Summary
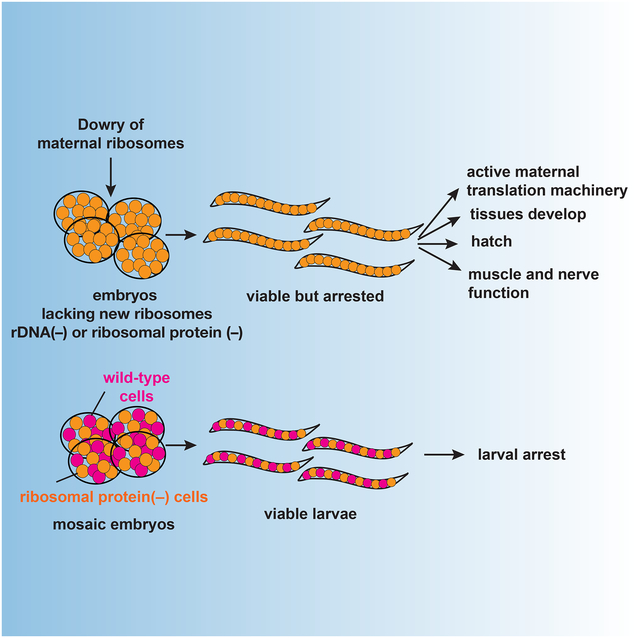
Caenorhabditis elegans provides an amenable system to explore whether newly composed ribosomes are required to progress through development. Despite the complex pattern of tissues that are formed during embryonic development, we found that null homozygotes lacking any of five different ribosomal proteins (RP) can produce fully functional first stage larvae, with similar developmental competence seen upon complete deletion of the multi-copy ribosomal RNA locus. These animals, relying on maternal but not zygotic contribution of ribosomal components, are capable of completing embryogenesis. In the absence of new ribosomal components, the resulting animals are arrested before progression from the first larval stage and fail in two assays for postembryonic plasticity of neuronal structure. Mosaic analyses of larvae that are a mixture of ribosome-competent and non-competent cells suggest a global regulatory mechanism in which ribosomal insufficiency in a subset of cells triggers organism-wide growth arrest.
Graphical Abstract
Cenik et al. find that Caenorhabditis elegans is able to complete embryogenesis with only maternal-deposited ribosomal components but arrest before progression from the first larval stage. Organism-wide arrest in larvae mosaic for zygotic ribosome production-competent and non-competent cells suggest ribosomal insufficiency in a subset of cells triggers whole organism arrest.
Introduction
Early in the study of molecular regulation, a realization of the complexity of translational machineries spurred speculation that certain global regulatory transitions might emanate from changes in composition of translating ribosomes (Waller and Harris, 1961, Moore et al., 1968, Kurland et al., 1969, Deusser and Wittmann, 1972).
Ribosome heterogeneity can potentially stem from variability in synthesis of ribosomal protein (RP) allelic variants, post-synthesis modifications of ribosomal RNAs (rRNAs) and RPs, or protein composition of the translating ribosomes (Waller and Harris, 1961, Lee et al., 2002, Slavov et al, 2015, Sharma and Lafontaine, 2015). Duplicated RP genes, prevalent in yeast and plants, are reported to have differential expression and associate with paralog-specific mutant phenotypes (Barakat et al., 2001, Ni and Snyder, 2001, Enyenihi and Saunders, 2003, Carroll, 2013, Falcone Ferreyra et al., 2010, Komili et al., 2007, Hummel et al., 2012). To explain the paralog-specific mutant phenotypes, a model has been proposed where functionally distinct ribosomes exist and regulate the translational activity of their target mRNAs (Komili et al., 2007) A complication with this argument comes from the rapid accumulation of suppressor mutations in paralog deletion strains due to slow growth, challenging the paralog-specific phenotype comparisons (Steffen et al. 2012).
Tissue, cell and/or stage specific ribosomal machinery (e.g., Ramagopal and Ennis, 1981, Gunderson et al., 1987) would provide a remarkable means for broad modulation of protein synthesis in order to diversify expression. Both ribosomal proteins and ribosomal RNA sequences could have message-dependent consequences for translation (Mauro and Edelman, 2002, Landry et al., 2009, Lee et al., 2013, Xue et al., 2015, Ferretti et al., 2017). The possibility of differential expression from ribosomal protein genes in different tissues has prompted a debate over whether translation of specific mRNAs in a tissue-dependent manner by a heterogeneous ribosome pool during development might play a role in proper tissue diversification (Bortoluzzi et al., 2001, Xue and Barna, 2012, Gupta and Warner, 2014). As a specific testable hypothesis, these studies raise the question of whether distinct ribosome compositions might be required for tissue diversification during embryonic development.
Complicating the functional identification of tissue-specific components of translational machinery in higher organisms is a series of apparently-tissue-specific dosage effects. Congenital abnormalities, bone marrow failure and tissue specific defects are characteristic phenotypes associated with germline mutations disrupting one copy of RPs or ribosome biogenesis factor genes. Such deficiencies lead to a number of human diseases including Diamond Blackfan Anemia (DBA) (Ellis and Lipton, 2008, Gazda et al., 2008). Tissue-specific haploinsufficient phenotypes could be explained by numerous factors: 1-Differential sensitivities of specific tissues to allelic feedback regulation, 2-Seemingly specific alteration of poorly initiated/translated mRNAs reflecting decreased ribosome availability, 3-Involvement of additional tissue-specific factors such as stress signaling pathway or ribosome recycling factors (Mills et al., 2016, Mills and Green, 2017, Narla and Ebert, 2010, Khajuria et al., 2018).
One of the confounding aspects of studying translational contributions to embryonic tissue diversification is the intertwinement of overall growth with tissue differentiation in most animal species. In such cases, we are limited in attempting to distinguish whether (i) specialized ribosomal properties are required during embryonic tissue diversification, or (ii) embryonic ribosome production is important. C. elegans is amenable to addressing such questions, because total organismal volume is constant from the pre-fertilization oocyte through the production of a fully differentiated ~550 cell L1 larvae.
A plethora of genetic and cytological tools in C. elegans facilitate addressing these fundamental questions. A well-defined cell lineage (Sulston and Horvitz, 1977, Sulston et al., 1983), a detailed cyto-anatomy (White et al., 1986), and a history of genetic screens and balancer chromosomes are foundations for this analysis. More recently, CRISPR-Cas-9 tools have been developed to readily introduce specific mutations to the C. elegans genome (e.g., Dickinson and Goldstein, 2016) permitting investigation of the phenotypic consequences of loss of function mutations in individual ribosomal components. As with all organisms that utilize embryonic development, C. elegans development involves a transition between early processes executed primarily by maternally encoded components and later events increasingly controlled and driven by zygotically encoded components (Newman-Smith and Rothman, 1998). This is manifested in genetic analysis of C. elegans embryogenesis in that homozygotes for loss of function in essential genes can generally progress to the point at which maternally encoded components are no longer sufficient for the relevant process and must be replaced by embryonically encoded (zygotic) products. Exemplary of differential maternal and zygotic requirements for C. elegans development, a single tandem repeat locus in C. elegans encodes both a common trans-spliced leader RNA and the C. elegans 5S rRNA (Nelson and Honda, 1985). Development proceeds through embryogenesis in the absence of new 5S rRNA synthesis, while early zygotic expression of the splice leader RNAs is required as early as gastrulation (Ferguson et al., 1996). Despite the evident lack of requirement for new 5S rRNA synthesis in embryonic development, the sufficiency of existing translation machinery is unclear since large amounts of 5S rRNA can be stored in ribosome-free particles in oocytes (Picard and Wegnez, 1979, Picard et al. 1980).
As in humans, C. elegans encodes most RPs from single copy genes distributed throughout the genome. In contrast, the ribosomal RNAs (rRNAs) are encoded in tandemly repeated clusters, with the major large and the small subunit RNAs (18S, 5.8 S and 28S) located in a cluster of ~100–250 repeats at the end of chromosome I (Files and Hirsh, 1981, Albertson, 1984, Bik et al., 2013). Presence of a single cluster encoding 18S, 5.8S and 28S rRNA provides a particular opportunity for genetic manipulation, in contrast to most organisms where rDNAs are positioned in multiple positions (5 different chromosomes for human) (e.g., Stults et al., 2008).
In this manuscript, we address the following questions: 1-In the absence of a requirement for growth in an embryo, how far can the maternally-encoded ribosome pool support tissue diversification? 2-Are certain tissues or cell types specifically dependent on the synthesis of new and qualitatively different ribosomes? 3-For cell types with no requirement for qualitatively different ribosomes, are there underlying mechanisms that respond to ribosomal quantity to maintain viability in the absence of new synthesis?
Results
Zygotic ribosomal RNA (rRNA) genes are transcribed from early stages but functional ribosomes remain predominantly maternal
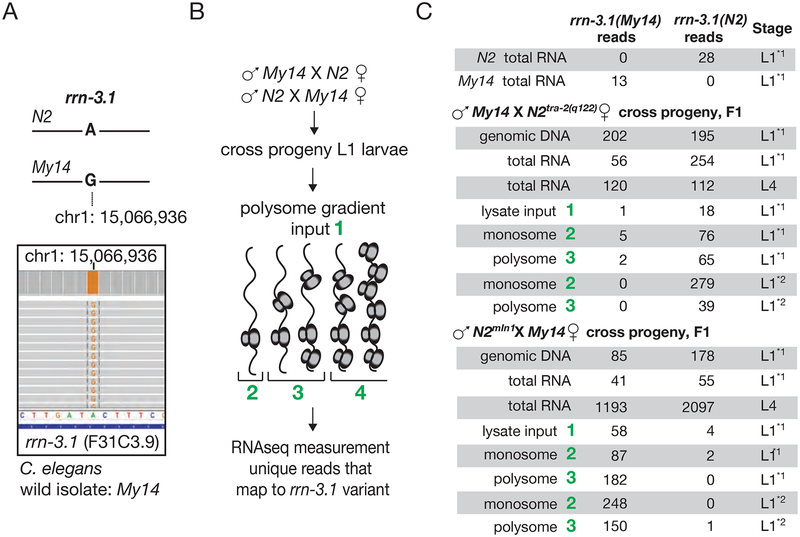
To understand the dynamics of zygotic ribosome expression, we analyzed rDNA sequences among wild strains of C. elegans with the intention of finding natural variants of rDNA to differentially track maternal and zygotic expression. Given that the rDNA locus is both conserved and extensively duplicated (Ellis et al., 1986), occurrence of a complete conversion is rare among wild strains. Despite the scarcity of such variation, we found a single nucleotide polymorphism in all copies of the rrn-3.1 gene in wild C. elegans isolate My14 at 28S position 15066936 on chromosome I (WBcel235/ce11 assembly, 2098th base on F31C3.9) (Figure 1A). My14 genome is sequenced among other wild isolates (Thompson et al., 2013). The observed rDNA variation allowed us to track parental rRNA expression in crosses between My14 and N2 strains, with a measurement of maternal versus zygotic rRNA prevalence deriving from RNA-seq of F1 progeny at different stages of development (L1 and L4). In L4 stage total RNA, both variants are represented at a similar ratio as their genomic DNA copies (Figure 1C). Both variants are also observed in L1 stage total RNA (Figure 1C) consistent with earlier observations that rRNA expression starts early in development (Korčeková et al., 2012).
Figure 1: Zygotic ribosomal RNA is transcribed but functional ribosomes remain predominantly maternal A.
C. elegans strain My14 harbors a single nucleotide variant in the rrn-3.1 gene, encoding for 28S rRNA. Genomic DNA sequencing of the canonical laboratory strain N2 (Brenner, 1974) shows only the reference allele at this site, while sequencing of the My14 strain reveals only copies containing the variant allele (Thompson et al., 2013).
B. RNA-seq of total RNA, monosome and polysome pools from cross progeny of either My14 males and N2 females (feminized strain: tra-2(q122)) or N2 males (mIn1) and My14 hermaphrodites (Goodwin et al., 1993). Total RNA, genomic DNA and ribosomes from cross progeny L1 or L4 larvae were extracted and uniquely mapping reads to each variant were counted.
C. Number of unique reads that map to each allele is shown for total RNA at L1 and L4 stages as well as monosome and polysome fractions. Comparing My14 rRNA read counts to all rRNA provides a measure of zygotic rRNA representation for total RNA, polysomes, and monosomes. At hatching [L12*], lower zygotic representation in monosome and polysome fractions is significant as compared to total RNA (p-values at hatching for monosome and polysome fractions p<1e-06, p = 0.0004 respectively for cross progeny of My14 males and N2 females. This significant difference persisted at 3 hours after hatching [L11*], (p = 0.016 and p = 0.00018 for the monosome and polysome fractions respectively for cross progeny of My14 males and N2 females, right). L1 progeny of the reciprocal cross of N2 (mIn1) sperm and My14 oocytes at hatching [L12*] and 3 hours after hatching [L11*], shows significantly lower zygotic N2 variant representation in monosome and polysome fractions as compared to total RNA. (p<1e-6)
We next investigated what fraction of the embryonic rRNA is in actively translating ribosome pools. We isolated monosome and polysome fractions from the L1 progeny of My14-derived males and N2-derived females (feminized strain: tra-2(q122), Goodwin et al., 1993) using sucrose gradient centrifugation (Figure 1B). We detected rRNA from the paternal rDNA allele in monosomes and polysomes only after hatching (Figure 1C). Comparing paternal My14 rRNA read counts to all rRNA provides us with a measure of zygotic rRNA representation for total RNA, polysomes, and monosomes. At hatching, we observe lower zygotic representation in monosome and polysome fractions as compared to total RNA (p-values at hatching for monosome and polysome fractions p<1e-06, p = 0.0004, respectively). This difference persisted for at least 3 hours after hatching (p = 0.016 and p = 0.00018 for the monosome and polysome fractions, respectively; Figure 1C). The reciprocal cross (N2-derived sperm (mIn1) and My14 oocytes) yielded a complementary pattern, with zygotic N2 rRNA underrepresented in monosomes and polysomes compared to L1 total RNA (p<1e-06 both at hatching and after hatching for monosome and polysome fractions).
From these results, we hypothesize that ribosome capacity for embryogenesis in C. elegans may be effectively derived from the maternal dowry, with post-fertilization synthesis contributing mainly to pools used postembryonically. Nonetheless, it remains possible that zygotically-derived ribosomes (despite their low number) might serve key developmental roles. To pursue this idea, we applied a genetic approach to investigate the requirement of zygotically-derived ribosomes in tissue diversification.
A shared larval arrest phenotype for ribosomal protein null mutants
C. elegans provides a unique genetic model for rapid introduction of mutations via gene editing. To study the effects of ribosomal machinery dysfunction we introduced early stop codons to generate loss of function mutation strains for the ribosomal protein (RP) genes: rpl-5, rps-30, rps-23, rps-10 and rpl-33 (in current ribosome nomenclature, these encode the proteins uL18, eS30, uS12, eS10 and eL33 (Figure 2A) (Ban et al., 2014) using CRISPR-Cas9 gene editing methods we have developed for C. elegans (Arribere et al., 2014). Rpl-5, rps-10 and rpl-33 (human homolog name: rpl35A) are among the single copy loss-of-function mutations observed in human Diamond Blackfan Anemia patients (Farrar et al., 2008, Gazda et al., 2008). Missense mutations in rps-23 were recently described in two individuals (Paolini et al., 2017). rps-30 is homologous to human FAU gene. Rps-30 is expressed as a fusion protein containing ubiquitin-like protein fubi at the N terminus and RPS30 at the C terminus in nematodes and humans; posttranslational cleavage liberates the two functional proteins (Michiels et al., 1993).
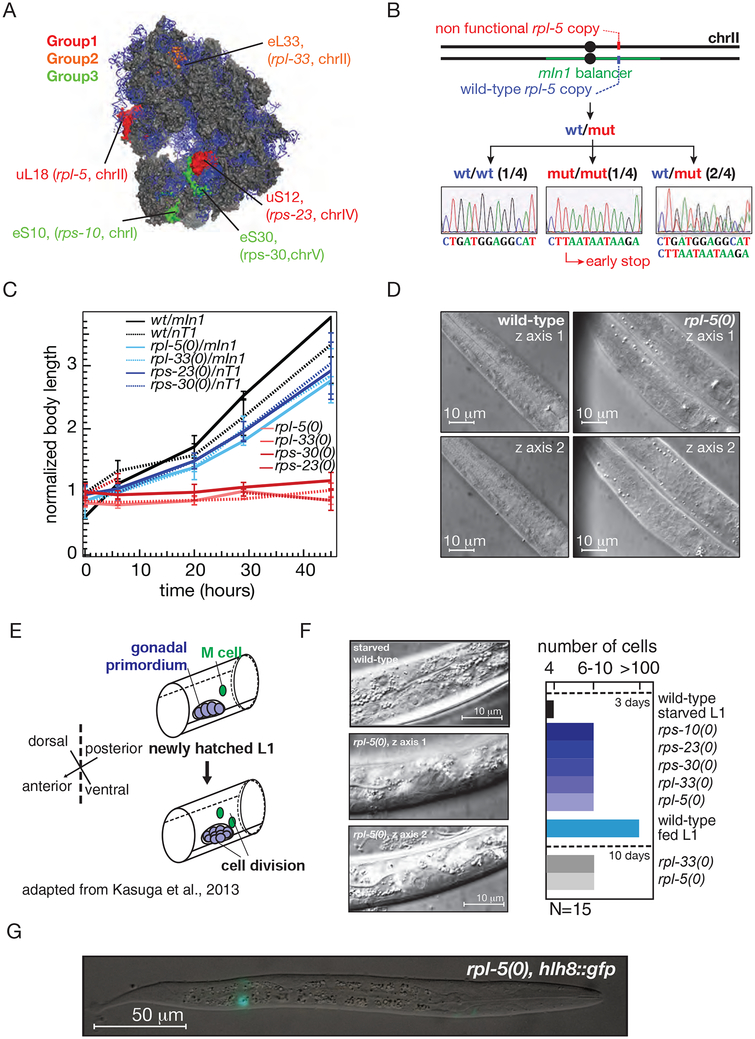
Figure 2: Complete embryonic development and partial post-embryonic development for ribosomal protein null mutants.
A. Mutated ribosomal proteins (RPs) with corresponding C. elegans gene names are shown in color on the human 80S ribosome structure (adapted from PDB ID: 4ug0) (Khatter et al., 2015). The three colors depict the extent of conservation of ribosomal proteins. Group-1 RPs have homologs in archaebacteria and eubacteria. Group-2 RPs have homologs only in archaebacteria, and Group-3 is unique to eukaryotes (Wool et al., 1995). Ribosomal RNA (rRNA) is labeled in blue.
B. Representative diagram for one of the mutations: rpl-5(0) depicts how the strain is maintained. The heterozygous mutation is maintained with the balancer mIn1 [dpy-10(e128)] (II). Representative Sanger sequencing traces in the mutated region is shown. All ribosomal protein mutations were made via CRISPR-Cas9 gene editing and via insertion of tandem early stop (TAA) mutations. All mutations were verified by PCR amplification of the target region and Sanger sequencing.
C. The body length of the newly hatched homozygous larvae matched the length of heterozygous and wild-type siblings in the same genetic background at hatching. Homozygous larvae failed to grow over time unlike its heterozygous or wild-type siblings.
D. Comparative differential interference microscopy images of pharynx for wild-type and rpl-5(0) larvae: different z-sections (top and bottom) of the pharynx of the wild-type larvae (left) and the representative rpl-5(0) homozygous larvae are shown (right). Scale bar, 10 μm.
E. Diagram depicts mesoblast (M) cell and gonadal primordium cellular divisions during L1 stage (adapted from Kasuga et al., 2013).
F. Gonadal primordium images of three-day-old rpl-5(0) in two different z-sections shows at least a single cellular division from the 4 cell gonadal primordium present canonically at hatching whereas wild-type starved L1 lacks post-embryonic cell division in gonadal primordium (left images). Brightness and contrast were individually adjusted. Right graph shows the number of cells in the gonadal primordium 72 hours after hatching at 16°C. For rpl-33 and rpl-5, gonadal primordium cells were also counted after 10 days. For all homozygous mutants, gonadal cells divides (N=15 for each mutant), whereas no cell division occurs in the gonadal primordium of starved L1 (N=15; also see Fukuyama et al., 2006). Scale bar, 10 μm.
G. GFP-based imaging of the mesodermal M lineage in arrested rpl-5(0) larvae. hlh-8::gfp labels the M cell and its descendants (Harfe et al., 1998). We observed that M lineage is arrested at a single-cell stage with no evidence of postembryonic divisions in larvae after 72 hours at 16°C.
For our genetic analysis, we chose RPs that have varying degrees of conservation amongst domains of life (Figure 2A) and with protein products incorporated into nascent ribosomes at different phases of the assembly process. RPL33, RPL5 and RPS23 have homologs in archaebacteria and eubacteria and are stably incorporated into nascent ribosomes at an early nucleolar stage. RPS30 has homologs only in archaebacteria, and assemble at a late nucleolar stage. RPS10 is unique to eukaryotes, with incorporation into nascent ribosomes reported to occur in the cytoplasm (Wool et al., 1995, de la Cruz et al., 2015).
We maintained RP mutations in heterozygous strains bearing a single copy loss of function mutation balanced with a chromosomal balancer [mIn1, nT1 or hT2] (McKim et al., 1993, Ferguson and Horvitz, 1985, Edgley and Riddle, 2001, Edgley et al., 2006) (Figure 2B). RP mRNAs containing an early stop mutation were detected at levels ~10–100 fold lower compared to wild-type mRNA in heterozygous strains (Figure S1A), consistent with known properties of the Nonsense-Mediated Decay system in C. elegans (Longman et al., 2008). We used these strains to follow the extreme case in which both genomic copies of the RPs are null. We observed that among all homozygous RP mutants [rpl-5(0), rps-23(0), rps-30(0), rps-10(0) and rpl-33(0)], early and late embryonic development proceeded similar to wild-type embryonic development (Table S1). Newly-hatched homozygous larvae were phenotypically indistinguishable from their heterozygous or wild-type counterparts (Figure S1B). Despite being phenotypically similar to wild-type first-stage (L1) larvae, the homozygous larvae arrest at L1 stage without any detectable growth but live up to 2 weeks at 16°C (Figure 2C). We likewise did not detect any significant difference in movement or mechanosensory response for the homozygous null newly hatched larvae, suggesting mechanosensory (touch receptor) neurons, interneurons, motor neurons, and muscles are functioning properly (Chalfie et al., 1985).
The C. elegans pharynx rhythmically contracts and relaxes (pharyngeal pumping) to deliver food into intestinal lumen. The rate of pharyngeal pumping determines the rate of ingestion and decreases with food availability. We observed that RP null larvae had pharyngeal pumping rates similar to wild-type larvae, suggesting absence of any abnormality in the pharynx development and function as well as food intake and ingestion (Avery and Horvitz, 1990). Differential interference contrast microscopy of pharyngeal (Figure 2D) and intestinal cells as well as structures of the gonadal primordium showed patterns indistinguishable from wild-type L1 larvae. Likewise, GFP markers for body wall muscle showed patterns that were similar to wild-type. These observations suggest that embryonic development proceeds normally in the absence of a zygotic contribution from any of these five different RP genes.
From a metabolic perspective, the ability to complete embryogenesis without synthesis of zygotically-encoded ribosomes is not necessarily unexpected: C. elegans embryogenesis proceeds rapidly once the oocyte has been fertilized and there is no overall volume/mass change from oocyte to newly hatched larva. Thus, by mass, persisting maternal ribosomes (and RNAs loaded into oocytes) might have been expected to prove sufficient in providing the translation machinery needed for normal embryonic processes. Indeed, our data demonstrate that the maternal ribosomes are sufficient.
Partial post-embryonic development in the absence of zygotic ribosomal protein (RP) genes
Given the lack of detectable size increase in the homozygous RP null mutant larvae, we wondered whether any postembryonic developmental programs continue in these animals. We have primarily focused on rpl-5(0) and rpl-33(0) in these analyses as mutation in either RP disrupts ribosome biogenesis at an early nucleolar stage (de la Cruz et al., 2015). To investigate the effect of homozygous mutations on the progression from L1 stage, we imaged newly hatched and ten day-old rpl-33(0) larvae using differential interference contrast microscopy. Consistent with previous observations of nucleolar hypertrophy as a result of certain ribosome assembly defects (Nicolas et al., 2016), we observed abnormally large nucleoli in ten day-old rpl-33(0) and rpl-5(0) larvae (Figure S1C).
One of the most dramatic changes during post-embryonic development is the proliferation of gonadal primordia from 4 to ~2500 cells in mature adults. L1 larvae have two primordial germ cells (Z2 and Z3) located between two somatic gonad precursors (Z1 and Z4) (Sulston and Horvitz, 1977). Primordial germ cells form in the early embryo, and migrate to their destination during late embryogenesis. Until larvae start feeding, these cells arrest at the G2 phase of the cell cycle in newly hatched L1 animals (Sulston et al. 1983, Fukuyama et al., 2006, Fukuyama et. al. 2012). After feeding, Z2 and Z3 proliferate around mid-L1 stage alongside somatic precursors Z1 and Z4, which also divide during L1 to generate 12 cells before the second molt (L2) (Figure 2E) (Kimble and Hirsh, 1979).
We inspected gonadal primordia of arrested homozygous mutant larvae at three or ten days after being laid as eggs. After three or ten days, gonadal primordia of homozygous RP null mutants contained 6–10 cells (Figure 2F, N=15 for each RP mutant). In an equivalent time frame, a robust proliferation of cells in the gonad (>100 cells) was observed for wild-type larvae (Hirsh et al., 1976). This observation distinguishes larval arrest in homozygote RP nulls from the starvation-dependent arrest of L1 larvae (Figure 2F, N=15), as the latter process blocks all gonadal divisions (Sulston et al. 1983, Fukuyama et al., 2006).
An additional proliferative population in C. elegans larva derives from a single mesoblast precursor called the M cell. Born next to the pharynx during embryonic development, the M cell migrates towards the posterior during embryogenesis. M cell divisions start in the mid-L1 stage and generate 18 cells by the end of the L1 stage (Figure 2E) (Sulston et al., 1983, Sulston and Horvitz, 1977). Taking advantage of specific labeling with hlh-8::GFP (Harfe et al., 1998), we observed the expected position of the M cell in newly-hatched homozygous rpl-5(0) larvae. The labeled M cell failed to divide and remained as a single cell in homozygous mutant larvae (Figure 2G, no M cell division, N=65, see Figure S2 for wild-type comparison).
Taken together, these results suggest the phasing in of a strong requirement for newly produced ribosomes as the animal commences postembryonic development.
Assessment of structure, function, and regrowth for embryonic-born mechanosensory neurons.
For a further analysis of embryonic development and the extent of post-embryonic larval arrest, we focused on an organ system known to involve substantial translational changes, localized translational heterogeneity, and translational control. Neuronal development and function are highly specialized processes with characteristics (such as local translation) that suggest the potential for greater dynamics and complexity in the translational machinery (Fernandez-Moya et al., 2014, Van Driesche and Martin, 2018, Glock et al., 2017, Wang et al., 2016). To assess the role of zygotic and/or specialized neuronal RP forms contributing to neuronal development, we examined the mechanosensory neurons (touch receptor neurons). We introduced the touch receptor neuron specific markers pmec-4::GFP(zdIs5) or pmec-7::GFP(muIs32) (Clark and Chiu 2003, Ch’ng et al., 2003) which label the embryonically-born bilaterally symmetric ALMs and PLMs, into the six RP null mutants and homozygous rDNA deleted larvae. In wild-type young L1 larvae, i.e. shortly after hatching, ALM and PLM extend their axons anteriorly, with the PLM anterior axon overlapping with ALM soma (Figure 3A, top left). In similar staged L1 larvae of all five RP mutants, we observed normal ALM and PLM position and axonal morphology (Figure 3A), indicating that the maternal ribosomes are sufficient for embryonic touch receptor neuron development. In addition, the mutant animals displayed normal patterns of sinusoidal locomotion and responded normally to a light touch. Following hatching, the movement of RP null mutants were observed to slow down, eventually becoming immobile before demise. Given that all the RP null mutants tested showed similar phenotypes, we focus on characterization of rpl-33(0).
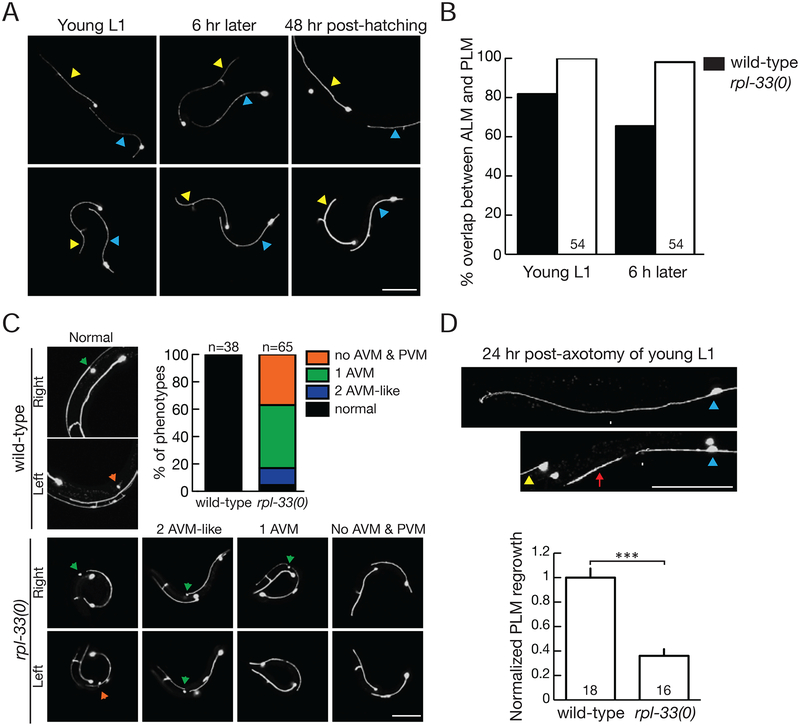
Figure 3: Ribosomal protein null mutants show normal development of embryonically born touch receptor neurons, but display impaired axonal tiling and regrowth after injury.
A. Confocal images of ALM (yellow arrowhead) and PLM (blue arrowhead) in young L1 larvae. Left panels: Normal positions and axon trajectory of ALM and PLM. PLM anterior axons overlap and extend beyond ALM soma (*), which are indistinguishable between wild-type and rpl-33(0). Middle panels: Positions of ALM and PLM in the same larva 6 h later. In wild-type, slow growth of PLM axons results in separation from ALM soma (process of tiling). In rpl-33(0) mutants, PLM axons remain overlapping with ALM soma. Right panels: 48 hours post-hatching, wild-type larvae reach L4 stage, PLM axons terminate posterior to ALM soma, but rpl-33(0) mutants remained as young larvae, and PLM axons remain overlapping with ALM soma.
B. Quantitative analysis of tiling defects of rpl-33(0). Larvae were mounted for observation (defined as time 0) from <6 hours post-hatching, and re-mounted 6 hours later.
C. Confocal images of AVM (green arrow) and PVM (orange arrow) in animals 48 hours post-hatching. Note that wild-type larvae reach L4 stage, whereas rpl-33(0) mutants remain as young larvae. Graph shows quantification of the phenotypes observed. N=38 for wild-type; N=65 for rpl-33(0).
D. PLM axons in ribosomal mutant L1 larvae do not regenerate. Representative images show regrowth of PLM in wild-type, but not in rpl-33(0). White arrows mark site of axotomy. The red arrow indicates the distal fragment of axotomized axon in rpl-33(0) mutants. Graph shows normalized PLM regrowth 24 h post-axotomy.
Over a next few hours after wild-type L1 larvae hatch, ALM and PLM axons underwent continued growth, but with slowed axon growth from PLM gradually reducing the overlapping of PLM axon with ALM soma (Figure 3A, top middle), a process known as “axon tiling” (Gallegos and Bargmann, 2004). This post-embryonic touch receptor neuron tiling process was blocked in rpl-33(0) L1 larvae (Figure 3A, bottom middle; quantification in Figure 3B), with sustained tiling defect 48 hours after hatching in arrested RP null mutants (Figure 3A, bottom right).
At late L1 larvae, two additional touch receptor neurons known as AVM and PVM are born as the result of Q neuroblast migration and cell division (Middelkoop and Korswagen, 2014). AVM is a descendant of QR following anterior migration, and PVM is a descendant of QL following posterior migration. In wild-type late larvae to young adults (~48 hours post-hatching), all animals had both AVM and PVM at the correct location (Figure 3C). In contrast, ~40% of the rpl-33(0) mutant larvae had only AVM by their anterior position on the right side, while another ~40% had no AVM or PVM (Figure 3C). <10% of the rpl-33(0) mutant larvae appeared to have two AVM-like neurons, based on their anterior location and axon morphology (Figure 3C). As the transgenic zdIs5 or muIs32 reporters do not label Q neuroblasts, a duplicate AVM-like cell could reflect a defective Q neuroblast migration, resulting in mislocalization of the PVM to the anterior part of the body.
To further examine the effect of RP deficit in growth-demanding conditions, we tested axon regrowth after injury. We severed the axon of PLM in L1 animals using laser axotomy (Wu et al., 2007). In wild-type animals, proximal axons of injured PLM were able to initiate rapid regrowth and extend for a long distance, when assessed at both 6 hours and 24 hours post-injury (Figure 3D). In contrast, in rpl-33(0) mutants, injured PLM axons showed no sign of regrowth (Figure 3D), supporting previous observations that axon regeneration was impaired upon translation inhibition (Noma et al., 2017).
In addition to the regrowth requirements, we examined degeneration of distal fragments of axotomized PLMs following microsurgery. Consistent with previous observation (Wu et al., 2007), these fragments underwent degeneration in wild-type animals 24 hour post-axotomy (“N=18; 100% degenerated distal axon fragments). In contrast, we observed morphologically normal distal axon fragments in 56% of rpl-33(0) mutants 24 hour post-axotomy (N=16), suggesting the degeneration of distal axon fragments was impaired in rpl-33(0) mutants.
In sum, these data indicate that maternally-derived RPs are sufficient for embryonic mechanosensory neuron development, but that additional ribosome synthesis is needed for a specialized set of post-embryonic capabilities: axon growth, axonal tiling, AVM and PVM positioning, and the ability of axons to undergo regenerative processes after injury.
Ribosomal protein null arrested larvae can translate new proteins
Can RP null homozygotes support new protein expression? To address this question, we constructed strains in which these mutations had been crossed into a background with GFP inserted into a heat shock inducible locus (hsp-16.2/hsp16.41). In both wild-type and RP null mutant backgrounds, we observed robust hsp-16 promoter induced GFP expression upon heat-shock. Negligible background activity was observed without heat-shock. GFP expression was also detected if homozygous arrested rpl-5(0) larvae were heat-shocked after ~100 hours of larval arrest (Figure 4A), indicating an ongoing ability of the homozygous mutant larvae to conduct translation resulting in new proteins. This result establishes that maternal ribosomes in addition to their functionality during embryonic development, persist in homozygous larvae and can actively translate new proteins upon induction.
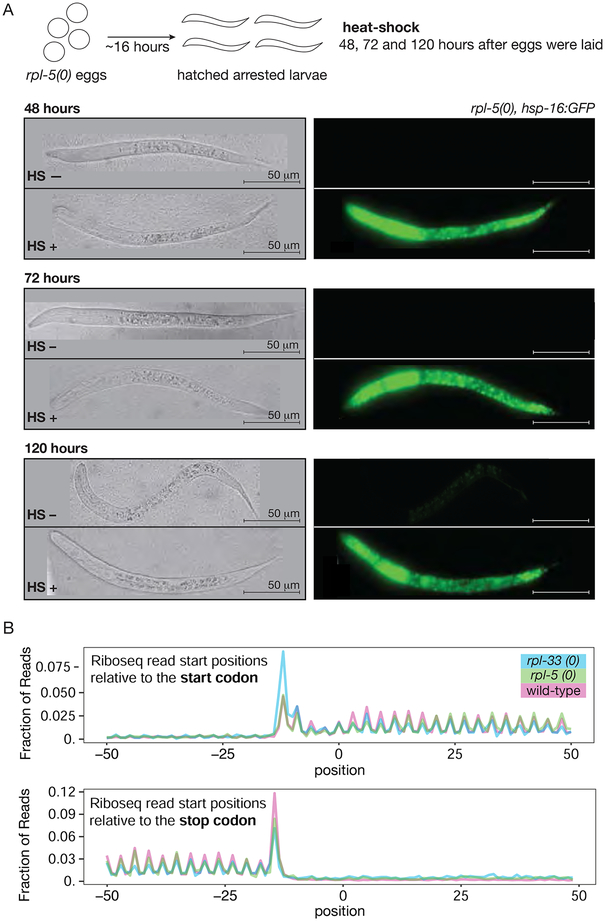
Figure 4: Maternal ribosomes are active in ribosomal protein null mutants.
A. Arrested rpl-5 null larvae express heat-shock inducible hsp-16 GFP. rpl-5(0), hsp16::GFP were kept at 16°C for 48, 72 and 120 hours (top, middle and bottom image panels respectively) after being laid as embryos, then heat-shocked at 34°C for 3 hours, followed by 3 hours incubation at 16°C before imaging (top diagram). First row of each image depicts the sibling larva in the absence of heat-shock (HS–) and second row shows the heat-shocked larva (HS+). Differential interference contrast (left), and GFP (right) images were captured sequentially with identical settings.
B. Metagene analysis of normalized ribosome footprint start site density in stop and start codon proximity in wild-type and rpl null larvae. 5’ end of mapped reads were plotted on their position relative to start and stop codons on protein coding transcripts in wild-type and rpl null larvae.
Riboseq methods provide an additional experimental window into mechanistic features of translation, with sequences of large numbers ribosome protected fragments allowing deductions related to ribosome positional information on specific mRNAs (Steitz, 1969, Ingolia et. al. 2012). In actively translated regions, ribosome-protected fragments generally follow three nucleotide periodicity and specifically pile up within coding regions. Global deviations in the utilization of the translational machinery in which the majority of mRNA-associated ribosomes are no longer actively involved in translation could result in ribosome pile-ups in non-coding regions or specific sequences of coding regions. For example, a change in ribosome recycling can result in increased number of reads in 3’UTRs (Mills et al., 2016). For a global analysis of translation in RP null larvae, we optimized Ribo-seq for a small-scale sample (arrested L1 larvae). Ribosome occupancy in RP null larvae was then compared to wild type immediately after hatching. Ribosome footprints in RP null larvae were observed in coding regions, had typical 3 nucleotide periodicity with no observable differential pile up near start or stop codon. We also did not detect an aggregate pile-up in 3’UTRs (Figure 4B). These results suggest that maternally rescued RP null mutant larvae retain substantial aspects of the global translation patterns.
Cell non-autonomous growth arrest in genetic mosaics suggests a global ribosome synthesis checkpoint
C. elegans genetics provides a tractable model for the study of developmental checkpoints, with mutant homozygotes derived from a heterozygote mother providing a valuable test for processes that arrest or progress under adverse cytological conditions (e.g., Albertson et al., 1978, Korzelius et al., 2011, Brauchle et al., 2003). It was conceivable that this arrest is “passive”, with homozygous RP null larvae arresting through a biosynthetic insufficiency – essentially lacking a sufficient number of ribosomes to grow. Alternatively there could be a specific (and possibly global) “checkpoint” engaged under such circumstances. In a passive arrest model, one might expect genetically mosaic animals with some normal and some RP deleted cells to exhibit mosaic growth patterns with RP(+) tissues growing while RP(–) tissues enter arrest states. By contrast, some checkpoint systems might have a systemic character whereby insufficiency in a subset of cells might arrest the entire animal.
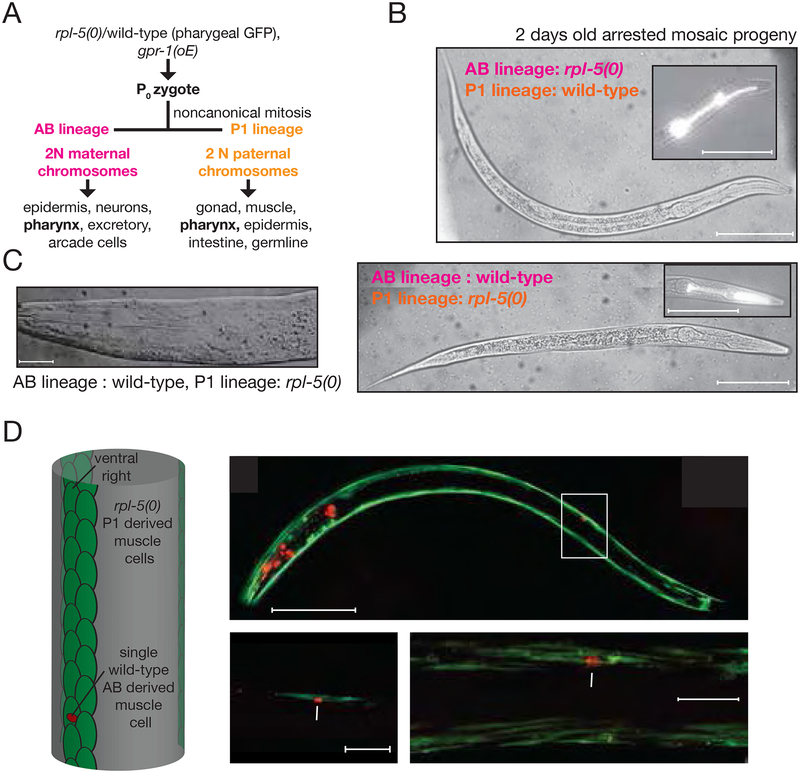
To test this hypothesis, we took advantage of a novel means to produce mosaics in which a viable two-cell C. elegans embryo is constructed with the anterior cell (‘AB’) having a completely maternally derived genome and the posterior cell (‘P1’) having a completely paternally-derived genome (Besseling and Bringmann, 2016, Artiles et al. 2018, co-submitted). In an otherwise wild-type genetic background (with no defects in RP genes), these mosaic embryos yield developmentally normal larvae and fertile adults. The production of mosaics in this background provides a tool for examining differential contributions of autonomous and non-autonomous signals to cellular and organismal development.
Using the rpl-5(0) mutation, we produced a series mosaics in which (i) all AB-derived cells were rpl-5(0) homozygous and all P1-derived cells were wild-type in genotype, or (ii) in which all AB-derived cells were wild-type and all P1 derived cells were rpl-5(0) homozygous. Lineage-specific derivations for the resulting larvae were confirmed based on precise lineage-specific patterns for a series of fluorescent protein markers that label muscle tissues that are split between the two lineages. Strikingly, both classes of mosaic larvae arrested at a stage similar to the rpl-5(0) mutant homozygote (L1 stage without any evident overall growth). We then examined the mosaic tissues in detail with the intention of determining whether there was differential growth for adjacent rpl-5(+) and rpl-5(−) cells. In each case we observed a near-normal overall anatomy suggesting no dramatic difference in growth between the mutant and wild-type cells (Figure 5A, 5B). Two different muscle types facilitate this comparison. For pharyngeal muscle tissue, the AB/P1 mosaics will contain a mixture of cells that are definitively rpl-5(0), that are wild-type, and that are heterokaryons of the two (a fraction of the pharyngeal muscle cells are multinucleate and derive from both P1 and AB lineages (Sulston et al., 1983). AB-derived cells are generally more anterior, with P1-derived cells more posterior. We observed that the mosaic pharynxes, with either AB or P1 being rpl-5(0) showed a normal morphology with no evident differential growth (Figure 5C).
Figure 5: Genetic mosaics with rpl-5 null and wild-type cells show normal embryonic development with a cell non-autonomous larval growth arrest.
A. gpr-1 overexpression (Artiles et al. 2018 co-submitted) led to a mosaic embryo with (i) rpl-5(0) in AB lineage and wild-type cells in P1 lineage and (i) wild-type cells in AB lineage and rpl-5(0) cells in P1 lineage because of non-canonical mitosis.
B. Pharyngeal musculature contains both AB and P1 lineage cells with anterior pharynx being predominantly AB-derived and posterior being predominantly P1-derived. GFP+ wild-type pharyngeal cells are P1-derived in the top image and AB-derived in the bottom image. GFP–pharynx cells are rpl-5(0). Both of these mosaic larvae were imaged 72 hours post egg laying at 16°C. Both have the typical anatomy and size of L1 arrested larvae. Scale bar, 50 μm.
C. Differential interference contrast image of the pharynx of a mosaic larva where the P1 lineage is wild-type and AB lineage is rpl-5(0)).
D. Left diagram: C. elegans body muscle tissue is predominantly P1-derived with the exception of a single AB-derived cell (adapted from (Sulston and Horvitz, 1977, Sulston et al., 1983, Moerman & Fire. C. elegans II, Chapter 16).). Right section, top image: Confocal GFP (unc-54::GFP, present in both lineages) and mCherry (myo2:mCherry:let858[3’UTR], only present in the AB lineage) image of a 48 hour old arrested mosaic larva whose P1 lineage is rpl-5(0) and AB lineage is wild-type. GFP in this animal labels muscle structure, with the heavy chain myosin gene unc-54 endogenously tagged with GFP at the C terminus. mCherry labels pharyngeal and body muscle cells from the AB lineage. Bottom left: In this image GFP and nuclear mCherry coincide for the single wild-type AB-derived body muscle cell. This cell doesn’t grow significantly over 3 days in the (AB: wild-type, P1: rpl-5(0)) mosaic larva; in contrast to a wild-type animal wherein the cell would have grown > 5 fold in length in a wild-type animal. Bottom right: A more detailed image of the muscle arrangement surrounding the nuclear mCherry labeled wild-type AB lineage cell. No defects in muscle structure were observed.
The body wall musculature provides a distinct partitioning with 80 cells of this type in the newly hatched larva being P1-derived and one cell AB-derived. The cells are closely interleaved in four quadrants and in wild-type animals there are no reported cytological differences between the single AB-derived cell and its many P1-derived cousins (Figure 5D, left diagram adapted from Sulston and Horvitz, 1977, Sulston et al., 1983, Moerman & Fire. C. elegans II, Chapter 16). In wild-type animals, all of these cells grow dramatically between the L1 and adult stages, with overall cell length increasing from about 20 microns in a newly hatched larva to over 100 microns two days later in adults. We first examined the mosaic progeny in which AB-derived cells were wild-type and all P1-derived cells were rpl-5(0) homozygous. We were able to label the AB-derived cell through the presence of a body-muscle-specific nuclear mCherry marker (pmyo2::mCherry) (Dibb et al., 1989), while labeling the full body wall muscle thick filament lattice with GFP-tagged functional MyosinB heavy chain (UNC54). The combined labeling allowed us to compare muscle filament structures in adjacent AB and P1-derived cells. We saw no difference in growth after two days, indicating that the AB-derived wild-type cells experience the same growth arrest as their RPL5-limited cousins (Figure 5D). Overall these results suggest the presence of a cell non-autonomous signal that inhibits postembryonic wild-type cell growth in mosaic larvae.
A common gene expression signature of zygotic ribosome deficiency
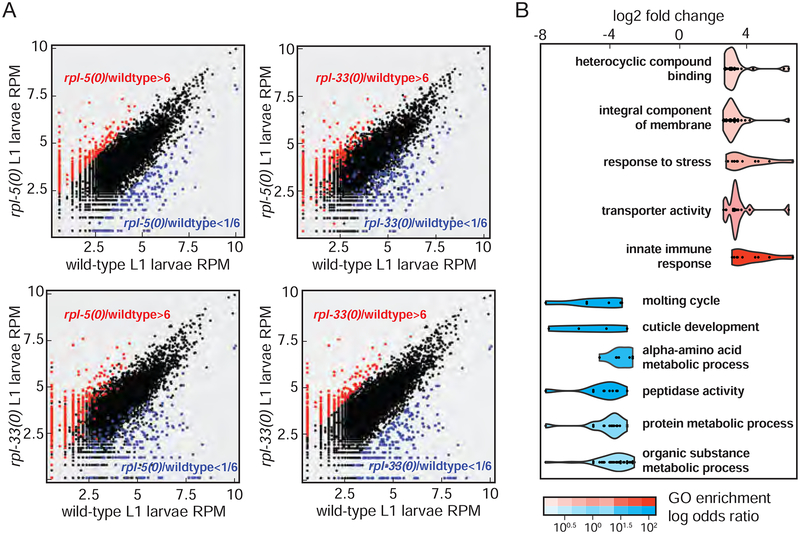
Toward exploring molecular processes accompanying cell non-autonomous signals, we applied RNA sequencing to wild-type and rpl null larvae. Homozygous rpl null mutants, rpl-5(0) and rpl-33(0), were analyzed, revealing considerable similarity between their global expression profiles, with numerous shared differences compared to wild-type. We identified an overlapping set of over- or under-expressed genes in these RP mutants (Figure 6A). Categories that were overexpressed in ribosome-limited conditions include stress response genes (e.g.; numr-1/2, gst-24, fbxa-78, dnj-15, zip-10, hil-1) (exemplary coverage plots in Figure S3), membrane transporters (e.g.; tat-6, unc-47, slcf-1) and genes that are involved in innate immunity (Figure 6B, Table S2). Under-expressed genes include molting cycle related genes (e.g.; myrf-1, qua-1, Figure 6B, and Table S2, exemplary coverage plots in Figure S4); suggesting an underlying arrest mechanism and lack of molting as a consequence of ribosome insufficiency.
Figure 6: A shared gene expression response in rpl-5 and rpl-33 null homozygous larvae.
A. Comparison of RNA abundance in wild type and RP mutant larvae. Read counts per gene in wild-type and rpl null conditions are shown. Each point on each graph indicates counts for a single gene in two conditions. Points along a single diagonal would indicate comparable read counts in the two samples; below-diagonal and above-diagonal points are indicative of genes which are outliers in their relative expression between the two conditions. Top two graphs: RNA-seq counts per million reads (RPM) compare rpl-5(0) on the y axis to wild-type L1 larvae on the x axis. Bottom two graphs highlight the same for rpl-33(0) on the y axis compared to wild-type L1 larvae on the x axis. Genes that were at least 6 fold up (red) or down (blue) in a mutant RP (rpl-5(0) on the left, rpl-33(0) on the right) compared to wild-type larvae are labeled (total minimum counts= 5 counts per million reads).
B. Gene Ontology analysis of differentially expressed transcripts. rpl null mutant larvae expression from two different datasets (rpl-5(0) and rpl-33(0)) were compared to wild-type larvae data using R limma package. RNA-seq libraries of each mutant and wild-type larval population were prepared and sequenced at separate times to ensure full biological replication. Genes that were significantly over or under expressed (>6-fold or <1/6-fold; (adjusted p-value cut-off = 0.002) were analyzed for significant gene ontology (GO) term enrichment (adjusted p-value cut-off = 0.05) using FuncAssociate 3.0 (Berriz et al., 2009). Selected categories are shown with a violin plot where the distribution of log2 fold change of each GO category is plotted. Points in each distribution represent the log2 fold change of each differentially expressed gene in the category. Complete list is in Table S2.
To understand whether some of the observed over- or under-expressed genes are differentially expressed due to developmental progression defects, we used public “ModEncode” data to assess gene-specific developmental profiles (E-MTAB-2812, Celniker et al. 2009). Among genes underexpressed in rpl null larvae, we observed a universal preferential expression at L2 stage compared to L1 (Figure S5). This result is suggestive of a failure to progress in larval development, concordant with the observed larval arrest phenotype. By contrast to the underexpressed genes, we did not note any universal stage specific patterns amongst genes overexpressed in rpl null genetic backgrounds, with some such genes elevated in earlier or later stages in comparison to L1 animals and some genes depleted in alternative stages (Figure S6). A number of the over and underexpressed genes suggested additional potential regulators of the ribosome depletion arrest (Figure S7).
One of the genes that is underexpressed in RP null larvae and over expressed in L2 larvae, elo-5, a long chain fatty acid elongation enzyme, is involved in the synthesis of monomethyl branched-chain fatty acids. RNAi knockdown of elo-5 results in L1 arrest that can be rescued by supplementing its product, 14-methyl hexadecanoic acid (Kniazeva et al., 2004). We tested the hypothesis that significant elo-5 downregulation in RP null larvae might underly the observed L1 arrest. To test this hypothesis, we supplemented animals with 14-methyl hexadecanoic acid and visualized postembryonic M cell division in RP null larvae. In a control experiment with elo-5 (gk208) strain, supplementation with 14-methyl hexadecanoic acid reduced larval arrest rate from 57% (17/30) to 14% (10/72)[2 sided bootstrap p value=1e-5] after 4 days. In contrast, no M cell division was observed in rpl-5 null arrested larvae when supplemented with 14-methyl hexadecanoic acid under similar conditions (observed arrest rate 100%(47/47) for supplemented rpl-5(0) and 100%(18/18) for non-supplemented DMSO control, p=1). These data suggest alternative or additional factors that play role in RP null mediated L1 arrest.
We next tested the hypothesis of similarity between the gene expression profiles of starvation induced L1 arrested and RP null L1 larvae. To test this hypothesis, we determined whether overlaps exist between the rpl(0) data and the published RNA expression data from starved animals (Stadler and Fire, 2013). Overlap between starved L1 and RP null L1 is limited such that a very small number of deregulated genes are shared between the RP null and starved animals (shared overexpressed genes: Y47H9A.1, Y38E10A.14, clec-75; shared underexpressed genes: spp-17, ilys-5), while most other gene expression changes are not shared.
Among the most significantly overexpressed genes in ribosome deficiency conditions–10 fold FDR 0.002–, we observed 5 genes that are specifically overexpressed in dauer larvae or post-dauer stages (Y45F10D.6, Y105C5A.8, fbxa–189, C29F9.6 and C07A4.2) as well as a set of (~21) genes that are not enriched in a specific developmental stage relative to L1 (eg. ZC266.1, nhr–102, unc–47, clec–75, tat–6, numr–1, tbb–6, gst–24). Among these genes, ZC266.1 is an ortholog of human RXFP4 (insulin-like family peptide receptor), nhr-102 is a nuclear receptor, unc-47 is a transmembrane vesicular GABA transporter, tat-6 is a subfamily IV P-type ATPse predicted to function as a flippase and gst-24 is an ortholog of human glutathione S-transfereases.
Insulin-like signaling plays an important role in the genetic regulation of starvation induced L1 quiescence in C. elegans (reviewed in Baugh, 2013). Two genes that have been identified in screens for downstream effectors of insulin-like signaling pathway are daf-16 and daf-18. We used alleles of the two genes, daf-16(mu86) and daf-18(ok480), that result in defective starvation induced L1 arrest. We reasoned that rpl-5(0), daf-16(mu86) or rpl-5(0), daf-18(ok480) larvae under fed conditions might exhibit incomplete M cell arrest, and tested this hypothesis by using the hlh8-GFP marker. We failed to observe any M cell divisions after 72 hours (23 and 44 observations for rpl-5(0), daf-18(ok480) and rpl-5(0), daf-16(mu86), respectively) (Figure S7). These results suggest that the daf-16/daf-18 insulin-like signaling pathway is insufficient to release limited ribosome mediated L1 arrest. In older starved plates (10 days of starvation), we observed rare instances with a single M cell division (observed in rpl-5(0), daf-18(ok480) and rpl-5(0), daf-16(mu86) animals).
L1 larval arrest phenotype in the absence of new 18S, 5.8S and 28S ribosomal RNA synthesis
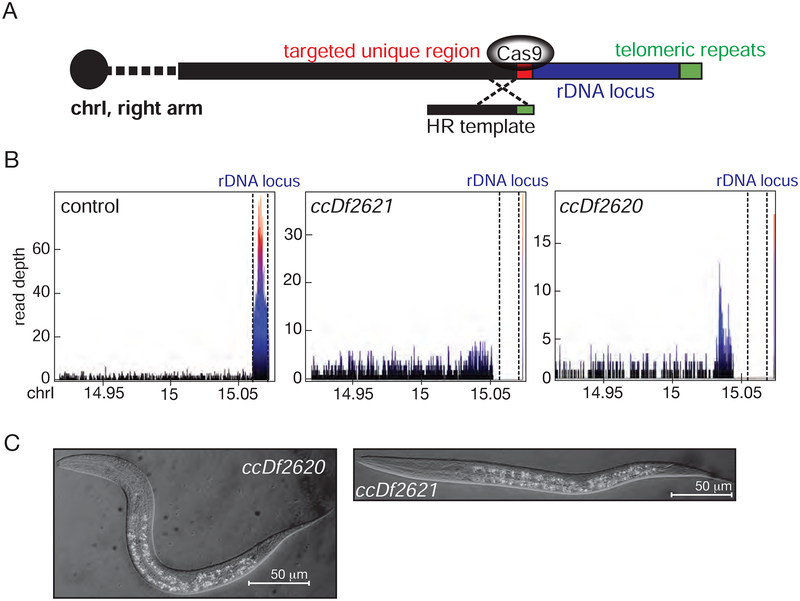
Deletions removing the entire 18S/5.8S/28S rDNA cluster offer an effective means to independently test the capacity of embryos to develop in the absence of new ribosome synthesis. Deletion of rRNA encoding loci at the DNA level will prevent new rRNA synthesis; given the requirements for nascent rRNA in ribosome assembly (Mougey et al., 1993), such deletions would be expected to illuminate on the issue of whether an embryo can fully avoid new ribosome synthesis. For the C. elegans rDNA loci, transcription is absent for a short time in the very early embryo (Korčeková et al., 2012). Later embryos reform nucleoli and show evidence for nascent rRNA chains (Korčeková et al., 2012, Henras et al., 2015). An advantage of C. elegans for such studies is the presence of all full copies of 18S, 5.8 and 28S rDNA at a single locus, transcribed as a single precursor from each copy in a tandem array of ~100–250 copy 7.2 kb repeat units (Files and Hirsh, 1981, Albertson, 1984, Ellis et al., 1986, Bik et al., 2013). To examine requirements for new ribosome synthesis in the embryo, we generated a full deletion of this locus (proximal to the right telomere of chromosome I of C. elegans). We used a CRISPR strategy that replaces the rDNA repeat array with a simple telomere (Figure 7A). Two independent chromosome I variants in which the entire rRNA locus is deleted were obtained (ccDf2620 and ccDf2621), with DNA sequencing confirming that both chromosomes have fully deleted the rDNA locus (Figure 7B). Homozygous progeny from both of the balanced rRNA deletion strains hatched and arrested as viable wild-type looking larvae (Figure 7C). The overall anatomy of the rDNA deletion larvae were similar to the RP mutations described above. Under differential interference contrast optics, numerous puncta within nuclei were visible but we couldn’t observe a distinct nucleolus in the cells of homozygous larvae.
Figure 7: Embryonic development can be completed in the absence of all functional copies of 18S, 5.8S, and 28S rDNA in the zygote.
A. The chromosomal locus containing all functional copies of 18S, 5.8S and 28S rDNA was deleted in two independent strains. The diagrammed Cas9-mediated break is repaired using a homologous recombination template that juxtaposes the desired 5’-deletion breakpoint to a set of putative subtelomeric sequences (annotated sequence from end of chromosome I).
B. Whole genome sequencing data from homozygous rDNA-deletion larvae and wild-type worms were generated and plotted using the R-Sushi Package (Phanstiel et al., 2014). The end of chromosome I where the rDNA locus is located is marked with dashed lines. No copies of the 18S/28S/5.8 S locus remain in homozygotes for both ccDf2620 and ccDf2621.
C. Differential interference contrast microscopy images of rRNA deletion homozygotes in whole larva.
Taken together these observations establish a sufficiency of maternally-produced ribosomal RNA in embryonic development, while confirming the role of the rRNA loci in the assembly of a functional nucleolus (Falahati et al., 2016) in this system.
Discussion
We find that new synthesis of distinct embryonic ribosomes is not required for tissue diversification during C. elegans embryonic development. We examined homozygous mutations eliminating six different ribosomal proteins (RPs) and constructed a chromosome eliminating the sole C. elegans locus encoding the 18S, 5.8S and 28S rRNAs. For all homozygous RPs, and for the full deletion of 18S/5.8S/28S cluster, we observe fully differentiated but arrested L1 larvae that appear surprisingly similar to wild-type larvae. Relying on maternally encoded ribosomal components, these larvae developed all tissue types that we analyzed (fully differentiated hypodermis and gut, a well formed cuticle, pharynx and body wall muscle, germline primordium, and differentiated neurons), and were capable of normal movement and behavioral responses. A subset of postembryonic cell divisions occurred before arrest suggesting that the arrest is distinct from starvation induced L1 quiescence. The arrested animals retain ribosomes composed of maternal components and are able to translate new proteins.
The capability of embryos to complete development in the absence of zygotically-encoded ribosomal components suggests a model where a sufficient number of maternal ribosomes are stockpiled in the oocyte and can subsequently be used for all embryonic processes. Our findings of complete tissue diversification and active translation in the absence of embryonic ribosomal components, as well as observation of minimal embryonic rRNA in active translation pools in the hatched larvae support this model. Storage of a maternal ribosome pool that would be used during embryonic development is prevalent in most animal eggs (Woodland, 1974, Bachvarova et al., 1981). Such prevalence evolutionarily makes sense given that ribosome maturation is a complicated and energy expensive process requiring stoichiometric and spatial-temporal coordination of ~80 different RPs and 4 rRNAs (Shajani et al., 2011, Panse and Johnson, 2010).
We find that ribosomal null and wild-type mosaic embryos arrest at a stage similar to ribosomal null mutant larvae suggesting the presence of a cell non-autonomous checkpoint. This addresses a key question of how organismal response to lack of new ribosomal components lead to L1 arrest. One possibility would have been a passive process in which insufficient capacity to meet each cells’ protein production demands causes a cell-autonomous arrest. Alternatively, coordination across various compartments of the organism might be involved in halting growth. We observed a consistent organism-wide arrest with both types of first-cleavage mosaic (P1-derived ribosomal deletion mixed with AB-derived wild-type, and AB-derived ribosomal deletion mixed with P1-derived wild-type). The consistent arrest phenotype seen with the two mosaic classes suggests the presence of a global cell non-autonomous checkpoint that inhibits further organismal development.
Embryonic development requires tight growth coordination between different parts of the organism. Thus, a systemic feedback, including cell non-autonomous growth regulation, could exist to maintain such synchrony in growth. Ribosome machinery mutations in C. elegans provide a distinct model to probe the coordination of cellular growth; given that ribosomes determine a cell’s capacity to make proteins required for growth (Lempiainen and Shore, 2009). The coordination of growth in response to different doses of RPs (and hence ribosomes) could also be of considerable importance in understanding human ribosomapathy disorders. These disorders result from haploinsufficiency of ribosomal components or ribosome maturation factors, but penetrance even amongst genetic heterozygotes is variable or incomplete. Healthy family members can carry the same mutation (e.g., 43% silent carriers, Moetter et al., 2011) as patients and certain tissues are affected more than others (shared phenotype of bone marrow failure in the case of Diamond Blackfan Anemia) (Carlston et al., 2017, Ellis and Lipton, 2008, Farrar and Dahl, 2011). C. elegans may provide a paradigm and model for studying genetic influences, checkpoint control, and other modulating factors in ribosomopathy.
Neuronal development and maintenance are highly specialized processes that rely on the dynamic regulation of protein synthesis, with dysfunction present in many neurodegenerative diseases (Kapur et al., 2017). As an intriguing possibility, could ribosome machinery specializations explain the dynamic regulation under physiological circumstances or deregulation in disease? Localized heterogeneity in translational output and kinetics is observed for distinct subcellular regions such as dendrites (Glock et al., 2017, Wang et al., 2016). Here, we investigated how a failure in new ribosome synthesis affects embryonic and postembryonic neuronal development, anatomy and repair. For a subset of neurons born during embryogenesis and readily labeled (mechanosensory neurons), we observed what appears to be normal development in the absence of embryonic ribosomes; further, homozygous RP null larvae had a normal touch response indicating normal function of these neurons (Chalfie et al., 1985). Overall these results suggest that embryonic ribosomes are dispensable for extensive embryonic neuronal development and for at least a subset of post-development neuronal functions. By contrast, postembryonic mechanosensory neuron development (axon tiling) and their response to injury (axotomy) were significantly impaired in the absence of new ribosome synthesis. At present, we do not know the degree to which these effects are specific–are these processes arrested due to compromised growth, due to systemic signals of ribosome insufficiency, or as a result of specific requirements for modified ribosomes for which new ribosomal RNA and RP synthesis might be needed.
Beyond the accessible C. elegans system, how general might the sufficiency of a pre-existing pool of ribosomes for embryonic development be? With mid-20th-century knowledge that the nucleolus is required for ribosome maturation (Warner, 1990), and that a class of haploinsufficient mutants–and deficiencies–(“minutes”) in Drosophila melanogaster affect RPs and ribosomal components (Kongsuwan et al., 1985, Lambertsson, 1998, Marygold et al., 2007) we can derive some suggestive observations from much earlier classic genetic literature. Indeed the possibility of embryonic development in the absence of embryonically-produced ribosomes arises from early 20th century studies in Xenopus embryos seemingly lacked nucleoli but had smaller nuclear blobs (Wallace, 1960). Similarly, a majority of Drosophila minute homozygotes develop as first instar larvae–despite the presence of homozygous minutes that are also embryonic lethal–suggesting a potential for extensive embryonic development in the absence of zygotic RPs (Brehme, 1939, Farnsworth, 1957a, Farnsworth, 1957b). Some of the limitations in drawing conclusions from these earlier observations are: (i) the presence of residual rDNA copies, (ii) generalized defects in the early mutant homozygotes, with overall lack of vitality in anucleolate Xenopus embryos (Steele et al., 1984, Wallace, 1960), and a small and sluggish character of Drosophila minute homozygotes (Brehme, 1939, Farnsworth, 1957a, Farnsworth, 1957b). Despite such limitations, these studies are suggestive of at least partial tissue diversification in the absence of embryonically-synthesized ribosomes in other systems. The combination of those observations with the remarkable developmental sufficiency of the maternal ribosome pool in C. elegans suggests that an ability to progress through development using an existing ribosomal pool may be a common feature in a variety of biological contexts. Thus, our results suggest that any potential function of ribosome heterogeneity in embryonic development does not depend on new ribosome synthesis. Ribosome heterogeneity that can potentially stem from “post-synthesis” modifications of rRNAs and RPs or post-assembly substitutions in protein composition of ribosomes might instead play functional roles in tissue diversification, as could the many translational control mechanisms that modulate activity of an existing ribosomal pool.
STAR Methods
Contact for reagent and resource sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Andrew Fire (afire@stanford.edu)
Experimental model and subject details
C. elegans strains were kept and grown at 16°C or 20°C on agar plates containing NGM growth media (RPI Research Products International, N81250–5000) seeded with E. coli strain OP-50 (Brenner, 1974). A subset of C. elegans strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). A complete list of strains is provided (Key Resources Table). To obtain newly hatched and fed L1 larvae, selected adult worms were transferred onto a fresh plate and removed after 2 hours of egg laying. The eggs were allowed to develop at 16°C for 16 hours in the presence of food (bacterial strain OP50; Brenner 1974).
Key Resources Table
| Reagent or Resource | Source | Identifier |
|---|---|---|
| Experimental Models: Organisms/Strains | ||
|
C. elegans strain:
rps-30[cc2530,5LX], unc-60(gk239)/nT1 [qIs51] (IV;V) |
This study | PD2530 |
|
C. elegans strain:
rpl-33[cc2558,R9X]/mIn1 [dpy-10(e128) mIs14] (II) |
This study | PD2558 |
|
C. elegans strain:
rps-23[cc5994,A67X]/nT1 [qIs51] (IV;V) |
This study | PD5994 |
|
C. elegans strain:
rps-23[cc5995,A67X]/nT1 [qIs51] (IV;V) |
This study | PD5995 |
|
C. elegans strain:
rpl-5 [cc5998,A166X]/mIn1 [dpy-10(e128) mIs14] (II) |
This study | PD5998 |
|
C. elegans strain:
rps-10[cc2557,T8X]/hT2 [bli-4(e937) let-? (q782) qIs48] (I;III) |
This study | PD2557 |
|
C. elegans strain:
ccDf2620/unc-54(e1152) |
This study | PD2620 |
|
C. elegans strain:
ccDf2621/unc-54(e1152) |
This study | PD2621 |
|
C. elegans strain:
hsp-16. 2(hsp-16. 2_del::GFP)V; hsp-16. 41(hsp-16.41_unknown) |
This study | PD6443 |
|
C. elegans strain:
zdIs5 [mec-4p::GFP + lin-15(+)] I; rpl-5 [cc5998,A166X]/mIn1 [dpy-10(e128) mIs14] (II) |
This study, from Yishi Jin, UCSD | CZ26400 |
|
C. elegans strain:
rps-10[cc2557,T8X]/hT2 [bli-4(e937) let-? (q782) qIs48] (I;III); muIs32(mec-7p::GFP) II |
This study, from Yishi Jin, UCSD | CZ26401 |
|
C. elegans strain:
muIs32(mec-7p::GFP) II; rps-23[cc5995,A67X]/nT1 [qIs51] (IV;V) |
This study, from Yishi Jin, UCSD | CZ26402 |
|
C. elegans strain:
muIs32(mec-7p::GFP) II; rps-30[cc2530,5LX], unc-60(gk239)/nT1 [qIs51] (IV;V) |
This study, from Yishi Jin, UCSD | CZ26403 |
|
C. elegans strain:
zdIs5 [mec-4p::GFP + lin-15(+)] I; rpl-33[cc2558,R9X]/mIn1 [dpy-10(e128) mIs14] (II) |
This study, from Yishi Jin, UCSD | CZ26404 |
|
C. elegans strain:
unc-54(unc-54::gfp)I |
Joshua Arribere, UCSC | PD2855 |
|
C. elegans strain:
ccTi1594 [mex-5p::GFP::gpr-1::smu-1 3’UTR + Cbr-unc-119(+), III: 680195] III. hjSi20 [myo-2p::mCherry::unc-54 3’UTR] IV. |
Karen Artiles, Stanford University; Christian Frøkjær-Jensen, KAUST |
PD2217 |
|
C. elegans strain:
ayIs6 [hlh-8::GFP fusion + dpy-20(+)] |
Andrew Fire, Stanford University | PD4666 |
|
C. elegans strain:
rpl-5 [cc5998,A166X]/mIn1 [dpy-10(e128) mIs14] (II); daf-16 (mu86) I. |
This study | PD2642 |
|
C. elegans strain:
rpl-5 [cc5998,A166X]/mIn1 [dpy-10(e128) mIs14] (II); daf-18 (ok480) IV. |
This study | PD2625 |
|
C. elegans strain:
ccTi1594[Pmex-5 GFP-gpr-1 smu-1UTR]; hjSi20 [myo-2p::mCherry::unc-54 3primeUTR] IV. |
Karen Artiles, Stanford University; Christian Frøkjær-Jensen, KAUST | PD2217 |
|
C. elegans strain: My14 Wild isolate from Hinrich Schulenburg |
Caenorhabditis Genetics Center | WB Cat# MY14, RRID:WB-STRAIN:MY14 |
|
C. elegans strain:
tra-2(q122) II (Goodwin et al., 1993) |
Caenorhabditis Genetics Center | WB Cat# JK726, RRID:WB-STRAIN:JK726 |
|
C. elegans strain:
ptp-3(mu245) muIs32 [mec-7p::GFP + lin-15(+)] II, from Cynthia J Kenyon |
Caenorhabditis Genetics Center | WB Cat# CF1756, RRID:WB-STRAIN:CF1756 |
|
C. elegans strain:
zdIs5 [mec-4p::GFP + lin-15(+)] I, from Andrew D Chisholm |
Caenorhabditis Genetics Center |
WB Cat# CZ10175, RRID:WB-STRAIN:CZ10175 |
| C. brenneri strain from Jonathan A Hodgkin | Caenorhabditis Genetics Center | WB Cat# CB5161, RRID:WB-STRAIN:CB5161 |
|
C. elegans strain:
hjSi20 [myo-2p::mCherry::unc-54 3’UTR], from Ho Yi Mak |
Caenorhabditis Genetics Center | WB Cat# VS21, RRID:WB-STRAIN:VS21 |
|
C. elegans strain:
rps-18(ok3353) IV/nT1 [qIs51] (IV;V) |
Caenorhabditis Genetics Center | WB Cat# VC2638, RRID:WB-STRAIN:VC2638 |
|
C. elegans strain:
daf-18 (ok480) IV from OMRF Knockout Group |
Caenorhabditis Genetics Center | WB Cat# RB712, RRID:WB-STRAIN:RB712 |
|
C. elegans strain:
elo-5 (gk208) IV from Vancouver KO Group |
Caenorhabditis Genetics Center | WB Cat# VC410, RRID:WB-STRAIN:VC410 |
|
C. elegans strain:
daf-16 (mu86) I from C. Kenyon and K. Kin |
Caenorhabditis Genetics Center | WB Cat# CF1038, RRID:WB-STRAIN:CF1038 |
|
C. elegans strain:
unc-54(e1152) I. from Robert H Waterston |
Caenorhabditis Genetics Center | WB Cat# CB1152, RRID:WB-STRAIN:CB1152 |
| E. coli strain: OP-50 | Caenorhabditis Genetics Center | WB Cat# OP50, RRID:WB-STRAIN:OP50 |
| Chemicals, Recombinant Proteins | ||
| TRIzol Reagent | ThermoFisher Scientific | 15596026 |
| Triton X-100 | Sigma-Aldrich | X100–500ML |
| Cycloheximide | Sigma-Aldrich | C7698–1G |
| Ribonucleoside vanadyl complexes | Sigma-Aldrich | 94740–250MG |
| 1-phenoxy-2-propanol | TCI America | P0118 |
| 14-methyl hexadecanoic acid | Sigma-Aldrich | M3164 |
| Levamisole | Sigma-Aldrich | L0380000 |
| Hybridase Thermostable RNaseH | Lucigen | H39500 |
| TURBO DNase | ThermoFisher Scientific | AM2238 |
| T4 polynucleotide kinase | NEB | MS201S |
| Proteinase K | Agilent | S3004 |
| Ambion RNase I | ThermoFisher Scientific | AM2294 |
| S.p. Cas9 Nuclease V3 | Integrated DNA Technologies | 1081058 |
|
Critical Commercial Assays |
||
| SMARTer Stranded RNA-Seq Kit | Clontech, Takara | 634838 |
| SMARTersmRNA-Seq Kit for Illumina | Clontech, Takara | 635029 |
| Agencourt AMPure XP | Beckman Coulter | A63880 |
| Qubit dsDNA HS Assay Kit | ThermoFisher Scientific | Q32851 |
| MiSeq Reagent Kit v3 (150-cycle) |
Illumina | MS-102–3001 |
| MiSeq Reagent Kit v2 (50-cycles) | Illumina | MS-102–2001 |
| Nextera DNA Library Preparation Kit | Illumina | FC-121–1030 |
| 10% TBE-Urea Gel | ThermoFisher Scientific | EC6875BOX |
| Deposited Data | ||
| RNAseq | This study | NCBI SRA, BioProject ID PRJNA509065 |
| Riboseq | This study | NCBI SRA, BioProject ID PRJNA509065 |
| Genomic DNA sequencing | This study | NCBI SRA, BioProject ID PRJNA509065 |
| C. elegans RNAseq from starved L1 larvae (Stadler & Fire 2013) | Gene Expression Omnibus | GSE48140 |
| C. elegans transcriptome from various developmental stages (Celniker et al. 2009) | ArrayExpress | E-MTAB-2812 |
| Structure of the Human 80S ribosome (Khatter et al., 2015) | Protein Data Bank | 4UG0 |
| Oligonucleotides | ||
| Oligonucleotides, guide RNAs, gBlocks Gene fragments | Integrated DNA Technologies | Table S3 |
| Recombinant DNA | ||
| Empty vector for gRNA cloning, pRB1017 | Addgene | 59936 |
| Software and Algorithms | ||
| ImageJ | Imagej.nih.gov | RRID:SCR_003070 |
| ZEN Digital Imaging for Light Microscopy | Zeiss | RRID:SCR_013672 |
| Axioskop | Zeiss | RRID:SCR_014587 |
| Integrative Genomics Viewer, IGV version 2.3.92 | https://software.broadinstitute.org/software/igv/RelNotes2.3.x | RRID:SCR_011793 |
| Tophat2 | https://ccb.jhu.edu/software/tophat/index.shtml | RRID:SCR_013035 |
| Bowtie 2 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml | RRID:SCR_016368 |
| Picard toolkit | http://broadinstitute.github.io/picard | RRID:SCR_006525 |
| SAMtools | http://samtools.sourceforge.net/ | RRID:SCR_002105 |
| featureCounts | http://subread.sourceforge.net/ | RRID:SCR_012919 |
| cutadapt 2.0 | https://cutadapt.readthedocs.io/en/stable/index.html | RRID:SCR_011841 |
| RStudio 0.98.501 | https://www.rstudio.com/ | RRID:SCR_000432 |
| limma, BioConductor | https://www.bioconductor.org/packages/release/bioc/html/limma.html | RRID:SCR_010943 |
| Sushi 3.8, Bioconductor (Phanstiel et al., 2014) | https://www.bioconductor.org/packages/release/bioc/html/Sushi.html | N/A |
| ggplot | https://cran.r-project.org/web/packages/ggplot2/index.html | N/A |
| FuncAssociate 3.0 (Berriz et al., 2009) | http://llama.mshri.on.ca/funcassociate/ | RRID:SCR_005768 |
| Gene Ontology | http://www.geneontology.org/ | RRID:SCR_002811 |
| Adobe Photoshop CS6 | Adobe | RRID:SCR_014199 |
| Adobe Illustrator CS6 | Adobe | RRID:SCR_010279 |
| Other | ||
| Nikon Eclipse E6000, 40× or 100× oil DIC objective | Nikon | N/A |
| Olympus BH-2, 40× or 100× oil DIC objective | Olympus | N/A |
| LSM 800, LD C-Apochromat 40× | Zeiss | N/A |
| LSM 510, 40× or 63× oil DIC objective | Zeiss | N/A |
| LSM 710, 40× or 63× oil DIC objective | Zeiss | N/A |
| Miseq System | Illumina | RRID:SCR_016379 |
| Density fractionation system | Brandel | BR-188–176 |
Method Details
Construction of C. elegans strains
Ribosomal protein loss of function mutations were generated via CRISPR-Cas9 genome editing in N2 background (PD1074) (Arribere et al., 2014) with the exception of rps-30 which was generated in a unc-60(gk239)/nT1 [qIs51] (IV;V) background (The C. elegans Deletion Mutant Consortium., 2012). The mutated strains were kept in a balanced heterozygote background (+/nT1 [qIs51] (IV;V); +/mIn1 [dpy-10(e128)] (II); +/hT2 [bli-4(e937) let-?(q782) qIs48] (I;III) (McKim et al., 1993, Ferguson and Horvitz, 1985, Edgley and Riddle, 2001, Edgley et al., 2006). Stop codons were introduced in the first exon to generate loss of function mutations. The presence of mutations was assessed by PCR of genomic DNA and Sanger sequencing. Oligonucleotides used for the generation and assessment of mutations are provided in Table S3. One-fourth of the progeny from simple heterozygous strains [balanced using the intrachromosomal balancer mIn1] are expected to be homozygous (Figure 2B). For the interchromosomal balancers (nT1 and hT2 in this case), the fraction of homozygotes amongst zygotes formed will be lower (1/16) (Edgley et al., 2006). For this reason, we counted the progeny of simple heterozygotes obtained as needed through outcross of the long-term-maintenance (balanced) strains (Table S1).
To construct a precise rDNA deletion, we targeted a unique region for CRISPR-Cas9 editing between coding genes and the repeated rDNA locus. After 20 uM Cas9 and crRNA:tracrRNA duplex was incubated for 5 minutes at room temperature, it was mixed with a single stranded repair template for a final concentration of 15 uM guide RNA/cas9 complex and 6 uM repair template. Resulting mix was injected into gonads of young hermaphrodites. Targeted DNA was repaired using a homologous template (single stranded repair template) that juxtaposes sequences just to the left of rDNA sequence to a subtelomeric sequence from the reference genome.
GFP was inserted at the C-terminus of unc-54. F1 animals were visually inspected for GFP expression in the body wall muscle. Integration events were confirmed by PCR and sequencing. hsp-16.2/41 locus was targeted with multiple guide RNAs and an HR template vector where hsp-16.2/41 genes were substituted with GFP. F1 animals were heat-shocked and animals expressing GFP were selected. Progeny were continuously selected until a population with true-breeding heat shock GFP was isolated. Oligonucleotides used for the construction of the strains as well as detection of the mutations are provided in Table S3.
gpr-1(overexpression) strains and the application of pharyngeal and body muscle-muscle-specific GFP markers to identify mosaic animals is described in detail (Artiles et al. 2018, co-submitted).
Sample and library preparation for RNA sequencing
Synchronized larvae were collected in 50 mM NaCl and were cleaned from OP-50 bacteria by sedimentation through a 5% sucrose cushion including 50 mM NaCl (<1g for 1 minute) After sucrose clean up of bacteria, >100 L1 larvae were lysed in 1 ml TRIzol (ThermoFisher Scientific), vortexed for 25 min in the cold room followed by a 5 min incubation at room temperature. To extract RNA, 200 μl volume of chloroform was added, then the sample was mixed and spun at 13K rpm for 10 min. Aqueous layer was used for further RNA precipitation. Isolated RNA was isopropanol precipitated and 80% ethanol washed. Thermostable RNAseH (Lucigen) and a pool of 94 DNA oligonucleotides anti-sense to C. elegans ribosomal RNA were used to deplete rRNA from 100 ng total C. elegans RNA (Arribere et al., 2016). RNA-seq libraries were prepared using SMARTer Stranded RNA-Seq kit (Clontech). Initially, RNA was alkaline fragmented at 95°C for 4 min followed by the protocol optimized <10 ng RNA input. 12–14 cycles of PCR were used to amplify the sequences. Library DNA was then purified using Agencourt AMPure XP beads (Beckman Coulter). The resulting libraries were quantified with Qubit dsDNA HS Assay Kit (ThermoFisher Scientific) and sequenced on a MiSeq system using the MiSeq reagent version 3 kit to yield 2×75 base-pair reads (Illumina).
Sample and library preparation for Riboseq
~100 newly hatched L1 larvae were flash frozen in 20 mM Tris-HCl pH 7.4, 150 mM NaCl, 5 mM MgCl2 and ground in liquid nitrogen with mortar and pestle. The frozen worm powder was thawed on ice and mixed with 5 mM DTT, 1% Triton X-100, 100 μg ml−1 cycloheximide (Sigma Aldrich) and 5 U ml−1 Turbo DNase (ThermoFisher Scientific). This lysate was vortexed briefly, and 1 U of RNAseI (ThermoFisher Scientific) is added per ug of RNA (measured by A260), followed by incubation for 30 min at room temperature. The RNAseI reaction was stopped by addition of 15 mM ribonucleoside vanadyl complexes (Sigma Aldrich). The lysate was loaded onto 34% sucrose in 20 mM Tris-HCl pH 7.4, 150 mM NaCl, 5 mM MgCl2 and 5 mM DTT, and spun at 70,000 rpm using TLA 120.2 rotor (Beckman Coulter, 357656) for 4 hours. The resulting pellet was solubilized in 1 ml of Trizol (ThermoFisher Scientific). To extract RNA, 200 μl chloroform was added, the sample was mixed and spun at 13K rpm for 10 min. The aqueous layer was used for further isopropanol precipitation in the presence of 30 mM Na-Acetate(pH 5.5), and 5 mM MgCl2, with an equal volume of isopropanol. Pellets were washed with 80% ethanol before solubilizing the RNA in nuclease free water. Resulting RNA was resolved on 10% Acrylamide TBE-urea gel (ThermoFisher Scientific), fragments between 26 and 34 nucleotide were cut. Gel pieces were nutated in 30 mM Na-Acetate (pH 5.5) solution overnight in the cold room, with the eluate then precipitated as described above. For 3’ dephosphorylation, 1 ul T4 polynucleotide kinase (PNK) (NEB) was added in a 15 ul reaction in the presence of 1× T4 PNK buffer without ATP. The reaction was incubated at 37°C for 1 hour. Resulting RNA was precipitated as described above and libraries were generated using SMARTer smRNA-Seq Kit (Clontech). The resulting libraries were quantified with Qubit dsDNA HS Assay Kit (ThermoFisher Scientific) and Agilent Bioanalyzer 2100. Libraries were sequenced on a MiSeq system using the MiSeq reagent version 3 kit to yield 1×75 base-pair reads or version 2 kit to yield 1×50 base-pair reads (Illumina).
RNA-seq data analysis
After trimming and de-multiplexing using standard Illumina software, the reads were further trimmed to remove the non-templated ~5-base G stretch that was introduced in the library preparation. A minimum read length of 20 base-pairs was enforced after trimming nucleotides with sequence quality less than 30 (cutadapt 2.0). Trimmed reads were mapped to C. elegans genome (Ensembl70–WBcel235) using Tophat2 paired end settings with the flag:--b2-sensitive. Reads containing the same start and stop mapping positions (possible PCR duplicates) were collapsed to single reads using the Picard toolkit. For additional analyses, reads with a mapping quality score (MAPQ >= 20) were selected from the alignment files using SAMtools. RStudio version 0.98.501 was used to visualize the data and for statistical analysis. Differentially expressed genes between ribosomal protein (RP) null larvae compared to wild-type were determined using the limma package. Gene ontology (GO) category enrichment analysis (adjusted p-value cut-off = 0.05) was carried out using FuncAssociate 3.0 (Berriz et al., 2009). Specifically, all genes that were used for gene expression analysis was provided as the genespace file. Differential expression criteria for the GO analysis were (i) adjusted p-value < 0.002, and (ii) at least 6-fold over- or under-expression relative to mean wild-type levels compared to all rpl null replicates as calculated by the limma Package. Starved and fed L1 larvae datasets were included (Stadler and Fire, 2013) and batch corrected for this comparison using R ComBat package. List of differently expressed genes and significantly enriched gene ontology terms for over and under expressed genes are provided in Table S2. RNA-Seq data from the modENCODE project was retrieved from ArrayExpress (E-MTAB-2812, Celniker et al. 2009). For each gene, the log2 of the ratio of expression between the different developmental states and L1 larvae was calculated. Genes that were over or underexpressed at least 10 fold with an adjusted p value of 0.002 were plotted. A heatmap representing these ratios was drawn in R using the gplots library heatmap.2 function. Darkest blue color indicates undetectable expression in the given stage (Figure S5, S6).
Ribo-seq data analysis
After de-multiplexing using standard Illumina software, the reads were further 3’ trimmed to remove polyA extension that was introduced in the library preparation. A minimum read length of 20 base-pairs was enforced after trimming nucleotides with sequence quality less than 30 (cutadapt 2.0). Trimmed reads were mapped to the C. elegans genome (Ensembl70–WBcel235) using Tophat2 paired end settings with the flag:--b2-sensitive. For metagene analysis, start and stop codon positions for each protein-coding transcript isoform was extracted from Ensembl70–WBcel235 annotation (Arribere & Fire 2018). Closest distance to the start or stop codon of a spliced transcript was calculated for start position of each ribo-seq read mapping to a given protein-coding transcript. Relative density of start positions of read counts were plotted in +50 and–50 nucleotide proximal window (Figure 4B).
Ribosomal RNA analysis of monosome and polysome fractions
L1 larvae were grown as described above. Animals were liquid nitrogen flash frozen in polysome lysis buffer as described (Arribere et al., 2016) and ground in liquid nitrogen (with mortar and pestle). L1 staged cross progeny of (My14 males and N2 (tra-2(q122) females or N2 males (mIn1) and My14 hermaphrodites)) (Goodwin et al., 1993) were mixed with a C. brenneri lysate (staged L4), which was used as a carrier. The frozen worm powder was thawed on ice and solubilized in polysome lysis buffer that was supplemented with 5 mM DTT, 15 mM ribonucleoside vanadyl complexes (Sigma Aldrich). Lysates were loaded onto 10–60% sucrose gradients and spun for 3.5 hours at 35,000 rpm using SW41 Ti rotor in an ultracentrifugation system (Beckman Coulter). RNA from monosome and polysome peaks was isolated using a density fractionation system (Brandel), and RNA-seq libraries were prepared as described above from equivalent amount of input RNA without an initial rRNA depletion step. The data was used for the analysis in Figure 1C.
DNA library preparation and data analysis
Larvae were synchronized as above. The collected worms were lysed at 65°C for 3 hours with Proteinase K (Agilent, S3004) in 10 mM Tris pH-8, 50 mM KCl, 2 mM MgCl2. DNA extraction was done using phenol-chloroform and ethanol precipitation. DNA was dissolved in TE buffer (10 mM Tris pH-8, 1 mM EDTA). Nextera DNA library Prep kits (Illumina) were used for genomic library preparation, starting with 50 ng genomic DNA. Final DNA libraries were gel extracted to yield a size distribution around 500 base pairs. Libraries were sequenced on a MiSeq machine as described above. The resulting 75 base-pair paired end reads were trimmed and de-multiplexed by the Illumina software. The reads were aligned to C. elegans genome (UCSC version ce10) using Bowtie2. Reads were visualized using Integrative Genomics Viewer (IGV version 2.3.92). The data was used for the colored chromosome histogram depicted in Figure 7B (R, Sushi Package, Phanstiel et al., 2014).
Microscopy and image analysis
Animals were immobilized on 1% agar slides with 50 mM NaCl and 1% 1-phenoxy-2-propanol. Images were obtained using Nikon Eclipse E6000 or Olympus BH-2 equipped with Differential interference contrast (DIC) optics (Figures 2D, 2F, 2G, 4A left panel, 5B, 5C 7C, S1B, S1C, S2, S7). Brightness and contrast of all DIC images were adjusted using Adobe Photoshop CS6 to enhance visible contrast. To image GFP (Figures 2G, 4A right panel, 5B, S1B, S2, S7,), a mercury lamp and Nikon filter cube 96342 were used. Images were taken with a 5.1 MP camera (OMAX, A3550U3). Body wall muscle GFP and mCherry images were captured using ZEISS LSM 800 on ZEISS Axio Examiner.Z1 (Figure 5D). The organism length was calculated by measuring the length of a segmented line that connects the mouth opening to the tail, images were processed and measured using ImageJ (Rueden et al., 2017). An “England finder” graticule slide was used to deduce absolute length on the images, and to provide scale bars.
Touch receptor neuron observation, confocal microscopy and Laser axotomy
Parallel staged worms were mounted onto 2% agarose pads in M9 solution (Figure 3A), 0.6 mM levamisole (Figure 3C) or 1% 1-phenoxy-2-propanol (Figure 3D) (TCI America), and touch receptor neuron morphology and position were scored under 63× lens on Zeiss Axioskop. Mounting media made no difference in TRN neuron development. Laser axotomy was performed as previously described (Wu, 2007). Images in Figure 3 were acquired using LSM510 (Figure 3D) or LSM710 (Figure 3A, 3C) (Zeiss Plan Apochromat 40× or 63× oil DIC objective) confocal microscope controlled by ZEN software (Zeiss).
Fatty acid complementation assay
Fatty acid phenotypic rescue was assessed by supplementing animals with 14–9methyl hexadecanoic acid (Kniazeva et al. 2004). Stock solution of 14-methyl hexadecanoic acid (Sigma Aldrich) was prepared in DMSO. A 10 mM solution was diluted to 1/10 ratio in 300 ul OP-50 bacterial culture in 2xTY media, and 30 ul of this mixture was plated onto NGM plates and incubated overnight. For controls, plates with same ratio of DMSO/OP-50 were used. Bleached eggs were placed onto fatty acid supplemented and control NGM plates. Growth or M cell division was monitored after 4 days.
Quantification and Statistical Analysis
We used a bootstrapping approach to test the statistical significance of the observed difference in My14 to N2 reads across the sucrose gradient fractions (Figure 1) as well as M cell division analysis. For sucrose gradient fractions, we resampled with replacement the same number of reads in each given fraction using the observed read counts in the total RNA sample (56 My14, and 254 N2 reads). We repeated the sampling one million times and calculated the p-value as the fraction of bootstrap samples with the same or less number of My14 reads than observed.
Statistical significance of differentially expressed genes between ribosomal protein (RP) null larvae compared to wild-type was determined using the R-limma package (Figure 6, Table S2). Significant Gene ontology (GO) category enrichment analysis was carried out using FuncAssociate 3.0 (Figure 6, Table S2) (Berriz et al., 2009).
Data Availability
The RNA-seq, Ribo-seq and DNA sequencing data are deposited to NCBI SRA with BioProject ID PRJNA509065.
Supplementary Material
Highlights.
Functional ribosomes are predominantly maternal during C. elegans embryogenesis.
Embryogenesis is complete in the absence of zygotic ribosomal component contributions.
Additional ribosome synthesis is needed for postembryonic plasticity of neuronal structure.
Ribosomal insufficiency in a subset of cells triggers organism-wide growth arrest.
Acknowledgements
We thank C. Frokjar-Jensen and K. Artiles for gpr-1(OE) strains, and Z. Wu for laser axotomy. We thank Fire lab members, A. Villeneuve, P. Sarnow, M. Kay, K. Sakomoto, V. Walbot, and A. Narla for critical reading of the manuscript and/or critical discussions. We thank B. Wang and P.Sarnow for letting us use their lab’s microscopy system, and gradient fractionator respectively. This work was supported by NIH (Grants GM37706/GM130366 to A.Z.F., NS035546 to Y.J.) and Walter and Idun Berry Foundation (to E.S.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- Albertson DG, Sulston JE & White JG 1978. Cell cycling and DNA replication in a mutant blocked in cell division in the nematode Caenorhabditis elegans. Dev Biol 63: 165–178. [DOI] [PubMed] [Google Scholar]
- Albertson DG 1984. Localization of the ribosomal genes in Caenorhabditis elegans chromosomes by in situ hybridization using biotin-labeled probes. EMBO J 3: 1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere JA, Bell RT, Fu BX, Artiles KL, Hartman PS & Fire AZ 2014. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198: 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere JA, Cenik ES, Jain N, Hess GT, Lee CH, Bassik MC & Fire AZ 2016. Translation readthrough mitigation. Nature 534: 719–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere JA & Fire AZ 2018. Nonsense mRNA suppression via nonstop decay. Elife 7: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L & Horvitz HR 1990. Effects of starvation and neuroactive drugs on feeding in Caenorhabditis elegans. J Exp Zool 253: 263–270. [DOI] [PubMed] [Google Scholar]
- Bachvarova R, De Leon V & Spiegelman I 1981. Mouse egg ribosomes: evidence for storage in lattices. J Embryol Exp Morphol 62: 153–164. [PubMed] [Google Scholar]
- Ban N, Beckmann R, Cate JH, Dinman JD, Dragon F, Ellis SR, Lafontaine DL, Lindahl L, Liljas A, Lipton JM, Mcalear MA, Moore PB, Noller HF, Ortega J, Panse VG, Ramakrishnan V, Spahn CM, Steitz TA, Tchorzewski M, Tollervey D, Warren AJ, Williamson JR, Wilson D, Yonath A & Yusupov M 2014. A new system for naming ribosomal proteins. Curr Opin Struct Biol 24: 165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat A, Szick-Miranda K, Chang IF, Guyot R, Blanc G, Cooke R, Delseny M & Bailey-Serres J 2001. The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genome. Plant Physiol 127: 398–415. [PMC free article] [PubMed] [Google Scholar]
- Baugh LR 2013. To grow or not to grow: nutritional control of development during Caenorhabditis elegans L1 arrest. Genetics 194: 539–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriz GF, Beaver JE, Cenik C, Tasan M & Roth FP 2009. Next generation software for functional trend analysis. Bioinformatics 25: 3043–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besseling J & Bringmann H 2016. Engineered non-Mendelian inheritance of entire parental genomes in C. elegans. Nat Biotechnol 34: 982–986. [DOI] [PubMed] [Google Scholar]
- Bik HM, Fournier D, Sung W, Bergeron RD & Thomas WK 2013. Intra-genomic variation in the ribosomal repeats of nematodes. PLoS One 8: e78230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortoluzzi S, d’alessi F, Romualdi C & Danieli GA 2001. Differential expression of genes coding for ribosomal proteins in different human tissues. Bioinformatics 17: 1152–1157. [DOI] [PubMed] [Google Scholar]
- Brauchle M, Baumer K & Gönczy P 2003. Differential activation of the DNA replication checkpoint contributes to asynchrony of cell division in C. elegans embryos. Curr Biol 13: 819–827. [DOI] [PubMed] [Google Scholar]
- Brehme KS 1939. A Study of the Effect on Development of “Minute” Mutations in Drosophila Melanogaster. Genetics 24: 131–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. ElegansDeletion Mutant Consortium. 2012. large-scale screening for targeted knockouts in the Caenorhabditis elegans genome. G3 (Bethesda) 2: 1415–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlston CM, Afify ZA, Palumbos JC, Bagley H, Barbagelata C, Wooderchak-Donahue WL, Mao R & Carey JC 2017. Variable expressivity and incomplete penetrance in a large family with non-classical Diamond-Blackfan anemia associated with ribosomal protein L11 splicing variant. Am J Med Genet A 173: 2622–2627. [DOI] [PubMed] [Google Scholar]
- Carroll AJ 2013. The Arabidopsis Cytosolic Ribosomal Proteome: From form to Function. Front Plant Sci 4: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CELNIKER SE, DILLON LA, GERSTEIN MB, GUNSALUS KC, HENIKOFF S, KARPEN GH, KELLIS M, LAI EC, LIEB JD, MACALPINE DM, MICKLEM G, PIANO F, SNYDER M, STEIN L, WHITE KP, WATERSTON RH & MODENCODE C 2009. Unlocking the secrets of the genome. Nature 459: 927–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ch’ng Q, Williams L, Lie YS, Sym M, Whangbo J & Kenyon C 2003. Identification of genes that regulate a left-right asymmetric neuronal migration in Caenorhabditis elegans. Genetics 164: 1355–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Sulston JE, White JG, Southgate E, Thomson JN & Brenner S 1985. The neural circuit for touch sensitivity in Caenorhabditis elegans. J Neurosci 5: 956–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- clark s.g. & chiu c. 2003. C. elegans ZAG-1, a Zn-finger-homeodomain protein, regulates axonal development and neuronal differentiation. Development 130: 3781–3794. [DOI] [PubMed] [Google Scholar]
- de la Cruz J, Karbstein K & Woolford JL 2015. Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo. Annu Rev Biochem 84: 93–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deusser E & Wittmann HG 1972. Ribosomal proteins: variation of the protein composition in Escherichia coli ribosomes as function of growth rate. Nature 238: 269–270. [DOI] [PubMed] [Google Scholar]
- Dibb NJ, Maruyama IN, Krause M & Karn J 1989. Sequence analysis of the complete Caenorhabditis elegans myosin heavy chain gene family. J Mol Biol 205: 603–613. [DOI] [PubMed] [Google Scholar]
- Dickinson DJ & Goldstein B 2016. CRISPR-Based Methods for Caenorhabditis elegans Genome Engineering. Genetics 202: 885–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgley ML, Baillie DL, Riddle DL & Rose AM 2006. Genetic balancers. WormBook 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgley ML & Riddle DL 2001. LG II balancer chromosomes in Caenorhabditis elegans: mT1(II;III) and the mIn1 set of dominantly and recessively marked inversions. Mol Genet Genomics 266: 385–395. [DOI] [PubMed] [Google Scholar]
- Ellis RE, Sulston JE & Coulson AR 1986. The rDNA of C. elegans: sequence and structure. Nucleic Acids Res 14: 2345–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis SR & Lipton JM 2008. Diamond Blackfan anemia: a disorder of red blood cell development. Curr Top Dev Biol 82: 217–241. [DOI] [PubMed] [Google Scholar]
- Enyenihi AH & Saunders WS 2003. Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics 163: 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falahati H, Pelham-Webb B, Blythe S & Wieschaus E 2016. Nucleation by rRNA Dictates the Precision of Nucleolus Assembly. Curr Biol 26: 277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcone Ferreyra ML, Pezza A, Biarc J, Burlingame AL & Casati P 2010. Plant L10 ribosomal proteins have different roles during development and translation under ultraviolet-B stress. Plant Physiol 153: 1878–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnsworth MW 1957a. Effects of Homozygous First, Second and Third Chromosome Minutes on the Development of Drosophila Melanogaster. Genetics 42: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnsworth MW 1957b. Effects of the Homozygous Minute-IV Deficiency on the Development of Drosophila Melanogaster. Genetics 42: 7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrar JE & Dahl N 2011. Untangling the phenotypic heterogeneity of Diamond Blackfan anemia. Semin Hematol 48: 124–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrar JE, Nater M, Caywood E, Mcdevitt MA, Kowalski J, Takemoto CM, Talbot CC, Meltzer P, Esposito D, Beggs AH, Schneider HE, Grabowska A, Ball SE, Niewiadomska E, Sieff CA, Vlachos A, Atsidaftos E, Ellis SR, Lipton JM, Gazda HT & Arceci RJ 2008. Abnormalities of the large ribosomal subunit protein, Rpl35a, in Diamond-Blackfan anemia. Blood 112: 1582–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti MB, Ghalei H, Ward EA, Potts EL & Karbstein K 2017. Rps26 directs mRNA-specific translation by recognition of Kozak sequence elements. Nat Struct Mol Biol 24: 700–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson EL & Horvitz HR 1985. Identification and characterization of 22 genes that affect the vulval cell lineages of the nematode Caenorhabditis elegans. Genetics 110: 17–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson KC, Heid PJ & Rothman JH 1996. The SL1 trans-spliced leader RNA performs an essential embryonic function in Caenorhabditis elegans that can also be supplied by SL2 RNA. Genes Dev 10: 1543–1556. [DOI] [PubMed] [Google Scholar]
- Fernandez-Moya SM, Bauer KE & Kiebler MA 2014. Meet the players: local translation at the synapse. Front Mol Neurosci 7: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Files JG & Hirsh D 1981. Ribosomal DNA of Caenorhabditis elegans. J Mol Biol 149: 223–240. [DOI] [PubMed] [Google Scholar]
- Fukuyama M, Rougvie AE & Rothman JH 2006. C. elegans DAF-18/PTEN mediates nutrient-dependent arrest of cell cycle and growth in the germline. Curr Biol 16: 773–779. [DOI] [PubMed] [Google Scholar]
- Fukuyama M, Sakuma K, Park R, Kasuga H, Nagaya R, Atsumi Y, Shimomura Y, Takahashi S, Kajiho H, Rougvie A, Kontani K & Katada T 2012. C. elegans AMPKs promote survival and arrest germline development during nutrient stress. Biol Open 1: 929–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallegos ME & Bargmann CI 2004. Mechanosensory neurite termination and tiling depend on SAX-2 and the SAX-1 kinase. Neuron 44: 239–249. [DOI] [PubMed] [Google Scholar]
- Gazda HT, Sheen MR, Vlachos A, Choesmel V, O’Donohue MF, Schneider H, Darras N, Hasman C, Sieff CA, Newburger PE, Ball SE, Niewiadomska E, Matysiak M, Zaucha JM, Glader B, Niemeyer C, Meerpohl JJ, Atsidaftos E, Lipton JM, Gleizes PE & Beggs AH 2008. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am J Hum Genet 83: 769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glock C, Heumüller M & Schuman EM 2017. mRNA transport & local translation in neurons. Curr Opin Neurobiol 45: 169–177. [DOI] [PubMed] [Google Scholar]
- Goodwin EB, Okkema PG, Evans TC & Kimble J 1993. Translational regulation of tra-2 by its 3’ untranslated region controls sexual identity in C. elegans. Cell 75: 329–339. [DOI] [PubMed] [Google Scholar]
- Gunderson JH, Sogin ML, Wollett G, Hollingdale M, de la Cruz VF, Waters AP & Mccutchan TF 1987. Structurally distinct, stage-specific ribosomes occur in Plasmodium. Science 238: 933–937. [DOI] [PubMed] [Google Scholar]
- Gupta V & Warner JR 2014. Ribosome-omics of the human ribosome. RNA 20: 1004–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, Vaz Gomes A, Kenyon C, Liu J, Krause M & Fire A 1998. Analysis of a Caenorhabditis elegans Twist homolog identifies conserved and divergent aspects of mesodermal patterning. Genes Dev 12: 2623–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras AK, Plisson-Chastang C, O’Donohue MF, Chakraborty A & Gleizes PE 2015. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev RNA 6: 225–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsh D, Oppenheim D & Klass M 1976. Development of the reproductive system of Caenorhabditis elegans. Dev Biol 49: 200–219. [DOI] [PubMed] [Google Scholar]
- Hummel M, Cordewener JH, de Groot JC, Smeekens S, America AH & Hanson J 2012. Dynamic protein composition of Arabidopsis thaliana cytosolic ribosomes in response to sucrose feeding as revealed by label free MSE proteomics. Proteomics 12: 1024–1038. [DOI] [PubMed] [Google Scholar]
- INGOLIA NT, BRAR GA, ROUSKIN S, MCGEACHY AM & WEISSMAN JS 2012. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc 7: 1534–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale A, Li W, Lee CH & Baker NE 2015. Apoptotic mechanisms during competition of ribosomal protein mutant cells: roles of the initiator caspases Dronc and Dream/Strica. Cell Death Differ 22: 1300–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur M, Monaghan CE & Ackerman SL 2017. Regulation of mRNA Translation in Neurons-A Matter of Life and Death. Neuron 96: 616–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga H, Fukuyama M, Kitazawa A, Kontani K & Katada T 2013. The microRNA miR-235 couples blast-cell quiescence to the nutritional state. Nature 497: 503–506. [DOI] [PubMed] [Google Scholar]
- Khajuria RK, Munschauer M, Ulirsch JC, Fiorini C, Ludwig LS, Mcfarland SK, Abdulhay NJ, Specht H, Keshishian H, Mani DR, Jovanovic M, Ellis SR, Fulco CP, Engreitz JM, Schütz S, Lian J, Gripp KW, Weinberg OK, Pinkus GS, Gehrke L, Regev A, Lander ES, Gazda HT, Lee WY, Panse VG, Carr SA & Sankaran VG 2018. Ribosome Levels Selectively Regulate Translation and Lineage Commitment in Human Hematopoiesis. Cell 173: 90–103.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatter H, Myasnikov AG, Natchiar SK & Klaholz BP 2015. Structure of the human 80S ribosome. Nature 520: 640–645. [DOI] [PubMed] [Google Scholar]
- Kimble J & Hirsh D 1979. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev Biol 70: 396–417. [DOI] [PubMed] [Google Scholar]
- Kniazeva M, Crawford QT, Seiber M, Wang CY & Han M 2004. Monomethyl branched-chain fatty acids play an essential role in Caenorhabditis elegans development. PLoS Biol 2: E257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komili S, Farny NG, Roth FP & Silver PA 2007. Functional specificity among ribosomal proteins regulates gene expression. Cell 131: 557–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kongsuwan K, Yu Q, Vincent A, Frisardi MC, Rosbash M, Lengyel JA & Merriam J 1985. A Drosophila Minute gene encodes a ribosomal protein. Nature 317: 555–558. [DOI] [PubMed] [Google Scholar]
- Korčekova D, Gombitová A, Raška I, Cmarko D & Lanctôt C 2012. Nucleologenesis in the Caenorhabditis elegans embryo. PLoS One 7: e40290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzelius J, The I, Ruijtenberg S, Portegijs V, Xu H, Horvitz HR & van den Heuvel S 2011. C. elegans MCM-4 is a general DNA replication and checkpoint component with an epidermis-specific requirement for growth and viability. Dev Biol 350: 358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland CG, Voynow P, Hardy SJ, Randall L & Lutter L 1969. Physical and functional heterogeneity of E. coli ribosomes. Cold Spring Harb Symp Quant Biol 34: 17–24. [DOI] [PubMed] [Google Scholar]
- Lambertsson A 1998. The minute genes in Drosophila and their molecular functions. Adv Genet 38: 69–134. [DOI] [PubMed] [Google Scholar]
- Landry DM, Hertz MI & Thompson SR 2009. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev 23: 2753–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SW, Berger SJ, Martinović S, Pasa-Tolić L, Anderson GA, Shen Y, Zhao R & Smith RD 2002. Direct mass spectrometric analysis of intact proteins of the yeast large ribosomal subunit using capillary LC/FTICR. Proc Natl Acad Sci U S A 99: 5942–5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AS, Burdeinick-Kerr R & Whelan SP 2013. A ribosome-specialized translation initiation pathway is required for cap-dependent translation of vesicular stomatitis virus mRNAs. Proc Natl Acad Sci U S A 110: 324–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempiäinen H & Shore D 2009. Growth control and ribosome biogenesis. Curr Opin Cell Biol 21: 855–863. [DOI] [PubMed] [Google Scholar]
- Longman D, Arrisi P, Johnstone IL & Cáceres JF 2008. Chapter 7. Nonsense-mediated mRNA decay in Caenorhabditis elegans. Methods Enzymol 449: 149–164. [DOI] [PubMed] [Google Scholar]
- Marygold SJ, Roote J, Reuter G, Lambertsson A, Ashburner M, Millburn GH, Harrison PM, Yu Z, Kenmochi N, Kaufman TC, Leevers SJ & Cook KR 2007. The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Biol 8: R216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro VP & Edelman GM 2002. The ribosome filter hypothesis. Proc Natl Acad Sci U S A 99: 12031–12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mckim KS, Peters K & Rose AM 1993. Two types of sites required for meiotic chromosome pairing in Caenorhabditis elegans. Genetics 134: 749–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michiels L, Van der Rauwelaert E, Van Hasselt F, Kas K & Merregaert J 1993. fau cDNA encodes a ubiquitin-like-S30 fusion protein and is expressed as an antisense sequence in the Finkel-Biskis-Reilly murine sarcoma virus. Oncogene 8: 2537–2546. [PubMed] [Google Scholar]
- Middelkoop TC & Korswagen HC 2014. Development and migration of the C. elegans Q neuroblasts and their descendants. WormBook 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills EW & Green R 2017. Ribosomopathies: There’s strength in numbers. Science 358: [DOI] [PubMed] [Google Scholar]
- Mills EW, Wangen J, Green R & Ingolia NT 2016. Dynamic Regulation of a Ribosome Rescue Pathway in Erythroid Cells and Platelets. Cell Rep 17: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerman DG, FIRE A. Muscle: Structure, Function, and Development In: Riddle DL, Blumenthal T, Meyer BJ, et al. , editors. C. elegans II. 2nd edition Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 1997. Chapter 16. [PubMed] [Google Scholar]
- Moetter MJ, Meerpohl J, Fischer A, Simon T, Urbaniak S, Hirabayashi S, Kapp F, Noellke P, Kratz C, Niemeyer CM, and Wlodarski MW 2011. Analysis of Ribosomal Protein Genes Associated with Diamond Blackfan Anemia (DBA) In German DBA Patients and Their Relatives. Blood 118:729. [Google Scholar]
- Moore PB, Traut RR, Noller H, Pearson P & Delius H 1968. Ribosomal proteins of Escherichia coli. II. Proteins from the 30 s subunit. J Mol Biol 31: 441–461. [DOI] [PubMed] [Google Scholar]
- Mougey EB, O’Reilly M, Osheim Y, Miller OL, Beyer A & Sollner-Webb B 1993. The terminal balls characteristic of eukaryotic rRNA transcription units in chromatin spreads are rRNA processing complexes. Genes Dev 7: 1609–1619. [DOI] [PubMed] [Google Scholar]
- Narla A & Ebert BL 2010. Ribosomopathies: human disorders of ribosome dysfunction. Blood 115: 3196–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DW & Honda BM 1985. Genes coding for 5S ribosomal RNA of the nematode Caenorhabditis elegans. Gene 38: 245–251. [DOI] [PubMed] [Google Scholar]
- Newman-Smith ED & Rothman JH 1998. The maternal-to-zygotic transition in embryonic patterning of Caenorhabditis elegans. Curr Opin Genet Dev 8: 472–480. [DOI] [PubMed] [Google Scholar]
- Ni L & Snyder M 2001. A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol Biol Cell 12: 2147–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas E, Parisot P, Pinto-Monteiro C, de Walque R, De Vleeschouwer C & Lafontaine DL 2016. Involvement of human ribosomal proteins in nucleolar structure and p53-dependent nucleolar stress. Nat Commun 7: 11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K, Goncharov A, Ellisman MH & Jin Y 2017. Microtubule-dependent ribosome localization in C. elegans neurons. Elife 6: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panse VG & Johnson AW 2010. Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem Sci 35: 260–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolini NA, Attwood M, Sondalle SB, Vieira CMDS, van Adrichem AM, di Summa FM, O’Donohue MF, Gleizes PE, Rachuri S, Briggs JW, Fischer R, Ratcliffe PJ, Wlodarski MW, Houtkooper RH, von Lindern M, Kuijpers TW, Dinman JD, Baserga SJ, Cockman ME & Macinnes AW 2017. A Ribosomopathy Reveals Decoding Defective Ribosomes Driving Human Dysmorphism. Am J Hum Genet 100: 506–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phanstiel DH, Boyle AP, Araya CL & Snyder MP 2014. Sushi.R: flexible, quantitative and integrative genomic visualizations for publication-quality multi-panel figures. Bioinformatics 30: 2808–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard B & Wegnez M 1979. Isolation of a 7S particle from Xenopus laevis oocytes: a 5S RNA-protein complex. Proc Natl Acad Sci U S A 76: 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard B, le Maire M, Wegnez M & Denis H 1980. Biochemical Research on oogenesis. Composition of the 42-S storage particles of Xenopus laevix oocytes. Eur J Biochem 109: 359–368. [DOI] [PubMed] [Google Scholar]
- RAMAGOPAL S & ENNIS HL 1981. Regulation of synthesis of cell-specific ribosomal proteins during differentiation of Dictyostelium discoideum. Proc Natl Acad Sci USA 78: 3083–3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shajani Z, Sykes MT & Williamson JR 2011. Assembly of bacterial ribosomes. Annu Rev Biochem 80: 501–526. [DOI] [PubMed] [Google Scholar]
- Sharma S & Lafontaine DL 2015. View From A Bridge’: A New Perspective on Eukaryotic rRNA Base Modification. Trends Biochem Sci 40: 560–575. [DOI] [PubMed] [Google Scholar]
- Slavov N, Semrau S, Airoldi E, Budnik B & van Oudenaarden A 2015. Differential Stoichiometry among Core Ribosomal Proteins. Cell Rep 13: 865–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler M & Fire A 2013. Conserved translatome remodeling in nematode species executing a shared developmental transition. PLoS Genet 9: e1003739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele RE, Thomas PS & Reeder RH 1984. Anucleolate frog embryos contain ribosomal DNA sequences and a nucleolar antigen. Dev Biol 102: 409–416. [DOI] [PubMed] [Google Scholar]
- Steffen KK, McCormick MA, Pham KM, MacKay VL, Delaney JR, Murakami CJ, Kaeberlein M & Kennedy BK 2012. Ribosome deficiency protects against ER stress in Saccharomyces cerevisiae. Genetics 191: 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz JA 1969. Polypeptide chain initiation: nucleotide sequences of the three ribosomal binding sites in bacteriophage R17 RNA. Nature 224: 957–964. [DOI] [PubMed] [Google Scholar]
- Stults DM, Killen MW, Pierce HH & Pierce AJ 2008. Genomic architecture and inheritance of human ribosomal RNA gene clusters. Genome Res 18: 13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE & Horvitz HR 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol 56: 110–156. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG & Thomson JN 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol 100: 64–119. [DOI] [PubMed] [Google Scholar]
- Thompson O, Edgley M, Strasbourger P, Flibotte S, Ewing B, Adair R, Au V, Chaudhry I, Fernando L, Hutter H, Kieffer A, Lau J, Lee N, Miller A, Raymant G, Shen B, Shendure J, Taylor J, Turner EH, Hillier LW, Moerman DG & Waterston RH 2013. The million mutation project: a new approach to genetics in Caenorhabditis elegans. Genome Res 23: 1749–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Driesche SJ & Martin KC 2018. New frontiers in RNA transport and local translation in neurons. Dev Neurobiol 78: 331–339. [DOI] [PubMed] [Google Scholar]
- Wallace H 1960. The development of anucleolate embryos of Xenopus laevis. J Embryol Exp Morphol 8: 405–413. [PubMed] [Google Scholar]
- Waller JP & Harris JI 1961. Studies on the composition of the protein from Escherichia coli ribosomes. Proc Natl Acad Sci U S A 47: 18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Han B, Zhou R & Zhuang X 2016. Real-Time Imaging of Translation on Single mRNA Transcripts in Live Cells. Cell 165: 990–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR 1990. The nucleolus and ribosome formation. Curr Opin Cell Biol 2: 521–527. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN & Brenner S 1986. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci 314: 1–340. [DOI] [PubMed] [Google Scholar]
- Woodland HR 1974. Changes in the polysome content of developing Xenopus laevis embryos. Dev Biol 40: 90–101. [DOI] [PubMed] [Google Scholar]
- Wool IG, Chan YL & Glück A 1995. Structure and evolution of mammalian ribosomal proteins. Biochem Cell Biol 73: 933–947. [DOI] [PubMed] [Google Scholar]
- Wu Z, Ghosh-Roy A, Yanik MF, Zhang JZ, Jin Y & Chisholm AD 2007. Caenorhabditis elegans neuronal regeneration is influenced by life stage, ephrin signaling, and synaptic branching. Proc Natl Acad Sci U S A 104: 15132–15137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S & Barna M 2012. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol 13: 355–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S, Tian S, Fujii K, Kladwang W, Das R & Barna M 2015. RNA regulons in Hox 5’ UTRs confer ribosome specificity to gene regulation. Nature 517: 33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq, Ribo-seq and DNA sequencing data are deposited to NCBI SRA with BioProject ID PRJNA509065.