Natural transformation is a broadly conserved mechanism of horizontal gene transfer in bacterial species that can shape evolution and foster the spread of antibiotic resistance determinants, promote antigenic variation, and lead to the acquisition of novel virulence factors. Surface appendages called competence pili promote DNA uptake during the first step of natural transformation1, however, their mechanism of action has remained unclear due to an absence of methods to visualize these structures in live cells. Here, using the model naturally transformable species Vibrio cholerae and a pilus labeling method, we define the mechanism for type IV competence pilus-mediated DNA uptake during natural transformation. First, we show that type IV competence pili bind to extracellular double-stranded DNA via their tip and demonstrate that this binding is critical for DNA uptake. Next, we show that type IV competence pili are dynamic structures and that pilus retraction brings tip-bound DNA to the cell surface. Finally, we show that pilus retraction is spatiotemporally coupled to DNA internalization and that sterically obstructing pilus retraction prevents DNA uptake. Together, these results indicate that type IV competence pili directly bind DNA via their tip and mediate DNA internalization through retraction during this conserved mechanism of horizontal gene transfer
It is widely believed that competence pili bind extracellular DNA and then actively translocate this DNA across the outer membrane via retraction2,3. In the predominant alternate model, competence pili act as gatekeepers to open the outer membrane secretin pore via their dynamic activity allowing for passive DNA diffusion across the outer membrane4. The main distinction between the proposed models for DNA uptake is the ability of pili to bind DNA. Previously, pilus-DNA binding was visually assessed by electron microscopy, which provides a static view and makes differentiating between coincidental co-localization and de facto binding difficult5. To test these models directly, we sought to observe both pili and DNA by fluorescence microscopy. Type IV competence pili are dynamic surface appendages that are polymerized and extended via the action of an ATPase, PilB, and retracted by depolymerization via the action of an antagonistic ATPase, PilT. We first labeled type IV competence pili in a hyperpiliated pilT mutant of V. cholerae6,7, using a technique8 in which an amino acid residue of the major pilin subunit PilA is replaced by a cysteine for subsequent labeling with a thiol-reactive fluorescent maleimide dye (AF488-mal) (Supplementary Fig. 1a–c). Neither the cysteine mutation nor labeling with AF488-mal substantially affected the transformation frequency either in wildtype cells or a constitutively competent strain where pilT is intact (Supplementary Fig. 1d–e). The constitutively competent strain maintained competence for an extended period of time (≥3 hours) (Supplementary Fig. 2). Because we sought to characterize the function of type IV competence pili downstream of competence induction, we used constitutively competent strains throughout the remainder of the study.
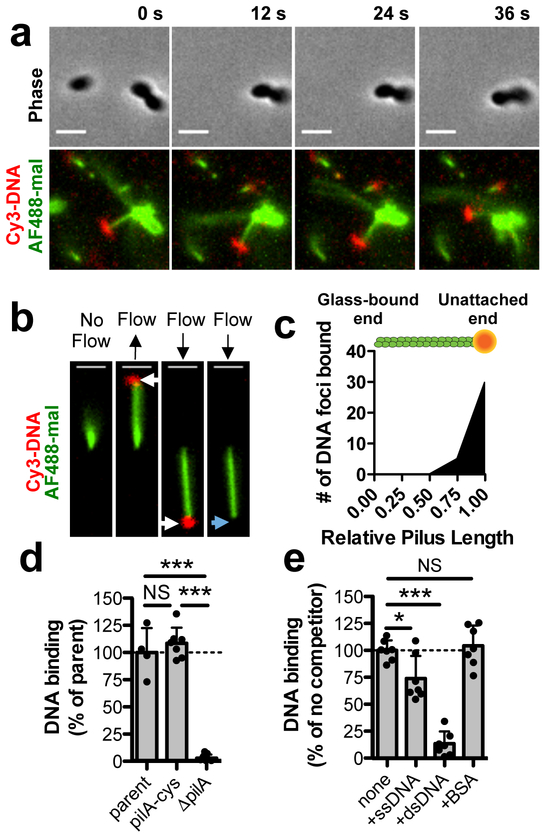
To directly observe pilus-DNA binding, we fluorescently labeled both pili and DNA and tracked them by microscopy in real time. Imaging revealed several cells harboring DNA-bound pili, and adherence was evident from the co-localized movement of DNA and pili (Fig. 1a, Supplementary Movie 1). We further confirmed DNA adherence using sheared pili bound to microfluidic channels by observing the correlated movement of DNA with single pilus fibers along with subsequent DNA detachment when flow was maintained (Fig. 1b, Supplementary Fig. 3, Supplementary Movies 2–3). We found that the majority of DNA interactions occurred at the pilus tip (Fig. 1c). DNA binding assays with fluorescently labeled DNA demonstrated that the piliated pilA-cys and parent strains bound DNA equally, while the non-piliated pilA mutant was deficient in DNA binding (Fig. 1d). Furthermore, competition assays with unlabeled competitors indicated that this binding was specifically inhibited by nucleic acids with a strong preference for double-stranded DNA (dsDNA) (Fig. 1e). This result is consistent with dsDNA binding to Streptococcus pneumoniae competence pili5 and the preferential uptake of dsDNA in Neisseria spp.9, although ssDNA can be transformed with similar efficiency in the latter10. Importantly, we found that DNA binding was dependent on the pilus fiber and could not be attributed to the outer membrane secretin11,12, which still localized properly in the pilA mutant (Supplementary Fig. 4a–c). Thus, type IV competence pili specifically bind to extracellular dsDNA.
Fig. 1. The tips of type IV competence pili directly bind DNA.
Because type IV competence pili are dynamically active and relatively few pili are extended at any given time, all pilus-DNA binding studies were performed in ΔpilT backgrounds to ensure that cells contained numerous extended pili as shown in Supplementary Fig. 1c. (a) Montage of time-lapse imaging of a pilA-cys ΔpilT cell labeled with AF488-mal after mixing with Cy3-labeled DNA in a wet mount. Scale bar, 2 μm. (b) Example of a sheared pilus labeled with AF488-mal bound to glass within a microfluidic device after the addition of fluorescently labeled DNA. Before flow is applied, the relatively long pilus fiber is out of the field of view, but after flow is applied, bound DNA becomes apparent (white arrows) and moves with the direction of flow. When flow is maintained, the DNA detaches from the pilus fiber (blue arrow). Black arrows above images represent the direction of flow. Scale bar, 2 μm. Data in a-b are representative of 3 independent experiments. (c) Localization of bound DNA along sheared pili in microfluidic devices. Pili were normalized in length to 1.00 (arbitrary units), and the position of DNA foci bound along the length for each fiber was plotted, n = 51 independent DNA-bound pili analyzed. Green rod above plot is a visual representation of a labeled pilus fiber bound to orange DNA. (d) Relative DNA binding assay using a fluorescently labeled 6 kb PCR product. parent n = 4, pilA-cys n = 7, ΔpilA, n = 7. (e) Relative DNA binding as in d except with the addition of the indicated non-labeled competitors at 50-fold excess. Competitors were ssDNA (PhiX174 virion), dsDNA (PhiX174 RFII), or bovine serum albumin (BSA). n = 7 for all samples. Each data point in d-e represents an independent biological replicate and bar graphs indicate the mean ± SD. Statistical comparisons were made by two-tailed Student’s t-test. NS = not significant, *P < 0.05, ***P < 0.001.
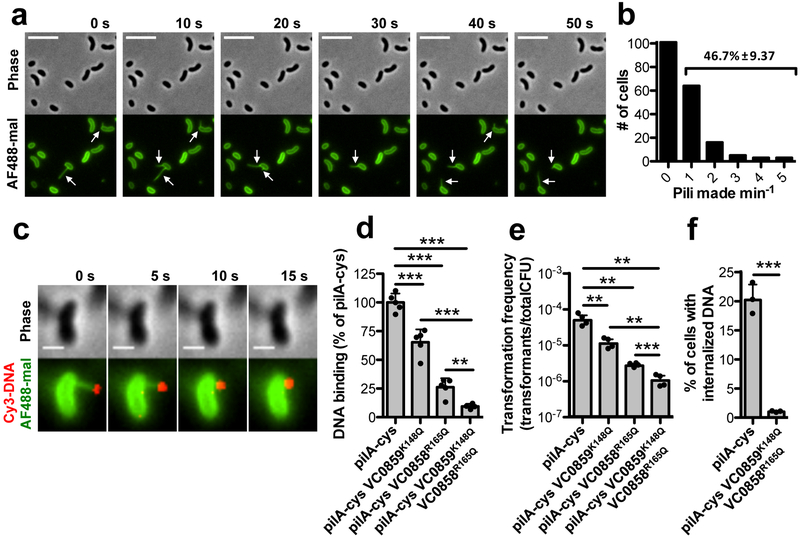
Because the majority of DNA binding occurs at the pilus tip, we hypothesized that retraction helps bring tip-bound DNA to the cell. Indeed, we found that type IV competence pili are highly active surface structures, with 47% of cells grown under competence inducing conditions making at least one pilus of an average length of ~1.0 μm within a one-minute period (Fig. 2a–b, Supplementary Fig. 5a–c, and Supplementary Movie 4). Extension and retraction rates of type IV competence pili were significantly different, with retraction occurring twice as fast (Supplementary Fig. 5b). Pilus dynamic activity, however, was not altered by the presence of extracellular DNA (Supplementary Fig. 6). In some cases, cells synthesized pili repeatedly at the same site, suggesting that the same assembly machinery may be reused (Fig. 2a, Supplementary Movie 4). These frequent extension and retraction events allow for repetitive interactions of type IV competence pili with the extracellular environment. Using fluorescently labeled DNA, we also found that retracting type IV competence pili brought tip-bound DNA to the cell surface (Fig. 2c, Supplementary Fig. 7, Supplementary Movies 5–6).
Fig. 2. Type IV competence pilus dynamic activity and DNA binding are critical for DNA internalization.
(a) Montage of time-lapse imaging of pilA-cys cells after labeling with AF488-mal dye. The white arrows indicate pili. Scale bar, 5 μm. (b) Number of cells making 0–5 pili within a one-minute period. Data are from three independent, biological replicates, n = 192 total pili observed. Percentage is the percent of cells within the population that make pili within the one-minute time-frame ± SD. (c) Montage of time-lapse imaging of a retracting pilA-cys strain after labeling with AF488-mal and incubation with fluorescently labeled DNA in a wet mount. Scale bar, 1 μm. Similar events were captured in three independent experiments. (d) Relative DNA binding assay of the indicated strains in ΔpilT mutant backgrounds using a fluorescently labeled 6 kb PCR product. (e) Natural transformation assays of the indicated strains. Cells were incubated with 5 ng of transforming DNA (tDNA) and DNase I was added to reactions after 10 mins to prevent additional DNA uptake. (f) DNA internalization assays of the indicated strains using a fluorescently labeled 6 kb PCR product. VC0858 and VC0859 encode minor pilins. Each data point in d-f represents an independent biological replicate (d-e, n = 4 for all samples; f, n = 3 for all samples) and bar graphs indicate the mean ± SD. All statistical comparisons were made by two-tailed Student’s t-test. **P < 0.01, ***P < 0.001.
To determine if pilus-DNA interactions were required for DNA uptake, we next sought to disrupt this interaction. Minor pilins can initiate pilus assembly and localize to the pilus tip13,14, which we hypothesized could promote non-specific DNA binding15 by ionic interactions. Consistent with this, we made targeted mutations of several positively charged residues within the major and minor pilins and identified Arg/Lys mutations in two independent minor pilins (VC0858R165Q and VC0859K148Q) that additively reduced DNA binding (Fig. 2d and Supplementary Fig. 5f–g), but did not substantially alter pilus dynamics (Supplementary Fig. 5a–c, h and Supplementary Movie 7). These minor pilin mutants had significantly reduced rates of natural transformation (Fig. 2e) and DNA internalization (Fig. 2f), consistent with pilus-DNA interactions playing an important role in DNA uptake. Additionally, a distinct minor pilin point mutant (VC0858R168Q) reduced DNA binding and natural transformation to the level of a pilA mutant (Supplementary Fig. 5d–e, g). While this mutant produced as many pili as the pilA-cys strain (Supplementary Fig. 5c), these pili displayed statistically significantly altered pilus dynamics (Supplementary Fig. 5a–b, h and Supplementary Movie 8). Despite this, many of the pilus events in the VC0858R168Q mutant were comparable to the rates of extension and retraction in the pilA-cys strain (Supplementary Fig. 5a–b), which further supports that DNA binding is critical for DNA uptake.
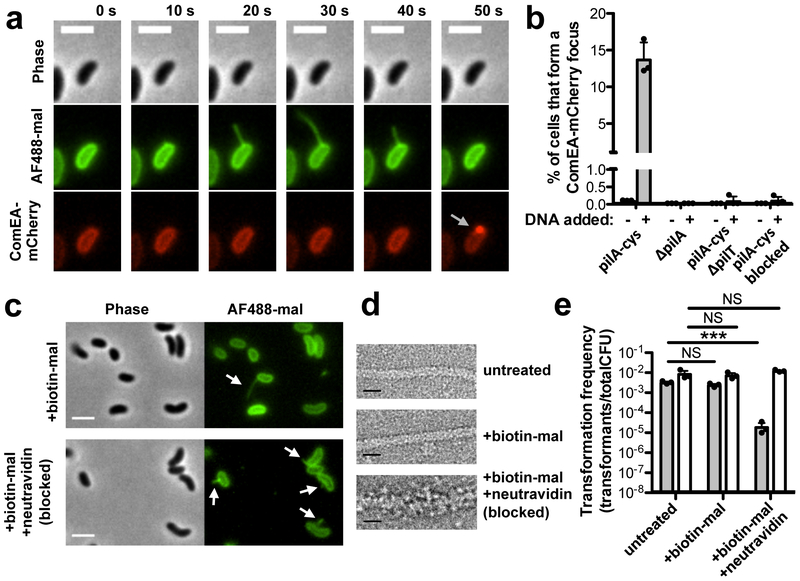
Next, we assessed the spatiotemporal relationship between pilus dynamic activity and DNA internalization. As a marker for DNA uptake into the periplasm we used a fluorescent fusion to ComEA, a critical competence protein that binds to DNA in the periplasm and results in the formation of readily observed foci upon DNA uptake16–18, a finding that we recapitulate here (Supplementary Fig. 8). We found that an average of 14% of cells formed new ComEA foci in the presence of DNA, and that of those events, 64 ± 3% were immediately preceded by a retracting pilus at the same location (Fig. 3a–b, Supplementary Movie 9). This value is likely an underestimate, as it does not account for short pili that cannot be resolved by light microscopy, pili occluded by the cell body, or pili that may form outside the plane of focus. For the events where readily observed pilus retraction precedes DNA uptake, ComEA focus formation only occurred after what appears to be the completion of pilus retraction, which is consistent with DNA binding to the tip of type IV competence pili (Fig. 1a–c). The non-piliated pilA mutant formed no ComEA foci either in the presence or absence of DNA (Fig. 3b). Thus, these data suggested that complete retraction of DNA bound pili is required for DNA uptake.
Fig 3. Pilus retraction is required for DNA uptake.
(a) Montage of time-lapse imaging of pilA-cys ComEA-mCherry strain labeled with AF488-mal showing ComEA focus formation after pilus retraction (gray arrow). Scale bar, 2 μm. (b) Percent of cells that formed a ComEA-mCherry focus within a five min window in the presence or absence of DNA. Cells that had already formed ComEA-mCherry foci at the start of imaging were excluded from the analysis. Data are from three independent experiments (n = 3 for each condition) and shown as the mean ± SD. (c) Static images of pilA-cys cells labeled with a 1:1 ratio of biotin-mal:AF488-mal with or without added neutravidin. White arrows indicate pili. Scale bar, 2 μm. (d) Transmission electron micrographs of pilA-cys pili with the indicated treatments. While untreated and biotin-mal treated pili exhibit a similar thickness, the biotin-mal + neutravidin treated pilus is approximately twice as thick. Scale bar, 10 nm. Images in c-d are representative of two independent experiments. (e) Natural transformation assays of pilA-cys (black bars) or parent (white bars) strains after the indicated treatment. Cells were incubated with 500 ng of tDNA and DNase I was added to reactions after 10 mins to prevent additional DNA uptake. Data are from three independent, biological replicates and shown as the mean ± SD. Statistical comparisons were made by two-tailed Student’s t-test. NS = not significant. ***P < 0.001.
To test this model further, we assessed whether retraction was required for DNA uptake. Because deletion of the retraction ATPase pilT alters the activity of type IV MSHA pili19 and may also have pleiotropic effects on gene expression20, we tested this hypothesis more directly by sterically blocking type IV competence pilus retraction. This was accomplished by co-labeling pilA-cys cells with biotin-maleimide and AF488-mal before treating with neutravidin (a high-affinity biotin binding protein), which specifically blocks retraction of extended pili through the outer membrane secretin likely by thickening the pilus fiber (Fig. 3c–d, Supplementary Fig. 9, Supplementary Movies 10–11). Obstruction of pilus retraction resulted in a ~100-fold decrease in transformation frequency, indicating that at least ~99% of all DNA uptake events require retraction of an extended pilus (Fig. 3e). Furthermore, blocking pilus retraction resulted in very few DNA internalization events (Fig. 3b). Importantly, blocking pilus retraction using this approach did not inhibit binding to dsDNA (Supplementary Fig. 10). Recent work demonstrates that ComEA acts as a molecular ratchet to internalize DNA across the outer membrane, but it is currently unclear how DNA uptake is initiated because ComEA is localized to the periplasm2,16,17. Our results support a previously proposed model21 whereby ComEA-dependent internalization is initiated by the dynamic activity of DNA-binding type IV competence pili. Consistent with this, we found that DNA was not internalized in a comEA mutant despite this strain maintaining normal type IV competence pilus dynamic activity (Supplementary Fig. 11). Together, these results indicate that retraction of DNA-bound type IV competence pili initiates the process of DNA uptake into the periplasm during natural transformation with concomitant uptake facilitated by ComEA-dependent molecular ratcheting.
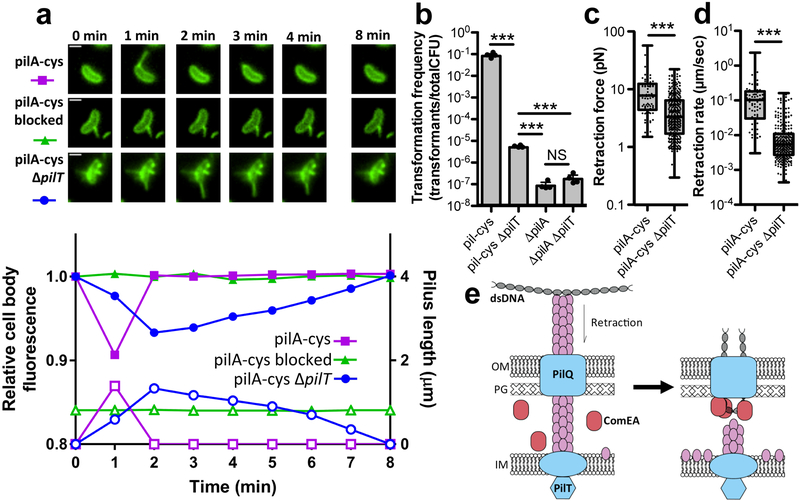
Previous studies have shown that type IV pili require a PilT retraction ATPase to retract22,23. Contrary to this, we found that pilT mutants were still capable of retraction as observed by correlated changes in cell body fluorescence that occur upon extension and retraction of labeled pili during time-lapse experiments (Fig. 4a, Supplementary Fig. 12, Supplementary Movie 12). The pilT mutant, however, exhibited a 10,000-fold reduction in transformation frequency and very little DNA internalization, despite exhibiting pilus retraction (Fig. 3b, Fig. 4a–d). Transformation of the ΔpilT mutant above the levels observed in the ΔpilA strain contrasts with a prior study7, which may be due to technical or strain differences (see Supplementary Discussion). Importantly, sterically blocking pili with biotin-mal and neutravidin as discussed above inhibited residual retraction events and transformation of the pilT mutant, indicating that retraction is still required for natural transformation in this background (Supplementary Fig. 13). We also found that PilT-independent natural transformation and retraction is not mediated by the PilT homolog PilU (Supplementary Fig. 14). Our data thus far indicate that tip bound DNA is internalized via retraction through the outer membrane secretin pore. Therefore, one possibility is that incomplete retraction in the pilT mutant may partially account for the highly reduced rate of transformation and DNA uptake observed.
Fig 4. Residual retraction in ΔpilT mutants allows for low rates of transformation.
(a) Montage (top) of cells measured for corresponding plots (bottom) showing the relative cell body fluorescence (closed symbols) and correlated pilus length (open symbols) over time for pilA-cys labeled with AF488-mal (circles), pilA-cys blocked for retraction by labeling with 1:1 ratio of biotin-mal:AF488-mal and neutravidin (triangles), or pilA-cys ΔpilT labeled with AF488-mal (squares). Scale bar, 1 μm. Data are representative of 3 independent experiments. (b) Natural transformation assays of the indicated strains using 500 ng of tDNA. Data are from four independent, biological replicates and shown as the mean ± SD. Micropillar assays were performed with the indicated strains to measure the retraction (c) force (pilA-cys n = 79, pilA-cys ΔpilT n = 339) and (d) speed (pilA-cys n = 76, pilA-cys ΔpilT n = 288). Box plots indicate the median and first and third quartiles, while the whiskers denote the range. Each data point in the overlay for c-d represents an independent retraction event. Statistical comparisons in b-d were made by two-tailed Student’s t-test. NS = not significant. ***P < 0.001. (e) Model of pilus retraction-mediated DNA uptake. Retraction of DNA-bound pili threads dsDNA across the outer membrane (left) followed by ComEA-dependent molecular ratcheting (right) to promote uptake; OM = outer membrane, PG = peptidoglycan, IM = inner membrane.
DNA could be thread through the PilQ outer membrane secretin pore from one end, or may undergo extreme bending to fit through this pore. We found that cells could internalize linear DNA and circular plasmids (i.e. DNA lacking a free end) equally well (Supplementary Fig. 15), suggesting that DNA is likely bent when threaded across the membrane through PilQ. Because dsDNA has a persistence length of ~50 nm (or 150 bp), the extreme bending required to bring DNA through the relatively small PilQ pore (diameter of 7–8 nm based on closely related systems)24,25 would require a high force of retraction. Accordingly, another explanation for lack of DNA uptake in pilT mutants may be a requirement for a high retraction force and speed to pull DNA through PilQ. To test whether the force and speed of retraction are altered in pilT mutants, we employed a previously described micropillar assay in which cells use pili to bind micropillars that bend as pilus retraction occurs, resulting in measurable retraction rates and forces26. The pilT mutant exhibited a significant reduction in both the speed and force of retraction (Fig. 4c–d). Consequently, reduced transformation in the pilT mutant may be explained by either a lack of full retraction and/or a reduction in the force or speed of retraction, which prevents translocation of DNA across the outer membrane. Coupled with recent reports that divergent pili lacking a pilT homologue can retract8,13, these data support a model whereby pilT-independent retraction may be conserved across diverse systems including competence pili from species that lack defined pilus retraction machinery (e.g. S. pneumoniae, B. subtilis, and H. influenzae). Whether additional canonical type IV pili also exhibit PilT-independent retraction, however, remains to be seen.
Binding and transport of large DNA molecules across the cell envelope is critical for horizontal gene transfer in diverse microbial species, and DNA taken up during this process can also be used as a nutrient. Here, we clarify the mechanism of action for type IV competence pili, the complex nanomachines that promote DNA uptake during natural transformation (Fig. 4e). DNA uptake in V. cholerae is sequence independent15, and we show that minor pilins likely promote DNA binding at the pilus tip. DNA binding to pili in Neisseria is also mediated by a minor pilin, ComP, which binds to a species-specific DNA uptake sequence (DUS)27,28. Despite minor pilins playing a role in DNA binding in both of these systems, the proteins involved lack homology, suggesting that distinct mechanisms for pilus DNA binding likely exist among diverse competence pilus systems.
Binding of DNA to the pilus tip as observed here is also compelling because the secretin pore through which the pilus extends and retracts is only wide enough to accommodate the pilus fiber and would likely exclude DNA bound along the pilus length24,25. Thus, retraction of tip bound DNA could allow for threading DNA through the secretin pore in the wake of a retracting pilus fiber. Our data indicate that pilus retraction, however, is not sufficient for DNA uptake into the periplasm, but likely works in conjunction with the molecular ratchet ComEA2,17 to mediate efficient DNA uptake.
We show that type IV competence pili in V. cholerae exhibit different rates of extension and retraction, which contrasts with other type IV pilus systems that have been characterized where rates of extension and retraction are equal8,29. Notably, the speed of extension and/or retraction were markedly slower than the rates observed for other canonical type IV pili as in Neisseria gonorrhoeae (retraction: ~1.2 μm/s30) and Pseudomonas aeruginosa (extension and retraction = ~0.5 μm/s29). The mechanism underlying this difference is unclear, however, the pilus from V. cholerae studied here is largely dedicated to competence, while the pili in N. gonorrhoeae and P. aeruginosa can mediate twitching motility, which could account for this difference.
While the type IV competence pili of V. cholerae studied here are long micrometer-scale surface appendages, many naturally competent species (such as Bacillus subtilis and Haemophilus influenzae) possess short “pseudopili” that are thought to simply span the cell envelope. Both long competence pili and short pseudopili, however, could engage in DNA uptake through a similar mechanism by binding DNA via their exposed tip and using retraction to pull bound DNA through the outer membrane in Gram-negative species or to pull this DNA through the thick peptidoglycan layer in Gram-positive species. Long extended pili involved in competence (as in V. cholerae, Neisseria spp., S. pneumoniae, and Acinetobacter baylyi) may not be critical for recruitment of freely diffusible DNA (since short pseudopili would suffice), but instead, these structures may be important for binding and uptake of DNA bound to surfaces as would likely be the case in bacterial biofilms.
Methods
Bacterial strains and culture conditions
All V. cholerae strains used throughout this study are derivatives of the El Tor isolate E794631. Competence induction in V. cholerae requires two cues – chitin oligosaccharides and quorum sensing which activate the expression of TfoX and HapR, respectively6,32–34. Thus, to constitutively activate competence, we ectopically expressed TfoX (via Ptac-tfoX) and constitutively activated quorum sensing via inactivation of luxO35,36. For a list of all strains used throughout this study, see Supplementary Table 1. Bacteria were routinely cultivated on LB Miller broth and agar supplemented with kanamycin (50 μg/mL), spectinomycin (200 μg/mL), trimethoprim (10 μg/mL), chloramphenicol (1 μg/mL), erythromycin (10 μg/mL), carbenicillin (100 μg/mL), and/or zeocin (50 μg/mL) as appropriate. Instant ocean medium (7 g/L – Aquarium Systems) was used throughout the study where indicated.
Construction of mutant strains
Mutants were constructed by MuGENT, Exo-MuGENT, and/or natural transformation exactly as previously described35,37. Briefly, mutant constructs for deletions or fluorescent fusions were generated via splicing-by-overlap (SOE) PCR to stitch (1) the upstream region of homology (aka the UP arm), (2) the mutation (aka the MIDDLE arm, which could represent an antibiotic resistance cassette or a fluorescent gene), and (3) the downstream region of homology (aka the DOWN arm). For a list of primers used to generate all mutant constructs, see Supplementary Table 2. The UP arm was amplified via an F1/R1 primer pair, while the DOWN arm was amplified with F2/R2 primer pair. All antibiotic resistance cassettes were amplified with ABD123 (ATTCCGGGGATCCGTCGAC) and ABD124 (TGTAGGCTGGAGCTGCTTC). Fluorescent genes were amplified with the primers indicated in Supplementary Table 2. SOE PCR reactions were carried out using a mixture of the UP, MIDDLE, and DOWN arms in a PCR reaction as template with the F1 and R2 primers. These constructs were then introduced into naturally competent strains as described below in natural transformation assays (for products with an antibiotic resistance marker) or by co-transformation (for constructs that lack a resistance marker – i.e. fluorescent fusions) as previously described35.
For point mutations, mutant constructs were generated via a single PCR reaction using an allele specific F primer that contained >35bp of homology upstream of the point mutation and an R2 primer to generate the downstream region of homology. This product was introduced into a recJ exoVII mutant of V. cholerae via co-transformation as previously described37. Subsequently, point mutations were amplified off of the gDNA of these strains with F1/R2 primers and then transferred to strains of interest by co-transformation35.
Mutants were confirmed by PCR and/or sequencing. The Ptac-tfoX and ΔlacZ::lacIq mutant constructs are from a previously published study35. To design cysteine replacement mutants and minor pilin lysine/arginine mutations, amino acid residues from pilin sequences were analyzed for solvent accessibility as described previously8 using solvent accessibility prediction software38.
Natural transformation assays
Chitin-dependent and chitin-independent transformation assays were conducted as previously described35,37. Unless otherwise noted, natural transformation assays were chitin-independent using strains containing IPTG-inducible Ptac-tfoX and ΔluxO mutations to induce competence. For chitin-independent transformation assays, the indicated strains were grown to late-log in LB + 100 μM IPTG (to induce TfoX expression) + 20 mM MgCl2 + 10 mM CaCl2. Then, ~108 CFUs of this culture were diluted into instant ocean medium. Next, ~500 ng of a transforming DNA was added to each reaction and allowed to incubate for an additional 5 hours at 30°C to allow for natural transformation. For each strain tested, negative controls were performed where no DNA was added. VC1807 is a frame-shifted transposase and its mutation does not impact the growth of V. cholerae35, thus, PCR products for ΔVC1807::AbR (AbR = CmR, ZeoR, TmR, or ErmR resistance cassettes depending on the resistance profiles of strains tested) containing 3 kb homology arms (required for maximum rates of transformation35) were used to test transformation frequency throughout this manuscript. Next, reactions were outgrown by addition of 1 mL of LB and shaking at 37°C for 2 hours followed by plating on media to select for transformants (i.e. media to select for integration of ΔVC1807::AbR) and on nonselective media to assess the total viable counts. Transformation frequency was defined as the number of transformants divided by the total viable counts. Where indicated, 25 μg/mL of Alexa Fluor 488 C5 Maleimide (AF488-mal)(Thermo Fisher) was added to the chitin-independent transformation assays 15 min before the addition of transforming DNA.
For chitin-dependent natural transformation assays (only used in Supplementary Fig. 1c), ~108 CFUs of a mid-log culture of the indicated strain were incubated on chitin powder from shrimp cells (Alfa Aesar) in instant ocean medium for 24 hours to allow for competence induction. Then, 500 ng of ΔVC1807::AbR transforming DNA was added, and reactions were incubated, outgrown, and plated exactly as described above for chitin-independent transformation assays to attain the transformation frequency.
For cotransformation assays, cells were incubated with two genetically unlinked markers. One marker was a ΔVC1807::ErmR mutant construct, while the other marker was a previously described ΔVCA1045 mutant construct39, which inactivates the sole mannitol transporter in V. cholerae. In these assays, 10ng of the ΔVC1807::ErmR PCR product was added to transformation assays and then ~3 μg of ΔVCA1045 PCR product was either added to cells immediately or after a 3 hour delay. Cell were then outgrown and plated onto Erm containing media to select for integration of the ΔVC1807::ErmR product. 24 colonies were then patched into M9 minimal medium containing mannitol as a sole carbon source to screen for integration of the ΔVCA1045 mutant construct. The cotransformation frequency was defined as the percent of the ΔVC1807::ErmR mutants that incorporated the ΔVCA1045 product.
To physically obstruct pilus retraction, ~108 CFUs of the indicated strains grown to late-log under competence inducing conditions were washed in instant ocean medium and incubated with 50 μg/mL EZ link Biotin-PEG11-maleimide (biotin-mal)(Thermo Fisher) for 45 min at room temperature. Then, cells were pelleted and washed twice in instant ocean medium. Next, ~107 CFUs were diluted into fresh instant ocean medium. Where indicated, neutravidin (Thermo Fisher) was then added to reactions at a final concentration of 1.32 mg/mL and incubated at room temperature for 30 min. Then, 500 ng of ΔVC1807::AbR transforming DNA was added and reactions were incubated at 30°C. For strains where pilT was intact, reactions were allowed to proceed for 10 min before the addition of 10 units of DNAse I (NEB) to prevent additional DNA uptake. This was performed because production of new pilins during the normal 5-hour DNA incubation would circumvent our method to block pilus retraction. For transformation assays in the ΔpilT mutant background, a 10 min incubation with DNA was insufficient to obtain any transformants. Thus, reactions were allowed to proceed for 1 hour at 30°C prior to DNAse I addition as described above. Following DNA addition, all reactions were incubated at 30°C for a total of 5 hours (regardless of the time of DNAse I addition) to allow for DNA integration. Reactions were then outgrown and plated exactly as described above to attain the transformation frequency.
For natural transformation assays of minor pilin DNA binding mutants, strains were incubated with 5 ng of ΔVC1807::AbR transforming DNA. After 5 mins, DNAse I was added to prevent additional DNA uptake, and reactions were then outgrown and plated as described above. This was performed to sensitize the assay to the effects of DNA binding on natural transformation.
Pilin labeling, imaging, and quantification
Pilin labeling was achieved using AF488-mal. Cultures were grown to late-log phase in LB supplemented with 20 mM MgCl2, 10 mM CaCl2, and 100 μM IPTG to induce competence. ~108 CFUs were then centrifuged at 16,000 × g for 1 min and then resuspended in instant ocean medium supplemented with 20 mM MgCl2 and 10 mM CaCl2 before labeling with 25 μg/ml of AF488-mal for 30 min. Labeled cells were centrifuged, washed once, and resuspended in instant ocean medium. Cell bodies were imaged using phase contrast microscopy while labeled pili were imaged using fluorescence microscopy on a Nikon Ti-2 microscope using a Plan Apo 60X objective, a GFP or DsRed filter cube, a Hamamatsu ORCAFlash4.0 camera and Nikon NIS Elements Imaging Software. Cell numbers were quantified using MicrobeJ40. To determine pilus length and rates of extension and retraction, labeled cells were imaged by time-lapse microscopy every second for 1 min. For pilus length measurements, pili that were already retracting when imaging began were excluded from the analysis. For extension and retraction rate calculations, only cells that made a single pilus that began extension and fully retracted within the 1 min window were analyzed. To determine the number of pili made per cell, the percent of cells that make pili, and the percent of cells that that form a ComEA-mCherry focus, labeled cells were imaged in the presence or absence of 1 μg DNA by time-lapse microscopy every 10 s for 5 min. Pilus length, extension and retraction rates, number of pili made per cell, the percent of cells that make pili, and the percent of cells that form a ComEA-mCherry focus were manually calculated using measurement tools of the NIS Elements analysis software. To block pilus retraction for fluorescence microscopy, cells were co-labeled with 25 μg/ml of AF488-mal and 25 μg/ml biotin-mal for 30 min before washing. Where indicated, 1.32 mg/mL neutravidin was then added to washed cells and incubated for 30 min at room temperature before imaging. All imaging was performed under 0.2% Gelzan (Sigma) pads made with instant ocean medium.
Fluorescent DNA binding and localization assays
Strains used for DNA binding and localization assays contained ΔpilT mutations to ensure the presence of extended pili for DNA binding and to diminish DNA uptake. Fluorescently labeled Cy3- and MFP488-DNA was generated using a 6 kb ΔVC1807::AbR PCR product (the same product used for transformation efficiency assays throughout this manuscript) and the LabelIT kit (Mirus Biosciences), which covalently adds a fluorescent label onto ~1 in every 20–60 bp. For DNA binding assays, the indicated strains were grown to late log in LB + 100 μM IPTG + 20 mM MgCl2 + 10 mM CaCl2. Then, ~108 CFUs were centrifuged and washed in instant ocean medium. Cells were then incubated with 100 ng of Cy3- or MFP488-labeled DNA at room temperature for 15 min (MFP488-labeled DNA was used for DNA pulldowns of minor pilin mutant strains because they contained comEA-mCherry, which has significant spectral overlap with Cy3). Cells were then pelleted and washed twice with fresh instant ocean to remove unbound DNA. Reactions were then transferred to a 96-well plate, and Cy3 or MFP488 fluorescence was determined on a Biotek H1M plate reader. Where indicated, 5 μg ssDNA (PhiX virion DNA, NEB), 5 μg dsDNA (PhiX RFII DNA, NEB), or 10 μg acetylated BSA (Promega) were added to DNA binding reactions as non-labeled competitors. Where indicated, strains were labeled with biotin-mal and/or neutravidin exactly as described above for natural transformation assays.
For microscopy localization assays on cell-associated pili, cells were labeled as described above before they were added to a coverslip with BSA (0.2 mg/mL) and 4 ng Cy3-labeled DNA. After incubation for 5–15 min, cells were imaged using a Cy3 filter cube every 3 s for 2 min. During imaging, we noticed the majority of DNA binding events occurred at the ends of pilus fibers. However, we experienced several limitations using this setup that prevented quantitative analysis. For example, the large number of pili in ΔpilT mutants obscured how many fibers were binding DNA puncta, the cell body fluorescence prevented observation of DNA binding to pili shorter than 1 μm, and Brownian motion in this setup resulted in pili and DNA movement outside of the focal plane.
For robust quantification of DNA localization along pilus filaments, we used sheared pili bound to surfaces within microfluidic channels to alleviate the limitations of using cell-associated pili and to further demonstrate that DNA binding was pilus-dependent. Microfluidic channel devices were constructed exactly as described previously8. Channels were pre-treated with BSA (0.2 mg/mL) in instant ocean medium to reduce nonspecific adherence. Then, ~108 AF488-mal labeled cells were vortexed to shear pili, and then cells and sheared pili were added to microchannels and incubated for 30 min at room temperature to allow for pilus attachment to the microfluidic channel surfaces. Next, 11 ng of Cy3-labeled DNA in a total volume of 400 μl instant ocean medium was added to the channel to both flush away unattached cells and pili and to introduce DNA to the microchannel. Then, the chamber was incubated at room temperature for 5–15 min with DNA to allow for DNA-pilus association before imaging. To confirm DNA-pilus binding, gravity flow was applied to microfluidic channels by altering the heights of the input and output tubing to bring both pili and bound DNA into the same field of view for imaging. Only DNA molecules that exhibited colocalization with pili during flow directional changes were analyzed, allowing for exclusion of glass-bound DNA from the analysis. Pili shorter than 0.5 μm were excluded from the analysis since their movement could not be resolved with directional flow changes. Pilus length and DNA localization was determined using MicrobeJ32. Pilus lengths were normalized where the glass bound end was fixed at 0 and the free end was 1.0, and relative DNA binding was then plotted along the normalized length.
To observe type IV competence pili pulling fluorescent DNA, pilA-cys cells harboring native comEA (TND0651) were grown to late-log in LB + 100 μM IPTG + 20 mM MgCl2 + 10 mM CaCl2, washed in instant ocean and then mixed with ~5 ng of Cy3-labeled DNA and acetylated BSA was added to 100 μg/mL. Next, a wet mount was prepared for microscopy on an inverted scope by placing 5–10 μL of sample onto a 22 × 50 mm coverslip and then covering the sample with a 22 × 22 mm coverslip. Time-lapse imaging was then performed as described above.
DNA internalization assay
~108 CFUs of cells grown to late-log in LB + 100 μM IPTG + 20 mM MgCl2 + 10 mM CaCl2 were pelleted and resuspended in instant ocean medium. Cells were then incubated with 10 ng of either an MFP488-labeled 6kb ΔVC1807::AbR PCR product or an ~6kb supercoiled plasmid (pBAD18Kan) at room temperature for 1 hour. Next, 10 units of DNase I was added to reactions and incubated at room temperature for 1 min to degrade any remaining extracellular DNA. Cells were then washed once with instant ocean medium to remove excess dye and imaged. The percent of cells with internalized DNA was quantified using MicrobeJ.
mCherry-PilQ localization
Cells were grown to late-log in LB + 100 μM IPTG + 20 mM MgCl2 + 10 mM CaCl2 and imaged. Localization of mCherry-PilQ foci was determined and plotted using MicrobeJ.
Cell body fluorescence quantification
Strains were labeled with AF488-mal as indicated above and imaged for 10 s intervals over a 20 min window. Integrated fluorescence intensity was measured for both the cell body and the pilus for each cell for each time point using ImageJ41. Fluorescence intensities were normalized for photobleaching over the time-lapse experiment by multiplying the measured fluorescence intensity by the percentage of fluorescence lost from the total fluorescence intensity (the sum of the cell body and pilus intensities) for each time point. After adjustment for photobleaching, the cell body fluorescence for each time point was divided by the cell body fluorescence measured at time 0. The relative cell body intensity changes were plotted with the measured pilus length for each time point.
Negative stain electron microscopy
To ensure that the pili imaged via EM were type IV competence pili, we inactivated the two other type IV pilus systems (MSHA and TCP) in V. cholerae yielding strain SAD2094 (see Supplementary Table 1 for additional strain details). We also generated a pilA-cys mutation in this background (SAD2093). Strains were gown to late-log in LB + 100 μM IPTG + 20 mM MgCl2 + 10 mM CaCl2 and then treated with biotin-mal and/or neutravidin exactly as described above for natural transformation assays.
To prepare the negative stain specimen, 4 μL of sample solution was applied on a glow-discharged, carbon-coated, 300-mesh copper grid for 30 s. The excess solution was removed by a piece of filter paper. The grid was washed with 4 μL of Milli-Q water, stained with 4 μL of 0.75% (w/v) uranyl formate for 25 s and blotted to dry. The images were acquired at a nominal magnification of 10,000x, 20,000x or 40,000x using JEOL JEM-1400 Plus transmission electron microscope operated at 120 kV and a Gatan 4k × 4k OneView Camera.
Micropillar retraction assay
To ensure that all measurements recorded were due to type IV competence pili, all the experiments were performed with strains lacking all external appendages other than type IV competence pili as well as exopolysaccharide production (i.e. mutants lacking flagella, MSHA pili, TCP pili, and VPS production). See Supplementary Table 1 for additional strain details. Strains were grown overnight in LB either directly from frozen stocks or from a single colony off of an LB agar plate. 50 μL of the overnight liquid culture was subcultured into 3 ml of LB + 100 μM IPTG + 20 mM MgCl2 + 10 mM CaCl2 and allowed to grow at 30 °C for 5 hours. Then, 100 μL of this culture was centrifuge for 5 min at 20,000 × g and the bacteria were resuspended in instant ocean medium. At different times, 10 μL of the resuspension was added to micropillars in an observation chamber as previously reported26. Briefly, silica molds were inverted on activated coverslips with polyacrylamide gels in between. The result is an array of flexible micropillars in a hexagonal array of 3 μm × 3 μm. Once the bacteria are in contact with the micropillars, 10Hz movies of the top of the pillars are recorded. The motion of the tips of the pillars is tracked using a cross-correlation algorithm in ImageJ41. The amplitude and speeds of the pillars’ motions were then analyzed using Matlab. Finally, we calibrated the pillars’ stiffness constant using optical tweezers as previously described26. The pillars used in this study have a stiffness constant of 17±4 pN/μm.
Statistics
Significance was calculated using statistical tests on the GraphPad (Prism) 5.0 software. Statistical differences between two groups were analyzed using two-tailed Student’s t-tests. Complete statistics can be found in Supplementary Table 3. Sample sizes were chosen based on historical data and no statistical methods were used to predetermine sample size.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon request.
Supplementary Material
Acknowledgments:
We thank A. Camilli, F. Yildiz, D. Kearns, N. Greene, C. Berne, and B. LaSarre for critical comments on the manuscript. We also thank members of the Biais lab, L. Khosla, R. Rayyan and A. Ratkiewicz for assistance with micropillar assays. This work was supported by grant R35GM122556 from the National Institutes of Health to YVB, by grant AI118863 from the National Institutes of Health to ABD, by National Science Foundation fellowship 1342962 to CKE, and by grant AI116566 from the National Institutes of Health to NB.
Footnotes
Competing interests:
The authors declare no competing interests.
Supplementary Materials:
Supplementary Discussion
Supplementary Figures 1–15
Supplementary Tables 1–3
Supplementary Movies 1–12
References:
- 1.Chen I & Dubnau D DNA uptake during bacterial transformation. Nat Rev Microbiol 2, 241–249 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Hepp C & Maier B Kinetics of DNA uptake during transformation provide evidence for a translocation ratchet mechanism. Proc Natl Acad Sci U S A 113, 12467–12472 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muschiol S, Balaban M, Normark S & Henriques-Normark B Uptake of extracellular DNA: competence induced pili in natural transformation of Streptococcus pneumoniae. Bioessays 37, 426–435 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graupner S, Weger N, Sohni M & Wackernagel W Requirement of novel competence genes pilT and pilU of Pseudomonas stutzeri for natural transformation and suppression of pilT deficiency by a hexahistidine tag on the type IV pilus protein PilAI. J Bacteriol 183, 4694–4701 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laurenceau R et al. A type IV pilus mediates DNA binding during natural transformation in Streptococcus pneumoniae. PLoS Pathog 9, e1003473 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meibom KL, Blokesch M, Dolganov NA, Wu CY & Schoolnik GK Chitin induces natural competence in Vibrio cholerae. Science 310, 1824–1827 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Seitz P & Blokesch M DNA-uptake machinery of naturally competent Vibrio cholerae. Proc Natl Acad Sci U S A 110, 17987–17992 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellison CK et al. Obstruction of pilus retraction stimulates bacterial surface sensing. Science 358, 535–538 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hepp C, Gangel H, Henseler K, Gunther N & Maier B Single-Stranded DNA Uptake during Gonococcal Transformation. J Bacteriol 198, 2515–2523 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duffin PM & Seifert HS Genetic transformation of Neisseria gonorrhoeae shows a strand preference. FEMS Microbiol Lett 334, 44–48 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Assalkhou R et al. The outer membrane secretin PilQ from Neisseria meningitidis binds DNA. Microbiology 153, 1593–1603 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burkhardt J, Vonck J & Averhoff B Structure and function of PilQ, a secretin of the DNA transporter from the thermophilic bacterium Thermus thermophilus HB27. J Biol Chem 286, 9977–9984 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ng D et al. The Vibrio cholerae Minor Pilin TcpB Initiates Assembly and Retraction of the Toxin-Coregulated Pilus. PLoS Pathog 12, e1006109 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen Y et al. Pseudomonas aeruginosa minor pilins prime type IVa pilus assembly and promote surface display of the PilY1 adhesin. J Biol Chem 290, 601–611 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suckow G, Seitz P & Blokesch M Quorum sensing contributes to natural transformation of Vibrio cholerae in a species-specific manner. J Bacteriol 193, 4914–4924 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gangel H et al. Concerted spatio-temporal dynamics of imported DNA and ComE DNA uptake protein during gonococcal transformation. PLoS Pathog 10, e1004043 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seitz P et al. ComEA is essential for the transfer of external DNA into the periplasm in naturally transformable Vibrio cholerae cells. PLoS Genet 10, e1004066 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borgeaud S, Metzger LC, Scrignari T & Blokesch M The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science 347, 63–67 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Watnick PI & Kolter R Steps in the development of a Vibrio cholerae El Tor biofilm. Mol Microbiol 34, 586–595 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dietrich M, Mollenkopf H, So M & Friedrich A Pilin regulation in the pilT mutant of Neisseria gonorrhoeae strain MS11. FEMS Microbiol Lett 296, 248–256 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hepp C & Maier B Bacterial Translocation Ratchets: Shared Physical Principles with Different Molecular Implementations: How bacterial secretion systems bias Brownian motion for efficient translocation of macromolecules. Bioessays 39, e201700099 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Burrows LL Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu Rev Microbiol 66, 493–520 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Craig L, Pique ME & Tainer JA Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol 2, 363–378 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Chang YW et al. Architecture of the type IVa pilus machine. Science 351, aad2001 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gold VA, Salzer R, Averhoff B & Kuhlbrandt W Structure of a type IV pilus machinery in the open and closed state. eLife 4, e07380 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biais N, Higashi D, So M & Ladoux B Techniques to measure pilus retraction forces. Methods Mol Biol 799, 197–216 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cehovin A et al. Specific DNA recognition mediated by a type IV pilin. Proc Natl Acad Sci U S A 110, 3065–3070 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berry JL et al. A Comparative Structure/Function Analysis of Two Type IV Pilin DNA Receptors Defines a Novel Mode of DNA Binding. Structure 24, 926–934 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skerker JM & Berg HC Direct observation of extension and retraction of type IV pili. Proc Natl Acad Sci U S A 98, 6901–6904 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maier B et al. Single pilus motor forces exceed 100 pN. Proc Natl Acad Sci U S A 99, 16012–16017 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller VL, DiRita VJ & Mekalanos JJ Identification of toxS, a regulatory gene whose product enhances toxR-mediated activation of the cholera toxin promoter. J Bacteriol 171, 1288–1293 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dalia AB, Lazinski DW & Camilli A Identification of a membrane-bound transcriptional regulator that links chitin and natural competence in Vibrio cholerae. MBio 5, e01028–01013 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lo Scrudato M & Blokesch M The regulatory network of natural competence and transformation of Vibrio cholerae. PLoS Genet 8, e1002778 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lo Scrudato M & Blokesch M A transcriptional regulator linking quorum sensing and chitin induction to render Vibrio cholerae naturally transformable. Nucleic Acids Res 41, 3644–3658 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dalia AB, McDonough E & Camilli A Multiplex genome editing by natural transformation. Proc Natl Acad Sci U S A 111, 8937–8942 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu J et al. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci U S A 99, 3129–3134 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dalia TN et al. Enhancing multiplex genome editing by natural transformation (MuGENT) via inactivation of ssDNA exonucleases. Nucleic Acids Res 45, 7527–7537 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petersen B, Petersen TN, Andersen P, Nielsen M & Lundegaard C A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct Biol 9, 51 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayes CA, Dalia TN & Dalia AB Systematic genetic dissection of PTS in Vibrio cholerae uncovers a novel glucose transporter and a limited role for PTS during infection of a mammalian host. Mol Microbiol 104, 568–579 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ducret A, Quardokus EM & Brun YV MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat Microbiol 1, 16077 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Collins TJ ImageJ for microscopy. Biotechniques 43, 25–30 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request.