Abstract
The DNA replication timing program is modulated throughout development and is also one of the main factors influencing the distribution of mutation rates across the genome. However, the relationship between the mutagenic influence of replication timing and its developmental plasticity remains unexplored. Here, we studied the distribution of copy number variations (CNVs) and single nucleotide polymorphisms across the zebrafish genome in relation to changes in DNA replication timing during embryonic development in this model vertebrate species. We show that CNV sites exhibit strong replication timing plasticity during development, replicating significantly early during early development but significantly late during more advanced developmental stages. Reciprocally, genomic regions that changed their replication timing during development contained a higher proportion of CNVs than developmentally constant regions. Developmentally plastic CNV sites, in particular those that become delayed in their replication timing, were enriched for the clustered protocadherins, a set of genes important for neuronal development that have undergone extensive genetic and epigenetic diversification during zebrafish evolution. In contrast, single nucleotide polymorphism sites replicated consistently early throughout embryonic development, highlighting a unique aspect of the zebrafish genome. Our results uncover a hitherto unrecognized interface between development and evolution.
Keywords: DNA replication timing, embryonic development, germline mutations, zebrafish
Introduction
Mutation rates are not constant across the genome. One of the main factors influencing mutation rate variation is DNA replication timing: genomic regions that replicate later in S phase have higher rates of mutations and harbor more single nucleotide polymorphisms (SNPs) compared with earlier-replicating regions. This has been observed in humans (Stamatoyannopoulos et al. 2009; Chen et al. 2010; Koren et al. 2012), mice (Pink and Hurst 2009; Chen et al. 2010), flies (Weber et al. 2012), yeast (Ito-Harashima et al. 2002; Lang and Murray 2011; Agier and Fischer 2012), archaea (Flynn et al. 2010), and bacteria (Deschavanne and Filipski 1995). DNA replication timing is also a major factor shaping the mutational landscape of cancer genomes, in a cancer-type-specific manner (Woo and Li 2012; Lawrence et al. 2013; Liu et al. 2013; Polak et al. 2015). The correlation of mutation rate across the genome with DNA replication timing appears to be at least in part due to diminished DNA repair capacity at late stages of DNA replication (Zheng et al. 2014; Supek and Lehner 2015). DNA copy number variations (CNVs) are also associated with replication timing and are generally enriched in later-replicating genomic regions (Cardoso-Moreira and Long 2010; De and Michor 2011; Koren et al. 2012; Donley and Thayer 2013). However, at least in the human genome, a GC-rich sequence motif (CCNCCNTNNCCNC) promotes the binding of the recombination protein PRDM9 (Myers et al. 2008) and influences the distribution of a subset of CNVs independently of DNA replication timing (Koren et al. 2012). Taken together, DNA replication timing could have important implications for germline and somatic mutations, and therefore development, genetic disease, cancer, and evolution.
DNA replication timing is also known to change significantly during development. Comparison of replication timing across mammalian cell types has shown that ∼20–30% of the genome, spanning hundreds of distinct genomic regions, varies in replication timing between any two cell types, with a total of up to ∼50% of the genome showing replication timing plasticity across all cell types (Desprat et al. 2009; Hansen et al. 2010). In addition, during differentiation from pluripotent to specialized human cell types, replication timing changes from early to late or vice versa across replication domains covering 30.5% of the genome (Rivera-Mulia et al. 2015). Although these developmental changes in replication timing in mammalian cells do not confound the genome-wide correlations of mutation rates with late replication, specific developmental changes in DNA replication timing could nonetheless alter the mutational landscape and/or its association with genes and gene regulation, potentially having important implications for somatic mutations and for development and cancer. In contrast to somatic mutations, germline mutations should only be influenced by cellular processes in the germline itself. Accordingly, no previous studies have addressed a potential cross-talk between the germline mutational landscape and development with regards to DNA replication timing. Bridging this gap would ideally require measurements of replication timing in the germline, or its nearest proxy—early embryonic development (Yehuda et al. 2018)—alongside more mature developmental stages.
Zebrafish (Danio rerio) serves as a powerful model organism for developmental research (Lele and Krone 1996). We recently reported the replication timing of the zebrafish genome in five stages of embryonic development: pre-MBT (mid-blastula transition), Dome, Shield, Bud, and 28 hpf (hours postfertilization) stages (Siefert et al. 2017) measured by sorting and sequencing G1 and S phase DNA. These stages encompass key events in embryonic development: rapid cell cycles consisting of alternating S phases and mitoses with repressed transcription in pre-MBT; germ layer determination at gastrulation, between the Shield and Bud stages; and the formation of a basic vertebrate body plan by 28hpf. Replication timing was also profiled in an adult cell line derived from a zebrafish tailfin fibroblast (ZTF). We identified numerous regions in the genome that change their replication timing during development, with some showing an advancement of replication timing along development, whereas others showing a delay of replication timing during development. These localized changes occurred on a background of progressive global structuring of the replication program, from partially structured during pre-MBT to highly structured in 28hpf and ZTF stages, with prominent, consistently active replication origins (Siefert et al. 2017).
Using replication timing maps along zebrafish embryonic development, we asked whether there is an overlap between sites of germline mutations and sites of developmental plasticity in DNA replication timing. We analyzed both CNVs and SNPs and found that CNVs are enriched in genomic regions that replicate earlier than expected during the first stages of development. Moreover, CNV sites were particularly prone to changes in replication timing, showing progressively later replication timing along development. In contrast, SNPs in zebrafish tended to replicate early in S phase. Our results provide a new dimension to the association between DNA replication timing and mutation rates and suggest a novel interface between the genome, the epigenome, and development.
Materials and Methods
DNA Replication Timing Data
DNA replication timing data were obtained from Siefert et al. (2017). Briefly, cells from whole embryos were sorted into G1 and S phase fractions, and genomic DNA was sequenced. S phase DNA copy number was normalized by G1 phase DNA copy number in windows of 200 G1 reads. DNA copy number values were then smoothed using a cubic smoothing spline and scaled to a z-score distribution, with a genome-wide mean of 0 and a standard deviation of 1. Chromosome 4 was divided into left and (“p”) and right (“q”) arms based on the developmental shift in replication timing at 27.7 Mb.
The replication timing data are defined in nonuniformly sized genomic windows. To determine replication timing at specific locations throughout the genome not directly represented in the data, replication timing was interpolated using linear interpolation (Matlab function interp1). Linear interpolation infers replication timing values for queried locations using flanking available locations and replication timing values.
Genetic Variation Data
CNV data were obtained from Brown et al. (2012) and Holden et al. (2018), which used microarrays to measure copy number across four common zebrafish strains. Randomly selected individuals were used as a reference. CNV locations were combined across all four zebrafish strains, and overlapping CNVs were merged as before (Brown et al. 2012). Because CNVs were called based on a randomly selected individual, deletions and amplifications could not be discriminated from each other. SNP data were based on high throughput sequencing of the hybrid strain NHGRI1 (LaFave et al. 2014) and the laboratory fish strains Tü, WIK, AB, and TLF (Bowen et al. 2012) or TL, WIK, and Tg (fli1a-eGFPy1) (Butler et al. 2015). SNPs located within exonic regions (RefSeq, GRCz10) and gene promoters (identified as nonmethylated DNA islands) (Long et al. 2013) were removed from analysis. Genomic coordinates were based on the GRCz10 zebrafish genome assembly and were matched across data sets using LiftOver when required.
Clustering of CNVs
Inter-CNV distances were calculated between the center locations of all CNVs on each chromosome, all distances across all chromosomes were combined and sorted by increasing distance. Expected distances were calculated by selecting random locations throughout the genome equal to the number of CNVs; this site permutation was repeated 100 times. Inter-CNV distances in the randomized locations were calculated for each of the 100 iterations, sorted by increasing distance as above, and then averaged across all iterations. In order to define CNV clusters, the center locations of CNVs on each chromosome were clustered using hierarchical clustering with a distance threshold of 500 kb. Only clusters containing three or more CNVs were considered further.
Aggregated Replication Timing Profiles at Genetic Variant Sites
Replication timing of the 5-Mb areas on each side of each SNP or CNV was interpolated to evenly spaced coordinates 1 kb apart along each chromosome. Replication timing values were then averaged across all variant sites (SNPs or CNVs) for each 1-kb coordinate. Randomized profiles were generated by choosing random locations across each chromosome equal to the number of variant sites on that chromosome, followed by interpolation to 1-kb-spaced coordinates as above; this was repeated 20 times. To correct for global background replication trends (figs. 2A and B and 6A and B ; see supplementary fig. 1, Supplementary Material online), the average replication timing profile was calculated from all randomized profiles and subtracted from the genetic variation site profile.
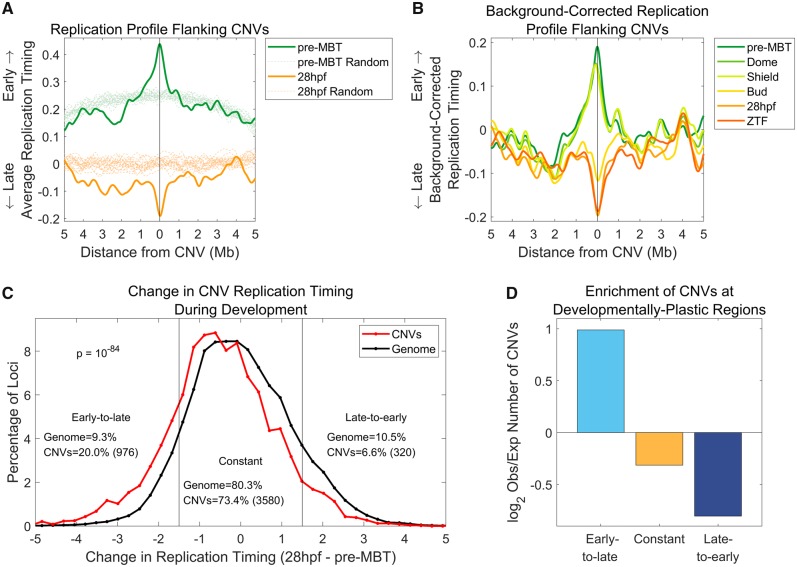
Fig. 2.
—CNV sites undergo replication timing shifts along development. (A) CNVs shift from early to late replication genome-wide. The replication timing profiles in 10-Mb regions surrounding all CNV sites were aggregated and averaged to produce a “meta-CNV” replication profile. In parallel, 20 sets of randomly permuted, chromosome-matched CNV locations were analyzed in a similar manner. (B) The CNV replication timing shift coincides with gastrulation, between the Shield and Bud embryonic stages. Same as (A), for all developmental time points, with the average permutation profile for each time point subtracted from the meta-CNV profile. (C) Genomic regions that shift from early to late replication are enriched for CNVs. The replication timing difference between pre-MBT and 28hpf was calculated for each CNV (excluding chromosome 4), and, separately, for replication timing values in 1-kb windows across the genome (excluding CNV locations and chromosome 4). CNVs are enriched in regions that change from early to late replication and are depleted from regions that change from late to early replication (Kolmogorov–Smirnov test P = 10−84). (D) Genomic regions that continuously change in their replication timing along the entire embryonic development time course were previously identified using a HMM (Siefert et al. 2017). Regions that become later replicating show a nearly 2-fold enrichment of CNVs compared with expectation, whereas regions that become earlier replicating or maintained constant replication timing are depleted of CNVs. Chi-square P ≪ 10−300 for all categories.
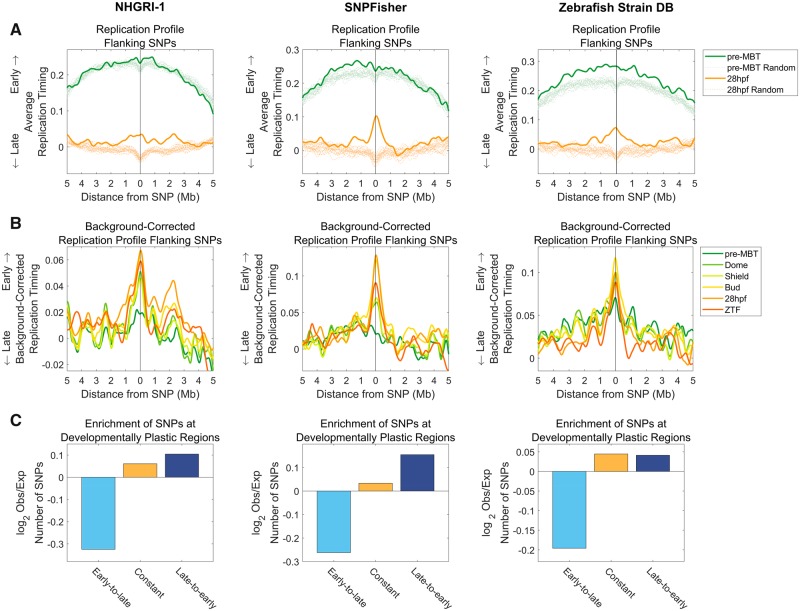
Fig. 6.
—SNP loci replicate early in zebrafish. (A) SNPs show a shift toward earlier replication at 28hpf when compared with genomic regions in general (excluding exonic and regulatory regions). This trend is consistent in all three independent data sets: NHGRI-1 (LaFave et al. 2014), SNPFisher (Butler et al. 2015), and Zebrafish Strain DB (Bowen et al. 2012). The replication timing profiles in 10-Mb regions surrounding all SNP sites were aggregated and averaged as described in figure 2A. In parallel, 20 sets of randomly permuted, chromosome-matched SNP locations were analyzed in a similar manner, also excluding exonic and regulatory regions. (B) SNPs replicate early in zebrafish and may possibly even shift to earlier replication along development. Same as (A) for all developmental time points, with the average permutation profile for each time point subtracted from the average SNP profile. (C) SNPs are enriched in late-to-early and constant replicating regions across all data sets, whereas being consistently depleted in early-to-late replicating regions. Replicating regions were classified according to HMM classification (Siefert et al. 2017). Chi-square P ≪ 10−300 for all categories.
Partial Correlations
In order to evaluate the contribution of different factors to the association between variant sites and replication timing, we performed a correlation and partial correlation analysis, the former evaluating the direct correlation between the density of genetic variants along chromosomes and DNA replication timing, whereas the latter repeats the correlation calculation taking into account one additional confounding factor at a time. To do this, the genome was divided into 100-kb-nonoverlapping windows. Replication timing was interpolated at the window center locations, whereas GC content was averaged within each window. The representation of all other genomic features was determined by counts within the 100-kb windows. Spearman rank correlations were calculated between CNVs or SNPs and one other variable (Matlab function corr), whereas Spearman partial correlations included an additional variable that was being controlled for (Matlab function partialcorr); this was repeated for all tested variables.
Multiple Linear Regression
The density of all genomic features was binned into 100-kb bins across the genome as described above. A linear model was fit using all genomic and epigenetic features (excluding replication timing) as input with CNV density as the observed output. Subsequently, a second linear model was fit with replication timing from a given developmental time point added as an additional input. The two linear models were compared using an ANOVA (R function anova) to determine if the fit of the linear model was improved with the addition of replication timing. This procedure was performed for replication timing at each developmental time point. Change in residual sum of squares (RSS) was calculated by subtracting the RSS from the first linear model (without replication timing) from the RSS from the second linear model (with replication timing). A negative change in RSS means the RSS decreased with the addition of replication timing to the model and the model better explains the variation in CNV density.
Enrichment of Variant Sites at Particular Genomic Regions
In order to determine whether CNV or SNP localization was biased to specific regions of the genome, such as particular chromosomes (fig. 3B) or regions with developmental replication timing plasticity (figs. 2D and 6C), we calculated the expected number of sites assuming an equal distribution throughout the genome. In the case of SNPs, the “genome” was defined as regions not classified as exonic or regulatory. To calculate the expected number of variants in a specific region of interest, the length of the region was divided by the total length of the genome and multiplied by the total number of variants (CNVs or SNPs) across the genome. A two proportions chi-square test was used to calculate the significance between the observed and the expected number of variant sites.
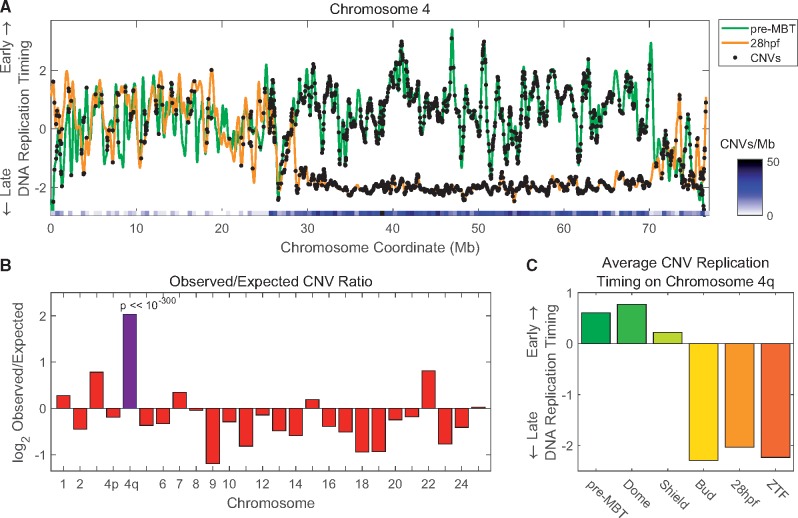
Fig. 3.
—The right arm of chromosome 4 shows a dramatic replication delay and the highest density of CNVs in the genome. (A) Replication timing profiles of chromosome 4 at pre-MBT (green) and at 28hpf (orange), with the locations of CNVs marked (black dots). Chromosome 4q shows the strongest replication timing delay in the genome (see Siefert et al. 2017) and also has an unusually high density of CNVs. The heat map shows the density of CNVs in 500-kb windows and highlights the sharp increase in CNV density coinciding with the developmental shift in replication timing. (B) Chromosome 4q has the highest density of CNVs throughout the genome. The ratios of observed versus expected numbers of CNVs were calculated per chromosome, with chromosome 4 divided into p and q arms (given their different properties). (C) The replication timing of CNVs on chromosome 4q shifts from early to late between shield and bud stages.
Gene Ontology Enrichment Analysis
Enrichment for gene annotations was analyzed using Gene Ontology (Ashburner et al. 2000; Gene Ontology Consortium 2016). CNVs were parsed using a Hidden Markov model (HMM) into early-to-late, late-to-early, or constant replication timing (Siefert et al. 2017), and genes overlapping with these CNVs were identified (Refseq gene annotation for GRCz10). Genes with more than one overlapping CNV in a given category were considered once for the enrichment analysis, and genes spanning more than one HMM category were excluded. Background gene lists for comparisons were generated based on all the genes in each HMM category, excluding genes spanning more than one HMM category. Gene enrichment was calculated using a Fisher’s Exact test with Bonferroni correction for multiple tests.
Chromosome-Level Replication Timing Trends
To reveal chromosome-scale replication timing patterns (supplementary fig. 1, Supplementary Material online), the replication timing of each chromosome was smoothed using a cubic smoothing spline (Matlab function csaps) with a parameter of 10−24. Each chromosome was binned into 1,000 evenly spaced intervals, and the smoothed replication timing for each interval was averaged across all chromosomes to produce a composite replication timing profile for each developmental stage.
Results
Sites of Germline CNVs Undergo Developmental Changes in DNA Replication Timing
Mutation rates are nonuniform across the genome and correlate with the time of DNA replication in a variety of species. To further understand these associations, we sought to characterize the loci that harbor genetic variants. We focus on a previously unexplored aspect of mutation sites: whether their replication timing remains constant or is plastic during organismal development. We previously showed that CNVs exhibit particularly strong associations with DNA replication timing (in contrast to SNPs, which have more subtle correlations; Koren et al. 2012). To begin to explore this question, we therefore analyzed the locations of CNVs in comparison to DNA replication timing at six stages of zebrafish embryonic development. Importantly, replication timing is not strongly correlated with GC content in zebrafish (Siefert et al. 2017), thus minimizing confounding effects on the genomic distribution of CNVs (Koren et al. 2012). We used a zebrafish CNV data set inferred from DNA copy number microarray (aCGH) analysis of 40 fish from four diverged strains (10 fish per strain) in comparison to a randomly selected reference fish from each strain, for a total of 5,855 nonoverlapping CNVs (Brown et al. 2012; Holden et al. 2018). Notably, because the data set is based on comparative microarray hybridization with an arbitrarily chosen reference, the inference of CNVs in this data set does not enable one to distinguish between deletions in one strain versus duplications in the other. Similarly, because microarrays use preselected sequence probes, they do not map CNVs to bp-resolution. As a result, the exact breakpoints of the CNVs are not known, precluding the search for microhomology at breakpoints and distinguishing between CNVs caused by homologous recombination or by nonhomologous end joining and related mechanisms.
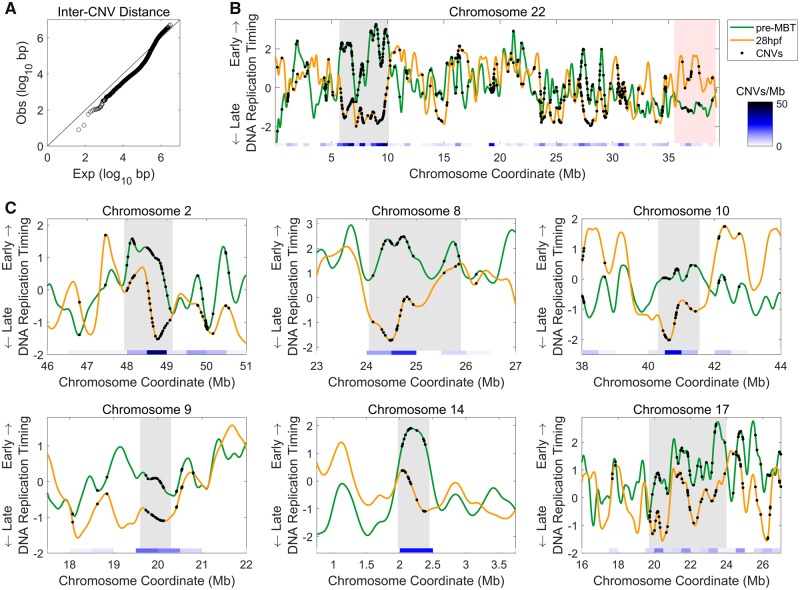
We first noted a tendency of CNVs to cluster along chromosomes, such that more CNVs were located within <∼500 kb from each other than expected if CNVs were randomly distributed along chromosomes (fig. 1A and B). Clustering of CNVs, previously observed in the human genome (Koren et al. 2012), suggests that regional factors may affect their formation at particular chromosomal regions. DNA replication timing could be one of these factors.
Fig. 1.
—Zebrafish CNVs cluster in genomic regions that shift from early to late replication. (A) CNVs tend to cluster within <500 kb from each other. Distances between adjacent CNVs were compared with the expected inter-CNV distances determined from 100 iterations of randomized locations equal to the number of CNVs assuming an even distribution of CNVs throughout the genome (Kolmogorov–Smirnov P = 10 − 145). (B) CNVs are concentrated at sites that change from early to late replication. An example of a chromosome replication profile showing early (pre-MBT) and late (28hpf) embryonic developmental time points together with the locations of CNVs along the chromosome (shown on both profiles). A large region at ∼5–10-Mb switches replication timing from early to late and has the highest density of CNVs on the chromosome (gray shading). Conversely, at ∼36 Mb to the end of the chromosome, the replication timing switches from late to early and has a low density of CNVs (red shading). Replication timing is presented in z-score units, that is, as standard deviations from the mean; positive values represent early replication and negative values represent late replication. Heatmap indicates the density of CNVs in 500-kb windows. (C) Genomic regions with high densities of CNVs tend to be regions that change replication timing from early to late. Representative examples are shown. Gray shading marks regions with high CNV densities.
Visualizing the locations of CNVs on the replication timing profiles of pre-MBT and of 28hpf embryos revealed an intriguingly high density of variant sites overlapping genomic regions that change in replication timing along development, particularly within regions that changed from early replicating (positive values) at pre-MBT to late replicating (negative values) at 28hpf (fig. 1B and C).
To more formally evaluate the association between structural variation loci and DNA replication timing, we aggregated CNV loci (excluding chromosome 4; see below) and measured the average replication timing at these sites and their flanking regions. Compared with the genomic average (empirically obtained by generating 20 sets of randomly chosen sites, each set matched to the number of actual genetic variants tested; see Materials and Methods), CNV sites replicated earlier than expected in pre-MBT embryos, but replicated later than expected in 28hpf embryos (fig. 2A). The regions of replication timing bias relative to the genomic expectation extended approximately one megabase to each side of the CNV sites. Of note, the random expectation—or average replication timing at randomly selected genomic sites—was itself different between early and late embryos, resembling an inverse-parabolic-like shift to early replication in early embryogenesis but a mostly uniform replication timing in late embryos (fig. 2A). These inverse-parabolic patterns were confirmed to represent the global replication timing pattern along each of the chromosomes in pre-MBT embryos and to be gradually reduced during development (supplementary fig. 1, Supplementary Material online). Importantly, the replication timing of CNV sites and their developmental shift remained significantly biased even after considering these global background patterns (fig. 2A).
To more systematically analyze the changes in replication timing along development at sites of genetic variation, we repeated the above analysis for all six developmental time points (five embryonic time points and somatic tailfin fibroblast cells). We subtracted the genomic average replication timing from the germline variant aggregate profiles to better evaluate the changes during development without the confounding influence of the background replication timing patterns described above (fig. 2B; see Materials and Methods). CNV sites showed a sharp shift from early to late replication between the Shield and Bud stages, the time at which gastrulation occurs (fig. 2B).
The developmental changes in replication timing at CNV sites were also evident as shifts in their replication timing distributions compared with the rest of the genome (supplementary fig. 2A, Supplementary Material online). This confirms that the shifts in the aggregated sites do not result from a minority of outliers but rather represent a general trend for CNVs. The replication timing differences between CNV sites and the rest of the genome were relatively modest, but highly significant (Kolmogorov–Smirnov P = 10−16–10−53 for the different developmental stages), and again, CNV sites showed the major shift from early to late replicating at the time of gastrulation (supplementary fig. 2A, Supplementary Material online).
CNVs Are Enriched in Regions of Replication Timing Plasticity across Development
The above results suggest that CNV sites change in their replication timing during development. As a complementary analysis, we asked what fraction of developmentally plastic genomic region corresponds to CNV sites. In other words, to what extent are CNVs enriched within genomic regions that change in their replication timing along development? To answer this, we performed two analyses. First, we calculated the change in replication timing from pre-MBT to 28hpf for each CNV site compared with all DNA replication timing values across the genome in 1 kb windows excluding CNV sites. Genomic regions in general showed a near normal distribution of replication timing changes, with a similar number of regions changing from early to late or from late to early replication. In contrast, CNVs showed a marked skew in favor of being delayed from early to late replication. Specifically, the replication of 976 CNVs (20.0% of the total) was delayed by more than 1.5 standard deviations along development, whereas only 320 CNVs (6.6%), or >3-fold less, showed the opposite change from late to early replication (fig. 2C). Overall, the distribution of replication timing changes for CNV sites was significantly different than other genomic sites (Kolmogorov–Smirnov test, P = 10−84). We also compared the distribution of changes in replication timing at CNV sites to the distribution of replication timing changes at random subsets of genomic region equal to the number of CNV sites (supplementary fig. 2B, Supplementary Material online). Random locations had a similarly shaped distribution of replication timing changes as the genome as a whole, confirming that the biased timing changes at CNV sites were not due to their smaller numbers compared with the remainder of genomic sites analyzed (P = 10−42). These results reinforce the conclusion that CNVs are enriched at sites with delayed replication along development.
A second analysis of the overlap between variant sites and developmental plasticity started with a list of genomic regions that we previously identified as developmentally plastic, based on a HMM of replication timing changes along development (Siefert et al. 2017). Genomic regions that changed from early to late replication were much more likely to contain CNVs than regions that changed from late to early replication (early-to-late CNVs: 1.96-fold enrichment of CNVs compared with expectation; chi-square P ≪ 10−300 and late-to-early CNVs: 1.72-fold depletion; P ≪ 10−300; fig. 2D). Taken together, these results indicate that CNVs sites are enriched within genomic regions that change in their replication timing from early to late during development.
Because CNVs tend to cluster along chromosomes (fig. 1), we repeated the above analyses (fig. 2) on CNV clusters instead of individual CNVs. By hierarchicaly clustering the locations of CNVs along each chromosome with a distance threshold of 500 kb, we identified 704 clusters containing at least three CNVs each (on average seven CNVs per cluster, mean cluster size = 402 kb). Despite the lower number (and hence reduced statistical power) of clusters compared with individual CNVs, all of the above results were reproduced with CNV clusters: cluster centers gradually changed from early to late replication along development (randomization-corrected average replication timing, pre-MBT: 0.15; Dome: 0.13; Shield: 0.14; Bud: −0.03; 28hpf: −0.11; and ZTF: −0.15); the replication timing distribution of CNV clusters was significantly earlier than the remainder of the genome in early development but significantly later than the remainder of the genome in late development (CNVs compared with the rest of the genome, pre-MBT: P = 10−4; Dome: P = 10−3; Shield: P = 10−3; Bud: P = 0.49; 28hpf: P = 0.038; and ZTF: P = 10−4); CNV clusters showed a marked skew toward replicating later in 28hpf than in pre-MBT compared with the rest of the genome (25.88% of CNV clusters, or 161 clusters, changed to later replicating compared with 10.03% of the genome; Kolmogorov–Smirnov P = 10−17); and CNV clusters were enriched 1.6-fold in regions of the genome classified as switching from early to late replication (chi-square P = 10−7 compared with an even distribution throughout the genome) and depleted 1.43-fold in regions of the genome classified as switching from late to early replication (chi-square P = 10−2). Thus, CNVs, individually or when found in clusters with other CNVs, show a robust change from early to late replication along embryonic development in zebrafish.
CNV Sites Correlate with DNA Replication Timing Plasticity Independently of Any Known Confounding Factors
The above results suggest a link between DNA replication timing changes during development and the locations of CNVs along the zebrafish genome. This link could indicate a common mechanism that influences both the generation of germline structural mutations and developmental changes in DNA replication. However, it remains possible that other factors influence both mutation generation and DNA replication timing and thus indirectly explain this association. To test this, we considered several genetic and epigenetic properties as potential confounders, including GC content, distance from the telomere, the locations of genes and repeat sequences, gene expression levels, and chromatin structure (supplementary fig. 2C, Supplementary Material online). We calculated the density of these properties as well as CNVs in 100-kb windows along the genome and inferred the average replication timing in the same windows at different developmental stages. The density of CNVs showed a weak (as expected given their sparsity) but significantly positive correlation with replication timing at early embryonic development (pre-MBT: Spearman ρ = 0.08; P = 10−19), which switched to a negative correlation at late stages of embryonic development (28hpf: Spearman ρ = −0.09; P = 10−22). This supports our conclusions above pointing to developmental changes in replication timing at CNV sites. Importantly, none of the tested potential confounding factors could explain these associations or the developmental trend, as the correlations remained almost identical when controlling for these factors using partial correlation analysis (supplementary fig. 2C, Supplementary Material online; Materials and Methods). Using linear regression, replication timing was significantly associated with CNV density (pre-MBT: regression coefficient = 0.068 ± 0.0156, P = 10−16; Dome: regression coefficient = 0.039 ± 0.0156, P = 10−7; Shield: regression coefficient = 0.044 ± 0.0156, P = 10−8; Bud: regression coefficient = −0.060 ± 0.0154, P = 10−15; 28hpf: regression coefficient = −0.100 ± 0.0154, P = 10−16; and ZTF: regression coefficient = −0.103 ± 0.0152, P = 10−16). Multiple linear regression with sequential ANOVA (Materials and Methods) indicated that replication timing associated with CNV density beyond the contribution of all 12 other tested variables, marked by a decrease in the RSS with the addition of replication timing to the linear model (pre-MBT ΔRSS = −173.3, P = 10−16; Dome ΔRSS = −113.6, P = 10−16; Shield ΔRSS = −93.8, P = 10−16; Bud ΔRSS = −4.3, P = 0.014; 28hpf ΔRSS = −47.6, P = 10−15; and ZTF ΔRSS = −49.4, P = 10−16), and as above, CNVs were positively associated with replication timing at pre-MBT, Dome, and Shield stages but negatively associated with replication timing at Bud and 28hpf stages, as well as in ZTF. We note, however, that all examined features cumulatively explained only 7.8% of the variance in CNV density, suggesting that other contributing factors remain to be identified.
Chromosome 4q Demonstrates the Tight Link between CNV Density and Developmental Delay in DNA Replication Timing
The most dramatic developmental change in DNA replication timing in the zebrafish genome involves the right arm of chromosome 4. The entire arm undergoes a shift from mid-to-late replication during gastrulation and becomes one of the latest replicating genomic regions (Siefert et al. 2017; fig. 3A). Strikingly, chromosome 4q was also highly enriched in CNVs (fig. 3A). When comparing the observed versus expected number of CNVs across all chromosomes, chromosome 4q showed the highest enrichment of CNVs in the genome, at 17.82 CNVs/Mb, compared with an average of 3.84 CNVs/Mb for the remainder of the genome (4.1-fold enrichment compared with the expected CNV density; chi-square P ≪ 10−300; fig. 3B). CNVs on chromosome 4q showed the strongest shift from early to late replication between the Shield and Bud stages (fig. 3C), mirroring the change in CNV replication timing in the remainder of the genome. Thus, the right arm of chromosome 4 provides a powerful demonstration of the association between CNV density and developmental changes in DNA replication timing.
CNVs That Change from Early to Late Replication Overlap the Clustered Protocadherin Genes
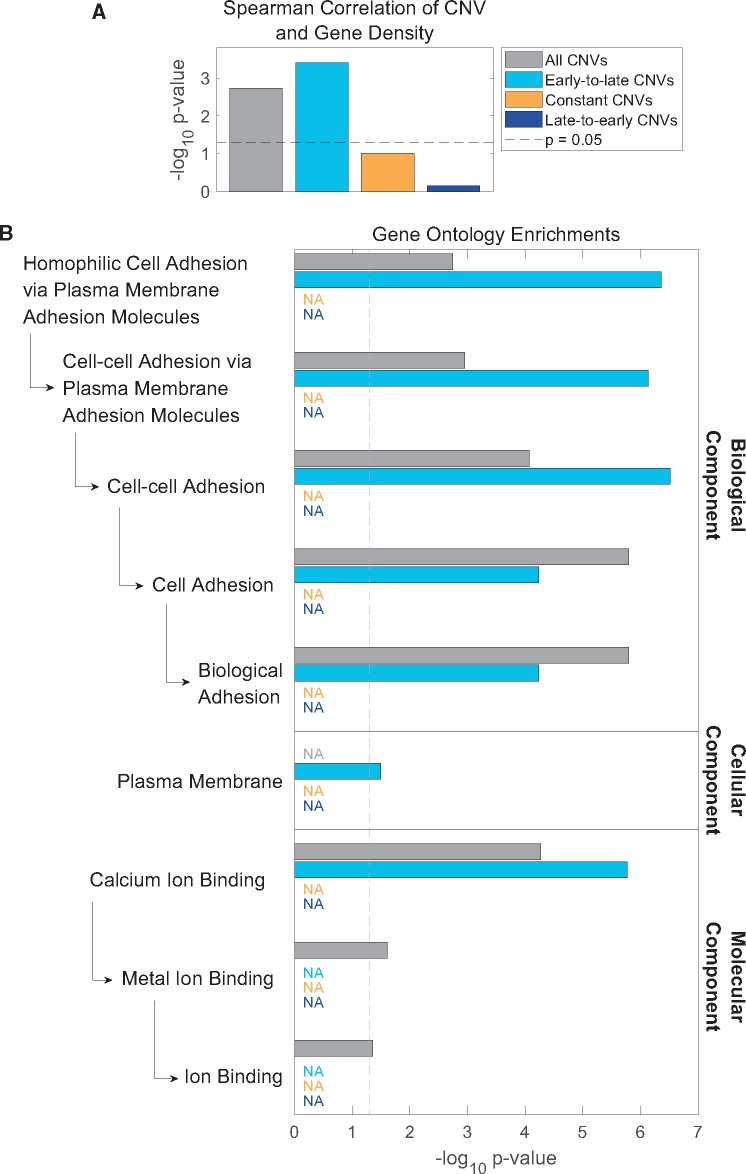
Although the association between DNA replication timing plasticity and CNV formation could point to a mechanism that jointly affects genetic and epigenetic processes, we also considered whether it could have any functional and/or evolutionary consequences. We first compared the locations of CNVs in the zebrafish genome to the locations of genes. CNVs tended to be depleted of genes (CNVs overlapped with 1,590 genes compared with 2,166 expected overlaps from a random distribution throughout the genome, chi-square P ≪ 10−300), consistent with previous findings that attributed this depletion to negative selection (Nguyen et al. 2006; Brown et al. 2012). Nonetheless, CNV density along chromosomes showed a weak yet positive correlation with gene density (supplementary fig. 2C, Supplementary Material online, P = 10−3). This correlation was independent of any tested potential confounders (supplementary fig. 2C, Supplementary Material online), indicating that the covariance of CNVs and genes does not result from their correlations with other genomic or epigenomic variables. To test whether a particular subset of CNVs explains the correlation with genes, we parsed the CNVs to CNVs within genomic regions that change during development from early to late, or from late to early replication, and CNVs in regions that do not significantly change their replication timing (as in fig. 2D). Intriguingly, the most significant correlation with gene density (P = 10−4) was found among CNVs that shift from early to late replication during development, despite this category not being the most abundant CNV category (representing 28.9% of CNVs), whereas constant and late-to-early CNVs (60.9% and 10.2% of CNVs, respectively) were not significantly correlated with gene density (fig. 4A).
Fig. 4.
—Early-to-late CNVs drive covariation of CNVs and genes and are enriched for cell adhesion genes. (A) Early-to-late CNV density covaries with gene density. Spearman rank correlation P values for gene density and CNVs parsed by change in replication timing throughout development (chromosome 4 excluded). Although all spearman correlation ρ values are modest (<0.1), early-to-late CNVs are the drivers of the correlation with gene density. Dashed gray line: P = 0.05. (B) CNVs that change from early- to late-replicating are enriched for genes related to homophilic cell adhesion and are the only CNV category with a significant gene annotation enrichment. Enrichments are in comparison to other genes within each replication timing change category. Late-to-early CNVs and constant CNVs had no significantly enriched genes (“NA”: category colorcode as in A).
To further ask if the covariance of genes and CNVs could have a functional significance, we tested CNVs for enrichment of specific gene annotations using GO Gene Ontology (Ashburner et al. 2000; Gene Ontology Consortium 2016). CNVs, as a whole, significantly overlapped genes with annotations related to homophilic cell adhesion (fig. 4B). When parsing CNVs by replication timing categories and comparing genes within CNVs to all other genes within that category, early-to-late CNVs emerged as the sole drivers of these gene enrichments. CNVs with late-to-early or constant replication timing showed no significant enrichment for any gene annotations. As a further control, we found no significant enrichments for randomly selected genes equal to the number of genes overlapping CNVs in each replication timing category.
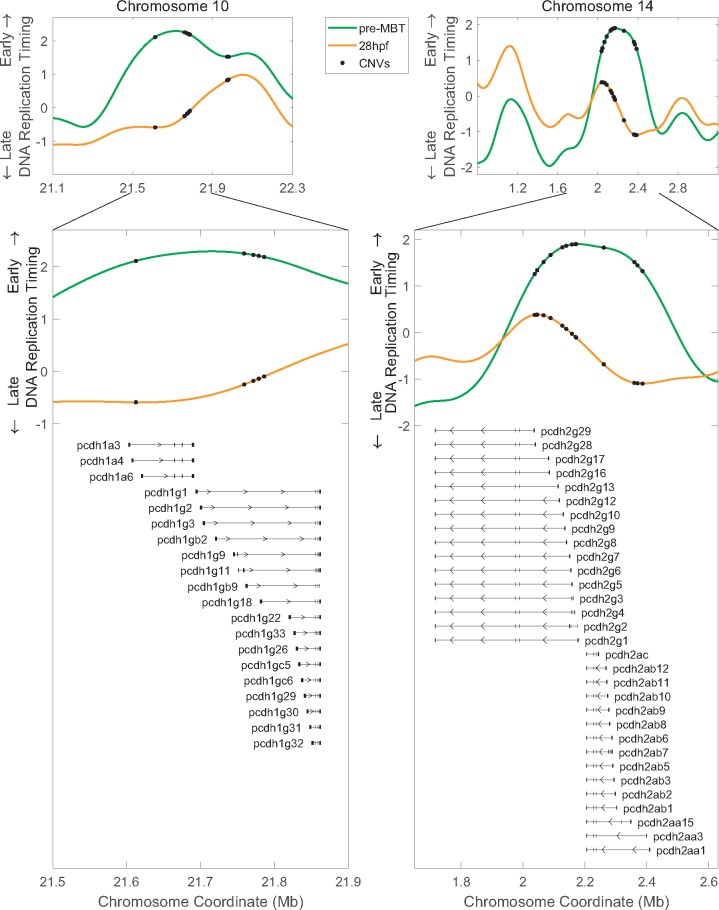
The enrichment for homophilic cell adhesion genes was entirely due to protocadherin genes. Early-to-late CNVs overlapped protocadherin 15a and the clustered protocadherin 1α (Pcdh1α), 1γ (Pcdh1γ), and 2α (Pcdh2α) genes (fig. 5 and supplementary fig. 3a and table 1, Supplementary Material online). Importantly, most of the clustered Pcdh2γ genes were excluded from the GO analyses because they spanned more than one replication timing category, yet they also overlapped CNVs that specifically changed from early- to late-replicating (fig. 5; the variable gene regions [see below] in particular overlapped early-to-late CNVs). Thus, all clustered protocadherin genes in zebrafish (four clusters in total) contain CNVs that change from early to late replication during embryonic development. The clustered protocadherins genes encode cell surface receptors that are expressed primarily in the nervous system in early embryos beginning at gastrulation (Emond and Jontes 2008). They have an unusual genomic organization in which multiple, variable first exons, each transcribed from its own promoter, are arranged in tandem upstream from three short, constant exons. Different isoforms of clustered Pcdh are expressed stochastically and combinatorically in single neurons, giving rise to an immense molecular diversity that is thought to serve as “molecular barcodes” for selecting appropriate synaptic partners and facilitating the establishment of complex neural circuits in the brain (Lefebvre et al. 2012; Chen and Maniatis 2013; Hirayama and Yagi 2013). Clustered Pcdh genes have undergone extensive evolutionary diversification, particularly in teleost fishes (including zebrafish), through tandem gene duplications, gene conversions, and lineage-specific degeneration (Noonan et al. 2004; Wu 2005; Yu et al. 2007). Taken together, clustered Pcdh genes are important for development and have undergone rapid structural evolution. At the same time, they conform to the pattern we observe of CNVs being progressively delayed in their replication timing during development. Thus, replication delays at CNVs sites may have broader functional implications for zebrafish biology, at Pcdh and potentially other genes.
Fig. 5.
—Early-to-late CNVs overlap the clustered protocadherin genes. Early-to-late CNV gene enrichment is driven by clustered protocadherin genes on chromosomes 10 and 14. Most Pcdh2γ genes were not included in the GO enrichment analysis because they span both an early-to-late and a constant replication timing region; however, their variable exons also overlap CNVs that shift from early to late replication. All clustered Pcdh genes are shown (not all overlapped CNVs, and several were not annotated by GO; supplementary table S1, Supplementary Material online).
SNPs Shift to Earlier Replication during Development
We next asked whether SNPs show replication timing changes during development similar to CNVs. SNPs are much more abundant in the genome than CNVs, and their rates across the genome vary in more subtle ways that represent the influences of mutational biases but also, to a large extent, of natural selection and random drift. Thus, the ability to relate their genomic distribution to DNA replication timing is more limited than for CNVs. Indeed, previous studies in humans have shown modest (but nonetheless significant) enrichments of SNPs in later-replicating genomic regions (Stamatoyannopoulos et al. 2009; Koren et al. 2012); this correlation is much stronger for de novo germline mutations (Francioli et al. 2015) and for somatic mutations in cell lines (Koren et al. 2012) and in cancer (Lawrence et al. 2013). In zebrafish, SNP data are currently the only available genomic data relating to point mutations (to our knowledge). Accordingly, we analyzed replication timing changes at SNP sites using three data sets obtained from whole-genome sequencing: 1) sites of heterozygosity, representing segregating SNPs, in the NHGRI-1 zebrafish strain that was derived from a cross between two divergent fish strains (“NHGRI-1”) (LaFave et al. 2014); 2) SNPs among three laboratory zebrafish strains (TL, WIK, and Tg; “SNPFisher”) (Butler et al. 2015); and 3) SNPs identified by sequencing four wild-type fish strains (Tü, WIK, AB, and TLF; “Zebrafish Strain DB”) (Bowen et al. 2012). To minimize the effects of selection and other population genetic processes, we removed SNPs located within exonic regions and in nonmethylated islands—markers of gene promoters in cold-blooded vertebrates (Long et al. 2013). We also excluded SNPs on chromosome 4, leaving a total of 14,945,106, 6,141,728, and 6,324,096 SNPs for the respective data sets.
We first compared the average replication timing of SNP sites and their flanking regions to randomized control sites. In this case, the random control, which also excluded exonic and regulatory regions to match the SNPs, showed a distinctive replication timing dip around tested sites that results from the relatively late replication of intergenic regions (consistently, exonic regions replicate early in S phase; Siefert et al. 2017; fig. 6A). Compared with this background, SNP sites replicated around the genomic mean at pre-MBT (or earlier than the mean in the case of the Zebrafish Strain DB data set) but replicated consistently earlier than expected at 28hpf (fig. 6A) and at other developmental time points (fig. 6B). There was even some evidence suggesting a gradual change to earlier replication along development (fig. 6A)—the opposite of the trend observed for CNVs. The early replication of SNP loci in zebrafish is surprising given that in previously studied species, SNP sites are typically enriched at late-replicating regions (Pink and Hurst 2009; Stamatoyannopoulos et al. 2009; Koren et al. 2012; Weber et al. 2012).
When analyzing the distribution of SNPs within genomic regions that changed from early to late replication during development, from late to early replication, or neither, we found that SNPs were weakly but significantly enriched in regions that shifted to earlier replication throughout development or that had a constant replication timing, whereas regions that changed from early to late replication were depleted of SNPs (fig. 6C). This was the opposite pattern from what was observed for CNVs, suggesting that SNPs and CNVs localize to distinct regions of the genome with different replication timing properties. Consistently, SNP and CNV densities were negatively correlated. This correlation and the correlation of SNP densities with DNA replication timing were not confounded by any known genomic or epigenetic features (supplementary fig. 4, Supplementary Material online). Taken together, these results suggest that replication timing affects the localization of both CNVs and SNPs, albeit in apparently opposite ways along zebrafish development. The early replication of SNP loci is unique to zebrafish among species studies thus far.
Human Genetic Variant Loci Demonstrate Replication Timing Plasticity
Because of the strong replication timing plasticity observed at genetic variant locations in zebrafish, we were interested to see whether replication timing plasticity at germline genetic variant locations was present in other species. To the best of our knowledge, zebrafish is the only species with both replication timing data during embryonic development as well as comprehensive catalogs of genetic variation. Therefore, as a proxy for embryonic development, we used replication timing measured during in vitro differentiation of human embryonic stem cells (ESCs) to the three germ cell layers: endoderm, mesoderm, or ectoderm, for a total of 14 cell lineages (Rivera-Mulia et al. 2015; Yehuda et al. 2018). We analyzed CNVs that were identified by whole-genome sequencing and have been parsed into those likely formed by homologous recombination (HR, n = 2,865) or nonhomologous end joining (NHEJ, n = 21,221) based on the presence or absence of microhomology at the break points (Sudmant et al. 2015). We also analyzed sites of de novo mutations (DNMs, n = 11,020), which represent single nucleotide mutation sites with minimal influences of natural selection (Francioli et al. 2015). Changes in replication timing were calculated between ESCs and each differentiated cell type at all genetic variant locations compared with all genomic DNA replication timing values in 1-kb windows. Modest, yet significant, delays in replication timing were observed in several differentiated cell types, most notably at locations with HR CNVs, for example during differentation to pancreatic cells (CNV replication timing was delayed on average by 0.15 standard deviations [sd] more than non-CNVs when comparing differentiated cells to ESCs; P = 10−29), mesothelial cells (average relative CNV delay = 0.3 sd; P = 10−63), smooth muscle (average relative CNV delay = 0.29 sd; P = 10−62), neural crest (average relative CNV delay = 0.15 sd; P = 10−16), and neural progenitor cells (average relative CNV delay = 0.24 sd; P = 10−43). More modest changes were observed for NHEJ CNVs and for de novo mutations (supplementary fig. 5, Supplementary Material online). Of note, the human clustered protocadherins genes, like their zebrafish counterparts, were located in a region with clear development shift from early to late replication. The same region also contains several CNVs, although these CNVs were not as locally clustered at the Pcdh genes as they were in zebrafish (supplementary fig. 3B, Supplementary Material online). We conclude that, while not as prevasive as in zebrafish (to the extent we could evaluate with existing data), some human genetic variation loci also show replication timing changes during development. Thus, the link between developmental replication timing plasticity and germline mutations may be present across a wide array of organisms but may play a larger role in the development of some species.
Discussion
The association of DNA replication timing with mutation rates and the developmental plasticity of DNA replication timing are both established phenomena. Here, we link the two together. The availability of high-resolution replication timing profiles along several time points of embryonic development makes zebrafish a particularly powerful model for comparing the replication timing influences on mutations with its developmental plasticity. We find that CNV locations correspond to regions that change from early- to late-replicating along development. Conversely, regions that are progressively delayed in their replication timing are enriched with CNVs. These observations hold for individual CNVs and for CNV clusters and could not be explained by other known confounding genetic or epigenetic factors. The right arm of chromosome 4 provides a dramatic example of this relationship between CNVs and developmental changes in replication timing—it is the most extreme instance of developmental delay in replication timing in zebrafish and also harbors the highest density of CNVs in the genome. Taken together, the genomic regions that experience developmental changes in replication timing are also more prone to the accumulation of structural changes that manifest as CNVs.
We hypothesize that an underlying property of the genome and/or epigenome influences both the plasticity of DNA replication timing and the formation of structural mutations. For instance, particular chromatin structures, DNA sequences, and/or DNA secondary structures may make certain genomic regions more amenable to replication timing plasticity but also more fragile. Further research would be required in order to understand the significance of replication timing changes at many of the same locations that tend to be structurally variant. It is possible that DNA replication timing serves different functions in the early embryo and in different somatic cell lineages, including the germline. For instance, particular replication times in the germline could facilitate specific rates and spectra of evolutionary mutations, whereas later in development (or in different cell lineages), somatic structural mutation patterns could be better tuned to preventing cancer and modulating the activity of certain sets of genes.
Although our study is based on early embryonic development, mutations with heritable and evolutionary significance occur specifically in the germline. Although replication timing is mostly conserved between cell types, and embryonic cells appear to be the closest to germ cells in terms of replication timing (Yehuda et al. 2018), it remains to be determined whether germline replication timing is more similar to pre- or post-gastrulation replication timing, and whether CNV-containing genomic loci replicate early or late in the zebrafish germline. Regardless, the pervasive correlation of DNA replication timing with CNV locations suggests that replication timing may be a central factor influencing the formation of CNVs during evolution.
CNV loci that were delayed in their replication timing during embryonic development coincided with 440 genes. It will be interesting to study the activity of these genes across different developmental lineages and link them to changes in replication timing on one hand, and to genome stability on the other hand. Of particular interest are the clustered protocadherin genes, virtually all of which coincide with CNVs that replicate early pregastrulation but late postgastrulation. The genomic organization of the Pcdh gene clusters resembles that of the immunoglobulin and T-cell receptor gene clusters, both of which generate enormous diversity in the antibody repertoire through a mechanism that involves somatic rearrangement and mutations. Clustered Pcdh have been suggested to generate diversity in the brain by a related, but distinct mechanism mostly based on combinatorial epigenetic regulation of promoter choice and alternative transcripts (Chen and Maniatis 2013; Hirayama and Yagi 2013). Our results raise the possibility that genomic rearrangements and their intersection with replication timing regulation may play an underappreciated role in Pcdh biology in either somatic cells or during evolution. Furthermore, zebrafish belong to the teleost infraclass, the largest and most diverse vertebrate clade that exhibits wide diversity in habitat, morphology, behavior, physiology, and adaptations. The dynamic Pcdh clusters have been proposed to have facilitated the diversification of neural circuitry among teleosts and potentially contribute to their behavioral and physiological diversity (Yu et al. 2007). It would be interesting to consider a role for DNA replication timing regulation in such phenotypic adaptations. More generally, it is conceivable that unique mechanisms of genome evolution related to developmental regulation are operating in these species and facilitate their rapid evolution. In line with this notion, a recent study (Xie et al. 2019) showed that recurrent evolutionary adaptations in stickleback fish (a teleost) could be facilitated by specific chromosomal structures and DNA replication timing programs.
Our analysis of CNVs was limited by the lack of sequencing-based CNV calls, which could inform regarding formation mechanism (i.e., homologous recombination vs. nonhomologous end joining; Mills et al. 2011) and allow deeper mechanistic insight. Future studies, utilizing better refined maps of zebrafish structural variation, will be instrumental in providing more detailed understanding of the links between replication timing changes along development and CNVs.
Although CNV loci showed developmental variation in DNA replication timing, zebrafish SNP loci also showed an unexpected localization with respect to the replication timing program. Single nucleotide variants are enriched in late replication regions in all species studies so far and in both germline and somatic cells. In contrast, we used three independent data sets to show that SNPs in zebrafish replicate early in all developmental time points studied. We propose at least three implications for the early replication of SNPs in zebrafish: first, because genes tend to replicate early (Siefert et al. 2017), zebrafish may harbor a larger fraction of genic diversity compared with other species in which germline mutations tend to localize to gene-poor, late replicating regions. As suggested above, this could conceivably contribute to the vast genetic diversity of the teleost infraclass. Second, in mammalian genomes, GC content varies across chromosomes in correlation to DNA replication timing, and this correlation has been suggested to result from mutagenic pressures exerted by germline DNA replication timing programs (Kenigsberg et al. 2016). Zebrafish is unique in this respect as its replication timing program is not correlated with genomic GC content (Siefert et al. 2017). We speculate that the weak, positive correlation between replication timing and SNP density (and by inference, mutation rate) in zebrafish may be the cause of the lack of strong correlation between replication timing and GC content. Finally, the accumulation of mutations in late replicating regions in humans is likely attributable to DNA repair pathways (Zheng et al. 2014; Supek and Lehner 2015), which presumably become less effective in late-S phase and enable the increased accumulation of mutations. The lack of increased SNP densities in late-replicating loci in zebrafish may point to differences in DNA repair mechanisms in zebrafish compared with other species. A comparative approach to DNA repair and mutagenesis could thus be enlightening for the understanding of the mechanisms affecting mutation accumulation across the genome.
Taken together, our results suggest that a complete understanding of the genetic and epigenetic factors influencing mutation rate distribution across the genome would require studies across species as well as different developmental stages. Future studies of DNA replication timing in different systems, together with ever-refined maps of mutation and genetic variation, will address the mechanisms that cause the developmental shifts in replication timing at sites of germline variation and illuminate this interface between the genome, the epigenome, development, and evolution.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Itamar Simon and Qiliang Ding, Dashiell Massey and other members of our labs for helpful discussions and comments on the manuscript. This work was supported by grants R01GM121703 (to C.L.S.) and DP2GM1234951 (to A.K.) from the National Institutes of Health. M.L.H. was supported by a National Science Foundation Graduate Research Fellowship DGE-1650441.
Author Contributions
J.C.S., C.L.S., and A.K. conceived of the study; M.L.H. and J.C.S. performed the study; and all authors interpreted data, wrote, and edited the manuscript.
Literature Cited
- Agier N, Fischer G.. 2012. The mutational profile of the yeast genome is shaped by replication. Mol Biol Evol. 29(3):905–913. [DOI] [PubMed] [Google Scholar]
- Ashburner M, et al. 2000. Gene Ontology: tool for the unification of biology. Nat Genet. 25(1):25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanovic O, et al. 2012. Dynamics of enhancer chromatin signatures mark the transition from pluripotency to cell specification during embryogenesis. Genome Res. 22(10):2043–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen ME, Henke K, Siegfried KR, Warman ML, Harris MP.. 2012. Efficient mapping and cloning of mutations in zebrafish by low-coverage whole-genome sequencing. Genetics 190(3):1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KH, et al. 2012. Extensive genetic diversity and substructuring among zebrafish strains revealed through copy number variant analysis. Proc Natl Acad Sci U S A. 109(2):529–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MG, et al. 2015. SNPfisher: tools for probing genetic variation in laboratory-reared zebrafish. Development 142(8):1542–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso-Moreira MM, Long M.. 2010. Mutational bias shaping fly copy number variation: implications for genome evolution. Trends Genet. 26(6):243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-L, et al. 2010. Impact of replication timing on non-CpG and CpG substitution rates in mammalian genomes. Genome Res. 20(4):447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WV, Maniatis T.. 2013. Clustered protocadherins. Development 140(16):3297–3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De S, Michor F.. 2011. DNA replication timing and long-range DNA interactions predict mutational landscapes of cancer genomes. Nat Biotechnol. 29(12):1103.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschavanne P, Filipski J.. 1995. Correlation of GC content with replication timing and repair mechanisms in weakly expressed E. coli genes. Nucleic Acids Res. 23(8):1350–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprat R, et al. 2009. Predictable dynamic program of timing of DNA replication in human cells. Genome Res. 19(12):2288–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donley N, Thayer MJ.. 2013. DNA replication timing, genome stability and cancer: late and/or delayed DNA replication timing is associated with increased genomic instability. Semin Cancer Biol. 23(2):80-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emond MR, Jontes JD.. 2008. Inhibition of protocadherin-alpha function results in neuronal death in the developing zebrafish. Dev Biol. 321(1):175–187. [DOI] [PubMed] [Google Scholar]
- Flynn KM, Vohr SH, Hatcher PJ, Cooper VS.. 2010. Evolutionary rates and gene dispensability associate with replication timing in the archaeon Sulfolobus islandicus. Genome Biol Evol. 2:859–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francioli LC, et al. 2015. Genome-wide patterns and properties of de novo mutations in humans. Nat Genet. 47(7):822.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium. 2016. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 45:D331–D38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen RS, et al. 2010. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc Natl Acad Sci U S A. 107(1):139–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T, Yagi T.. 2013. Clustered protocadherins and neuronal diversity. Prog Mol Biol Transl Sci. 116:145–167. [DOI] [PubMed] [Google Scholar]
- Holden LA, Wilson C, Heineman Z, Dobrinski KP, Brown KH.. 2018. An interrogation of shared and unique copy number variants across genetically distinct zebrafish strains. Zebrafish 16(1):29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito-Harashima S, Hartzog PE, Sinha H, McCusker JH.. 2002. The tRNA-Tyr gene family of Saccharomyces cerevisiae: agents of phenotypic variation and position effects on mutation frequency. Genetics 161(4):1395–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenigsberg E, et al. 2016. The mutation spectrum in genomic late replication domains shapes mammalian GC content. Nucleic Acids Res. 44(9):4222–4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren A, et al. 2012. Differential relationship of DNA replication timing to different forms of human mutation and variation. Am J Hum Genet. 91(6):1033–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFave MC, Varshney GK, Vemulapalli M, James CM, Shawn MB.. 2014. A defined zebrafish line for high-throughput genetics and genomics: nHGRI-1. Genetics 198(1):167–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GI, Murray AW.. 2011. Mutation rates across budding yeast chromosome VI are correlated with replication timing. Genome Biol Evol. 3:799–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, et al. 2013. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499(7457):214.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, et al. 2015. Developmental enhancers revealed by extensive DNA methylome maps of zebrafish early embryos. Nat Commun. 6:6315.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre JL, Kostadinov D, Chen WV, Maniatis T, Sanes JR.. 2012. Protocadherins mediate dendritic self-avoidance in the mammalian nervous system. Nature 488(7412):517–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lele Z, Krone PH.. 1996. The zebrafish as a model system in developmental, toxicological and transgenic research. Biotechnol Adv. 14(1):57–72. [DOI] [PubMed] [Google Scholar]
- Liu L, De S, Michor F.. 2013. DNA replication timing and higher-order nuclear organization determine single-nucleotide substitution patterns in cancer genomes. Nat Commun. 4:1502.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long HK, et al. 2013. Epigenetic conservation at gene regulatory elements revealed by non-methylated DNA profiling in seven vertebrates. eLife 2:e00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills RE, et al. 2011. Mapping copy number variation by population-scale genome sequencing. Nature 470(7332):59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S, Freeman C, Auton A, Donnelly P, McVean G.. 2008. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat Genet. 40:1124–1129. [DOI] [PubMed] [Google Scholar]
- Nguyen D-Q, Webber C, Ponting CP.. 2006. Bias of selection on human copy-number variants. PLoS Genet. 2:e20.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan JP, Grimwood J, Schmutz J, Dickson M, Myers RM.. 2004. Gene conversion and the evolution of protocadherin gene cluster diversity. Genome Res. 14:354–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pink CJ, Hurst LD.. 2009. Timing of replication is a determinant of neutral substitution rates but does not explain slow Y chromosome evolution in rodents. Mol Biol Evol. 27:1077–1086. [DOI] [PubMed] [Google Scholar]
- Polak P, et al. 2015. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature 518(7539):360.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Mulia JC, et al. 2015. Dynamic changes in replication timing and gene expression during lineage specification of human pluripotent stem cells. Genome Res. 25(8):1091–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siefert JC, Georgescu C, Jonathan DW, Koren A, Christopher LS.. 2017. DNA replication timing during development anticipates transcriptional programs and parallels enhancer activation. Genome Res. 27(8):1406–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos JA, et al. 2009. Human mutation rate associated with DNA replication timing. Nat Genet. 41(4):393.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, et al. 2015. An integrated map of structural variation in 2,504 human genomes. Nature 526(7571):75.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Lehner B.. 2015. Differential DNA mismatch repair underlies mutation rate variation across the human genome. Nature 521(7550):81.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP.. 2011. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147(7):1537–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber CC, Pink CJ, Hurst LD.. 2012. Late-replicating domains have higher divergence and diversity in Drosophila melanogaster. Mol Biol Evol. 29(2):873–882. [DOI] [PubMed] [Google Scholar]
- Woo YH, Li W-H.. 2012. DNA replication timing and selection shape the landscape of nucleotide variation in cancer genomes. Nat Commun. 3:1004.. [DOI] [PubMed] [Google Scholar]
- Wu Q. 2005. Comparative genomics and diversifying selection of the clustered vertebrate protocadherin genes. Genetics 169(4):2179–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie KT, et al. 2019. DNA fragility in the parallel evolution of pelvic reduction in stickleback fish. Science 363(6422):81–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehuda Y, et al. 2018. Germline DNA replication timing shapes mammalian genome composition. Nucleic Acids Res. 46(16):8299–8310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu WP, Yew K, Rajasegaran V, Venkatesh B.. 2007. Sequencing and comparative analysis of fugu protocadherin clusters reveal diversity of protocadherin genes among teleosts. BMC Evol Biol. 7(1):49.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng CL, et al. 2014. Transcription restores DNA repair to heterochromatin, determining regional mutation rates in cancer genomes. Cell Rep. 9(4):1228–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.