Abstract
Monoacylglycerol lipase (MAGL) hydrolyzes monoacylglycerol, producing free fatty acid and glycerol. Although this enzyme has been shown to play important roles in mammal, its potential function in plants remains poorly understood. In a survey of the MAGL genes in Brassica napus, we found tapetal expression of BnaC.MAGL8.a, a homolog of AtMAGL8, results in male sterility in Arabidopsis thaliana. Retarded tapetal PCD and defective pollen wall were observed in the transgenic plants. The tapetal cells became vacuolated at stage 9, and then degenerated at stage 11. Most microspores degenerated with the tapetal cells, and only few pollen grains with an irregular-shaped exine layer were produced in the transgenic plants. Transcriptome analysis identified 398 differentially expressed genes. Most of them are involved in pollen development and stress response. ABORTED MICROSPORES and its downstream pollen wall biosynthesis genes were down-regulated, but genes related with reactive oxygen species homeostasis and jasmonates signaling were up-regulated in the transgenic plants. These results suggest that expression of BnaC.MAGL8.a in tapetum invokes stress response and impairs pollen development. The apparent phenotypic similarity between atgpat1 mutant and BnA9::BnaC.MAGL8.a transgenic plants lead us to propose a role for monoacylglycerol (MAG) in pollen development in Arabidopsis. Our study provides insights on not only the biological function of plant MAGL genes but also the role of MAG in pollen development.
Keywords: Arabidopsis thaliana, Brassica napus, MAGL, male sterility, pollen wall, tapetum, AMS, jasmonates
Introduction
In mammals, triacylglycerol (TAG) breakdown proceeds through sequential hydrolysis reactions catalyzed by adipose tissue triacylglycerol lipase, hormone sensitive lipase, and monoacylglycerol lipase (MAGL) (Labar et al., 2010). In addition to participate in fat mobilization, MAGLs also regulate the endocannabinoid signaling system by controlling the level of 2-arachidonoylglycerol, which is a key messenger of the endocannabinoid system (Labar et al., 2010). The first MAGL gene was cloned from mice (Karlsson et al., 1997), and then many MAGL identifications followed from various species (Karlsson et al., 2001; Heier et al., 2010). MAGLs belong to the alpha/beta hydrolase superfamily, containing the GXSXG motif and the catalytic triad of Ser, Asp, and His.
In Arabidopsis, a total of 16 putative MAGL genes were identified (Kim et al., 2016). MAG hydrolytic activity were measured with 11 recombinant proteins, among which, AtMAGL6 and AtMAGL8 exhibited the highest MAGL activities (Kim et al., 2016). The involvement of AtMAGL8 in lipid mobilization is supported by its preferential expression in developing pollen and germinating seeds, the hydrolytic activity of AtMAGL8 toward the MAG containing eicosenoic acid (20:1), and its oil body associated localization (Kim et al., 2016). AtMAGL3 has two different enzymatic roles: lysophospholipase and monoacylglycerol acyltransferase (Gao et al., 2010; Vijayaraj et al., 2012). It is also named as lysoPL2 after its hydrolytic activity toward lysophosphatidylcholine and protein sequence similarity with lysoPL1 (Gao et al., 2010). AtMAGL3 was proposed to protect the plant from heavy metal ions by replacing oxidized membrane lipids with its protein partner acyl-CoA-binding protein 2 (ACBP2) (Gao et al., 2010).
The tapetum supports pollen development in an altruistic way, providing nutrients, enzymes, lipids and other materials. In the bicellular pollen stage, tapetal cells undergo programmed cell death (PCD) to convert their contents to building blocks for pollen development. Tapetal PCD is regulated by a number of transcription factors including ABORTED MICROSPORES (AMS) (Sorensen et al., 2003; Xu et al., 2010), MALE STERILE 188/MYB103/MYB80 (MS188/MYB103/MYB80) (Higginson et al., 2003; Phan et al., 2012; Xiong et al., 2016) and DYSFUNCTIONAL TAPETUM1 (DYT1) (Feng et al., 2012). Absence of these genes results in premature or retarded tapetal PCD and male sterility. AMS acts as a master regulator in pollen wall development, regulating 8 sporopollenin synthesis genes with MS188 (Xiong et al., 2016; Wang et al., 2018). In addition to transcription factors, reactive oxygen species (ROS) also mediate tapetal PCD. It has been reported that the ROS levels peaked at stage 8 and 9 and then decreased at stage 11 in the rice anther (Hu et al., 2011). Similar ROS patterns were also observed in Arabidopsis, tobacco and tomato (Xie et al., 2014; Yu et al., 2017). RESPIRATORY BURST OXIDASE HOMOLOG E (RBOHE) encodes a NADPH oxidase producing ROS (Xie et al., 2014). Tapetal PCD was delayed in the rbohe mutant, and RBOHE overexpression resulted in premature tapetum degeneration (Xie et al., 2014).
Lipids are important for pollen development. The pollen wall comprises two layers, the exine and the intine (Ariizumi and Toriyama, 2011; Zhang et al., 2016). Lipid derived compounds are the major constitutes of the pollen coat and the exine (Ariizumi and Toriyama, 2011; Zhang et al., 2016). The pollen wall not only protects gamete from environmental stresses, but also plays an important role in pollen-stigma communication (Wang et al., 2017; Nasrallah, 2019). Defective pollen wall frequently results in male sterility. Many lipid related genes are important for pollen wall development, such as MALE STERILITY2 (Aarts et al., 1997; Chen et al., 2011), CYTOCHROME P450 FAMILY 703 SUBFAMILY A POLYPEPTIDE 2 (CYP703A2) (Morant et al., 2007), CYP704B1 (Dobritsa et al., 2009), and ACYL-COA SYNTHETASE 5 (ACOS5) (de Azevedo Souza et al., 2009). Moreover, loss of genes involved in glycerolipid synthesis also leads to male sterility. Glycerol-3-phosphate acyltransferases (GPATs) mediate the production of lysophosphatidic acids from glycerol-3-phosphate and acyl-CoA. gpat1 and gpat6 mutants in Arabidopsis (Zheng et al., 2003; Li et al., 2012) and gpat3 mutant in rice (Men et al., 2017) are characterized with defective tapetum development and male sterility. Knockout of GPAT9 also leads to male gametophytic lethality in Arabidopsis (Shockey et al., 2016).
In this work, we cloned BnaC.MAGL8.a, which encodes a MAG lipase, from an oilseed rape cultivar (Zhongshuang 11). AtMAGL8 is a promising candidate gene related with lipid mobilization, and it is preferentially expressed in developing pollen and germinating seeds. In this study, we used the BnA9 promoter and the cauliflower mosaic virus (CaMV) 35S promoter to drive BnaC.MAGL8.a expression in Arabidopsis to explore its potential biological function. Though the 35S::BnaC.MAGL8.a transgenic plants exhibited similar phenotype as wild type, overexpression of BnaC.MAGL8.a in tapetum impaired pollen development. The tapetal cells became vacuolated at stage 10 and degenerated with microspores at later stages in the BnA9::BnaC.MAGL8.a transgenic plants. Transcriptome analysis uncovered 398 differentially expressed genes in comparison with the wild type, mainly involved in pollen development and stress response. Based on these results, we discussed the potential mechanism underlying male sterility in the transgenic plants.
Materials and Methods
Plant Materials and Growth Condition
Zhongshuang 11 (ZS 11), a Brassica napus cultivar, was grown in the experimental field of Huazhong Agricultural University, Wuhan, China. Wild-type Arabidopsis thaliana Columbia (ecotype Col-0) and BnA9::BnaC.MAGL8.a transgenic plants were grown in a greenhouse (8 h dark and 16 h light, 23°C).
Protein Sequence Alignment
The deduced amino acid sequence of AtMAGL8 and its four homologous genes were aligned using the ClustalW method in MEGA 7 (Kumar et al., 2016).
Vector Construction and Plant Transformation
Total RNA was extracted from ZS 11 flower buds using RNeasy Plant Mini Kit (Qiagen) and reverse transcription was performed using RevertAid First Strand cDNA Synthesis Kit from Thermo Scientific. Then, the coding sequence of BnaC.MAGL8.a was amplified with BnaC.MAGL8.a_LP and BnaC.MAGL8.a_RP. The BnA9 promoter was amplified from ZS 11 genomic DNA with BnA9_LP and BnA9_RP. The NOS terminator was amplified using NosT_LP and NosT_RP from the pCAMBIA2300 vector. Primers used in this study are listed in Supplementary Table S1. The produced fragments were assembled in the pCAMBIA2300 vector. Then, the vector was transferred into Agrobacterium tumefaciens strain GV3101 and transformed into Arabidopsis via the floral dip method (Zhang X. et al., 2006).
Recombinant Protein Purification and in vitro Enzyme Assay
The coding sequences from BnaC.MAGL8.a and AtMAGL8 were inserted into the pMAL-c5X vector from the MBP purification system kit (NEB). After confirmed by sequencing, the vectors were introduced into E. coli strain ER2523. Protein purification and western blot were performed according to the manufacturer’s instructions. After dialysis against 50 mM phosphate sodium solution (pH 8.0), the maltose binding protein (MBP):MAGL recombinant proteins were quantified using Bradford protein assay kit (Tiangen). The recombinant proteins were incubated with the substrates (Supplementary Table S2), MAGs with various fatty acids, including palmitic acid (16:0), palmitoleic acid (16:1), stearic acid (18:0), oleic acid (18:1), and linoleic acid (18:2), at sn-1 position and the MAG with 18:1 at sn-2 position in 100 μl reaction solution containing 50 mM sodium phosphate buffer (pH 8.0), 0.2% Triton X-100 (Kim et al., 2016). All substrates were bought from Sigma-Aldrich and were emulsified in 0.2% Triton X-100 solution before use. After 30 min incubation, the reaction was stopped by 90°C treatment for 5 min and the released free fatty acid was measured by NEFA kit (Wako).
Alexander’s Staining of Pollen
Anthers were stained with Alexander solution (Alexander, 1969) and examined microscopically.
Light and Electron Microscopy
The semi-thin sections were prepared and analyzed as previously described (Chen et al., 2009). Transmission electron microscopy (TEM) analysis and scanning electron microscopy (SEM) analysis were conducted as previously described (Yi et al., 2010).
Transcriptome Analysis
Total RNA was extracted from young flower buds (stage 6–9) with RNAprep pure Plant Kit (Tiangen) from the wild type and transgenic lines. Three biological replicates for the wild type and BnA9::BnaC.MAGL8.a plants were included. Libraries were sequenced with HiSeq X Ten system. Hisat2 was used to map the reads to the Arabidopsis genome (Kim et al., 2015). During this process, BnaC.MAGL8.a coding sequence was added to the reference genome to get its expression value. Raw read counts were estimated by featureCount, and differentially expressed genes was detected via DESeq2 using P-value <0.01 as the threshold (Liao et al., 2014; Love et al., 2014). The data was deposited in GEO database (GSE124918). The Gene Ontology classification was conducted using GO annotation tool from TAIR1 (Berardini et al., 2004). The MAPMAN annotation was conducted according to the user’s manual (Thimm et al., 2004). DAVID was used for functional classification and KEGG pathway enrichment (Huang et al., 2009).
Real-Time PCR
Total RNA was extracted from flower buds (stage 6–9) from the wild type and BnA9::BnaC.MAGL8.a plants. First-strand cDNA was synthesized using RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher). Real-time PCR was performed using TOYOBO SYBR GREEN mix and Bio-Rad cycler. Experiments were conducted in triplicate. Primers were listed in Supplementary Table S1. PP2AA3 (PROTEIN PHOSPHATE 2A SUBUNIT 3, AT1G13320) was used for standardization (Singer et al., 2016) and the relative expression levels were calculated with CFX Manager Software (Bio-Rad) according to the 2−ΔΔCt method.
Results
Discovery of the BnA9::BnaC.MAGL8.a Male Sterile Plants
To explore the MAGL gene family in Brassica napus, 47 genes were identified from the Darmor-bzh genome database (Chalhoub et al., 2014) based on the protein sequence similarity with 16 AtMAGLs (Supplementary Figure S1). We have successfully cloned 23 genes. Then, these genes were inserted into an ectopic expression vector containing CaMV35S or BnA9 promoter and introduced into Arabidopsis. Stable transgenic lines harboring 15 genes were obtained (Supplementary Table S3). The CaMV35S promoter is commonly used for ectopically expressing genes in transgenic plants, but it does not drive a sufficiently high level of expression in the tapetum. BnA9 promoter drives gene expression in tapetum from stage 6 to stage 11 (Konagaya et al., 2008; Song et al., 2016). Lipid metabolism plays an important role in pollen development. To explore the affect of ectopic expression of MAGL genes in pollen development, BnA9 promoter were used to drives tapetum specific gene expression. Among the transgenic lines, BnA9::BnaC.MAGL8.a and BnA9::BnaA.MAGL10.a exhibited male sterility (Supplementary Table S3 and Supplementary Figure S2). As we were unable to purify MBP:BnaA.MAGL10.a recombinant protein for enzyme assay, this study is focused on the BnA9::BnaC.MAGL8.a transgenic plants.
In the BnA9::BnaC.MAGL8.a plants, the siliques were stunt and failed to set seeds (Figure 1A,B). Among the 12 BnA9::BnaC.MAGL8.a transgenic lines, 9 showed similar male-sterile phenotypes (Supplementary Table S4). The anthers from transgenic plants turned brown and no pollen grains were observed on the stigma or style (Figure 1D). In contrast, abundant pollen grains were observed in the wild type (Figure 1C). Alexander’s staining of anthers from the transgenic plants revealed that degenerated pollen grains adhered with round pollen grains in the locule (Figure 1E,F). Pollinating the BnA9::BnaC.MAGL8.a plants with wild type pollen grains led to normal siliques, suggesting the male sterility is caused by defective pollen grains (Supplementary Table S4).
FIGURE 1.
Phenotype of the BnA9::BnaC.MAGL8.a plants. (A) The wild type plant with long siliques. (B) The BnA9::BnaC.MAGL8.a plant with stunted siliques. Open flowers from the wild type (C) and the BnA9::BnaC.MAGL8.a plants (D). The anthers of transgenic plants turn brown. Bar = 1 mm. Alexander’s staining of anthers from the wild type (E) and the BnA9::BnaC.MAGL8.a plants (F). Bar = 100 μm.
BnaC.MAGL8.a Is a Homologous Gene of AtMAGL8 and Encodes a MAG Lipase
Based on phylogenetic analysis (Supplementary Figure S1), there are four homologous genes of AtMAGL8 in Brassica napus, designated as BnaC.MAGL8.a, BnaC.MAGL8.b, BnaA.MAGL8.a, and BnaA.MAGL8.b. Alignment of the deduced protein sequences from these genes with AtMAGL8 indicated that the GXSXG motif and the catalytic triad are conserved (Figure 2). The BnaC.MAGL8.a shows the highest identity (89%) with AtMAGL8. BnaC.MAGL8.b, BnaA.MAGL8.a, and BnaA.MAGL8.b exhibit 87.7, 87.7, and 80% identity with AtMAGL8, respectively.
FIGURE 2.
Alignment of deduced amino acid sequences of AtMAGL8 and four homologous genes. These deduced amino acid sequences were aligned by ClustalW in MEGA 7. The identical and conserved amino acids residues are shaded in black and gray. The Gly-X-Ser-X-Gly (X = any residue) lipase motif is marked with red box and the catalytic triad (Ser, Asp, and His) is highlighted with blue arrows.
In vitro enzyme assay indicated the MBP:BnaC.MAGL8.a recombinant protein possessed biochemical properties similar to MBP:AtMAGL8. The recombinant proteins were purified with amylose resin (Supplementary Figure S3). The production rate of non-esterified fatty acid increased with the MBP:BnaC.MAGL8.a recombinant protein content in the reaction and reached a maximum rate with 1.4 μg or more recombinant protein (Figure 3A), suggesting that BnaC.MAGL8.a and AtMAGL8 have a similar hydrolytic activity of MAG (Kim et al., 2016). We further investigated regio-specificity and substrate specificity of MBP:BnaC.MAGL8.a to different MAG species. Same as MBP:AtMAGL8, the MBP:BnaC.MAGL8.a recombinant protein preferred MAG with 18:1 at sn-1 position to sn-2 position. In addition, MBP:BnaC.MAGL8.a also showed a higher hydrolase activity with MAG containing unsaturated fatty acid (16:1) than saturated fatty acid (16:0) (Figure 3B). Hence, MBP:BnaC.MAGL8.a recombinant protein showed a similar regio-specificity and substrate specificity as MBP:AtMAGL8.
FIGURE 3.

In vitro enzymatic assays of MBP:BnaC.MAGL8.a recombinant protein. (A) Protein-dependent MAGL assay were conducted with various amounts of recombinant protein (0.03–0.18 μg). The recombinant protein was incubated with 400 μM MAG containing 18:2 at the sn-1 position. (B) Substrate-dependent MAGL assay were conducted with 0.1 μg recombinant protein. The substrates were added to 400 μM in the reactions and released non-esterified fatty acid (NEFA) were measured using a NEFA assay kit. The values are means ± standard errors of three independent experiments.
Defective Pollen Development in the BnA9::BnaC.MAGL8.a Plants
To unravel the mechanism of defective pollen formation, sections of anthers from the wild type and BnA9::BnaC.MAGL8.a plants were examined under light microscope. Normal microsporogenesis was observed in the transgenic plants (Figure 4A,B,G,H and Supplementary Figures S4A,E). In the BnA9::BnaC.MAGL8.a plants, microspores were released from the tetrads as the wild type at stage 8 (Figure 4H and Supplementary Figure S4E). At stage 9, the wild type tapetal cells initiated programmed cell death, evidenced by their reduced size (Figure 4C and Supplementary Figures S4B,C). However, in the transgenic plants, the tapetum became vacuolated and abundant small vacuoles were presented in the tapetal cells (Figure 4I and Supplementary Figure S4F). In addition, some of the microspores were degraded at this stage (Figure 4I and Supplementary Figure S4G). At stage 11, tapetum was degenerated in the wild type (Figure 4D,E and Supplementary Figure S4D). However, vacuolated tapetal cells and microspores filled the locules and large vacuoles were present in the tapetum of transgenic plants (Figure 4J and Supplementary Figure S4H). Some of the tapetal cells were degraded, leaving cell debris among the microspores. Most microspores were degraded, presented as empty exine ring in the section, and only a few still viable (Figure 4J). At stage 12, tapetal PCD were completed and pollen grains were freely distributed in the wild type (Figure 4E). In the BnA9::BnaC.MAGL8.a plants, the degenerated pollen grains were attached to normal ones and all of them appeared stuck to the locule wall (Figure 4K). At stage 13, only pollen debris and irregular pollen grains could be observed in the BnA9::BnaC.MAGL8.a plants (Figure 4L), when mature pollen grains were observed in the locules of wild type (Figure 4F).
FIGURE 4.
Transverse semi-thin sections of anthers from the wild type and BnA9::BnaC.MAGL8.a plants. (A–F) Anther from the wild type at stage 7 (A), stage 8 (B), stage 9 and 10 (C), stage 11 (D), stage 12 (E), and stage 13 (F); (G–L): anther from the BnA9::BnaC.MAGL8.a plants at stage 7 (G), stage 8 (H), stage 9 and 10 (I), stage 11 (J), stage 12 (K), and stage 13 (L). BnM8, BnA9::BnaC.MAGL8.a; dPG, degenerated pollen grain; E, epidermis; En, endothecium; Msp, microspore; PG, pollen grain; T, tapetum; Tds, tetrads; WT, the wild type. Bars = 25 μm.
Transmission electron microscope analysis was employed to examine the defective pollen development at subcellular level. In the wild type, layers of ER were presented beneath the plasma membrane facing the locule, indicating an active secretory function; in addition, the cell walls were degraded (Figure 5A). In the BnA9::BnaC.MAGL8.a plants, the outer tangential cell walls of tapetal cells were degraded and layers of ER were also observed (Figure 5B), but the cell wall between adjacent tapetal cells remained visible till stage 9 (Figure 5D). At stage 9, the wild type tapetum contained large vacuoles as well as many small vesicles, releasing fibrillar materials into the locule (Figure 5C). In the BnA9::BnaC.MAGL8.a plants, in addition to the large vacuoles, a great number of small vesicles were present in the tapetal cells, giving them a spongy appearance (Figure 5D). At stage 10, elaioplasts and tapetosomes were observed in the wild type and BnA9::BnaC.MAGL8.a plants (Figure 5E,F). Some of the tapetal cells were degenerated in the transgenic plants, leaving cell remnants in the locule (Figure 5F). Degenerated pollen grains were observed in the transgenic plants, featured with detached cytoplasm (Figure 5F).
FIGURE 5.
Transmission electron micrographs of the tapetum. (A,C,E) Tapetal cells from the wild type at stage 8 (A), stage 9 (C), and stage 10 (E); (B,D,F): tapetal cells from the BnA9::BnaC.MAGL8.a plants at stage 8 (B), stage 9 (D), and stage 10 (F). BnM8, BnA9::BnaC.MAGL8.a; CW, cell wall; dPG, degenerated pollen grain; dT, degenerated tapetum; El, elaioplast; ER, endoplasmic reticulum; Ex, exine; In, intine; Mt, mitochondria; N, nucleus; Ts, tapetosome; V, vacuole; WT, the wild type. Arrow in (C) points to the tapetal cell releasing fibrillar materials. Arrow in (D) points to the small vesicle inside the tapetal cells. Bars = 2 μm.
Pollen wall formation was also affected in the BnA9::BnaC.MAGL8.a plants, especially the exine. Although some pollen grains from BnA9::BnaC.MAGL8.a appeared normal at stage 10, others were mostly degenerated in the locule (Figure 6B). At stage 11, the wild type pollen accumulated lipid droplets and starch granules (Figure 6C). On the other hand, large vacuoles presented in the pollen cytoplasm and the defect in exine became obvious in transgenic plants (Figure 6D). Furthermore, round pollen grains and degenerated pollen grains were attached to each other in the locule (Figure 6D). As the tapetal PCD was perturbed in the transgenic lines, limited pollen coat was uploaded onto the pollen wall (Figure 6F). Finally, after tapetum and pollen degradation, the collapsed pollen grains and tapetum remnants attached to the locule (Figure 6F). Notably, the locule fluid in the transgenic lines contained more fibrillar materials than the wild type at stage 10 and stage 11 (Figure 6B,D), which may came from degenerated tapetum and caused pollen grains aggregation.
FIGURE 6.

Transmission electron micrographs of microspores and pollen grains. (A,C,E) Microspores and pollen grains from the wild type in stage 10 (A), stage 11 (C), and stage 13 (E); (B,D,F) microspores and pollen grains from the BnA9::BnaC.MAGL8.a plants in stage 10 (B), stage 11 (D), and stage 13 (F). BnM8, BnA9::BnaC.MAGL8.a; dPG, degenerated pollen grain; dT, degenerated tapetum; Ex, exine; In, intine; PC, pollen coat; PG, pollen grain; VN, vegetative nucleus; WT, the wild type. Arrows in (A,B,C,D) point to the fibrillar materials in the locule. Bars = 2 μm.
Scanning electron microscopy analysis showed defective pollen wall in shrunken pollen grains. The exine was practically smooth and had small or no lacunae (Figure 7B). In addition, no intine or cytoplasm could be observed under the exine layer (Figure 7D). The round pollen grains were decorated with intine, exine and pollen coat (Figure 7E), and storage organelles were presented in the cytoplasm. However, the round pollen grains showed defective exine structure and limited pollen coat (Figure 7E).
FIGURE 7.
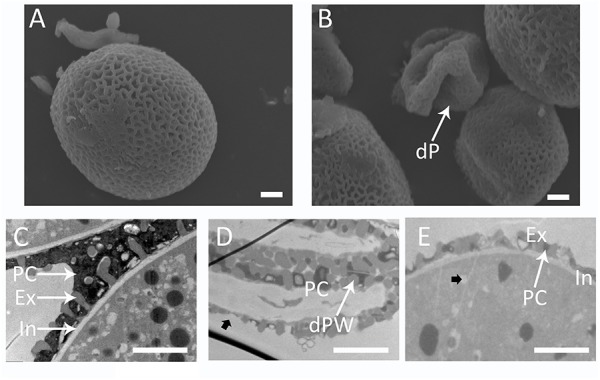
Defective pollen wall in the BnA9::BnaC.MAGL8.a plants. Scanning electron micrographs of pollen grains from the wild type (A) and the BnA9::BnaC.MAGL8.a plants (B). Transmission electron micrographs emphasizing pollen wall of the wild type pollen (C), and shrunken pollen grains (D) and round pollen grains (E) from the BnA9::BnaC.MAGL8.a plants. dP, degenerated pollen; dPW, degenerated pollen wall; Ex, exine; In, intine; PC, pollen coat. Arrow in (D) points to the thin exine layer in degenerated pollen. Arrow in (E) points to a storage organelle in the pollen cytoplasm. Bars = 2 μm.
Transcriptome Analysis of the BnA9::BnaC.MAGL8.a Flower Buds
To assess the genes associated with male sterility in the BnA9::BnaC.MAGL8.a plants, transcriptome analysis was performed with young flower buds (stage 6–9) from the wild type and transgenic plants. In addition to BnaC.MAGL8.a, 397 differentially expressed genes (DEGs) were identified, composing 157 up-regulated genes and 240 down-regulated genes (Supplementary Table S5). The result of transcriptome analysis was validated with real-time PCR conducted on 6 selected genes (Supplementary Figure S5). Then, the DEGs were annotated with GO and MAPMAN (Berardini et al., 2004; Thimm et al., 2004). GO enrichment analysis showed that up-regulated genes were mainly enriched in response to stress, while down-regulated genes were mainly involved in developmental processes (Table 1). KEGG pathway enrichment analysis indicated that the DEGs were mainly localized in four pathways: biosynthesis of secondary metabolites, glucosinolate biosynthesis, phenylpropanoid biosynthesis and 2-oxocarboxylic acid metabolism (Table 2).
Table 1.
Gene Ontology classification of DEGs.
| GO term Biological process | Number of differentially expressed genes |
||
|---|---|---|---|
| Total | Down-regulated | Up-regulated | |
| Response to stress | 86 | 41 | 45 |
| Response to abiotic or biotic stimulus | 75 | 35 | 40 |
| Developmental processes | 58 | 44 | 14 |
| Cell organization and biogenesis | 53 | 34 | 19 |
| Protein metabolism | 47 | 33 | 14 |
| Transcription, DNA-dependent | 44 | 31 | 13 |
| Transport | 40 | 25 | 15 |
| Signal transduction | 30 | 23 | 7 |
| DNA or RNA metabolism | 6 | 5 | 1 |
| Electron transport or energy pathways | 6 | 2 | 4 |
| Other cellular processes | 202 | 121 | 81 |
| Other metabolic processes | 192 | 114 | 78 |
| Other biological processes | 66 | 34 | 32 |
| Unknown biological processes | 61 | 40 | 21 |
Table 2.
Enriched KEGG pathways for differentially expressed genes.
| KEGG_ID | Pathway name | Count | Total | % | Fold enrichment | P-value | |
|---|---|---|---|---|---|---|---|
| Up regulated | ath01110 | Biosynthesis of secondary metabolites | 20 | 38 | 12.99 | 2.34 | 0.0001 |
| ath00966 | Glucosinolate biosynthesis | 3 | 38 | 1.95 | 31.47 | 0.0037 | |
| ath00940 | Phenylpropanoid biosynthesis | 6 | 38 | 3.90 | 4.81 | 0.0067 | |
| ath01210 | 2-Oxocarboxylic acid metabolism | 4 | 38 | 2.60 | 6.81 | 0.0189 | |
| Down regulated | ath00940 | Phenylpropanoid biosynthesis | 6 | 38 | 2.50 | 4.81 | 0.0067 |
Notably, six up-regulated genes and six down-regulated genes are involved in the phenylpropanoid biosynthesis pathway (Supplementary Table S6 and Supplementary Figure S6). The up-regulated genes are PEROXIDASE 15 (PRX15), PRX22, PRX49, PRX52, PRX72, and AT5G20940 (glycosyl hydrolase family protein). PRX49, PRX52, and PRX72 participate in lignin synthesis progress and their up-regulated expression suggested an enhancement in lignin formation (Herrero et al., 2013a,b; Fernandez-Perez et al., 2015a,b). CYP98A8 and TAPETUM-SPECIFIC METHYLTRANSFERASE 1 (TSM1) are involved in spermidine synthesis, which is an important constituent for the pollen wall. The down-regulation of CYP98A8 and TSM1 in transgenic plants indicated that the phenylpropanoid polyamine conjugates synthesis was repressed in the transgenic plants.
Phenylpropanoid biosynthesis and glucosinolate biosynthesis are regulated by jasmonates signals as wound responses (Gaquerel et al., 2014; Wasternack and Strnad, 2019). VEGETATIVE STORAGE PROTEIN1 (VSP1), a jasmonates response gene, was induced in transgenic plants. We compared our DEGs list with jasmonates regulated genes (JRGs) and 12-oxo-phytodienoic acid (OPDA) regulated genes (ORGs) (Taki et al., 2005). In total, 26 JRGs and five ORGs were differentially expressed in the transgenic plants (Supplementary Table S7). Of the 26 JRGs, 23 were also induced in wound response (Taki et al., 2005). Moreover, 11 of them were up-regulated in jasmonates treated stage 12 flowers (Supplementary Table S7) (Mandaokar et al., 2006). Based on this result, we propose that jasmonates, instead of OPDA, induced stress response in the transgenic plants.
Reactive oxygen species homeostasis genes showed altered expression in the transgenic plants. 19 ROS homeostasis genes were differentially expressed in the transgenic plants, and only three of them were down-regulated (Supplementary Table S8). Six ROS scavengers were up-regulated, including FERRITIN 1 (FER1), FER3, FER4, GLUTAREDOXIN 5, THIOREDOXIN-DEPENDENT PEROXIDASE 2 (TPX2), and FE SUPEROXIDE DISMUTASE 1 (Mittler et al., 2004). FER1, FER3, and FER4 function as a link between iron homeostasis and oxidative stress (Briat et al., 2010). As free iron induces harmful free radical production via the Fenton reaction, ferritins reduce ROS production by binding free irons in the cytoplasm (Ravet et al., 2009). GLUTATHIONE S-TRANSFERASE PHI 9 and GLUTATHIONE S-TRANSFERASE TAU 20 also reduce ROS levels (Noctor et al., 2011). The up-regulation of ROS scavenger genes suggested an unbalanced ROS homeostasis and activated ROS response in the transgenic plants.
Histology analysis indicated that pollen wall development was impaired in the transgenic plants. Consistent with this, 21 down regulated genes are among the 98 genes essential for pollen wall development (Table 3; Xu et al., 2014). As a master regulator in pollen wall formation, AMS directly associate with the promoters of QUARTET3 (QTR3), 3-KETOACYL-COA SYNTHASE7 (KCS7), KCS21, CYP98A8, CYP704B1, and EXTRACELLULAR LIPASE 5 (EXL5) (Xu et al., 2014). In the BnA9::BnaC.MAGL8.a plants, AMS and its six down-stream genes were down-regulated (Table 3). Moreover, AMS, working with MS188, regulates eight sporopollenin biosynthesis genes during pollen wall formation, including ACOS5, CYP703A2, CYP704B1, MS2, POLYKETIDE SYNTHASE A (PKSA), PKSB, TETRAKETIDE α-PYRONE REDUCTASE1 (TKPR1), and TKPR2 (Wang et al., 2018). According to transcriptome analysis result, only CYP704B1 was significantly down-regulated, but all of them tend to express at lower levels in the BnA9::BnaC.MAGL8.a transgenic plants (Supplementary Table S9). Real-time PCR indicated that all of the eight genes were differentially expressed in the wild type and transgenic plants. CYP703A2, PKSA, and TKPR1 were up-regulated and CYP704B1, ACOS5, MS2, PKSB, and TKPR2 were down-regulated in the transgenic plants. Although the real-time PCR result and transcriptome analysis result are not perfectly matched, they suggested that CYP704B1 and several sporopollenin biosynthesis genes were down regulated in transgenic plants and may lead to the irregular exine layer.
Table 3.
Down-regulated gene involved in pollen wall formation.
| TAIR IDa | Name | Description | Log2foldchange | P-value |
|---|---|---|---|---|
| AT2G16910 | AMS | bHLH transcription factor | −0.76 | 0.0004 |
| AT3G26125 | CYP86C2 | Cytochrome P450 86C2 | −1.58 | 0.0001 |
| AT1G69500 | CYP704B1b | Cytochrome P450 704B1 | −1.54 | 0.0013 |
| AT1G28430 | CYP705A24 | Cytochrome P450 705A24 | −1.53 | 0.0012 |
| AT1G75920 | EXL5b | Family II extracellular lipase 5 | −1.44 | 0.0002 |
| AT5G53190 | SWEET3 | Nodulin MtN3 family protein | −1.36 | 0.0049 |
| AT1G33430 | KNS4/UPEX1 | Galactosyltransferase family protein | −1.28 | <0.0001 |
| AT4G14815 | Lipid transfer protein family protein | −1.26 | <0.0001 | |
| AT1G22015 | DD46 | Galactosyltransferase | −1.04 | 0.0042 |
| AT4G29250 | Transferase family protein | −1.00 | 0.0001 | |
| AT5G07530 | GRP17 | Glycine-rich protein 17 | −0.99 | 0.0012 |
| AT5G16960 | NADP-dependent oxidoreductase, putative | −0.92 | 0.0001 | |
| AT1G26710 | Transmembrane protein | −0.90 | 0.0038 | |
| AT1G71160 | KCS7b | β-Ketoacyl-CoA synthase 7 | −0.90 | 0.0005 |
| AT4G20050 | QRT3b | Polygalacturonase | −0.87 | 0.0003 |
| AT1G74540 | CYP98A8b | Cytochrome P450 98A8 | −0.74 | 0.0066 |
| AT1G75940 | ATA27 | Hydrolase, hydrolyzing O-glycosyl compounds | −0.69 | 0.0020 |
| AT1G61110 | NAC25 | NAC transcription factor related with GA signal | −0.68 | 0.0037 |
| AT5G49070 | KCS21b | β-Ketoacyl-CoA synthase 21 | −0.57 | 0.0049 |
| AT5G48210 | Unknown protein | −0.57 | 0.0077 | |
| AT1G67990 | TSM1 | Tapetum-specific O-methyltransferase | −0.56 | 0.0096 |
aLocus identifier from TAIR (www.arabidopsis.org). bGenes regulated by AMS (Xu et al., 2014).
Discussion
Since the discovery of mice MAGL in 1960, many other species have been proved to have multiple MAGL genes (Karlsson et al., 1997; Labar et al., 2010; Li-Beisson et al., 2013; Kim et al., 2016). MAGLs not only participate in fat mobilization, but also play important roles in endocannabinoid signaling in mammals (Labar et al., 2010). As a widespread gene family, the MAGL genes may be involved with many biological processes in plants. In Arabidopsis, 16 MAGL genes were characterized biochemically and molecularly (Kim et al., 2016). However, besides AtMAGL3, the potential functions of the MAGL genes have not been investigated in mutants or transgenic lines (Gao et al., 2010; Vijayaraj et al., 2012; Vanholme et al., 2013). In this study, we showed that expressing BnaC.MAGL8.a in tapetum affected tapetal PCD and microspore development, leading to male sterility in Arabidopsis.
Retarded Tapetal PCD Was Correlated With Altered Expression of AMS and ROS Homeostasis Genes in the BnA9::BnaC.MAGL8.a Plants
An obvious difference between the BnA9::BnaC.MAGL8.a plants and the wild type is the vacuolated tapetum. The tapetum of transgenic plants contained many small vacuoles at stage 9 and 10. Meanwhile, the cytoplasm of several microspores receded from their pollen wall. The tapetum vacuolation became more severe at stage 11. Tapetal PCD is controlled by DYT1, AMS, MALE STERILE1 and other transcription factors (Zhang W. et al., 2006; Ito et al., 2007; Yang et al., 2007; Xu et al., 2010). Their mutation caused defective tapetal PCD and tapetum vacuolation. The reduced AMS expression in the BnA9::BnaC.MAGL8.a plants was confirmed by transcriptome analysis and real-time PCR. DYT1, AMS, and MS188 constitute a regulation pathway tuning tapetal PCD (Wang et al., 2018). The real-time PCR result indicated that MS188 was significantly down-regulated in the transgenic plants and DYT1 showed similar expression level in the wild type and transgenic plants. BASIC HELIX LOOP HELIX PROTEIN 10 (bHLH010), bHLH089, and bHLH091 are needed for DYT1 to regulate anther development (Zhu et al., 2015). bHLH010, bHLH089, and bHLH091 were significantly down-regulated in the transgenic plants, confirmed by the real-time PCR analysis. Moreover, down-regulated bHLH091 expression was also supported by transcriptome analysis result (Supplementary Table S9). The down-regulation of bHLH010, bHLH089 and bHLH091 may repressed AMS expression.
In addition to transcription factors, tapetal PCD is also related with the ROS levels (Hu et al., 2011; Xie et al., 2014; Yu et al., 2017). The similar developmental changes in ROS profiles were found in Arabidopsis, tobacco and tomato (Xie et al., 2014; Yu et al., 2017). In addition, the manipulation of RESPIRATORY-BURST OXIDASE HOMOLOG E (RBOHE) changed anther ROS levels and resulted in defective tapetal PCD (Xie et al., 2014). BnaC.MAGL8.a may produce excessive free fatty acid (FFA) in the tapetum, same as AtMAGL3 hydrolyzing membrane lipids (Gao et al., 2010). There are two possible ways for FFA to affect ROS homeostasis, in the form of FFA or lipid derived oxylipins (Bonaventure, 2014). FFA may interact with respiratory chain, change the mitochondrial membrane fluidly and oxidizes glutathione inside the mitochondria (Schonfeld and Wojtczak, 2008). Under stress conditions, FFA could be transformed into oxylipins, including OPDA and jasmonates (Wasternack, 2007; Wasternack and Strnad, 2019). Based on our transcriptome data, large numbers of jasmonates-regulated genes were differentially expressed in the transgenic plants, suggesting that it is jasmonates, not OPDA, caused altered genes expression. TPX2 and GLUTATHIONE S-TRANSFERASE PHI 9 are regulated by jasmonates and function as ROS scavengers. The up-regulated ROS homeostasis genes in transgenic plants supported a ROS levels change in the transgenic plants. Notably, RBOHE is regulated by AMS and MS188. Hence, the regulator in tapetal PCD may initiate PCD process via regulating ROS levels (Xie et al., 2014). FFA, jasmonates and AMS regulated ROS levels in transgenic plants and the change in ROS level may also lead to a delayed tapetal PCD.
Defective Pollen Wall Formation Was Correlated With Reduced Sporopollenin Synthesis Genes Expression in the BnA9::BnaC.MAGL8.a Plants
Many sporopollenin synthesis and pollen wall formation related genes were down-regulated in the transgenic plants (Table 3). The sporopollenin precursor is synthesized in the tapetum and then transported to the surface of microspores for exine wall formation. The sporopollenin is mainly composed of biopolymers derived from fatty acids and phenolic compounds (Ariizumi and Toriyama, 2011; Quilichini et al., 2015; Zhang et al., 2016). CYP704B1 mediates long chain fatty acids hydroxylation (Dobritsa et al., 2009). ACOS5, PKSA, PKSB, and TKPR1 are localized in the ER of tapetal cells and also in the locule (Grienenberger et al., 2010; Kim et al., 2010; Wang et al., 2018). It was proposed that these genes function as a metabolon for sporopollenin synthesis (Lallemand et al., 2013). Our real-time PCR result showed that ACOS5, CYP704B1, MS2, TKPR2, and PKSB were down-regulated in the transgenic plants (Supplementary Table S9), indicating a reduced sporopollenin synthesis. In phenylpropanoid biosynthesis pathway, CYP98A8 and TSM1 are involved in phenylpropanoid spermidine conjugates synthesis (Fellenberg et al., 2009) and the peroxidase genes, PRX49, PRX52, and PRX72, participate in lignin synthesis. The down-regulated CYP98A8 and TSM1 and up-regulated PRX49, PRX52, and PRX72 in the transgenic plants may drive more phenylpropanoid compounds to lignin synthesis and reduced spermidine compounds production. The expression changes in sporopollenin related genes may resulted in irregular exine structure in the transgenic plants.
As the tapetal PCD was delayed in the transgenic lines, only limited pollen coat was loaded onto the pollen wall. In consistency with this, transcriptome analysis showed that some pollen coat related genes were down-regulated in the transgenic plants. GRP17 is the most abundant pollen coat protein in Arabidopsis and EXL5 is characterized as an extra cellular lipase in the pollen coat (Mayfield and Preuss, 2000; Mayfield et al., 2001). Down-regulation of GRP17 and EXL5 during early pollen stage may also affect pollen coat formation. Tunicamycin induced 1 mutant shows altered pollen surface structure and the pollen grains are sticky (Iwata et al., 2012). Down-regulation of TUNICAMYCIN INDUCED 1 may contribute to the defective pollen wall in transgenic plants (Supplementary Table S5).
In the BnA9::BnaC.MAGL8.a plants, the sticky pollen grains may caused by the defective pollen wall and the thick locule fluid. Mutation of sporopollenin synthesis genes results defective pollen grains. cyp704b1 produces sticky pollen grains (Dobritsa et al., 2011). cyp703a2 and tkpr1 are characterized with thin-exine pollen wall (Dobritsa et al., 2011). The repressed sporopollenin synthesis genes expression correlated with thin and destructed exine layer in transgenic plants. As tapetum degenerated with microspores during stage 10 and stage 11, the locule fluid contained cell debris and cellular components, indicated as fibrillar materials in Figure 6. This change promoted pollen grains adhesion. As the microspores from BnA9::BnaC.MAGL8.a plants coalesced with each other in stage 11, when the pollen coat substrates was still enclosed in tapetum, the sticky pollen phenotype was more likely caused by defective pollen wall (Figure 6D). This notion was further supported by the fact that limited pollen coat was loaded on round or shrunken pollen grains (Figure 7).
The Potential Role of MAG in Pollen Development
At stage 8, microspores are separated from the tapetum and obtain nutrients from the locule fluid. The exact composition of the locule fluid remains poorly understood. MAG, as a transportable metabolite, may serve as a locule fluid component. Arburscular mycorrhizal fungi and its host plant evolve a symbiosis relationship: the fungi transports mineral nutrients to the plant and receives MAGs and sugars as feedback (Jiang et al., 2017; Luginbuehl et al., 2017). ram2 mutant in Medicago truncatula is defective in mycorrhizal fungal colonization and addition of C16 fatty acids complements the mutation (Wang et al., 2012). Further analysis indicated that RAM2 encodes a GPAT synthesizing 2-monoacylglycerol, which likely exported to the fungi (Wang et al., 2012). Pollen development requires plenty of lipids (Zhang et al., 2016). As a high concentration of free fatty acid is toxic to plant cells, transporting MAG from tapetum could be a safe and efficient way to deliver lipid derived compounds to microspores. In the atgpat1 mutant, the tapetum became hypertrophic and even occupied half of the locule (Zheng et al., 2003). Then, the microspores and tapetum degenerated, leaving collapsed pollen walls and several round pollen grains attached to the locule (Zheng et al., 2003). The similar phenotype of the atgpat1 mutant and the BnA9::BnaC.MAGL8.a transgenic plants suggested a common mechanism for defect pollen development. We speculate that MAG could be the link, as the mutation of AtGPAT1 and the BnaC.MAGL8.a overexpression may cause a MAG shortage. Further investigation is needed to prove the importance of MAG for microspore development.
Author Contributions
JG, JZ, and JS contributed to the conception and design of the study. JG, QL, NW, and BT conducted the plant transformation and histology analysis. JG performed the enzyme assay and transcriptome analysis. JG wrote the first draft of the manuscript. BY, CM, JT, TF, and QL provided the valuable suggestions for this research. All authors contributed to the manuscript revision, read and approved the submitted version.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Kaining Hu for his help in transcriptome analysis. We also thank Nesma Shalby and Fawad Khan for revising the manuscript. We greatly acknowledge the reviewers for their valuable comments.
Funding. This work was supported by the National Natural Science Foundation of China (Grant No. 31871654), the National Key Research and Development Program of China (Grant No. 2016YFD0101300), and the Program for Modern Agricultural Industrial Technology System (Grant No. CARS-12).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00763/full#supplementary-material
References
- Aarts M. G., Hodge R., Kalantidis K., Florack D., Wilson Z. A., Mulligan B. J., et al. (1997). The arabidopsis male sterility 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J. 12 615–623. 10.1046/j.1365-313x.1997.00615.x [DOI] [PubMed] [Google Scholar]
- Alexander M. P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44 117–122. 10.3109/10520296909063335 [DOI] [PubMed] [Google Scholar]
- Ariizumi T., Toriyama K. (2011). Genetic regulation of sporopollenin synthesis and pollen exine development. Annu. Rev. Plant Biol. 62 437–460. 10.1146/annurev-arplant-042809-112312 [DOI] [PubMed] [Google Scholar]
- Berardini T. Z., Mundodi S., Reiser L., Huala E., Garcia-Hernandez M., Zhang P., et al. (2004). Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol. 135 745–755. 10.1104/pp.104.040071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaventure G. (2014). Lipases and the biosynthesis of free oxylipins in plants. Plant Signal. Behav. 9:e28429. 10.4161/psb.28429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briat J. F., Ravet K., Arnaud N., Duc C., Boucherez J., Touraine B., et al. (2010). New insights into ferritin synthesis and function highlight a link between iron homeostasis and oxidative stress in plants. Ann. Bot. 105 811–822. 10.1093/aob/mcp128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalhoub B., Denoeud F., Liu S., Parkin I. A., Tang H., Wang X., et al. (2014). Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345 950–953. 10.1126/science.1253435 [DOI] [PubMed] [Google Scholar]
- Chen W., Yu X. H., Zhang K., Shi J., De Oliveira S., Schreiber L., et al. (2011). Male Sterile2 encodes a plastid-localized fatty acyl carrier protein reductase required for pollen exine development in Arabidopsis. Plant Physiol. 157 842–853. 10.1104/pp.111.181693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Lei S., Zhou Z., Zeng F., Yi B., Wen J., et al. (2009). Analysis of gene expression profile in pollen development of recessive genic male sterile Brassica napus L. line S45A. Plant Cell Rep. 28 1363–1372. 10.1007/s00299-009-0736-9 [DOI] [PubMed] [Google Scholar]
- de Azevedo Souza C., Kim S. S., Koch S., Kienow L., Schneider K., McKim S. M., et al. (2009). A novel fatty Acyl-CoA synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. Plant Cell 21 507–525. 10.1105/tpc.108.062513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa A. A., Geanconteri A., Shrestha J., Carlson A., Kooyers N., Coerper D., et al. (2011). A large-scale genetic screen in Arabidopsis to identify genes involved in pollen exine production. Plant Physiol. 157 947–970. 10.1104/pp.111.179523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa A. A., Shrestha J., Morant M., Pinot F., Matsuno M., Swanson R., et al. (2009). CYP704B1 is a long-chain fatty acid omega-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant Physiol. 151 574–589. 10.1104/pp.109.144469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellenberg C., Bottcher C., Vogt T. (2009). Phenylpropanoid polyamine conjugate biosynthesis in Arabidopsis thaliana flower buds. Phytochemistry 70 1392–1400. 10.1016/j.phytochem.2009.08.010 [DOI] [PubMed] [Google Scholar]
- Feng B., Lu D., Ma X., Peng Y., Sun Y., Ning G., et al. (2012). Regulation of the Arabidopsis anther transcriptome by DYT1 for pollen development. Plant J. 72 612–624. 10.1111/j.1365-313X.2012.05104.x [DOI] [PubMed] [Google Scholar]
- Fernandez-Perez F., Pomar F., Pedreno M. A., Novo-Uzal E. (2015a). Suppression of Arabidopsis peroxidase 72 alters cell wall and phenylpropanoid metabolism. Plant Sci. 239 192–199. 10.1016/j.plantsci.2015.08.001 [DOI] [PubMed] [Google Scholar]
- Fernandez-Perez F., Pomar F., Pedreno M. A., Novo-Uzal E. (2015b). The suppression of AtPrx52 affects fibers but not xylem lignification in Arabidopsis by altering the proportion of syringyl units. Physiol. Plantarum 154 395–406. 10.1111/ppl.12310 [DOI] [PubMed] [Google Scholar]
- Gao W., Li H. Y., Xiao S., Chye M. L. (2010). Acyl-CoA-binding protein 2 binds lysophospholipase 2 and lysoPC to promote tolerance to cadmium-induced oxidative stress in transgenic Arabidopsis. Plant J. 62 989–1003. 10.1111/j.1365-313X.2010.04209.x [DOI] [PubMed] [Google Scholar]
- Gaquerel E., Gulati J., Baldwin I. T. (2014). Revealing insect herbivory-induced phenolamide metabolism: from single genes to metabolic network plasticity analysis. Plant J. 79 679–692. 10.1111/tpj.12503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienenberger E., Kim S. S., Lallemand B., Geoffroy P., Heintz D., Souza Cde A., et al. (2010). Analysis of TETRAKETIDE alpha-PYRONE REDUCTASE function in Arabidopsis thaliana reveals a previously unknown, but conserved, biochemical pathway in sporopollenin monomer biosynthesis. Plant Cell 22 4067–4083. 10.1105/tpc.110.080036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heier C., Taschler U., Rengachari S., Oberer M., Wolinski H., Natter K., et al. (2010). Identification of Yju3p as functional orthologue of mammalian monoglyceride lipase in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1801 1063–1071. 10.1016/j.bbalip.2010.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero J., Esteban-Carrasco A., Zapata J. M. (2013a). Looking for Arabidopsis thaliana peroxidases involved in lignin biosynthesis. Plant Physiol. Biochem. 67 77–86. 10.1016/j.plaphy.2013.02.019 [DOI] [PubMed] [Google Scholar]
- Herrero J., Fernandez-Perez F., Yebra T., Novo-Uzal E., Pomar F., Pedreno M. A., et al. (2013b). Bioinformatic and functional characterization of the basic peroxidase 72 from Arabidopsis thaliana involved in lignin biosynthesis. Planta 237 1599–1612. 10.1007/s00425-013-1865-5 [DOI] [PubMed] [Google Scholar]
- Higginson T., Li S. F., Parish R. W. (2003). AtMYB103 regulates tapetum and trichome development in Arabidopsis thaliana. Plant J. 35 177–192. 10.1046/j.1365-313x.2003.01791.x [DOI] [PubMed] [Google Scholar]
- Hu L., Liang W., Yin C., Cui X., Zong J., Wang X., et al. (2011). Rice MADS3 regulates ROS homeostasis during late anther development. Plant Cell 23 515–533. 10.1105/tpc.110.074369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Lempicki R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Ito T., Nagata N., Yoshiba Y., Ohme-Takagi M., Ma H., Shinozaki K. (2007). Arabidopsis MALE STERILITY1 encodes a PHD-type transcription factor and regulates pollen and tapetum development. Plant Cell 19 3549–3562. 10.1105/tpc.107.054536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y., Nishino T., Iwano M., Takayama S., Koizumi N. (2012). Role of the plant-specific endoplasmic reticulum stress-inducible gene TIN1 in the formation of pollen surface structure in Arabidopsis thaliana. Plant Biotechnol. 29 51–56. 10.5511/plantbiotechnology.11.1228a [DOI] [Google Scholar]
- Jiang Y., Wang W., Xie Q., Liu N., Liu L., Wang D., et al. (2017). Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 356 1172–1175. 10.1126/science.aam9970 [DOI] [PubMed] [Google Scholar]
- Karlsson M., Contreras J. A., Hellman U., Tornqvist H., Holm C. (1997). cDNA cloning, tissue distribution, and identification of the catalytic triad of monoglyceride lipase. Evolutionary relationship to esterases, lysophospholipases, and haloperoxidases. J. Biol. Chem. 272 27218–27223. 10.1074/jbc.272.43.27218 [DOI] [PubMed] [Google Scholar]
- Karlsson M., Reue K., Xia Y. R., Lusis A. J., Langin D., Tornqvist H., et al. (2001). Exon-intron organization and chromosomal localization of the mouse monoglyceride lipase gene. Gene 272 11–18. 10.1016/s0378-1119(01)00559-5 [DOI] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim R. J., Kim H. J., Shim D., Suh M. C. (2016). Molecular and biochemical characterizations of the monoacylglycerol lipase gene family of Arabidopsis thaliana. Plant J. 85 758–771. 10.1111/tpj.13146 [DOI] [PubMed] [Google Scholar]
- Kim S. S., Grienenberger E., Lallemand B., Colpitts C. C., Kim S. Y., Souza Cde A., et al. (2010). LAP6/polyketide synthase a and LAP5/polyketide synthase b encode hydroxyalkyl alpha-pyrone synthases required for pollen development and sporopollenin biosynthesis in Arabidopsis thaliana. Plant Cell 224045–4066. 10.1105/tpc.110.080028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konagaya K., Ando S., Kamachi S., Tsuda M., Tabei Y. (2008). Efficient production of genetically engineered, male-sterile Arabidopsis thaliana using anther-specific promoters and genes derived from Brassica oleracea and B. rapa. Plant Cell Rep. 27 1741–1754. 10.1007/s00299-008-0598-6 [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labar G., Wouters J., Lambert D. M. (2010). A review on the monoacylglycerol lipase: at the interface between fat and endocannabinoid signalling. Curr. Med. Chem. 17 2588–2607. 10.2174/092986710791859414 [DOI] [PubMed] [Google Scholar]
- Lallemand B., Erhardt M., Heitz T., Legrand M. (2013). Sporopollenin biosynthetic enzymes interact and constitute a metabolon localized to the endoplasmic reticulum of tapetum cells. Plant Physiol. 162 616–625. 10.1104/pp.112.213124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. C., Zhu J., Yang J., Zhang G. R., Xing W. F., Zhang S., et al. (2012). Glycerol-3-phosphate acyltransferase 6 (GPAT6) is important for tapetum development in Arabidopsis and plays multiple roles in plant fertility. Mol. Plant 5 131–142. 10.1093/mp/ssr057 [DOI] [PubMed] [Google Scholar]
- Liao Y., Smyth G. K., Shi W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Li-Beisson Y., Shorrosh B., Beisson F., Andersson M. X., Arondel V., Bates P. D., et al. (2013). Acyl-lipid metabolism. Arabidopsis Book 11:e0161. 10.1199/tab.0161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luginbuehl L. H., Menard G. N., Kurup S., Van Erp H., Radhakrishnan G. V., Breakspear A., et al. (2017). Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 356 1175–1178. 10.1126/science.aan0081 [DOI] [PubMed] [Google Scholar]
- Luo W. J., Brouwer C. (2013). Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 291830–1831. 10.1093/bioinformatics/btt285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandaokar A., Thines B., Shin B., Lange B. M., Choi G., Koo Y. J., et al. (2006). Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J. 46 984–1008. 10.1111/j.1365-313X.2006.02756.x [DOI] [PubMed] [Google Scholar]
- Mayfield J. A., Fiebig A., Johnstone S. E., Preuss D. (2001). Gene families from the Arabidopsis thaliana pollen coat proteome. Science 292 2482–2485. 10.1126/science.1060972 [DOI] [PubMed] [Google Scholar]
- Mayfield J. A., Preuss D. (2000). Rapid initiation of Arabidopsis pollination requires the oleosin-domain protein GRP17. Nat. Cell Biol. 2 128–130. 10.1038/35000084 [DOI] [PubMed] [Google Scholar]
- Men X., Shi J., Liang W., Zhang Q., Lian G., Quan S., et al. (2017). Glycerol-3-phosphate acyltransferase 3 (OsGPAT3) is required for anther development and male fertility in rice. J. Exp. Bot. 68 513–526. 10.1093/jxb/erw445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Gollery M., Van Breusegem F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9 490–498. 10.1016/j.tplants.2004.08.009 [DOI] [PubMed] [Google Scholar]
- Morant M., Jorgensen K., Schaller H., Pinot F., Moller B. L., Werck-Reichhart D., et al. (2007). CYP703 is an ancient cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. Plant Cell 19 1473–1487. 10.1105/tpc.106.045948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrallah J. B. (2019). Self-incompatibility in the brassicaceae: regulation and mechanism of self-recognition. Curr. Top. Dev. Biol. 131 435–452. 10.1016/bs.ctdb.2018.10.002 [DOI] [PubMed] [Google Scholar]
- Noctor G., Queval G., Mhamdi A., Chaouch S., Foyer C. H. (2011). Glutathione. Arabidopsis Book 9:e0142. 10.1199/tab.0142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan H. A., Li S. F., Parish R. W. (2012). MYB80, a regulator of tapetal and pollen development, is functionally conserved in crops. Plant Mol. Biol. 78 171–183. 10.1007/s11103-011-9855-0 [DOI] [PubMed] [Google Scholar]
- Quilichini T. D., Grienenberger E., Douglas C. J. (2015). The biosynthesis, composition and assembly of the outer pollen wall: a tough case to crack. Phytochemistry 113 170–182. 10.1016/j.phytochem.2014.05.002 [DOI] [PubMed] [Google Scholar]
- Ravet K., Touraine B., Boucherez J., Briat J. F., Gaymard F., Cellier F. (2009). Ferritins control interaction between iron homeostasis and oxidative stress in Arabidopsis. Plant J. 57 400–412. 10.1111/j.1365-313X.2008.03698.x [DOI] [PubMed] [Google Scholar]
- Schonfeld P., Wojtczak L. (2008). Fatty acids as modulators of the cellular production of reactive oxygen species. Free Radic. Biol. Med. 45 231–241. 10.1016/j.freeradbiomed.2008.04.029 [DOI] [PubMed] [Google Scholar]
- Shockey J., Regmi A., Cotton K., Adhikari N., Browse J., Bates P. D. (2016). Identification of Arabidopsis GPAT9 (At5g60620) as an essential gene involved in triacylglycerol biosynthesis. Plant Physiol. 170 163–179. 10.1104/pp.15.01563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer S. D., Chen G., Mietkiewska E., Tomasi P., Jayawardhane K., Dyer J. M., et al. (2016). Arabidopsis GPAT9 contributes to synthesis of intracellular glycerolipids but not surface lipids. J. Exp. Bot. 67 4627–4638. 10.1093/jxb/erw242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L. P., Zhou Z. F., Tang S., Zhang Z. Q., Xia S. Q., Qin M. M., et al. (2016). Ectopic expression of BnaC.CP20.1 results in premature tapetal programmed cell death in arabidopsis. Plant Cell Physiol. 57 1972–1984. 10.1093/pcp/pcw119 [DOI] [PubMed] [Google Scholar]
- Sorensen A. M., Krober S., Unte U. S., Huijser P., Dekker K., Saedler H. (2003). The arabidopsis aborted microspores (AMS) gene encodes a MYC class transcription factor. Plant J. 33 413–423. 10.1046/j.1365-313x.2003.01644.x [DOI] [PubMed] [Google Scholar]
- Taki N., Sasaki-Sekimoto Y., Obayashi T., Kikuta A., Kobayashi K., Ainai T., et al. (2005). 12-oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiol. 139 1268–1283. 10.1104/pp.105.067058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O., Blasing O., Gibon Y., Nagel A., Meyer S., Kruger P., et al. (2004). MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37 914–939. 10.1111/j.1365-313x.2004.02016.x [DOI] [PubMed] [Google Scholar]
- Vanholme R., Cesarino I., Rataj K., Xiao Y. G., Sundin L., Goeminne G., et al. (2013). Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in arabidopsis. Science 341 1103–1106. 10.1126/science.1241602 [DOI] [PubMed] [Google Scholar]
- Vijayaraj P., Jashal C. B., Vijayakumar A., Rani S. H., Venkata Rao D. K., Rajasekharan R. (2012). A bifunctional enzyme that has both monoacylglycerol acyltransferase and acyl hydrolase activities. Plant Physiol. 160 667–683. 10.1104/pp.112.202135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E. T., Schornack S., Marsh J. F., Gobbato E., Schwessinger B., Eastmond P., et al. (2012). A common signaling process that promotes mycorrhizal and oomycete colonization of plants. Curr. Biol. 22 2242–2246. 10.1016/j.cub.2012.09.043 [DOI] [PubMed] [Google Scholar]
- Wang K., Guo Z. L., Zhou W. T., Zhang C., Zhang Z. Y., Lou Y., et al. (2018). The regulation of sporopollenin biosynthesis genes for rapid pollen wall formation. Plant Physiol. 178 283–294. 10.1104/pp.18.00219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. D., Clarke L. A., Eason R. J., Parker C. C., Qi B. X., Scott R. J., et al. (2017). PCP-B class pollen coat proteins are key regulators of the hydration checkpoint in Arabidopsis thaliana pollen-stigma interactions. New Phytol. 213 764–777. 10.1111/nph.14162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C. (2007). Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 100 681–697. 10.1093/aob/mcm079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C., Strnad M. (2019). Jasmonates are signals in the biosynthesis of secondary metabolites - pathways, transcription factors and applied aspects - a brief review. New Biotechnol. 48 1–11. 10.1016/j.nbt.2017.09.007 [DOI] [PubMed] [Google Scholar]
- Xie H. T., Wan Z. Y., Li S., Zhang Y. (2014). Spatiotemporal production of reactive oxygen species by NADPH oxidase is critical for tapetal programmed cell death and pollen development in arabidopsis. Plant Cell 26 2007–2023. 10.1105/tpc.114.125427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong S. X., Lu J. Y., Lou Y., Teng X. D., Gu J. N., Zhang C., et al. (2016). The transcription factors MS188 and AMS form a complex to activate the expression of CYP703A2 for sporopollenin biosynthesis in Arabidopsis thaliana. Plant J. 88 936–946. 10.1111/tpj.13284 [DOI] [PubMed] [Google Scholar]
- Xu J., Ding Z., Vizcay-Barrena G., Shi J., Liang W., Yuan Z., et al. (2014). Aborted microspores acts as a master regulator of pollen wall formation in arabidopsis. Plant Cell 26 1544–1556. 10.1105/tpc.114.122986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Yang C., Yuan Z., Zhang D., Gondwe M. Y., Ding Z., et al. (2010). The aborted microspores regulatory network is required for postmeiotic male reproductive development in Arabidopsis thaliana. Plant Cell 22 91–107. 10.1105/tpc.109.071803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Vizcay-Barrena G., Conner K., Wilson Z. A. (2007). Male sterility1 is required for tapetal development and pollen wall biosynthesis. Plant Cell 19 3530–3548. 10.1105/tpc.107.054981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi B., Zeng F., Lei S., Chen Y., Yao X., Zhu Y., et al. (2010). Two duplicate CYP704B1-homologous genes BnMs1 and BnMs2 are required for pollen exine formation and tapetal development in Brassica napus. Plant J. 63 925–938. 10.1111/j.1365-313X.2010.04289.x [DOI] [PubMed] [Google Scholar]
- Yu S. X., Feng Q. N., Xie H. T., Li S., Zhang Y. (2017). Reactive oxygen species mediate tapetal programmed cell death in tobacco and tomato. BMC Plant Biol. 17:76. 10.1186/s12870-017-1025-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Shi J., Yang X. (2016). Role of lipid metabolism in plant pollen exine development. Subcell. Biochem. 86 315–337. 10.1007/978-3-319-25979-6_13 [DOI] [PubMed] [Google Scholar]
- Zhang W., Sun Y., Timofejeva L., Chen C., Grossniklaus U., Ma H. (2006). Regulation of Arabidopsis tapetum development and function by dysfunctional tapetum1 (DYT1) encoding a putative bHLH transcription factor. Development 133 3085–3095. 10.1242/dev.02463 [DOI] [PubMed] [Google Scholar]
- Zhang X., Henriques R., Lin S. S., Niu Q. W., Chua N. H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1 641–646. 10.1038/nprot.2006.97 [DOI] [PubMed] [Google Scholar]
- Zheng Z., Xia Q., Dauk M., Shen W., Selvaraj G., Zou J. (2003). Arabidopsis AtGPAT1, a member of the membrane-bound glycerol-3-phosphate acyltransferase gene family, is essential for tapetum differentiation and male fertility. Plant Cell 15 1872–1887. 10.1105/tpc.012427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu E., You C., Wang S., Cui J., Niu B., Wang Y., et al. (2015). The DYT1-interacting proteins bHLH010, bHLH089 and bHLH091 are redundantly required for Arabidopsis anther development and transcriptome. Plant J. 83 976–990. 10.1111/tpj.12942 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.






