Abstract
Background
Although immunotherapy has recently achieved clinical successes in a variety of cancers, thus far there is no immunotherapeutic strategy for breast cancer (BC). Thus, it is important to discover biomarkers for identifying BC patients responsive to immunotherapy. TP53 mutations were often associated with worse clinical outcomes in BC whose triple-negative subtype has a high TP53 mutation rate (approximately 80%). To explore a potentially promising therapeutic option for the TP53-mutated BC subtype, we studied the association between TP53 mutations and immunogenic activity in BC.
Methods
We compared the enrichment levels of 26 immune signatures that indicated activities of diverse immune cells, functions, and pathways between TP53-mutated and TP53-wildtype BCs based on two large-scale BC multiomics datasets. Moreover, we explored the molecular cues associated with the differences in immunogenic activity between TP53-mutated and TP53-wildtype BCs. Furthermore, we performed experimental validation of the findings from bioinformatics analysis.
Results
Bioinformatics analysis showed that almost all analyzed immune signatures showed significantly higher enrichment levels in TP53-mutated BCs than in TP53-wildtype BCs. Moreover, in vitro experiments confirmed that mutant p53 could increase BC immunogenicity. Both computational and experimental results demonstrated that TP53 mutations could promote BC immunogenicity via regulation of the p53-mediated pathways including cell cycle, apoptosis, Wnt, Jak-STAT, NOD-like receptor, and glycolysis. Furthermore, we found that elevated immune activity was likely associated with a better survival prognosis in TP53-mutated BCs, but not necessarily in TP53-wildtype BCs.
Conclusions
TP53 mutations may promote immunogenic activity in BC, suggesting that the TP53 mutation status could be a useful biomarker for stratifying BC patients responsive to immunotherapy.
1. Introduction
The tumor suppressor p53 plays an important role in the regulation of cell-cycle, apoptosis, DNA repair, cellular senescence, and autophagy [1]. Accordingly, TP53 mutations and dysfunction are importantly involved in carcinogenesis due to the disturbance of these biologic processes it functions in. In fact, TP53 mutations occur in more than half of all human cancer cases [2] and are independent markers of poor prognosis in a variety of cancers [3]. p53 also plays an important role in immune regulation, e.g., the control of immune responses to infection, autoimmunity, and cancer [4]. p53 functions in immunity by induction of apoptosis, removal of apoptotic cells, antiviral defense, induction of type I IFN, enhanced pathogen recognition, cytokine production, and immune checkpoint regulation [4]. Several studies have explored the association of p53 and tumor immune regulation [5–8]. For example, the p53 activation in the tumor microenvironment (TME) might overcome tumor immune suppression and enhance antitumor immunity [8]. p53 could transactivate many tumor immunosuppressive genes such as PD-L1, VISTA, and FOXP3 [5]. p53 functioned in both tumor suppression and anticancer immunosurveillance via regulation of VISTA [5].
Recently, cancer immunotherapy has shown successes in treating various cancers [9]. In particular, the blockade of immune checkpoints has achieved rapid clinical successes in multiple cancers, including skin, lung, kidney, bladder, head and neck cancers, lymphoma, and the cancers with deficient DNA mismatch repair (dMMR) [10]. Unfortunately, current immunotherapies are only propitious to a subset of cancer patients [11]. Some molecular biomarkers associated with cancer immunotherapy response have been identified, e.g., tumor mutation burden (TMB) [12], neoantigens [13], dMMR [14], and PD-L1 expression [15]. However, few studies have correlated the TP53 mutation status with cancer immunotherapy response, although a recent clinical trial (phase II) data showed that patients with mutated-p53 metastatic breast cancer had better overall survival (OS) when treated with the immunooncology viral agent REOLYSIN® in combination with paclitaxel [16].
Breast cancer (BC) is the most common cancer and the second leading cause of cancer death in women [17]. The triple-negative BC (TNBC) is the BC subtype which does not express estrogen receptor (ER) and progesterone receptor (PR) and lacks overexpression of the human epidermal growth factor receptor 2 (HER2) [18]. TNBC has a high TP53 mutation rate (80% in TNBC versus 33% in general BC) [19] and has a poor prognosis due to its aggressive clinical behavior and lack of response to hormonal or HER2 receptor-targeted therapy. Although there is currently no immunotherapeutic drug clinically used for BC therapy, several studies have indicated that TNBC might be propitious to immunotherapy [20–22].
To explore the association of TP53 mutations with tumor immunity in BC, we compared the activity of 26 immune signatures between TP53-mutated and TP53-wildtype BCs based on the Cancer Genome Atlas (TCGA) [23] and METABRIC [24] BC genomic data. We found that these immune signatures exhibited significantly higher activity in TP53-mutated BCs than in TP53-wildtype BCs. Furthermore, we explored the molecular cues correlated with the differences in immune activities between TP53-mutated and TP53-wildtype BCs. Finally, we performed experimental validation of the findings from bioinformatics analysis.
2. Methods
2.1. Datasets
We downloaded TCGA BC RNA-Seq gene expression profiles (Level 3), gene somatic mutations (Level 3), somatic copy number alterations (SCNAs) (Level 3), protein expression profiles (Level 3), and clinical data from the genomic data commons data portal (https://portal.gdc.cancer.gov/), and the METABRIC gene expression profiles, gene somatic mutations, SCNAs, and clinical data from cBioPortal (http://www.cbioportal.org). We obtained 26 gene sets representing 26 different immune signatures from several publications, including 15 immune cell types and functions [25], tumor-infiltrating lymphocytes (TILs) [26], proinflammatory [27], parainflammation (PI) [28], cytokine and cytokine receptor (CCR) [29], human leukocyte antigen (HLA) [22], cancer testis (CT) antigen [30], regulatory T (Treg) cells [31], immune checkpoint [31], metastasis-promoting, and metastasis-inhibiting [32] (Supplementary Table S1). The sample sizes of breast cancers are presented in Supplementary Table S2. We performed computational and statistical analyses using R programming (https://www.r-project.org/).
2.2. Comparisons of Gene Expression Levels, Gene-Set Enrichment Levels, and Protein Expression Levels between Two Classes of Samples
We normalized the TCGA BC gene expression values by base-2 log transformation and used the downloaded normalized METABRIC gene expression data. For the TCGA BC protein expression profiles data, we used the downloaded normalized data. We quantified the activity of an immune signature in a sample by the single-sample gene-set enrichment analysis (ssGSEA) (“GSVA” version 1.24.2, R package) score [33, 34] of the gene set representing the immune signature (a higher ssGSEA score indicated a higher activity) (Supplementary Table S3). We compared gene or protein expression levels between two classes of samples using Student's t test and compared immune signature enrichment levels (ssGSEA scores) between two classes of samples using the Mann-Whitney U test. The false discovery rate (FDR) was utilized to adjust for multiple tests by the Benjamini and Hochberg (BH) method [35]. The threshold of FDR < 0.05 was used to identify the statistical significance. The comparisons involving normal tissue were performed only in TCGA since METABRIC had no normal tissue related data available.
2.3. Comparison of the Immune Cell Infiltration Degree between TP53-Mutated and TP53-Wildtype BCs
We evaluated the immune cell infiltration degree in BC using ESTIMATE (“estimate” version 1.0.13, R package) [36]. For each BC sample, ESTIMATE output an immune score that quantified its immune cell infiltration degree based on the BC gene expression data. In addition, we obtained the lymphocyte infiltration percentage data for BC from the TCGA BC clinical data. We compared immune scores or lymphocyte infiltration percentage between TP53-mutated and TP53-wildtype BCs using the Mann-Whitney U test.
2.4. Gene-Set Enrichment Analysis
We used GSEA [37] to identify differentially expressed KEGG pathways between TP53-mutated and TP53-widtype BCs with the threshold of FDR < 0.05.
2.5. Comparison of the Proportions of Leukocyte Cell Subsets between TP53-Mutated and TP53-Wildtype BCs
We used CIBERSORT [38] to calculate the proportions of 22 human leukocyte cell subsets and compared the proportions of the leukocyte cell subsets between TP53-mutated and TP53-wildtype BCs using the Mann-Whitney U test.
2.6. Correlation of Pathway or Protein Activity with Immune Activity in BCs
We obtained the gene-set collections for p53-mediated pathways from KEGG [39] and quantified the pathway activity with the ssGSEA score of the set of genes included in the pathway. To correct for the strong correlation between the p53 pathway and the other p53-mediated pathways, we evaluated the correlation between a pathway activity and an immune activity using the first-order partial correlation method (“ppcor” version 1.1, R package) [40] with the significance level of FDR<0.05. The correlation between a protein and an immune signature was evaluated by the Spearman correlation coefficient (“rho”) of the protein expression levels and the immune signature enrichment levels.
2.7. Survival Analyses
We compared overall survival (OS) and disease-free survival (DFS) between two classes of BC patients divided by the TP53 mutation status (TP53-mutated versus TP53-wildtype), or the median values of gene expression levels, immune signature enrichment levels, and immune scores. Kaplan-Meier survival curves were used to exhibit the 20-year survival differences between both groups, and the log-rank test was used to evaluate the significance of survival-time differences with a threshold of P < 0.05.
2.8. Comparison of Mutation Counts between TP53-Mutated and TP53-Wildtype BCs
We compared mutation counts (defined as total number of somatic point mutations and indels) between TP53-mutated and TP53-wildtype BCs using the Mann-Whitney U test. This comparison was performed only in TCGA since somatic mutation data in TCGA were generated by whole exome sequencing while in METABRIC they were generated by targeted exome sequencing.
2.9. In Vitro Experiments
2.9.1. Cell Lines and Cell Culture
Human cells from breast cancer, MCF-7 (ER+/HER2-), and natural killer cells NK-92 were from the American Type Culture Collection (ATCC). MCF-7 was cultured in RPMI-1640 (GIBCO, USA) supplemented with 10% fetal bovine serum (FBS, GIBCO, USA). NK92 cells were incubated in α-MEM (GIBCO, USA) with 2 mM L-glutamine, 0.2 mM inositol, 0.02 mM folic acid, 0.01 mM 2-mercaptoethanol, 10 ng/ml IL-2, 12.5% FBS, and 12.5% horse serum (GIBCO, USA). These cells were cultured in a humidified incubator at 37°C and a 5% CO2 atmosphere and were harvested in logarithmic growth phase.
2.9.2. Cell Transfection
The MCF-7 cells without antibiotic were maintained in the medium with TP53-mutated (c.596 G > T, c.818 A > G, and c.925 T > C) virus stock solution and polybrene (5 ug/mL) for 24h. The transfected MCF-7 cells were cultured in a humidified incubator at 37°C and a 5% CO2 atmosphere for 48h.
2.9.3. Coculture of MCF-7 and NK-92 Cells
The transwell chamber (Corning Inc., Corning, NY, USA) was inserted into a 6-well plate to construct a coculture system. MCF-7 cells were seeded on the 6-well plate at a density of 5×104 cells/well, and NK-92 cells were seeded on the membrane (polyethylene terephthalate, pore size, 0.4μm) of the transwell chamber at a density of 5×104 cells/chamber. NK-92 and MCF-7 cells were cocultured in a humidified incubator at 37°C and a 5% CO2 atmosphere for 24h.
2.9.4. Transwell Migration Assay
After coculture of 24h, NK-92 cells were harvested and resuspended in the upper transwell chambers (8-μm pores, Corning), and MCF-7 cells in the lower 24-well plates. Both NK-92 and MCF-7 cells were incubated at 37°C for 24h. The membrane was removed and its upper surface was wiped away with a cotton swab to remove the unmigrated NK-92 cells. The membrane was fixed in neutral formalin and air-dried at a room temperature and was stained with 0.1% crystal violet at 37°C for 30 min. The number of NK-92 cells that migrated to the lower surface of the membrane was counted under light microscope. Each assay was performed in triplicate wells.
2.9.5. EdU Proliferation Assay
After coculture of 24h, an EdU (5-ethynyl-2′-deoxyuridine, Invitrogen, CA, USA) proliferation assay [41] was performed to measure the proliferation ability of NK-92 cells. NK-92 cells were plated in 96-well plates at a density of 2 × 103 cells/well for 24h. The cells were incubated with 10 μM EdU for 24h at 37°C before fixation, permeabilization, and EdU staining according to manufacturer's protocol. The cell nuclei were stained with DAPI (Sigma) at a concentration of 1 μg/ml for 20 min. The proportion of the cells incorporated EdU was determined with fluorescence microscopy. Each assay was performed in triplicate wells.
2.9.6. CCL4 and CCL5 Enzyme-Linked Immunosorbent Assay
After coculture of 24h, supernatants from NK-92 cells were collected and assayed. CCL4 and CCL5 protein levels were evaluated by ELISA, according to manufacturer's protocol (Shanghai Enzyme-Linked Biotechnology Co., Ltd. China). Study samples and standard dilutions of the chemokines/cytokines were assayed in triplicate. The absorbance was read at 450 nm with the correction set to 570 nm by a microplate reader (BioTek, USA).
2.9.7. Reverse Transcription Quantitative PCR (qPCR) Analysis
Z-DEVD-FMK, abemaciclib, and MPP were purchased from Selleck, Haoyuan Chemexpress, and Cayman, respectively. MCF-7 cells were harvested after being treated by drugs (Z-DEVD-FMK, 50 μM, 72h; abemaciclib, 250 nM, 7 days; MPP, 0.1 nM, 72h). The total RNA was isolated by Trizol (Invitrogen, USA) and was reversely transcribed into cDNA using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher, USA). Primer sequences used for qPCR were presented in the Supplementary Table S4. Primers were diluted in nuclease-free water with the Real time PCR Master Mix (SYBR Green) (TOYOBO Co., LTD, JAPAN). Relative copy number was determined by calculating the fold-change difference in the gene of interest relative to β-actin. The qPCR was performed on an ABI 7500 FAST and Applied Biosystems StepOnePlus Real Time PCR machine.
2.9.8. Western Blotting
MCF-7 cells were washed twice with cold PBS and were lysed in SDS buffer (1% SDS, 0.1 M Tris pH 7.4, 10% glycerol) supplemented with protease inhibitors. The protein concentration was determined by Bradford Protein Assay (Bio-Rad). After normalization of the total protein content, samples were resolved by standard SDS-PAGE. After Western blotting transfer, NC membranes (Millipore) were incubated with antibodies ER-alpha (21244-1-AP, Proteintech Group, INC.) and cleaved-caspase 3 (KGYC0004-6, KeyGEN Biotech, China). After 2h incubation with the HRP-labeled secondary antibody (KGAA002-1, KeyGEN Biotech, China), proteins were visualized by enhanced chemiluminescence using a G: BOX chemiXR5 digital imaging system.
2.9.9. Knockdown of p53 with Small Interfering RNA (siRNA)
MCF-7 cells were seeded in 6-well plates and grown until they attained 70% confluency. Transfection of siRNA was performed using Lipofectamine 3000 (Invitrogen, CA) according to the manufacturer's instructions. p53 siRNA and control siRNA were synthesized by KeyGEN BioTECH. The sequence of siRNA against p53 was sense (5′-3′): CCAUCCACUACAACUACAUdTdT, and antisense (5′-3′): AUGUAGUUGUAGUGGAUGGdTdG.
2.9.10. Statistical Analyses
All experimental data were expressed as mean ± SD and were analyzed by t test using Prism 5.0 software (GraphPad). P < 0.05 was considered statistically significant.
3. Results
3.1. TP53-Mutated BCs Exhibit Significantly Stronger Immune Signatures than TP53-Wildtype BCs
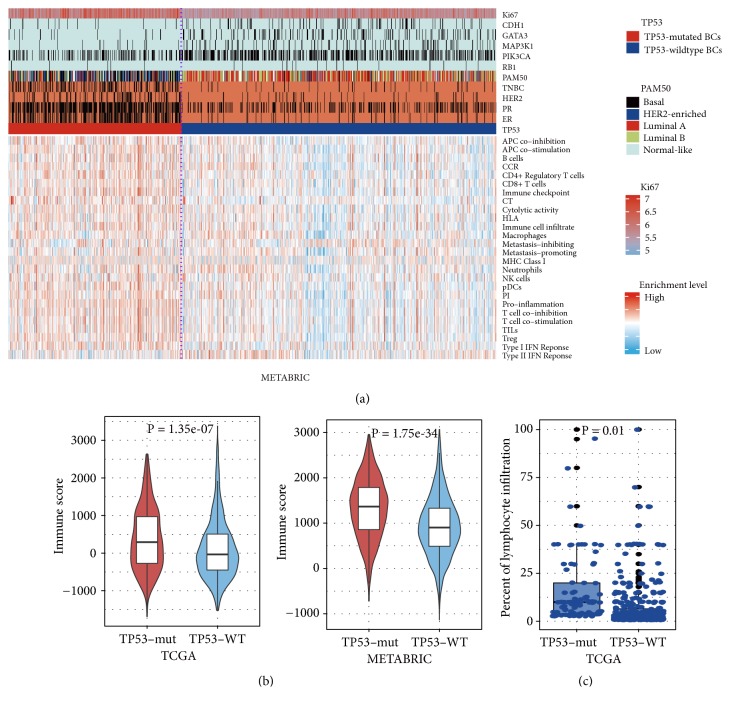
Strikingly, we found that almost all 26 immune signatures analyzed showed significantly higher enrichment levels in TP53-mutated BCs than in TP53-wildtype BCs consistently in both TCGA and METABRIC datasets (Mann-Whitney U test, P<0.05; Figure 1(a), Supplementary Figure S1A). Moreover, TP53-mutated BCs had significantly higher immune scores than TP53-wildtype BCs in both datasets (Mann-Whitney U test; P=1.35∗10−7. 1.75∗10−34 for TCGA and METABRIC, respectively) (Figure 1(b)). On the basis of the TCGA BC pathological slides data, we found that TP53-mutated BCs had markedly higher percentages of lymphocyte infiltration compared to TP53-wildtype BCs (Mann-Whitney U test, P=0.01) (Figure 1(c)). Altogether, these data indicate that TP53 mutations are associated with elevated immune activity in BC.
Figure 1.
TP53-mutated breast cancers (BCs) have increased immune activities compared to TP53-wildtype BCs. (a) Heatmap showing the ssGSEA scores of 26 immune gene-sets in TP53-mutated and TP53-wildtype BCs (METABRIC). ssGSEA: single-sample gene-set enrichment analysis. TNBC: triple-negative breast cancer. Red color indicates higher enrichment levels (ssGSEA scores) of gene-sets, and blue color indicates lower enrichment levels of gene-sets in the heatmap. RB1 are more frequently mutated in TP53-mutated BCs while CDH1, GATA3, MAP3K1, and PIK3CA are more frequently mutated in TP53-wildtype BCs (Fisher's exact test, P<0.05). The black vertical lines in the horizontal bars beside gene symbols indicate that the genes are mutated in corresponding samples. The black vertical lines in the horizontal bar beside “TNBC” indicate that the sample is a TNBC. The black vertical lines in the horizontal bars beside “ER”, “PR,” and “HER2” indicate that the sample is ER-, PR-, or HER2-. (b) TP53-mutated BCs have significant higher degree of immune infiltration than TP53-wildtype cancers evaluated by ESTIMATE [29]. (c) The TCGA BC pathological slides data show that TP53-mutated BCs had markedly higher percent of lymphocyte infiltration than TP53-wildtype BCs. TP53-mut: TP53-mutated BCs. TP53-WT: TP53-wildtype BCs. It applies to all the other figures.
Of the 15 immune cell type and function signatures [25], 14 showed significantly higher enrichment levels in TP53-mutated BCs than in TP53-wildtype BCs (Mann-Whitney U test, FDR<0.05) (Supplementary Table S5). Moreover, numerous marker genes of immune cells and function had significantly higher expression levels in TP53-mutated BCs than in TP53-wildtype BCs, e.g., CD8A (CD8+ T cell), B2M, HLA-A, and TAP1 (MHC Class I), and GZMA and PRF1 (cytolytic activity) (Supplementary Table S5). The TILs signature composed of 122 genes [26] showed significantly higher enrichment levels in TP53-mutated BCs than in TP53-wildtype BCs (Mann-Whitney U test; P=2.84∗10−8, 4.22∗10−33 for TCGA and METABRIC, respectively) (Figure 1(a), Supplementary Figure S1B). Moreover, 112 (92%) TIL signature genes were more highly expressed in TP53-mutated BCs than in TP53-wildtype BCs in at least 1 dataset (88 in both datasets) (Supplementary Table S6; Figure S1C). Altogether, these data indicate that TP53-mutated BCs have higher degree of immune cell infiltration than TP53-wildtype BCs.
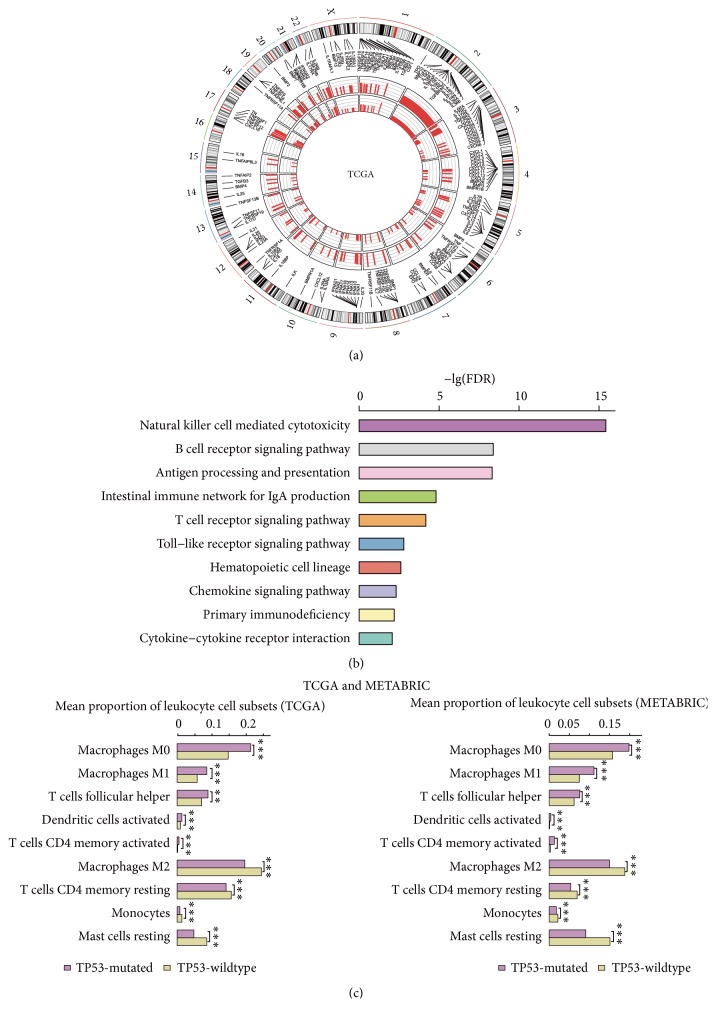
Cytokines are important constituents of the immune system and play crucial roles in the immune regulation of cancer [42]. The enrichment levels of the CCR signature [29] were significantly higher in TP53-mutated BCs than in TP53-wildtype BCs in both datasets (Mann-Whitney U test; P=1.9∗10−10, 5.32∗10−39 for TCGA and METABRIC, respectively) (Figure 1(a); Supplementary Figure S1D; Table S7). Of the 261 CCR genes, 158 (61%) were more highly expressed in TP53-mutated BCs and 230 (88%) showed more frequent SCNAs in TP53-mutated BCs compared to TP53-wildtype BCs (Fisher's exact test, FDR < 0.05; Figure 2(a)). These data suggest that TP53 mutations are associated with higher cytokine activity in BC.
Figure 2.
TP53-mutated breast cancers (BCs) have higher activity of cytokines, immune pathways, and immune-promoting leukocyte cell subsets than TP53-wildtype BCs. (a) Cytokine and cytokine receptor (CCR) genes show more frequent somatic copy number alterations (SCNAs) in TP53-mutated BCs than in TP53-wildtype BCs (Fisher's exact test, FDR<0.05). The outmost circle indicates 23 human chromosomes. The bars in both inner circles (outside and inside) indicate the frequency of SCNAs of CCR genes in TP53-mutated and TP53-wildtype BCs, respectively. A longer bar indicates a higher frequency of SCNAs. (b) Immune-related KEGG pathways upregulated in TP53-mutated BCs relative to TP53-wildtype BCs (FDR q-value<0.05). (c) TP53-mutated breast cancers (BCs) have significantly different leukocyte cell subset infiltrates estimated by CIBERSORT [30] compared to TP53-wildtype BCs.
The proinflammatory signature was more enriched in TP53-mutated BCs than in TP53-wildtype BCs in both datasets (Mann-Whitney U test; P=2.07∗10−17, 1.81∗10−61 for TCGA and METABRIC, respectively). Strikingly, all 16 proinflammatory genes [27] were upregulated in TP53-mutated BCs relative to TP53-wildtype BCs in at least 1 dataset (13 in both datasets) (Supplementary Figure S1E, S1F; Table S8). Remarkably, STAT1 (signal transducer and activator of transcription 1) was upregulated in TP53-mutated BCs compared to both TP53-wildtype BCs and normal tissue. This gene has been shown to interact with p53 [43] and enhance immunosuppression in BC [44]. Another proinflammatory gene GZMB (granzyme B) was upregulated in TP53-mutated BCs compared to both TP53-wildtype BCs and normal tissue. This gene and GZMA (one of the cytolytic activity marker genes upregulated in TP53-mutated BCs) are associated with immune cytolytic activity as their protein products are mainly secreted by NK cells and cytotoxic T lymphocytes [25]. These data suggest that TP53 mutations may promote inflammatory and immune cytolytic activities in BC. In addition, we found that another inflammatory signature parainflammation (PI) [28], a low-grade inflammatory reaction associated with carcinogenesis [28], was more enriched in TP53-mutated BCs than in TP53-wildtype BCs (Mann-Whitney U test; P=1.03∗10−9, 5.03∗10−37 for TCGA and METABRIC, respectively) and was also more enriched in both TP53-mutated BCs and TP53-wildtype BCs than in normal tissue (Mann-Whitney U test; P=6.89∗10−12, 0.003 for TP53-mutated and TP53-wildtype BCs, respectively) (Supplementary Table S8). These observations are in line with a previous study showing that PI significantly correlated with the p53 status in cancer [28].
GSEA [37] identified significantly upregulated pathways in TP53-mutated BCs compared to TP53-wildtype BCs, many of which were immune-related, including natural killer cell mediated cytotoxicity, B cell receptor signaling, antigen processing and presentation, intestinal immune network for IgA production, T cell receptor signaling, toll-like receptor signaling, hematopoietic cell lineage, chemokine signaling, primary immunodeficiency, and cytokine-cytokine receptor interaction (Figure 2(b)). In contrast, only the cytokine-cytokine receptor interaction pathway was significantly enriched in TP53-wildtype BCs. These results again demonstrate that TP53 mutations are associated with elevated immune activity in BC.
Furthermore, we compared the proportions of 22 human leukocyte cell subsets that were evaluated by CIBERSORT [38] between TP53-mutated and TP53-wildtype BCs. We found that TP53-mutated BCs harbored higher proportions of activated dendritic cells, M0 macrophages, M1 macrophages, activated T cells CD4 memory, and T cells follicular helper cell subsets (Mann-Whitney U test; FDR<0.05; Figure 2(c)). In contrast, TP53-wildtype BCs harbored higher proportions of resting dendritic cells, M2 macrophages, resting mast cells, monocytes, and resting T cells CD4 memory cell subsets (Mann-Whitney U test; FDR<0.05; Figure 2(c)). This further demonstrates that TP53 mutations are associated with stronger immune activity in BC. Intriguingly, M1 macrophages that incite inflammation had higher proportions in TP53-mutated BCs than in TP53-wildtype BCs, while M2 macrophages that repress inflammation and encourage tissue repair had lower proportions in TP53-mutated BCs. It suggests that TP53 mutations may promote inflammatory behavior and inhibit tissue repair in BC, thereby contributing to higher invasiveness of TP53-mutated BCs [45].
3.2. TP53 Mutations Are Associated with Elevated HLA Activity in BC
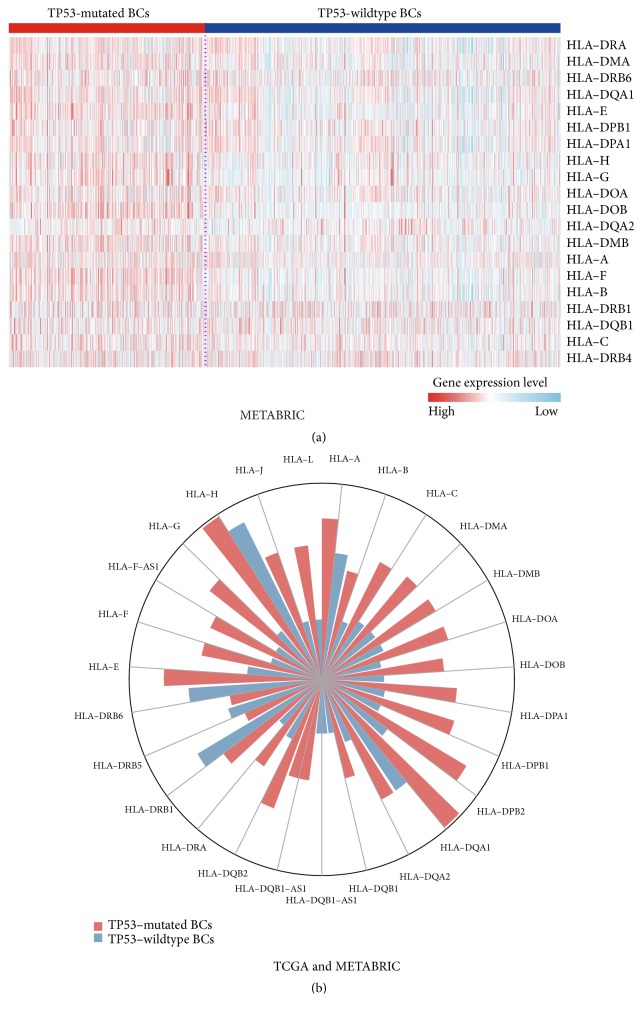
The products of HLA genes MHC proteins play important roles in the regulation of the immune system [46]. We found that most HLA genes showed significantly higher expression levels in TP53-mutated BCs than in TP53-wildtype BCs (Figure 3(a); Supplementary Table S9; Figure S1G). Moreover, HLA genes were more frequently amplified in TP53-mutated BCs compared to TP53-wildtype BCs (Figure 3(b)). TP53-mutated BCs had lower somatic mutation rates of HLA genes than TP53-wildtype BCs in TCGA (Fisher's exact test, P=0.02, OR=0.6), while METABRIC had no somatic mutation data available for HLA genes. These data suggest that TP53 mutations may promote HLA activity in BC. This finding appears not to be consistent with a previous study showing that p53 increased expression of MHC proteins in cancer [47]. This inconsistency supports the notion that the p53 function is context-dependent and largely depends on the cell type [48, 49].
Figure 3.
TP53-mutated breast cancers (BCs) have more elevated expression of HLA genes than TP53-wildtype BCs. (a) Heatmap shows that TP53-mutated BCs likely more highly express HLA genes than TP53-wildtype BCs (METABRIC). (b) HLA genes are more frequently amplified in TP53-mutated BCs than in TP53-wildtype BCs. The length of the bars in the rose diagram is proportional to the frequency of HLA gene amplification in TP53-mutated or TP53-wildtype BCs.
Gene mutations may yield neoantigens that are associated with antitumor immune response [11]. Although TP53-mutated BCs had markedly higher total mutation counts than TP53-wildtype BCs in TCGA (Mann-Whitney U test; P=4.18∗10−25), the numbers of gene mutations yielding predicted HLA-binding peptides [25] showed no significant differences between TP53-mutated and TP53-wildtype BCs (Mann-Whitney U test; P=0.4). It suggests that TMB or neoantigens may not be the essential factor explaining the differential immune activities between TP53-mutated and TP53-wildtype BCs.
3.3. Immune Activities Are Associated with Activities of p53-Regulated Pathways in BC
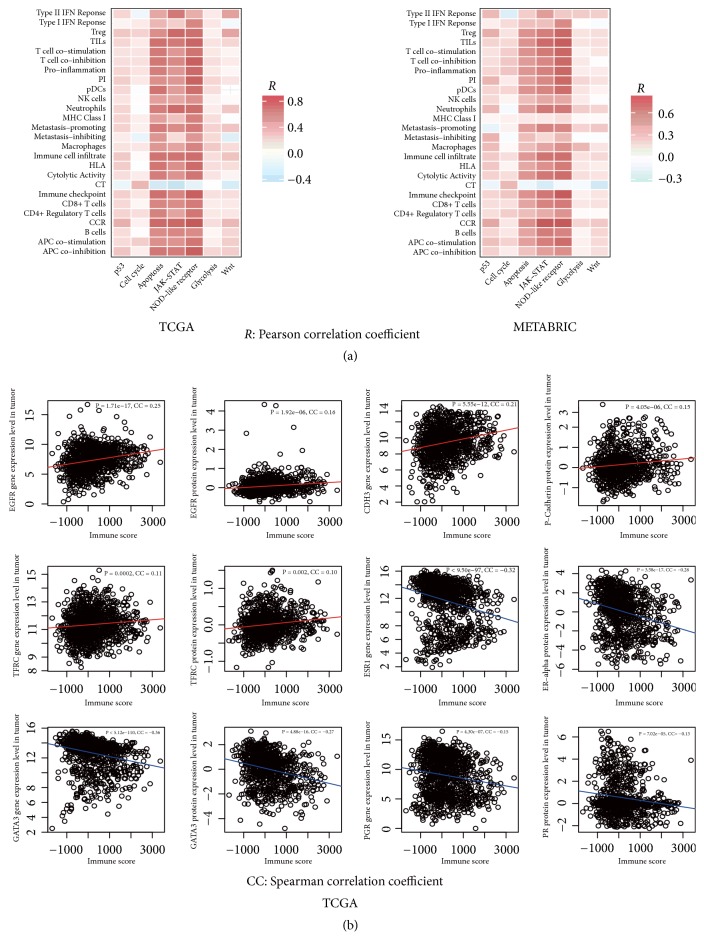
p53 plays important roles in regulating the cancer-associated pathways, e.g., cell cycle, apoptosis, DNA damage repair, autophagy, metabolism, inflammation, epithelial–mesenchymal transition (EMT), angiogenesis, and metastasis [48]. Accordingly, TP53 mutations often result in the disturbance of the p53-mediated pathways [3]. Indeed, we found that a number of p53-mediated pathways showed significantly differential activity between TP53-mutated and TP53-wildtype BCs, such as the p53, cell cycle, apoptosis, Jak-STAT, NOD-like receptor, glycolysis, and Wnt pathways showing significantly higher activity in TP53-mutated BCs than in TP53-wildtype BCs (Mann-Whitney U test; P<0.05). Moreover, these pathways tended to positively correlate with the immune signatures analyzed (Figure 4(a)). These results indicate that the altered immune activity in TP53-mutated BCs could be associated with the disturbance of the p53-mediated pathways.
Figure 4.
Immune signatures significantly correlate with p53-regulated pathways, genes, and proteins in BC. (a) Immune signatures likely positively correlate with the p53-mediated pathways that show higher activity in TP53-mutated BCs than in TP53-wildtype BCs. (b) Immune signatures positively correlate with EGFR, CDH3, and TFRC, and their protein products that are upregulated in TP53-mutated BCs, while they negatively correlate with ESR1, GATA3, and PCR, and their protein products that are downregulated in TP53-mutated BCs relative to TP53-wildtype BCs.
3.4. Identification of Genes and Proteins Differentially Expressed between TP53-Mutated and TP53-Wildtype BCs and Significantly Correlating with Immune Activity in BC
Based on the gene and protein expression data in TCGA, we identified the genes and proteins that were differentially expressed between TP53-mutated and TP53-wildtype BCs (Student's t test; FDR<0.05). Of these, 10 genes (EGFR, CDH3, TFRC, CCNE1, CDK1, CDKN2A, CHEK1, FOXM1, NDRG1, and STMN1) and their protein products had significantly higher expression levels in TP53-mutated BCs and 8 genes (ESR1, GATA3, PGR, AR, ERBB3, BCL2, IGF1R, and CCND1) and their protein products had significantly lower expression levels in TP53-mutated BCs. We termed the 10 genes and their protein products upregulated in TP53-mutated BCs as GPU and the 8 genes and their protein products downregulated in TP53-mutated BCs as GPD. Interestingly, GPU had a significant positive expression correlation with almost all 26 immune signatures and immune scores, while GPD had a significant negative expression correlation with them (Spearman correlation, FDR<0.05; Figure 4(b); Supplementary Figures S2A, S2B, S2C). These results showed that the expression of these molecules correlated with elevated or depressed immune activity in BC.
3.5. Association of Immune Activity with Clinical Outcomes in BC
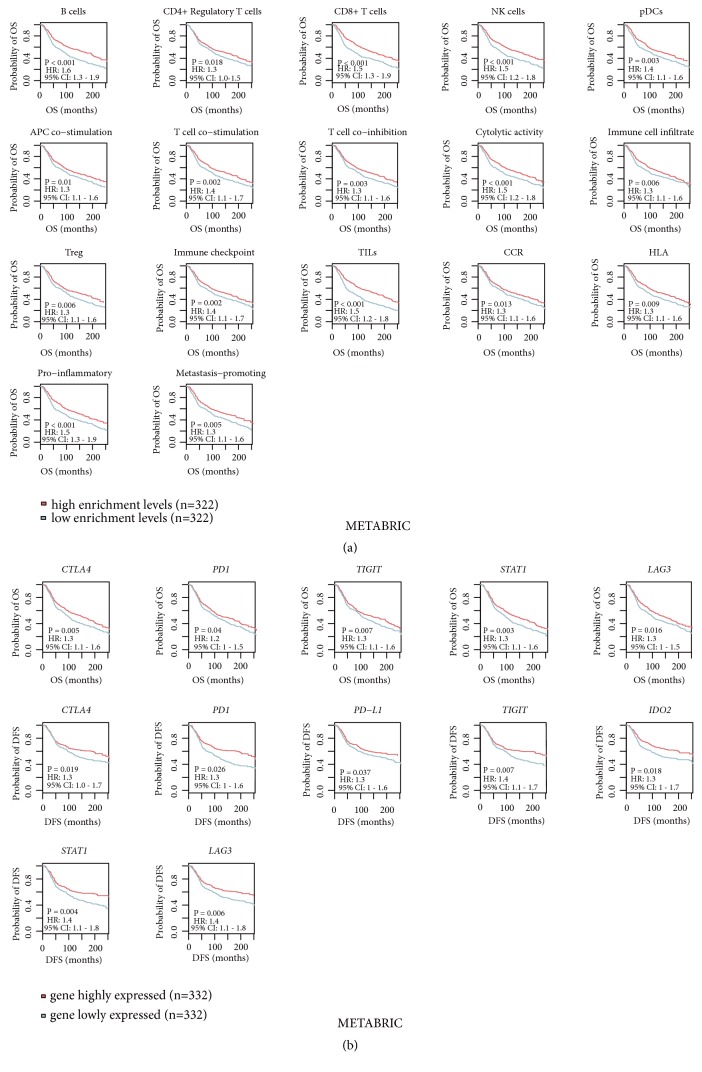
Among 26 immune signatures, 17 and 15 showed a significant correlation with survival (OS and/or DFS) prognosis in TP53-mutated and TP53-wildtype BCs, respectively (log-rank test; P<0.05) (Figure 5(a); Supplementary Figures S3A, S3B). Strikingly, elevated enrichment of the 17 immune signatures consistently correlated with a more favorable prognosis in TP53-mutated BCs. In contrast, 10 and 5 immune signatures were positively and negatively associated with survival in TP53-wildtype BCs, respectively. The B cell, cytolytic activity, T cell coinhibition, immune checkpoint, TILs, CCR, HLA, and proinflammatory signatures showed a positive correlation with survival consistently in both TP53-mutated and TP53-wildtype BCs. However, the CD4+ regulatory T cell signature showed a positive correlation with DFS in TP53-mutated BCs while showing a negative correlation in TP53-wildtype BCs (Supplementary Figures S3A, S3B; Table S10).
Figure 5.
Immune activities are positively associated with a 20-year survival prognosis in TP53-mutated BCs. (a) Kaplan-Meier survival curves show that the elevated enrichment of immune signatures is associated with a better 20-year survival in TP53-mutated BCs. (b) Kaplan-Meier survival curves show that higher expression levels of immune genes are associated with a better 20-year survival in TP53-mutated BCs. The log-rank test P<0.05 indicates the significance of survival-time differences between two classes of patients. HR: hazard ratio; CI: confidence interval.
Furthermore, we found numerous immune-related genes whose expression was associated with survival prognosis in TP53-mutated and/or TP53-wildtype BCs. For example, the immune checkpoint genes CTLA4, PD1, PD-L1, PD-L2, and TIGIT, and the CD8+ T cell marker gene CD8A were positively associated with prognosis in both TP53-mutated and TP53-wildtype BCs (Figure 5(b); Supplementary Figure S3C). In addition, some genes were positively associated with prognosis exclusively in TP53-mutated or TP53-wildtype BCs, e.g., IL10, CD247, GZMA, GZMB, CD276, CCR4, and CCR7 (Supplementary Table S11). Interestingly, some genes had a positive correlation with survival in TP53-mutated BCs while having a negative correlation in TP53-wildtype BCs, e.g., IDO2, STAT1, and LAG3 (Figure 5(b); Supplementary Table S11; Figure S3C). The mechanism underlying these discrepancies may lie in that TP53 mutations alter the tumor immune microenvironment (TIM) in BC.
3.6. In Vitro Experiments Validate that TP53 Mutations Promote Immune Activity in BC
3.6.1. TP53 Mutations Increase the Expression of MHC Class I Genes in MCF-7 Cells
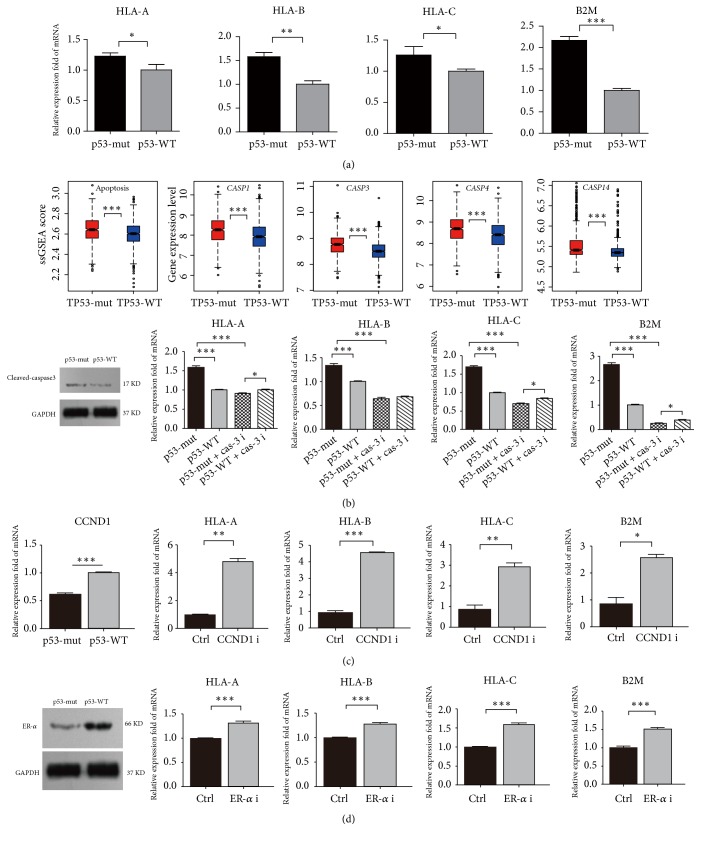
We used a pair of isogenic BC cell lines with different p53 status (MCF-7 p53-wildtype versus MCF-7 p53-mutant) and evaluated MHC class I gene expression levels in both cell lines. The MHC Class I genes (HLA-A, HLA-B, HLA-C, and B2M) had significantly higher expression levels in p53-mutant MCF-7 cells than in p53-wildtype MCF-7 cells, demonstrated by real-time qPCR (Figure 6(a)). These experimental results verified that TP53 mutations increased the expression of HLA molecules in BC.
Figure 6.
The expression of MHC Class I genes is significantly upregulated in p53-mutant MCF-7 cells versus p53-wildtype MCF-7 cells and is regulated by the cell cycle, apoptosis, and estrogen receptor (ER) activities. (a) MHC Class I genes (HLA-A, HLA-B, HLA-C, and B2M) have significantly higher mRNA expression levels in p53-mutant MCF-7 cells than in p53-wildtype MCF-7 cells, evident by real-time quantity PCR. (b) Promotion of apoptosis increases the expression of MHC Class I genes, evident by both computational and experimental analyses. (c) Inhibition of cell cycle increases the expression of MHC Class I genes. (d) Inhibition of ER alpha increases the expression of MHC Class I genes. p53-WT: p53-wildtype; p53-mut: p53-mutant; CCND1 i: CCND1 inhibitor; ERa i: ER alpha inhibitor.
3.6.2. TP53 Mutations Increase the Expression of MHC Class I Genes via Regulation of Apoptosis in BC
p53 plays an important role in regulation of apoptosis [50]. Surprisingly, our bioinformatics analysis showed that TP53-mutated BCs had significantly higher activity of the apoptosis pathway than TP53-wildtype BCs and that TP53-mutated BCs more highly expressed apoptosis-inducing caspases such as CASP1, CASP3, CASP4, and CASP14 (Figure 6(b)). Furthermore, our experiments verified that caspase-3 expression markedly increased in p53-mutant MCF-7 cells versus p53-wildtype MCF-7 cells (Figure 6(b)). We treated both p53-mutant and p53-wildtype MCF-7 cells with the caspase-3 inhibitor Z-DEVD-FMK and found that the MHC Class I genes had markedly decreased expression in both p53-mutant and p53-wildtype MCF-7 cells (Figure 6(b)). Interestingly, we observed that p53-mutant MCF-7 cells more lowly expressed three out of the four MHC Class I genes than p53-wildtype MCF-7 cells after they were treated with Z-DEVD-FMK. These data indicate that apoptosis may have an appreciable effect on tumor immunity and that TP53 mutations alter tumor immunity via regulation of apoptosis.
3.6.3. TP53 Mutations Increase the Expression of MHC Class I Genes via Regulation of Cell Cycle in BC
Our bioinformatics analysis showed that CCND1 (cyclin D1), a regulator of cyclin-dependent kinases, was downregulated in TP53-mutated BCs versus TP53-wildtype BCs. Furthermore, our experiments verified that CCND1 had significantly lower mRNA expression levels in p53-mutant MCF-7 cells than in p53-wildtype MCF-7 cells (Figure 6(c)). We treated p53-wildtype MCF-7 cells with the cyclin D1 inhibitor abemaciclib and observed a substantial increase in the expression of MHC Class I genes in MCF-7 cells (Figure 6(c)). Thus, the alteration of the p53-mediated cell cycle pathway may contribute to the differential tumor immunity between TP53-mutated and TP53-wildtype BCs.
3.6.4. TP53 Mutations Increase the Expression of MHC Class I Genes via Downregulation of Estrogen Receptor Alpha
Our bioinformatics analysis showed that both ESR1 and its protein product estrogen receptor alpha (ERα) were downregulated in TP53-mutated BCs versus TP53-wildtype BCs (Supplementary Table S12). This is consistent with previous studies showing that p53 upregulated ERα expression in BC and that TP53 mutations downregulated ERα expression [51, 52]. Furthermore, our experiments verified that ERα was more lowly expressed in p53-mutant MCF-7 cells than in p53-wildtype MCF-7 cells (Figure 6(d)). We treated p53-wildtype MCF-7 cells with the ERα inhibitor MPP and observed a marked increase in the expression of MHC Class I genes in MCF-7 cells (Figure 6(d)). These results indicate that the downregulation of ERα may contribute to the elevated tumor immunity in TP53-mutated BCs.
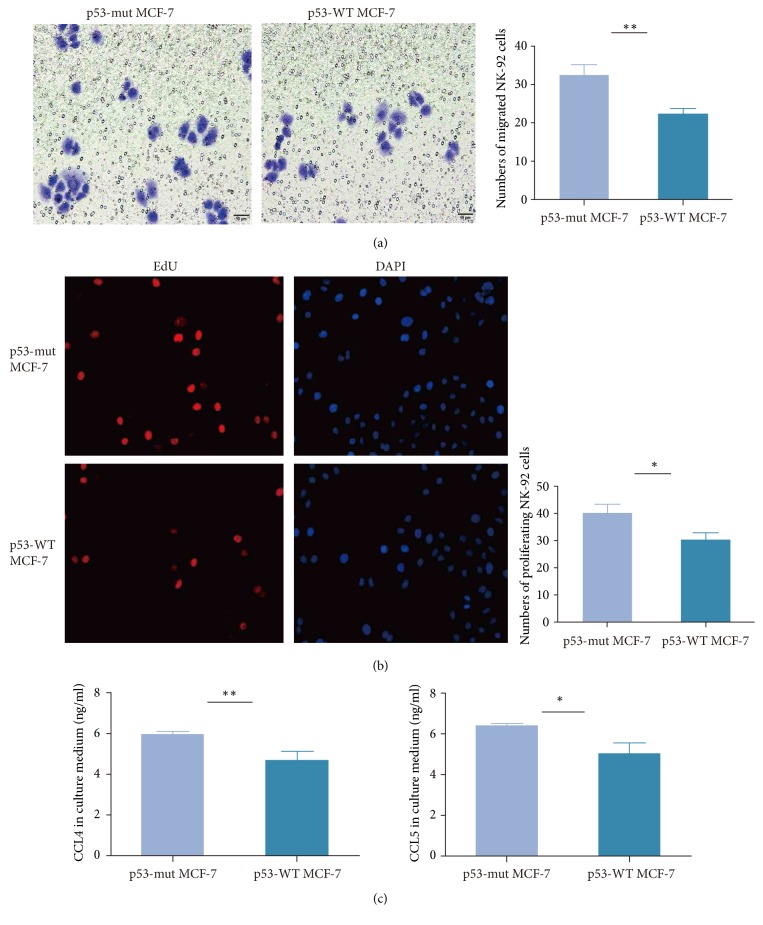
3.6.5. Mutant p53 Promotes Migration and Proliferation of NK Cells Cocultured with MCF-7 Cells
We used the transwell migration and EdU proliferation assay to observe the migration and proliferation of NK92 cells cocultured with p53-mutant and p53-wildtype MCF-7 cells for 24h, respectively. We found that the number of migrated NK92 cells cocultured with p53-mutant MCF-7 cells far exceeded the number of migrated NK92 cells cocultured with p53-wildtype MCF-7 cells (Figure 7(a)). Moreover, the NK92 cells cocultured with p53-mutant MCF-7 cells showed significantly stronger proliferation ability compared to the NK92 cells cocultured with p53-wildtype MCF-7 cells (Figure 7(b)). Furthermore, we observed that the cytokines CCL4 and CCL5 had markedly higher levels in the serum containing NK92 cells cocultured with p53-mutant MCF-7 cells than in the serum containing NK92 cells cocultured with p53-wildtype MCF-7 cells (Figure 7(c)). These observations verified our computational results that TP53-mutated BCs had stronger activities of immune cells including NK cells and more highly expressed a number of CCR genes including CCL4 and CCL5 than TP53-mutated BCs. These findings are also consistent with previous studies showing that cytokines such as CCL4 could induce NK cells migration [53] and that activated NK cells could secrete cytokines to mediate immune response [54].
Figure 7.
Mutant p53 promotes migration and proliferation of NK cells cocultured with MCF-7 cells. (a) NK92 cells cocultured with p53-mutant MCF-7 cells show stronger migration ability than NK92 cells cocultured with p53-wildtype MCF-7 cells, evident by transwell migration assay. (b) NK92 cells cocultured with p53-mutant MCF-7 cells show stronger proliferation ability than NK92 cells cocultured with p53-wildtype MCF-7 cells, evident by EdU proliferation assay. (c) Cytokines CCL4 and CCL5 have markedly higher levels in the serum containing NK92 cells cocultured with p53-mutant MCF-7 cells than in the serum containing NK92 cells cocultured with p53-wildtype MCF-7 cells, evident by quantitative enzyme-linked immunosorbent assay (ELISA).
4. Discussion
We performed a comprehensive portrait of the association between TP53 mutations and immune signatures in BC. We found that TP53-mutated BCs showed significantly higher levels of immune infiltration and higher activity of various immune cells, function, and pathways than TP53-wildtype BCs (Figure 1(a)). TP53-mutated BCs had higher proportions of activated immune cell subsets and lower proportions of resting immune cell subsets compared to TP53-wildtype BCs within the TME. TP53-mutated BCs have significant differences in clinical features compared to TP53-wildtype BCs. Typically, TP53-mutated BCs contain a higher proportion of ER-, PR-, HER2+, or triple-negative/basal-like BCs. Previous studies have shown that the ER- and HER2+ features were associated with stronger immunogenic activity in BC [22, 55]. Thus, both features may contribute to the higher immune activity in TP53-mutated BCs as compared to TP53-wildtype BCs. However, when comparing the enrichment levels of the 26 immune signatures between TP53-mutated and TP53-wildtype BCs within the ER+ subtype of BC, we obtained similar results that almost all these immune signatures were more enriched in TP53-mutated ER+ BCs than in TP53-wildtype ER+ BCs (Supplementary Table S13). Similarly, TP53-mutated HER2- BCs had significantly higher enrichment levels of immune signatures than TP53-wildtype HER2- BCs (Supplementary Table S14). These results indicate that the TP53 mutation itself is capable of contributing to the elevated immune activity in BC as was further verified by in vitro experiments. Our computational and experimental results suggest that TP53 mutations may alter immune activity in BC via regulation of the p53-mediated pathways, including cell cycle, apoptosis, Wnt, Jak-STAT, NOD-like receptor, and glycolysis. It should be noted that previous studies have demonstrated that p53 could increase immune activity in various cancers, e.g., colon cancer [47], gastric cancer [56], and lymphoma [8], as appears not to be in line with the present findings. Nevertheless, a number of studies have shown that p53 functions in a context-dependent fashion [8, 48, 49]. Thus, these distinct effects of p53 in regulating tumor immunity could be attributed to the different cellular contexts. Numerous studies have shown that TP53 mutations may result in not only loss of the wildtype p53 tumor-suppressive function but also gain of oncogenic function of mutant p53 [57]. A recent study showed that TP53 gain of function mutation could promote inflammatory activity in glioblastoma [58]. To explore whether the elevated immune/inflammatory activity in TP53-mutated BC is attributed to TP53 gain of function mutations, we silenced TP53 expression in MCF-7 cells by siRNA. Interestingly, we observed a significant increase in the expression levels of MHC Class I genes in p53-knockdown MCF-7 cells compared with p53-wildtype MCF-7 cells (Supplementary Figure S5). This indicates that the elevated immune/inflammatory activity in TP53-mutated BC is likely caused by TP53 loss of function mutations.
One tumor sample may contain a certain percentage of nontumor cells such as normal cells and stromal cells. To exclude the impact of nontumor associated cells on the present results, we selected the BC samples composed of 100% tumor cells based on the TCGA BC pathological slides data. We observed the similar results that almost all 26 immune signatures exhibited significantly higher enrichment levels in the TP53-mutated class than in the TP53-wildtype class of these samples (Mann-Whitney U test; FDR<0.05) (Supplementary Table S15). Thus, the differential immune activity between TP53-mutated and TP53-wildtype BCs referred to the actual difference in tumor immunity.
Cancer-testis (CT) antigens are a group of immunogenic proteins overexpressed in many cancers [59]. We found that the CT antigen signature [30] was more active in TP53-mutated BCs than in TP53-wildtype BCs in both datasets (Mann-Whitney U test; P=9.52∗10−35, 9.46∗10−24 for TCGA and METABRIC, respectively) (Figure 1(a); Supplementary Figure S4A). Many CT antigen genes were upregulated in TP53-mutated BCs and encode the CT antigens that are potential targets for developing cancer vaccines, e.g., MAGEA, NY-ESO-1, and PRAME (Supplementary Figure S4B; Table S16). It suggests that p53 could inhibit the expression of many CT antigens, a finding in line with a prior study [60].
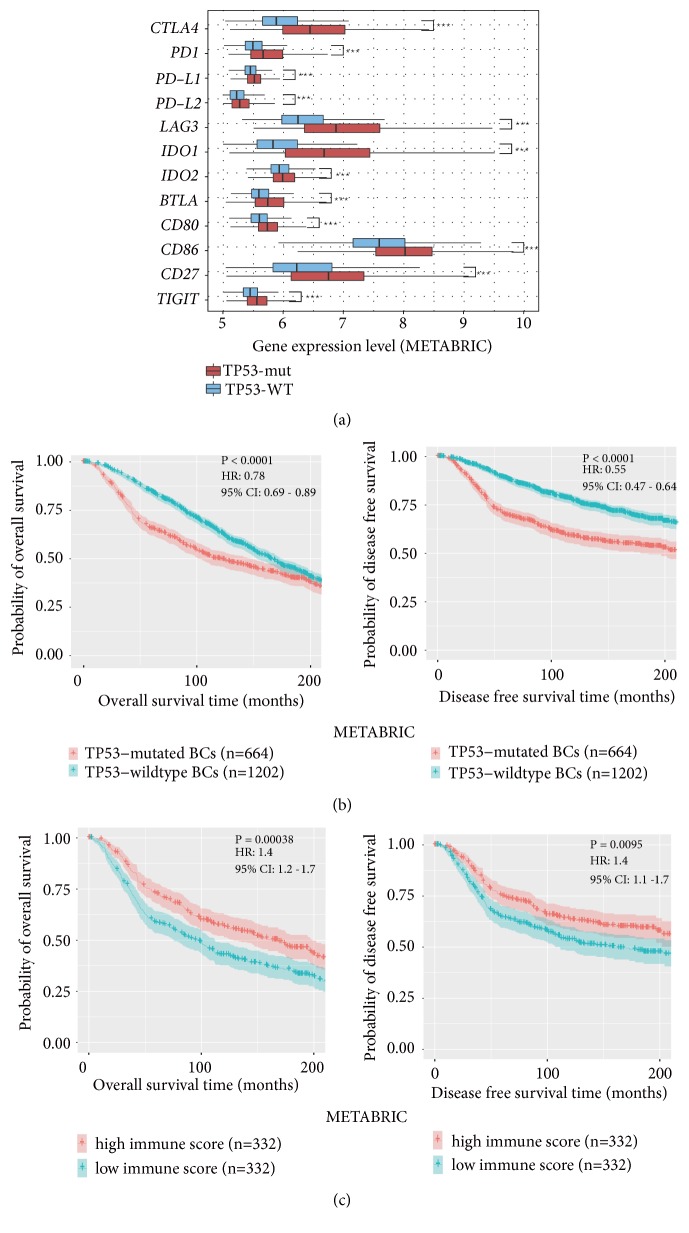
Interestingly, we found that TP53-mutated BCs had remarkably higher enrichment levels of Treg signature and immune checkpoint signature [31] than TP53-wildtype BCs in both datasets (Mann-Whitney U test; P<10−10) (Figure 1(a); Supplementary Figure S4C; Table S17). In particular, numerous notable immune checkpoint genes were upregulated in TP53-mutated BCs, including CTLA4, PD1, PD-L1, PD-L2, LAG3, IDO1/2, BTLA, CD80, CD86, CD27, and TIGIT (Figure 8(a); Supplementary Table S18; Figure S4D). These results suggest that p53 may play a role in inhibiting tumor immunosuppression in BC. Weyden et al. [32] identified 19 genes which function in immune regulation of cancer metastasis, of which 12 promoted tumor metastasis and 7 inhibited tumor metastasis. The enrichment levels of the metastasis-promoting signature were markedly higher in TP53-mutated BCs than in TP53-wildtype BCs in both datasets (Mann-Whitney U test; P=3.62∗10−6, 0.003 for TCGA and METABRIC, respectively) (Supplementary Figure 4E). In contrast, the metastasis-inhibiting signature exhibited significantly lower enrichment levels in TP53-mutated BCs than in TP53-wildtype BCs in TCGA (Mann-Whitney U test; P=0.01) (Supplementary Figure 4E). Notably, SPNS2 (sphingolipid transporter 2) which most incited tumor metastasis by regulating lymphocyte trafficking [32], had higher expression levels in TP53-mutated BCs than in TP53-wildtype BCs in TCGA (Student's t test; FDR=1.34∗10−6; SPNS2 expression data was lacking in METABRIC). These results suggest that TP53-mutated BCs are metastasis-prone and that this characteristic may be attributed to the defect in p53 immune regulation of BC and its TME.
Figure 8.
TP53-mutated breast cancers (BCs) more highly express immune checkpoint genes and have a worse 20-year survival than TP53-wildtype BCs and associated with unfavorable survival prognosis in BC, while higher degree of immune cell infiltration is associated with a 20-year better survival prognosis in BC. (a) A number of important immune checkpoint genes are upregulated in TP53-mutated BCs versus TP53-wildtype BCs. (b) Kaplan-Meier survival curves show that TP53-mutated BCs have a worse 20-year survival prognosis than TP53-wildtype BCs. (c) Kaplan-Meier survival curves show that higher degree of immune cell infiltration is associated with a better 20-year survival prognosis in TP53-mutated BCs. The log-rank test P<0.05 indicates the significance of survival-time differences between two classes of patients.
The elevated expression of immunosuppressive, proinflammatory, and metastasis-promoting signatures in TP53-mutated BCs may promote tumor invasion and lead to a worse prognosis in BC. Indeed, previous studies have shown that p53 mutations were associated with unfavorable clinical outcomes in BC [61, 62]. The METABRIC data also showed that TP53-mutated BCs had worse OS and DFS compared to TP53-wildtype BCs (Figure 8(b)). Moreover, TP53-mutated BCs more highly expressed Ki67 (a marker for cell proliferation) than TP53-wildtype BCs (Figure 1(a)), again indicating the higher aggressiveness of TP53-mutated BCs. Interestingly, the activities of different immune cell types, function, and pathways, and the immune cell infiltration degree were consistently positively associated with survival prognosis in TP53-mutated BCs (Figures 5(a), 5(b), and 8(c)). It is sensible that the elevated enrichment of CD8+ T cell, B cell, NK cell, cytolytic activity, HLA, immune cell infiltrate, TILs, and CCR is associated with favorable clinical outcomes in cancer since these immune signatures can promote anticancer immune response. Furthermore, the observation that the elevated enrichment of Treg, immune checkpoint, proinflammatory, and metastasis-promoting immune signatures was associated with better survival in TP53-mutated BCs may be due to the fact that the elevated immunosuppressive activity is likely to promote chemotherapy sensitivity of TP53-mutated BCs [63]. Thus, to achieve successes in immunotherapy of TP53-mutated BCs, the effective combination of chemotherapy with immunotherapy may represent a promising direction [64].
Interestingly, compared to TP53-wildtype BCs, TP53-mutated BCs more highly express a majority of the gene targets for immunotherapy agents that are currently used in the clinic or clinical trials [65] (Supplementary Table S19). It indicates that these immunotherapy agents may be more effective against TP53-mutated BCs than TP53-wildtype BCs. In fact, several clinical trials [20, 21] have shown that immune checkpoint blockade was effective against TNBC, a BC subtype with a high TP53 mutation rate.
5. Conclusions
TP53 mutations promote immune activity in BC. This finding suggests that the TP53 mutation status could be a useful biomarker for stratifying BC patients responsive to immunotherapy.
Acknowledgments
This work was supported by the China Pharmaceutical University (grant numbers 3150120001 and 2632018YX01 to Xiaosheng Wang). The authors would like to thank Dr. Jiaqi Ge for his valuable suggestions on their experimental design.
Data Availability
The TCGA data can be downloaded from the genomic data commons data portal (https://portal.gdc.cancer.gov/), and the METABRIC data can be downloaded from cBioPortal (http://www.cbioportal.org).
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Supplementary Materials
Supplementary Tables. Table S1: the list of 26 immune signatures and related gene sets. Table S2: sample sizes of breast cancers. Table S3: ssGSEA scores of immune signature in TCGA and METABRIC. Table S4: primer sequences used for real time quantity PCR. Table S5: comparison of the enrichment levels of 15 immune cell types and function signatures between two classes of samples. Table S6: comparison of the enrichment levels of the tumor-infiltrating lymphocytes signature between two classes of samples. Table S7: comparison of the enrichment levels of the cytokine and cytokine receptor signature between two classes of samples. Table S8: comparison of the enrichment levels of the inflammation-promoting and parainflammation (PI) signatures between two classes of samples. Table S9: comparison of the enrichment levels of the HLA signature between two classes of samples. Table S10: comparisons of the ssGSEA scores of immune signatures between TP53-mutated and TP53-wildtype BCs and their associations with survival prognosis in BC. Table S11: comparisons of the expression levels of immune genes between TP53-mutated and TP53-wildtype BCs and their associations with survival prognosis in BC. Table S12: comparisons of the expression levels of genes and their protein products between TP53-mutated and TP53-wildtyped BCs. Table S13: comparisons of the enrichment levels of immune signatures between TP53-mutated and TP53-wildtype BCs within the ER+ subtype of BC. Table S14: comparisons of the enrichment levels of immune signatures between TP53-mutated and TP53-wildtype BCs within the HER2- subtype of BC. Table S15: comparisons of the enrichment levels of immune signatures between TP53-mutated and TP53-wildtype BCs within the 100% tumor purity of BC. Table S16: comparisons of the enrichment levels of the cancer-testis signature between two classes of samples. Table S17: comparisons of the enrichment levels of the Treg signature between two classes of samples. Table S18: comparisons of the enrichment levels of the immune checkpoint signature between two classes of samples. Table S19: comparisons of the expression levels of the genes targeted by immunotherapy agents in clinical use or trials or in preclinical development between TP53-mutated and TP53-wildtype BCs. Supplementary Figures Legends. Figure S1: TP53-mutated breast cancers (BCs) have increased immune activity compared to TP53-wildtype BCs. A: heatmap shows the ssGSEA scores of 26 immune signatures in TP53-mutated and in TP53-wildtype BCs (TCGA). ssGSEA: single-sample gene-set enrichment analysis; TNBC: triple-negative breast cancer. RB1 is more frequently mutated in TP53-mutated BCs while CDH1, GATA3, MAP3K1, and PIK3CA are more frequently mutated in TP53-wildtype BCs (Fisher's exact test; P<0.05). The black vertical lines in the horizontal bars beside gene symbols indicate that the genes are mutated in corresponding samples. The black vertical lines in the horizontal bar beside “TNBC” indicate that the samples are TNBC. The black vertical lines in the bars beside “ER”, “PR,” and “HER2” indicate that the samples are ER-, PR-, or HER2-. B: TP53-mutated BCs have higher levels of tumor-infiltrating lymphocytes (TILs) infiltration than TP53-wildtype BCs (Mann-Whitney U test; p<0.001). C: heatmaps show that TP53-mutated BCs likely more highly express TILs genes than TP53-wildtype BCs. D: TP53-mutated BCs have higher enrichment levels of the cytokine and cytokine receptor (CCR) signature than TP53-wildtype BCs. E: heatmaps show that TP53-mutated BCs likely more highly express proinflammatory and parainflammation (PI) gene signatures than TP53-wildtype BCs. F: most of the proinflammatory genes are upregulated in TP53-mutated BCs relative to TP53-wildtype BCs. G: heatmaps show that TP53-mutated BCs likely more highly express HLA genes than TP53-wildtype BCs in TCGA. ∗, P<0.05. ∗∗, P<0.01. ∗∗∗, P< 0.001. It also applies to the following figures. Figure S2: the genes and their protein products have a significant expression correlation with immune activities in BC that are differentially expressed between TP53-mutated and TP53-wildtype breast cancers (BCs). A: the genes TFRC, CDH3, and EGFR show a positive expression correlation with immune activities in BC. B: the genes GATA3, IGF1R, PGR, ESR1, ERBB3, BCL2, and AR show a negative expression correlation with immune activities in BC. C: AR, ERBB3, BCL2, and IGF1R, and their protein products consistently show a negative expression correlation with immune scores in BC. Figure S3: association of immune signatures with clinical outcomes in breast cancers (BCs). A: Kaplan-Meier survival curves show that the upregulation of immune signatures is consistently associated with better survival in TP53-mutated BCs. B: Kaplan-Meier survival curves show that the upregulation of immune signatures is associated with better or worse survival in TP53-wildtype BCs. C: the genes IDO2, STAT1, and LAG3 have a negative expression correlation with survival prognosis in TP53-wildtype BCs. Figure S4: TP53-mutated breast cancers (BCs) have higher expression levels of cancer-testis (CT) antigens, Treg, immune checkpoint, metastasis-promoting, and metastasis-inhibiting immune signatures than TP53-wildtype BCs. A: a number of genes encoding the CT antigens that are potential targets for developing cancer vaccines are upregulated in TP53-mutated BCs relative to TP53-wildtype BCs. B: TP53-mutated BCs have higher enrichment levels of the CT antigen signature than TP53-wildtype BCs. C: TP53-mutated BCs have higher enrichment levels of the Treg and immune checkpoint signatures than TP53-wildtype BCs. D: a number of important immune checkpoint genes are more highly expressed in TP53-mutated BCs than in TP53-wildtype BCs in TCGA. Treg: regulatory T cell. E: the metastasis-promoting and the metastasis-inhibiting immune signatures are significantly upregulated and downregulated in TP53-mutated BCs versus TP53-wildtype BCs. Figure S5: knockdown of p53 by siRNA significantly increases the expression of MHC Class I genes in MCF-7 cells.
References
- 1.Zilfou J. T., Lowe S. W. Tumor suppressive functions of p53. Cold Spring Harbor Perspectives in Biology. 2009;1(5) doi: 10.1101/cshperspect.a001883.a001883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hollstein M., Sidransky D., Vogelstein B., Harris C. C. p53 Mutations in human cancers. Science. 1991;253(5015):49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 3.Wang X., Sun Q. TP53 mutations, expression and interaction networks in human cancers. Oncotarget . 2017;8(1):624–643. doi: 10.18632/oncotarget.13483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muñoz-Fontela C., Mandinova A., Aaronson S. A., Lee S. W. Emerging roles of p53 and other tumour-suppressor genes in immune regulation. Nature Reviews Immunology. 2016;16(12):741–750. doi: 10.1038/nri.2016.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zitvogel L., Kroemer G. CANCER. A p53-regulated immune checkpoint relevant to cancer. Science. 2015;349(6247):476–477. doi: 10.1126/science.aac8475. [DOI] [PubMed] [Google Scholar]
- 6.Textor S., Fiegler N., Arnold A., Porgador A., Hofmann T. G., Cerwenka A. Human NK cells are alerted to induction of p53 in cancer cells by upregulation of the NKG2D ligands ULBP1 and ULBP2. Cancer Research. 2011;71(18):5998–6009. doi: 10.1158/0008-5472.CAN-10-3211. [DOI] [PubMed] [Google Scholar]
- 7.Shatz M., Menendez D., Resnick M. A. The human TLR innate immune gene family is differentially influenced by DNA stress and p53 status in cancer cells. Cancer Research. 2012;72(16):3949–3957. doi: 10.1158/0008-5472.CAN-11-4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo G., Yu M., Xiao W., Celis E., Cui Y. Local activation of p53 in the tumor microenvironment overcomes immune suppression and enhances antitumor immunity. Cancer Research. 2017;77(9):2292–2305. doi: 10.1158/0008-5472.CAN-16-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Del Paggio J. C. Immunotherapy: cancer immunotherapy and the value of cure. Nature Reviews Clinical Oncology. 2018;15(5):268–269. doi: 10.1038/nrclinonc.2018.27. [DOI] [PubMed] [Google Scholar]
- 10.Li X., Shao C., Shi Y., Han W. Lessons learned from the blockade of immune checkpoints in cancer immunotherapy. Journal of Hematology & Oncology. 2018;11(1):p. 31. doi: 10.1186/s13045-018-0578-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braun D. A., Burke K. P., Van Allen E. M. Genomic approaches to understanding response and resistance to immunotherapy. Clinical Cancer Research. 2016;22(23):5642–5650. doi: 10.1158/1078-0432.CCR-16-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodman A. M., Kato S., Bazhenova L., et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Molecular Cancer Therapeutics. 2017;16(11):2598–2608. doi: 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Efremova M., Finotello F., Rieder D., Trajanoski Z. Neoantigens generated by individual mutations and their role in cancer immunity and immunotherapy. Frontiers in Immunology. 2017;8:p. 1679. doi: 10.3389/fimmu.2017.01679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le D. T., Uram J. N., Wang H., et al. PD-1 blockade in tumors with mismatch-repair deficiency. The New England Journal of Medicine. 2015;372(26):2509–2520. doi: 10.1056/nejmoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel S. P., Kurzrock R. PD-L1 expression as a predictive biomarker in cancer immunotherapy. Molecular Cancer Therapeutics. 2015;14(4):847–856. doi: 10.1158/1535-7163.mct-14-0983. [DOI] [PubMed] [Google Scholar]
- 16.Inc. OB. Oncolytics Biotech® Inc.'s REOLYSIN® More than Doubles Overall Survival in Patients with Mutated p53 Metastatic Breast Cancer. 2017.
- 17.Siegel R. L., Miller K. D., Jemal A. Cancer statistics, 2017. CA: A Cancer Journal for Clinicians. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 18.Anders C., Carey L. A. Understanding and treating triple-negative breast cancer. Oncology. 2008;22(11):1233–1239. [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X., Guda C. Integrative exploration of genomic profiles for triple negative breast cancer identifies potential drug targets. Medicine. 2016;95(30) doi: 10.1097/MD.0000000000004321.e4321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nanda R., Chow L. Q. M., Dees E. C., et al. Pembrolizumab in patients with advanced triple-negative breast cancer: phase Ib keynote-012 study. Journal of Clinical Oncology. 2016;34(21):2460–2467. doi: 10.1200/JCO.2015.64.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emens L. A., Braiteh F. S., Cassier P., et al. Inhibition of PD-L1 by MPDL3280A leads to clinical activity in patients with metastatic triple-negative breast cancer. Proceedings of the AACR Annual Meeting; 2015; Pennsylvania, Pa, USA. [DOI] [Google Scholar]
- 22.Liu Z., Li M., Jiang Z., Wang X. A comprehensive immunologic portrait of triple-negative breast cancer. Translational Oncology. 2018;11(2):311–329. doi: 10.1016/j.tranon.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Curtis C., Shah S. P., Chin S.-F., et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rooney M. S., Shukla S. A., Wu C. J., Getz G., Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160(1-2):48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Massink M. P. G., Kooi I. E., Martens J. W. M., Waisfisz Q., Meijers-Heijboer H. Genomic profiling of CHEK2⁎1100delC-mutated breast carcinomas. BMC Cancer. 2015;15:p. 877. doi: 10.1186/s12885-015-1880-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bedognetti D., Hendrickx W., Marincola F. M., Miller L. D. Prognostic and predictive immune gene signatures in breast cancer. Current Opinion in Oncology. 2015;27(6):433–444. doi: 10.1097/CCO.0000000000000234. [DOI] [PubMed] [Google Scholar]
- 28.Aran D., Lasry A., Zinger A., et al. Widespread parainflammation in human cancer. Genome Biology. 2016;17(1):p. 145. doi: 10.1186/s13059-016-0995-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong H. S., Chang C., Liu X., Huang W., Chang W. Characterization of cytokinome landscape for clinical responses in human cancers. OncoImmunology. 2016;5(11) doi: 10.1080/2162402X.2016.1214789.e1214789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Almeida L. G., Sakabe N. J., de Oliveira A. R., et al. CTdatabase: a knowledge-base of high-throughput and curated data on cancer-testis antigens. Nucleic Acids Research. 2009;37(1):D816–D819. doi: 10.1093/nar/gkn673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Simone M., Arrigoni A., Rossetti G., et al. Transcriptional landscape of human tissue lymphocytes unveils uniqueness of tumor-infiltrating T regulatory cells. Immunity. 2016;45(5):1135–1147. doi: 10.1016/j.immuni.2016.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Der Weyden L., Arends M. J., Campbell A. D., et al. Genome-wide in vivo screen identifies novel host regulators of metastatic colonization. Nature. 2017;541(7636):233–236. doi: 10.1038/nature20792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barbie D. A., Tamayo P., Boehm J. S., et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462(7269):108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics. 2013;14:p. 7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society B: Methodological. 1995;57(1):289–300. [Google Scholar]
- 36.Yoshihara K., Shahmoradgoli M., Martínez E., et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nature Communications. 2013;4, article no. 2612 doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subramanian A., Tamayo P., Mootha V. K., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Acadamy of Sciences of the United States of America. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newman A. M., Liu C. L., Green M. R., et al. Robust enumeration of cell subsets from tissue expression profiles. Nature Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Research. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim S. ppcor: an R package for a fast calculation to semi-partial correlation coefficients. Communications for Statistical Applications and Methods. 2015;22(6):665–674. doi: 10.5351/CSAM.2015.22.6.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salic A., Mitchison T. J. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proceedings of the National Acadamy of Sciences of the United States of America. 2008;105(7):2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lippitz B. E. Cytokine patterns in patients with cancer: a systematic review. The Lancet Oncology. 2013;14(6):e218–e228. doi: 10.1016/S1470-2045(12)70582-X. [DOI] [PubMed] [Google Scholar]
- 43.Townsend P. A., Scarabelli T. M., Davidson S. M., Knight R. A., Latchman D. S., Stephanou A. STAT-1 interacts with p53 to enhance DNA damage-induced apoptosis. The Journal of Biological Chemistry. 2004;279(7):5811–5820. doi: 10.1074/jbc.M302637200. [DOI] [PubMed] [Google Scholar]
- 44.Hix L. M., Karavitis J., Khan M. W., Shi Y. H., Khazaie K., Zhang M. Tumor STAT1 transcription factor activity enhances breast tumor growth and immune suppression mediated by myeloid-derived suppressor cells. The Journal of Biological Chemistry. 2013;288(17):11676–11688. doi: 10.1074/jbc.m112.441402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Overgaard J., Yilmaz M., Guldberg P., Hansen L. L., Alsner J. TP53 mutation is an independent prognostic marker for poor outcome in both node-negative and node-positive breast cancer. Acta Oncologica. 2000;39(3):327–333. doi: 10.1080/028418600750013096. [DOI] [PubMed] [Google Scholar]
- 46.Munro A., Bright S. Products of the major histocompatibility complex and their relationship to the immune response. Nature. 1976;264(5582):145–152. doi: 10.1038/264145a0. [DOI] [PubMed] [Google Scholar]
- 47.Wang B., Niu D., Lai L., Ren E. C. P53 increases MHC class i expression by upregulating the endoplasmic reticulum aminopeptidase ERAP1. Nature Communications. 2013;4:p. 2359. doi: 10.1038/ncomms3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kastenhuber E. R., Lowe S. W. Putting p53 in context. Cell. 2017;170(6):1062–1078. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andrysik Z., Galbraith M. D., Guarnieri A. L., et al. Identification of a core TP53 transcriptional program with highly distributed tumor suppressive activity. Genome Research. 2017;27(10):1645–1657. doi: 10.1101/gr.220533.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen J. The cell-cycle arrest and apoptotic functions of p53 in tumor initiation and progression. Cold Spring Harbor Perspectives in Medicine. 2016;6(3) doi: 10.1101/cshperspect.a026104.a026104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Angeloni S. V., Martin M. B., Garcia-Morales P., Castro-Galache M. D., Ferragut J. A., Saceda M. Regulation of estrogen receptor-α expression by the tumor suppressor gene p53 in MCF-7 cells. Journal of Endocrinology. 2004;180(3):497–504. doi: 10.1677/joe.0.1800497. [DOI] [PubMed] [Google Scholar]
- 52.Rasti M., Arabsolghar R., Khatooni Z., Mostafavi-Pour Z. P53 binds to estrogen receptor 1 promoter in human breast cancer cells. Pathology & Oncology Research. 2012;18(2):169–175. doi: 10.1007/s12253-011-9423-6. [DOI] [PubMed] [Google Scholar]
- 53.Taub D. D., Sayers T. J., Carter C. R. D., Ortaldo J. R. α and β chemokines induce NK cell migration and enhance NK-mediated cytolysis. The Journal of Immunology. 1995;155(8):3877–3888. [PubMed] [Google Scholar]
- 54.Dorner B. G., Smith H. R. C., French A. R., et al. Coordinate expression of cytokines and chemokines by NK cells during murine cytomegalovirus infection. The Journal of Immunology. 2004;172(5):3119–3131. doi: 10.4049/jimmunol.172.5.3119. [DOI] [PubMed] [Google Scholar]
- 55.Safonov A., Jiang T., Bianchini G., et al. Immune gene expression is associated with genomic aberrations in breast cancer. Cancer Research. 2017;77(12):3317–3324. doi: 10.1158/0008-5472.CAN-16-3478. [DOI] [PubMed] [Google Scholar]
- 56.Jiang Z., Liu Z., Li M., Chen C., Wang X. Immunogenomics analysis reveals that TP53 mutations inhibit tumor immunity in gastric cancer. Translational Oncology. 2018;11(5):1171–1187. doi: 10.1016/j.tranon.2018.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oren M., Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harbor Perspectives in Biology. 2010;2(2) doi: 10.1101/cshperspect.a001107.a001107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ham S. W., Jeon H., Jin X., et al. TP53 gain-of-function mutation promotes inflammation in glioblastoma. Cell Death & Differentiation. 2019;26(3):409–425. doi: 10.1038/s41418-018-0126-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Caballero O. L., Chen Y.-T. Cancer/testis (CT) antigens: potential targets for immunotherapy. Cancer Science. 2009;100(11):2014–2021. doi: 10.1111/j.1349-7006.2009.01303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Renaud S., Pugacheva E. M., Delgado M. D., et al. Expression of the CTCF-paralogous cancer-testis gene, brother of the regulator of imprinted sites (BORIS), is regulated by three alternative promoters modulated by CpG methylation and by CTCF and p53 transcription factors. Nucleic Acids Research. 2007;35(21):7372–7388. doi: 10.1093/nar/gkm896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gasco M., Shami S., Crook T. The p53 pathway in breast cancer. Breast Cancer Research. 2002;4(2):70–76. doi: 10.1186/bcr426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jansson T., Inganäs M., Sjögren S., et al. p53 status predicts survival in breast cancer patients treated with or without postoperative radiotherapy: a novel hypothesis based on clinical findings. Journal of Clinical Oncology. 1995;13(11):2745–2751. doi: 10.1200/JCO.1995.13.11.2745. [DOI] [PubMed] [Google Scholar]
- 63.Bracci L., Schiavoni G., Sistigu A., Belardelli F. Immune-based mechanisms of cytotoxic chemotherapy: implications for the design of novel and rationale-based combined treatments against cancer. Cell Death & Differentiation. 2014;21(1):15–25. doi: 10.1038/cdd.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoshida Y., Naito M., Yamada T., et al. Clinical study on the medical value of combination therapy involving adoptive immunotherapy and chemotherapy for stage IV colorectal cancer (COMVI Study) Anticancer Reseach. 2017;37(7):3941–3946. doi: 10.21873/anticanres.11777. [DOI] [PubMed] [Google Scholar]
- 65.Smyth M. J., Ngiow S. F., Ribas A., Teng M. W. L. Combination cancer immunotherapies tailored to the tumour microenvironment. Nature Reviews Clinical Oncology. 2016;13(3):143–158. doi: 10.1038/nrclinonc.2015.209. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables. Table S1: the list of 26 immune signatures and related gene sets. Table S2: sample sizes of breast cancers. Table S3: ssGSEA scores of immune signature in TCGA and METABRIC. Table S4: primer sequences used for real time quantity PCR. Table S5: comparison of the enrichment levels of 15 immune cell types and function signatures between two classes of samples. Table S6: comparison of the enrichment levels of the tumor-infiltrating lymphocytes signature between two classes of samples. Table S7: comparison of the enrichment levels of the cytokine and cytokine receptor signature between two classes of samples. Table S8: comparison of the enrichment levels of the inflammation-promoting and parainflammation (PI) signatures between two classes of samples. Table S9: comparison of the enrichment levels of the HLA signature between two classes of samples. Table S10: comparisons of the ssGSEA scores of immune signatures between TP53-mutated and TP53-wildtype BCs and their associations with survival prognosis in BC. Table S11: comparisons of the expression levels of immune genes between TP53-mutated and TP53-wildtype BCs and their associations with survival prognosis in BC. Table S12: comparisons of the expression levels of genes and their protein products between TP53-mutated and TP53-wildtyped BCs. Table S13: comparisons of the enrichment levels of immune signatures between TP53-mutated and TP53-wildtype BCs within the ER+ subtype of BC. Table S14: comparisons of the enrichment levels of immune signatures between TP53-mutated and TP53-wildtype BCs within the HER2- subtype of BC. Table S15: comparisons of the enrichment levels of immune signatures between TP53-mutated and TP53-wildtype BCs within the 100% tumor purity of BC. Table S16: comparisons of the enrichment levels of the cancer-testis signature between two classes of samples. Table S17: comparisons of the enrichment levels of the Treg signature between two classes of samples. Table S18: comparisons of the enrichment levels of the immune checkpoint signature between two classes of samples. Table S19: comparisons of the expression levels of the genes targeted by immunotherapy agents in clinical use or trials or in preclinical development between TP53-mutated and TP53-wildtype BCs. Supplementary Figures Legends. Figure S1: TP53-mutated breast cancers (BCs) have increased immune activity compared to TP53-wildtype BCs. A: heatmap shows the ssGSEA scores of 26 immune signatures in TP53-mutated and in TP53-wildtype BCs (TCGA). ssGSEA: single-sample gene-set enrichment analysis; TNBC: triple-negative breast cancer. RB1 is more frequently mutated in TP53-mutated BCs while CDH1, GATA3, MAP3K1, and PIK3CA are more frequently mutated in TP53-wildtype BCs (Fisher's exact test; P<0.05). The black vertical lines in the horizontal bars beside gene symbols indicate that the genes are mutated in corresponding samples. The black vertical lines in the horizontal bar beside “TNBC” indicate that the samples are TNBC. The black vertical lines in the bars beside “ER”, “PR,” and “HER2” indicate that the samples are ER-, PR-, or HER2-. B: TP53-mutated BCs have higher levels of tumor-infiltrating lymphocytes (TILs) infiltration than TP53-wildtype BCs (Mann-Whitney U test; p<0.001). C: heatmaps show that TP53-mutated BCs likely more highly express TILs genes than TP53-wildtype BCs. D: TP53-mutated BCs have higher enrichment levels of the cytokine and cytokine receptor (CCR) signature than TP53-wildtype BCs. E: heatmaps show that TP53-mutated BCs likely more highly express proinflammatory and parainflammation (PI) gene signatures than TP53-wildtype BCs. F: most of the proinflammatory genes are upregulated in TP53-mutated BCs relative to TP53-wildtype BCs. G: heatmaps show that TP53-mutated BCs likely more highly express HLA genes than TP53-wildtype BCs in TCGA. ∗, P<0.05. ∗∗, P<0.01. ∗∗∗, P< 0.001. It also applies to the following figures. Figure S2: the genes and their protein products have a significant expression correlation with immune activities in BC that are differentially expressed between TP53-mutated and TP53-wildtype breast cancers (BCs). A: the genes TFRC, CDH3, and EGFR show a positive expression correlation with immune activities in BC. B: the genes GATA3, IGF1R, PGR, ESR1, ERBB3, BCL2, and AR show a negative expression correlation with immune activities in BC. C: AR, ERBB3, BCL2, and IGF1R, and their protein products consistently show a negative expression correlation with immune scores in BC. Figure S3: association of immune signatures with clinical outcomes in breast cancers (BCs). A: Kaplan-Meier survival curves show that the upregulation of immune signatures is consistently associated with better survival in TP53-mutated BCs. B: Kaplan-Meier survival curves show that the upregulation of immune signatures is associated with better or worse survival in TP53-wildtype BCs. C: the genes IDO2, STAT1, and LAG3 have a negative expression correlation with survival prognosis in TP53-wildtype BCs. Figure S4: TP53-mutated breast cancers (BCs) have higher expression levels of cancer-testis (CT) antigens, Treg, immune checkpoint, metastasis-promoting, and metastasis-inhibiting immune signatures than TP53-wildtype BCs. A: a number of genes encoding the CT antigens that are potential targets for developing cancer vaccines are upregulated in TP53-mutated BCs relative to TP53-wildtype BCs. B: TP53-mutated BCs have higher enrichment levels of the CT antigen signature than TP53-wildtype BCs. C: TP53-mutated BCs have higher enrichment levels of the Treg and immune checkpoint signatures than TP53-wildtype BCs. D: a number of important immune checkpoint genes are more highly expressed in TP53-mutated BCs than in TP53-wildtype BCs in TCGA. Treg: regulatory T cell. E: the metastasis-promoting and the metastasis-inhibiting immune signatures are significantly upregulated and downregulated in TP53-mutated BCs versus TP53-wildtype BCs. Figure S5: knockdown of p53 by siRNA significantly increases the expression of MHC Class I genes in MCF-7 cells.
Data Availability Statement
The TCGA data can be downloaded from the genomic data commons data portal (https://portal.gdc.cancer.gov/), and the METABRIC data can be downloaded from cBioPortal (http://www.cbioportal.org).