of C. jejuni, in particular to fluoroquinolones, in recent years is alarming. As a result, in 2017 the World Health Organization listed fluoroquinolone-resistant Campylobacter spp. as one of the six high priority pathogens for research and development of new antibiotics [11].
Multilocus sequence typing (MLST), based on sequence comparison of seven housekeeping genes defined as sequence types (STs) and clonal complexes (CCs), has been an essential tool in studying of C. jejuni phylogeny and epidemiology [12]. However, MLST does not include medically relevant information such as the virulence or antibiotic resistance determinants, also known as virulome and resistome [12, 13]. In addition, since C. jejuni is genetically variable pathogen with high level of horizontal gene exchange and recombination, even strains representing the same STs may possess distinct virulence patterns [9, 14]. At present, whole-genome sequencing (WGS) is considered as the most informative and discriminative typing method of bacterial pathogens [14–16], allowing for comprehensive phylogenetic analyses of numerous traits associated with virulence [17, 18] or antibiotic resistance [16, 19].
In this study we applied WGS in order (i) to characterize C. jejuni strains isolated from children with acute diarrhea in northeastern Poland, and (ii) to compare their virulence and antibiotic resistance patterns with phylogenetically related, i.e. representing the same STs, C. jejuni strains from other parts of the world.
Results
Sequence types (STs), serotypes, phylogenetic relatedness and pan-genome
Four C. jejuni isolates from children with diarrhea were classified into three STs, ST50 (strains KF017 and KF042), ST51 (strain KF070), and ST257 (strain KF045). According to the Campylobacter PubMLST database involving 6977 distinct C. jejuni STs, ST50, ST257, and ST51 were among the top ten STs (Table 1). Likewise, among 139 various STs from Poland (289 records), ST257 (8.6%) and ST50 (3.5%) occupied the second and third place respectively, outdistanced by ST6411 (10.0%). Also, in the PATRIC genome database ST50, ST257 and ST51 represented twelve the most commonly sequenced C. jejuni STs (Additional file 1: Table S3).
Table 1.
The ten most common C. jejuni STs collected in the Campylobacter PubMLST database comprising 58,179 records
| ST | Number of strains | % of the total STsa | Clonal Complex |
|---|---|---|---|
| 21 | 3517 | 6.0 | CC-21 |
| 45 | 3141 | 5.4 | CC-45 |
| 50 | 3064 | 5.3 | CC-21 |
| 257 | 2623 | 4.5 | CC-257 |
| 48 | 2478 | 4.3 | CC-48 |
| 53 | 1483 | 2.5 | CC-21 |
| 61 | 1296 | 2.2 | CC-61 |
| 19 | 1270 | 2.2 | CC-21 |
| 51 | 1270 | 2.2 | CC-443 |
| 354 | 1171 | 2.0 | CC-354 |
| 42 | 1053 | 1.8 | CC-42 |
a6977 is the total number of STs, profiles without ascribed ST (n = 53) were excluded from the comparison
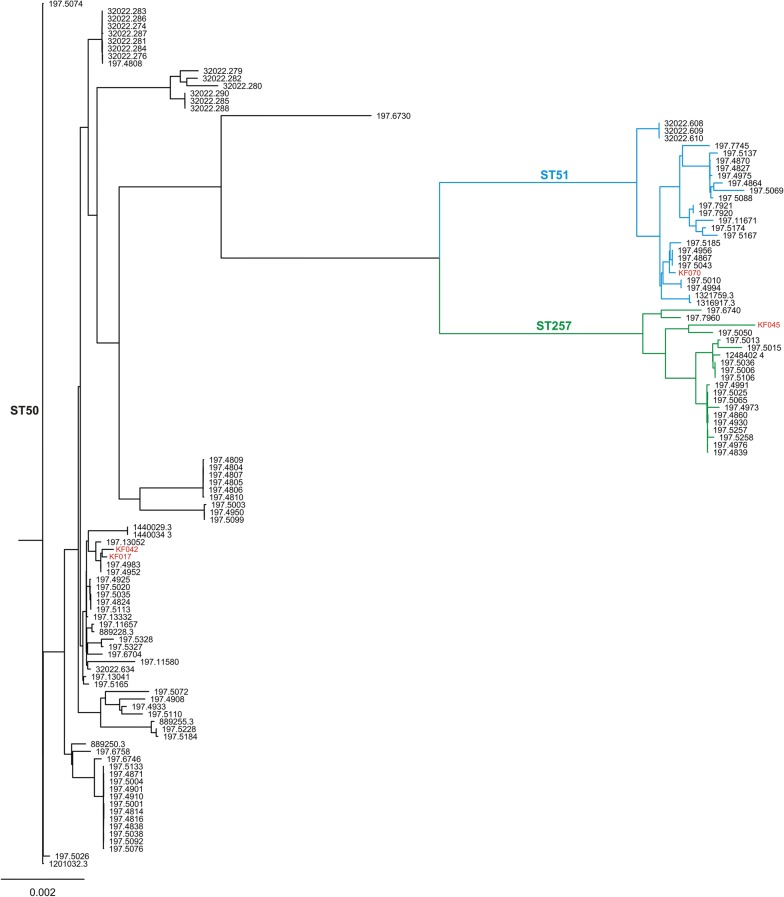
All strains classified as ST51 and ST257 belonged to the HS37 and HS11 serotypes, respectively. On the contrary, ST50 strains were represented by five serotypes, where HS8c predominated (49.3%), including our two isolates KF042 and KF017, followed by HS1 (33.8%), HS2 (8.4%), HS10 (4.2%), and HS5c (4.2%) (Additional file 1: Table S1). This diversity was reflected by the phylogenetic analysis that revealed several discrete clusters formed by the ST50 strains (Fig. 1).
Fig. 1.
Phylogenetic tree of C. jejuni ST50, ST51 and ST257 constructed based on core genes comparison using Roary and RAxML software. Black, blue and green colors of the tree branches indicate ST50, ST51 and ST257 strains, respectively. The isolates from northeastern Poland are in red
The pan-genome analysis revealed a total of 5961 genes, including 907 and 376 of the core and the shell core genes, respectively (Additional file 1: Fig. S1). Additional data regarding the pan-genome analysis, i.e. frequency of genes and their presence or absence in genomes, were included in Additional file 1: Fig. S2 and S3.
Resistome
Overall, based on in silico analysis in C. jejuni isolates from northeastern Poland we identified resistance determinants to three groups of antibiotics, beta-lactams (blaoxa-61 or blaoxa-184), tetracyclines (tetO), and fluoroquinolones (point mutation T86I in gyrA) (Table 2, Fig. 2), which was consistent with their in vitro antibiotic resistance patterns. The same resistance markers were noted in the remaining C. jejuni genomes, which only sporadically possessed aminoglycoside resistance genes (three strains). In addition, in one genome (C. jejuni BIGS0022) tetW was present instead of tetO. No mutations in 23S rRNA or other genes (cmeR, rplV, rpsL) associated with macrolide or spectinomycin resistance were detected. Therefore, altogether twelve antibiotic resistance patterns were distinguished (Table 2).
Table 2.
Antibiotic resistance determinants and profiles in genomes (n = 116) of C. jejuni ST50, ST51 and ST257
| Profile | Resistance determinants | ST number | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| bla oxa-61 | bla oxa-184 | tetO | tetW | gyrA (T86I) | aph(2′)-If | aph(2′')-If | aph(3′)-IIIa | ant9 | aadE | ST50 | ST51 | ST257 | |
| 1 | + | 34 | 14 | ||||||||||
| 2 | + | + | + | 21 | 1 | 2a | |||||||
| 3 | + | + | 10 | 7 | 1 | ||||||||
| 4 | + | + | 7 | ||||||||||
| 5 | + | + | + | 6 | |||||||||
| 6 | + | + | 4a | 1 | |||||||||
| 7 | + | 3 | |||||||||||
| 8 | + | + | + | + | + | 1 | |||||||
| 9 | + | + | + | + | + | + | + | 1 | |||||
| 10 | + | + | + | + | + | + | 1 | ||||||
| 11 | + | + | 1 | ||||||||||
| 12 | + | + | 1a | ||||||||||
aProfiles present in C. jejuni strains from northeastern Poland
Fig. 2.
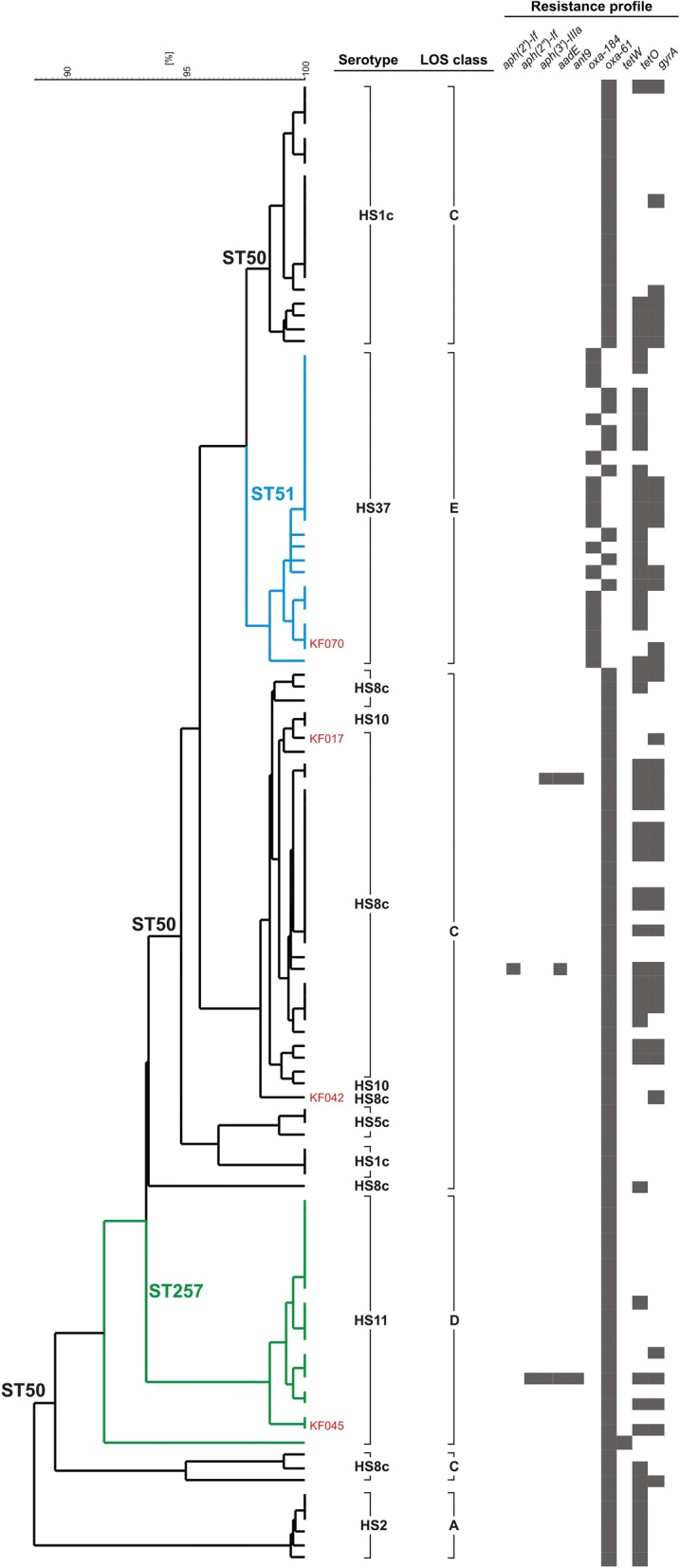
Dendrogram representing similarity of virulence patterns of C. jejuni ST50, ST51 and ST257 strains. The dendrogram was constructed based on presence/absence of virulence genes (n = 143) using UPGMA method and Dice coefficient in NTSYS-pc software. Black, blue and green colors of the tree branches indicate ST50, ST51 and ST257 strains, respectively. The isolates from northeastern Poland are shown in red
In total, tetracycline and fluoroquinolone resistance determinants were present in 51.3% and 31.8% of the genomes, respectively. Conversely, tetO was detected only in one of our strains (KF045), but all have T86I mutation in gyrA. Furthermore, in 58.6% of C. jejuni strains tetO has plasmid origin, since it is located on contigs sharing similarity with pTet plasmid of C. jejuni 81–176 or similar plasmids (Additional file 1: Fig. S4). In addition, 14 variants of TetO protein were noted with amino acid sequence identity varied from 93 to 99% (Additional file 1: Fig. S5). Interestingly, in C. jejuni KF045 tetO is localized in unique chromosomal position in comparison to other strains representing ST257, ST50, and ST51 (Additional file 1: Fig. S6).
As aforementioned, aminoglycoside resistance genes were noted only in three genomes, two representing ST50 (aadE, ant9, and aph(3′)-IIIa or aadE and aph(2′)-If) and one ST257 (aadE, ant9, aph(3′)-IIIa and aph(2′')-If) (Table 2). In all cases the genes seem to be located on plasmids, e.g. similar to plasmid pGMI16-00 from C. jejuni strain CFSAN054107 (Additional file 1: Fig. S7).
Although the resistance patterns were not connected with a particular ST, tetO was more common in genomes belonging to ST51 (84.0%) than in ST50 (47.8%) or ST257 (20%). Furthermore, 32.0% of ST51 genomes possess blaoxa-61, like all ST50 and ST257, instead of blaoxa-184 (Table 2).
Virulome
In general, a majority of the virulence genes, with rare exceptions such as flaA, flaB or flaE, were evenly distributed throughout the genomes of all STs under study (Additional file 1: Table S4). However, the greatest number of virulence genes was noted in strains representing ST50 (mean = 95.1, range = 88–115), followed by ST257 (mean = 90.9, range = 80–93;) and ST51 (mean = 88.4, range = 86–90) strains. The differences were statistically significant between ST50 and ST51 (p = 0.000147). Similarly, the prevalence of the 25 genes recognized as markers of human pathogenic C. jejuni strains [17] was significantly higher (p = 0.000114) in the ST50 strains (mean = 23.2, range = 20–24) than ST257 (mean and range = 17) and ST51 (mean = 13.5, range = 11–17) (Additional file 1: Table S5). The differences in virulence patterns (Fig. 2) resulted mainly from a variation in the polysaccharide capsule and LOS biosynthesis loci. The former is also responsible for serotype specificity, and the latter is used to classify C. jejuni strains into so-called LOS classes. Hence, all ST51 and ST257 were classified into class E or class D, respectively. Whereas class C was recognized in 91.3% of the ST50 strains, including KF017 and KF042, and class A-specific genes in the remaining genomes. Noteworthy, the latter are characterized also by distinct HS2 serotype (Fig. 2). Finally, plasmids carrying T4SS or T6SS secretion systems were detected only in three genomes, all belonging to ST50.
Discussion
Campylobacter jejuni is a highly diverse pathogen represented currently by around 7000 distinct STs, which are distributed over 44 clonal complexes (CCs) and 2606 singletons (STs without assignment to any CCs). The relative frequency of particular C. jejuni genotypes and their diversity may vary between countries, and possibly is influenced by multiple factors, including food sources, animal reservoirs, seasons, levels of zoonotic transmissions, as well as the rate of horizontal gene transfer [7, 20]. Nevertheless, the three STs found among C. jejuni Polish isolates, ST50 (CC-21), ST257 (CC-257), and ST51 (CC-443), were in the top ten of C. jejuni STs noted in Europe (data from 46,237 records gathered in 28 countries). In fact, ST50 belongs to the largest CC-21, which clusters 23.1% of all STs. Similarly, in Poland the prevalence of ST257 and ST50 is outdistanced by ST6411 only [21]. However, the data from Poland might be skewed by the fact that all strains classified into ST50, ST51 and ST257 were isolated from animals, mostly chickens (86.1%). On the other hand, an isolation of the same STs from animal sources and humans, likely indicates their zoonotic transmission frequently reported by other studies [18, 22]. Actually, CC-257 (ST257) represents so-called ‘specialists’ C. jejuni lineage, i.e. strongly associated with certain hosts, chickens in this case for instance [7, 23]. Whereas, CC-21 (ST50) and CC-443 (ST51) are frequently isolated from various animal species, hence considered as ‘generalists’.
Nevertheless, C. jejuni pathogenic characteristics, with rare exceptions [24], generally is not attributed to particular phylogenetic lineages [7]. However, we noted that ST50 genomes, followed by ST257, possessed overall the highest number of virulence genes, including those ones considered as typical for human pathogenic C. jejuni isolates [17]. Interestingly, Harvala et al. [25] showed that patients infected with C. jejuni ST-50 or ST-257 strains were slightly more likely to be hospitalized than those infected with other STs.
Additionally, genetic boundaries between C. jejuni genotypes might be readily blurred by horizontal gene transfer and recombination, which are the major driving forces of variability in this species [26, 27]. Indeed, we found a notable variation of ST50 strains in regard to serotype (HS1c, HS2, HS5c, HS8c and HS10) and the LOS class (A and C), the traits mediated via gene diversity in capsular and lipooligosaccharide biosynthesis loci, respectively. Similar observations were made by Skarp et al. [28], who noted microevolution in ST50 strains resulting in their diversification into clusters distinguished by differences in the capsule loci and the distribution of accessory genetic content. Furthermore, based on the core genome analysis the authors revealed tendency for C. jejuni ST-50 strains to partially cluster according to their isolation site, i.e. blood vs faeces [28].
Noteworthy, certain serotypes and LOS classes are considered as important risk factors in development of post-infectious neurological sequels such as GBS and MFS [4, 9]. Briefly, the molecular mimicry between sialylated LOS components and gangliosides present on human peripheral nerves explains this relationship. However, genes involved in the synthesis of sialylated LOS are present only in a few (A, B, C, M, and R) among numerous LOS classes (A to S) [29]. Generally, only C. jejuni strains with LOS classes A, B and C are frequently isolated from stools of patients with GBS, and the LOS classes A and B are associated with GBS and MFS, respectively [4]. However, this connection is not universal and recently Heikema et al. [9] have recognized the serotypes HS1/44c, HS2, HS4c, HS19, HS23/36c and HS41 as an additional risk factors in GBS. Although we have no information with regard to a potential link between our C. jejuni isolates and GBS/MFS, the two ST50 isolates (KF017 and KF042), which partially correspond to this characteristics, i.e. LOS class C but serotype HS8c, seem to pose some risk in GBS/MFS. In contrast, six other ST50 genomes perfectly fit to this LOS/serotype-related GBS/MFS characteristics, i.e. LOS class A and serotype HS2 (Fig. 2, Additional file 1: Table S1). Therefore, MLST-based epidemiology of C. jejuni might be insufficient to recognize or to assess all risks associated with Campylobacter-related diseases, e.g. GBS or MFS, albeit a connection between these neurologic conditions and infections caused by specific C. jejuni strains definitely requires more studies. Despite, C. jejuni is a very common enteropathogen in children, it seems that Polish epidemiological data is underestimated and this pathogen is not routinely detected by laboratories in Poland [30], since each year 2–4 GBS cases are noted in children in Podlasie Province and their cause is unknown (personal communication; Department of Neurology, University Children’s Hospital in Bialystok). In our opinion this fact deserves to be publicized and should impact further investigation.
Another remarkable observation is the presence of vitamin B5 biosynthesis pathway (panBCD operon) in all C. jejuni ST50 and ST257 as well as in 32% of ST51 strains. The panBCD operon has been recently recognized as a marker of human pathogenic C. jejuni strains [17]. In addition, it seems to be a cattle-specific feature and the gain or loss of this locus was suggested as mechanism of rapid C. jejuni host switching from cattle to chickens [31]. Although chickens are generally considered as the ultimate vehicle responsible for transmission of C. jejuni to humans, then cattle may be an important source of strains contaminating the chicken production system, finally leading to human campylobacteriosis [17]. Furthermore, the panBCD operon is co-localized with the blaoxa-61 gene, that is also considered as a trait specific for strains causing infections in humans [17]. However, it is unclear whether this association is coincidental or somewhat linked with the agriculture niche and ability of C. jejuni to colonize ruminants. On the other hand, the study focused on zoonotic transmission of C. jejuni between birds, primates and livestock in US revealed that the presence of another beta-lactamase gene, blaoxa-184, is specific for C. jejuni lineage limited to American crows and not associated with disease [32]. Since we detected blaoxa-184 in 68% of C. jejuni ST51 genomes, including one our carrier isolate (KF070), it is possible that their primary reservoir is not related to agriculture-associated animals or their transmission chain may differ from the used by strains possessing blaoxa-61. Moreover, since this group is characterized by the lowest number (mean = 11.9) of the 25 genes identified as determinants of C. jejuni strains causing infections in humans [17], then its pathogenic potential may be lower than the remaining ST51 or ST50 and ST257 strains.
The antimicrobial resistance patterns identified in our C. jejuni ST50, ST51 and ST257 strains, i.e. resistance to fluoroquinolones only or fluoroquinolones and tetracyclines, in general are consistent with the patterns observed in other C. jejuni genomes representing these STs. Since aminoglycoside resistance genes were recognized only in three genomes, and no macrolide or other antimicrobial resistance determinants were noted. These results are in line with epidemiological data from Poland (for details see an excellent paper by Wieczorek et al. [33]) showing low frequency of resistance to macrolides (from 0% to 3.3%) and aminoglycosides. On the other hand, level of resistance to fluoroquinolones either in our isolates (100%) and other Polish isolates ( > 90%) is critical on the background of European Union ( ~ 55%) [33–36]. Recently, some reports have shown that clonal spreading of C. jejuni fluoroquinolone resistant strains, for example of ST50 clone, is responsible for an emergence of resistance to fluoroquinolones in certain European countries [37]. However, its ubiquitous presence in phylogenetically unrelated STs in our country, indicates rather multiple independent mutation events as the primary cause. This situation is likely connected with a selective pressure associated with a broad use of fluoroquinolones in animal husbandry [33]. In contrast, resistance of C. jejuni to tetracyclines in Poland seems to be variable or locally dependent, and ranging from < 10% even to up 100% [33]. Although, resistance to tetracyclines in C. jejuni is generally considered as plasmid mediated [10], we noted various chromosomal localization of the tetO gene in 41.4% of genomes.
Conclusions
Campylobacter jejuni ST50, ST51 and ST257 are among the top ten of STs isolated in Europe. However, WGS analysis revealed a notable diversity in occurrence of certain clinically relevant genes, serotypes and LOS classes even in C. jejuni strains representing the same STs, ST50 in particular. This observation deserves further clinical and epidemiological investigations as it might be related to risk associated with post-infectious development of GBS or MSF. In addition, the presence or lack of certain host-specific determinants, such as the vitamin B5 biosynthesis pathway or the blaoxa-184 gene may implicate various transmission chains or reservoirs of C. jejuni ST51 strains responsible for campylobacteriosis in northeastern Poland.
Methods
Three predominant sequence types (STs), namely ST50 (CC-21), ST257 (CC-257) and ST51 (CC-443), were identified among C. jejuni isolates collected in our previous study on the etiology of acute diarrhea in northeastern Poland, ie. in Podlasie Province [30]. In detail, the C. jejuni strains were isolated from stool samples of children hospitalized with acute diarrhea in the Department of Pediatric Infectious Diseases of University Children's Hospital in Bialystok (Poland) [30]. The written informed consent from the children's parents or legal guardians for research studies were obtained, and the study was approved by the Bioethics Commission of the Medical University of Bialystok (Consent Number: R-I-002/31/2010 from 28 January 2010).
WGS of four C. jejuni isolates representing the most common STs: diarrheal isolates KF017 and KF042 (ST50), and KF045 (ST257) as well as KF070 strain (ST51) isolated from a control patient (without diarrhea), was performed on Ion PGM Machine (Life Technologies, USA) strictly followed the procedure developed in earlier study [38]. Sequences were assembled with Newbler v2.9 software (Roche, Germany) and deposited in the GenBank database under accession numbers: RDSQ00000000 (KF017), RDSP00000000 (KF042), RDSO00000000 (KF045), and RDSN00000000 (KF070).
In total, 1260 C. jejuni genomes collected in the PATRIC database (Pathosystems Resource Integration Center; accessed in November 2018) were screened for STs using mlst script; https://github.com/tseemann/mlst). In the next step, the genomes representing ST50 (n = 71), ST51 (n = 25) and ST257 (n = 20) (Additional file 1: Table S1) were annotated with Prokka v. 1.3.13 tool (with –usegenus and –genus Campylobacter parameters) [39] followed by pan-genome based phylogenetic analysis performed using Roary v. 3.11.2 software (with minimum percentage identity for blastp of 95% [-i parameter], and PRANK aligner v. 170427 to create a multiFASTA alignment of core genes [-e parameter]) [40, 41]. The Roary output, i.e. alignment of core genes, was used to built maximum-likelihood based on phylogenetic tree with bootstrap value of 500 iterations using RAxML tool v. 8.2.10 and the following parameters: [-m] GTRGAMMA [-p] 12,345 [-f] a [-x] 12,345 [-N] 500 [42].
Antibiotic resistance determinants were detected using ABRicate v0.8.5 software (https://github.com/tseemann/abricate) equipped with the CARD (Comprehensive Antibiotic Resistance Database) database [43]. In addition, PointFinder tool [44] was used to identify point mutations in gyrA (A70T, D85T, T86I, T86A, T86K, T86V, D90A, D90N, D90T, P104S), 23S rRNA (A2074G, A2074T, A2074C, A2075G), cmeR (A86G), rplV (A103C), and rpsL (K88E, K88R, K88Q) responsible for resistance to quinolones, macrolides and spectinomycin, respectively. Resistance patterns were verified by in vitro tests based on the disk diffusion method according to the EUCAST guidelines. The software tools were run with the default settings, except minimum DNA % coverage parameter [–mincov, default ‘0′] in ABRicate which was set to 30.
Virulence patterns were determined with ABRicate software using in-house created virulence gene database involving genes (n = 143) associated with motility, chemotaxis, adhesion, invasion, iron acquisition, capsule and LOS biosynthesis, general stress as well as T4SS and T6SS secretion systems (Additional file 1: Table S2). Additionally, we analyzed the prevalence of 25 genes identified as markers of human pathogenic C. jejuni strains [17]. Furthermore, C. jejuni serotypes were determined based on in silico multiplex PCR with primers described by Poly et al. [45] and simulate_pcr script [46]. Finally, Campylobacter spp. PubMLST database (https://pubmlst.org/campylobacter/; accessed in November 2018), was screened (58 179 records) for the prevalence of the STs under study.
NTSYSpc 2.2 (Exeter Software) and MEGA7 [47] were used to create dendrograms. Genome regions were compared and visualized with Easyfig v2.1 tool [48].
The Tukey's HSD for unequal N test (Spjotvolla/Stoline) was used to compare differences in average number of the virulence genes among the STs with Statistica v7 (StatSoft) software and a p-value 0.01 was considered statistically significant.
Additional file
Additional file 1: Table S1. C. jejuni ST50, ST51 and ST257 genomes (n = 116) collected in the PATRIC genome database. Table S2. C. jejuni virulence genes (n = 143) under the study. Table S3. The twelve most common STs among C. jejuni genomes (n = 116) deposited in the PATRIC genome database. Table S4. The occurrence of virulence genes (n = 143) in C. jejuni ST50, ST51 and ST257 strains under the study. Table S5. The occurence of 25 genes recognized by Buchanan et al. [17] as markers of human pathogenic C. jejuni strains in genomes of C. jejuni ST50, ST51 and ST257. Figure S1. The distribution C. jejuni pan-genome genes and the number of isolates possessing them (visualized using roary_plots.py script). Figure S2. The frequency of C. jejuni genes versus the number of genomes (visualized using roary_plots.py script). Figure S3. The phylogenetic tree compared to a matrix with the presence and absence of C. jejuni core and accessory genes (visualized using roary_plots.py script). Figure S4. Similarity of C. jejuni 81-176 plasmid pTet (A) and C. jejuni CFSAN054107 plasmid pGMI16-002 (B) with C. jejuni strains under the study carrying tetO on plasmid contigs. Figure S5. UPGMA cluster tree illustrating identity of amino acid sequences of TetO variants detected in C. jejuni strains. The evolutionary distances were computed using the Poisson correction method, and are in the units of the number of amino acid substitutions per site. The tree was built in MEGA7 software. Figure S6. Chromosomal locations of the tetO gene in C. jejuni genomes under the study. Figure S7. Similarity of C. jejuni CFSAN054107 plasmid pGMI16-002 with C. jejuni genomes under the study carrying aminoglycoside resistance genes.
Acknowledgments
Authors thank Emilia Murawska and Paulina Marjanska (Department of Microbiology, University of Bialystok) for their involvement in the preparation of genetic libraries into WGS.
Authors’ contributions
KF developed the study concept and elaborated the general notion of the manuscript, performed WGS experiments, accomplished selection of bioinformatics tools and analyzed data, prepared all graphic materials, and wrote the final version of the manuscript; TD performed in vitro antibiotic susceptibility tests; DR and EO involved in critical revision of the manuscript; SP participated in bioinformatics databases analysis; IS involved in the development of the study concept and revised the final version of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by projects N/ST/ZB/18/002/1122 (KF) and N/ST/ZB/18/001/1122 (TD) from the Medical University of Bialystok. WGS equipment is maintained from the ‘Specific Scientific Equipment’ grant of the Ministry of Science and Higher Education in Poland (Decision No. 8636/E-342/SPUB/2018/2, IS).
Availability of data and materials
The authors confirm that the data supporting the findings of this study are available within the article and/or its additional materials.
Ethics approval and consent to participate
The study was approved by the Bioethics Commission of the Medical University of Bialystok (Consent Number: R-I-002/31/2010 from 28 January 2010).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Krzysztof Fiedoruk, Email: krzysztof.fiedoruk@umb.edu.pl.
Tamara Daniluk, Email: tamara.daniluk@umb.edu.pl.
Dorota Rozkiewicz, Email: dorota.rozkiewicz@umb.edu.pl.
Elzbieta Oldak, Email: elzbieta.oldak@umb.edu.pl.
Suhanya Prasad, Email: suhanyavp@gmail.com.
Izabela Swiecicka, Email: izabelas@uwb.edu.pl.
References
- 1.Kirk MD, Pires SM, Black RE, Caipo M, Crump JA, Devleesschauwer B, et al. Correction: World Health Organization Estimates of the Global and Regional Disease Burden of 22 Foodborne Bacterial, Protozoal, and Viral Diseases, 2010: a data synthesis. PLoS Med. 2015;12(12):e1001940. doi: 10.1371/journal.pmed.1001940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2015. EFSA J.14(12):4634. [DOI] [PMC free article] [PubMed]
- 3.Kaakoush NO, Castano-Rodriguez N, Mitchell HM, Man SM. Global epidemiology of campylobacter infection. Clin Microbiol Rev. 2015;28(3):687–720. doi: 10.1128/CMR.00006-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Godschalk PC, Heikema AP, Gilbert M, Komagamine T, Ang CW, Glerum J, et al. The crucial role of Campylobacter jejuni genes in anti-ganglioside antibody induction in Guillain-Barre syndrome. J Clin Invest. 2004;114(11):1659–1665. doi: 10.1172/JCI200415707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poropatich KO, Walker CL, Black RE. Quantifying the association between Campylobacter infection and Guillain-Barre syndrome: a systematic review. J Health Popul Nutr. 2010;28(6):545–552. doi: 10.3329/jhpn.v28i6.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burnham PM, Hendrixson DR. Campylobacter jejuni: collective components promoting a successful enteric lifestyle. Nat Rev Microbiol. 2018;16(9):551–565. doi: 10.1038/s41579-018-0037-9. [DOI] [PubMed] [Google Scholar]
- 7.Wu Z, Mou K, Sahin O, Zhang Q. Genomic insights into Campylobacter jejuni virulence and population genetics. Infect Dis Transl Med. 2016;2(3):109–119. [Google Scholar]
- 8.Dasti JI, Tareen AM, Lugert R, Zautner AE, Gross U. Campylobacter jejuni: a brief overview on pathogenicity-associated factors and disease-mediating mechanisms. Int J Med Microbiol. 2010;300(4):205–211. doi: 10.1016/j.ijmm.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Heikema AP, Islam Z, Horst-Kreft D, Huizinga R, Jacobs BC, Wagenaar JA, et al. Campylobacter jejuni capsular genotypes are related to Guillain-Barre syndrome. Clin Microbiol Infect. 2015;21(9):852. doi: 10.1016/j.cmi.2015.05.031. [DOI] [PubMed] [Google Scholar]
- 10.Wieczorek K, Osek J. Antimicrobial resistance mechanisms among Campylobacter. Biomed Res Int. 2013;2013:340605. doi: 10.1155/2013/340605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tacconelli E, Carrara E, Savoldi A, Kattula D, Burkert F. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. 2018;18(3):318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 12.Colles FM, Maiden MC. Campylobacter sequence typing databases: applications and future prospects. Microbiology. 2012;158(Pt 11):2695–2709. doi: 10.1099/mic.0.062000-0. [DOI] [PubMed] [Google Scholar]
- 13.Drewnowska JM, Swiecicka I. Eco-genetic structure of Bacillus cereus sensu lato populations from different environments in northeastern Poland. PLoS ONE. 2013;8(12):e80175. doi: 10.1371/journal.pone.0080175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Llarena AK, Taboada E, Rossi M. Whole-genome sequencing in epidemiology of Campylobacter jejuni infections. J Clin Microbiol. 2017;55(5):1269–1275. doi: 10.1128/JCM.00017-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nadon C, Van Walle I, Gerner-Smidt P, Campos J, Chinen I, Concepcion-Acevedo J, et al. PulseNet international: vision for the implementation of whole genome sequencing (WGS) for global food-borne disease surveillance. Euro Surveill. 2017;22:23. doi: 10.2807/1560-7917.ES.2017.22.23.30544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whitehouse CA, Young S, Li C, Hsu CH, Martin G, Zhao S. Use of whole-genome sequencing for Campylobacter surveillance from NARMS retail poultry in the United States in 2015. Food Microbiol. 2018;73:122–128. doi: 10.1016/j.fm.2018.01.018. [DOI] [PubMed] [Google Scholar]
- 17.Buchanan CJ, Webb AL, Mutschall SK, Kruczkiewicz P, Barker DOR, Hetman BM, et al. A genome-wide association study to identify diagnostic markers for human pathogenic Campylobacter jejuni strains. Front Microbiol. 2017;8:1224. doi: 10.3389/fmicb.2017.01224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rokney A, Valinsky L, Moran-Gilad J, Vranckx K, Agmon V, Weinberger M. Genomic epidemiology of Campylobacter jejuni transmission in Israel. Front Microbiol. 2018;9:2432. doi: 10.3389/fmicb.2018.02432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao S, Tyson GH, Chen Y, Li C, Mukherjee S, Young S, et al. Whole-Genome Sequencing Analysis Accurately Predicts Antimicrobial Resistance Phenotypes in Campylobacter spp. Appl Environ Microbiol. 2016;82(2):459–466. doi: 10.1128/AEM.02873-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dearlove BL, Cody AJ, Pascoe B, Meric G, Wilson DJ, Sheppard SK. Rapid host switching in generalist Campylobacter strains erodes the signal for tracing human infections. ISME J. 2016;10(3):721–729. doi: 10.1038/ismej.2015.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wieczorek K, Denis E, Lachtara B, Osek J. Distribution of Campylobacter jejuni multilocus sequence types isolated from chickens in Poland. Poult Sci. 2017;96(3):703–709. doi: 10.3382/ps/pew343. [DOI] [PubMed] [Google Scholar]
- 22.Ramonaite S, Tamuleviciene E, Alter T, Kasnauskyte N, Malakauskas M. MLST genotypes of Campylobacter jejuni isolated from broiler products, dairy cattle and human campylobacteriosis cases in Lithuania. BMC Infect Dis. 2017;17(1):430. doi: 10.1186/s12879-017-2535-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheppard SK, Colles FM, McCarthy ND, Strachan NJ, Ogden ID, Forbes KJ, et al. Niche segregation and genetic structure of Campylobacter jejuni populations from wild and agricultural host species. Mol Ecol. 2011;20(16):3484–3490. doi: 10.1111/j.1365-294X.2011.05179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sahin O, Plummer PJ, Jordan DM, Sulaj K, Pereira S, Robbe-Austerman S, et al. Emergence of a tetracycline-resistant Campylobacter jejuni clone associated with outbreaks of ovine abortion in the United States. J Clin Microbiol. 2008;46(5):1663–1671. doi: 10.1128/JCM.00031-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harvala H, Rosendal T, Lahti E, Engvall EO, Brytting M, Wallensten A, et al. Epidemiology of Campylobacter jejuni infections in Sweden, November 2011-October 2012: is the severity of infection associated with C. jejuni sequence type? Infect Ecol Epidemiol. 2016;6:31079. doi: 10.3402/iee.v6.31079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karlyshev AV, Champion OL, Churcher C, Brisson JR, Jarrell HC, Gilbert M, et al. Analysis of Campylobacter jejuni capsular loci reveals multiple mechanisms for the generation of structural diversity and the ability to form complex heptoses. Mol Microbiol. 2005;55(1):90–103. doi: 10.1111/j.1365-2958.2004.04374.x. [DOI] [PubMed] [Google Scholar]
- 27.Wilson DJ, Gabriel E, Leatherbarrow AJ, Cheesbrough J, Gee S, Bolton E, et al. Rapid evolution and the importance of recombination to the gastroenteric pathogen Campylobacter jejuni. Mol Biol Evol. 2009;26(2):385–397. doi: 10.1093/molbev/msn264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skarp CPA, Akinrinade O, Kaden R, Johansson C, Rautelin H. Accessory genetic content in Campylobacter jejuni ST21CC isolates from feces and blood. Int J Med Microbiol. 2017;307(4–5):233–240. doi: 10.1016/j.ijmm.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 29.Parker CT, Gilbert M, Yuki N, Endtz HP, Mandrell RE. Characterization of lipooligosaccharide-biosynthetic loci of Campylobacter jejuni reveals new lipooligosaccharide classes: evidence of mosaic organizations. J Bacteriol. 2008;190(16):5681–5689. doi: 10.1128/JB.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fiedoruk K, Daniluk T, Rozkiewicz D, Zaremba ML, Oldak E, Sciepuk M, et al. Conventional and molecular methods in the diagnosis of community-acquired diarrhoea in children under 5 years of age from the north-eastern region of Poland. International Journal of Infectious Diseases. 2015;37:145–151. doi: 10.1016/j.ijid.2015.06.028. [DOI] [PubMed] [Google Scholar]
- 31.Sheppard SK, Didelot X, Meric G, Torralbo A, Jolley KA, Kelly DJ, et al. Genome-wide association study identifies vitamin B5 biosynthesis as a host specificity factor in Campylobacter. Proc Natl Acad Sci USA. 2013;110(29):11923–11927. doi: 10.1073/pnas.1305559110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weis AM, Storey DB, Taff CC, Townsend AK, Huang BC, Kong NT, et al. Genomic comparison of Campylobacter spp. and their potential for zoonotic transmission between birds, primates, and livestock. Appl Environ Microbiol. 2018;82(24):7165–7175. doi: 10.1128/AEM.01746-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wieczorek K, Wolkowicz T, Osek J. Antimicrobial resistance and virulence-associated traits of Campylobacter jejuni isolated from poultry food chain and humans with diarrhea. Front Microbiol. 2018;9:1508. doi: 10.3389/fmicb.2018.01508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wozniak-Biel A, Bugla-Ploskonska G, Kielsznia A, Korzekwa K, Tobiasz A, Korzeniowska-Kowal A, et al. High prevalence of resistance to fluoroquinolones and tetracycline Campylobacter spp. isolated from poultry in Poland. Microb Drug Resist. 2018;24(3):314–322. doi: 10.1089/mdr.2016.0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szczepanska B, Andrzejewska M, Spica D, Klawe JJ. Prevalence and antimicrobial resistance of Campylobacter jejuni and Campylobacter coli isolated from children and environmental sources in urban and suburban areas. BMC Microbiol. 2017;17(1):80. doi: 10.1186/s12866-017-0991-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anonymous The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2016. EFSA J. 2018;16:5182. doi: 10.2903/j.efsa.2018.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kovac J, Cadez N, Stessl B, Stingl K, Gruntar I, Ocepek M, et al. High genetic similarity of ciprofloxacin-resistant Campylobacter jejuni in central Europe. Front Microbiol. 2015;6:1169. doi: 10.3389/fmicb.2015.01169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fiedoruk K, Daniluk T, Swiecicka I, Murawska E, Sciepuk M, Leszczynska K. First Complete Genome Sequence of Escherichia albertii Strain KF1, a New Potential Human Enteric Pathogen. Genome announcements. 2014;2(1):e00004–14. doi: 10.1128/genomeA.00004-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 40.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31(22):3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loytynoja A. Phylogeny-aware alignment with PRANK. Methods Mol Biol. 2014;1079:155–170. doi: 10.1007/978-1-62703-646-7_10. [DOI] [PubMed] [Google Scholar]
- 42.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, et al. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 2013;57(7):3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zankari E, Allesoe R, Joensen KG, Cavaco LM, Lund O, Aarestrup FM. PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J Antimicrob Chemother. 2017;72(10):2764–2768. doi: 10.1093/jac/dkx217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poly F, Serichantalergs O, Kuroiwa J, Pootong P, Mason C, Guerry P, et al. Updated Campylobacter jejuni capsule PCR multiplex typing system and its application to clinical isolates from South and Southeast Asia. PLoS ONE. 2015;10(12):e0144349. doi: 10.1371/journal.pone.0144349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gardner SN, Slezak T. Simulate_PCR for amplicon prediction and annotation from multiplex, degenerate primers and probes. BMC Bioinf. 2014;15:237. doi: 10.1186/1471-2105-15-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumar S, Stecher S, Tamura K. MEGA7: molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27(7):1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. C. jejuni ST50, ST51 and ST257 genomes (n = 116) collected in the PATRIC genome database. Table S2. C. jejuni virulence genes (n = 143) under the study. Table S3. The twelve most common STs among C. jejuni genomes (n = 116) deposited in the PATRIC genome database. Table S4. The occurrence of virulence genes (n = 143) in C. jejuni ST50, ST51 and ST257 strains under the study. Table S5. The occurence of 25 genes recognized by Buchanan et al. [17] as markers of human pathogenic C. jejuni strains in genomes of C. jejuni ST50, ST51 and ST257. Figure S1. The distribution C. jejuni pan-genome genes and the number of isolates possessing them (visualized using roary_plots.py script). Figure S2. The frequency of C. jejuni genes versus the number of genomes (visualized using roary_plots.py script). Figure S3. The phylogenetic tree compared to a matrix with the presence and absence of C. jejuni core and accessory genes (visualized using roary_plots.py script). Figure S4. Similarity of C. jejuni 81-176 plasmid pTet (A) and C. jejuni CFSAN054107 plasmid pGMI16-002 (B) with C. jejuni strains under the study carrying tetO on plasmid contigs. Figure S5. UPGMA cluster tree illustrating identity of amino acid sequences of TetO variants detected in C. jejuni strains. The evolutionary distances were computed using the Poisson correction method, and are in the units of the number of amino acid substitutions per site. The tree was built in MEGA7 software. Figure S6. Chromosomal locations of the tetO gene in C. jejuni genomes under the study. Figure S7. Similarity of C. jejuni CFSAN054107 plasmid pGMI16-002 with C. jejuni genomes under the study carrying aminoglycoside resistance genes.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and/or its additional materials.