Message
Computer-aided diagnosis using deep learning (CAD-DL) may be an instrument to improve endoscopic assessment of Barrett’s oesophagus (BE) and early oesophageal adenocarcinoma (EAC). Based on still images from two databases, the diagnosis of EAC by CAD-DL reached sensitivities/specificities of 97%/88% (Augsburg data) and 92%/100% (Medical Image Computing and Computer-Assisted Intervention [MICCAI] data) for white light (WL) images and 94%/80% for narrow band images (NBI) (Augsburg data), respectively. Tumour margins delineated by experts into images were detected satisfactorily with a Dice coefficient (D) of 0.72. This could be a first step towards CAD-DL for BE assessment. If developed further, it could become a useful adjunctive tool for patient management.
In more detail
The incidence of BE and EAC in the West is rising significantly, and because of its close association with the metabolic syndrome this trend is expected to continue.1–3 Reports of CAD in BE analysis have used mainly handcrafted features based on texture and colour.4–7 In our study, two databases (Augsburg data and the ‘Medical Image Computing and Computer Assisted-Intervention’ [MICCAI] data) were used to train and test a CAD system on the basis of a deep convolutional neural net (CNN) with a residual net (ResNet) architecture.8 Images included 148 high-definition (1350×1080 pixels) WL and NBI of 33 early EAC and 41 areas of non-neoplastic Barrett’s mucosa in the Augsburg data set, while the MICCAI data set contained 100 high-definition (1600×1200 pixels) WL images—17 early EAC and 22 areas of non-neoplastic Barrett’s mucosa. All images were pathologically validated and this served as the ground truth for the classification task. Manual delineation of tumour margins by experts was the reference standard for the segmentation task.
The ResNet consisted of 100 layers. Training and subsequent testing were done completely independent of each other using the principle of ‘leave-one-patient-out cross-validation’. For training, small patches were generated from the endoscopic colour images and augmented to simulate similar instances of the same class (figure 1). The class probability of each patch of the test image was then estimated during the classification task, after which the class decision for the full image was compiled from the patch class probabilities.
Figure 1.
Illustration of the deep learning system. The training of the Augsburg data (top row) is done for three classes (EAC: orange; background: blue; and Barrett: green) after patch extraction and augmentation. The patch sampling for the test image is done equidistantly (bottom row) in contrast to the training patch extraction (top row). The probability of EAC class is stored in the image result for the size of the patch sampling offset. EAC, early oesophageal adenocarcinoma.
Using Augsburg data, our CAD-DL system diagnosed EAC with a sensitivity of 97% and a specificity of 88% for WL images, and a sensitivity and specificity of 94% and 80% for NBI images, respectively. CAD-DL achieved a sensitivity and specificity of 92% and 100%, respectively, for the MICCAI images. Thirteen endoscopists, who were shown the same WL images, achieved a mean sensitivity and specificity of 76% and 80% for the Augsburg data and 99% and 78% for the MICCAI data set, respectively. The McNemar test revealed statistically significant outperformance of the CAD-DL system for 11 of the 13 endoscopists for the Augsburg data either for sensitivity or specificity or for both.
The measure of overlap (Dice coefficient D) between the segmentation of CAD-DL and that of experts was computed for images correctly classified by CAD-DL as cancerous (figure 2). A mean value of D=0.72 was computed for the Augsburg data, equally for WL and NBI images. In the MICCAI data, D was 0.56 on average.
Figure 2.
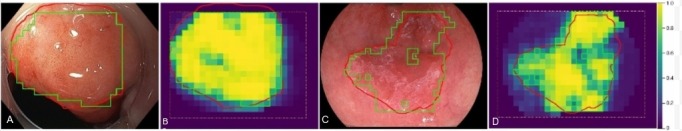
Automatic tumour segmentations on Augsburg image (A, B) and MICCAI image (C, D) are shown by green contours overlaid on the original images and the pseudocoloured, patch-based probability maps. For comparison, the manual segmentations of an expert are drawn in red. Note that the CAD-DL segmentation is restricted to the area indicated by the orange dashed line. CAD-DL, computer-aided diagnosis using deep learning; MICCAI, Medical Image Computing and Computer-Assisted Intervention.
Find more details on the methods and results in the online supplementary material.
Comments
In this manuscript, we extend on our prior study on CNN in BE analysis,9 and demonstrate excellent performance scores (sensitivity and specificity) for the computer-aided diagnosis of early EAC using a deep learning approach. In addition to the classification task, an automated segmentation of the tumour region based on the cancer probability provided by the deep learning system showed promising results.
However, the results of this study are preliminary and represent only the first step towards implementation in real life. The experimental setting involved assessment of optimal endoscopic images. Furthermore, video sequences were not part of the evaluation, and images were limited to flat or elevated EAC, while high-grade/low-grade dysplasia was not included in the analysis. For this reason, future work should focus on further development of the CAD system to assist endoscopists immediately onsite and on real-life endoscopic video sequences while including low-grade/high-grade dysplasia.
A further limitation of our evaluation involves the histological reference standard used for the classification task, whereby we assumed that cancer was spread uniformly across the lesion. However, the idea of one uniform region of cancer is not always the case. Very often, multifocal patches of cancer, dysplasia and normal Barrett’s mucosa are scattered across the surface of the lesion. However, because the image was classified as cancer depending on the patch-specified probability, the authors do not think that the histological pattern described above had a decisive influence on the performance of the CAD-DL system.
Finally, the reference standard used for the segmentation task was based on the macroscopic delineations provided by experts and not on the histological margins. In the Augsburg data set, these delineations were always controlled by a second expert, while in the MICCAI data set the intersection area of five experts was taken as the ground truth. For this reason, we assume that the reference standard used for the segmentation task still had a high level of validity. However, even if the results of tumour segmentation by our CAD-DL system are viewed critically, it still holds a lot of promise as a tool for better visualisation of tumour margins, but may need further improvement and enhancement.
In conclusion, we have shown that CAD-DL on the basis of a deep neural network can be trained to classify lesions in BE with a high level of accuracy. Furthermore, a rough segmentation of the tumour region is possible automatically. However, the diagnostic ability of CAD-DL on less optimal real-life images as well as video sequences needs to be evaluated before its implementation in the clinical setting can be considered. At least, this pilot study may show that CAD with deep learning has the potential to become an important add-on for endoscopists facing the challenge of tumour detection and characterisation in BE.
gutjnl-2018-317573supp001.pdf (16.3MB, pdf)
Footnotes
AE and RM contributed equally.
Contributors: AE: study concept and design, drafting of the manuscript, analysis and interpretation of data, critical revision of the manuscript. RM: study concept and design, software implementation, drafting of the manuscript, analysis and interpretation of data, critical revision of the manuscript. AP: study concept and design, acquisition of data, drafting of the manuscript, critical revision of the manuscript, study supervision. JM: study concept and design, acquisition of data, critical revision of the manuscript. LAdS: statistical analysis, critical revision of the manuscript. JPP: statistical analysis, critical revision of the manuscript, study supervision. CP: study concept and design, drafting of the manuscript, analysis and interpretation of data, statistical analysis, critical revision of the manuscript, administrative and technical support, study supervision. HM: study concept and design, acquisition of data, drafting of the manuscript, critical revision of the manuscript, administrative and technical support, study supervision.
Funding: The authors thank Capes/Alexander von Humboldt Foundation (grant number #BEX 0581-16-0) and the Deutsche Forschungsgemeinschaft (grant number PA 1595/3-1).
Competing interests: None declared.
Patient consent: Not required.
Ethics approval: Beratungskommission für Klinische Forschung am Klinikum Augsburg.
Provenance and peer review: Not commissioned; internally peer reviewed.
References
- 1. Lagergren J, Lagergren P. Oesophageal cancer. BMJ 2010;341:c6280 10.1136/bmj.c6280 [DOI] [PubMed] [Google Scholar]
- 2. Coleman HG, Xie SH, Lagergren J. The Epidemiology of Esophageal Adenocarcinoma. Gastroenterology 2018;154:390–405. 10.1053/j.gastro.2017.07.046 [DOI] [PubMed] [Google Scholar]
- 3. Drahos J, Ricker W, Parsons R, et al. Metabolic syndrome increases risk of Barrett esophagus in the absence of gastroesophageal reflux: an analysis of SEER-Medicare Data. J Clin Gastroenterol 2015;49:282–8. 10.1097/MCG.0000000000000119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Klomp S, van der Sommen F , Swager A-F, et al. Evaluation of image features and classification methods for barrett’s cancer detection using vle imaging In Proceedings of the SPIE Medical Imaging. 2017;10134:101340D. [Google Scholar]
- 5. van der Sommen F, Zinger S, Curvers WL, et al. Computer-aided detection of early neoplastic lesions in Barrett’s esophagus. Endoscopy 2016;48:617–24. 10.1055/s-0042-105284 [DOI] [PubMed] [Google Scholar]
- 6. de Souza LA, Hook C, Papa JP, et al. Barrett’s Esophagus Analysis Using SURF Features: Bildverarbeitung für die Medizin: Springer, 2017:141–6. [Google Scholar]
- 7. de Souza LA, Afonso LCS, Palm C, et al. ; Barrett’s Esophagus Identification Using Optimum-Path Forest, 30th SIBGRAPI Conference on Graphics, Patterns and Images (SIBGRAPI, 2017:308–14. [Google Scholar]
- 8. He K, Zhang X, Ren S, et al. Deep Residual Learning for Image Recognition: In Proceedings of the IEEE conference on computer vision and pattern recognition, 2016:770–8. [Google Scholar]
- 9. Mendel R, Ebigbo A, Probst A, et al. Barrett’s Esophagus Analysis Using Convolutional Neural Networks: Bildverarbeitung für die Medizin: Springer, 2017:80–5. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2018-317573supp001.pdf (16.3MB, pdf)



