Abstract
Structural characterization of sulfated glycosaminoglycans (GAGs) by mass spectrometry has long been a formidable analytical challenge owing to their high structural variability and the propensity for sulfate decomposition upon activation with low-energy ion activation methods. While derivatization and complexation workflows have aimed to generate informative spectra using low-energy ion activation methods, alternative ion activation methods present the opportunity to obtain informative spectra from native GAG structures. Both electron- and photon-based activation methods, including electron detachment dissociation (EDD), negative electron transfer dissociation (NETD), and extreme ultraviolet photon activation, have been explored previously to overcome the limitations associated with low-energy activation methods for GAGs and other sulfated oligosaccharides. Further, implementation of such methods on high-resolution mass spectrometers has aided the interpretation of the complex spectra generated. Here, we explore ultraviolet photodissociation (UVPD) implemented on an Orbitrap mass spectrometer as another option for structural characterization of GAGs. UVPD spectra for both dermatan and heparan sulfate structures display extensive fragmentation including both glycosidic and cross-ring cleavages with the extent of sulfate retention comparable to that observed by EDD and NETD. In addition, the relatively short activation time of UVPD makes it promising for higher throughput analysis of GAGs in complex mixtures.
Graphical Abstract
Glycosaminoglycans (GAGs), linear polysaccharides composed of repeating hexosamine and uronic acid disaccharides, are complex molecules that participate in a number of biological processes ranging from tissue development to inflammation.1–3 GAG structural complexity arises from variations in their degree of polymerization, extent of saccharide sulfation, and uronic acid stereochemistry. Such modifications are responsible for the specific interactions GAGs have with proteins, with misregulation of modifications implicated in a number of genetic diseases, including Alzheimer’s and cancer.4–7 Development and quality control of GAG therapeutics also relies on a detailed knowledge of GAG structures.1,8 In 2008, heparin, an anticoagulant drug, contaminated with oversulfated chondroitin sulfate resulted in 81 deaths.8 Detailed structural characterization of GAGs is therefore essential yet still impeded by a number of technical challenges. Advances in modern mass spectrometry, including the development of high-resolution/high-mass accuracy mass analyzers and hybrid instrumentation, have made it an indispensable tool for the structural characterization of GAGs.9–13 Owing to their acidic nature, GAGs are most frequently analyzed in the negative ion mode, and deprotonation of the sulfate half-ester leads to stabilization of this labile modification. If these moieties remain protonated, sequencing attempts with low-energy tandem mass spectrometry methods, including collision-induced dissociation (CID) and infrared multiphoton dissociation (IRMPD), or by using improper tuning conditions in high-pressure regions of the mass spectrometer can result in preferential sulfate decomposition, especially for highly modified heparin and heparan sulfate. The prevalence of sulfate decomposition impedes complete saccharide sequencing and sulfate localization.14,15 Previous studies have shown that informative CID spectra can be obtained after deprotonation of all sites of sulfation.16,17 Metal adduction using sodium or calcium ions effectively allows deprotonation of acidic sites that would otherwise lead to SO3 loss and enables informative spectra to be obtained from low-energy collisional activated dissociation.14,18–20 Derivatization strategies, while requiring more sample preparation, have similarly been beneficial for obtaining information spectra upon low-energy collisional activated dissociation.21,22 High-energy collisional activation has also been found to produce informative MS/MS spectra with some fragments resulting from electron detachment.23
As a result of the limitations of low-energy activation methods, a number of alternative ion activation methods have successfully been applied to analyze GAGs. Electron-based activation methods include electron detachment dissociation (EDD),24–33 negative electron transfer dissociation (nETD),33–37 and electron-induced dissociation (EID).38 EDD produces an extensive array of fragment ions, including sulfate-retaining glycosidic and cross-ring cleavage products for both free acid and sodium-proton exchanged species with observed fragments resulting from both direct dissociation and electron detachment.24–27 Both EDD and NETD has also shown promise for distinguishing hexuronic acid epimers based on both the presence of unique fragments and fragment ion abundances.28–31 While long activation times and low abundance product ions have prevented coupling of EDD with online separations, successful interfacing of field asymmetric-waveform ion mobility spectrometry (FAIMS) with an EDD-equipped Fourier transform ion cyclotron resonance (FTICR) instrument has enabled gas-phase separation and detection of GAG epimers.32 NETD has also shown great promise as an alternative activation method for negative ions, exhibiting similar fragmentation to EDD and the potential to be performed on any mass spectrometer capable of gas phase ion-ion reactions.33,35,36 The recent application of NETD to GAG analysis on an Orbitrap platform confirmed that NETD can produce highly informative spectra with activation times amenable to a chromatographic or electro-phoretic time scale.36
Ultraviolet photon-based activation methods have also been considered as alternatives to low-energy methods for the analysis of GAGs anions.39–46 The use of UV photons for oligosaccharide fragmentation was first reported by Reilly.47 In their work, both underivatized and derivatized oligosaccharides were fragmented on a home-build MALDI TOF/TOF equipped with a 157 nm excimer laser.47–49 Since then, others have investigated a range of wavelengths to activate oligosaccharides on a number of platforms. Racaud et al. performed ultraviolet photodissociation (UVPD) in the 220–290 nm range in an ion trap mass spectrometer on a set of heparin-derived disaccharides and observed informative cross-ring cleavage fragments.40 UVPD favored a large number of cross ring cleavages in addition to unique electron-photo-detachment ions with corresponding charge-reduced neutral loss products. Racaud et al. also investigated the influence of charge on the preference for fragmentation versus electron-photodetachment using UVPD.41 Similar to low-energy and electron-based activation methods, deprotonation of sulfate modification greatly influenced the fragmentation patterns with full deprotonation achieved through sodium adduction.40,41 Extreme UVPD (X-UVPD) at a photon energy of 18 eV was performed with synchrotron radiation on an ion trap mass spectrometer and provided detailed structure characterization of highly sulfated oligosaccharides.43–45 While synchrotrons provide a high-energy and tunable source of radiation, their limited accessibility make them less feasible for routine usage and broad applications. Moreover, while UVPD produces rich fragmentation patterns, it is apparent with increasingly large and complex biomolecules that interpretation of UVPD spectra becomes challenging, if not impossible, when using low-resolution ion trap mass spectrometers.
Excimer lasers have been successfully interfaced to a number of mass spectrometers and likewise provide access to high-energy photons.50 Integration of 193 nm excimer lasers with high-accuracy Orbitrap mass analyzers has greatly expanded the utility of UVPD as an activation method for larger biomolecules, enabling successful interpretation of complex fragmentation patterns.50–52 The successful structural characterization of acidic saccharides53 and localization of sulfate modifications on peptides54 using 193 nm UVPD suggest it as a viable tool for GAG analysis. Additionally, the recent commercialization of UVPD with a 213 nm Nd:YAG laser has further increased its accessibility.55,56 The present study evaluates the use of UVPD for the structural characterization of GAGs. UVPD was found to perform comparably to collision-based activation methods for heparan and dermatan sulfate GAGs with low degrees of sulfation. More significantly, UVPD produced an extensive array of cross-ring cleavage fragments for fondaparinux, a highly sulfated heparin-like anticoagulant drug, enabling more detailed characterization than collision-based methods and comparable fragmentation to electron-based methods such as NETD.
EXPERIMENTAL SECTION
Materials and Reagents.
HPLC grade methanol (MeOH) and water (H2O) were acquired from EMD Millipore (Billerica, MA). Fondaparinux sodium was purchased from Sigma (St. Louis, MO). For data collection of fondaparinux with salt adduction, fondaparinux sodium was desalted on a PD MiniTrap G-10 column (GE Healthcare, Buckinghamshire, U.K.). Heparan sulfate tetrasaccharide standards were prepared by chemical synthesis using a modular approach.57 Dermatan sulfate oligosaccharides were prepared by partial enzymatic depolymerization of porcine intestinal mucosa dermatan sulfate (Celsus Laboratories, Cincinnati, OH). A detailed protocol can be found in prior reports.30 The structures of all the GAGs analyzed in the present study are shown in Table S1.
Mass Spectrometry.
Higher-energy collisional dissociation (HCD) and UVPD spectra for all samples were collected on a Thermo Fisher Orbitrap Fusion Lumos mass spectrometer (San Jose, CA) equipped with a Coherent Excistar 193 nm excimer laser (Santa Clara, CA).52 UVPD at 213 nm was performed on the same mass spectrometer used for 193 nm UVPD with a 213 nm CryLaS solid state laser (Berlin, Germany) in place of the 193 nm excimer laser. Samples were diluted in 50:50 MeOH/H2O to 50 μg/mL and sprayed from a nanoESI static source with a spray voltage of 0.8−1.2 kV. The spray voltage and distance between the nanoESI source and the mass spectrometer inlet were adjusted to manipulate the observed charge state distribution. Spectra were collected in the negative ion mode at a resolving power of 120 000 at m/z 200 with the instrument in full-profile mode. The ion funnel rf was set to 10% to minimize sulfate decomposition in the MS1 spectra. Precursor ions were isolated in the ion trap with an isolation width of 3 m/z. HCD was performed with a normalized collision energy (NCE) of 15−25, 193 nm UVPD was performed with 8 pulses at 4 mJ per pulse resulting in a 16 ms activation period, and 213 nm UVPD was performed with 500 pulses at 3 μJ per pulse resulting in a 200 ms activation period. All presented HCD and UVPD spectra are an average of 50 transients. Each spectrum took approximately 30–60 s to collect. EDD and NETD spectra of IdoA-GlcNAc6S-GlcA-GlcNAc6S-(CH2)5NH2 were collected on a 9.4 T Bruker Apex Ultra Qe Fourier transform mass spectrometer (FTMS, Billerica, MA) and a Thermo Scientific Orbitrap Elite mass spectrometer (Bremen, Germany), respectively. EDD was performed on quadrupole mass selected precursor ions by irradiation with 19 eV electrons for a pulse duration of 1 s. A total of 24 spectra were averaged and internally calibrated. NETD was enabled by software modifications and performed on mass-selected precursors by ion-ion reaction with radical fluoranthene cations for 150 ms. A total of 36 spectra were summed with a specified resolution of 120 000. Fragment ions are labeled using Domon and Costello nomenclature (Figure S1).58 Spectra were interpreted in a semiautomated fashion through the use of GlycoWork-bench.59 Fragment ions maps are depicted using dashed lines drawn through chemical structures with hash marks at the ends of dashed lines to indicate the presence of a fragment ion. An open circle at the end of a hash mark indicates a fragment ion with loss of a single SO3. A filled circle at the end of a hash mark indicates a fragment ion with loss of more than one SO3. Donut plots represent percentages of the summed ion abundances for each ion type (glycosidic, cross-ring, glycosidic-SO3, cross-ring-SO3) relative to summed abundances of all ion types. Precursor neutral losses and ions resulting from electron photodetachment are reported as percentages of the total ion current.
RESULTS AND DISCUSSION
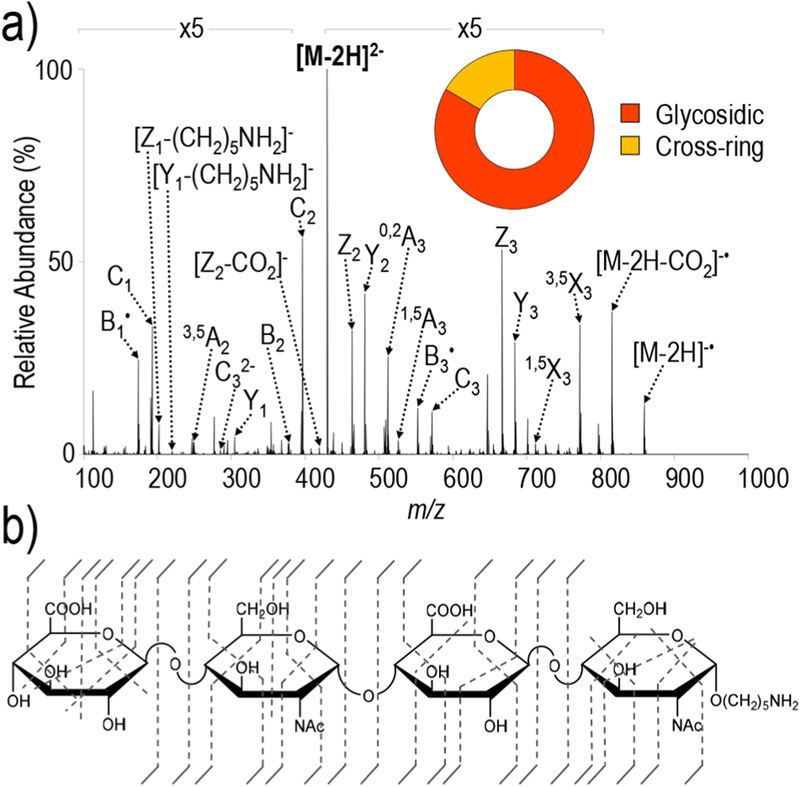
Both synthetic and enzymatically depolymerized GAG standards were analyzed to evaluate the capabilities of UVPD for structural characterization, along with comparisons to HCD, EDD, and NETD spectra. Unless specified, all UVPD spectra were collected using 193 nm. Figure 1 and Figure S2 show the UVPD and HCD spectra, respectively, of the [M − 2H]2− precursor ion of the unsulfated tetrasaccharide GlcA-GlcNAc-GlcA-GlcNAc-(CH2)5NH2. The UVPD spectrum displays cleavage of every glycosidic bond in addition to many cross-ring cleavages distributed across the four monosaccharide residues. The UVPD spectrum also contains a characteristic charge-reduced, electron-photodetachment product at m/z 859.31 which accounts for 2.6% of the total ion current, in addition to the loss of CO2 from that species (m/z 815.32) which accounts for 7.6% of the total ion current. HCD of the same tetrasaccharide primarily yielded glycosidic cleavages in addition to a few cross-ring cleavages, as illustrated in Figure S2. The distribution of glycosidic and cross-ring fragments between HCD (glycosidic = 86.7%; cross-ring = 13.2%) and UVPD (glycosidic = 83.5%; cross-ring = 16.5%), as depicted in the donut plots of Figure 1 and Figure S2, respectively, is quite similar owing to the lack of labile sulfate modifications and ionization of both carboxylates. Comparison of fragment ions that are observed in both the UVPD and HCD spectra reveals that although B/Y and C/Z are prominent in both spectra, additional odd-electron species are generated exclusively upon UVPD. For example, in the HCD spectrum the even-electron B3 ion of m/z 554.14 is observed, while both even- and odd-electron B3 and B3● ions appear (m/z 554.14 and m/z 553.13) in the UVPD spectrum (Figure S3 and Tables S2 and S3). These results are analogous to prior findings reported for the electron-based activation method EDD and suggest radical-based fragmentation as one of the contributing pathways of UVPD.24,25 Moreover, the broad array of cross-ring cleavages observed upon UVPD, including fragmentation across residues without ionizable sites, suggests that radical migration may play an active role in the fragmentation processes akin to prior observations for EDD and NETD of GAGs.25
Figure 1.
(a) UVPD spectrum of [M − 2H]2− (m/z 429) of GlcA-GlcNAc-GlcA-GlcNAc-(CH2)5NH2 with donut plot depicting the fragment ion composition (based on summed abundances of products from glycosidic versus cross-ring cleavages) and (b) UVPD fragment ion map. Table S2 contains a list of identified fragment ions.
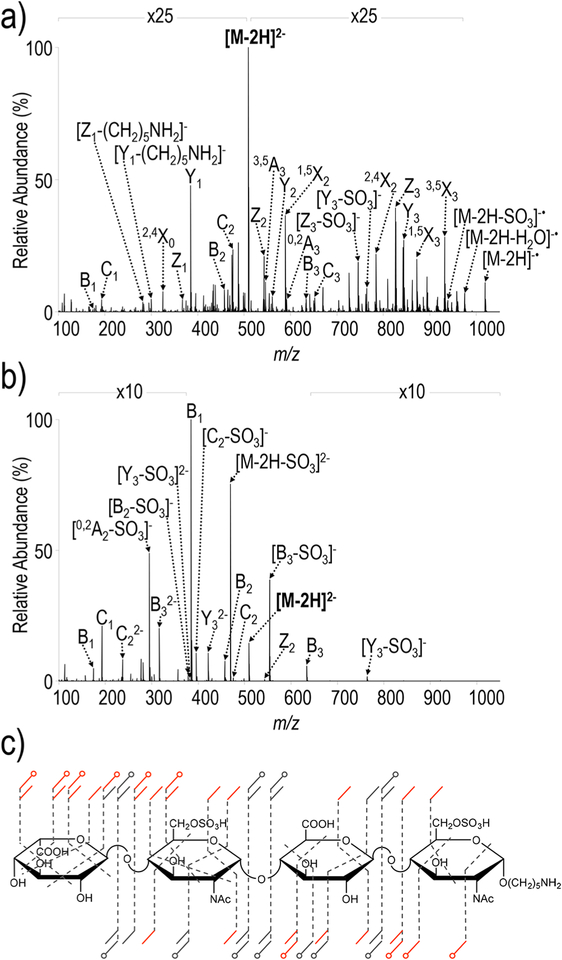
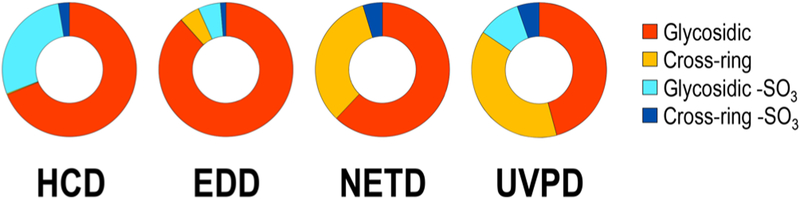
Figure 2 shows the UVPD and HCD spectra of the [M − 2H]2− precursor ion of the tetrasaccharide IdoA-GlcNAc6S-GlcA-GlcNAc6S-(CH2)5NH2, a synthetic heparan sulfate with a low degree of sulfation. While the HCD spectrum does contain informative glycosidic fragments (primarily B, C, Y, and Z ions), sulfate decomposition is a predominant fragmentation pathway. Sulfate decomposition is possible when sites of sulfation are protonated, a process that can result from either protonation during ionization or possible proton migration in the gas phase.24,60 With increasingly sulfated heparan sulfate species that have more than one sulfate per disaccharide, complete deprotonation is unlikely owing to charge repulsion, and incomplete deprotonation contributes to excessive sulfate decomposition upon collisional activation. In contrast, the UVPD spectrum exhibits cleavage of every glycosidic bond in addition to cross-ring cleavages with considerably less SO3 loss. Cross-ring cleavages allow determination of the saccharide linkage pattern and enable localization of 6-O sulfation to the glucosamine residues. Similar to GlcA-GlcNAc-GlcA-GlcNAc-(CH2)5NH2, UVPD of IdoA-GlcNAc6S-GlcA-GlcNAc6S-(CH2)5NH2 (2−) generates an electron-photodetachment fragment of m/z 1019.22, in addition to a series of secondary electron-photodetachment products (m/z 975.23 and m/z 939.26) originating from neutral losses of CO2 and SO3, respectively. A comparative summary of fragmentation pathways for this compound in HCD, EDD, NETD, and UVPD is visualized in the donut plots in Figure 3. The predominant ion of m/z 469.63 corresponding to sulfate decomposition from the precursor (Figure 2b) accounts for 18.1% of the total ion current in the HCD spectrum. Based on the summed abundances of glycosidic and cross-ring fragments in the HCD spectrum, 28.2% and 2.7% of the glycosidic and cross-ring fragments, respectively, undergo sulfate decomposition. For EDD, NETD, and UVPD, 5.4%, 0.0%, and 10.3% of glycosidic, and 1.3%, 4.7%, and 5.2% of cross-ring fragments, respectively, undergo sulfate decomposition relative to the total summed abundances of glycosidic and cross-ring fragments. EDD, NETD, and UVPD all exhibit less sulfate decomposition than HCD and generally produce spectra that are more structurally informative. While the distribution of glycosidic and cross-ring cleavages is quite comparable between NETD and UVPD, EDD appears to favor production of glycosidic cleavages. Based on the summed abundances of all cross-ring fragment ions, 33.2% and 38.6% are sulfate-retaining cross-ring fragments for NETD and UVPD, respectively, while only 4.9% are sulfate-retaining cross-ring fragments for EDD.
Figure 2.
(a) UVPD and (b) HCD spectra of [M − 2H]2− (m/z 509) of IdoA-GlcNAc6S-GlcA-GlcNAc6S-(CH2)5NH2. (c) Fragment ion maps contain fragment ions for both UVPD and HCD (NCE 25). Fragment ions only produced exclusively by UVPD are in red. Tables S4 and S5 contain lists of identified fragment ions.
Figure 3.
Donut plots depicting fragment ion compositions for HCD, EDD, NETD, and UVPD for [M – 2H]2− of IdoA-GlcNAc6S-GlcA-GlcNAc6S-(CH2)5NH2 based on summed abundances of fragment types. Tables S6 and S7 contain lists of identified fragment ions for EDD and NETD, respectively.
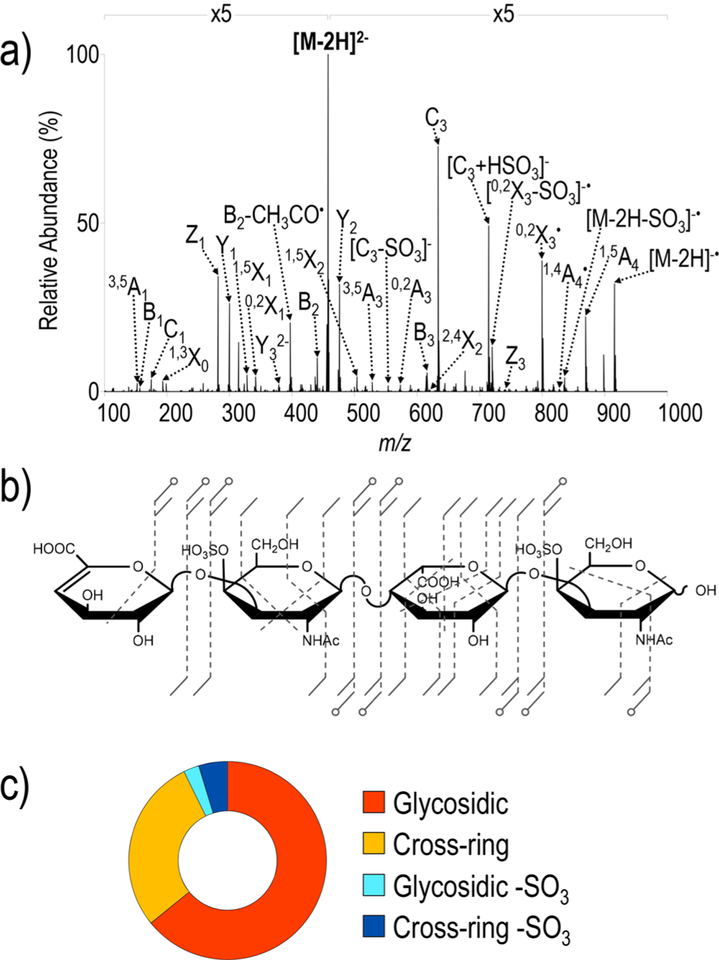
Dermatan sulfate (DS) is a sulfated GAG composed of repeating β−1,4 linked disaccharide units each comprised of β1,3-linked iduronic acid and 4-O-sulfated N-acetyl galactosamine (GalNAc) residues. Figure 4 shows the UVPD spectrum for a doubly sulfated DS tetrasaccharide (DS dp4). Although similar in length and sulfation extent, it contains a different linkage pattern and therefore different sulfate positions and differing uronic acid stereochemistry than IdoA-GlcNAcS6-GlcA-GlcNAcS6-(CH2)5NH2. These subtle structural differences have resulted in challenges to determine sequence and epimer content of chondroitin sulfate (CS)/DS GAGs de novo.25,30 The UVPD spectrum of [M − 2H]2− contains a complete series of glycosidic cleavages in addition to cross-ring cleavages that again enable localization of sulfate modifications. For example, pairs of fragment ions, Z3 and 2,4X2, and C3 and 1,4A4, confirm 4-O sulfation of each GalNAc. These cross-ring cleavages do not arise during EDD or NETD of the same precursor ion. The symmetry of this GAG precludes differentiation of C2/Z2 ions as these fragment ions are isobaric with the precursor ion. UVPD again causes a relatively low degree of sulfate decomposition, thus minimizing the congestion of the spectrum with uninformative ions. In general, a larger number of cross-ring fragments were identified for UVPD of DS dp4 compared to EDD and NETD, which is perhaps related in part to the increased signal-to-noise of the UVPD spectrum.25,34 We estimate that approximately 20—30% of ions in the UVPD spectra are currently unassigned, whereas this value is 10−20% in EDD/NETD spectra. Interestingly, there is a prominent fragment ion of m/z 715.05 (1−). Further interrogation of this UVPD fragment ion by subsequent fragmentation with either HCD or UVPD in an MS3 event results in a predominant neutral loss of 80.96 Da (HSO3●) to form an abundant C3 ion of m/z 634.09 (Figure S4). We speculate that this product arises during MS2 from a gas-phase rearrangement that entails loss of GalNAc by glycosidic bond cleavage in conjunction with the transfer of the sulfate modification to the reducing end of the incipient product. This fragment ion occurs in the 1− charge state and contains all sites of ionization; therefore, this fragment is likely an odd-electron ion resulting from electron-photodetachment. The MS3 spectrum also shows an ion of m/z 272.99, corresponding to a sulfate-containing uronic acid, further suggesting retention of the reducing end 4-O GalNAc sulfate group on uronic acid. This unusual ion of m/z 715.05 has also been observed in EDD and NETD spectra of [M − 2H]2− of DS dp4 but has not been previously assigned.25,30 UVPD of [M − 3H]3− does not generate the ion at m/z 715.05, indicating that the number and sites of deprotonation influence gas-phase rearrangement processes (Figure S5a,b). As expected, [M − 3H]3− yields lower percentages of sulfate decomposition in relation to the summed abundances of the glycosidic and cross-ring fragments; [M − 3H]3− has 0.7% and 0.02% sulfate decomposition for glycosidic and cross-ring fragments, respectively, whereas [M − 2H]2− displays 2.6% and 4.7% sulfate decomposition for glycosidic and cross-ring fragment, respectively.
Figure 4.
(a) UVPD spectrum of [M − 2H]2− (m/z 458) of DS dp4, (b) UVPD fragment ion map, and (c) UVPD donut plot depicting the fragment ion composition based on summed abundances of fragment types. Table S8 contains a list of identified fragment ions.
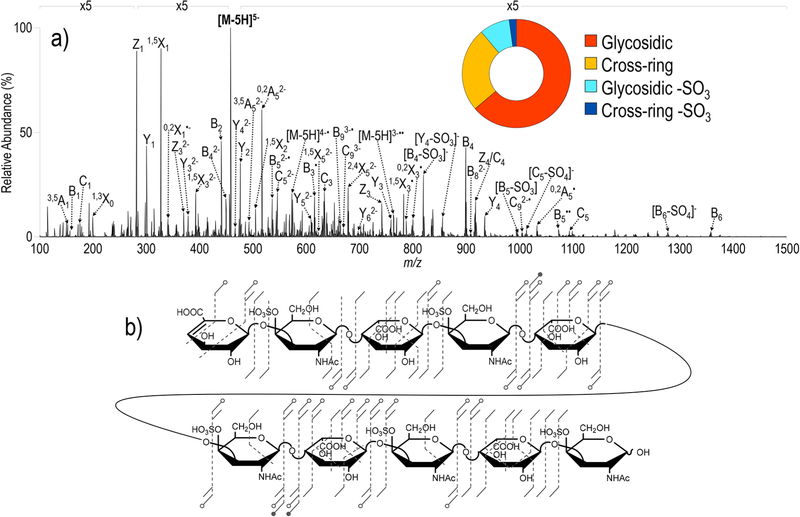
While many structural characterization studies have focused on relatively small GAGs (dp2−dp4), analysis of larger oligosaccharides offers the potential for correlation of modification motifs with biological activity, analogous to analysis of combinatorial post-translation modification of proteins. Typical GAG-protein binding epitopes range from dp4 to dp10; the previous analytes represent the lower boundary for this regime. DS dp10 represents a GAG standard with a higher degree of polymerization. The UVPD spectrum of the 5− charge state of DS dp10 is shown in Figure 5. The rich spectrum contains a full set of glyosidic cleavages and a number of cross-ring cleavages with a relatively low degree of sulfate decomposition. In addition, ions originating from sequential photodetachment events, analogous to multiple electron transfer events that are possible with NETD, are observed.61 For example, ions of m/z 763.43 and m/z 1145.15 correspond to [M − 5H]3−●● and [M − 5H]2−●●●, representing doubly and triply charge-reduced species. The observed UVPD fragmentation pattern for DS dp10 is quite similar to that of EDD, with UVPD affording more identified cross-ring fragments.25 With increasingly large GAG structures like DS dp10, high-resolution and high-accuracy mass analysis for assignment of fragment ions is necessary. The fragmentation caused by UVPD for both DS dp4 (2−) and DS dp10 (5−) is consistent with the fragmentation observed from EDD25,30 and NETD,30,34 including abundant uronic acid cross-ring cleavages, albeit production of more abundant fragment ions by UVPD than EDD with a significantly shorter ion activation period.
Figure 5.
(a) UVPD spectrum of [M − 5H]5− (m/z 458) of DS dp10 with inset donut plot depicting the fragment ion composition based on summed abundances of fragment types and (b) UVPD fragment ion map. Table S12 contains a list of identified fragment ions.
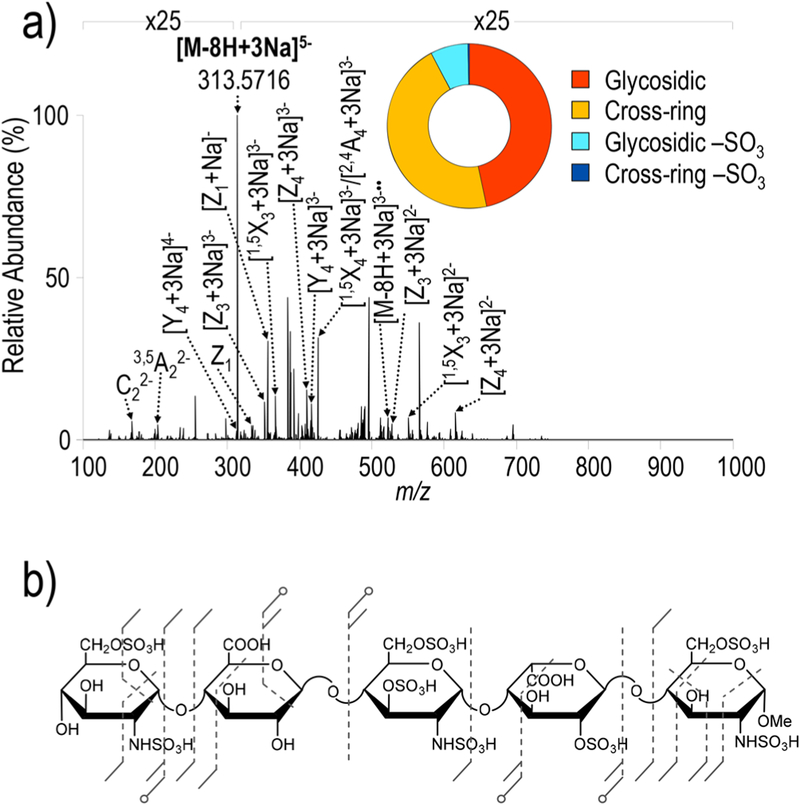
The prior examples have demonstrated the utility of UVPD in analytes with varied sulfation density (0−1 per dp2) and oligomer length yet were all homopolymers. Fondaparinux (trade name Arixtra) is a synthetic, highly sulfated pentasaccharide with anticoagulant activity.62 The sulfation pattern of fondaparinux is based on a naturally occurring pentasaccharide motif in heparin that binds antithrombin III and is therefore representative of the complex GAG structures that occur naturally in heteropolymers. Fondaparinux has thus served as a benchmark for appraisal of new GAG ion activation methods. As stated previously, with increasingly sulfated GAGs, charge repulsion limits the number of ionized sites during ESI and promotes sulfate decomposition via residual protons. Assessing the impact of precursor charge state in conjunction with the use of sodium adduction to achieve a higher degree of deprotonation is a key milestone in the evaluation of performance metrics of MS/MS methods. While deprotonation of highly sulfated GAGs via sodium adduction increases the effectiveness of collisional activation, benefits are only observed when all sites of ionization are deprotonated.14 The limited utility of collisional activation for highly sulfated GAGs has led to exploration of other ion activation methods, including NETD. For comparison to previously reported collisional activation and NETD data, UVPD was used to characterize two ionization states of sodiated fondaparinux ([M − 8H + 3Na]5−and [M − 10H + 5Na]5−).14,36 Figure 6 shows the UVPD mass spectrum acquired for the [M − 8H + 3Na]5−. This charge state represents one in which the number of ionized sites matches the number of sulfate groups in an attempt to minimize sulfate decomposition. The fragment ion map in Figure 6b reflects cleavage between all saccharides while the donut plot in Figure 6a confirms a relatively low degree of SO3 loss (glycosidic-SO3 = 7.3%; cross-ring-SO3 = 0.3%). Figure S6 shows the UVPD fragmentation spectrum for [M − 10H + 5Na]5− and corresponding fragment ion map and donut plot. This charge state exhibits an even lower degree of SO3 loss (glycosidic-SO3 = 5.8%; cross-ring-SO3 = 0.0%) than [M − 8H + 3Na]5− but overall produces fewer informative fragments. This observation is consistent with NETD and EDD results for GAGs in which SO3 loss is minimized when the number of ionized sites is greater than the number of sulfate groups.26,36 However, less overall fragmentation is observed when all ionizable sites are deprotonated, as sites that remain protonated are hypothesized to facilitate radical-based reactions that provide informative fragment ions.
Figure 6.
(a) UVPD spectrum of [M − 8H + 3Na]5− (m/z 313) of fondaparinux with donut plot depicting the fragment ion composition based on summed abundances of fragment types and (b) UVPD fragment ion map. Table S13 contains a list of identified fragment
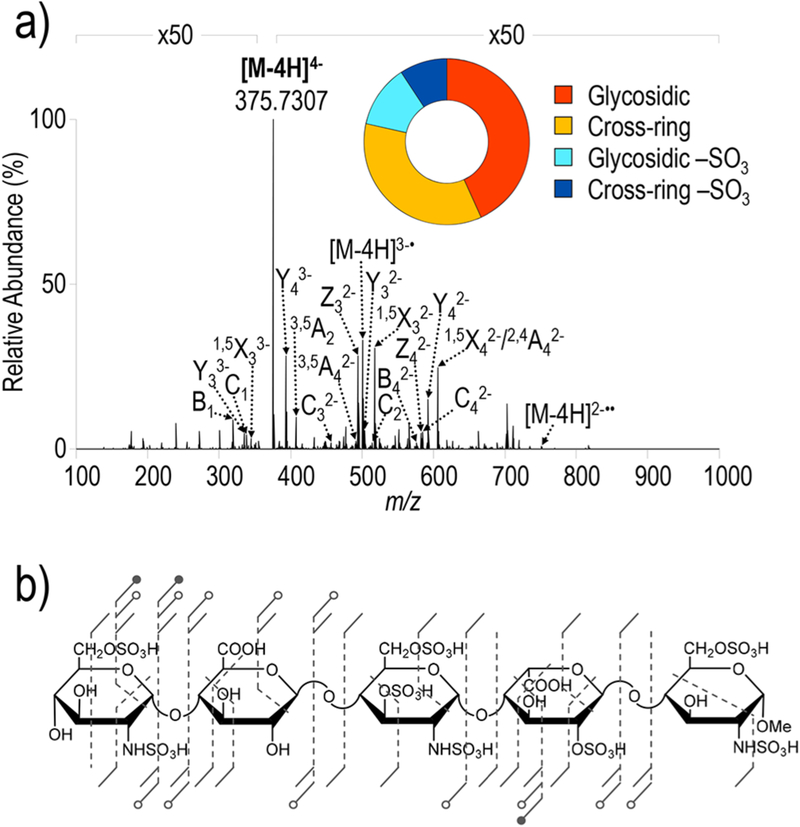
While deprotonation via sodium adduction represents one strategy to sequence highly sulfated GAGs, sodium adduction increases the number of possible ion types through generation of ions both with and without sodium adducts, which has the potential to increase spectral complexity, convolute interpretation, and increase the likelihood of false identifications. In addition, fragment ion signals can be split across various adduction states and can lead to reduced ion abundances. Conversely, highly sulfated GAG ions without salt adducts often contain protonated sulfate moieties, which are particularly problematic during collisional activation as they promote sulfate decomposition and preclude modification localization. EDD and NETD have previously shown success for activation of fondaparinux with protonated sulfate groups, generating spectra that contain informative fragment ions with minimal sulfate loss.33 The UVPD mass spectrum of the [M − 4H]4− ion of fondaparinux is displayed in Figure 7. In contrast to collisional activation (Table S16), UVPD generates a complete series of glycosidic fragments that retain sulfate modifications to a much larger extent (glycosidic-SO3 = 12.2%; cross-ring-SO3 = 9.2%). The informative spectrum reconfirms that sulfate decomposition is further minimized with UVPD even when sulfate groups are likely protonated, consistent with the prevailing understanding of the different mechanism of UVPD compared to collisional activation.63
Figure 7.
a) UVPD spectrum of [M − 4H]4− (m/z 375) of fondaparinux with donut plot depicting the fragment ion composition based on summed abundances of fragment type and (b) UVPD fragment ion map. Table S15 contains a list of identified fragment.
While the advantages of alternative ion activation methods for GAG analysis are apparent, many methods have limited accessibility and require instrument hardware modifications. Addition of a 213 nm solid state laser to an Orbitrap mass spectrometer was commercialized in 2017, thus making UVPD a more widely available technique.55,56 We assessed the capabilities of 213 nm UVPD for GAG structural characterization, and one example is illustrated in Figure S5c,d for DS dp4, [M − 3H]3−. While the 213 nm Nd:YAG laser is considerably less powerful than the 193 nm excimer laser, comparison of the UVPD fragment ion maps in Figure S5 suggests that 213 nm UVPD results in fragmentation that is similar to that obtained by 193 nm UVPD. However, the activation period for 213 nm UVPD (200 ms) is substantially greater than that used for 193 nm UVPD (16 ms). Future improvements to 213 nm UVPD are anticipated to decrease the required activation period. Variations between the 193 nm and 213 nm UVPD spectra in Figure S5 are attributed to differences in the laser powers (which influence the distribution of multiphoton absorption events and accumulation of energy) and the different wavelengths which may access different excited electronic states.
The fast, high-energy deposition of UVPD enhances cleavage of C–C and C–O bonds while minimizing loss of labile modifications. Moreover, UV photoabsorption can cause electron detachment, particularly for multiply charged negative ions, potentially leading to both radical and hydrogen migration in the course of fragmentation. Considering the informative spectra that can be generated with short activation times, UVPD appears to be a natural fit for coupling to online or gas-phase separations of GAGs.64,65 Recent coupling of NETD with trapped ion mobility spectrometry demonstrates the advantage of coupling alterative ion activation methods with separation techniques for GAG analysis.65 For GAGs with a higher degree of sulfation, it is worthwhile to investigate separation techniques that can accommodate sodium cationization of GAGs for increased sulfate retention with spectra collected on high-resolution mass spectrometers to alleviate concerns regarding spectral interpretation. These spectra are also amenable to automatic spectral interpretation which further enables the incorporation of this sequencing method for increased throughput.66
CONCLUSIONS
The work presented here confirms that UVPD demonstrates great promise for sequencing of GAGs by tandem mass spectrometry. Similar to electron-based activation methods, UVPD produces sulfate-retaining glycosidic and cross-ring fragment ions, with activation times comparable to NETD and much shorter than EDD. In general, the high-resolution capabilities of the Orbitrap mass analyzer enabled confident assignment of fragment ions and permitted interpretation of the complex tandem mass spectra typical of GAGs. While informative fragment ions were generated for all charge states, the degree of sulfate decomposition and extent of fragmentation showed some dependence on the ionization state, reiterating the important impact of both ionization conditions and precursor ion selection. In addition, the ability to modulate fragmentation patterns based on the particular ionization state of the precursor provides another strategic option when developing a workflow for high-throughput analysis of GAGs in mixtures.
Supplementary Material
ACKNOWLEDGMENTS
Funding from the NIH (Grant RO1 GM103655 to J.S.B.) and the Welch Foundation (Grant F-1155 to J.S.B.) are gratefully acknowledged. I.J.A. and F.E.L. are grateful for generous support from the National Institutes of Health and the Common Fund for Glycoscience, Grants R21HL136271, U01CA231074, and P41GM103390. Funding from the UT System for support of the UT System Proteomics Core Facility Network is gratefully acknowledged.
Footnotes
Notes
The authors declare no competing financial interest.
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.anal-chem.9b00521.
Figure S1, scheme of Domon and Costello nomenclature with sulfate retention/decomposition key; Figure S2, HCD spectrum of GlcA-GlcNAc-GlcA-GlcNAc-(CH2)NH2 Figure S3, expanded spectra of GlcA-GlcNAc-GlcA-GlcNAc-(CH2)5NH2; Figure S4, MS3 spectra of m/z 715.05 from DS dp4; Figure S5, 193 nm UVPD and 213 nm spectra of [M − 3H]3− of DS dp4; Figure S6, UVPD spectrum, fragment ion maps, and donut plot for [M − 10H + 5Na]5− for fondaparinux; Table S1, GAG structures; and Tables S2—S17, lists of identified fragment ions for all analyzed GAGs (PDF)
REFERENCES
- (1).Lever R; Page CP Nat. Rev. Drug Discovery 2002, 1, 140–148. [DOI] [PubMed] [Google Scholar]
- (2).Hacker U; Nybakken K; Perrimon N Nat. Rev. Mol. Cell Biol 2005, 6, 530. [DOI] [PubMed] [Google Scholar]
- (3).Parish CR Nat. Rev. Immunol 2006, 6, 633–643. [DOI] [PubMed] [Google Scholar]
- (4).Sasisekharan R; Shriver Z; Venkataraman G; Narayanasami U Nat. Rev. Cancer 2002, 2, 521. [DOI] [PubMed] [Google Scholar]
- (5).Soares da Costa D; Reis RL; Pashkuleva I Annu. Rev. Biomed. Eng 2017, 19, 1–26. [DOI] [PubMed] [Google Scholar]
- (6).Turnbull JE; Linhardt RJ Nat. Chem. Biol 2006, 2, 449–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Afratis N; Gialeli C; Nikitovic D; Tsegenidis T; Karousou E; Theocharis AD; Pavao MS; Tzanakakis GN; Karamanos NK FEBS J 2012, 279, 1177–1197. [DOI] [PubMed] [Google Scholar]
- (8).Liu H; Zhang Z; Linhardt RJ Nat. Prod. Rep 2009, 26, 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Mormann M; Zamfir AD; Seidler DG; Kresse H; Peter-Katalinic J J. Am. Soc. Mass Spectrom 2007, 18, 179–187. [DOI] [PubMed] [Google Scholar]
- (10).Laremore TN; Zhang F; Dordick JS; Liu J; Linhardt RJ Curr. Opin. Chem. Biol 2009, 13, 633–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Laremore TN; Leach FE III; Solakyildirim K; Amster IJ; Linhardt RJ Glycosaminoglycan Characterization by Electrospray Ionization Mass Spectrometry Including Fourier Transform Mass Spectrometry In Glycomics; Methods in Enzymology, Vol. 478; Abelson JN, Simon MI, Eds.; Academic Press, 2010; pp 79–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Chi L; Wolff JJ; Laremore TN; Restaino OF; Xie J; Schiraldi C; Toida T; Amster IJ; Linhardt RJ J. Am. Chem. Soc 2008, 130, 2617–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Ly M; Leach FE III; Laremore TN; Toida T; Amster IJ; Linhardt RJ Nat. Chem. Biol 2011, 7, 827–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Kailemia MJ; Li L; Ly M; Linhardt RJ; Amster IJ Anal. Chem 2012, 84, 5475–5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Wolff JJ; Laremore TN; Leach FE III; Linhardt RJ; Amster IJ Eur. J. Mass Spectrom 2009, 15, 275–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Zaia J; McClellan JE; Costello CE Anal. Chem 2001, 73, 6030–6039. [DOI] [PubMed] [Google Scholar]
- (17).Zaia J; Costello CE Anal. Chem 2001, 73, 233–239. [DOI] [PubMed] [Google Scholar]
- (18).Zaia J; Costello CE Anal. Chem 2003, 75, 2445–2455. [DOI] [PubMed] [Google Scholar]
- (19).Naggar EF; Costello CE; Zaia J J. Am. Soc. Mass Spectrom 2004, 15, 1534–1544. [DOI] [PubMed] [Google Scholar]
- (20).Kailemia MJ; Li L; Xu Y; Liu J; Linhardt RJ; Amster IJ Mol. Cell. Proteomics 2013, 12, 979–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Huang R; Pomin VH; Sharp JS J. Am. Soc. Mass Spectrom 2011, 22, 1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Huang R; Liu J; Sharp JS Anal. Chem 2013, 85, 5787–5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Taylor CJ; Burke RM; Wu B; Panja S; Nielsen SB; Dessent CEH Int. J. Mass Spectrom 2009, 285, 70–77. [Google Scholar]
- (24).Wolff JJ; Amster IJ; Chi L; Linhardt RJ J. Am. Soc. Mass Spectrom 2007, 18, 234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Wolff JJ; Laremore TN; Busch AM; Linhardt RJ; Amster IJ J. Am. Soc. Mass Spectrom 2008, 19, 294–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Wolff JJ; Laremore TN; Busch AM; Linhardt RJ; Amster IJ J. Am. Soc. Mass Spectrom 2008, 19, 790–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Leach FE III; Wolff JJ; Laremore TN; Linhardt RJ; Amster IJ Int. J. Mass Spectrom 2008, 276, 110–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Wolff JJ; Chi L; Linhardt RJ; Amster IJ Anal. Chem 2007, 79, 2015–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Oh HB; Leach FE III; Arungundram S; Al-Mafraji K; Venot A; Boons GJ; Amster IJ J. Am. Soc. Mass Spectrom 2011, 22, 582–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Leach FE III; Ly M; Laremore TN; Wolff JJ; Perlow J; Linhardt RJ; Amster IJ J. Am. Soc. Mass Spectrom 2012, 23, 1488–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Agyekum I; Zong C; Boons GJ; Amster IJ J. Am. Soc. Mass Spectrom 2017, 28, 1741–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Kailemia MJ; Park M; Kaplan DA; Venot A; Boons GJ; Li L; Linhardt RJ; Amster IJ J. Am. Soc. Mass Spectrom 2014, 25, 258–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Huang Y; Yu X; Mao Y; Costello CE; Zaia J; Lin C Anal. Chem 2013, 85, 11979–11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Wolff JJ; Leach FE III; Laremore TN; Kaplan DA; Easterling ML; Linhardt RJ; Amster IJ Anal. Chem 2010, 82, 3460–3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Leach FE III; Wolff JJ; Xiao Z; Ly M; Laremore TN; Arungundram S; Al-Mafraji K; Venot A; Boons GJ; Linhardt RJ; Amster IJ Eur. J. Mass Spectrom 2011, 17, 167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Leach FE III; Riley NM; Westphall MS; Coon JJ; Amster IJ J. Am. Soc. Mass Spectrom 2017, 28, 1844–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Wu J; Wei J; Hogan JD; Chopra P; Joshi A; Lu W; Klein J; Boons GJ; Lin C; Zaia JJ Am. Soc. Mass Spectrom 2018, 29, 1262–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Wolff JJ; Laremore TN; Aslam H; Linhardt RJ; Amster IJ J. Am. Soc. Mass Spectrom 2008, 19, 1449–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Ropartz D; Li P; Fanuel M; Giuliani A; Rogniaux H; Jackson GP J. Am. Soc. Mass Spectrom 2016, 27, 1614–1619. [DOI] [PubMed] [Google Scholar]
- (40).Racaud A; Antoine R; Joly L; Mesplet N; Dugourd P; Lemoine J J. Am. Soc. Mass Spectrom 2009, 20, 1645–1651. [DOI] [PubMed] [Google Scholar]
- (41).Racaud A; Antoine R; Dugourd P; Lemoine J J. Am. Soc. Mass Spectrom 2010, 21, 2077–2084. [DOI] [PubMed] [Google Scholar]
- (42).Ortiz D; Enjalbert Q; MacAleese L; Dugourd P; Salpin J-Y Rapid Commun. Mass Spectrom 2015, 29, 1135–1144. [DOI] [PubMed] [Google Scholar]
- (43).Ropartz D; Lemoine J; Giuliani A; Bittebiere Y; Enjalbert Q; Antoine R; Dugourd P; Ralet M-C; Rogniaux H Anal. Chim. Acta 2014, 807, 84–95. [DOI] [PubMed] [Google Scholar]
- (44).Ropartz D; Giuliani A; Herve C; Geairon A; Jam M; Czjzek M; Rogniaux H Anal. Chem 2015, 87, 1042–1049. [DOI] [PubMed] [Google Scholar]
- (45).Ropartz D; Giuliani A; Fanuel M; Herve C; Czjzek M; Rogniaux H Anal. Chim. Acta 2016, 933, 1–9. [DOI] [PubMed] [Google Scholar]
- (46).Compagnon I; Schindler B; Renois-Predelus G; Daniel R Curr. Opin. Struct. Biol 2018, 50, 171–180. [DOI] [PubMed] [Google Scholar]
- (47).Devakumar A; Thompson MS; Reilly JP Rapid Commun. Mass Spectrom 2005, 19, 2313–2320. [DOI] [PubMed] [Google Scholar]
- (48).Cui W; Thompson MS; Reilly JP J. Am. Soc. Mass Spectrom 2005, 16, 1384–1398. [DOI] [PubMed] [Google Scholar]
- (49).Reilly JP Mass Spectrom. Rev 2009, 28, 425–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Brodbelt JS Chem. Soc. Rev 2014, 43, 2757–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Shaw JB; Li W; Holden DD; Zhang Y; Griep-Raming J; Fellers RT; Early BP; Thomas PM; Kelleher NL; Brodbelt JS J. Am. Chem. Soc 2013, 135, 12646–12651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Klein DR; Holden DD; Brodbelt JS Anal. Chem 2016, 88, 1044–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Ko BJ; Brodbelt JS Anal. Chem 2011, 83, 8192–8200. [DOI] [PubMed] [Google Scholar]
- (54).Robinson MR; Moore KL; Brodbelt JS J. Am. Soc. Mass Spectrom 2014, 25, 1461–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Fornelli L; Srzentic K; Huguet R; Mullen C; Sharma S; Zabrouskov V; Fellers RT; Durbin KR; Compton PD; Kelleher NL Anal. Chem 2018, 90, 8421–8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Brodie NI; Huguet R; Zhang T; Viner R; Zabrouskov V; Pan J; Petrotchenko EV; Borchers CH Anal. Chem 2018, 90, 3079–3082. [DOI] [PubMed] [Google Scholar]
- (57).Arungundram S; Al-Mafraji K; Asong J; Leach FE III; Amster IJ; Venot A; Turnbull JE; Boons GJ J. Am. Chem. Soc 2009, 131, 17394–17405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Domon B; Costello CE Glycoconjugate J 1988, 5, 397–409. [Google Scholar]
- (59).Ceroni A; Maass K; Geyer H; Geyer R; Dell A; Haslam SM J. Proteome Res 2008, 7, 1650–1659. [DOI] [PubMed] [Google Scholar]
- (60).Shi X; Huang Y; Mao Y; Naimy H; Zaia J J. Am. Soc. Mass Spectrom 2012, 23, 1498–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Brunet C; Antoine R; Dugourd P; Canon F; Giuliani A; Nahon L J. Chem. Phys 2013, 138, 064301. [DOI] [PubMed] [Google Scholar]
- (62).Walenga JM; Jeske WP; Frapaise FX; Bick RL; Fareed J; Samama MM Expert Opin. Invest. Drugs 2002, 11, 397–407. [DOI] [PubMed] [Google Scholar]
- (63).Julian RR J. Am. Soc. Mass Spectrom 2017, 28, 1823–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Sanderson P; Stickney M; Leach FE; Xia Q; Yu Y; Zhang F; Linhardt RJ; Amster IJ J. Chromatogr. A 2018, 1545, 75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Wei J; Wu J; Tang Y; Ridgeway ME; Park MA; Costello CE; Zaia J; Lin C Anal. Chem 2019, 91, 2994–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Duan J; Jonathan Amster I J. Am. Soc. Mass Spectrom 2018, 29, 1802–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.