Abstract
Cancer stem cells (CSCs) are rare types of cells responsible for tumor development, relapse, and metastasis. However, current research in CSC biology is largely limited by the difficulty of obtaining sufficient CSCs. Single-cell analysis techniques are promising tools for CSC-related studies. Here, we used the Single-probe mass spectrometry (MS) technique to investigate the metabolic features of live colorectal CSCs at the single-cell level. Experimental data were analyzed using statistical analysis methods, including the t-test and partial least squares discriminant analysis. Our results indicate that the overall metabolic profiles of CSCs are distinct from non-stem cancer cells (NSCCs). Specifically, we demonstrated that tricarboxylic acid (TCA) cycle metabolites are more abundant in CSCs compared to NSCCs, indicating their major energy production pathways are different. Moreover, CSCs have relatively higher levels of unsaturated lipids. Inhibiting the activities of stearoyl-CoA desaturase-1 (SCD1), nuclear factor κB (NF-κB), and aldehyde dehydrogenases (ALDH1A1) in CSCs significantly reduced the abundances of unsaturated lipids and hindered the formation of spheroids, resulting in reduced stemness of CSCs. Our techniques and experimental protocols can be potentially used for metabolomic studies of other CSCs and rare types of cells and provide a new approach to discovering functional biomarkers as therapeutic targets.
Graphical Abstract
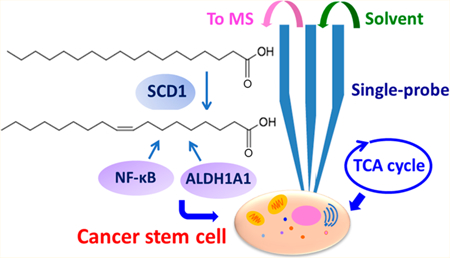
Cancer is one of the most common causes of death, and it threats the health of millions of people.1 Cancer therapy suffers from drug resistance, which leads to tumor recurrence and metastasis.2 Cancer stem cells (CSCs) are a rare subset of cancer cells possessing the ability to self-renew and initiate tumors.3 Recent studies indicate that CSCs are the main source of therapy resistance, and they are responsible for tumor recurrence and metastasis.4 In particular, the heterogeneity of CSCs forcefully impedes anticancer therapies.5 Unfortunately, the current understanding of the characteristics of CSCs and their roles in drug resistance is largely lacking, hindering the development of novel tumor diagnostic and therapeutic strategies.
One of the biggest challenges in CSC studies is to obtain sufficient cells, especially from patients, for analysis.6 However, traditional approaches for studying cell metabolomics, such as HPLC/MS, typically require a large number of cells for sample preparation.7 Therefore, techniques performing meaningful biological research from individual cells would provide a great advantage for the study of CSCs.8 A number of single-cell analysis techniques, such as single-cell sorting and single-cell sequencing, have been developed to effectively utilize the limited resources of CSCs.9,10 As the final downstream products, metabolites reflect gene regulation, pathway interactions, and environmental perturbations, and they directly indicate cell status. Therefore, the single cell metabolomics study, particularly based on live single cells, can truly provide chemical information on individual cells to effectively investigate their biological phenotypes.11,12 Due to its high detection sensitivity and broad range of molecular analysis, mass spectrometry (MS) has become a predominant technique for metabolomics research. However, the single cell MS (SCMS) analysis of single CSCs has been rarely performed. To the best of our knowledge, only one MS-based metabolomics study of single CSCs has been reported,13 in which the time-of-flight secondary ion mass spectrometry (TOF-SIMS) has been utilized. Due to complex background at low mass range and high degree of ion fragmentation, only a few molecules were identified. In addition, because cell metabolites can rapidly change upon the alteration of a cell living environment,14,15 any method requiring a vacuum environment (e.g., for sampling and ionization) and nontrivial sample preparation, such as SIMS and MALDI (matrix assisted laser desorption/ionization), precludes the capability of obtaining molecular information from live cells. Thus, metabolomics of live single CSCs is largely unexplored.16 Current methods used to detect the CSCs are mainly based on a number of common cell surface biomarkers, such as CD133, CD24, and CD44, and the activity of aldehyde dehydrogenase (ALDH1).17 Because CSCs are highly heterogeneous, using novel techniques to discover new candidates for CSC markers will provide additional approaches to identify target CSCs.18 Moreover, many CSCs markers are selected without knowing their functional roles, leading to limited reliability as biomarkers.19 Therefore, characteristic molecules identified using SCMS techniques can be potentially used as novel function-based biomarkers of CSCs.
Colorectal cancer (CRC) is the fourth leading cause of cancer-related death, and CSCs play a critical role in tumor relapse and metastasis;20 however, the metabolomic characteristics of colon CSCs in their living status are largely unknown. In this study, we used live CSCs derived from the HCT-116 (colorectal cancer) cells as the model system and conducted metabolomic analysis at the single-cell level. In the comparison study, we utilized the regular HCT-116 as the model of nonstem cancer cells (NSCCs) to illustrate the metabolic differences between CSCs and NSCCs. All SCMS experiments were conducted using the Single-probe MS techniques (Figure 1A,B), which have been previously applied in live single-cell analysis,21–24 mass spectrometry imaging of tissues,25 and metabolomic analysis of extracellular molecules in live multicellular spheroids.26
Figure 1.
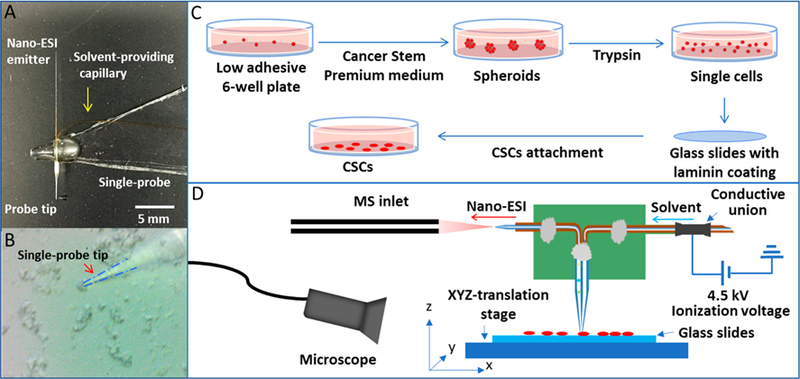
Single-probe SCMS experimental setup for the analysis of individual CSCs and NSCCs. (A) Photograph of a Single-probe device; (B) zoomed-in photograph of the Single-probe tip probing a single cancer cell; (C) workflow of CSC sample preparation; (D) schematic diagram of the Single-probe SCMS setup for the analysis of live single cells.
EXPERIMENTAL SECTION
Cell Lines and Cell Culture.
Enriched human colon cancer HCT-116 CSCs were purchased from ProMab Biotechnology (Richmond, CA, USA), and HCT-116 cancer cells (NSCCs) were originally obtained from American Type Culture Collection (ATCC; Rockville, MD, USA). The HCT-116 CSCs were cultured in Cancer Stem Premium medium (Probmab Biotechnology) to maintain their stemness (Figure 1C),27 and the passage number of cells was limited within three to maintain the population of CSCs during cell culture. The HCT-116 NSCCs were cultured in McCoy’s 5A cell culture medium containing 10% FBS (fetal bovine serum) and 1% Pen Strep (Life Technologies, Grand Island, NY, USA). All cells were cultured at 37 °C in an incubator with 5% CO2 supply (HeraCell, Heraeus, Germany).
Reagents for the Treatment of CSCs.
CAY10566 (SCD1 inhibitor, ≥98%), (dimethylamino)parthenolide (DMAPT; NF-κB inhibitor, ≥99%), CM037 (ALD1H1 inhibitor, ≥98%), and retinoic acid (≥98%) were purchased from Cayman Chemical Co. (Ann Arbor, MI, USA). The stock solution of these compounds were prepared in dimethyl sulfoxide (DMSO; Sigma-Aldrich, St. Louis, MO, USA) and further diluted into McCoy’s 5A cell culture medium to prepare inhibitor solutions. Based on previous studies, CSCs were treated by CAY10566 (1 μM), DMAPT (5 μM), CM037 (4.6 μM), or retinoic acid (1 μM) for 7 days prior to SCMS measurement.28,29
CSC Sorting.
To purify CSCs for our SCMS experiments, we utilized florescence-activated cell sorting (FACS; BD FACS Jazz flow cytometer, BD Biosciences, San Jose, CA, USA) under sterile conditions for cell isolation. Briefly, CSC spheroids formed during culture were dissociated using TrypLE reagents (Life Technologies, Grand Island, NY, USA) and then resuspended in the solution containing 0.5% bovine serum albumin (BSA; Sigma-Aldrich) and antibodies on ice. According to previous studies,18 CD133+/CD24+ population is recognized as CSCs, whereas CD133−/CD24− cells (HCT-116 cells) are regarded as NSCCs. In our experiments, antibodies of CD133 conjugated with phycoerythrin (PE) and CD24 conjugated with allophycocyanin (APC; Biolegend, San Diego, CA, USA) were added into the cell suspension. CD133 and CD24 double-positive cells (CD133+/CD24+) were gated using control cells that were incubated with IgG1 isotype controls (APC− and PE-conjugated antibodies, Biolegend, San Diego, CA, USA).
Fabrication of the Single-Probe.
The fabrication details of the Single-probe have been described in previous studies.21 Briefly, there are three components in a single probe (Figure 1A): a needle pulled from dual-bore quartz tubing (outer diameter (o.d.), 500 μm; inner diameter (i.d.), 127 μm; Friedrich & Dimmock, Inc., Millville, NJ, USA) using a laser pipet puller (P-2000 micropipette puller, Sutter Instrument, Novato, CA, USA), a fused silica capillary (o.d., 105 μm; i.d., 40 μm; Polymicro Technologies, Phoenix, AZ, USA), and a nano-ESI emitter produced using the same type of fused silica capillary. A Single-probe is fabricated by embedding a laserpulled dual-bore quartz needle with a fused silica capillary and a nano-ESI emitter.
Single-Probe SCMS Setup.
CSCs and NSCCs were attached onto the laminin-coated glass slides through overnight culture (Figure 1C), and the slides were placed on a motorized XYZ-translation stage system (Figure 1D), which was controlled by a LabView software package.30 A syringe (250 μL; Hamilton Co., Reno, NV, USA) was used to continuously provide the sampling solvent (acetonitrile; Sigma-Aldrich), and a stable liquid junction was formed at the single-probe tip to extract cellular contents during the experiment. Using a microscope as a guide, a cell of interest was selected by controlling the stage system, and the selected cell was penetrated by lifting the Z-stage (Figure 1B). Cellular components were extracted by sampling solvent at the Single-probe tip, withdrawn toward the nano-ESI emitter through a self-aspiration process, and ionized for analysis using a Thermo LTQ Orbitrap XL mass spectrometer (Thermo Scientific, Waltham, MA, USA; Figure 1D). MS scans were acquired for individual cells, whereas MS2 experiments were conducted for ions of interest. MS analysis parameters are listed as follows: mass resolution, 60,000; ionization voltage, +4.5 kV (positive ion mode) or −4 kV (negative ion mode); 1 microscan; 100 ms max injection time; AGC (automatic gain control), on.
Data Analysis.
We performed the pretreatment of SCMS data prior to statistical analysis. We exported MS data (m/z values with their relative intensities) as tab-delimited data files using Thermo Xcalibur Qual Browser (Thermo Scientific, Waltham, MA, USA). Only relatively abundant peaks (intensity > 103) were exported, whereas background signals, such as peaks from solvent and cell culture medium, were subtracted from MS data.31 To minimize the influence induced by fluctuations of ion signals during experiments, we normalized ion intensities to the total ion current (TIC). We used the Geena 2 online software tool (http://bioinformatics.hsanmartino.it/geena2/) to preform peak alignment. The pretreated data were subjected to Levene’s test to assess the equality of variance of data, and Student’s t-test (for data with equal variance) or Welch’s t-test (for data with unequal variance) was then applied to obtain ions with significantly different abundance between two groups (p < 0.05). Metaboanalyst 4.0 (http://www.metaboanalyst.ca/) was used to conduct partial least squares discriminant analysis (PLS-DA) to illustrate the overall differences of metabolomics profiles between CSCs and NSCCs. Finally we use online database METLIN (https://metlin.scripps.edu/) and human metabolome database (HMDB; http://www.hmdb.ca) to tentatively label all ions based on their accurate m/z values, whereas more confident identification of ions of interest was proposed based on tandem MS measurements. It is worth noting that, due to the lack of separation of cellular contents in the real-time SCMS measurement, we cannot completely exclude the coexistence of isomers producing common fragment ions in tandem MS.
RESULTS AND DISCUSSION
Metabolic Profiles of CSCs and NSCCs Significantly Different.
We used florescence-activated cell sorting (FACS) to isolate CD133+/CD24+ population from HCT-116 CSCs18 prior to SCMS experiments and utilized the regular HCT-116 cells as NSCCs for comparison studies. For SCMS experiments, CSCs and NSCCs were attached onto the laminincoated glass sides through overnight culture (Figure 1C), and the slides were placed on a motorized XYZ-translation stage system (Figure 1D). Using a microscope as a guide, a cell of interest was selected by controlling the stage system. The selected cell was penetrated by the single probe (tip size < 10 μm) for sampling of cellular contents followed by real-time MS analysis (Figure 1A,B). To obtain a broader coverage of cellular metabolites, we measured 60 and 40 cells from each group in the positive and negative ion modes, respectively. Our experimental results indicate that the positive ion mode is suitable for MS detection of the majority of cellular metabolites (e.g., lipids), whereas the negative ion mode is desired for certain organic acids (e.g., TCA cycle metabolites and fatty acids). Mass spectrum patterns obtained from CSCs and NSCCs are distinct (Supporting Information Figures S1 and S2), and PLS-DA results confirmed that their overall metabolomic features are significantly different as shown in Figure 2 (p = 0.0015 from the permutation test).
Figure 2.
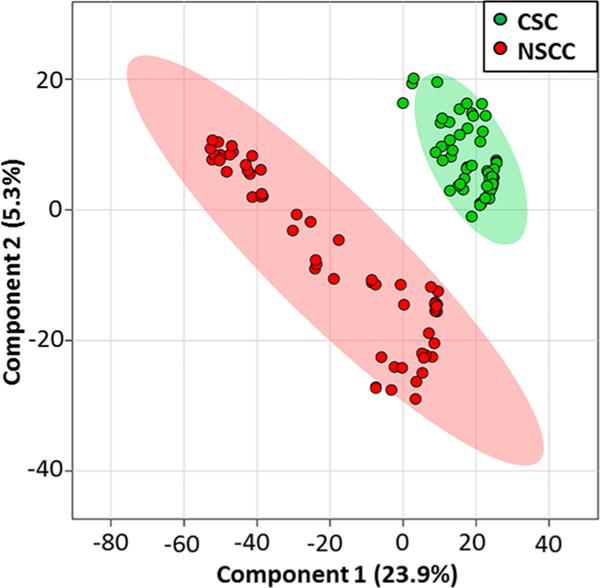
Results from partial least squares discriminant analysis (PLS-DA) of SCMS data illustrating the overall difference of metabolites between CSCs and NSCCs (positive ion mode, n = 60 in each group).
Higher Abundances of Metabolites of TCA Cycle in CSCs Than in NSCCs.
We further investigated the specific metabolites that are different between CSCs and NSCCs. TCA cycle, a step in the aerobic oxidative respiration, plays a critical role in the energetic metabolism in normal mammalian cells (Figure 3A). We found that multiple metabolites of the TCA cycle, including malic acid, citric acid, succinic acid, and pyruvate, have significantly higher abundances in CSCs than NSCCs (Figures 3B and S3). However, other relevant metabolites, including fumaric acid, α-ketoglutarate, and oxoglutaric acid, are present with comparable abundances (Figure 3B). All these metabolites of the TCA cycle were identified by MS2 experiments at the single-cell level.
Figure 3.
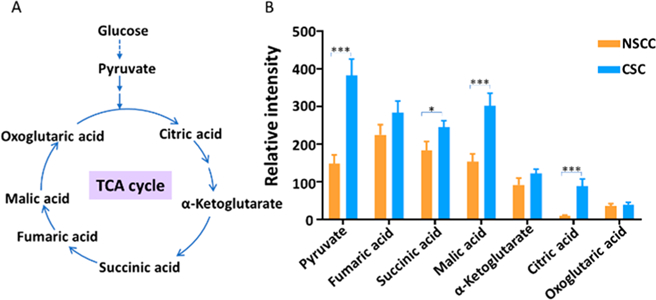
Comparison of abundances of metabolites in TCA cycle between CSCs and NSCCs (negative ion mode, n = 40 in each group). (A) TCA cycle pathway and the associated key metabolites; (B) bar graph showing metabolites present at significantly higher levels in CSCs than in NSCCs (labeled in the panel). All metabolites were identified from MS2 and MS2 experiments at single-cell level. (From t-test: *, p < 0.05; ***, p < 0.001.)
Our results demonstrate that the activities of the TCA cycle are different between colonic CSCs and NSCCs at the singlecell level. According to previous studies, cancer cells have a distinct energy metabolism pathway compared to normal cells.32 Aerobic glycolysis is the major energetic source for most cancer cells even under sufficient oxygen environment (known as “the Warburg effect”), while normal cells rely on the mitochondrial oxidative phosphorylation (OXPHOS) to provide most energy.33 The TCA cycle, a cellular process supplying the material to OXPHOS,34 is downregulated in cancer cells. Studies based on bulk analysis (LC-MS) found that intermediates of the TCA cycle are decreased in human ovarian cancer and cervical cancer cells,35 whereas ovarian and cervical squamous CSCs obtain energy through OXPHOS by activating the TCA cycle.36 In fact, CSCs can utilize different energy metabolism pathways based on their phenotypes,36,37 and they maintain the homeostasis by switching their energy metabolism pathways between OXPHOS and glycolysis.38 In the present study, we identified multiple intermediates of the TCA cycle (including pyruvate, citric acid, malic acid, and succinic acid), and demonstrated they are significantly upregulated in CSCs than NSCCs. Our results may suggest that the dominant energy metabolism pathways are different for HCT-116 CSCs and NSCCs under our experimental conditions (Figure 3A,B).
Higher Levels of Unsaturated Lipids in CSCs Than in NSCCs.
In addition to the detection of small metabolites such as species involved in the TCA cycle, we observed a large number of lipids in both CSCs and NSCCs. Our experimental results indicate the levels of 13 unsaturated lipids are significantly higher in CSCs compared to NSCCs (in positive ion mode, Figure 4A). All of these lipids were confirmed through MS2 analysis (Figure S4) from single cells. As the building blocks of lipids, fatty acids were also detected in both CSCs and NSCCs in the negative ion mode. The ratios of monounsaturated fatty acids (MUFAs) to saturated fatty acids (SFAs), e.g., palmitic acid/palmitoleic acid (C16:0/C16:1) and stearic acid/oleic acid (C18:0/C18:1), are significantly higher in CSCs compared to NSCCs (Figures 4B,C and S5).
Figure 4.
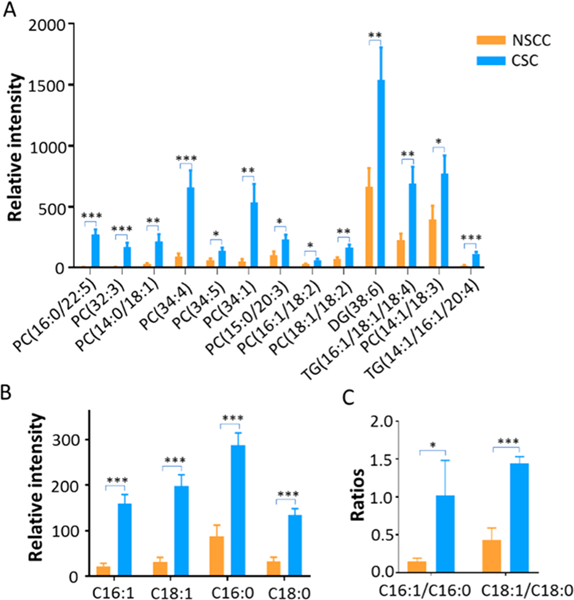
Saturation levels of lipids and fatty acids in CSCs and NSCCs. All species were identified from MS2 experiments at singlecell level. (A) CSCs contain relatively higher abundances of unsaturated lipids compared to NSCCs (positive ion mode, n = 60 in each group); (B) relative abundances of fatty acids (C16:0, C16:1, C18:0, and C18:1) detected in CSCs and NSCCs (negative ion mode, n = 40 in each group); (C) ratios of unsaturated fatty acid (UFA) to saturated fatty acid (SFA) in CSCs and NSCCs. (From t-test: *, p < 0.05; **, p < 0.01; ***, p < 0.001.)
We observed drastically different levels of unsaturated lipids and fatty acids between CSCs and NSCCs through SCMS analysis (Figure 4A,B). Relatively higher abundances of unsaturated lipids in CSCs are likely related to the formation of lipid droplets.39 Previous studies found that in CSCs de novo fatty acid synthesis pathway is upregulated, and the lipids storage compartment, lipid droplets (LDs), accumulates.40 An increasing amount of evidence indicates that lipid molecules and lipid droplets are essential for the stemness and tumorigenicity of CSCs.40 For example, a recent study reported a higher degree of LDs accumulation in different patient-derived colon CSCs than normal cancer cells using hyperspectral-stimulated Raman spectroscopy and LC/MS, and relatively more abundant LDs are positively correlated with the tumorigenic potential of CSCs.41
Interestingly, correlations between metabolites of TCA cycle and unsaturated lipids were also discovered in CSCs (Figure S6). We conducted correlation analysis of species measured from same cells, and found positive correlations (correlation coefficient > 0.6, p < 0.01) between pyruvate and each of these fatty acids: C16:0, C16:1, C18:0, and C18:1. Because pyruvate can be converted into acetyl-CoA, which is directly involved in the sythesis of fatty acids, by pyruvate dehydrogenase,42 higher levels of pyruvate likely lead to more production of fatty acids, including unsaturated fatty acids.
In addition to those species reported in previous LC-MS studies (i.e., palmitic (C16:0), palmitoleic (C16:1), stearic (C18:0), oleic (C18:1), linoleic (C18:2), arachidonic (C20:4), and docosahexaenoic acid (C22:6)), we discovered additional unsaturated lipids with drastically different abundances between CSCs and NSCCs. For example, phosphatidylcholines (PC(16:0/22:5), PC(32:3), PC(14:0/18:1), PC(34:4), PC(34:5), PC(34:1), PC(15:0/20:3), PC(16:1/18:2), PC-(18:1/18:2), PC(14:1/18:3)), diglyceride (DG(38:6)), and triglycerides (TG(16:1/18:1/18:4), TG(14:1/16:½0:4)) are present at significantly higher abundances in CSCs than NSCCs (Figures 4A,B and S7). Higher levels of unsaturated lipids and fatty acids are mainly produced by stearoyl-CoA desaturase-1 (SCD1) enzyme, and they are directly associated with the stemness of CSCs.28
Reduction of CSC Stemness Due to Inhibition of SCD1, NF-κB, and ALDH1A1.
Previous studies indicate that SCD1, nuclear factor κB (NF-κB), and aldehyde dehydrogenases (ALDH1A1) play critical roles in maintaining the abundances of unsaturated lipids in CSCs.28 We conducted SCMS experiments to investigate their functions regulating the saturation level of lipids and fatty acids, and the consequential influence on the stemness of CSCs. Overall, PLS-DA indicates that metabolomic profiles of CSCs were significantly altered (p < 0.05 from permutatoin test) by these inhibitors (Figure S8). Because inhibitors used in our experiments can induce cell apoptosis,43–45 it is unclear if changes of cell metabolites are related to cell apoptosis, although cells are still alive prior to the SCMS measurements.
Stearoyl-CoA desaturase-1 (SCD1) is one of the major lipid desaturases catalyzing the conversion of SFAs to MUFAs in mammalian cells, and the major products of SCD1 are palmitoleic (C16:1) and oleic acid (C18:1).46 A recent study shows that SCD1 is a key factor mediating the expression of desaturated lipids and fatty acids in ovarian cancer cells.28 To determine whether SCD1 plays a role in regulating the expression of unsaturation lipids and fatty acids in colorectal CSCs, we treated CSCs with CAY10566, a small molecule inhibitor of SCD1, to suppress the activity of SCD1.47 As shown in Figures 5 and 6A, the ratios of MUFAs to SFAs (C16:1/C16:0 and C18:1/C18:0) and the abundances of numerous unsaturated lipids are drastically decreased in CSCs treated with CAY10566, indicating the expression of unsaturated lipids and fatty acids can be regulated by SCD1. Interestingly, PC(16:0/22:5) exhibits an opposite trend: its abundance is increased after CAY10566 treatment. A reasonable explanation follows: inhibiting SCD1 can reduce the synthesis of C16:1 from C16:0, result in the accumulation of C16:0, and further increase the synthesis of lipids using C16:0, such as PC(16:0/22:5), through competative pathways. Moreover, we found that CSCs treated with CAY10566 possess significantly lower tendencies to form spheroids during cell culture (Figures S8A and S9B). Because forming spheroids in serum-free medium is an important feature of CSCs,48 our results suggest CAY10566 impacts the formation of speroids and reduces the stemness of CSCs.28 We conclude that levels of unsaturated lipids and fatty acids can be regulated by SCD1, and their abundances are important for the maintenance of the stemness of CSCs, which is consistent with the previous studies.28
Figure 5.
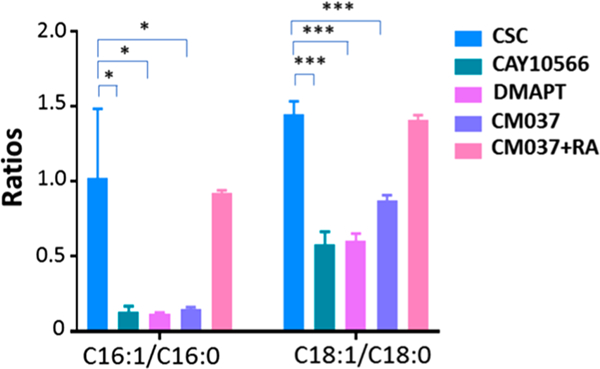
Ratios of monounsaturated fatty acid (MUFA) to its saturated fatty acid (SFA) in CSCs under different treatment conditions (negative ion mode, n = 40 in each group. (From t-test: *, p < 0.05; ***, p < 0.001.)
Figure 6.
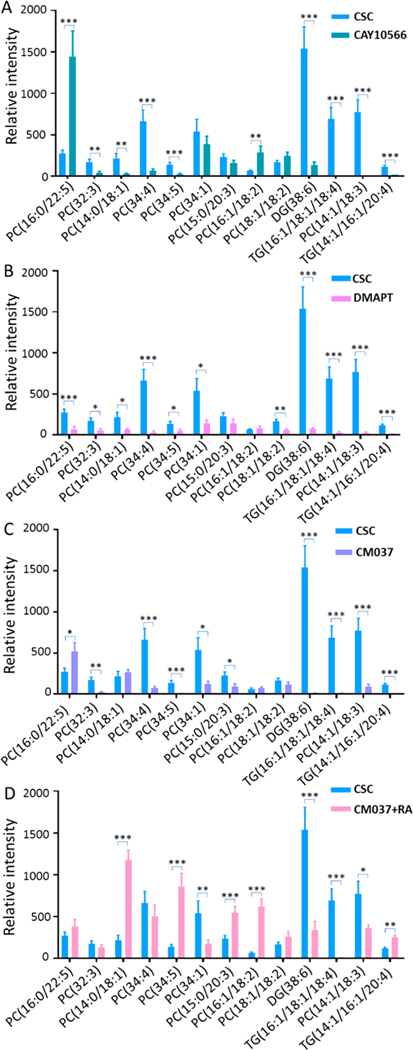
Investigation of roles of SCD1, NF-κB, and ALDH1A1 in regulating the saturation levels of lipids and fatty acids in CSCs (positive ion mode, n = 60 in each group). Expression of unsaturated lipids is decreased by inhibitors of (A) SCD1 (CAY10566), (B) NF-κB (DMART), and (C) ALDH1A1 (CM037). (D) Retinoic acid (RA) rescues the inhibition by CM037 and results in increased levels of unsaturated lipids. (From t-test: *, p < 0.05; **, p < 0.01; ***, p < 0.001.)
In addition to directly suppressing the activity of SCD-1, we investigated the influence of NF-κB and ALDH1A1, which are crucial for regulating SCD1 activities and maintaining the stemness of ovarian CSCs,28 on metabolites of CSCs. NF-κB is a protein complex that is essential for DNA transcription, cytokine production, and cell survival.49,50 NF-kB is extensively tied to cancer biology and tightly related to cancer stem cell.51,52 Particularly, NF-κB signaling is highly sensitive to the inhibition of desaturase, which is related to the stemness of CSCs.28 ALDH1A1 is one of the major enzymes producing retinoic acid (RA), which is also a well-known biomarker of colon CSCs.53 To investigate whether NF-κB and ALDH1A1 are associated with lipid saturation and the stemness of CSCs, we treated CSCs with inhibitors of NF-κB and ALDH1A1 (i.e., DMAPT and CM037,29,54 respectively). Experimental results show that these two inhibitors significantly suppressed the expression of unsaturated lipids and fatty acids (Figures 5, 6B,C), and drastically hindered the formation of CSC spheroids (Figures S9C,D), i.e., reduced stemness of CSCs. We conclude that the NF-κB and ALDH1A1 affect the growth of CSCs.
Because RA is an important product of ALDH1A1,55 inhibiting the activity of ALDH1A1 will apparently lead to a reduced production of RA. This consequence is likely related to the altered properties of CSCs such as the reduced stemness. It is reasonable to hypothesize that increasing RA supply during the cell culture will likely alleviate the inhibition by CM037. We then treated CM037-inhibited cells with RA and observed that the levels of unsaturated fatty acids and lipids were significantly restored in CM037-inhibited CSCs (Figures 5 and 6D). Taken together, we demonstrate SCD1, NF-κB, and ALDH1A1 are important regulating factors in CSCs, and they are responsible for the expression of higher levels of unsaturated lipids and fatty acids to maintain the stemness of CSCs.
Regulation of CSC Stemness by Activities of SCD1, NF-κB, and ALDH1A1.
As summarized in Figure 7, demonstrate that SCD1, NF-κB, and ALDH1A1 participate in the regulation of the saturation levels of lipids and fatty acids, and further affect the stemness of CSCs. First, we found SCD1 inhibitor (CAY10566) can reduce the levels of unsaturated lipids in CSCs and inhibit the formation of CSC spheroids (Figures 6A, S8A, and S9B), indicating that SCD1 activity is important for the stemness and tumorigenicity of CSCs. Second, we investigated the influence of inhibiting ALDH1A1, which is a known biomarker of CSCs.56 ALDH1A1 converts retinaldehyde to RA, which functions as a ligand of transcription factor, i.e., retinoic acid receptor (RAR). RA we regulates the activation of numerous nuclear transcription factors and plays a central role in regulating lipid metabolism.57 Treatment using ALDH1A1 inhibitor (CM037) significantly decreased levels of unsaturated lipids and suppressed spheroid forming of CSCs (Figures 6C, S8B, and S9D). Our results indicate that ALDH1A1 is a mediating factor of the stemness and tumorigenicity of CSCs, which is consistent with a previous study of ovarian CSCs.28 Last, we proved that the function of NF-κB is cruicial for the maintenance of the stemness of CSCs. NF-κB signaling pathway is responsible for maintaining the stemness of CSCs,58 and it is a crucial regulator of inflammation and immune responses as well as multiple cancer-associated processes such as proliferation, apoptosis, angiogenesis, and metastasis.59 The hyperactivation of NF-κB signaling in CSCs provides a proper niche for the survival and expansion of CSCs and contributes to their capabilities of invasion, metastasis, and self-renewal.51,58,60 In this study, we found that, similar to the treatment effects of using inhibitors of SCD1 and ALD1H1, inhibiting the activity of NF-κB using DMAPT changed the metabolomic profiles, as well as decreased the level of unsaturated lipid and the spheroid formation of colon CSCs (Figures 6B, S8C, and S9C), indicating this transcription factor may be important for maintaining the stemness of CSCs.
Figure 7.
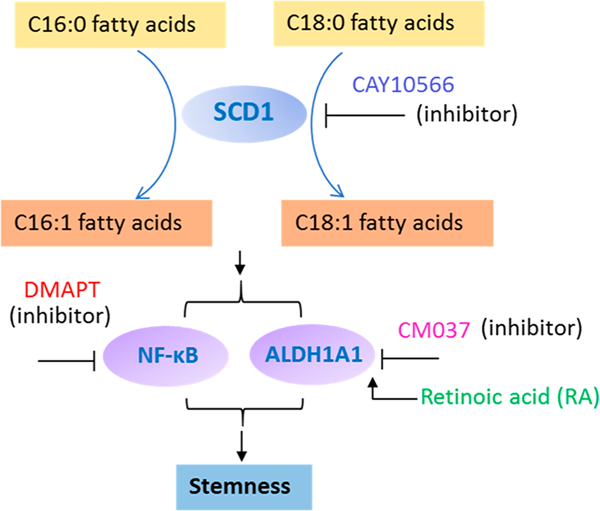
Potential mechanisms showing the regulation of unsaturated lipids and fatty acids by SCD1, NF-κB, and ALDH1A1, and their relationship to the stemness of CSCs.
CONCLUSIONS
In this study we successfully applied the SCMS technique to detecting a number of key metabolic features of colon CSCs, including higher levels of metabolites in the TCA cycle and unsaturated fatty acids and lipids. The high abundances of unsaturated lipids and fatty acids are the metabolic features of CSCs. SCD1 is a key factor mediating the expression of unsaturated fatty acids and lipids such as palmitoleic and oleic acid. Inhibiting the activity of SCD1 could significantly suppress the expression of unsaturated fatty acids and lipids. In addition, NF-κB and ALDH1A1 play critical roles in maintaining stemness of CSCs and also regulate the expression of unsaturated lipids. Together, we demonstrate that SCD1, NF-κB, and ALDH1A1 regulate the metabolism of unsaturated lipids and fatty acids, which are associated with the stemness of CSCs, at the single-cell level. Our current study indicates that the Single-probe SCMS technique can be applied to analyze live single CSCs, and our findings can promote the understanding of the biological characteristics of CSCs. Moreover, because metabolic features and regulatory factors detected here are deeply associated with the stemness and tumorigenicity of colon CSCs, they can be potentially used as the functional biomarkers and new therapeutic targets for colon CSCs.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Anthony Burgett (The University of Oklahoma) for providing the HCT-116 cells and suggestions for our studies. We thank the Laboratory for Molecular Biology and Cytometry Research at OUHSC (The University of Oklahoma Health Sciences Center) for the use of the Flow Cytometry and Imaging facility to isolate cancer stem cells. This research was supported by grants from National Institutes of Health (R01GM116116 and R21CA204706).
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.anal-chem.8b05166.
Relative abundances of fatty acids in CSCs and NSCCs, MS2 analysis of TCA cycle components, unsaturated lipids, and fatty acids, and images showing that inhibitors prevent the formation of CSC spheroids (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Torre LA; Bray F; Siegel RL; Ferlay J; Lortet-Tieulent J; Jemal A CA Ca-Cancer J. Clin 2015, 65, 87–108. [DOI] [PubMed] [Google Scholar]
- (2).Housman G; Byler S; Heerboth S; Lapinska K; Longacre M; Snyder N; Sarkar S Cancers 2014, 6, 1769–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Reya T; Morrison SJ; Clarke MF; Weissman IL Nature 2001, 414, 105. [DOI] [PubMed] [Google Scholar]
- (4).Lawson DA; Bhakta NR; Kessenbrock K; Prummel KD; Yu Y; Takai K; Zhou A; Eyob H; Balakrishnan S; Wang C-Y; Yaswen P; Goga A; Werb Z Nature 2015, 526, 131–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Tang DG Cell Res 2012, 22, 457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Bao B; Ahmad A; Azmi AS; Ali S; Sarkar FH Curr. Protoc. Pharmacol 2013, 61, 14.25.1–14.25. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Dunn WB; Erban A; Weber RJ; Creek DJ; Brown M; Breitling R; Hankemeier T; Goodacre R; Neumann S; Kopka J; Viant MR Metabolomics 2013, 9, 44–66. [Google Scholar]
- (8).Hu P; Zhang W; Xin H; Deng G Front. Cell Dev. Biol 2016, 4, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Gruber W; Scheidt T; Aberger F; Huber CG Cell Commun. Signaling 2017, 15, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Davey HM; Kell DB Microbiol. Rev 1996, 60, 641–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Vermeersch KA; Styczynski MP J. Carcinog 2013, 12, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).O’Connell TM Bioanalysis 2012, 4, 431–451. [DOI] [PubMed] [Google Scholar]
- (13).Waki M; Ide Y; Ishizaki I; Nagata Y; Masaki N; Sugiyama E; Kurabe N; Nicolaescu D; Yamazaki F; Hayasaka T; et al. Biochimie 2014, 107, 73–77. [DOI] [PubMed] [Google Scholar]
- (14).Turner BM Philos. Trans. R. Soc., B 2009, 364, 3403–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Villas-Bôas SG; Højer-Pedersen J; Åkesson M; Smedsgaard J; Nielsen J Yeast 2005, 22, 1155–1169. [DOI] [PubMed] [Google Scholar]
- (16).Peiris-Pages̀ M; Martinez-Outschoorn UE; Pestell RG; Sotgia F; Lisanti MP Breast Cancer Res 2016, 18, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Zhou Y; Wang Y; Ju X; Lan J; Zou H; Li S; Qi Y; Jia W; Hu J; Liang W; Zhang W; Pang L; Li F Biomarkers Med 2015, 9, 777–790. [DOI] [PubMed] [Google Scholar]
- (18).Langan RC; Mullinax JE; Raiji MT; Upham T; Summers T; Stojadinovic A; Avital IJ Cancer 2013, 4, 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Visvader JE; Lindeman GJ Nat. Rev. Cancer 2008, 8, 755. [DOI] [PubMed] [Google Scholar]
- (20).Mitra A; Mishra L; Li S Oncotarget 2015, 6, 10697–10711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Pan N; Rao W; Kothapalli NR; Liu R; Burgett AW; Yang Z Anal. Chem 2014, 86, 9376–9380. [DOI] [PubMed] [Google Scholar]
- (22).Pan N; Rao W; Liu R; Kothapalli N; Burgett A; Yang Z Planta Med 2015, 81, IL55. [Google Scholar]
- (23).Pan N; Rao W; Standke SJ; Yang Z Anal. Chem 2016, 88, 6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Sun M; Yang Z; Wawrik B Front. Plant Sci 2018, 9, 571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Rao W; Pan N; Tian X; Yang Z J. Am. Soc. Mass Spectrom 2016, 27, 124–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Sun M; Tian X; Yang Z Anal. Chem 2017, 89, 9069–9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Bielecka ZF; Maliszewska-Olejniczak K; Safir IJ; Szczylik C; Czarnecka AM Biological Reviews 2017, 92, 1505–1520. [DOI] [PubMed] [Google Scholar]
- (28).Li J; Condello S; Thomes-Pepin J; Ma X; Xia Y; Hurley TD; Matei D; Cheng J-X Cell stem cell 2017, 20, 303–314 e305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Condello S; Morgan CA; Nagdas S; Cao L; Turek J; Hurley TD; Matei D Oncogene 2015, 34, 2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Lanekoff I; Heath BS; Liyu A; Thomas M; Carson JP; Laskin J Anal. Chem 2012, 84, 8351–8356. [DOI] [PubMed] [Google Scholar]
- (31).Liu R; Pan N; Zhu Y; Yang Z Anal. Chem 2018, 90, 11078–11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Dang CV Cancer Res 2010, 70, 859–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Hsu PP; Sabatini DM Cell 2008, 134, 703–707. [DOI] [PubMed] [Google Scholar]
- (34).Saraste M Science 1999, 283, 1488–1493. [DOI] [PubMed] [Google Scholar]
- (35).Sato M; Kawana K; Adachi K; Fujimoto A; Yoshida M; Nakamura H; Nishida H; Inoue T; Taguchi A; Takahashi J; et al. Oncotarget 2016, 7, 33297–33305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Sancho P; Barneda D; Heeschen C Br. J. Cancer 2016, 114, 1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Ye XQ; Li Q; Wang GH; Sun FF; Huang GJ; Bian XW; Yu SC; Qian GS Int. J. Cancer 2011, 129, 820–831. [DOI] [PubMed] [Google Scholar]
- (38).Snyder V; Reed-Newman TC; Arnold L; Thomas SM; Anant S Front. Oncol 2018, 8, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Mukherjee A; Kenny HA; Lengyel E Cell stem cell 2017, 20, 291–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Tirinato L; Pagliari F; Limongi T; Marini M; Falqui A; Seco J; Candeloro P; Liberale C; Di Fabrizio E Stem Cells Int 2017, 2017, 1656053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Gökmen-Polar Y; Nakshatri H; Badve S Biomarkers Med 2011, 5, 661–671. [DOI] [PubMed] [Google Scholar]
- (42).Pietrocola F; Galluzzi L; Bravo-San Pedro JM; Madeo F; Kroemer G Cell Metab 2015, 21, 805–821. [DOI] [PubMed] [Google Scholar]
- (43).Chen L; Ren J; Yang L; Li Y; Fu J; Li Y; Tian Y; Qiu F; Liu Z; Qiu Y Sci. Rep 2016, 6, 19665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Deraska PV; O’Leary C; Reavis HD; Labe S; Dinh TK; Lazaro J-B; Sweeney C; D’Andrea AD; Kozono D Cell Death Discov 2018, 4, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Nakshatri H; Appaiah H; Anjanappa M; Gilley D; Tanaka H; Badve S; Crooks P; Mathews W; Sweeney C; Bhat-Nakshatri P Cell Death Dis 2015, 6, No. e1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Matsui H; Yokoyama T; Sekiguchi K; Iijima D; Sunaga H; Maniwa M; Ueno M; Iso T; Arai M; Kurabayashi M PLoS One 2012, 7, No. e33283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Mohammadzadeh F; Mosayebi G; Montazeri V; Darabi M; Fayezi S; Shaaker M; Rahmati M; Baradaran B; Mehdizadeh A; Darabi M Breast Cancer Res 2014, 17, 136–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Bielecka ZF; Maliszewska-Olejniczak K; Safir IJ; Szczylik C; Czarnecka AM BIOL REV 2017, 92, 1505–1520. [DOI] [PubMed] [Google Scholar]
- (49).Beg AA; Baltimore D Science 1996, 274, 782–784. [DOI] [PubMed] [Google Scholar]
- (50).Jia D; Yang W; Li L; Liu H; Tan Y; Ooi S; Chi L; Filion LG; Figeys D; Wang L Cell Death Differ 2015, 22, 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Gonzalez-Torres C; Gaytan-Cervantes J; Vazquez-Santillan K; Mandujano-Tinoco EA; Ceballos-Cancino G; Garcia-Venzor A; Zampedri C; Sanchez-Maldonado P; Mojica-Espinosa R; Jimenez-Hernandez LE; Maldonado V Arch. Med. Res 2017, 48, 343–351. [DOI] [PubMed] [Google Scholar]
- (52).Zakaria N; Mohd Yusoff N; Zakaria Z; Widera D; Yahaya BH Front. Oncol 2018, 8, 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Langan RC; Mullinax JE; Raiji MT; Upham T; Summers T; Stojadinovic A; Avital IJ Cancer 2013, 4, 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Shanmugam R; Kusumanchi P; Appaiah H; Cheng L; Crooks P; Neelakantan S; Peat T; Klaunig J; Matthews W; Nakshatri H; Sweeney CJ Int. J. Cancer 2011, 128, 2481–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Phoenix K; Hong X; Tannenbaum S; Claffey K Cancer Res 2009, 69, 1135.19155302 [Google Scholar]
- (56).Tomita H; Tanaka K; Tanaka T; Hara A Oncotarget 2016, 7, 11018–11032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Zhang R; Wang Y; Li R; Chen G Int. J. Mol. Sci 2015, 16, 14210–14244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Rinkenbaugh AL; Baldwin AS Cells 2016, 5, 16. [Google Scholar]
- (59).Xia Y; Shen S; Verma IM Cancer Immunol. Res 2014, 2, 823–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Zhou J; Zhang H; Gu P; Bai J; Margolick JB; Zhang Y Breast Cancer Res. Treat 2008, 111, 419–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


