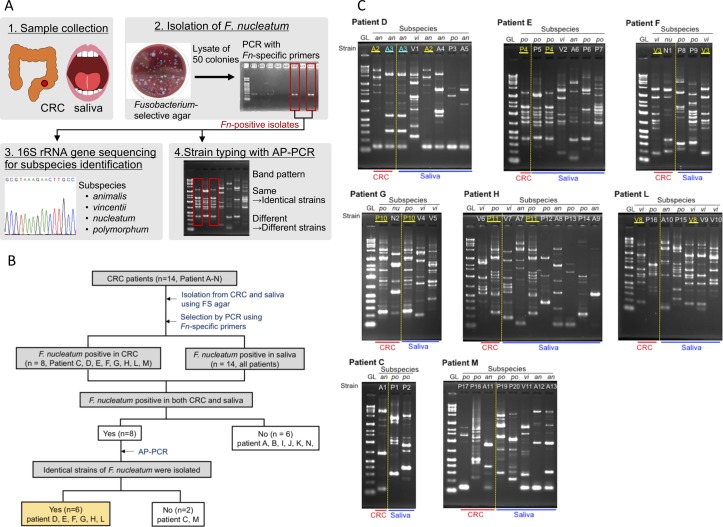
We read with great interest the article by Flemer et al, which suggests that analysis of the oral microbiota could potentially be used as a screening method for colorectal cancer (CRC) and polyp detection.1 Fusobacterium (F.) nucleatum is one of the most densely colonised bacterial species in the oral cavity and is known to be associated with periodontitis.2 Recently, many researchers have demonstrated that F. nucleatum is related to CRC development and pathogenicity.3 4 However, the relationship between F. nucleatum in CRC and the oral cavity is not well understood. For this purpose, we examined whether identical strains of F. nucleatum could be isolated from CRC and saliva specimens obtained from the same patient. The approach used in this study is detailed in figure 1A (see online supplementary information for details). We collected CRC and saliva samples from 14 patients (online supplementary table 1) and isolated bacteria from the specimens on Fusobacterium-selective agar. All colonies (1,351 in total) were analysed by PCR using F. nucleatum-specific primer sets, and 361 F. nucleatum isolates were obtained. F. nucleatum was detected in 8 of 14 patients (57.1%) from CRC samples and in all patients (100%) from saliva samples (figure 1B). The F. nucleatum subspecies identified by 16S rRNA gene sequencing and the number of isolates from each specimen are shown in table 1.
Figure 1.
Detection of Fusobacterium nucleatum subspecies in paired colorectal cancer and saliva samples. (A) Schematic of the experimental procedures. AP-PCR, arbitrarily primed PCR; CRC, colorectal cancer; Fn, Fusobacterium nucleatum. See online supplementary information for more details. (B) Flowchart of the study process. FS agar, Fusobacterium-selective agar. (C) AP-PCR patterns detected with primer D11344. Data are representative of at least two independent experiments. Identical pairs are highlighted in yellow or blue. GL, gene ladder (0.1, 0.2, 0.3, 0.4, 0.5, 0.7, 1.0, 1.3, 1.5, 2.0, 3.0, 4.0, 5.0, 7.0, 10 and 20 kbp). Subspecies, an, nu, po and vi are F. nucleatum subsp. animalis, F. nucleatum subsp. nucleatum, F. nucleatum subsp. polymorphum and F. nucleatum subsp. vincentii, respectively.
Table 1.
Subspecies and strains detected in each patient
| Fusobacterium nucleatum subspecies | Number of isolates | |||||||||||||||
| Patient C | Patient D | Patient E | Patient F | Patient G | Patient H | Patient L | Patient M | |||||||||
| CRC | Saliva | CRC | Saliva | CRC | Saliva | CRC | Saliva | CRC | Saliva | CRC | Saliva | CRC | Saliva | CRC | Saliva | |
| animalis | ||||||||||||||||
| Strain A1 | 21 | 0 | ||||||||||||||
| Strain A2 | 13* | 3* | ||||||||||||||
| Strain A3 | 1* | 5* | ||||||||||||||
| Strain A4 | 0 | 1 | ||||||||||||||
| Strain A5 | 0 | 2 | ||||||||||||||
| Strain A6 | 0 | 1 | ||||||||||||||
| Strain A7 | 0 | 2 | ||||||||||||||
| Strain A8 | 0 | 1 | ||||||||||||||
| Strain A9 | 0 | 1 | ||||||||||||||
| Strain A10 | 0 | 3 | ||||||||||||||
| Strain A11 | 1 | 0 | ||||||||||||||
| Strain A12 | 0 | 1 | ||||||||||||||
| Strain A13 | 0 | 1 | ||||||||||||||
| nucleatum | ||||||||||||||||
| Strain N1 | 11 | 0 | ||||||||||||||
| Strain N2 | 1 | 0 | ||||||||||||||
| polymorphum | ||||||||||||||||
| Strain P1 | 0 | 1 | ||||||||||||||
| Strain P2 | 0 | 1 | ||||||||||||||
| Strain P3 | 0 | 1 | ||||||||||||||
| Strain P4 | 1* | 5* | ||||||||||||||
| Strain P5 | 0 | 3 | ||||||||||||||
| Strain P6 | 0 | 1 | ||||||||||||||
| Strain P7 | 0 | 1 | ||||||||||||||
| Strain P8 | 0 | 3 | ||||||||||||||
| Strain P9 | 0 | 1 | ||||||||||||||
| Strain P10 | 47* | 3* | ||||||||||||||
| Strain P11 | 1* | 3* | ||||||||||||||
| Strain P12 | 0 | 5 | ||||||||||||||
| Strain P13 | 0 | 1 | ||||||||||||||
| Strain P14 | 0 | 2 | ||||||||||||||
| Strain P15 | 0 | 7 | ||||||||||||||
| Strain P16 | 1 | 0 | ||||||||||||||
| Strain P17 | 43 | 0 | ||||||||||||||
| Strain P18 | 3 | 0 | ||||||||||||||
| Strain P19 | 0 | 26 | ||||||||||||||
| Strain P20 | 0 | 8 | ||||||||||||||
| Strain P21 | 0 | 1 | ||||||||||||||
| vincentii | ||||||||||||||||
| Strain V1 | 0 | 3 | ||||||||||||||
| Strain V2 | 0 | 2 | ||||||||||||||
| Strain V3 | 2* | 2* | ||||||||||||||
| Strain V4 | 0 | 3 | ||||||||||||||
| Strain V5 | 0 | 1 | ||||||||||||||
| Strain V6 | 48 | 0 | ||||||||||||||
| Strain V7 | 0 | 21 | ||||||||||||||
| Strain V8 | 32* | 1* | ||||||||||||||
| Strain V9 | 0 | 1 | ||||||||||||||
| Strain V10 | 0 | 1 | ||||||||||||||
| Strain V11 | 0 | 6 | ||||||||||||||
Strain P21 did not grow from stock.
*Strains detected from both specimens.
gutjnl-2018-316661supp001.pdf (205KB, pdf)
gutjnl-2018-316661supp003.pdf (393.7KB, pdf)
Four subspecies, F. nucleatum subsp. animalis, F. nucleatum subsp. nucleatum, F. nucleatum subsp. polymorphum and F. nucleatum subsp. vincentii were isolated from the samples. To identify F. nucleatum isolates from CRC and saliva at the strain level, we performed arbitrarily primed PCR (AP-PCR) as the strain typing method, which can be applied without genome information or specialised techniques and equipment.5–7 We performed AP-PCR targeting the F. nucleatum isolates from the 8 patients whose CRC and saliva samples were both F. nucleatum-positive and analysed the detected AP-PCR patterns (figure 1C and online supplementary figure 1). Focusing on patient C (left, bottom), there were no common isolates between their CRC and saliva samples (figure 1C). However, patient D (left, top) had two and four strains of F. nucleatum subsp. animalis detected in their CRC and saliva, respectively. Furthermore, strains A2 and A3 (highlighted in yellow and blue) were indicated as identical strains by the AP-PCR patterns (figure 1C). We detected identical F. nucleatum strains in both CRC and saliva from 42.9% (6/14) of the patients. Notably, an identical strain was detected in 75% (6/8) of patients who were both F. nucleatum-positive in CRC and saliva specimens. From our results, there were no significant differences in the detection rate of F. nucleatum among each lesion site from the 8 patients. F. nucleatum was detected from stages 0 to IV (online supplementary table 1), indicating that F. nucleatum could adhere to CRC tissue from an early stage of tumorigenesis, as previously reported.8 9 From our results, more than 40% of CRC patients exhibited identical strains of F. nucleatum in their CRC and saliva specimens. This suggests that F. nucleatum in CRC originates in the oral cavity. Our findings support that targeting F. nucleatum in the oral cavity may provide insights for further studies in the field of human microbiome research and CRC.
gutjnl-2018-316661supp002.pdf (393.7KB, pdf)
Acknowledgments
We thank Christina Croney, PhD, and H Nikki March, PhD, from Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript.
Footnotes
Contributors: YK and YSh contributed equally to this paper. MM and AN share senior authorship. TH and MM conceived this study. YK, YSh and YSu performed the experiments. YK, TH, JA, SUm and SUc provided clinical specimens. YK, YSh, TH, YSu and MM interpreted the data. YK wrote the first draft, and TH, YSu, MM and AN contributed to the completion of the manuscript. All authors read, critically revised for important intellectual content and approved the final manuscript.
Competing interests: None declared.
Patient consent: Obtained.
Ethics approval: Ethics Committee of Yokohama City University Hospital.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. Flemer B, Warren RD, Barrett MP, et al. The oral microbiota in colorectal cancer is distinctive and predictive. Gut 2018;67:1454–63. 10.1136/gutjnl-2017-314814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Signat B, Roques C, Poulet P, et al. Fusobacterium nucleatum in periodontal health and disease. Curr Issues Mol Biol 2011;13:25–36. [PubMed] [Google Scholar]
- 3. Mima K, Nishihara R, Qian ZR, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut 2016;65:1973–80. 10.1136/gutjnl-2015-310101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kostic AD, Chun E, Robertson L, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013;14:207–15. 10.1016/j.chom.2013.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Haraldsson G, Holbrook WP, Könönen E. Clonal persistence of oral Fusobacterium nucleatum in infancy. J Dent Res 2004;83:500–4. 10.1177/154405910408300613 [DOI] [PubMed] [Google Scholar]
- 6. George KS, Reynolds MA, Falkler WA. Arbitrarily primed polymerase chain reaction fingerprinting and clonal analysis of oral Fusobacterium nucleatum isolates. Oral Microbiol Immunol 1997;12:219–26. 10.1111/j.1399-302X.1997.tb00382.x [DOI] [PubMed] [Google Scholar]
- 7. Li W, Raoult D, Fournier PE. Bacterial strain typing in the genomic era. FEMS Microbiol Rev 2009;33:892–916. 10.1111/j.1574-6976.2009.00182.x [DOI] [PubMed] [Google Scholar]
- 8. Sobin LH, Gospodarowicz MK, Wittekind C, et al. : Hoboken NJ, TNM classification of malignant tumours. 7th edn Chichester, West Sussex, UK: Wiley-Blackwell, 2009:94–105. [Google Scholar]
- 9. Tahara T, Yamamoto E, Suzuki H, et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res 2014;74:1311–8. 10.1158/0008-5472.CAN-13-1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2018-316661supp001.pdf (205KB, pdf)
gutjnl-2018-316661supp003.pdf (393.7KB, pdf)
gutjnl-2018-316661supp002.pdf (393.7KB, pdf)