Abstract
Accurate and efficient transcription termination is an important step for cells to generate functional RNA transcripts. In bacteria, two mechanisms are responsible for terminating transcription: intrinsic (Rho-independent) termination and Rho-dependent termination. Growing examples suggest that neither type of transcription termination is static, but instead are highly dynamic and regulated. Regulatory small RNAs (sRNAs) are key players in bacterial stress responses, are frequently expressed under specific growth conditions, and are predominantly terminated through the intrinsic termination mechanism. Once made, sRNAs can base-pair with mRNA targets and regulate mRNA translation and stability. Recent findings suggest that alterations in the efficiency of intrinsic termination for sRNAs under various growth conditions may affect the availability of a given sRNA and the ability of the sRNA to function properly. Moreover, alterations of mRNA structure, translation, and accessibility by sRNAs have the potential to impact the access of Rho factor to mRNAs and thus termination of the mRNA. Indeed, recent studies have revealed that some sRNAs can modulate target gene expression by stimulating or inhibiting Rho-dependent termination, thus expanding the regulatory power of bacterial sRNAs. Here we review the current knowledge on intrinsic termination of sRNAs and sRNA-mediated Rho-dependent termination of protein coding genes in bacteria.
Keywords: SgrS, ChiX, SraL, Hfq, CsrA, Rho
Bacterial sRNA Function and the Role of HFQ
Critical processes in bacteria, including those necessary for pathogenesis, are frequently regulated at multiple levels. Small regulatory RNAs play important roles in both bacteria and hosts. The bacterial small RNAs (sRNAs) function by base-pairing to target mRNAs, resulting in stimulation or inhibition of mRNA stability and translation. These sRNAs are usually around 100 nt long, and use a short seed region (8–15 nt) to form an RNA duplex with mRNA for regulation (Storz et al., 2011; Wagner and Romby, 2015). In E. coli and many related bacteria, sRNAs often act in concert with the RNA chaperone Hfq. Mutations in hfq are associated with attenuated virulence in many pathogens, suggesting critical roles for sRNAs in pathogenesis (Chao and Vogel, 2010), although, in a few instances, Hfq can act independently of sRNAs [see, for instance (Chen and Gottesman, 2017)].
Hfq, an Lsm/Sm family RNA binding protein, forms a ring-shaped homohexamer. Hfq binds to and stabilizes sRNAs and promotes their pairing with mRNA targets (Vogel and Luisi, 2011; Updegrove et al., 2016). Three different surfaces on the hexamer have been shown to be important for RNA binding. The proximal face binds polyU sequences, and mutations on this surface disrupt sRNA binding in vitro and sRNA stability in vivo (Otaka et al., 2011; Sauer and Weichenrieder, 2011). The distal face binds AAN repeats (Robinson et al., 2014), frequently found on mRNA targets of sRNAs. Some RNAs bind to the rim of Hfq through AU rich regions; the rim has also been implicated in helping bring sRNA and mRNA together (Panja et al., 2013). Most sRNAs are quite stable in the cell when bound to Hfq, but are presumably displaced from Hfq and degraded after pairing to mRNA targets (Massé et al., 2003; Schu et al., 2015). Thus, sRNA function and stability depend on the ability to bind Hfq properly.
Here we focus on sRNAs in Escherichia coli, one of the species in which these regulators have been well studied. sRNAs are usually expressed from dedicated and well-regulated promoters, but in some cases, sRNAs are processed from the 3′ end of mRNAs, and are thus dependent on the upstream mRNA promoters for expression (Miyakoshi et al., 2015b; Kavita et al., 2018). Regulation of sRNA promoters is often in response to specific cellular stresses, contributing to the cells' ability to adapt to changing environments. For instance, the iron-responsive RyhB sRNA is induced in response to iron deficiency and negatively regulates multiple iron binding proteins, thus helping the cell save iron for critical proteins (Massé and Gottesman, 2002). SgrS, on the other hand, is an sRNA made in response to accumulation of toxic sugar phosphates; it negatively regulates genes involved in the import of the sugar phosphates into the cell, while upregulating a phosphatase gene critical for detoxification of phosphosugar stress (Papenfort et al., 2013; Bobrovskyy and Vanderpool, 2014).
Regardless of whether an sRNA is processed from a longer transcript or not, a critical feature that enables sRNAs to bind Hfq is the presence of a Rho-independent terminator at its 3′ end (Otaka et al., 2011; Morita et al., 2017). This requirement suggests that transcription termination is crucial for both sRNA biogenesis and function. Recent work highlighting this process of sRNA termination, ways in which intrinsic terminators of sRNAs may be distinct from other intrinsic terminators, and the ways in which termination may be regulated are discussed in the first part of this review.
sRNAs, by binding to mRNAs, can affect many aspects of mRNA folding, translation, and decay, as well as access of RNA binding proteins. While sRNAs frequently act to alter ribosome binding and translation initiation, recent findings highlight how sRNAs can affect gene expression by blocking or facilitating premature Rho-dependent transcription termination within the genes of target mRNAs. We review the characteristics of these interactions in the second part of this review.
Rho-independent Termination
Transcription termination that is independent of termination factors is known as Rho-independent termination, also called intrinsic termination [reviewed in Roberts (in press)]. Essential elements of a Rho-independent terminator consist of a GC-rich dyad repeat that forms a stem-loop (hairpin) structure followed by a T-rich stretch, generating a U-rich tail in the RNA after termination (Adhya and Gottesman, 1978). Rho-independent termination is achieved by formation of the stem-loop structure, which is facilitated by RNA polymerase pausing during transcription of the T-rich tract (Ray-Soni et al., 2016). The T-rich stretch is highly conserved among Rho-independent terminators in bacteria, while sequences of the stem-loop seem not to be conserved except for their GC-rich characteristic. Interestingly, T-rich stretches can be found in terminators for both eukaryotic RNA polymerase III and archaeal RNA polymerase (Arimbasseri et al., 2013; Maier and Marchfelder, 2019), suggesting that the intrinsic termination within a T-stretch is a fundamental characteristic of termination pathways. Although many of the T-rich stretches contain four to eight Ts, they are frequently disrupted with other nucleotides (d'Aubenton Carafa et al., 1990; Chen et al., 2013). Intrinsic termination can be directly measured in vitro by appearance of properly terminated transcripts, and in vivo by reporters to measure termination read-through [see, for instance (Morita et al., 2017)].
Characteristics of sRNA Rho-Independent Terminators
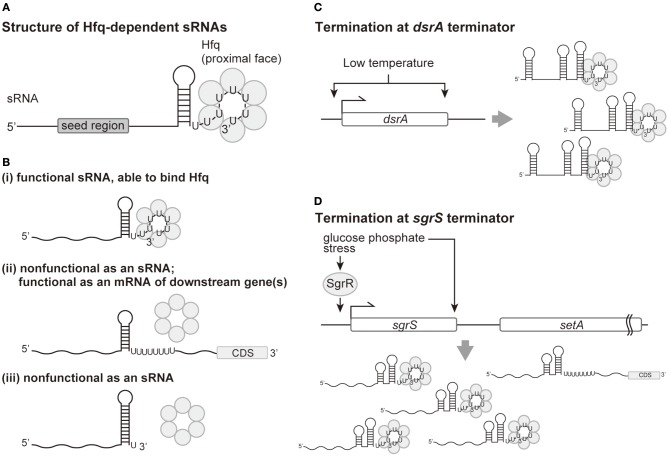
Transcription of genes encoding sRNAs are generally terminated by Rho-independent termination (Livny and Waldor, 2007). A notable feature of Rho-independent terminators of sRNAs is a consecutive T stretch longer than seven nucleotides, which is not necessarily found in all Rho-independent terminators (Otaka et al., 2011; Morita et al., 2017). The fact that discontinuous and relatively short T-rich stretches are found at many Rho-independent terminators implies that seven or more Ts found in sRNA terminators is not required for transcription termination. The transcribed long T stretch, i.e., a polyU tail of seven or more Us, is the primary element responsible for the binding of sRNAs to Hfq (Figure 1A). Studies of sRNA SgrS and others demonstrated that shortening the polyT stretch to four Ts in terminators of sRNA genes no longer produced functional sRNAs in vivo (Otaka et al., 2011). Additionally, the individual consecutive six uridines, including the last U, bind to the proximal face of the Hfq hexamer in vitro (Sauer and Weichenrieder, 2011). Consistent with these findings, deep sequencing analyses of Hfq-binding sRNAs verified that polyU tails of sRNAs longer than six Us are primary binding sites for Hfq in vivo (Holmqvist et al., 2016; Melamed et al., 2016) (Figure 1Bi). It should be noted that even shortening an sgrS variant from eight Ts to six Ts generated less functional SgrS, implying that six Us is not long enough to produce a fully functional sRNA (Otaka et al., 2011). Because, from a structural perspective, the six U tail seems to be sufficient for binding to Hfq, a question remains what the role of extra U(s) plays in sRNA function in vivo. In addition, some sRNAs possess a discontinuous U-rich tail disrupted with other nucleotides (Otaka et al., 2011; Morita et al., 2017). Effects of these discontinuities on the function and/or production of sRNAs also remain to be studied.
Figure 1.
Production of the polyU tail of sRNAs by Rho-independent termination and the role of a polyU tail in Hfq binding and sRNA function. (A) Functional structure of an Hfq-dependent sRNA. The polyU tail of seven or longer is responsible for binding to the Hfq proximal face. The seed region is defined as the region which pairs with target mRNAs and is generally located 5′ to the terminator. (B) (i): Termination at the seventh or longer position within the T stretch is necessary for functional sRNAs. (ii): Transcripts with 3′ extensions resulting from termination site read-through bind poorly to Hfq, resulting in non-functional sRNAs. (iii): The transcripts with the U tail shortened by premature termination are no longer able to bind Hfq, resulting in non-functional sRNAs. (C) Low temperature increases both the level of transcription initiation and the termination efficiency for the gene encoding the DsrA sRNA. (D) Termination efficiency at the sgrS terminator is improved by glucose-phosphate stress, which also induces transcription initiation of sgrS by SgrR, a transcriptional regulator. Increased transcription termination will result in more production of SgrS and less expression of the downstream setA mRNA.
In contrast to a conserved polyT stretch, nucleotide sequences for the stem-loop structure seem not to be conserved between intrinsic terminators (Ishikawa et al., 2012; Morita et al., 2017). In fact, the SgrS sRNA was found to be fully functional with a heterologous terminator with a long polyU tail and hairpin stability approximately equal to that of the native terminator (Otaka et al., 2011). The stem-loop structure itself is critical for sRNA function, likely because without this structure termination efficiency is disrupted, resulting in 3′-extended transcripts. Nucleotide substitutions in the DsrA and RybB terminators that reduce thermodynamic stability of the stem were isolated as mutations affecting their function (Sledjeski and Gottesman, 1995; Balbontín et al., 2010). Consistent with the structure of Hfq specifically binding to the hydroxyl group at the 3′-end of the RNA in vitro (Sauer and Weichenrieder, 2011), a recent study found that read-through products of SgrS and RyhB with a polyU tract internal to the transcript did not bind Hfq in vivo (Morita et al., 2015). These findings suggest that 3′-extended transcripts resulting from read-through no longer function as sRNAs (Figure 1Bii). One sRNA, DicF, was reported to be produced both via termination and via 3′-processing by RNase III from a longer read-through transcript (Faubladier et al., 1990). However, a dicF coding region expressed from a plasmid and lacking the downstream processing site still generated functional DicF in vivo (Balasubramanian et al., 2016), suggesting that the intrinsic termination pathway is sufficient for the production of DicF, as with other sRNAs.
Another important feature of the terminator stem-loop structure is the strength of the stem, which affects the position of termination. A moderate but not too strong stem is needed to generate a polyU tail of seven nucleotides or longer. Extension of SgrS and RyhB terminator stems with additional G-C pairs resulted in premature termination and the generation of non-functional transcripts with polyU tails shorter than six (Morita et al., 2017; Figure 1Biii).
Based on the sRNAs studied thus far, the intrinsic terminators of sRNAs are expected to have unique features, forming a subset of the Rho-independent terminators in E. coli and possibly other bacteria. These features—a polyT stretch of seven or more and a moderate-strength stem-loop that enables termination after a stretch of seven or more Ts—can help in identifying potential Hfq-dependent sRNAs from genomic sequence analysis, and possibly help in distinguishing RNA transcripts that encode short proteins from those that have the potential to act as sRNAs. This would also be useful for the design and engineering of synthetic sRNAs for the control of specific gene expression. The absence of the critical characteristics for Hfq binding in the intrinsic terminators for mRNAs might prevent these mRNAs from binding to and blocking the proximal face of Hfq, where sRNAs must bind.
Regulation of Rho-Independent Termination of sRNAs
A growing number of studies have found that sRNAs can be encoded upstream of ORFs, where the promoter upstream of the sRNA is responsible for expression of the downstream ORF. In these cases, expression of the ORF requires that transcription continue through the sRNA termination sequences, leading to a mRNA in which the embedded sRNA is non-functional. SgrS is co-transcribed with the downstream gene setA encoding a SET (sugar efflux transporter) family protein, although SgrS requires transcription termination at its own terminator for function (Sun and Vanderpool, 2011; Morita et al., 2015). SroC, one of the 3′ derived Hfq-binding sRNAs, is encoded within the gltIJKL operon encoding a Glu/Asp ABC transporter and likely needs to terminate at its own Rho-independent terminator for function (Vogel et al., 2003; Miyakoshi et al., 2015a). Given that Rho-independent termination impacts functional sRNA production, the downstream genes are assumed to be expressed discordantly from these sRNAs. Although the biological significance of the discordant expression of sRNAs and the downstream genes remains to be determined, a recent study revealed that such discordant expression by internal Rho-independent termination occurs frequently within operons and contributes to preferred expression levels for individual proteins (Lalanne et al., 2018).
Intriguingly, growing evidence suggests that the efficiency of transcription termination at Rho-independent terminators of sRNAs can be regulated by the same physiological and/or stress signals that induce initiation of sRNA transcription. Termination efficiency at the terminator for the DsrA sRNA, a positive regulator of RpoS translation, is increased at low temperature, where the dsrA promoter is most active (Sledjeski et al., 1996; Figure 1C). Similarly, an increase in termination efficiency at the SgrS and RyhB terminators was observed under conditions of both the cognate stress for transcriptional induction of these sRNAs and non-cognate stresses (Morita et al., 2015; Figure 1D). One can envision that this increase in termination efficiency would result in more effective production of sRNAs under specific conditions.
Previous studies on Rho-independent termination provide clues to the molecular mechanism by which Rho-independent termination is regulated. Lower levels of UTP nucleotide were found to improve transcription termination at several Rho-independent terminators in vitro (Farnham et al., 1982; McDowell et al., 1994). The density of RNA polymerase on an mRNA was found to influence the rate of transcription elongation/termination at Rho-independent terminators in vivo and in vitro, with higher transcription (more RNA polymerases) decreasing pausing and termination (Jacquet and Reiss, 1992; Epshtein and Nudler, 2003). Therefore, this linkage between promoter strength (transcription initiation) and termination provides a way in which stresses that decrease overall transcription activity on a given gene may result in increases in the termination efficiency. In addition, specific protein factors can be involved in regulation of Rho-independent termination. NusA is an essential protein which affects termination efficiency at Rho-independent terminators in multiple ways, by contributing to RNA polymerase pausing and by helping form and stabilizing RNA structures upon termination (Nudler and Gottesman, 2002; Guo et al., 2018; Holmqvist and Vogel, 2018), and thus could regulate termination in response to stresses. Hfq can be excluded as a candidate factor because it seems not to be involved in the termination at sRNA terminators (Morita et al., 2015).
Most in vivo experiments on Rho-independent termination have been carried out with protein-coding genes as templates. The situation at the sRNA terminators might be different from that at the mRNA terminators because the protein-coding genes are typically longer than sRNA genes. For instance, for short sRNA-encoding transcripts, dissociation of the sigma factor from the RNA polymerase, usually assumed to occur soon after transcription initiation, might not occur before the polymerase reaches the intrinsic terminator, possibly changing the efficiency of termination. A recent long-read RNA sequencing strategy enabled analysis of intact transcripts and revealed that the degree of readthrough of several mRNA terminators also varied between growth conditions (Yan et al., 2018). A critical question for future investigation will be how Rho-independent terminators are modulated and whether these mechanisms of modulation are specific to sRNA terminators, or also affect those for mRNAs.
Rho-dependent Transcription Termination
In E. coli, 20–30% of transcription events are terminated in a Rho-dependent manner [(Peters et al., 2009, 2012); reviewed in Roberts (in press)]. While Rho is generally not used to form the 3′ ends of regulatory sRNAs, its ability to terminate mRNAs can be regulated by sRNAs.
Transcription termination factor Rho is an ATP-dependent RNA helicase/translocase, which can bind to a sequence motif called the Rho utilization (rut) site on the nascent transcript, translocate along the naked RNA and dissociate the elongation complex to terminate transcription (Grylak-Mielnicka et al., 2016; Ray-Soni et al., 2016). A typical rut site is a single-stranded, ribosome-free, cytosine-rich/low-guanine RNA sequence with a length of ~60–80 nucleotides. Thus, molecular or cellular processes that change ribosome occupancy on mRNA or the single-stranded nature of an RNA stretch at or adjacent to the rut site may regulate Rho's function. Rho forms a homohexamer ring, and uses its N-terminal OB-like protein fold to bind cytosine-rich sequences, while a region near the C-terminal part of the protein binds to the RNA threaded in the channel; the latter binding event activates Rho's ATPase activity, driving its translocation (Skordalakes and Berger, 2006). Several models of Rho-dependent transcription termination have been proposed (Peters et al., 2011; Ray-Soni et al., 2016), but the molecular details in terms of the mechanism of Rho translocation and RNA polymerase dissociation on Rho-dependent terminators remain to be fully determined. In vitro approaches to study Rho-dependent termination and its regulation by sRNAs have been recently described (Nadiras et al., 2018). In vivo approaches primarily depend on defining Rho-dependent termination with Rho-specific inhibitors such as bicyclomycin, and evaluating effects of sRNAs on mRNA expression by measuring read-through products with reporter assays, quantitative PCR or deep sequencing (Hussein et al., 2015; Elgamal et al., 2016; Sedlyarova et al., 2016).
Rho-Dependent Transcription Termination Affected by sRNAs and RNA-Binding Proteins
Rho-dependent termination at the 3′ end of protein-coding genes is well documented, and this process is not believed to be significantly regulated. Rho can also terminate transcription inside genes and within operons, and this is likely a highly regulated process given the fact that the translation status of mRNAs can directly affect Rho access to rut sites inside coding sequences (Adhya and Gottesman, 1978). Genome-wide analysis of RNA Polymerase redistribution in the presence of the Rho-specific inhibitor bicyclomycin (BCM) found that Rho terminates transcription intragenically within ~100 genes (Peters et al., 2009, 2012). The observation of such intragenic termination sites raised the possibility that factors that can affect translation, such as sRNAs or RNA-binding proteins, may regulate Rho-dependent termination at these sites.
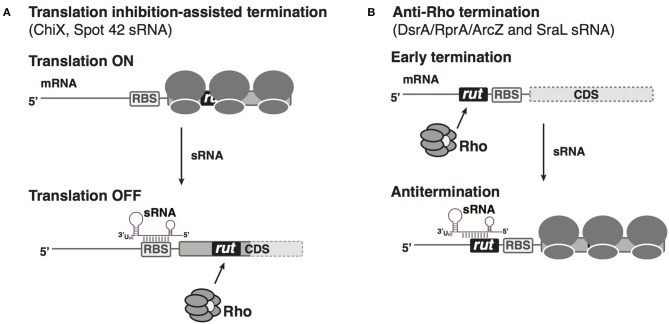
A seminal study documented sRNA-mediated regulation of Rho-dependent termination in Salmonella (Bossi et al., 2012). ChiX is an Hfq-dependent sRNA that can bind to the chiP mRNA, encoding the outer membrane channel (porin) for chitosugars, at its ribosome binding site, inhibit translation and trigger chiP mRNA decay (Figueroa-Bossi et al., 2009). Interestingly, ChiX-mediated translation inhibition has a secondary effect. By blocking entry of ribosomes on chiP mRNA, ChiX binding exposes an intragenic rut site for Rho loading, leading to premature termination of transcription inside the chiP coding sequence (Figure 2A). This regulation not only reinforces the inhibition of chiP translation but also leads to a polarity effect, inhibiting expression of the downstream chiQ gene in the same operon (Bossi et al., 2012). When chitosugars are available and thus the transporter is needed, ChiX is destroyed by interaction with an RNA decoy, induced dependent on chitosugar sensing (Figueroa-Bossi et al., 2009; Overgaard et al., 2009). Therefore, regulation coordinates with other transcriptional circuits to ensure genes responsible for utilization of chitin-derived oligosaccharides are switched on only when chitosugars are present. In a second example, the E. coli Spot 42 sRNA base pairs with the galK leader, the third gene in the galETKM operon, resulting in translational inhibition of galK as well as an increased Rho-dependent transcription termination, possibly by interfering with translational coupling between ribosomes translating through galT with those initiating translation of galK, and thus allowing Rho access (Wang et al., 2015; Figure 2A). This type of regulation allows discoordinate expression of operon genes in response to metabolic needs of the cells.
Figure 2.
sRNA-based negative or positive regulation of Rho-dependent transcription attenuation in bacteria. (A) Translation inhibition by sRNA assists Rho-dependent termination. When the accumulation of Salmonella ChiX or E. coli Spot 42 sRNA is low, the mRNAs chiPQ and galETKM are translated well. When ChiX or Spot42 sRNA is produced at high levels, under specific environmental conditions, sRNA annealing blocks the ribosome binding site of the corresponding mRNA target (chiP or galK, respectively); reduced translation allows Rho loading onto a cryptic rut site to terminate transcription inside the genes (dotted pale portion of box represents untranscribed portion of the mRNA). (B) Antagonization of Rho-dependent attenuation by sRNAs. In genes with long 5′ UTRs, rut sites can allow Rho termination, blocking expression of the downstream gene (dotted box represents untranscribed coding region). sRNAs binding within the 5′ UTR can block Rho access to the rut sites, thus acting as anti-terminators for the downstream CDS.
In the cases described above, sRNAs facilitate Rho access to rut sites by interfering with mRNA translation, consistent with the consensus that mRNA sequences devoid of translating ribosomes are preferable targets for Rho binding and regulation. However, sRNAs can also positively regulate genes, raising the possibility that in some cases, sRNAs might antagonize Rho function by blocking Rho binding and termination. In addition to the 3′ end of genes and the intragenic regions containing rut sites, long 5′ UTRs (>80 nt) of mRNAs are another reservoir of potential sites for Rho-dependent termination. Transcriptomic analyses of RNA samples from E. coli treated with sublethal concentrations of Rho inhibitor BCM (Sedlyarova et al., 2016) suggested that, out of the 1,200 5′ UTRs longer than 80 nts, at least 250 were targets for Rho-dependent termination. One such 5′ UTR is the 567 nt leader of rpoS, an mRNA known to be positively regulated by sRNAs (Battesti et al., 2011). In-depth analysis of regulation in the rpoS 5′ UTR revealed that the three base-pairing sRNAs (RprA, DsrA, and ArcZ) that activate rpoS mRNA translation can antagonize Rho-dependent premature termination in the rpoS leader (Sedlyarova et al., 2016; Figure 2B). Such a regulatory mechanism is likely not limited to rpoS, but may apply to many other putative targets of these three sRNAs, as well as other positively regulated mRNAs. As optimal growth of E. coli in the stationary stress condition relies on expression of the stationary sigma factor RpoS, the sRNA-mediated upregulation of RpoS production both via increasing ribosome entry and via anti-Rho-dependent premature termination provides an important way for sRNAs to contribute to the response to stress.
In another example of an sRNA preventing Rho-dependent termination, the Salmonella SraL sRNA was reported to base-pair with the leader for rho mRNA, protecting it from transcription attenuation and thus increasing levels of full-length rho mRNA (Silva et al., 2019; Figure 2B). While the physiological role of this regulatory interaction is not known, it raises the possibility that under certain conditions the cell might modulate Rho levels and thus Rho-dependent termination via the SraL sRNA. For both the rpoS and rho leader, it is still not clear how sRNA binding prevents Rho-dependent premature termination. In both cases, the sRNA binds upstream of the RBS site, and in the absence of corresponding sRNAs, Rho can prematurely terminate transcription in the leader. Therefore, it is very likely that sRNA blocks Rho loading by occluding a rut site, either directly by overlapping the site, or indirectly via RNA structure rearrangements. The latter regulatory mechanism is exploited by the E. coli CsrA RNA-binding protein in regulating pgaABCD operon expression (Figueroa-Bossi et al., 2014). pgaA mRNA is prematurely terminated in its leader in a Rho-dependent manner, and this regulation relies on CsrA binding to a region in the long pgaA leader; CsrA binding refolds the pgaA leader, exposing an intragenic rut site for Rho binding to prematurely terminate transcription.
In addition to the role of Hfq in chaperoning sRNA stability and function, sRNA-independent roles of Hfq also have been reported (Wagner and Romby, 2015; Chen and Gottesman, 2017). Rabhi et al. (2011) elegantly demonstrated that Hfq, as a general RNA-binding protein, can antagonize Rho-dependent transcription termination at a prototypical terminator (λtR1) both in vitro and in vivo. The antitermination activity of Hfq depends on (1) its distal face binding to an A-rich sequence motif upstream of rut sites in λtR1, and (2) physical interaction of Hfq with Rho protein; this interaction can block Rho binding to other proteins, such as NusG, which has been shown to stimulate Rho termination at some terminators (Mitra et al., 2017). In this case, Hfq directly modulates Rho's RNA-DNA unwinding activity, rather than simply blocking access of Rho to RNA. Such a regulation is distinct from the roles that sRNAs and CsrA play in regulating Rho access to RNAs. Nonetheless, these reports of both negative and positive regulation of Rho-dependent termination by Hfq-dependent sRNAs or RNA-binding proteins reinforce the idea that RNA-based regulation of termination is widely exploited by bacteria.
Autoregulation of Rho
Rho is an abundant protein in many bacteria, including E. coli (Grylak-Mielnicka et al., 2016). It is autoregulated through a transcription attenuation mechanism at its own leader in E. coli and in other bacteria (Barik et al., 1985; Matsumoto et al., 1986; Ingham et al., 1999), emphasizing the importance of cells maintaining an appropriate level of Rho and presumably of Rho-dependent termination. In Rhodobacter capsulatus, Rho is significantly induced under anaerobic growth (Jäger et al., 2004), suggesting a role of Rho in response to changing environments. The recent finding of an sRNA, SraL, that regulates Rho expression, described above (Silva et al., 2019), further highlights the likelihood that cells adjust termination under some conditions by adjusting Rho availability.
Summary and Issues for the Future
sRNA research in the last 20 years have led to a growing appreciation of the ways in which small regulatory RNAs participate in stress circuits in bacteria. In the simplest terms, a sRNA will be induced in response to a stress, and it will pair and regulate the expression of mRNAs encoding proteins that help the cell deal with stress, and will thus contribute in significant ways to the recovery from the stress (Holmqvist and Wagner, 2017; Fröhlich and Gottesman, 2018). Here we have reviewed recent studies that highlight the complexity of this process and its intersection with transcription termination. In particular:
While transcription initiation remains the most important contributor to sRNA synthesis and accumulation, proper termination of the sRNA transcript is equally important for its function. The requirements for efficient termination and binding of the nascent sRNA to Hfq have likely selected for particular characteristics of intrinsic terminators for sRNA-encoding genes.
Recent work shows that the efficiency of proper intrinsic termination at sRNA terminators can be changed by cellular stress, by mechanisms still to be explored. It remains to be determined if these changes in termination affect intrinsic termination in general or are specific to sRNA terminators. For sRNAs, however, stress that changes the efficiency of termination will affect the fraction of transcripts that become functional sRNAs. This provides new insight into the mechanism of Rho-independent termination, which has been considered a simple, unregulated process.
Rho-dependent termination requires access of Rho to single-stranded RNA, and thus sRNAs that block translation may open up Rho entry sites within genes, reinforcing negative regulation of the mRNAs. sRNA binding to otherwise untranslated RNAs (for instance, in 5′ UTRs) can positively regulate downstream ORFs by blocking Rho entry.
The extent and importance of these levels of regulation are just beginning to be realized. Much still remains to be addressed to fully understand the effect of sRNAs on transcription termination. Unlike translational repression and mRNA degradation, effects on termination must occur rapidly, during the transcription process. This suggests that sRNA pairing with mRNAs must be happening co-transcriptionally; how often does that happen? Are there properties of the particular mRNAs, sRNAs, or transcriptional machinery that will favor this? In addition to the trans-acting sRNAs discussed here, the cis-regulatory riboswitches can also control gene expression through modulation of Rho-dependent transcription termination (Hollands et al., 2012; Proshkin et al., 2014; Bastet et al., 2018). It seems likely that, in addition to uncovering new levels of stress regulation, these studies may lead to a deeper understanding of transcription termination itself.
Author Contributions
All authors contributed equally and directly to the preparation of this review, and all approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank H. Aiba, M. Miyakoshi, T. Updegrove, K. Kavita, K. Ramamurthi, G. Storz, and S. Melamed for their comments on the manuscript. Preparation of this review was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and by the Japan Society for the Promotion of Science KAKENHI (grant number 17KK0146) to TM. JC acknowledges support from NCI Director's Intramural Innovation Awards 479434 and 602936.
Footnotes
Funding. Funding for preparation of this review: Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research funding to SG and JC. Japan Society for the Promotion of Science KAKENHI (grant number 17KK0146) to TM.
References
- Adhya S., Gottesman M. (1978). Control of transcription termination. Annu. Rev. Biochem. 47, 967–996. 10.1146/annurev.bi.47.070178.004535 [DOI] [PubMed] [Google Scholar]
- Arimbasseri A. G., Rijal K., Maraia R. J. (2013). Transcription termination by the eukaryotic RNA polymerase III. Biochim. Biophys. Acta 1829, 318–330. 10.1016/j.bbagrm.2012.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian D., Ragunathan P. T., Fei J., Vanderpool C. K. (2016). A prophage-encoded small rna controls metabolism and cell division in Escherichia coli. mSystems 1:e00021-15. 10.1128/mSystems.00021-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbontín R., Fiorini F., Figueroa-Bossi N., Casadesús J., Bossi L. (2010). Recognition of heptameric seed sequence underlies multi-target regulation by RybB small RNA in Salmonella enterica. Mol. Microbiol. 78, 380–394. 10.1111/j.1365-2958.2010.07342.x [DOI] [PubMed] [Google Scholar]
- Barik S., Bhattacharya P., Das A. (1985). Autogenous regulation of transcription termination factor Rho. J. Mol. Biol. 182, 495–508. 10.1016/0022-2836(85)90236-0 [DOI] [PubMed] [Google Scholar]
- Bastet L., Turcotte P., Wade J. T., Lafontaine D. A. (2018). Maestro of regulation: riboswitches orchestrate gene expression at the levels of translation, transcription and mRNA decay. RNA Biol. 15, 679–682. 10.1080/15476286.2018.1451721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battesti A., Majdalani N., Gottesman S. (2011). The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 65, 189–213. 10.1146/annurev-micro-090110-102946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrovskyy M., Vanderpool C. K. (2014). The small RNA SgrS: roles in metabolism and pathogenesis of enteric bacteria. Front. Cell. Infect. Microbiol. 4:61. 10.3389/fcimb.2014.00061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi L., Schwartz A., Guillemardet B., Boudvillain M., Figueroa-Bossi N. (2012). A role for Rho-dependent polarity in gene regulation by a noncoding small RNA. Genes. Dev. 26, 1864–1873. 10.1101/gad.195412.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Y., Vogel J. (2010). The role of Hfq in bacterial pathogens. Curr. Opin. Microbiol. 13, 24–33. 10.1016/j.mib.2010.01.001 [DOI] [PubMed] [Google Scholar]
- Chen J., Gottesman S. (2017). Hfq links translation repression to stress-induced mutagenesis in E. coli. Genes. Dev. 31, 1382–1395. 10.1101/gad.302547.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. J., Liu P., Nielsen A. A., Brophy J. A., Clancy K., Peterson T. (2013). Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat. Methods 10, 659–664. 10.1038/nmeth.2515 [DOI] [PubMed] [Google Scholar]
- d'Aubenton Carafa Y., Brody E., Thermes C. (1990). Prediction of rho-independent Escherichia coli transcription terminators. A statistical analysis of their RNA stem-loop structures. J. Mol. Biol. 216, 835–858. 10.1016/S0022-2836(99)80005-9 [DOI] [PubMed] [Google Scholar]
- Elgamal S., Artsimovitch I., Ibba M. (2016). Maintenance of transcription-translation coupling by elongation factor P. mBio 7:e01373-16. 10.1128/mBio.01373-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epshtein V., Nudler E. (2003). Cooperation between RNA polymerase molecules in transcription elongation. Science 300, 801–805. 10.1126/science.1083219 [DOI] [PubMed] [Google Scholar]
- Farnham P. J., Greenblatt J., Platt T. (1982). Effects of NusA protein on transcription termination in the tryptophan operon of Escherichia coli. Cell 29, 945–951. 10.1016/0092-8674(82)90457-3 [DOI] [PubMed] [Google Scholar]
- Faubladier M., Cam K., Bouché J. P. (1990). Escherichia coli cell division inhibitor DicF-RNA of the dicB operon. Evidence for its generation in vivo by transcription termination and by RNase III and RNase E-dependent processing. J. Mol. Biol. 212, 461–471. 10.1016/0022-2836(90)90325-G [DOI] [PubMed] [Google Scholar]
- Figueroa-Bossi N., Schwartz A., Guillemardet B., D'Heygère F., Bossi L., Boudvillain M. (2014). RNA remodeling by bacterial global regulator CsrA promotes Rho-dependent transcription termination. Genes. Dev. 28, 1239–1251. 10.1101/gad.240192.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa-Bossi N., Valentini M., Malleret L., Fiorini F., Bossi L. (2009). Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes. Dev. 23, 2004–2015. 10.1101/gad.541609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich K. S., Gottesman S. (2018). Small Regulatory RNAs in the enterobacterial response to envelope damage and oxidative stress. Microbiol. Spectr. 6:22. 10.1128/microbiolspec.RWR-0022-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grylak-Mielnicka A., Bidnenko V., Bardowski J., Bidnenko E. (2016). Transcription termination factor Rho: a hub linking diverse physiological processes in bacteria. Microbiology 162, 433–447. 10.1099/mic.0.000244 [DOI] [PubMed] [Google Scholar]
- Guo X., Myasnikov A. G., Chen J., Crucifix C., Papai G., Takacs M. (2018). Structural basis for NusA stabilized transcriptional pausing. Mol. Cell 69, 816–827. 10.1016/j.molcel.2018.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollands K., Proshkin S., Sklyarova S., Epshtein V., Mironov A., Nudler E. (2012). Riboswitch control of Rho-dependent transcription termination. Proc. Natl. Acad. Sci. U.S.A. 109, 5376–5381. 10.1073/pnas.1112211109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmqvist E., Vogel J. (2018). RNA-binding proteins in bacteria. Nat. Rev. Microbiol. 16, 601–615. 10.1038/s41579-018-0049-5 [DOI] [PubMed] [Google Scholar]
- Holmqvist E., Wagner E. G. H. (2017). Impact of bacterial sRNAs in stress responses. Biochem. Soc. Trans. 45, 1203–1212. 10.1042/BST20160363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmqvist E., Wright P. R., Li L., Bischler T., Barquist L., Reinhardt R. (2016). Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO. J. 35, 991–1011. 10.15252/embj.201593360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein R., Lee T. Y., Lim H. N. (2015). Quantitative characterization of gene regulation by Rho dependent transcription termination. Biochim. Biophys. Acta 1849, 940–954. 10.1016/j.bbagrm.2015.05.003 [DOI] [PubMed] [Google Scholar]
- Ingham C. J., Dennis J., Furneaux P. A. (1999). Autogenous regulation of transcription termination factor Rho and the requirement for Nus factors in Bacillus subtilis. Mol. Microbiol. 31, 651–663. 10.1046/j.1365-2958.1999.01205.x [DOI] [PubMed] [Google Scholar]
- Ishikawa H., Otaka H., Maki K., Morita T., Aiba H. (2012). The functional Hfq-binding module of bacterial sRNAs consists of a double or single hairpin preceded by a U-rich sequence and followed by a 3' poly(U) tail. RNA 18, 1062–1074. 10.1261/rna.031575.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquet M. A., Reiss C. (1992). In vivo control of promoter and terminator efficiencies at a distance. Mol. Microbiol. 6, 1681–1691. 10.1111/j.1365-2958.1992.tb00893.x [DOI] [PubMed] [Google Scholar]
- Jäger S., Hebermehl M., Schiltz E., Klug G. (2004). Composition and activity of the Rhodobacter capsulatus degradosome vary under different oxygen concentrations. J. Mol. Microbiol. Biotechnol. 7, 148–154. 10.1159/000078658 [DOI] [PubMed] [Google Scholar]
- Kavita K., de Mets F., Gottesman S. (2018). New aspects of RNA-based regulation by Hfq and its partner sRNAs. Curr. Opin. Microbiol. 42, 53–61. 10.1016/j.mib.2017.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalanne J. B., Taggart J. C., Guo M. S., Herzel L., Schieler A., Li G. W. (2018). Evolutionary convergence of pathway-specific enzyme expression stoichiometry. Cell 173, 749–761. 10.1016/j.cell.2018.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livny J., Waldor M. K. (2007). Identification of small RNAs in diverse bacterial species. Curr. Opin. Microbiol. 10, 96–101. 10.1016/j.mib.2007.03.005 [DOI] [PubMed] [Google Scholar]
- Maier L. K., Marchfelder A. (2019). It's all about the T: transcription termination in archaea. Biochem. Soc. Trans. 47, 461–468. 10.1042/BST20180557 [DOI] [PubMed] [Google Scholar]
- Massé E, Escorcia F. E., Gottesman S. (2003). Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes. Dev. 17, 2374–2383. 10.1101/gad.1127103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Gottesman S. (2002). A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 99, 4620–4625. 10.1073/pnas.032066599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y., Shigesada K., Hirano M., Imai M. (1986). Autogenous regulation of the gene for transcription termination factor Rho in Escherichia coli: localization and function of its attenuators. J. Bacteriol. 166, 945–958. 10.1128/jb.166.3.945-958.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell J. C., Roberts J. W., Jin D. J., Gross C. (1994). Determination of intrinsic transcription termination efficiency by RNA polymerase elongation rate. Science 266, 822–825. 10.1126/science.7526463 [DOI] [PubMed] [Google Scholar]
- Melamed S., Peer A., Faigenbaum-Romm R., Gatt Y. E., Reiss N., Bar A., Altuvia Y., Argaman L., Margalit H. (2016). Global mapping of Small RNA-target interactions in bacteria. Mol. Cell 63, 884–897. 10.1016/j.molcel.2016.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra P., Ghosh G., Hafeezunnisa M., Sen R. (2017). Rho protein: roles and mechanisms. Annu. Rev. Microbiol. 71, 687–709. 10.1146/annurev-micro-030117-020432 [DOI] [PubMed] [Google Scholar]
- Miyakoshi M., Chao Y., Vogel J. (2015a). Cross talk between ABC transporter mRNAs via a target mRNA-derived sponge of the GcvB small RNA. EMBO. J. 34, 1478–1492. 10.15252/embj.201490546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakoshi M., Chao Y., Vogel J. (2015b). Regulatory small RNAs from the 3' regions of bacterial mRNAs. Curr. Opin. Microbiol. 24, 132–139. 10.1016/j.mib.2015.01.013 [DOI] [PubMed] [Google Scholar]
- Morita T., Nishino R., Aiba H. (2017). Role of the terminator hairpin in the biogenesis of functional Hfq-binding sRNAs. RNA 23, 1419–1431. 10.1261/rna.060756.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T., Ueda M., Kubo K., Aiba H. (2015). Insights into transcription termination of Hfq-binding sRNAs of Escherichia coli and characterization of readthrough products. RNA 21, 1490–1501. 10.1261/rna.051870.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadiras C., Schwartz A., Delaleau M., Boudvillain M. (2018). Evaluating the effect of small RNAs and associated chaperones on Rho-dependent termination of transcription in vitro. Methods Mol. Biol. 1737, 99–118. 10.1007/978-1-4939-7634-8_7 [DOI] [PubMed] [Google Scholar]
- Nudler E., Gottesman M. E. (2002). Transcription termination and anti-termination in E. coli. Genes Cells :Devoted Mol. Cell. Mechan. 7, 755–768. 10.1046/j.1365-2443.2002.00563.x [DOI] [PubMed] [Google Scholar]
- Otaka H., Ishikawa H., Morita T., Aiba H. (2011). PolyU tail of rho-independent terminator of bacterial small RNAs is essential for Hfq action. Proc. Natl. Acad. Sci. U.S.A. 108, 13059–13064. 10.1073/pnas.1107050108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overgaard M., Johansen J., Møller-Jensen J., Valentin-Hansen P. (2009). Switching off small RNA regulation with trap-mRNA. Mol. Microbiol. 73, 790–800. 10.1111/j.1365-2958.2009.06807.x [DOI] [PubMed] [Google Scholar]
- Panja S., Schu D. J., Woodson S. A. (2013). Conserved arginines on the rim of Hfq catalyze base pair formation and exchange. Nucleic Acids Res. 41, 7536–7546. 10.1093/nar/gkt521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K., Sun Y., Miyakoshi M., Vanderpool C. K., Vogel J. (2013). Small RNA-mediated activation of sugar phosphatase mRNA regulates glucose homeostasis. Cell 153, 426–437. 10.1016/j.cell.2013.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J. M., Mooney R. A., Grass J. A., Jessen E. D., Tran F., Landick R. (2012). Rho and NusG suppress pervasive antisense transcription in Escherichia coli. Genes Dev. 26, 2621–2633. 10.1101/gad.196741.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J. M., Mooney R. A., Kuan P. F., Rowland J. L., Keles S., Landick R. (2009). Rho directs widespread termination of intragenic and stable RNA transcription. Proc. Natl. Acad. Sci. U.S.A. 106, 15406–15411. 10.1073/pnas.0903846106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J. M., Vangeloff A. D., Landick R. (2011). Bacterial transcription terminators: the RNA 3'-end chronicles. J. Mol. Biol. 412, 793–813. 10.1016/j.jmb.2011.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proshkin S., Mironov A., Nudler E. (2014). Riboswitches in regulation of Rho-dependent transcription termination. Biochim. Biophys. Acta 1839, 974–977. 10.1016/j.bbagrm.2014.04.002 [DOI] [PubMed] [Google Scholar]
- Rabhi M., Espéli O., Schwartz A., Cayrol B., Rahmouni A. R., Arluison V. (2011). The Sm-like RNA chaperone Hfq mediates transcription antitermination at Rho-dependent terminators. EMBO J. 30, 2805–2816. 10.1038/emboj.2011.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray-Soni A., Bellecourt M. J., Landick R. (2016). Mechanisms of bacterial transcription termination: all good things must end. Annu. Rev. Biochem. 85, 319–347. 10.1146/annurev-biochem-060815-014844 [DOI] [PubMed] [Google Scholar]
- Roberts J. W. (in press). Mechanisms of bacterial transcription termination. J. Mol. Biol. 10.1016/j.jmb.2019.04.003. [DOI] [PubMed] [Google Scholar]
- Robinson K. E., Orans J., Kovach A. R., Link T. M., Brennan R. G. (2014). Mapping Hfq-RNA interaction surfaces using tryptophan fluorescence quenching. Nucleic Acids Res 42, 2736–2749. 10.1093/nar/gkt1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer E., Weichenrieder O. (2011). Structural basis for RNA 3'-end recognition by Hfq. Proc. Natl. Acad. Sci. U.S.A. 108, 13065–13070. 10.1073/pnas.1103420108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schu D. J., Zhang A., Gottesman S., Storz G. (2015). Alternative Hfq-sRNA interaction modes dictate alternative mRNA recognition. EMBO J. 34, 2557–2573. 10.15252/embj.201591569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlyarova N., Shamovsky I., Bharati B. K., Epshtein V., Chen J., Gottesman S., Schroeder R., Nudler E. (2016). sRNA-mediated control of transcription termination in E. coli. Cell 167, 111–121. 10.1016/j.cell.2016.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva I. J., Barahona S., Eyraud A., Lalaouna D., Figueroa-Bossi N., Mass,é E. (2019). SraL sRNA interaction regulates the terminator by preventing premature transcription termination of rho mRNA. Proc. Natl. Acad. Sci. U.S.A. 116, 3042–3051 10.1073/pnas.1811589116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skordalakes E., Berger J. M. (2006). Structural insights into RNA-dependent ring closure and ATPase activation by the Rho termination factor. Cell 127, 553–564. 10.1016/j.cell.2006.08.051 [DOI] [PubMed] [Google Scholar]
- Sledjeski D., Gottesman S. (1995). A small RNA acts as an antisilencer of the H-NS-silenced rcsA gene of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 92, 2003–2007. 10.1073/pnas.92.6.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledjeski D. D., Gupta A., Gottesman S. (1996). The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 15, 3993–4000. 10.1002/j.1460-2075.1996.tb00773.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G., Vogel J., Wassarman K. M. (2011). Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell 43, 880–891. 10.1016/j.molcel.2011.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Vanderpool C. K. (2011). Regulation and function of Escherichia coli sugar efflux transporter A (SETA) during glucose-phosphate stress. J. Bacteriol. 193, 143–153. 10.1128/JB.01008-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updegrove T. B., Zhang A., Storz G. (2016). Hfq: the flexible RNA matchmaker. Curr. Opin. Microbiol. 30, 133–138. 10.1016/j.mib.2016.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J., Bartels V., Tang T. H., Churakov G., Slagter-Jäger J. G., Hüttenhofer A. (2003). RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 31, 6435–6443. 10.1093/nar/gkg867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J., Luisi B. F. (2011). Hfq and its constellation of RNA. Nat. Rev. Microbiol. 15, 578–589. 10.1038/nrmicro2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E. G. H., Romby P. (2015). Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv. Genet. 90, 133–208. 10.1016/bs.adgen.2015.05.001 [DOI] [PubMed] [Google Scholar]
- Wang X., Ji S. C., Jeon H. J., Lee Y., Lim H. M. (2015). Two-level inhibition of galK expression by Spot 42: degradation of mRNA mK2 and enhanced transcription termination before the galK gene. Proc. Natl. Acad. Sci. U.S.A. 112, 7581–7586. 10.1073/pnas.1424683112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B., Boitano M., Clark T. A., Ettwiller L. (2018). SMRT-Cappable-seq reveals complex operon variants in bacteria. Nat. Commun. 9:3676. 10.1038/s41467-018-05997-6 [DOI] [PMC free article] [PubMed] [Google Scholar]