Abstract
Background
Atopic dermatitis is a chronic inflammatory disease of the skin. It has a high prevalence worldwide and affected persons are prone to recurrent attacks, seriously affecting the physical and mental of patients. The exact etiology of the disease is still unclear.
Material/Methods
There are 7 datasets on atopic dermatitis in the Gene Expression Omnibus database, including 142 lesional and 134 non-lesional skin biopsy samples. Differential analysis was performed after datasets were integrated by robust multi-array average method. Functional modules of GSE99802 were explored by weighted gene co-expression network analysis. The 4 most important modules were enriched into the pathways by Metascape.
Results
Significantly differentially expressed genes included 41 upregulated and 10 downregulated genes. The following 5 of the most important upregulated genes had the strongest association with atopic dermatitis. SERPINB3&4 promote inflammation and impaired skin barrier function in the early stage of atopic dermatitis. S100A9 aggravates the inflammatory response by inducing the activation of toll-like receptor 4, neutrophil chemotaxis, neutrophilic inflammation, and the amplification of interleukin-8. MMP1 is the key protease of skin collagen degradation, keeping the extracellular matrix in dynamic balance. MMP12 induces the aggregation of various inflammatory cells into inflammatory tissue. The enriched pathways of each module mainly include Cellular responses to external stimuli, Metabolism of RNA and Translation, and Infectious disease.
Conclusions
The associated pathways and genes not only help us understand the molecular mechanism of the disease, but also provide research directions or targets for accurate diagnosis and treatment.
MeSH Keywords: Cluster Analysis; Critical Pathways; Dermatitis, Atopic; Genetic Association Studies
Background
Atopic dermatitis (AD), also known as atopic eczema, is a chronic inflammatory skin disease characterized by severe itching and recurrent, relapsing eczema-like skin lesions [1]. AD is a complex multifactorial disease, and its exact etiology and pathogenesis have not been fully elucidated. Moreover, clinical diagnosis of the disease still depends on the clinical features, since no specific tests and histopathology have been reported as diagnostic criteria [2]. Although AD as a skin disease is not fatal, it imposes a serious psychological burden on patients, and increases the risk of food allergies, asthma, allergic arthritis, and other autoimmune diseases [3]. AD is a very common chronic disease, affecting up to one-fifth of the population in developed countries. The disease has shown a global increase over the past 3 decades. The prevalence rate in developed regions is stable at 10–20%, while in developing countries the prevalence is low but increasing [4,5]. Roughly 60% of cases have their first episode within the first year of life, but the first episode can occur at any age [6,7]. The current treatment methods are mainly the external use of glucocorticoid and immunosuppressive agents, and phototherapy or systemic immunosuppressive therapy for severe cases, but they cannot achieve the goal of complete cure [8]. Nowadays, when medical research has advanced into the era of genes and molecules, research on the molecular basis of AD also made great progress. Previous studies have found that functional mutations, polymorphisms, and abnormal expressions of genes related to skin barrier, such as FLG, LOR, IVL, SPINK5, and TMEM79, are often closely related to the incidence of AD [9]. In addition, genes of key components of innate immunity, such as NOD1, NOD2, TLR2, CD14, and DEFB1, are associated with the pathogenesis of AD.
The Gene Expression Omnibus (GEO) is a database containing a large number of datasets on gene expression of various diseases. These datasets provide very valuable information that can be reused to gain new insights through different methods and perspectives.
Bioinformatics is an increasingly popular and reliable way to process existing data [10–12]. In this study, 2 major bioinformatics approaches – robust multi-array average (RMA) analysis and weighted gene co-expression network analysis (WGCNA) – were used. RMA is a strict systems biology tool that can integrate data from different datasets of the same platform for the same disease, so as to not only improve the sample size, but also reduce the resulting deviation caused by data combination [13]. WGCNA divides all selected genes into several biological functional modules, which are clusters of genes with biological significance, and can reflect the pathogenesis of disease [14,15].
The aim of our research was to identify the pathways and genes that may play an important role in the pathogenesis of AD, so as to narrow the target range and provide new targets for further research.
Material and Methods
Datasets search and eligibility criteria
The gene expression data of “Atopic Dermatitis Skin Biopsy” were downloaded from the Gene Expression Omnibus (GEO) database. The inclusion criteria used in our research were: (a) Gene expression profiling datasets that contained microarray chip technology; (b) Studies comparing expression data between lesional skin and non-lesional skin in atopic dermatitis patients; and (c) Sample size should be greater than 15. Studies that did not meet these eligibility criteria were excluded.
Data processing and differential analysis
Processing steps was conducted using R and Bioconductor Software [16]. Background correction, quantile normalization, and determination of expression levels were performed using an RMA analysis method [13]. The surrogate variable analysis (SVA) package was used to adjust for batch effects [17,18]. Differential expression analysis was performed with the Limma package [19]. |log Fold Change (FC)|>1 and P-value <0.05 were set as the cut-offs to screen differentially expressed genes (DEGs). A heat map was plotted by R package function heat map.
WGCNA
A gene co-expression network was constructed by WGCNA package of R software [14]. GSE99802 had the largest sample size, including lesional skin gene expression data of 59 patients, and was thus used in the WGCNA analysis. We calculated mean expression value for each gene, and only those with Fragments Per Kilobase Million (FPKM) greater than 8.5 were selected. We defined the soft-threshold power as 5 to make the scale-free topology index greater than 0.85. The cut height=0.3 and min size=100 were used to identify key modules. Each module was labeled with a random color. Genes that cannot be clustered into any given module were assigned to the grey module.
Functional enrichment analysis
Functional enrichment analyses were carried out using the online software Metascape (http://metascape.org) [20]. The top 20 clusters with the representative enriched terms were chosen.
Statistical analysis
All statistical analyses were performed using R software version 3.5.0. P<0.05 was considered to be statistically significant.
Results
AD datasets
Based on the inclusion criteria, 7 datasets from GPL570 were finally selected for study: GSE32924, GSE120899, GSE27887, GSE36842, GSE120721, GSE58558, and GSE99802. The description of datasets is provided in Table 1, such as GSE number, authors, participants, and platforms. The number of biopsy samples at lesional skin ranged from 8 to 59, and the number of biopsy samples at non-lesional skin ranged from 8 to 53. Finally, the research included 142 lesional and 134 non-lesional skin biopsy samples.
Table 1.
Summary of those 7 genome-wide gene expression datasets involving AD patients.
| Dataset ID | GSE number | Samples | Source types | Platform | Authors |
|---|---|---|---|---|---|
| 1 | GSE32924 | 13 lesional and 12 non-lesional | Skin tissues in AD patients | GPL570 | Suárez-Fariñas M, Tintle SJ, Shemer A et al. |
| 2 | GSE27887 | 9 lesional and 8 non-lesional | Skin tissues in AD patients | GPL570 | Tintle S, Shemer A, Suárez-Fariñas M et al. |
| 3 | GSE120899 | 20 lesional and 21 non-lesional | Skin tissues in AD patients | GPL570 | Simpson EL, Imafuku S, Poulin Y et al. |
| 4 | GSE120721 | 15 lesional and 15 non-lesional | Skin tissues in AD patients | GPL570 | Esaki H, Ewald DA, Ungar B et al. |
| 5 | GSE99802 | 59 lesional and 53 non-lesional | Skin tissues in AD patients | GPL570 | Brunner PM, Pavel AB, Khattri S et al. |
| 6 | GSE58558 | 18 lesional and 17 non-lesional | Skin tissues in AD patients | GPL570 | Khattri S, Shemer A, Rozenblit M, et al. |
| 7 | GSE36842 | 8 lesional and 8 non-lesional | Skin tissues in AD patients | GPL570 | Gittler JK, Shemer A, Suárez-Fariñas M et al. |
Identification of differentially expressed genes
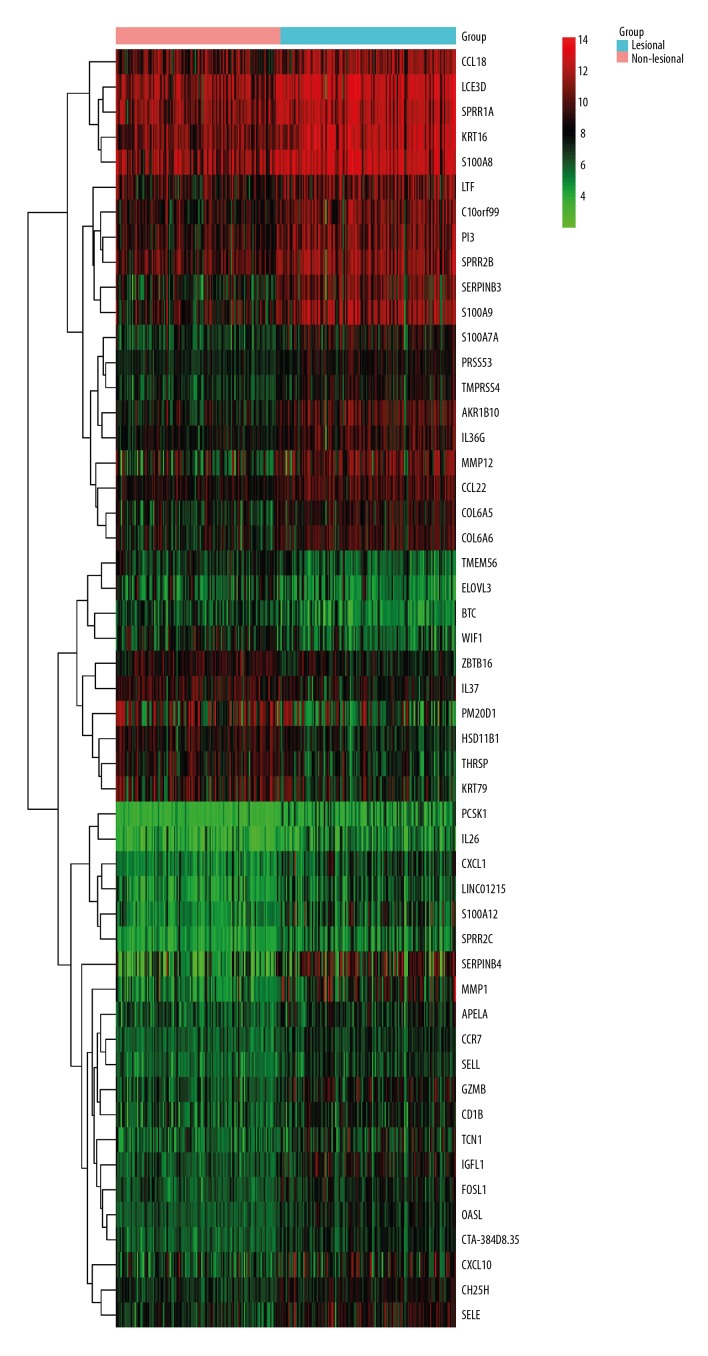
According to the criteria of |log FC| >1 and P<0.05, a total of 51 DEGs were identified (Supplementary Table 1), of which 41 were upregulated and 10 were downregulated. A larger number of upregulated genes were identified as compared with the downregulated genes, which indicated that the DEGs had a tendency to be upregulated in AD. These 51 DEGs are displayed by heatmap visualization in Figure 1.
Figure 1.
Heatmap of the top 51 DEGs according to the value of |logFC|. The color in the heatmap from green to red shows the progression from low expression to high expression. The heatmap can roughly distinguish the lesional skin group from the non-lesional skin group of AD.
WGCNA
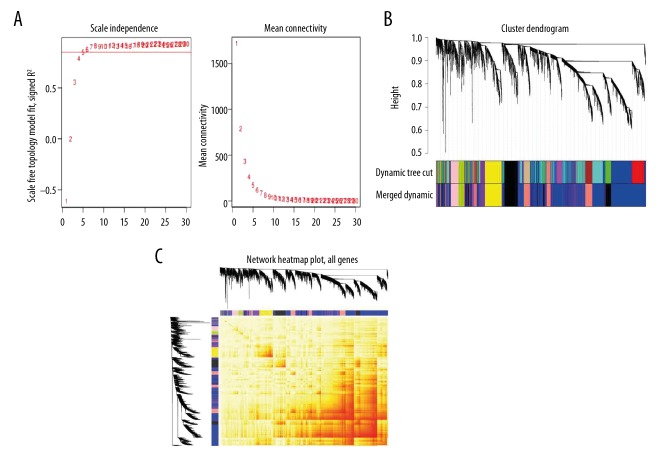
In total, 5481 genes were filtered out based on the requirement and used in the WGCNA analysis. The dimensions of the sample tree are obtained by defining sample clustering height=60. We adjusted the soft-threshold power to 5, and the index of scale-free topology was >0.85 (Figure 2A). When the minimum module size was set to 100 genes and a threshold setting for merging modules of 0.3, 7 coexpressed gene modules were identified (Figure 2B). The number of genes in modules ranged from 176 to 2694. The largest module, which had 2694 genes, was the blue module, and 919 genes were in the second largest module, the salmon module. Interaction relationship among the 7 co-expression modules of genes was further analyzed. As seen in Figure 2, the modules were independent of each other.
Figure 2.
Plots in the WGCNA using gene expression profiles of biopsy samples from lesional skin of 59 AD patients in the GSE99802. (A) We selected an appropriate threshold power to make the connections between genes in the network conform to the scale-free networks. The panel on the left shows that the appropriate soft-threshold power meets the topological index >0.85; the panel on the right shows the correspondence between soft-threshold and mean connectivity. (B) Gene dendrogram of co-expression genes obtained by hierarchical clustering. The underlying color row represents the functional modules resulting from the dynamic merge. When the threshold of merging similar modules was set to 0.3, 7 main modules with different colors were obtained. (C) Heatmap of topological overlap in the gene network. The gradually saturated red color indicates a high degree of overlap between functional modules.
Functional enrichment analysis
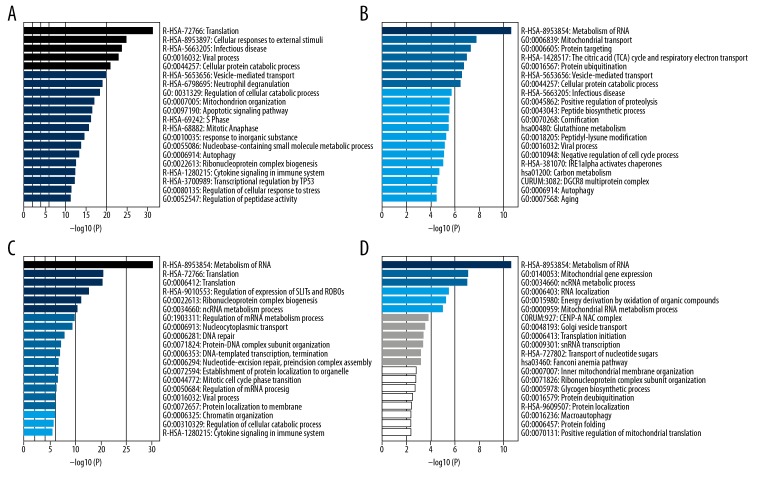
The main results in the functional enrichment analysis are displayed in Figure 3. The genes in the blue module are involved in many biological processes, such as Translation, Cellular responses to external stimuli, Infectious disease, and Viral process (Figure 3A). The genes in the salmon module are mainly involved in Metabolism of RNA, Mitochondrial transport, and Protein targeting (Figure 3B). The genes in the black and yellow modules are mainly involved in Metabolism of RNA (Figure 3C, 3D). Genes in the pink module are mainly involved in extracellular matrix organization, while the green-yellow module genes are mainly enriched in processes of the immune response, such as lymphocyte activation.
Figure 3.
Functional enrichment analysis of the top 4 modules which have the most genes. When the p-value is smaller, the bar’s color is darker and longer, and is more significant statistically. (A) Significant biological processes in blue module. (B) Significant biological processes in salmon module. (C) Significant biological processes in black module. (D) Significant biological processes in yellow module.
Discussion
AD has a high incidence and is prone to long-term recurrence; at present, there is no effective treatment. In recent years, there have been more and more reports on genes correlated with AD. Our research results have some similarities with previous research conclusions, and also has some new findings. First, we selected 7 datasets on AD from the same platform and integrated them by using the RMA method. Then, differential expression analysis was performed comparing lesional skin and non-lesional skin of AD patients.
According to the results, among the upregulated genes of DEGs, SERPINB3&4, S100A9, and MMP1&12 showed the most significant differences. For SERPINB3&4, there have been a number of previous studies that have confirmed its increased expression in the injured skin tissues and serum of AD patients [21–23]. However, the role of SERPINB3&4 in AD remains unclear. In a recent experiment, it was found that mouse SERPINB3a, corresponding to human SERPINB3&4, plays a pro-inflammatory role in the early stage of AD after allergen exposure, and plays an important role in the early stage of skin barrier dysfunction. However, there is no significant effect on the chronic inflammatory duration of AD. Moreover, the study also found that the expression of SERPINB3&4 directly promoted the expression of S100A8 gene in both mice and human patients [24]. Among the significantly different genes we found, S100A9, S100A12, and S100A8 all belong to the S100A family, which has been proved to be closely related to chronic inflammation and proliferation of bone marrow cells [25]. The one with the largest logFC of the 3 genes is S100A9. S100A9 is a calcium-binding protein that is an endogenous danger signal released by stress cells. S100A9 promotes and aggravates the inflammatory response by inducing the activation of Toll-like receptor 4, neutrophil chemotaxis, neutrophilic inflammation, and the amplification of interleukin-8 [26–-29]. It has been reported that S100A9 can directly, or in collaboration with inflammatory factors, to lead to the occurrence and progression of asthma neutrophilic inflammation [30]. The present study is the first to show that S100A9 is likely to be a hub gene in the pathogenesis of AD, which is also a multifactorial disease, along with asthma. Among the significantly different genes, there are 2 Matrix metalloproteinases (MMPs) family genes – MMP1 and MMP12. MMPs is closely related to the pathophysiological processes of allergic inflammation, tissue damage and repair, and host defense against pathogens [31,32].
Using skin-wash sampling assay technique, Harper et al. confirmed that the MMPs family has a very important role in the pathogenesis of AD [33]. MMP1 is the key protease of skin collagen degradation [34], which keeps the extracellular matrix in dynamic balance. It is probably activated during the process of skin damage in the chronic inflammatory phase of AD to inhibit the formation of skin scarring. MMP12 has been found to induce the aggregation of various inflammatory cells into inflammatory tissue, which is involved in the pathogenesis of asthma [35], and we believe it also plays an important role in the pathogenesis of AD.
In addition, BTC and PM20D1 genes showed the most significant difference among the downregulated genes. BTC, also known as Betacellulin, encodes a member of the epidermal growth factor (EGF) protein family [36]. The downregulated expression of BTC may weaken the ability of skin regeneration and repair, thereby damaging the skin barrier function and contributing to the occurrence of AD. However, there are still few studies on the function of PM20D1, and there are no reports on the correlation between PM20D1 and AD, which need further studies to clarify. The discovery of downregulated genes could provide a new perspective for the treatment of diseases.
WGCNA is an advanced analytical method with great biological importance and has been used in the study of molecular mechanisms of more and more diseases. It can cluster highly correlated genes into modules, and through the enrichment analysis of these modules, the general pathogenesis and core pathways of diseases can be observed [37]. To make the results reflect the overall picture of disease occurrence and to make the results more reliable, we selected the dataset (GSE99802) that has the largest sample size and is the most representative. We selected the whole-gene expression matrix of AD patients and deleted the genes with FPKM values that were too low. In the end, 7 modules were identified, among which the blue module contained the largest number of genes, followed by the salmon module, and the black module and the yellow module also contained a large number of genes. We applied Metascape to carry out enrichment analysis of the modules mentioned above. The genes of the blue module are significantly enriched in ‘Translation, Cellular responses to external stimuli, Infectious disease, and Viral process’, and the genes of the salmon module are significantly enriched in ‘Metabolism of RNA, Mitochondrial transport, and Protein targeting’. ‘Metabolism of RNA and Translation’ are the most important pathways enriched by the genes of the black module. Meanwhile, pathways such as ‘Metabolism of RNA, mitochondrial gene expression, and ncRNA metabolic process’ were found to be significantly enriched in the yellow module. Other smaller modules were enriched into pathways involving extracellular matrix and immune response.
The pathways described above are mainly reflected in 2 aspects. First of all, when the cells at the lesion site of AD are exposed to external stimuli, frequency or extent of the chemical reactions involving RNA is advanced. This further leads to the activation of biological processes such as the expression of inflammatory factors, cell repair, and proliferation. Secondly, infectious factors have once again been proved to play an important role in the occurrence and development of AD. In the initial stage of the disease, viruses or other pathogens may act as a trigger. In the chronic stage of AD, the skin microecosystem is out of balance due to skin barrier dysfunction, topical antibiotic application, and other factors. Antimicrobial peptides (AMPs) in the skin of patients decrease, and the overproliferation of colonized dominant bacteria, such as Staphylococcus aureus, stimulate the epidermis to develop persistent inflammation [38].
In the present study, the functions of the genes and pathways obtained are mostly consistent with the current consensus about the mechanism of AD, which reflects the reliability of our research methods and results. However, we are also aware of some limitations of the study. First, some data sets have large sample sizes, but the timing of biopsy is confusing or took place after treatment. To ensure diagnostic accuracy and reduce the impact of different experimental designs on this study, only 7 datasets were selected, which led to a small sample size. Second, we regret that the raw data from almost all datasets do not provide information about patient ethnicity, which may have some impact on the results. Third, it is insufficient to conduct secondary analysis of existing data only through bioinformatics, and we hope to verify these results through further experiments in the future.
Conclusions
By using RMA method, we selected hub genes based on the DEGs obtained from 7 datasets. Then, we used WGCNA to cluster the genes of AD samples of GSE99802 into 7 modules, and performed functional enrichment analysis by Metascape on the first 4 modules containing the most genes. These pathways and associated genes warrant more exploration and demonstration of their potential in diagnosis, prognosis, and therapeutic targets.
Supplementary Table
Supplementary Table 1.
51 genes with significant differences between lesional skin and non-lesional (|log FC|>1, P-value <0.05).
| Types | logFC | AveExpr | P. value | adj. P. value | |
|---|---|---|---|---|---|
| SERPINB4 | Up-regulated | 2.806586 | 6.991864 | 4.92E-18 | 5.32E-15 |
| MMP1 | Up-regulated | 2.185797 | 6.912649 | 1.35E-19 | 4.12E-16 |
| SERPINB3 | Up-regulated | 2.003999 | 9.04055 | 4.01E-17 | 2.48E-14 |
| S100A9 | Up-regulated | 2.002726 | 9.772449 | 6.06E-16 | 2.13E-13 |
| MMP12 | Up-regulated | 1.998675 | 8.810077 | 9.22E-15 | 2.18E-12 |
| AKR1B10 | Up-regulated | 1.671834 | 8.79022 | 8.69E-18 | 8.97E-15 |
| S100A12 | Up-regulated | 1.580258 | 6.285105 | 4.07E-15 | 1.06E-12 |
| GZMB | Up-regulated | 1.551569 | 7.411098 | 1.43E-18 | 2.06E-15 |
| KRT16 | Up-regulated | 1.493313 | 11.30241 | 1.52E-19 | 4.12E-16 |
| IGFL1 | Up-regulated | 1.467074 | 7.332853 | 2.72E-18 | 3.10E-15 |
| S100A7A | Up-regulated | 1.446888 | 7.948126 | 1.30E-14 | 2.86E-12 |
| CXCL1 | Up-regulated | 1.423856 | 6.295399 | 2.37E-18 | 3.02E-15 |
| C10orf99 | Up-regulated | 1.405349 | 9.630901 | 5.67E-13 | 6.33E-11 |
| PI3 | Up-regulated | 1.359249 | 9.837105 | 2.37E-12 | 2.06E-10 |
| SELE | Up-regulated | 1.318033 | 8.106887 | 3.55E-14 | 6.51E-12 |
| S100A8 | Up-regulated | 1.284684 | 11.63387 | 2.99E-12 | 2.48E-10 |
| LINC01215 | Up-regulated | 1.281348 | 5.859495 | 5.24E-16 | 1.92E-13 |
| TMPRSS4 | Up-regulated | 1.277667 | 7.941901 | 2.88E-16 | 1.27E-13 |
| COL6A5 | Up-regulated | 1.269852 | 8.345767 | 3.99E-12 | 3.08E-10 |
| COL6A6 | Up-regulated | 1.238226 | 8.666585 | 9.70E-12 | 6.36E-10 |
| SPRR2B | Up-regulated | 1.234246 | 10.32728 | 1.44E-10 | 6.08E-09 |
| TCN1 | Up-regulated | 1.223949 | 6.845003 | 3.96E-11 | 2.03E-09 |
| CD1B | Up-regulated | 1.183159 | 7.068287 | 3.71E-12 | 2.92E-10 |
| LCE3D | Up-regulated | 1.180626 | 11.62643 | 9.22E-10 | 2.96E-08 |
| OASL | Up-regulated | 1.166058 | 7.017667 | 4.66E-24 | 1.01E-19 |
| CCL18 | Up-regulated | 1.12904 | 10.45926 | 2.33E-09 | 6.58E-08 |
| IL36G | Up-regulated | 1.106151 | 8.760932 | 9.51E-13 | 9.74E-11 |
| FOSL1 | Up-regulated | 1.10205 | 7.13091 | 2.85E-17 | 2.05E-14 |
| APELA | Up-regulated | 1.100046 | 6.975293 | 2.54E-12 | 2.19E-10 |
| CTA-384D8.35 | Up-regulated | 1.078971 | 6.899799 | 3.76E-21 | 4.07E-17 |
| SELL | Up-regulated | 1.077479 | 6.611732 | 4.14E-16 | 1.57E-13 |
| CXCL10 | Up-regulated | 1.073656 | 7.540534 | 9.86E-07 | 1.21E-05 |
| CH25H | Up-regulated | 1.062305 | 7.836583 | 3.13E-16 | 1.28E-13 |
| PCSK1 | Up-regulated | 1.060591 | 4.703752 | 4.61E-16 | 1.72E-13 |
| SPRR2C | Up-regulated | 1.057754 | 5.470469 | 2.98E-12 | 2.48E-10 |
| IL26 | Up-regulated | 1.051377 | 4.916004 | 5.97E-12 | 4.28E-10 |
| CCR7 | Up-regulated | 1.040097 | 6.782151 | 1.04E-19 | 3.75E-16 |
| SPRR1A | Up-regulated | 1.034975 | 11.39261 | 1.05E-09 | 3.24E-08 |
| LTF | Up-regulated | 1.03336 | 9.690571 | 6.59E-07 | 8.59E-06 |
| CCL22 | Up-regulated | 1.010977 | 9.235341 | 1.67E-13 | 2.33E-11 |
| PRSS53 | Up-regulated | 1.001162 | 8.122699 | 3.57E-17 | 2.27E-14 |
| IL37 | Down-regulated | −1.00327 | 8.700774 | 7.08E-10 | 2.38E-08 |
| TMEM56 | Down-regulated | −1.01936 | 6.964349 | 8.33E-12 | 5.62E-10 |
| ZBTB16 | Down-regulated | −1.06223 | 8.465374 | 9.64E-14 | 1.43E-11 |
| KRT79 | Down-regulated | −1.0656 | 8.43783 | 1.92E-06 | 2.16E-05 |
| ELOVL3 | Down-regulated | −1.116 | 6.431317 | 2.60E-08 | 5.47E-07 |
| HSD11B1 | Down-regulated | −1.27707 | 8.309902 | 5.62E-16 | 2.03E-13 |
| WIF1 | Down-regulated | −1.29782 | 7.133747 | 1.98E-11 | 1.11E-09 |
| THRSP | Down-regulated | −1.30307 | 8.167562 | 1.73E-12 | 1.58E-10 |
| BTC | Down-regulated | −1.34965 | 6.20669 | 5.02E-19 | 9.87E-16 |
| PM20D1 | Down-regulated | −1.53753 | 8.192321 | 3.21E-07 | 4.62E-06 |
Footnotes
Source of support: Departmental sources
References
- 1.Weidinger S, Novak N. Atopic dermatitis. Lancet. 2016;387(10023):1109–22. doi: 10.1016/S0140-6736(15)00149-X. [DOI] [PubMed] [Google Scholar]
- 2.David Boothe W, Tarbox JA, Tarbox MB. Atopic dermatitis: Pathophysiology. Adv Exp Med Biol. 2017;1027:21–37. doi: 10.1007/978-3-319-64804-0_3. [DOI] [PubMed] [Google Scholar]
- 3.Eckert L, Gupta S, Amand C, et al. Impact of atopic dermatitis on health-related quality of life and productivity in adults in the United States: An analysis using the National Health and Wellness Survey. J Am Acad Dermatol. 2017;77(2):274–79.e3. doi: 10.1016/j.jaad.2017.04.019. [DOI] [PubMed] [Google Scholar]
- 4.Deckers IA, McLean S, Linssen S, et al. Investigating international time trends in the incidence and prevalence of atopic eczema 1990–2010: A systematic review of epidemiological studies. PLoS One. 2012;7(7):e39803. doi: 10.1371/journal.pone.0039803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams H, Stewart A, von Mutius E, et al. International Study of Asthma and Allergies in Childhood (ISAAC) Phase One and Three Study Groups. Is eczema really on the increase worldwide? J Allergy Clin Immunol. 2008;121(4):947–54.e15. doi: 10.1016/j.jaci.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Illi S, von Mutius E, Lau S, et al. The natural course of atopic dermatitis from birth to age 7 years and the association with asthma. J Allergy Clin Immunol. 2004;113(5):925–31. doi: 10.1016/j.jaci.2004.01.778. [DOI] [PubMed] [Google Scholar]
- 7.Garmhausen D, Hagemann T, Bieber T, et al. Characterization of different courses of atopic dermatitis in adolescent and adult patients. Allergy. 2013;68(4):498–506. doi: 10.1111/all.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Maria Diaz Granados L, Quijano MA, et al. Quality assessment of atopic dermatitis clinical practice guidelines in </=18 years. Arch Dermatol Res. 2018;310(1):29–37. doi: 10.1007/s00403-017-1791-7. [DOI] [PubMed] [Google Scholar]
- 9.Jurakic Toncic R, Marinovic B. What is new and hot in genetics of human atopic dermatitis: Shifting paradigms in the landscape of allergic skin diseases. Acta Dermatovenerol Croat. 2014;22(4):313–15. [PubMed] [Google Scholar]
- 10.Marques FZ, Campain AE, Yang YH, Morris BJ. Meta-analysis of genome-wide gene expression differences in onset and maintenance phases of genetic hypertension. Hypertension. 2010;56(2):319–24. doi: 10.1161/HYPERTENSIONAHA.110.155366. [DOI] [PubMed] [Google Scholar]
- 11.Rung J, Brazma A. Reuse of public genome-wide gene expression data. Nat Rev Genet. 2013;14(2):89–99. doi: 10.1038/nrg3394. [DOI] [PubMed] [Google Scholar]
- 12.Seifuddin F, Pirooznia M, Judy JT, et al. Systematic review of genome-wide gene expression studies of bipolar disorder. BMC Psychiatry. 2013;13:213. doi: 10.1186/1471-244X-13-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 14.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prom-On S, Chanthaphan A, Chan JH, Meechai A. Enhancing biological relevance of a weighted gene co-expression network for functional module identification. J Bioinform Comput Biol. 2011;9(1):111–29. doi: 10.1142/s0219720011005252. [DOI] [PubMed] [Google Scholar]
- 16.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 18.Chen C, Grennan K, Badner J, et al. Removing batch effects in analysis of expression microarray data: An evaluation of six batch adjustment methods. PLoS One. 2011;6(2):e17238. doi: 10.1371/journal.pone.0017238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wettenhall JM, Smyth GK. limmaGUI: A graphical user interface for linear modeling of microarray data. Bioinformatics. 2004;20(18):3705–6. doi: 10.1093/bioinformatics/bth449. [DOI] [PubMed] [Google Scholar]
- 20.Tripathi S, Pohl MO, Zhou Y, et al. Meta- and orthogonal integration of influenza „OMICs” data defines a role for UBR4 in virus budding. Cell Host Microbe. 2015;18(6):723–35. doi: 10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Broccardo CJ, Mahaffey SB, Strand M, et al. Peeling off the layers: Skin taping and a novel proteomics approach to study atopic dermatitis. J Allergy Clin Immunol. 2009;124(5):1113–15.e1-11. doi: 10.1016/j.jaci.2009.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu ZR, Park D, Lee KA, et al. Profiling the dysregulated genes of keratinocytes in atopic dermatitis patients: cDNA microarray and interactomic analyses. J Dermatol Sci. 2009;54(2):126–29. doi: 10.1016/j.jdermsci.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 23.Saaf AM, Tengvall-Linder M, Chang HY, et al. Global expression profiling in atopic eczema reveals reciprocal expression of inflammatory and lipid genes. PLoS One. 2008;3(12):e4017. doi: 10.1371/journal.pone.0004017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sivaprasad U, Kinker KG, Ericksen MB, et al. SERPINB3/B4 contributes to early inflammation and barrier dysfunction in an experimental murine model of atopic dermatitis. J Invest Dermatol. 2014;135(1):160–69. doi: 10.1038/jid.2014.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kovacic M, Mitrovic-Ajtic O, Beleslin-Cokic B, et al. TLR4 and RAGE conversely mediate pro-inflammatory S100A8/9-mediated inhibition of proliferation-linked signaling in myeloproliferative neoplasms. Cell Oncol (Dordr) 2018;41(5):541–53. doi: 10.1007/s13402-018-0392-6. [DOI] [PubMed] [Google Scholar]
- 26.Vogl T, Tenbrock K, Ludwig S, et al. Mrp8 and Mrp14 are endogenous activators of Toll-like receptor 4, promoting lethal, endotoxin-induced shock. Nat Med. 2007;13(9):1042–49. doi: 10.1038/nm1638. [DOI] [PubMed] [Google Scholar]
- 27.Ryckman C, Vandal K, Rouleau P, et al. Proinflammatory activities of S100: Proteins S100A8, S100A9, and S100A8/A9 induce neutrophil chemotaxis and adhesion. J Immunol. 2003;170(6):3233–42. doi: 10.4049/jimmunol.170.6.3233. [DOI] [PubMed] [Google Scholar]
- 28.Yano J, Kolls JK, Happel KI, et al. The acute neutrophil response mediated by S100 alarmins during vaginal Candida infections is independent of the Th17-pathway. PLoS One. 2012;7(9):e46311. doi: 10.1371/journal.pone.0046311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simard JC, Noel C, Tessier PA, Girard D. Human S100A9 potentiates IL-8 production in response to GM-CSF or fMLP via activation of a different set of transcription factors in neutrophils. FEBS Lett. 2014;588(13):2141–46. doi: 10.1016/j.febslet.2014.04.027. [DOI] [PubMed] [Google Scholar]
- 30.Lee TH, Chang HS, Bae DJ, et al. Role of S100A9 in the development of neutrophilic inflammation in asthmatics and in a murine model. Clin Immunol. 2017;183:158–66. doi: 10.1016/j.clim.2017.08.013. [DOI] [PubMed] [Google Scholar]
- 31.Coussens LM, Shapiro SD, Soloway PD, Werb Z. Models for gain-of-function and loss-of-function of MMPs. Transgenic and gene targeted mice. Methods Mol Biol. 2001;151:149–79. doi: 10.1385/1-59259-046-2:149. [DOI] [PubMed] [Google Scholar]
- 32.Ishii Y, Ogura T, Tatemichi M, et al. Induction of matrix metalloproteinase gene transcription by nitric oxide and mechanisms of MMP-1 gene induction in human melanoma cell lines. Int J Cancer. 2003;103(2):161–68. doi: 10.1002/ijc.10808. [DOI] [PubMed] [Google Scholar]
- 33.Harper JI, Godwin H, Green A, et al. A study of matrix metalloproteinase expression and activity in atopic dermatitis using a novel skin wash sampling assay for functional biomarker analysis. Br J Dermatol. 2009;162(2):397–403. doi: 10.1111/j.1365-2133.2009.09467.x. [DOI] [PubMed] [Google Scholar]
- 34.Scharffetter-Kochanek K, Brenneisen P, et al. Photoaging of the skin from phenotype to mechanisms. Exp Gerontol. 2000;35(3):307–16. doi: 10.1016/s0531-5565(00)00098-x. [DOI] [PubMed] [Google Scholar]
- 35.Zimmermann N, King NE, Laporte J, et al. Dissection of experimental asthma with DNA microarray analysis identifies arginase in asthma pathogenesis. J Clin Invest. 2003;111(12):1863–74. doi: 10.1172/JCI17912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rush JS, Peterson JL, Ceresa BP. Betacellulin (BTC) biases the EGFR to dimerize with ErbB3. Mol Pharmacol. 2018;94(6):1382–90. doi: 10.1124/mol.118.113399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan S, Wang W, Gao G, et al. Key genes and functional coexpression modules involved in the pathogenesis of systemic lupus erythematosus. J Cell Physiol. 2018;233(11):8815–25. doi: 10.1002/jcp.26795. [DOI] [PubMed] [Google Scholar]
- 38.Kong HH, Oh J, Deming C, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012;22(5):850–59. doi: 10.1101/gr.131029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1.
51 genes with significant differences between lesional skin and non-lesional (|log FC|>1, P-value <0.05).
| Types | logFC | AveExpr | P. value | adj. P. value | |
|---|---|---|---|---|---|
| SERPINB4 | Up-regulated | 2.806586 | 6.991864 | 4.92E-18 | 5.32E-15 |
| MMP1 | Up-regulated | 2.185797 | 6.912649 | 1.35E-19 | 4.12E-16 |
| SERPINB3 | Up-regulated | 2.003999 | 9.04055 | 4.01E-17 | 2.48E-14 |
| S100A9 | Up-regulated | 2.002726 | 9.772449 | 6.06E-16 | 2.13E-13 |
| MMP12 | Up-regulated | 1.998675 | 8.810077 | 9.22E-15 | 2.18E-12 |
| AKR1B10 | Up-regulated | 1.671834 | 8.79022 | 8.69E-18 | 8.97E-15 |
| S100A12 | Up-regulated | 1.580258 | 6.285105 | 4.07E-15 | 1.06E-12 |
| GZMB | Up-regulated | 1.551569 | 7.411098 | 1.43E-18 | 2.06E-15 |
| KRT16 | Up-regulated | 1.493313 | 11.30241 | 1.52E-19 | 4.12E-16 |
| IGFL1 | Up-regulated | 1.467074 | 7.332853 | 2.72E-18 | 3.10E-15 |
| S100A7A | Up-regulated | 1.446888 | 7.948126 | 1.30E-14 | 2.86E-12 |
| CXCL1 | Up-regulated | 1.423856 | 6.295399 | 2.37E-18 | 3.02E-15 |
| C10orf99 | Up-regulated | 1.405349 | 9.630901 | 5.67E-13 | 6.33E-11 |
| PI3 | Up-regulated | 1.359249 | 9.837105 | 2.37E-12 | 2.06E-10 |
| SELE | Up-regulated | 1.318033 | 8.106887 | 3.55E-14 | 6.51E-12 |
| S100A8 | Up-regulated | 1.284684 | 11.63387 | 2.99E-12 | 2.48E-10 |
| LINC01215 | Up-regulated | 1.281348 | 5.859495 | 5.24E-16 | 1.92E-13 |
| TMPRSS4 | Up-regulated | 1.277667 | 7.941901 | 2.88E-16 | 1.27E-13 |
| COL6A5 | Up-regulated | 1.269852 | 8.345767 | 3.99E-12 | 3.08E-10 |
| COL6A6 | Up-regulated | 1.238226 | 8.666585 | 9.70E-12 | 6.36E-10 |
| SPRR2B | Up-regulated | 1.234246 | 10.32728 | 1.44E-10 | 6.08E-09 |
| TCN1 | Up-regulated | 1.223949 | 6.845003 | 3.96E-11 | 2.03E-09 |
| CD1B | Up-regulated | 1.183159 | 7.068287 | 3.71E-12 | 2.92E-10 |
| LCE3D | Up-regulated | 1.180626 | 11.62643 | 9.22E-10 | 2.96E-08 |
| OASL | Up-regulated | 1.166058 | 7.017667 | 4.66E-24 | 1.01E-19 |
| CCL18 | Up-regulated | 1.12904 | 10.45926 | 2.33E-09 | 6.58E-08 |
| IL36G | Up-regulated | 1.106151 | 8.760932 | 9.51E-13 | 9.74E-11 |
| FOSL1 | Up-regulated | 1.10205 | 7.13091 | 2.85E-17 | 2.05E-14 |
| APELA | Up-regulated | 1.100046 | 6.975293 | 2.54E-12 | 2.19E-10 |
| CTA-384D8.35 | Up-regulated | 1.078971 | 6.899799 | 3.76E-21 | 4.07E-17 |
| SELL | Up-regulated | 1.077479 | 6.611732 | 4.14E-16 | 1.57E-13 |
| CXCL10 | Up-regulated | 1.073656 | 7.540534 | 9.86E-07 | 1.21E-05 |
| CH25H | Up-regulated | 1.062305 | 7.836583 | 3.13E-16 | 1.28E-13 |
| PCSK1 | Up-regulated | 1.060591 | 4.703752 | 4.61E-16 | 1.72E-13 |
| SPRR2C | Up-regulated | 1.057754 | 5.470469 | 2.98E-12 | 2.48E-10 |
| IL26 | Up-regulated | 1.051377 | 4.916004 | 5.97E-12 | 4.28E-10 |
| CCR7 | Up-regulated | 1.040097 | 6.782151 | 1.04E-19 | 3.75E-16 |
| SPRR1A | Up-regulated | 1.034975 | 11.39261 | 1.05E-09 | 3.24E-08 |
| LTF | Up-regulated | 1.03336 | 9.690571 | 6.59E-07 | 8.59E-06 |
| CCL22 | Up-regulated | 1.010977 | 9.235341 | 1.67E-13 | 2.33E-11 |
| PRSS53 | Up-regulated | 1.001162 | 8.122699 | 3.57E-17 | 2.27E-14 |
| IL37 | Down-regulated | −1.00327 | 8.700774 | 7.08E-10 | 2.38E-08 |
| TMEM56 | Down-regulated | −1.01936 | 6.964349 | 8.33E-12 | 5.62E-10 |
| ZBTB16 | Down-regulated | −1.06223 | 8.465374 | 9.64E-14 | 1.43E-11 |
| KRT79 | Down-regulated | −1.0656 | 8.43783 | 1.92E-06 | 2.16E-05 |
| ELOVL3 | Down-regulated | −1.116 | 6.431317 | 2.60E-08 | 5.47E-07 |
| HSD11B1 | Down-regulated | −1.27707 | 8.309902 | 5.62E-16 | 2.03E-13 |
| WIF1 | Down-regulated | −1.29782 | 7.133747 | 1.98E-11 | 1.11E-09 |
| THRSP | Down-regulated | −1.30307 | 8.167562 | 1.73E-12 | 1.58E-10 |
| BTC | Down-regulated | −1.34965 | 6.20669 | 5.02E-19 | 9.87E-16 |
| PM20D1 | Down-regulated | −1.53753 | 8.192321 | 3.21E-07 | 4.62E-06 |