Abstract
With improved screening methods, the numbers of abnormal breast lesions diagnosed in women have been increasing over time. However, it remains unclear whether these breast lesions will develop into invasive cancers. To more effectively predict the outcome of breast lesions and determine a more appropriate course of treatment, it is important to understand the underlying mechanisms that regulate progression of non-invasive lesions to invasive breast cancers. A hallmark of invasive breast cancers is the accumulation of fibroblasts. Fibroblast proliferation and activation in the mammary gland is in part regulated by the Transforming Growth Factor beta1 pathway (TGF-β). In animal models, TGF-β suppression of CCL2 and CXCL1 chemokine expression is associated with metastatic progression of mammary carcinomas. Here, we show that transgenic overexpression of the Polyoma middle T viral antigen in the mouse mammary gland of C57BL/6 mice results in slow growing non-invasive lesions that progress to invasive carcinomas in a stage dependent manner. Invasive carcinomas are associated with accumulation of fibroblasts that show decreased TGF-β expression and high levels of CXCL1, but not CCL2. Using co-transplant models, we show that decreased TGF-β signaling in fibroblasts contribute to mammary carcinoma progression through enhancement of CXCL1/CXCR2 dependent mechanisms. Using cell culture models, we show that CXCL1 mediated mammary carcinoma cell invasion through NF-κB, AKT, Stat3 and p42/44MAPK dependent mechanisms. These studies provide novel mechanistic insight into the progression of pre-invasive lesions and identify new stromal biomarkers, with important prognostic implications.
Keywords: breast carcinoma, invasion, chemokine, TGF-β, fibroblast, MMTV-PyVmT, C57BL/6
Introduction:
Improved imaging approaches combined with core needle biopsies have led to earlier detection of abnormal breast lesions in women over time, with most lesions diagnosed as non-cancerous [1–3]. Non-cancerous breast lesions are highly variable, and include non-proliferative diseases such as breast cysts, and proliferative diseases such as hyperplasia and intraductal papilloma [4, 5]. Due to heterogeneity of breast disease, it currently remains difficult to assess whether some lesions are associated with breast cancers. Risk for breast cancers varies among lesions; for example, typical hyperplasia carries a lower risk than atypical hyperplasia [6, 4, 5]. Some high risk lesions also exhibit features of ductal carcinoma in situ [7], or may be accompanied by breast carcinoma detected in subsequent surgery [8, 9, 3]. As such, there is no uniform agreement on how to manage early stage breast lesions [10, 11]. To more effectively predict the outcome of breast lesions and determine appropriate course of treatment, it is important to understand the underlying molecular mechanisms that regulate progression of non-invasive lesions to invasive breast cancers.
A hallmark of invasive breast cancers is the accumulation of fibroblasts [12, 13]. Fibroblasts are a major cellular component of the breast stroma, and exert profound effects on epithelial cell growth and survival during wound healing and mammary gland development through secretion of growth factors, matrix proteins and proteases [14, 15]. Fibroblast proliferation and activation in the mammary gland is in part regulated by the Transforming Growth Factor beta1 pathway (TGF-β). TGF-β signals through cell surface type I transmembrane receptors, which heterodimerize with the type II receptors, leading to activation of canonical Smad2/3/4 pathways and Smad independent pathways including: Rho, p38MAPK and PI3 kinase. These signaling pathways regulate the actin cytoskeleton and gene expression of cell cycle regulators, cytokines matrix proteins and proteases [16, 17]. The role of TGF-β signaling in breast fibroblasts during breast cancer progression is complex. Mice with stromal specific knockout of Tgfbr2 die prior to 8 weeks of age, and exhibit defects in mammary ductal morphogenesis and increased fibrosis [18]. Fibroblasts isolated from homozygous Tgfbr2 knockout mice (Tgfbr2FspKO) promote mammary tumor growth and invasion. These phenotypes are associated with impairment of TGF-β signaling and increased expression of chemokines [19–21].
Chemokines are small soluble molecules (8kda) in size that form molecular gradients to attract cells to sites of inflammation [22, 23]. Chemokines are classified into C, C-C, C-X-C or CX3C groups depending on the composition of a conserved cysteine motif. While CXCL1 and CCL2 belong to different chemokine subgroups, they both regulate angiogenesis and recruitment of bone marrow derived cells in acute chronic inflammation [24–28]. CXCL1 and CCL2 signal to seven transmembrane spanning receptors expressed on the cell surface, which activate G protein dependent and independent signaling pathways to regulate cell migration and differentiation. CXCL1 binds with highest affinity to CXCR2, while CCL2 binds primarily to CCR2 [29, 30]. Studies have indicated an important role for these chemokine signaling pathways in cancer. In animal models of melanoma and mammary carcinoma, CXCL1 regulates tumor growth and metastasis through recruitment of CXCR2 expressing neutrophils and myeloid immune suppressor cells to tumor tissues [31–33]. CXCL1 also signals to CXCR2 expressing ovarian, lung and breast cancer cells to enhance cell growth and motility [34–38]. In animal models of prostate and mammary carcinoma, CCL2 enhances recruitment of CCR2 expressing macrophages and neutrophils to promote tumor growth and metastasis [39–41]. CCL2 also signals to CCR2 expressing prostate and breast cancer cells to enhance survival and growth [42, 43]. These studies indicate multiple roles for CXCL1/CXCR2 and CCL2/CCR2 signaling in cancer, which remain poorly understood.
In breast carcinomas, increased stromal CXCL1 and CCL2 expression in the stroma correlate with disease recurrence and poor patient prognosis [44, 45]. In animal models, TGF-β suppression of CCL2 and CXCL1 chemokine expression is associated with metastatic progression of mammary carcinomas in individual studies [20, 46]. The mechanisms that regulate the progression of pre-invasive breast lesions to invasive breast cancer have remained poorly understood. Here, we show that transgenic overexpression of the Polyoma middle T viral antigen in the mouse mammary gland of C57BL/6 mice results in slow growing benign lesions that progress to invasive carcinomas in a stage dependent manner. Invasive carcinomas are associated with accumulation of fibroblasts that show decreased TGF-β expression and high levels of CXCL1, but not CCL2. Using co-transplant models, we show that decreased TGF-β signaling in fibroblasts contribute to mammary carcinoma progression through enhancement of CXCL1/CXCR2 dependent mechanisms. Using cell culture models, we show that CXCL1 mediated mammary carcinoma cell invasion through NF-κB, AKT, Stat3 and p42/44MAPK dependent mechanisms. These studies provide novel mechanistic insight into the progression of pre-invasive lesions and identify new stromal biomarkers, with important prognostic implications.
Materials and methods
Maintenance of transgenic mice
MMTV-PyVmT (C57BL/6N) mice were obtained from Dr. Harold L. Moses (Vanderbilt University). PyVmT positive animals were generated by crossing PyVmT positive males with Wildtype (WT) females. Animals were genotyped for the PyVmT gene as described [47]. C57BL/6N female mice (8–10) weeks of age were purchased from Envigo. Animals were maintained under an approved IACUC protocol at the University of Kansas Medical Center under AALAAC accredited facilities.
Cell culture
All cell lines were cultured in DMEM/10% FBS unless otherwise specified. Raw264.7 cells (cat no. TIB-71), 4T1 (cat no. CRL-2539) and NMuMG (cat no. CRL-1636) mammary epithelial cells were obtained from American Type Culture Collection. Immortalized lung microvascular endothelial cells [48] were kindly provided by Dana Brantley-Sieders (Vanderbilt University, Nashville, TN) and cultured in EGM-2 Bullet Kit media (cat no. CC-3162 Lonza). Tgfbr2FspKO fibroblasts were isolated and characterized as reported [19]. Normal mammary fibroblasts (NAFs) were isolated from tumor free C57BL/6 female mice 19 weeks old. Carcinoma associated fibroblasts (P-CAFs) were isolated from 29-week-old MMTV-PyVmT (C57BL/6) female mice, which formed invasive carcinomas characterized by extensive fibrosis as determined by H&E stain. Mammary epithelial cells (144epi) were isolated from mammary tissues of 15-week-old MMTV-PyVmT female mice (C57BL/6), in which mammary lesions showed accumulation of stroma and appearance of few invasive carcinoma cells. NAFs, P-CAFs, and 144epi cells were isolated and cultured using methods described previously [49]. Briefly, tissues were minced with surgical scissors, and digested overnight on ice in 10 ml of PBS containing: 5 mg/ml collagenase A (Sigma-Aldrich, cat no. 234153–1GM), 2 mg/ml trypsin (Sigma-Aldrich cat no. T3924–100ml),1000 units/ml hyaluronidase (Sigma-Aldrich cat no. H3884), 4 mg/ml DNAse (Sigma-Aldrich cat no. D5025). Cells were pelleted at 1000 g for 5 minutes at 4°C and washed with PBS/10% FBS 3 times, and plated on 10 cm dishes coated with rat tail collagen I (Fisher Scientific, cat no. CB 40236) in DMEM/10% FBS. To coat plates with collagen, 1 ml of 1.5 mg/ml collagen was diluted in 39 ml of 0.02 N acetic acid. Plates were incubated with 5 ml of collagen working solution at room temperature for a minimum for 10 minutes, and excess collagen was aspirated. After 24 hours, fibroblasts were separated from epithelial cells through differential trypsinization. Cells were incubated with 2 ml of 0.25% trypsin/0.54 mM EDTA at room temperature for approximately 2 minutes, fibroblasts were transferred to a 15 ml conical tube, quenched in DMEM/10% FBS, and pelleted. The remaining mammary epithelial cells were trypsinized at 37°C for an additional 5 minutes, quenched in DMEM/10% FBS and pelleted. Fibroblasts and epithelial cells were plated on 10 cm dishes coated with rat tail collagen I in DMEM/10% FBS. All cell lines were maintained in culture no longer than 3 months at a time. Cells were tested after each freeze/thaw using the MyCoAlert Mycoplasm Detection kit (Lonza cat no LT07).
Immunocytochemistry
Cells were seeded in a 96 well plate at 10,000/well in growth media. After 24 hours, cells were fixed in 10% neutral formalin buffer (NBF) for an additional 24 hours, permeabilized with methanol for 10 minutes at −20°C. For primary incubation with mouse monoclonal antibodies, cells were blocked in PBS containing 3.4% mouse FAB fragment for (Jackson Laboratory cat no. 715 001 003) for 1 hour. For all other antibodies, cells were blocked in PBS containing 3% FBS. Cells were incubated with primary antibodies at a 1: 3 dilution to Fibroblast Specific Protein 1 (Fsp1) (Abcam cat no. ab75550), or at a 1:100 dilution to: pan-cytokeratin (Santa Cruz Biotechnology cat no. sc 8018), α−smooth muscle actin (α-sma) (Abcam cat no. 7187), N-cadherin (Santa Cruz Biotechnology cat no. 7339) or Von Willebrand Factor 8 (VWF8, Millipore cat no. Ab7356). Pan-cytokeratin and α-sma were detected with secondary anti-mouse-hrp at 1:500 dilution for 2 hours in PBS/3% FBS and incubation with 3,3’-diaminobenzidine substrate (DAB, Vector Laboratories cat no. SK-4100). FSP1, N-cadherin and VWF8 were detected with secondary anti-rabbit-hrp at a 1:500 dilution and DAB substrate. Cells were counterstained with Mayer’s hematoxylin for 2 minutes. Images were captured using the EVOS FL Auto Imaging system.
Lentiviral transduction
For stable mCherry expression in cells, pSicoR-Ef1a-mCherry-Puro vectors [50] were obtained from Addgene (cat no. 31845). To generate lentivirus, 10 μg target vectors were co-transfected with 3μg PMD2G (Addgene cat no.12260) and 6 μg PDPAX2 (Addgene cat no.12259) in HEK 293T cells using Lipofectamine 2000 (ThermoFisher cat no. 11668027). Lentivirus conditioned medium was removed 48 hours after transfection and used to transduce fibroblasts two times. Transduced cells were sorted for mCherry expression by fluorescence activated cell sorting (FACS).
Soft agar colony formation assay
Colony formation assays were performed using previously reported procedures [51]. Briefly, 6 well plates were coated with 1.5 ml/well of solution containing 0.5% agar in DMEM/10% FBS, and allowed to solidify for 30 minutes at room temperature. 5,000 cells were resuspended in 0.75ml 2× DMEM/20% FBS, mixed with 0.75 ml 0.6% agarose, and then plated on top of the base agar layer, and allowed to solidify at 30 minutes at room temperature. 250 μl DMEM/10% FBS or fibroblast conditioned medium was added on top of the agar-cell layer. Plates were incubated for 14 days at 37°C based on growth of 4T1 mammary carcinoma cells, a positive control. Media was replaced twice a week. Colonies were then stained for 24 hours with 200 μl of 1mg/ml nitroblue tetrazolium chloride solution (Sigma cat no. N6876) at 37°C, and imaged using a dissecting microscope with an OMAX 18.0MP digital camera attachment. 4 fields/well were captured to cover the whole well. Colonies were counted using Image J software. To generate fibroblast conditioned medium, 500,000 fibroblasts were seeded overnight in a 10 cm dish. Cells were incubated with 5 ml of DMEM/10% FBS for 24 hours. Medium was collected and used for soft agar assays.
Mammary transplantation
Wildtype female C57BL/6NHsd mice, 6–8 weeks of age were obtained from Envigo (stock no. 044). Co-transplantation of mammary fibroblasts with 144epi cells were performed using methods previously described[49]. Briefly, 250,000 fibroblasts were embedded with or without 100,000 carcinoma cells in 50 μl of type I collagen overnight. C57BL/6 mice were anesthetized using 2% isoflurane. A Y incision was made in the abdomen to expose the left and right inguinal mammary glands. Using spring scissors, a 1–2 mm pocket was made in intact mammary fat pads, under the lymph node. Using a glass pipet, one collagen plug containing cells was inserted into the pocket in the left and right mammary glands. Wounds were closed using gut absorbable sutures. Animals were monitored and palpated twice weekly for tumor formation. Tumors were measured by caliper. Mice were sacrificed 60 days post-transplantation, when control tumors reached 1.5 mm in diameter, the maximum allowable size under IACUC guidelines. Animals were maintained under an approved IACUC protocol in AALAAC accredited facilities.
shRNA knockdown
pSM2c retroviral shRNA vectors were developed by the laboratory of Greg Hannon [52], and obtained from Open Biosystems (now part of GE Healthsystems). The CXCR2 shRNA sequences for F-6 are 5’-CAGTGTACTTACATATAAT-3′ and for G-1: 5′ GTAACATTTGAAATGTAA-3.′ The eGFP control shRNA sequence is 5’ GCTGACCCTGAAGTTCATC-3.’ Plasmids were transfected into Phoenix cells by Lipofectamine 2000 (Invitrogen, Carlsbad, CA). 48 hours after transfection, media was collected and used to treat 144epi cells in the presence of 5 μg/ml polybrene. 144epi cells were subject to three rounds of transduction before selection with 1.5 μg/ml puromycin.
Flow cytometry
500,000 cells were seeded in 10 cm dishes overnight. Cells were detached by 3 mM EDTA, washed in DMEM/10% FBS and fixed in 10% neutral formalin buffer. Cells were permeabilized in Triton X-100 at 37°C for 10 minutes, and incubated at a 1:100 dilution of anti-CXCR2 (Abcam cat no.14935) for 1 hour. Cells were washed in PBS 3 times and incubated with goat anti-Rabbit-Alexa-Fluor-488 (Invitrogen cat no.A11008) for 1 hour. Samples were read using an LSRII Flow Cytometer and normalized to unstained and secondary antibody only controls.
Histology/Immunohistochemistry/Immunofluorescence
Tissues were fixed in 10% neutral formalin buffer for 24 hours and processed for paraffin embedding as described [53]. For H&E stain, slides were incubated in Mayer’s hematoxylin for 1 minute and eosin for 2 minutes prior to dehydration and mounting in Cytoseal.
For DAB immunostaining, slides were subject to antigen retrieval by heating under low pressure for 2 minutes in 1 M urea for CCL2, or 10 mM Sodium Citrate pH 6.0 for all other antigens. Endogenous peroxidases were quenched by incubation in PBS buffer containing 20% methanol/3% H202. Slides were blocked in PBS/3% FBS and then incubated overnight in blocking buffer with primary antibodies at a 1:100 to: Ki67 (Santa Cruz Biotechnology cat no. sc 1307), cleaved caspase-3 (Cell Signaling Technology, cat no. 9579), α-sma (Spring Biosciences cat no. M4712), F4/80, Gr-1 (RnD Systems, cat no. mab1037), VWF8, CD45 (Biolegend cat no. 103101), CXCL1 (Santa Cruz Biotechnology cat no. 16961), CCL2 (Santa Cruz Biotechnology cat no.1784), CXCR2 (Abcam cat no. 14935), CCR2 (Santa Cruz Biotechnology cat no. 300393). For detection of Ki67, α-sma, cleaved caspase-3, VWF8, CXCR2 or CCR2, slides were incubated with secondary rabbit biotinylated antibodies at a 1:1000 dilution for 2 hours. To detect Gr-1 and F4/80, slides were incubated with rat-biotinylated secondary antibodies at a 1:1000 dilution for 2 hours. To detect CCL2 or CXCL1, slides were incubated with secondary anti-goat biotinylated antibodies at a 1:1000 dilution. Secondary antibodies were bound to streptavidin-peroxidase, and incubated with DAB substrate. Slides were counterstained in Mayer’s hematoxylin for 1 minute, dehydrated and. mounted with Cytoseal. 5 images per slide per sample were captured at 10 × magnification using an FL Auto Imaging System (Invitrogen). Immunostaining was quantified by Image J using methods previously described [53].
For immunofluorescence, slides were subject to antigen retrieval in 10 mM sodium citrate pH 6.0, blocked for 1 hour in PBS containing 3% FBS or 3.4% mouse FAB fragment for (Jackson Laboratory cat no. 715 001 003) and incubated with primary antibodies at a 1:50 to CK5/6 (Cell Marque, cat no. 356M-14) or p63 (Boster, cat no. P2056) and α-sma. α-sma was detected with goat anti-rabbit-AlexaFluor488 diluted 1:250 (Invitrogen cat no. A-11034). p63 was detected with donkey anti-mouse-AlexaFluor568 diluted 1:250 (Invitrogen cat no. A10037). CK5/6 was detected with goat anti-mouse-AlexaFluor488 diluted 1:250 (Invitrogen cat no. A11001). For co-staining of mCherry and α-sma, sections were blocked with PBS/3.4% mouse FAB, incubated with anti-mCherry (Rockland cat no. 600–404-P16) diluted 1:500, and anti-α-sma (Abcam cat no. 7187), diluted 1:100 for 24 hours. mCherry was detected with goat anti-rabbit-AlexaFluor647 (Invitrogen cat no. A-21244) diluted 1:250. α-sma was detected with goat anti-rabbit-AlexaFluor488 diluted 1:250. After a 2 hour incubation in the dark, slides were washed with PBS three times and counterstained with 4,6-Diamidino-2-phenylindole (DAPI), and mounted with 1:1 PBS: glycerol.
Scoring for tumor invasion
To quantify the extent of tumor invasion into the mammary fat pad, three serial sections per slide were stained by H&E. Using a numerical scoring system, tissues were scored in a blinded fashion. 0 indicated a non-invasive phenotype, with papillary structures, ductal hyperplasia or DCIS. Little to no stromal reactivity was observed. 1 (lowly invasive) indicated the presence of a few invasive carcinoma cells. Increased stromal reactivity was observed. 2 (highly invasive) indicated the absence of papillary structures and the presence of invasive carcinoma with stromal reactivity.
siRNA transfection
30,000 cells were seeded per well in a 24 well plate for 24 hours. For each well, 24 pmol control siRNAs or siRNAs targeting CXCL1 (Santa Cruz Biotechnology) were complexed to 2.4 ul lipofectamine 2000 (Invitrogen) in 100 ul Opti-MEM medium for 20 minutes at room temperature. Cells were incubated in 400 ul Opti-MEM with the siRNA/lipofectamine complexes for 24 hours, and recovered in Opti-MEM/10% medium for an additional 24 hours. Silencer® Negative Control #2 siRNA was obtained from Ambion (cat no. AM4613). 2 different CXCL1 siRNAs were generated through Ambion. CXCL1 si1 (sense): 5’-GGACUUGUUACAAAUGAAGUtt-3’, CXCL1 si2 (sense) 5’-GGGUCGUAUUUAUUUAUGUtt-3’.
ELISA
30,000 cells were seeded per well in a 24 well plate for 24 hours. To generate conditioned medium, cells were washed with PBS and incubated with 500 μl of serum free medium DMEM in the presence or absence of 10 ng/ml TGF-β1 (Peprotech cat no. 11021) for 24 hours. Media was assayed by ELISA for CXCL1 (Peprotech cat no. 250–11), CCL2 (Peprotech cat no. 250–10) or TGF−β (RnD Systems cat no. DY1679). Reactions were read at OD450nM using a Biotek plate reader.
Invasion assay
Invasion assays were adapted from previous studies [54]. Briefly, transwells (8 micron pores) were coated with Growth Factor Reduced Matrigel (BD Pharmingen, cat no 356230), diluted ¼ in PBS for 30 minutes at 37°C. 144epi cells were serum starved for 24 hours. 75,00 cells then seeded on top of Matrigel coated transwells in the presence or absence of: 60 ng/mLCXCL1 (Peprotech cat no. 250–11), 5 μM Bay11–7085 (Enzo Life Sciences cat no. 196309–76), 1 μM U0126 (Cell Signaling Technology cat no. 9903), 0.28 μM MK2206 (Selleck Chemicals cat no. S1078) or 1 mM Stat3 peptide inhibitors (Calbiochem cat no. 573096). Transwells were incubated at 37°C for 24 hours. The cells were fixed in 10% neutral formalin buffer for 10 min and stained with 0.1% crystal violet for 5 minutes. Cells on the topside of the filter were removed by cotton swab. The transwell filter was removed and mounted on glass slides in aqua-based mounting medium. Brightfield images of crystal violet stained cells were captured at 20 × magnification, 4 fields per well using an EVOS FL Auto Imaging System (Invitrogen). Quantitation of tumor cells was determined by measuring pixel density of using Image J (NIH) software (arbitrary units). For co-culture experiments, 100,000 fibroblasts were seeded on the underside of the Matrigel coated transwells. CMTMR-labeled tumor cells (75,000) were then seeded on top in DMEM/10% FBS and incubated for 24 hours. The cells were fixed in 10% neutral formalin buffer for 10 min. Tumor cells on the topside of the filter were removed by cotton swab. The transwell filter was removed and mounted on glass slides in aqua-based mounting medium. Invaded tumor cells were visualized by fluorescence microscopy and imaged at 20 × magnification, four fields per sample.
Immunoblot
Cells were seeded in 6 well plates at 250,000/well for 24 hours, serum starved for 24 hours, and stimulated with 60 ng/ml mouse recombinant CXCL1 (RnD Systems) for up to 24 hours. Cells were lysed in RIPA buffer containing protease inhibitors (Sigma cat no. P8340) and 10 μM sodium orthovanadate and sonicated. 50 μg protein were resolved on 10% SDS-PAGE and transferred to nitrocellulose membranes. Blots were blocked in PBS containing 0.05% Tween-20 (PBS-T) containing 3% dry milk for 1 hour and immunoblotted with primary antibodies at a 1:1000 overnight in PBS-T/3% BSA. With the exception of FAK, antibodies were obtained from Cell Signaling Technology: phospho-Src (Tyr416 cat no. 6943), Src (cat no. 2123), phospho-FAK (Tyr397 cat no.8856), FAK (Santa Cruz Biotechnology, cat no. sc558), phospho-p38MAPK (Thr180/Tyr182, cat no. 4511), p38MAPK (cat no. 9212), phospho-IκB (Ser32/36, cat no. 9246), IκB (cat no. 2678), phospho-p42/44MAPK (Thr202/Tyr204, cat no. 9101), p42/44MAPK (cat no. 9102), phospho-AKT (Ser473, cat no. 4060), AKT (cat no. 2965), phospho-Stat3 (Tyr705, cat no. 9145), Stat3 (cat no. 9132). Immunoblots were washed in PBS-T 3 times for 10 minutes each and incubated with secondary anti-rabbit conjugated to horse radish peroxidase at a 1:1000 dilution in PBS-T/3% milk for 2 hours. Reactions were catalyzed with West Femto ECL substrate.
Statistical analysis
Data were analyzed using Graphpad Prism software using Student’s Two Tailed Tests for 2 groups and One Way ANOVA with Bonferroni’s post-hoc comparison for more than 2 groups. In vitro experiments were conducted with triplicate samples, and performed a minimum of 3 times. *p<0.05, **p<0.01, ***p<0.001, ns= not significant. Data are presented as Mean±SEM.
Results
Carcinoma associated fibroblasts enhance progression of in situ mammary lesions to invasiveness
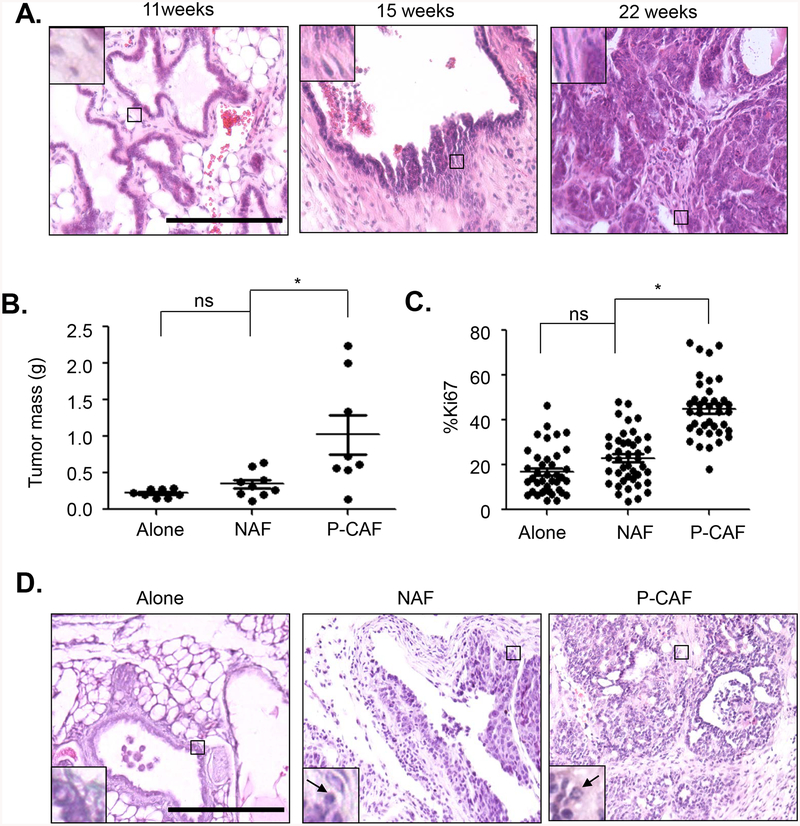
MMTV-PyVmT mice have been a reliable model for breast cancer progression. MMTV-PyVmT mice bred on an FVB genetic background form in situ lesions between 4–6 weeks of age that quickly progress to multi-focal invasive adenocarcinomas by 10 weeks of age [55–57]. C57BL/6 mice are more resistant to tumor formation [58, 59]; however mammary tumor development on this background has not been fully characterized. When MMTV-PyVmT mice were bred onto the C57BL/6 background, these mice formed benign mammary lesions between 7–11 weeks of age, with a papillary architecture, as confirmed by a clinical breast pathologist. The mammary lesions were characterized by non-invasive fern-like structures with cysts and few fibroblasts (Fig 1A). Between 13–18 weeks of age, lesions were characterized by increased numbers of fibroblasts, and presence of invasive carcinoma cells in the surrounding stroma. Myoepithelial cells were distinguished from stromal derived fibroblasts by overlapping expression of p63 and α-sma; fibroblasts expressed α-sma only (Supplemental Fig 1). Epithelial CK5/6 expression in MMTV-PyVmT lesions were characteristic of cytokeratin expression patterns detected in papillary lesions [60, 61]. By 29 weeks of age, lesions had progressed to invasive carcinomas with extensive fibrosis (Fig 1A). These data indicate that transgenic overexpression of MMTV-PyVmT on the C57BL/6 background results in slow growing mammary lesions that progress to invasive carcinomas, correlating with accumulation of fibroblasts.
Fig 1. P-CAFs enhance progression of 144epi mammary carcinoma cells.
A. H&E stain of mammary lesions from PyVmT/C57BL/6 mice. N=12 for mice 7–11 weeks, n=13 for mice 12–19 weeks old and n=12 for mice 25–29 weeks old. Magnified inset shows level of fibroblastic stroma. B-D. 144epi mammary epithelial cells were transplanted alone, with NAF or P-CAFs for 60 days. Mammary tissues were measured for tumor mass (B), Ki67 immunostaining (C) and H&E stain (D). Magnified inset in (D) shows mammary carcinoma cells. Arrowhead points to invasive carcinoma cells. Statistical analysis was performed using One Way ANOVA with Bonferroni post-hoc comparison. Statistical significance was determined by p<0.05. *p<0.05, **p<0.01. Scale bar=200 microns. n=8 for 144epi alone, n=9 for 144epi: NAF, n=8 for 144epi: P-CAF. Mean±SEM are shown.
We analyzed the role of fibroblasts on progression of in situ lesions in co-transplant studies. Fibroblasts were isolated from normal mammary tissues (NAFs), or from 29-week-old MMTV-PyVmT mice (P-CAFs), which formed invasive mammary tumors. Mammary epithelial cells (144epi) were isolated from mammary tissues of 15-week-old mice, at early stage invasion. 144epi cells were orthotopically transplanted alone, or embedded in NAFs or P-CAFs in collagen and transplanted directly into the mammary fat pad. Because breast cancers tend to develop in adult women, past the age of 55 [62], we transplanted cells directly into uncleared mammary fat pads of adult mice. Using this approach, we had previously demonstrated that donor fibroblasts enhanced mammary tumor growth compared to carcinoma cells transplanted alone in adult mice [49].
Mice were monitored for tumor formation on a twice weekly basis, until 60 days post-transplantation, when tumors reached approximately 1.5 cm in diameter, the maximum allowable tumor size allowed by the institutional animal review board. Mammary tissues were harvested and analyzed for changes in growth and invasiveness of lesions. Compared to 144 epi cells grafted alone, 144 epi cells co-grafted with NAFs resulted in a small but not statistically significant increase in tumor mass and tumor cell proliferation, as indicated by Ki67 immunostaining (Fig 1B–C). Compared to co-grafting with NAFs, co-grafting with P-CAFs significantly increased tumor mass from an average of 0.32 g to 0.89 g, and enhanced tumor cell proliferation by 1.9-fold (Fig 1B–C). By H&E stain, at 60 days (8 weeks), 144epi cells grafted alone formed non-invasive lesions like those seen in transgenic mice 7–11 weeks old, with slender frond-like structures and low stromal reactivity. Compared to 144 epi grafted alone, 144epi grafted with NAFs formed thicker frond-like structures, with a few invasive carcinoma cells, resembling transgenic lesions from mice 12–19 weeks old (Fig 1D). Co-grafting with P-CAFs resulted in invasive carcinoma characterized by loss of papillary structure and extensive stromal reactivity, resembling lesions from transgenic mice 25–29 weeks old (Fig 1D). In blinded scoring of H&E stains, co-transplantation with NAFs resulted in a few lowly and highly invasive lesions. Co-transplantation with P-CAFs significantly increased the number of highly invasive lesions compared to grafting with 144epi alone or 144epi with NAFs (Table 1). These data indicate that P-CAFs enhance progression of 144epi lesions over NAFs co-grafted with 144epi cells or 144epi cells transplanted alone.
Table 1. P-CAFs fibroblasts co-grafted with 144epi mammary carcinoma cells promotes tumor invasion.
144epi cells were transplanted alone, or with NAFs or P-CAFs for 60 days. Tumor samples stained by H&E were scored for invasiveness. Statistical analysis was performed by Chi-square test for trend. Statistical significance was determined by p<0.05. p<0.0001. n=8 for 144epi alone, n=9 for 144epi: NAF, n=8 for 144epi: P-CAF.
| non- invasive |
lowly- invasive |
highly invasive |
|
|---|---|---|---|
| Alone | 8 | 0 | 0 |
| NAF | 5 | 2 | 2 |
| P-CAF | 1 | 1 | 6 |
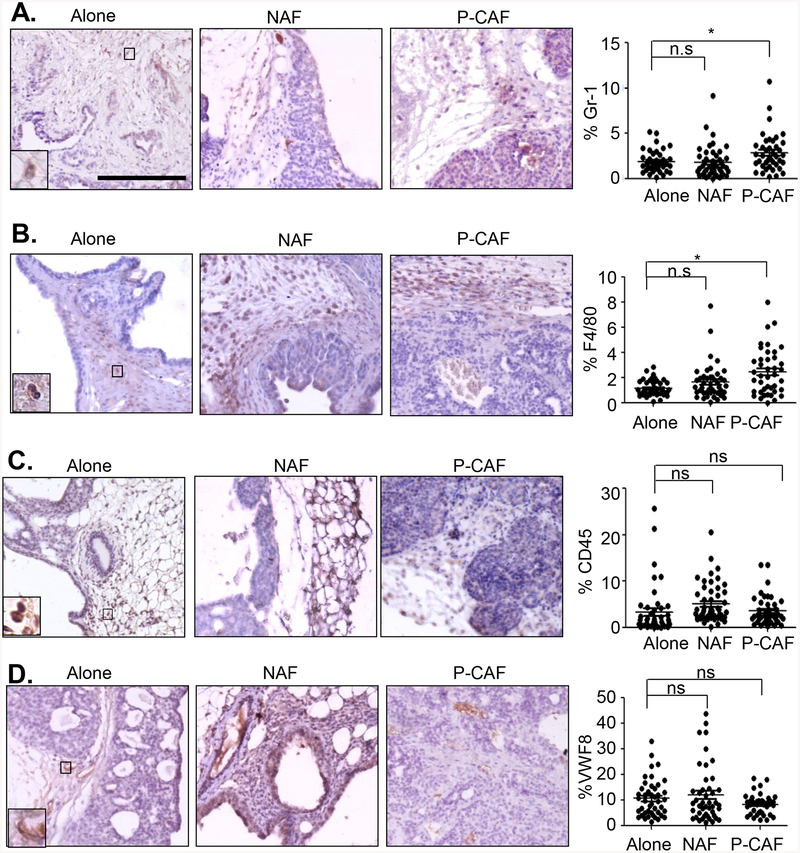
As analysis of H&E stains indicated changes in stromal reactivity with co-grafting of 144epi cells with P-CAFs, primary tumor tissues further analyzed for changes in immune cell recruitment and tumor and angiogenesis. Tissues were immunostained for F4/80, a macrophage marker, Gr-1, a neutrophil marker, CD45, a lymphocyte marker, and VWF8, an endothelial marker [63–66], and expression levels were quantified. 144epi cells co-grafted with P-CAFs showed a 1.6-fold increased expression of Gr-1 compared to 144epi co-grafted with NAFs (Fig 2A). Compared to 144epi cells grafted alone, F4/80 expression was increased in lesions co-grafted with NAFs by 1.4-fold, and in lesions co-grafted with P-CAFs by 1.9-fold (Fig 2B). There were no significant differences in CD45 or VWF8 expression among groups (Fig 2C–D). These data indicate that NAFs and P-CAFs enhance recruitment of F4/80+ cells and only P-CAFs enhance recruitment of Gr-1+ cells.
Fig 2. P-CAFs regulate immune cell recruitment to 144epi mammary lesions.
Mammary lesions were immunostained for: A. Gr-1, B. F4/80, C. CD45 or D. Von Willebrand Factor 8 (VWF8). Statistical analysis was performed using One Way ANOVA with Bonferroni post-hoc comparison. Statistical significance was determined by p<0.05. *p<0.05, n.s=not significant. Scale bar=200 microns. n=8 for 144epi alone, n=9 for 144epi: NAF, n=8 for 144epi: P-CAF. Mean±SEM are shown.
Mammary lesions show increased expression of CXCL1 and CXCR2 over CCL2 and CCR2
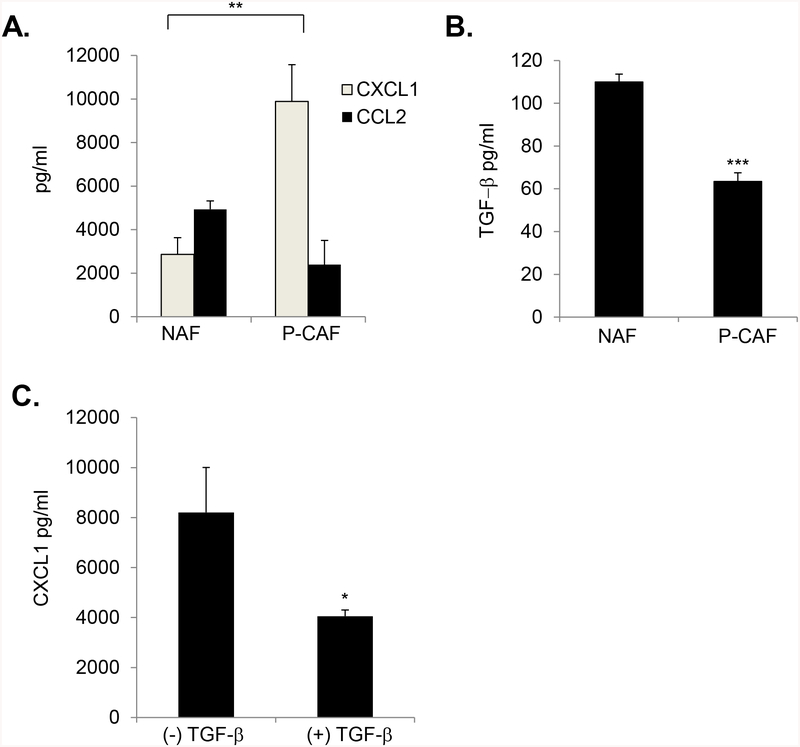
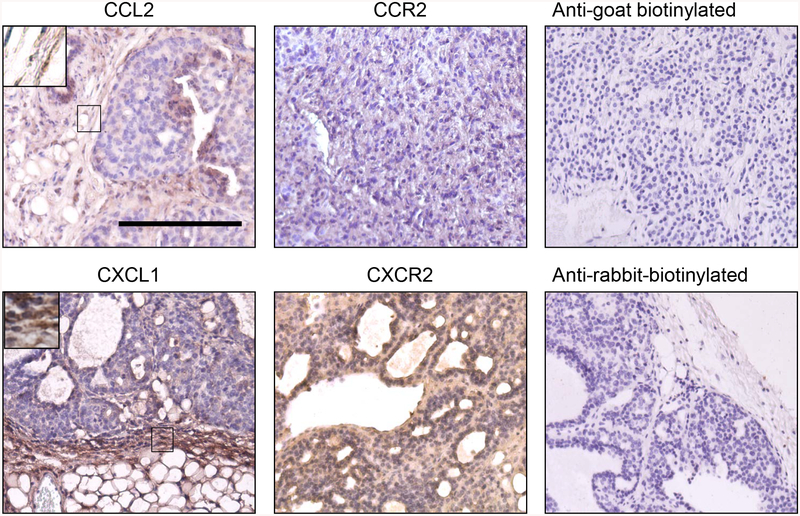
Previous studies have demonstrated that progression of invasive ductal carcinomas is associated with increased expression of CCL2 and CXCL1 chemokines and decreased expression of TGF-β in stromal derived fibroblasts [44, 20]. By ELISA analysis, CXCL1 was more highly expressed in P-CAFs than CCL2 and P-CAFs showed elevated levels of CXCL1 compared to NAFs (Fig 3A). Furthermore, elevated CXCL1 levels corresponded to decreased expression of TGF-β in P-CAFs (Fig 3B). TGF-β treatment of P-CAFs decreased CXCL1 expression (Fig 3C). These data indicate that P-CAFs elevate CXCL1 expression, which is negatively regulated by TGF-β signaling. By immunostaining, CCL2 was lowly expressed in the stroma and its primary receptor, CCR2 was expressed weakly in the mammary tumor epithelium. CXCL1 appeared strongly expressed in the stroma, and its primary receptor, CXCR2 was strongly expressed in the tumor epithelium (Fig 4). Overall, CXCL1 and CXCR2 were strongly expressed in mammary tumor tissues.
Fig 3. P-CAFs show increased CXCL1 and decreased TGF-β expression.
ELISA analysis of A. CXCL1 and CCL2 in NAFs vs. P-CAFs, B. TGF-β in NAFs vs. P-CAFs, C. CXCL1 in P-CAFs treated with 10 ng/ml of TGF-β for 24 hours. Statistical analyzed was performed using Two Tailed T-test. Samples were plated in triplicate per group. Experiments were performed three times. Statistical significance was determined by p<0.05. *p<0.05, **p<0.01, ***p<0.001. Mean±SEM are shown.
Fig 4. CXCL1 and CXCR2 are more highly expressed than CCL2 and CCR2 in 144epi mammary lesions.
Mouse mammary lesions from 144epi cells were co-transplanted with P-CAFs and immunostained for CCL2, CCR2, CXCL1 or CXCR2. Magnified insets show staining in fibroblastic cells. Scale bar=200 microns. n=8 per group.
CXCR2 knockdown inhibits invasive progression of mammary lesions mediated by TGF-β signaling deficient fibroblasts
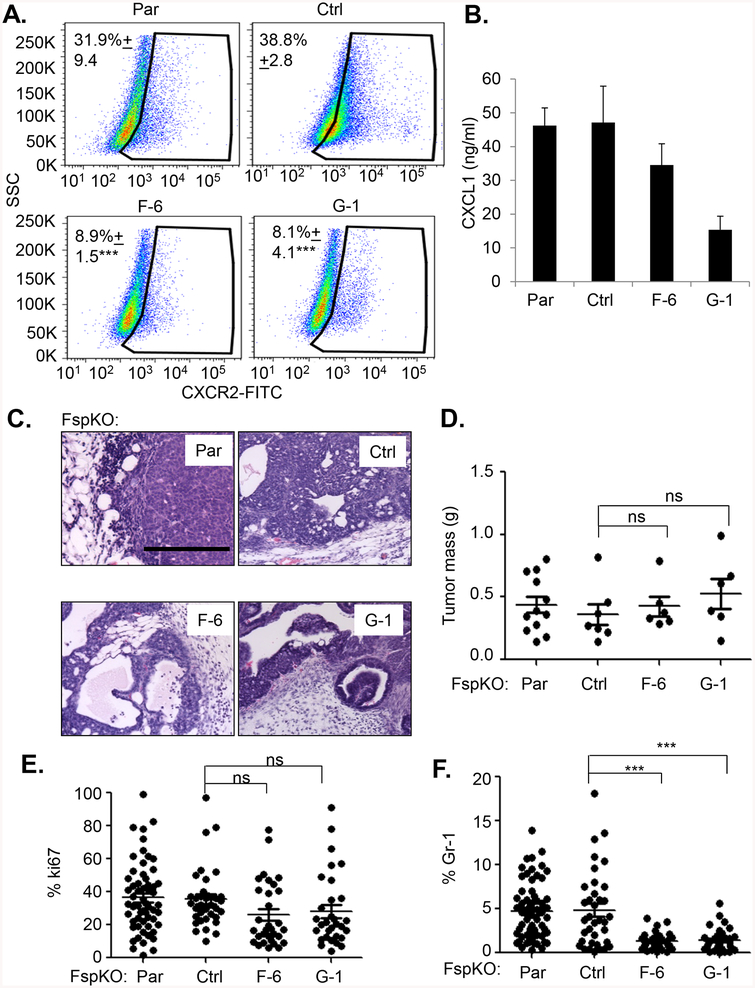
We determined the possibility that CXCL1 may signal to CXCR2+ epithelial cells to regulate progression of in situ lesions. CXCR2 was knocked down in 144epi mammary epithelial cells by stable shRNA expression to generate 2 different CXCR2 deficient cell lines (F-6, G-1). By flow cytometry CXCR2 was reduced in F-6 cells by 4.3-fold, and 4.7-fold in G-1 cells, relative to control shRNA expressing cells (Fig 5A). As CXCR2 knockdown decreased CXCL1 expression in F-6 cells by 1.3-fold and in G-1 cells 3-fold (Fig 5B), CXCR2 knockdown alone could affect progression of 144epi lesions through decreased autocrine CXCR2 signaling. To determine this possibility, we orthotopically transplanted 144epi cells expressing control or CXCR2 shRNAs (F-6, G-1) in C57BL/6 mice. At 60 days, F-6 and G-1 144epi cells grafted alone did not show significant changes to tumor growth, invasion or recruitment of Gr-1+ cells, compared to control 144epi cells, (Supplemental Fig 2). These data indicate that autocrine CXCR2 signaling in 144epi cells was not sufficient for invasive progression.
Fig 5. CXCR2 knockdown in 144epi cells inhibits Tgfbr2FspKO fibroblasts mediated progression of 144epi mammary lesions.
A. Flow cytometry analysis of CXCR2 in Parental (Par) or 144epi cells stably expressing control shRNA (Ctrl) or CXCR2 shRNAs (F-6, G-1). Percentage of positive cells are shown. B. CXCL1 ELISA of cultured 144epi cells. C-F. Tgfbr2FspKO fibroblasts were co-transplanted with control or CXCR2 deficient 144epi cells. Mammary tissues were analyzed by H&E stain (C), tumor mass (D), Ki67 (E), or Gr-1 expression (F). Statistical analysis was determined by One Way ANOVA followed by Bonferroni post-hoc comparison of F-6 and G-1 with control shRNA. Statistical significance was determined by p <0.05. *p<0.05; **p<0.01, ***p<0.001, ns=not significant. Scale bar=200 microns. n=12 for parental 144epi: Tgfbr2FspKO, n=7 for 144epi: Control shRNA, n=6 for 144epi:F-6, n=6 for 144epi:G-1. Flow cytometry experiments were performed 3 times. Mean±SEM are shown.
Given that P-CAFs show elevated CXCL1 expression, which is negatively regulated by TGF-β signaling, we determined the relevance of stromal derived TGF-β to CXCL1/CXCR2 signaling in in situ mammary PyVmT lesions. TGF-β signaling deficient fibroblasts (Tgfbr2FspKO) showed elevated CXCL1 expression [44, 19]. These cells were used as a model to determine how elevated CXCL1 expression and down-regulation of TGF-β signaling in stroma would affect progression of in situ lesions. At 60 days, Tgbr2FspKO fibroblasts co-grafted with parental 144epi cells resulted in invasive lesions similar to co-transplants with P-CAFs (Fig 5C, Table 2). To determine the relevance of paracrine CXCR2 signaling to progression of in situ mammary lesions, Tgfbr2FspKO fibroblasts were co-grafted with control or CXCR2 deficient 144epi cells (F-6, G-1). Compared to control shRNA expressing 144epi cells, CXCR2 knockdown in 144epi cells resulted in fewer cases of invasive lesions, but did not significantly affect tumor growth or cell proliferation mediated by Tgfbr2FspKO fibroblasts (Fig 5C–E, Table 2). Compared to control shRNA expressing 144epi cells, CXCR2 knockdown also inhibited recruitment of Gr1+ cells mediated by Tgfbr2FspKO fibroblasts by 3.6-fold with F-6 cells and 3.45-fold with G-1 cells (Fig 5F). Compared to control shRNA expressing 144epi cells, CXCR2 knockdown in 144epi cells did not affect recruitment of F4/80+ cells mediated by Tgfbr2FspKO fibroblasts (Supplemental Fig 3). These data indicate that CXCR2 expression is required for progression of 144epi cells mediated by Tgfbr2FspKO fibroblasts.
Table 2. CXCR2 knockdown inhibits Tgbr2FspKOmediated invasion of 144epi mammary carcinomas.
Parental (Par), Control (Ctrl) or CXCR2 deficient 144epi cells were co-transplanted with Tgfbr2FspKO fibroblasts for 60 days. Tumor samples stained by H&E were scored for invasiveness. Statistical analysis was performed by Chi-square test for trend. Statistical significance was determined by p<0.05. p<0.003. n=12 for parental 144epi: Tgfbr2FspKO, n=7 for 144epi:Control shRNA, n=6 for 144epi:F-6, n=6 for 144epi:G-1.
| Tgfbr2FspKO | non-invasive | lowly- invasive |
highly invasive |
|---|---|---|---|
| Par | 0 | 1 | 11 |
| Ctrl | 2 | 1 | 4 |
| F-6 | 0 | 5 | 1 |
| G-1 | 5 | 0 | 1 |
Oncogenic potential of mouse mammary fibroblasts
Our data indicated that P-CAFs and Tgfbr2FspKO fibroblasts co-grafted with 144epi mammary epithelial cells enhance mammary tumor progression compared to 144epi mammary epithelial cells grafted alone or co-grafted with NAFs. However, it was unclear whether the fibroblasts themselves were tumorigenic and if they could affect normal or benign mammary epithelial cells. Therefore, we determined the purity of the fibroblast cell lines used in this study. Immunocytochemistry staining indicated that NAFs, P-CAFs and Tgfbr2FspKO fibroblasts expressed FSP1, α-sma and N-cadherin (Supplemental Figure 4), mesenchymal markers typically expressed in fibroblasts [67–69]. Expression of FSP1 and N-cadherin appeared slightly weaker in Tgfbr2FspKO fibroblasts, possibly reflecting defects in TGF-β signaling, which regulate expression of Fsp1 and N-cadherin [70, 71]. All fibroblast cell lines were absent for expression of endothelial and epithelial markers (Supplemental Fig 4). By contrast, 144epi cells expressed pan-cytokeratin, an epithelial marker [72], and were absent for Fsp1, α-sma and N-cadherin expression. These data demonstrate that NAFs, P-CAFs and Tgfbr2FspKO cells were fibroblastic and devoid of potential contaminating carcinoma cells.
We next compared the oncogenic potential of fibroblasts with 144epi cells through soft agar assay. 144epi cells but not NAFs, P-CAFs or Tgfbr2FspKO fibroblasts formed colonies in soft agar (Supplemental Fig 5). 144epi cells formed fewer colonies in soft agar compared to 4T1 cells, a metastatic mammary carcinoma cell line [73]. We analyzed the effects of fibroblast conditioned medium on soft agar growth of 144epi cells and on NMuMG cells, a benign mammary epithelial cell line [74, 75]. NMuMG cells treated with conditioned medium from P-CAFs, but not control DMEM/10% FBS or NAF conditioned medium showed increased growth in soft agar to an average of 1.3 colonies per 5000 cells (Supplemental Fig 6). 144epi cells showed increased colony formation in soft agar when treated with conditioned medium from P-CAFs, compared to NAF conditioned medium or control medium. Similar to P-CAF conditioned medium, Tgfbr2FspKO fibroblast conditioned medium enhanced colony formation of 144epi cells in soft agar, and induced anchorage independent growth of NMuMG mammary epithelial cells, with an average of 1.4 colonies per 5000 cells (Supplemental Fig 6). These data indicate that fibroblasts do not form colonies in soft agar; however, conditioned medium from P-CAFs and Tgfbr2FspKO fibroblasts enhance anchorage independent growth of benign mammary epithelial cells.
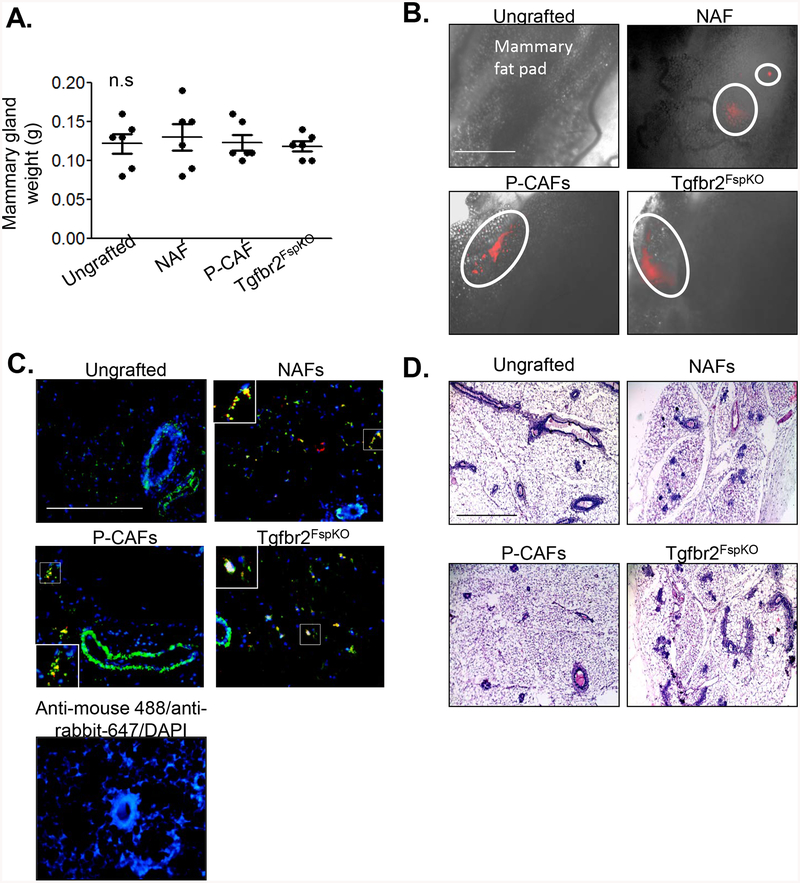
Could fibroblasts form tumors or transform the normal mammary gland, contributing to the mammary tumor phenotypes observed in this study? To determine these possibilities, NAFs, P-CAFs and Tgfbr2FsPKO fibroblasts were labeled with mCherry fluorochrome, embedded in collagen and implanted into the mouse mammary fat pad. Mice were palpated twice weekly for tumor formation, and sacrificed for tissue harvest 60 days post-transplantation. Compared to ungrafted tissues, there were no significant differences in mammary gland weight with mice transplanted with fibroblasts (Fig 6A). By fluorescence microscopy, mCherry positive cells were detected in mammary tissues, indicating retention of transplanted fibroblasts (Fig 6B). By co-immunofluorescence staining, mCherry positive cells overlapped with α-sma (Fig 6C), a marker expressed in activated breast fibroblasts [68, 76]. A few mCherry positive cells were α-sma negative, reflecting possible differences in the activation state and heterogeneity of fibroblasts. To determine the effects of transplanted fibroblasts on the mammary gland, mammary tissues were serial sectioned and stained by H&E. Compared to ungrafted mammary glands, fibroblasts did not significantly affect mammary gland architecture, which appeared normal (Fig 6D). Taken together, these data indicate that NAFs, P-CAFs and Tgfbr2FspKO fibroblasts are not transformed and do not transform normal mammary epithelial cells when orthotopically transplanted into mice.
Fig 6. Fibroblasts transplanted alone do not promote tumor formation in mammary glands.
NAFs, P-CAFs or Tgbr2FspKO fibroblasts were labeled with mcherry and transplanted alone in mammary fat pads of C57Bl/6 mice for up to 60 days. A. Mammary tissues were harvested and imaged by using the FL Auto EVOS imaging system for detection of mCherry positive cells. mCherry positive areas are circled. Scale bar=1000 microns. B. Co-immunofluorescence staining for mCherry (red) or α-sma expression (green) in mammary tissues. Magnified inset shows α-sma/mCherry co-expressing cells. Scale bar=200 microns. C. Mammary tissues were serial sectioned and stained by H&E. Representative of 200 sections per sample, n=6 mice per group. Scale bar=200 microns. Statistical analysis was performed using One Way ANOVA with Bonferroni post-hoc comparison. Statistical significance was determined by p<0.05. ns=not significant. Mean+SEM are shown.
CXCL1/CXCR2 signaling regulates invasion of 144epi mammary carcinoma cells through NF-κB, p42/44MAPK, AKT and Stat3 dependent mechanisms
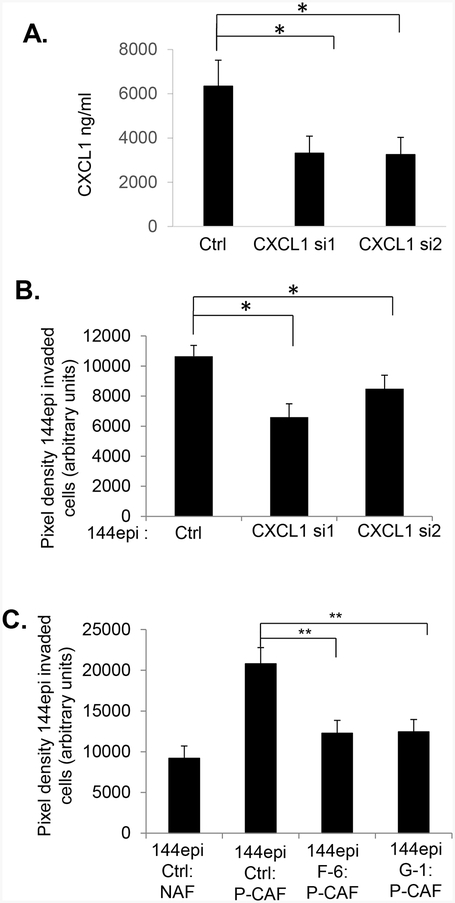
As data indicated that mammary tumor progression was mediated through fibroblast signaling to mammary carcinoma cells, we further investigated the role of CXCL1/CXCR2 signaling in regulating mammary tumor cell invasion. To further determine the contribution of CXCL1 derived from P-CAFs to invasion of 144epi cells, we performed Matrigel transwell invasion assays. P-CAFs were transfected with control or 2 different CXCL1 siRNAs (CXCL1 si1, CXCL1 si2), coated on the underside of Matrigel coated transwells, and examined for invasion of fluorochrome labeled 144epi cells to the underside. By ELISA, CXCL1 expression was reduced in P-CAFs by 1.9-fold with CXCL1 si1, and 2-fold reduction of CXCL1 expression with CXCL1 si2 (Fig 7A). CXCL1 knockdown in P-CAFs significantly decreased 144epi invasion 1.6-fold with CXCL1 si1, and 1.3-fold with CXCL1 si2 (Fig 7B). Control siRNA transfection in P-CAFs did not significantly affect CXCL1 expression or 144epi cell invasion compared to parental 144epi cells (Supplemental Fig 7A–B), indicating the reduction in 144epi invasion was specific to CXCL1 siRNA knockdown. These data indicate that CXCL1 expression is required for P-CAF mediated invasion of 144epi cells. To further determine the relevance of CXCR2 expression to tumor cell invasion mediated by P-CAFs, control or CXCR2 deficient 144epi cells were co-cultured with P-CAFs and analyzed for invasion by Matrigel transwell assay. Compared to control 144epi cells, CXCR2 deficient 144epi cells (F-6, G-1) showed a significant 1.7-fold reduction in invasion mediated by P-CAFs (Fig 7C). These data indicate that CXCR2 expression is necessary for P-CAF mediated invasion of 144epi cells.
Fig 7. Stromal derived CXCL1 promotes 144epi cell invasion through CXCR2 dependent mechanisms.
A-B. P-CAFs were transfected with control (Ctrl) or CXCL1 siRNAs, analyzed for CXCL1 expression by ELISA (A) and 144epi cell invasion by Matrigel transwell assay (B). C. Control (Ctrl) or CXCR2 shRNA expressing144epi cells (G-1, F-6) were co-cultured with control or CXCL1 siRNA transfected P-CAFs and analyzed for changes in invasion by Matrigel transwell assay. Statistical analysis was determined by One Way ANOVA followed by Bonferroni post-hoc analysis. Triplicate samples per group were plated; experiments were performed three times. Statistical significance was determined by p <0.05. *p<0.05; **p<0.01, ns=not significant. Mean±SEM are shown.
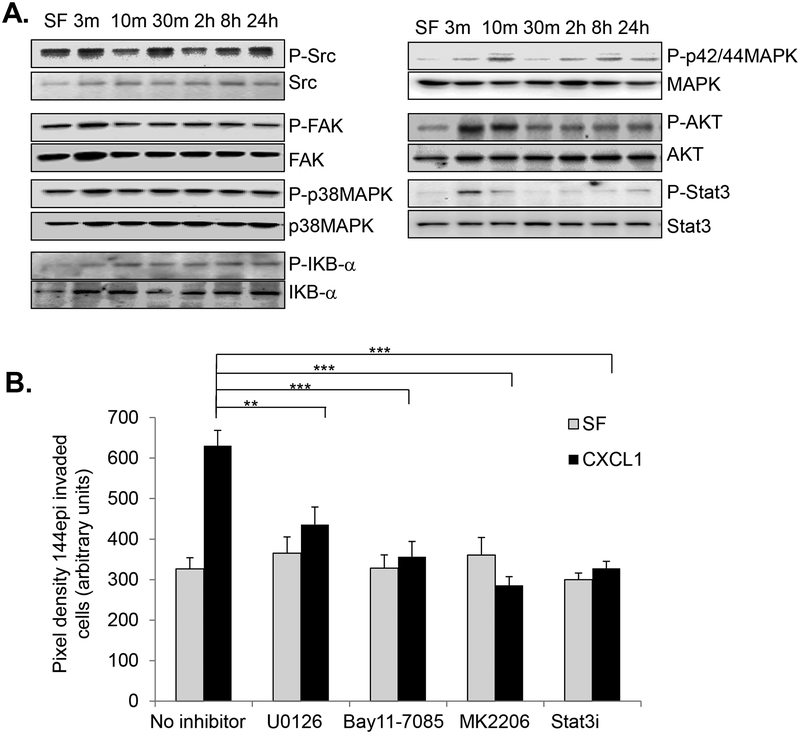
To identify the downstream mechanisms responsible for CXCL1 mediated invasion, we employed a candidate approach, examining for phosphorylation of downstream kinases related to invasion and migration. CXCL1 treatment did not significantly affect phosphorylation of Src, FAK or p38MAPK proteins in 144epi cells. CXCL1 enhanced phosphorylation of IκB, p42/44MAPK, AKT and Stat3 (Fig 8A). To determine the functional contribution of these pathways to CXCL1 induced invasion, we used pharmacologic inhibitors to block p42/44MAPK, NF-κB, AKT and Stat3 activity. U0126 non-competitively binds to and inhibits MEK1/2, upstream kinases of p42/44MAPK [77, 43]. Bay11–7085 irreversibly binds to IκB-α, preventing phosphorylation and NF-κB activation [78, 79]. MK2206 is an allosteric inhibitor to AKT1/2/3[78, 80]. Cell permeable Stat3 phospho-peptides (Stat3i) bind to the Stat3 SH2 domain to prevent Stat3 dimerization and activation [81, 82]. Treatment of 144epi cells alone with U0126, Bay11–7085, 0 MK2206 or Stat3i did not affect cellular invasion. CXCL1 induced invasion of 144epi cells was reduced by 1.4-fold with U0126 treatment, 1.8-fold with Bay11–7085 treatment, 2.2-fold with MK2206 treatment and 1.9-fold with Stat3i treatment (Fig 8B). These data indicate that p42/44MAPK, NF-κB, AKT and Stat3 activity are required for CXCL1 induced 144epi cell invasion.
Fig 8. CXCL1 regulates invasion of 144epi cells through NF-κB, MAPK, AKT and Stat3 dependent pathways.
A. 144epi cells were treated with 60 ng/ml of CXCL1 over time and analyzed by immunoblot for expression of the indicated proteins. B. 144epi cells were treated with 60 ng/ml CXCL1 in the presence or absence of: 5 μM NF-κB inhibitor (Bay11–7082), 1 μM MEK inhibitor (U0126),0.28 μM AKT inhibitor (MK2206) and 1mM Stat3 peptide inhibitors (Stat3i), and analyzed for changes in transwell invasion over 24 hours. Statistical analysis was determined by One Way ANOVA followed by Bonferroni post-hoc analysis. Transwell assays were plated in triplicate per group. Experiments were performed three times. Statistical significance was determined by p <0.05. **p<0.05, ***p<0.001, ns=not significant. Mean±SEM are shown.
Discussion
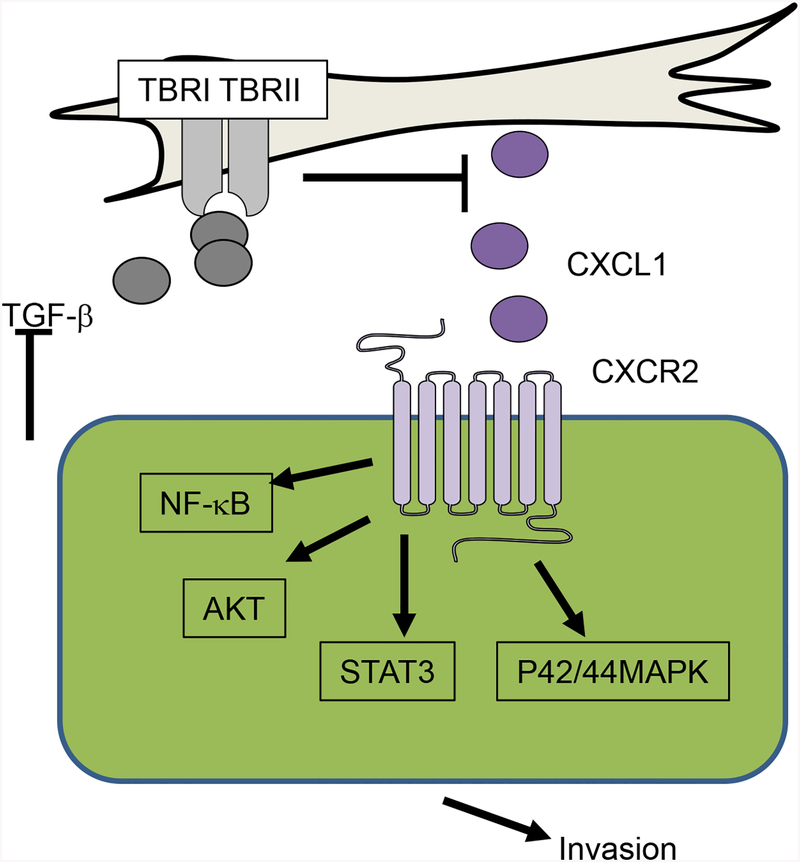
In these studies, we demonstrate that PyVmT overexpression in C57BL/6 mice form slow growing in situ lesions that progress to invasive carcinomas in a stage dependent manner. Furthermore, we demonstrate an important role for fibroblasts in regulating the invasive progression of mammary lesions through paracrine interactions with epithelial cells. Our studies suggest decreased TGF-β expression upregulates CXCL1 expression in P-CAFs. CXCL1 acts on CXCR2 expressing tumor epithelial cells to enhance cell invasion through NF-κB, p42/44MAPK, AKT and Stat3 dependent mechanisms (Fig 9).
Fig 9. Role of fibroblasts in invasive progression of breast lesions.
Decreased expression of TGF-β in stromal fibroblasts increase expression of CXCL1, which acts on CXCR2 expressed on mammary epithelial cells. CXCL1/CXCR2 signaling increases activation of: NF-κB, AKT, Stat3 and p42/44MAPK to enhance cellular invasion.
PyVmT transgenic mice on a C57BL/6 background initially formed non-invasive lesions with a papillary architecture. The physiologic relevance of the papillary phenotype in MMTV-PyVmT transgenic mice is unclear. Transgenic expression of cyclin E in the mammary epithelium of mice in the B6C3F2 genetic background result in hyperplastic lesions with a papillary architecture that progress to adenocarcinomas in a subset of mice [83]. MMTV-driven expression of c-Myc in FVB mice results in mammary lesions with papillary histology, a subset of which progress to metastatic mammary carcinomas [84]. Transgenic expression of WNT1 also form mammary tumors with a papillary architecture [85]. These suggest that papillary lesions in the mammary gland are common in transgenic mice.
A breast clinical pathologist indicated that the early transgenic lesions resembled intraductal papillomas diagnosed in patients. Papillomas are benign lesions that only carry a low risk of breast cancer in patients [86–88]. Yet, all of the female mice carrying the PyVmT transgene developed invasive mammary carcinomas with loss of papillary architecture, indicating that the papillary phenotype in mice are not stable lesions. The C57Bl/6 genetic background is tumor resistant [58], while the PyVmT is a potent oncogene [56]. While transgenic expression of PyVmT on a C57BL/6 background resulted in slow growing mammary lesions, these lesions eventually progressed to invasive mammary carcinomas indicating a dominant effect of the PyVmT oncogene to mammary carcinoma progression.
At this time, we cannot conclude that the lesions formed in PyVmT transgenic mice on C57Bl/6 background accurately represent papillary lesions diagnosed in patients. Yet, several aspects of the transgenic model may be considered advantageous to studying breast cancer. MMTV-PyVmT mice bred on to C67BL/6 genetic background formed slow growing in situ lesions that progressed to invasive carcinoma over time. Because breast cancers often take decades to develop in women, the MMTV-PyVmT (C57BL/6) model may be a useful model to study progression of early stage breast cancers. Accumulation of fibroblasts correlates with invasive mammary carcinoma progression, similar to a desmoplastic phenotype observed in IDC in humans [89, 90]. There are currently few transgenic models to mimic the various molecular subtypes of breast cancers diagnosed in women. MMTV-PyVmT mammary tumors show increased ErbB2 expression and signaling, mimicking luminal/ErbB2(Her2) overexpressing breast cancers [85, 91, 92, 57]. The WNT pathway may also be important in mediating mammary carcinoma progression tumors [85]. In our studies, we have demonstrated that fibroblasts mediate CXCL1/CXCR2 signaling to regulate mammary carcinoma progression in this model. Profiling studies have revealed that PyVmT tumors from different genetic backgrounds such as FVB and AKXD show significant differences in predicted pathway activity [59, 85]. It would be interest to further analyze how CXCL1/CXCR2 cooperates with other oncogenic pathways to regulate development of invasive mammary carcinomas.in PyVmT/C57BL/6 mice. In summary, studies in this model provide mechanistic understanding of how fibroblasts enhance invasive progression of mammary carcinomas in part through TGF-β and CXCR2 dependent mechanisms.
We demonstrate several molecular and functional differences in fibroblasts derived from tumor free mice and mice bearing PyVmT mammary lesions on a C57Bl/6 background. P-CAFs enhance tumor growth and invasion compared to NAFs. Our studies found that NAFs exerted a small increase in tumor growth and invasion of 144epi cells, similar to some studies on NAFs in breast cancer [93, 94]. The tumor promoting activities of P-CAFs are consistent with previous studies showing that breast derived CAFs enhance tumor progression of MCF7, MDA-MB-231 breast cancer cells and PDX models, [93, 95–97]. P-CAFs showed elevated CXCL1 expression and decreased TGF-β expression similar to other CAFs [44, 21]. Our studies suggest that a hallmark of some mammary carcinoma associated fibroblasts is increased CXCL1 expression, which is negatively regulated by TGF-β signaling. P-CAFs exhibit several phenotypes similar to Tgfbr2FspKO fibroblasts including: elevated CXCL1 expression and enhancement of 144epi cell invasion and anchorage independent growth. While our studies indicate that CXCL1 derived from CAFs is biologically significant, we cannot conclude at this time that loss of TGF-β signaling in fibroblasts is necessary for the tumor promoting phenotype of P-CAFs. Further studies would need to be performed to investigate for changes in TGF-β signaling in P-CAFs, and how these possible changes may functionally contribute to mammary carcinoma progression. In addition, it would be of interest to further examine the patterns in stromal CXCL1 and TGF-β expression in patient samples with pre-invasive lesions. These additional studies would advance our understanding on how pre-invasive lesions progress on a molecular level, and lead to a signature to predict invasion of in situ breast cancers.
Our studies show that P-CAFs enhance progression of 144epi mammary carcinoma cells in mice, corresponding to increased in vitro invasion and anchorage independent growth in soft agar. Orthotopic transplantation of fibroblasts alone for 60 days did not result in visible tumor formation. These cells were retained, but did not significantly expand. These data indicate that fibroblasts were not transformed themselves, and did not transform normal mammary epithelium within that timeframe. Thus, the mammary tumors observed in this study originated from PyVmT expressing mammary epithelial cells. Interestingly, conditioned medium from P-CAFs and Tgfbr2FspKO fibroblasts induced anchorage independent growth of NMuMG mammary epithelial cells. But orthotopic transplantations of NAFs, P-CAFs and Tgfbr2FspKO fibroblasts had no effect on the normal mammary gland. NMuMG cells are non-transformed but immortalized [74, 75]. Therefore, heritable alterations in this cell line combined with fibroblasts could have contributed to the soft agar colony growth of NMuMG cells in vitro.
Bone marrow derived cells including macrophages, neutrophils, myeloid derived suppressor cells (MDSC) play an integral role in regulating breast cancer progression [15, 98, 99]. Here, we show that P-CAFs mediated Gr-1+ recruitment is associated with CXCR2 mediated tumor invasion. These data are consistent with previous studies indicating a tumor promoting role for Gr-1+ myeloid cells in breast cancer [33, 32, 100]. While CXCL1 may attract Gr-1+ cells [33], the reduction in CXCL1 expression in 144epi cells caused by CXCR2 knockdown was not sufficient to affect Gr1+ recruitment. Other factors such as CXCL17, GM-CSF1 or IL-1α [101–103] may be expressed by 144epi cells, and attract and/or regulate differentiation of Gr-1+ cells in 144epi lesions. Interestingly, both NAFs and P-CAFs enhanced recruitment of F4/80+ macrophages over 144epi cells alone. Fibroblast mediated macrophage recruitment may be important in the early stages of progression of in situ breast lesions. Fibroblast accumulation was initially observed in transgenic mice 13–18 weeks of age during early invasion. Co-grafting with NAFs slightly increased tumor growth and invasion over 144epi cells grafted alone. Furthermore, studies have detected accumulation of macrophages and fibroblasts in in situ lesions associated with invasive breast cancer progression [104]. It would be of interest to further investigate the contributions of Gr-1+ and F4/80+ myeloid cells in progression of in situ breast lesions.
We show that paracrine but not autocrine CXCL1 signaling to CXCR2 signaling in 144epi cells is an important mechanism for cellular invasion. Previous studies have shown that autocrine CXCL1 signaling in PyVmT (FVB) mammary carcinoma cells and LM2–4175 breast cancer cells was important for tumor growth [33]. In contrast to these studies, which showed high epithelial CXCL1 expression [33], we observed high stromal expression and low epithelial expression of CXCL1. These differences in CXCL1 expression may explain why CXCR2 knockdown inhibited tumor invasion mediated by P-CAFs, but had no effect on 144epi cells transplanted alone. Epithelial cells from in situ breast lesions may be dependent on paracrine signaling interactions with stromal cells. It would of further interest to study the functional contribution of fibroblasts to invasive progression of breast lesions.
A few studies have investigated the CXCR2 mediated pathways in breast cancer. In MCF-7 breast cancer cells [105]. CXCR2 mediated MAPK is important for epithelial to mesenchymal transition while CXCR1/2 activates Src to enhance AKT and MAPK signaling, and enhance cancer stem cell activity [106]. In other cancers, CXCL1/CXCR2 signaling enhances growth, motility and invasion of hepatocellular, ovarian, gastric and prostate carcinoma cells, and melanoma through a common set of pathways including NF-κB, MAPK, and AKT [107, 34, 108–110]. Here, CXCL1 regulated invasion of 144epi cells through AKT and MAPK but not Src dependent mechanisms. Furthermore, we report for the first time that CXCL1-mediated Stat3 is important for invasion of mammary epithelial cells. Stat3 is more notable as a downstream transcriptional regulator of EGF, HGF and IL6 signaling mediated cancer growth and invasion [111–114]. The activation of NF-κB and Stat3 suggests that CXCL1/CXCR2 signaling may regulate gene expression in 144epi cells. Furthermore, Stat3 and P42/44MAPK phosphorylation appear transitory, and IκB and AKT phosphorylation is sustained long-term in CXCL1 stimulated cells. It is possible that Stat3 and p42/44MAPK may act upstream of IκB and AKT to regulate CXCL1 induced invasion. It would be of interest to further understand how these pathways function cooperatively to regulate CXCL1 induced cellular invasion at the molecular level.
In summary, we demonstrate an important role for stromal TGF-β and CXCL1/CXCR2 signaling in enhancing progression of in situ lesions to invasion through a genetically engineered mouse model. These studies reveal mechanistic insight into the progression of slow growing lesions with important implications for improving prognosis and treatment of patients with in situ breast carcinomas.
Supplementary Material
Acknowledgements:
We thank Iman Jokar for technical assistance.
Funding: These studies were supported by funds from KU Endowment and the National Institutes of Health/National Cancer Institute: R01CA172764 to N. Cheng.
References
- 1.Hussain A, Gordon-Dixon A, Almusawy H, Sinha P, Desai A. The incidence and outcome of incidental breast lesions detected by computed tomography. Annals of the Royal College of Surgeons of England. 2010;92(2):124–6. doi: 10.1308/003588410X12518836439083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falomo E, Strigel RM, Bruce R, Munoz Del Rio A, Adejumo C, Kelcz F. Incidence and outcomes of incidental breast lesions detected on cross-sectional imaging examinations. Breast J. 2018. doi: 10.1111/tbj.13040. [DOI] [PubMed] [Google Scholar]
- 3.Menes TS, Rosenberg R, Balch S, Jaffer S, Kerlikowske K, Miglioretti DL. Upgrade of high-risk breast lesions detected on mammography in the Breast Cancer Surveillance Consortium. Am J Surg. 2014;207(1):24–31. doi: 10.1016/j.amjsurg.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schnitt SJ. Benign breast disease and breast cancer risk: morphology and beyond. The American journal of surgical pathology. 2003;27(6):836–41. [DOI] [PubMed] [Google Scholar]
- 5.Dyrstad SW, Yan Y, Fowler AM, Colditz GA. Breast cancer risk associated with benign breast disease: systematic review and meta-analysis. Breast Cancer Res Treat. 2015;149(3):569–75. doi: 10.1007/s10549-014-3254-6. [DOI] [PubMed] [Google Scholar]
- 6.Hartmann LC, Degnim AC, Santen RJ, Dupont WD, Ghosh K. Atypical hyperplasia of the breast--risk assessment and management options. N Engl J Med. 2015;372(1):78–89. doi: 10.1056/NEJMsr1407164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tozbikian G, Brogi E, Vallejo CE, Giri D, Murray M, Catalano J et al. Atypical Ductal Hyperplasia Bordering on Ductal Carcinoma In Situ. International journal of surgical pathology. 2017;25(2):100–7. doi: 10.1177/1066896916662154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim J, Han W, Go EY, Moon HG, Ahn SK, Shin HC et al. Validation of a scoring system for predicting malignancy in patients diagnosed with atypical ductal hyperplasia using an ultrasound-guided core needle biopsy. J Breast Cancer. 2012;15(4):407–11. doi: 10.4048/jbc.2012.15.4.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagoner MJ, Laronga C, Acs G. Extent and histologic pattern of atypical ductal hyperplasia present on core needle biopsy specimens of the breast can predict ductal carcinoma in situ in subsequent excision. Am J Clin Pathol. 2009;131(1):112–21. doi: 10.1309/AJCPGHEJ2R8UYFGP. [DOI] [PubMed] [Google Scholar]
- 10.Nizri E, Schneebaum S, Klausner JM, Menes TS. Current management practice of breast borderline lesions--need for further research and guidelines. Am J Surg. 2012;203(6):721–5. doi: 10.1016/j.amjsurg.2011.06.052. [DOI] [PubMed] [Google Scholar]
- 11.Georgian-Smith D, Lawton TJ. Variations in physician recommendations for surgery after diagnosis of a high-risk lesion on breast core needle biopsy. AJR American journal of roentgenology. 2012;198(2):256–63. doi: 10.2214/AJR.11.7717. [DOI] [PubMed] [Google Scholar]
- 12.Ahn S, Cho J, Sung J, Lee JE, Nam SJ, Kim KM et al. The prognostic significance of tumor-associated stroma in invasive breast carcinoma. Tumour Biol. 2012;33(5):1573–80. doi: 10.1007/s13277-012-0411-6. [DOI] [PubMed] [Google Scholar]
- 13.Cid S, Eiro N, Fernandez B, Sanchez R, Andicoechea A, Fernandez-Muniz PI et al. Prognostic Influence of Tumor Stroma on Breast Cancer Subtypes. Clin Breast Cancer. 2018;18(1):e123–e33. doi: 10.1016/j.clbc.2017.08.008. [DOI] [PubMed] [Google Scholar]
- 14.Bussard KM, Mutkus L, Stumpf K, Gomez-Manzano C, Marini FC. Tumor-associated stromal cells as key contributors to the tumor microenvironment. Breast Cancer Res. 2016;18(1):84. doi: 10.1186/s13058-016-0740-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komohara Y, Takeya M. CAFs and TAMs: maestros of the tumour microenvironment. The Journal of pathology. 2017;241(3):313–5. doi: 10.1002/path.4824. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Alexander PB, Wang XF. TGF-beta Family Signaling in the Control of Cell Proliferation and Survival. Cold Spring Harb Perspect Biol. 2017;9(4). doi: 10.1101/cshperspect.a022145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kahata K, Dadras MS, Moustakas A. TGF-beta Family Signaling in Epithelial Differentiation and Epithelial-Mesenchymal Transition. Cold Spring Harb Perspect Biol. 2017. doi: 10.1101/cshperspect.a022194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhowmick N, Chytiil A, Plieth D, Gorska A, Dumont N, Shappel S et al. TGF-ß signaling in fibroblasts modulates the oncogenic potential of adjacent epithelia. Science. 2004;303:847–51. [DOI] [PubMed] [Google Scholar]
- 19.Cheng N, Bhowmick NA, Chytil A, Gorksa AE, Brown KA, Muraoka R et al. Loss of TGF-beta type II receptor in fibroblasts promotes mammary carcinoma growth and invasion through upregulation of TGF-alpha-, MSP- and HGF-mediated signaling networks. Oncogene. 2005;24(32):5053–68. doi: 10.1038/sj.onc.1208685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hembruff SL, Jokar I, Yang L, Cheng N. Loss of transforming growth factor-beta signaling in mammary fibroblasts enhances CCL2 secretion to promote mammary tumor progression through macrophage-dependent and -independent mechanisms. Neoplasia. 2010;12(5):425–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang WB, Mafuvadze B, Yao M, Zou A, Portsche M, Cheng N. TGF-beta Negatively Regulates CXCL1 Chemokine Expression in Mammary Fibroblasts through Enhancement of Smad2/3 and Suppression of HGF/c-Met Signaling Mechanisms. PLoS One. 2015;10(8):e0135063. doi: 10.1371/journal.pone.0135063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rees PA, Greaves NS, Baguneid M, Bayat A. Chemokines in Wound Healing and as Potential Therapeutic Targets for Reducing Cutaneous Scarring. Advances in wound care. 2015;4(11):687–703. doi: 10.1089/wound.2014.0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ridiandries A, Tan JT, Bursill CA. The Role of CC-Chemokines in the Regulation of Angiogenesis. International journal of molecular sciences. 2016;17(11). doi: 10.3390/ijms17111856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strieter RM, Belperio JA, Burdick MD, Keane MP. CXC chemokines in angiogenesis relevant to chronic fibroproliferation. Current drug targets Inflammation and allergy. 2005;4(1):23–6. [DOI] [PubMed] [Google Scholar]
- 25.Stillie R, Farooq SM, Gordon JR, Stadnyk AW. The functional significance behind expressing two IL-8 receptor types on PMN. J Leukoc Biol. 2009;86(3):529–43. doi: 10.1189/jlb.0208125. [DOI] [PubMed] [Google Scholar]
- 26.Balkwill FR. The chemokine system and cancer. The Journal of pathology. 2012;226(2):148–57. doi: 10.1002/path.3029. [DOI] [PubMed] [Google Scholar]
- 27.White GE, Iqbal AJ, Greaves DR. CC chemokine receptors and chronic inflammation--therapeutic opportunities and pharmacological challenges. Pharmacol Rev. 2013;65(1):47–89. doi: 10.1124/pr.111.005074. [DOI] [PubMed] [Google Scholar]
- 28.Greaves NS, Ashcroft KJ, Baguneid M, Bayat A. Current understanding of molecular and cellular mechanisms in fibroplasia and angiogenesis during acute wound healing. J Dermatol Sci. 2013;72(3):206–17. doi: 10.1016/j.jdermsci.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 29.Lowman HB, Slagle PH, DeForge LE, Wirth CM, Gillece-Castro BL, Bourell JH et al. Exchanging interleukin-8 and melanoma growth-stimulating activity receptor binding specificities. J Biol Chem. 1996;271(24):14344–52. [DOI] [PubMed] [Google Scholar]
- 30.Kurihara T, Bravo R. Cloning and functional expression of mCCR2, a murine receptor for the C-C chemokines JE and FIC. J Biol Chem. 1996;271(20):11603–7. [DOI] [PubMed] [Google Scholar]
- 31.Singh S, Varney M, Singh RK. Host CXCR2-dependent regulation of melanoma growth, angiogenesis, and experimental lung metastasis. Cancer Res. 2009;69(2):411–5. doi: 10.1158/0008-5472.CAN-08-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma B, Nannuru KC, Varney ML, Singh RK. Host Cxcr2-dependent regulation of mammary tumor growth and metastasis. Clin Exp Metastasis. 2015;32(1):65–72. doi: 10.1007/s10585-014-9691-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Acharyya S, Oskarsson T, Vanharanta S, Malladi S, Kim J, Morris PG et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell. 2012;150(1):165–78. doi: 10.1016/j.cell.2012.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dong YL, Kabir SM, Lee ES, Son DS. CXCR2-driven ovarian cancer progression involves upregulation of proinflammatory chemokines by potentiating NF-kappaB activation via EGFR-transactivated Akt signaling. PLoS One. 2013;8(12):e83789. doi: 10.1371/journal.pone.0083789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saintigny P, Massarelli E, Lin S, Ahn YH, Chen Y, Goswami S et al. CXCR2 expression in tumor cells is a poor prognostic factor and promotes invasion and metastasis in lung adenocarcinoma. Cancer Res. 2013;73(2):571–82. doi: 10.1158/0008-5472.CAN-12-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang B, Khachigian LM, Esau L, Birrer MJ, Zhao X, Parker MI et al. A key role for early growth response-1 and nuclear factor-kappaB in mediating and maintaining GRO/CXCR2 proliferative signaling in esophageal cancer. Mol Cancer Res. 2009;7(5):755–64. doi: 10.1158/1541-7786.MCR-08-0472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang B, Hendricks DT, Wamunyokoli F, Parker MI. A growth-related oncogene/CXC chemokine receptor 2 autocrine loop contributes to cellular proliferation in esophageal cancer. Cancer Res. 2006;66(6):3071–7. doi:66/6/3071 [pii] 10.1158/0008–5472.CAN-05–2871. [DOI] [PubMed] [Google Scholar]
- 38.Dhawan P, Richmond A. A novel NF-kappa B-inducing kinase-MAPK signaling pathway up-regulates NF-kappa B activity in melanoma cells. J Biol Chem. 2002;277(10):7920–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim SY, Yuzhalin AE, Gordon-Weeks AN, Muschel RJ. Targeting the CCL2-CCR2 signaling axis in cancer metastasis. Oncotarget. 2016;7(19):28697–710. doi: 10.18632/oncotarget.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borsig L, Wolf MJ, Roblek M, Lorentzen A, Heikenwalder M. Inflammatory chemokines and metastasis--tracing the accessory. Oncogene. 2014;33(25):3217–24. doi: 10.1038/onc.2013.272. [DOI] [PubMed] [Google Scholar]
- 41.Granot Z, Henke E, Comen EA, King TA, Norton L, Benezra R. Tumor entrained neutrophils inhibit seeding in the premetastatic lung. Cancer Cell. 2011;20(3):300–14. doi: 10.1016/j.ccr.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu Y, Chen Q, Corey E, Xie W, Fan J, Mizokami A et al. Activation of MCP-1/CCR2 axis promotes prostate cancer growth in bone. Clin Exp Metastasis. 2009;26(2):161–9. doi: 10.1007/s10585-008-9226-7. [DOI] [PubMed] [Google Scholar]
- 43.Fang WB, Jokar I, Zou A, Lambert D, Dendukuri P, Cheng N. CCL2/CCR2 chemokine signaling coordinates survival and motility of breast cancer cells through Smad3 protein- and p42/44 mitogen-activated protein kinase (MAPK)-dependent mechanisms. J Biol Chem. 2012;287(43):36593–608. doi: 10.1074/jbc.M112.365999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zou A, Lambert D, Yeh H, Yasukawa K, Behbod F, Fan F et al. Elevated CXCL1 expression in breast cancer stroma predicts poor prognosis and is inversely associated with expression of TGF-beta signaling proteins. BMC Cancer. 2014;14:781. doi: 10.1186/1471-2407-14-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yao M, Yu E, Staggs V, Fan F, Cheng N. Elevated expression of chemokine C-C ligand 2 in stroma is associated with recurrent basal-like breast cancers. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2016:810–23. doi: 10.1038/modpathol.2016.78. [DOI] [PubMed] [Google Scholar]
- 46.Bierie B, Chung CH, Parker JS, Stover DG, Cheng N, Chytil A et al. Abrogation of TGF-beta signaling enhances chemokine production and correlates with prognosis in human breast cancer. J Clin Invest. 2009;119(6):1571–82. doi: 10.1172/JCI37480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bugge TH, Lund LR, Kombrinck KK, Nielsen BS, Holmback K, Drew AF et al. Reduced metastasis of Polyoma virus middle T antigen-induced mammary cancer in plasminogen-deficient mice. Oncogene. 1998;16(24):3097–104. doi: 10.1038/sj.onc.1201869. [DOI] [PubMed] [Google Scholar]
- 48.Langley RR, Ramirez KM, Tsan RZ, Van Arsdall M, Nilsson MB, Fidler IJ. Tissue-specific microvascular endothelial cell lines from H-2K(b)-tsA58 mice for studies of angiogenesis and metastasis. Cancer Res. 2003;63(11):2971–6. [PubMed] [Google Scholar]
- 49.Lambert D, Cheng N. Mammary transplantation of stromal cells and carcinoma cells in C57BL/6 mice. Journal of Visualized Experiments. 2011. (54). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salomonis N, Schlieve CR, Pereira L, Wahlquist C, Colas A, Zambon AC et al. Alternative splicing regulates mouse embryonic stem cell pluripotency and differentiation. Proc Natl Acad Sci U S A. 2010;107(23):10514–9. doi: 10.1073/pnas.0912260107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Borowicz S, Van Scoyk M, Avasarala S, Karuppusamy Rathinam MK, Tauler J, Bikkavilli RK et al. The soft agar colony formation assay. Journal of visualized experiments : JoVE. 2014(92):e51998. doi: 10.3791/51998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paddison PJ, Cleary M, Silva JM, Chang K, Sheth N, Sachidanandam R et al. Cloning of short hairpin RNAs for gene knockdown in mammalian cells. Nat Methods. 2004;1(2):163–7. doi: 10.1038/nmeth1104-163. [DOI] [PubMed] [Google Scholar]
- 53.Fang WB, Yao M, Brummer G, Acevedo D, Alhakamy N, Berkland C et al. Targeted gene silencing of CCL2 inhibits triple negative breast cancer progression by blocking cancer stem cell renewal and M2 macrophage recruitment. Oncotarget. 2016. doi: 10.18632/oncotarget.9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheng N, Chytil A, Shyr Y, Joly A, Moses HL. Transforming growth factor-beta signaling-deficient fibroblasts enhance hepatocyte growth factor signaling in mammary carcinoma cells to promote scattering and invasion. Mol Cancer Res. 2008;6(10):1521–33. doi: 10.1158/1541-7786.MCR-07-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin E, Jones J, Zhu L, Whitney K, Muller W, Pollard J. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J of Path. 2003;163(5):2113–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maglione JE, Moghanaki D, Young LJ, Manner CK, Ellies LG, Joseph SO et al. Transgenic Polyoma middle-T mice model premalignant mammary disease. Cancer Res. 2001;61(22):8298–305. [PubMed] [Google Scholar]
- 57.Guy C, Cardiff R, Muller W. Induction of mammary tumors by expression a polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12:954–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davie SA, Maglione JE, Manner CK, Young D, Cardiff RD, MacLeod CL et al. Effects of FVB/NJ and C57Bl/6J strain backgrounds on mammary tumor phenotype in inducible nitric oxide synthase deficient mice. Transgenic Res. 2007;16(2):193–201. doi: 10.1007/s11248-006-9056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qiu TH, Chandramouli GV, Hunter KW, Alkharouf NW, Green JE, Liu ET. Global expression profiling identifies signatures of tumor virulence in MMTV-PyMT-transgenic mice: correlation to human disease. Cancer Res. 2004;64(17):5973–81. doi: 10.1158/0008-5472.CAN-04-024264/17/5973 [pii]. [DOI] [PubMed] [Google Scholar]
- 60.Tse GM, Tan PH, Lui PC, Gilks CB, Poon CS, Ma TK et al. The role of immunohistochemistry for smooth-muscle actin, p63, CD10 and cytokeratin 14 in the differential diagnosis of papillary lesions of the breast. Journal of clinical pathology. 2007;60(3):315–20. doi: 10.1136/jcp.2006.036830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reisenbichler ES, Balmer NN, Adams AL, Pfeifer JD, Hameed O. Luminal cytokeratin expression profiles of breast papillomas and papillary carcinomas and the utility of a cytokeratin 5/p63/cytokeratin 8/18 antibody cocktail in their distinction. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2011;24(2):185–93. doi: 10.1038/modpathol.2010.197. [DOI] [PubMed] [Google Scholar]
- 62.Fouad TM, Kogawa T, Liu DD, Shen Y, Masuda H, El-Zein R et al. Overall survival differences between patients with inflammatory and noninflammatory breast cancer presenting with distant metastasis at diagnosis. Breast Cancer Res Treat. 2015;152(2):407–16. doi: 10.1007/s10549-015-3436-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weidner N Current pathologic methods for measuring intratumoral microvessel density within breast carcinoma and other solid tumors. Breast Cancer Res Treat. 1995;36(2):169–80. [DOI] [PubMed] [Google Scholar]
- 64.Sacks GP, Clover LM, Bainbridge DR, Redman CW, Sargent IL. Flow cytometric measurement of intracellular Th1 and Th2 cytokine production by human villous and extravillous cytotrophoblast. Placenta. 2001;22(6):550–9. doi: 10.1053/plac.2001.0686. [DOI] [PubMed] [Google Scholar]
- 65.Mori M, Sadahira Y, Kawasaki S, Hayashi T, Awai M. Macrophage heterogeneity in bone marrow culture in vitro. J Cell Sci. 1990;95 (Pt 3):481–5. [DOI] [PubMed] [Google Scholar]
- 66.Cooper KD, Duraiswamy N, Hammerberg C, Allen E, Kimbrough-Green C, Dillon W et al. Neutrophils, differentiated macrophages, and monocyte/macrophage antigen presenting cells infiltrate murine epidermis after UV injury. J Invest Dermatol. 1993;101(2):155–63. [DOI] [PubMed] [Google Scholar]
- 67.Sharon Y, Alon L, Glanz S, Servais C, Erez N. Isolation of normal and cancer-associated fibroblasts from fresh tissues by Fluorescence Activated Cell Sorting (FACS). Journal of visualized experiments : JoVE. 2013(71):e4425. doi: 10.3791/4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sugimoto H, Mundel TM, Kieran MW, Kalluri R. Identification of fibroblast heterogeneity in the tumor microenvironment. Cancer Biol Ther. 2006;5(12):1640–6. [DOI] [PubMed] [Google Scholar]
- 69.Matsuyoshi N, Imamura S. Multiple cadherins are expressed in human fibroblasts. Biochem Biophys Res Commun. 1997;235(2):355–8. doi: 10.1006/bbrc.1997.6707. [DOI] [PubMed] [Google Scholar]
- 70.Bhowmick NA, Ghiassi M, Bakin A, Aakre M, Lundquist CA, Engel ME et al. Transforming growth factor-beta1 mediates epithelial to mesenchymal transdifferentiation through a RhoA-dependent mechanism. Mol Biol Cell. 2001;12(1):27–36. doi: 10.1091/mbc.12.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li Q, Zhang D, Wang Y, Sun P, Hou X, Larner J et al. MiR-21/Smad 7 signaling determines TGF-beta1-induced CAF formation. Sci Rep. 2013;3:2038. doi: 10.1038/srep02038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lenicek T, Szerda F, Demirovic A, Mijic A, Kruslin B, Tomas D. Pleomorphic ductal carcinoma of the breast with predominant micropapillary features. Pathol Int. 2007;57(10):694–7. doi: 10.1111/j.1440-1827.2007.02159.x. [DOI] [PubMed] [Google Scholar]
- 73.Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992;52(6):1399–405. [PubMed] [Google Scholar]
- 74.Owens RB. Glandular epithelial cells from mice: a method for selective cultivation. J Natl Cancer Inst. 1974;52(4):1375–8. [DOI] [PubMed] [Google Scholar]
- 75.Bandyopadhyay A, Cibull ML, Sun LZ. Isolation and characterization of a spontaneously transformed malignant mouse mammary epithelial cell line in culture. Carcinogenesis. 1998;19(11):1907–11. [DOI] [PubMed] [Google Scholar]
- 76.Lazard D, Sastre X, Frid MG, Glukhova MA, Thiery JP, Koteliansky VE. Expression of smooth muscle-specific proteins in myoepithelium and stromal myofibroblasts of normal and malignant human breast tissue. Proc Natl Acad Sci U S A. 1993;90(3):999–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Duncia JV, Santella JB 3rd, Higley CA, Pitts WJ, Wityak J, Frietze WE et al. MEK inhibitors: the chemistry and biological activity of U0126, its analogs, and cyclization products. Bioorg Med Chem Lett. 1998;8(20):2839–44. [DOI] [PubMed] [Google Scholar]
- 78.Pierce JW, Schoenleber R, Jesmok G, Best J, Moore SA, Collins T et al. Novel inhibitors of cytokine-induced IkappaBalpha phosphorylation and endothelial cell adhesion molecule expression show anti-inflammatory effects in vivo. J Biol Chem. 1997;272(34):21096–103. [DOI] [PubMed] [Google Scholar]
- 79.Jeon M, Han J, Nam SJ, Lee JE, Kim S. Elevated IL-1beta expression induces invasiveness of triple negative breast cancer cells and is suppressed by zerumbone. Chem Biol Interact. 2016;258:126–33. doi: 10.1016/j.cbi.2016.08.021. [DOI] [PubMed] [Google Scholar]
- 80.Sangai T, Akcakanat A, Chen H, Tarco E, Wu Y, Do KA et al. Biomarkers of response to Akt inhibitor MK-2206 in breast cancer. Clin Cancer Res. 2012;18(20):5816–28. doi: 10.1158/1078-0432.CCR-12-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Turkson J, Ryan D, Kim JS, Zhang Y, Chen Z, Haura E et al. Phosphotyrosyl peptides block Stat3-mediated DNA binding activity, gene regulation, and cell transformation. J Biol Chem. 2001;276(48):45443–55. doi: 10.1074/jbc.M107527200. [DOI] [PubMed] [Google Scholar]
- 82.Zhao W, Jaganathan S, Turkson J. A cell-permeable Stat3 SH2 domain mimetic inhibits Stat3 activation and induces antitumor cell effects in vitro. J Biol Chem. 2010;285(46):35855–65. doi: 10.1074/jbc.M110.154088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bortner DM, Rosenberg MP. Induction of mammary gland hyperplasia and carcinomas in transgenic mice expressing human cyclin E. Mol Cell Biol. 1997;17(1):453–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Andrechek ER, Cardiff RD, Chang JT, Gatza ML, Acharya CR, Potti A et al. Genetic heterogeneity of Myc-induced mammary tumors reflecting diverse phenotypes including metastatic potential. Proc Natl Acad Sci U S A. 2009;106(38):16387–92. doi: 10.1073/pnas.0901250106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rosner A, Miyoshi K, Landesman-Bollag E, Xu X, Seldin DC, Moser AR et al. Pathway pathology: histological differences between ErbB/Ras and Wnt pathway transgenic mammary tumors. Am J Pathol. 2002;161(3):1087–97. doi: 10.1016/S0002-9440(10)64269-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nassar H, Qureshi H, Adsay NV, Visscher D. Clinicopathologic analysis of solid papillary carcinoma of the breast and associated invasive carcinomas. The American journal of surgical pathology. 2006;30(4):501–7. [DOI] [PubMed] [Google Scholar]
- 87.Collins LC, Schnitt SJ. Papillary lesions of the breast: selected diagnostic and management issues. Histopathology. 2008;52(1):20–9. doi: 10.1111/j.1365-2559.2007.02898.x. [DOI] [PubMed] [Google Scholar]
- 88.Tan PH, Schnitt SJ, van de Vijver MJ, Ellis IO, Lakhani SR. Papillary and neuroendocrine breast lesions: the WHO stance. Histopathology. 2015;66(6):761–70. doi: 10.1111/his.12463. [DOI] [PubMed] [Google Scholar]
- 89.Yamashita M, Ogawa T, Zhang X, Hanamura N, Kashikura Y, Takamura M et al. Role of stromal myofibroblasts in invasive breast cancer: stromal expression of alpha-smooth muscle actin correlates with worse clinical outcome. Breast Cancer. 2012;19(2):170–6. doi: 10.1007/s12282-010-0234-5. [DOI] [PubMed] [Google Scholar]
- 90.Cardone A, Tolino A, Zarcone R, Borruto Caracciolo G, Tartaglia E. Prognostic value of desmoplastic reaction and lymphocytic infiltration in the management of breast cancer. Panminerva medica. 1997;39(3):174–7. [PubMed] [Google Scholar]
- 91.Webster M, Hutchinson JN, Rauh MJ, Muthuswamy SK, Anton A, Tortorice CG, Cardiff RD, Graham FL, Hassell JA, Muller WJ Requirement of both Shc and Phosphatidyl 3’ kinase signaling pathways in polyomavirus middle T-mediated mammar tumorigenesis. Mol Cell Biol. 1998;18(4):2344–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schwab LP, Peacock DL, Majumdar D, Ingels JF, Jensen LC, Smith KD et al. Hypoxia-inducible factor 1alpha promotes primary tumor growth and tumor-initiating cell activity in breast cancer. Breast Cancer Res. 2012;14(1):R6. doi: 10.1186/bcr3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121(3):335–48. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 94.Gache C, Berthois Y, Martin PM, Saez S. Positive regulation of normal and tumoral mammary epithelial cell proliferation by fibroblasts in coculture. In Vitro Cell Dev Biol Anim. 1998;34(4):347–51. doi: 10.1007/s11626-998-0012-2. [DOI] [PubMed] [Google Scholar]
- 95.Studebaker AW, Storci G, Werbeck JL, Sansone P, Sasser AK, Tavolari S et al. Fibroblasts isolated from common sites of breast cancer metastasis enhance cancer cell growth rates and invasiveness in an interleukin-6-dependent manner. Cancer Res. 2008;68(21):9087–95. doi: 10.1158/0008-5472.CAN-08-0400. [DOI] [PubMed] [Google Scholar]
- 96.Horgan K, Jones DL, Mansel RE. Mitogenicity of human fibroblasts in vivo for human breast cancer cells. Br J Surg. 1987;74(3):227–9. [DOI] [PubMed] [Google Scholar]
- 97.Ray P, Stacer AC, Fenner J, Cavnar SP, Meguiar K, Brown M et al. CXCL12-gamma in primary tumors drives breast cancer metastasis. Oncogene. 2015;34(16):2043–51. doi: 10.1038/onc.2014.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gabrilovich DI. Myeloid-Derived Suppressor Cells. Cancer Immunol Res. 2017;5(1):3–8. doi: 10.1158/2326-6066.CIR-16-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jorch SK, Kubes P. An emerging role for neutrophil extracellular traps in noninfectious disease. Nat Med. 2017;23(3):279–87. doi: 10.1038/nm.4294. [DOI] [PubMed] [Google Scholar]
- 100.Tabaries S, Ouellet V, Hsu BE, Annis MG, Rose AA, Meunier L et al. Granulocytic immune infiltrates are essential for the efficient formation of breast cancer liver metastases. Breast Cancer Res. 2015;17:45. doi: 10.1186/s13058-015-0558-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Matsui A, Yokoo H, Negishi Y, Endo-Takahashi Y, Chun NA, Kadouchi I et al. CXCL17 expression by tumor cells recruits CD11b+Gr1 high F4/80-cells and promotes tumor progression. PLoS One. 2012;7(8):e44080. doi: 10.1371/journal.pone.0044080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Morales JK, Kmieciak M, Knutson KL, Bear HD, Manjili MH. GM-CSF is one of the main breast tumor-derived soluble factors involved in the differentiation of CD11b-Gr1-bone marrow progenitor cells into myeloid-derived suppressor cells. Breast Cancer Res Treat. 2010;123(1):39–49. doi: 10.1007/s10549-009-0622-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lin D, Lei L, Zhang Y, Hu B, Bao G, Liu Y et al. Secreted IL-1alpha promotes T-cell activation and expansion of CD11b(+) Gr1(+) cells in carbon tetrachloride-induced liver injury in mice. European journal of immunology. 2015;45(7):2084–98. doi: 10.1002/eji.201445195. [DOI] [PubMed] [Google Scholar]
- 104.Sharma M, Beck AH, Webster JA, Espinosa I, Montgomery K, Varma S et al. Analysis of stromal signatures in the tumor microenvironment of ductal carcinoma in situ. Breast Cancer Res Treat. 2010;123(2):397–404. doi: 10.1007/s10549-009-0654-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sobolik T, Su YJ, Wells S, Ayers GD, Cook RS, Richmond A. CXCR4 drives the metastatic phenotype in breast cancer through induction of CXCR2 and activation of MEK and PI3K pathways. Mol Biol Cell. 2014;25(5):566–82. doi: 10.1091/mbc.E13-07-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Singh JK, Farnie G, Bundred NJ, Simoes BM, Shergill A, Landberg G et al. Targeting CXCR1/2 significantly reduces breast cancer stem cell activity and increases the efficacy of inhibiting HER2 via HER2-dependent and -independent mechanisms. Clin Cancer Res. 2013;19(3):643–56. doi: 10.1158/1078-0432.CCR-12-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dhawan P, Richmond A. Role of CXCL1 in tumorigenesis of melanoma. J Leukoc Biol. 2002;72(1):9–18. [PMC free article] [PubMed] [Google Scholar]
- 108.Wang Z, Wang Z, Li G, Wu H, Sun K, Chen J et al. CXCL1 from tumor-associated lymphatic endothelial cells drives gastric cancer cell into lymphatic system via activating integrin beta1/FAK/AKT signaling. Cancer Lett. 2017;385:28–38. doi: 10.1016/j.canlet.2016.10.043. [DOI] [PubMed] [Google Scholar]
- 109.Kuo PL, Shen KH, Hung SH, Hsu YL. CXCL1/GROalpha increases cell migration and invasion of prostate cancer by decreasing fibulin-1 expression through NF-kappaB/HDAC1 epigenetic regulation. Carcinogenesis. 2012;33(12):2477–87. doi: 10.1093/carcin/bgs299. [DOI] [PubMed] [Google Scholar]
- 110.Kemp DM, Pidich A, Larijani M, Jonas R, Lash E, Sato T et al. Ladarixin, a dual CXCR1/2 inhibitor, attenuates experimental melanomas harboring different molecular defects by affecting malignant cells and tumor microenvironment. Oncotarget. 2017;8(9):14428–42. doi: 10.18632/oncotarget.14803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chang Q, Bournazou E, Sansone P, Berishaj M, Gao SP, Daly L et al. The IL-6/JAK/Stat3 feed-forward loop drives tumorigenesis and metastasis. Neoplasia. 2013;15(7):848–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Song L, Turkson J, Karras JG, Jove R, Haura EB. Activation of Stat3 by receptor tyrosine kinases and cytokines regulates survival in human non-small cell carcinoma cells. Oncogene. 2003;22(27):4150–65. doi: 10.1038/sj.onc.1206479. [DOI] [PubMed] [Google Scholar]
- 113.Cheng N, Chytil A, Shyr Y, Joly A, Moses HL. Enhanced hepatocyte growth factor signaling by type II transforming growth factor-beta receptor knockout fibroblasts promotes mammary tumorigenesis. Cancer Res. 2007;67(10):4869–77. doi: 10.1158/0008-5472.CAN-06-3381. [DOI] [PubMed] [Google Scholar]
- 114.Elliott BE, Hung WL, Boag AH, Tuck AB. The role of hepatocyte growth factor (scatter factor) in epithelial-mesenchymal transition and breast cancer. Can J Physiol Pharmacol. 2002;80(2):91–102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.