Abstract
Lung cancer is the leading cause of cancer deaths in the United States. Patients with early stage lung cancer have the best prognosis with surgical removal of the tumor, but the disease is often asymptomatic until advanced disease develops, and there are no effective blood-based screening methods for early detection of lung cancer in at-risk populations. We have explored the lipid profiles of blood plasma exosomes using ultra high-resolution Fourier transform mass spectrometry (UHR-FTMS) for early detection of the prevalent non-small cell lung cancers (NSCLC). Exosomes are nanovesicles released by various cells and tumor tissues to elicit important biofunctions such as immune modulation and tumor development. Plasma exosomal lipid profiles were acquired from 39 normal and 91 NSCLC subjects (44 early stage and 47 late stage). We have applied two multivariate statistical methods, Random Forest (RF) and Least Absolute Shrinkage and Selection Operator (LASSO) to classify the data. For the RF method, the Gini importance of the assigned lipids was calculated to select 16 lipids with top importance. Using the LASSO method, 7 features were selected based on a grouped LASSO penalty. The Area Under the Receiver Operating Characteristic curve for early and late stage cancer versus normal subjects using the selected lipid features was 0.85 and 0.88 for RF and 0.79 and 0.77 for LASSO, respectively. These results show the value of RF and LASSO for metabolomics data-based biomarker development, which provide robust an independent classifiers with sparse data sets. Application of LASSO and Random Forests identifies lipid features that successfully distinguish early stage lung cancer patient from healthy individuals.
Keywords: non-small cell lung cancer, Ultrahigh Resolution Fourier Transform Mass Spectrometry, exosomal lipid profiling, Random Forest, LASSO
Graphical abstract
1. Introduction
1.1 Lung cancer is difficult to diagnose at early stage
Lung cancer is by far the leading cause of cancer deaths in the U.S. [1, 2] with an estimated 224,390 new cases and 158,080 deaths in 2016 [3]. Kentucky now leads the nation both in terms of lung cancer incidence and mortality, with the Appalachian population showing even higher incidence and mortality rates [4]. Most lung cancer patients are diagnosed at advanced stages due to the silent nature of the early stage disease. Although the five-year survival rate of localized lung cancer is ~55% with proper surgical intervention [5, 6], that of advanced stage disease drops to ~4%. Presently, there is no robust low-cost blood-based screening method for detecting asymptomatic early stage lung cancer. Current imaging or cytology-based methods are impractical for screening at-risk populations for lung cancer, as they are not sufficiently accurate, cost-effective or non-invasive [7, 8]. Although low dose helical CT screens have recently been reported to decrease lung cancer mortality by 20% in comparison to chest x-ray screening, there remains a high false positive rate [9]. Thus, techniques to detect and reliably screen lung cancer at its earliest stage in at-risk populations are urgently needed to improve survival and quality of life for lung cancer patients.
Non-small cell lung cancer (NSCLC) is the dominant form (ca. 85%) of lung cancer, and comprises many subtypes with different sets of oncogenic drivers such as mutant KRAS, EGFR, LKB1, EML4-ALK (adenocarcinomas), PIK3CA, NRF2 (squamous cell carcinomas), cMYC overexpression and inactivation of TP53 via mutations (both subtypes) [10–14], and numerous other genetic aberrations yet to be functionally defined. It is becoming clear that one of the key functions of these oncogenic drivers lies in reprogramming specific metabolic events in cancer cells to promote their proliferation, survival and metastasis. Thus, metabolic reprogramming in cancer has been recently recognized as a hallmark of cancer [15]. However, the global metabolic networks, and lipidomics in particular, modulated by these drivers and/or other undefined genetic aberrations are poorly characterized in NSCLC.
1.2 Lung cancer lipid metabolism is a rich ground for biomarker discovery
We have performed lipid profiling of paired CA and NC lung tissues using UHR-FTMS, “UHR” defined here as MS with sufficient resolving power to resolve the many hundreds of lipid species and their 13C isotopologues (same lipid differing only in the number of 13C atoms). A large fraction of our NSCLC tissue collections analyzed are classified as early stage adenocarcinoma (AdC) or squamous cell carcinoma (SqCC) [16]. We noticed consistent differences in the lipid profiles of paired CA versus NC lung tissues e.g. sphingomyelins (SM), ceramides, phosphatidylserines (PS), and cholesterol esters (Fan, Higashi & Lane unpublished data), which could reflect the altered expression of many lipid metabolic genes evident in lung [17] and other tumors [18].
1.3 Exosomes and microvesicles carry tumor cell-derived bioactive materials
Interestingly, both SM and PS have been linked to lipid microparticles (MP) shed from cells [19]. MP such as exosomes (EXO) and microvesicles (MV) can be shed from many different cell types, most notably immune cells and tumor cells, into the circulating blood. EXO are multivesicular bodies originating from the endosomal membrane, and are released upon fusion with the plasma membrane while MV are formed by outward budding and fission of the plasma membrane. Both types of lipidic MP are thought to mediate extracellular communications such as immune activation or suppression [19, 20]. MP derived from cancer cells including lung cancer cells can carry a variety of bioactive proteins (e.g. epidermal growth factor receptor, EGFR; vascular endothelial growth factor, VEGF; integrins; Fas ligand; latent membrane protein, LMP-1; angiogenic factor tetraspanin; macrophage migration inhibitory factor or MIF) and microRNAs to promote tumor growth/invasion/metastasis as well as to enact immune evasion [19, 21–31] and drug resistance [32–35]. Although largely unexplored, exosomal lipids derived from cancer cells have been shown to elicit apoptosis in sensitive cells via inhibition of the Notch-1 pathway [36] but activate the Akt survival pathway via promoting the NFκB-SDF1-CXCR4 axis in resistant cells [37]. Melanoma cells cultured under acidic conditions released EXO with a higher SM content, and were shown to have a higher capacity for cell fusion and delivery of caveolin-1 (tumor promoting) to less aggressive melanoma cells than neutral EXO [38]. Moreover, blocking CE buildup interferes with exosomal uptake [39] and has anti-cancer effects [18], while ceramide buildup is important for exosomal biogenesis [31] and triggers cancer cell death [40]. Thus, there are vital functions of lipids in exosomal biogenesis and interactions with the tumor microenvironment (TME) to influence tumor development and progression.
Recently, exosomal components such as microRNA and proteins have been shown to be promising diagnostic tools in human cancers including lung cancer [41–45]. However, it is unclear if these components can be generally useful in classifying lung cancer, as the microRNA signatures did not differ qualitatively between lung cancer and normal subjects [46] while the accuracy of protein markers for advanced stage NSCLC detection was only 75%. Such limitations do not meet the specificity and sensitivity requirements for lung cancer screening at early stages.
We have procured blood plasma samples from 39 normal and 91 NSCLC subjects (44 early stage and 47 late stage) for EXO isolation and lipid profiling using UHR-FTMS. We also applied two advanced multivariate statistical methods, Random Forest (RF) and Least Absolute Shrinkage and Selection Operator (LASSO) to perform supervised clustering analysis of the EXO lipid profiles. The Area Under the Receiver Operating Characteristic curve (AUROC) of normal versus early and late stage NSCLC using the top 16 (for RF) or top 6 (for LASSO) lipid features was 0.85 and 0.88 or 0.79 and 0.77, respectively. These results showed that selected lipid species of plasma EXO discriminated normal from early and late stage NSCLC and demonstrate the value of RF and LASSO for metabolomics-based biomarker development.
2. Materials and Methods
2.1 Materials
2.1.1 Blood collection
A total of 130 blood samples were collected prospectively with informed consent under University of Kentucky IRB-approved protocols from 39 healthy volunteers, 44 patients undergoing surgery for early stage (I, II) lung cancer and 47 patients with advanced NSCLC (stages III, IV). The age range was 40–85 y and there were similar number of males and females, and overall the population was >95% Caucasian.
Ten mL samples of blood were drawn into a purple top vacutainer containing K2-EDTA (Becton-Dickson), inverted twice to ensure dissolution of the EDTA, and kept on ice immediately after blood draw. The whole blood was separated into packed red cells, buffy coat, and plasma within 30 minutes of collection by centrifuging at 3,500 g for 15 min at 4 °C in a swing out rotor. All blood processing procedures were performed in a class II biosafety cabinet housed in a BSL category 2 laboratory. Plasma (0.7 mL) was aliquotted into 1.5 mL screw cap vials, flash frozen in liq. N2, and stored at −80 ° C until exosomal isolation. These collection and processing procedures were designed to minimize variations in plasma and exosome quality.
2.1.2 Exosome preparation
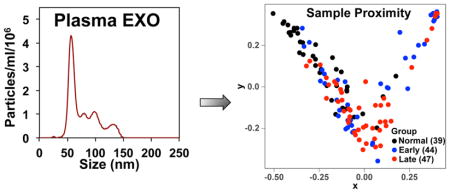
Exosomes were isolated from plasma by differential ultracentrifugation adapted from [47, 48]. 0.7 mL cleared plasma (see above) were placed in 5×41 mm polyallomer ultraclear ultracentrifuge tubes on ice, and centrifuged for 1 h at 70,000 g at 4 °C in a SWTi55 swing out rotor (Beckman). The supernatant was recentrifuged at 100,000 g for 1 h at 4°C, and the pellet was drained and resuspended in 0.7 mL cold PBS, and recentrifuged at 100,000 g for 1 h at 4°C. The washed exosomal pellets were resuspended in 100 μL nanopure water, vortexed for 30 sec and transferred to a fresh microcentrifuge tube. The ultracentrifuge tube was washed with another 100 μL of nanopure water, vortexed for 30 sec and the wash was transferred into same microcentrifuge tube, using the same pipet tip. The combined exosome suspensions were then lyophilized except for a small portion that was used for characterization by particle size distribution analysis (see below). These nanoparticles are operationally defined as exosomes.
2.1.3 Lipid extraction for exosomes
The lyophilized EXO preparations were extracted for lipidic metabolites using a solvent partitioning method with CH3CN:H2O:CHCl3 (2:1.5:1, v/v) as described previously [49]. The resulting lipid extracts were vacuum-dried in a vacuum centrifuge (Eppendorf), redissolved in 200 μL CHCl3:CH3OH (2:1) with 1 mM butylated hydroxytoluene, which was further diluted 1:20 in isopropanol/CH3OH/CHCl3 (4:2:1) with 20 mM ammonium formate for UHR-FTMS analysis.
2.2 Methods
2.2.1 Microparticle characterization
A small fraction (<1%) of each exosome preparation was characterized by size distribution analysis using a Nanosight 300 (Malvern Instruments), which provided the distribution of the Stokes’ radius (mean 60–66 nm) and the number density of the particles. A typical analysis is shown in Figure S1. The method eliminates very small particles, and provides a strongly peaked, narrow distribution at the expected size for exosomes (40–100 nm, observed mode of 60–65 nm for the main peaks in Fig S1A, B).
2.2.2 UHR-FTMS analysis of exosomal lipids
High sample throughput (≤ 16 min total cycle time per sample, <7 min for MS1 portion) was achieved using the nanoelectrospray TriVersa NanoMate (Advion Biosciences, Ithaca, NY, USA) with 1.5 kV electrospray voltage and 0.4 psi head pressure. UHR-FTMS data were acquired from an Orbitrap Fusion Tribrid (Thermo Scientific, San Jose, CA, USA) set at a resolving power of 450,000 (at 200 m/z) for MS1 full scans using 10 microscans per scan in the m/z range of 150 – 1,600, achieving sub ppm mass accuracy through > 800 m/z in positive mode. AGC (Automatic Gain Control) target was set to 1e5 and maximal injection time was set to 100 ms. During the MS1 run, the top 500 most intense monoisotopic precursor ions were isolated via quadrupole using 1 m/z isolation window and HCD (Higher Energy Collisional Dissociation) set at 25 % collision energy was performed in positive mode for data-dependent MS2 at a resolving power of 120,000 (at 200 m/z) to obtain fragments for acyl chain assignment and neutral loss of specific head groups. The AGC target was set to 5e4 with maximal injection time of 500 ms. MS2 does not distinguish the sn1 and sn2 acyl positions of glycerolipids, nor the position of unsaturations in acyl chains and acyl branching. Representative full scan MS along with an example MS2 spectrum are shown in Figure S2.
2.2.3 Lipid Assignment
The UHRMS raw data were assigned by our (CESB) in-house software PREMISE (PRecalculated Exact Mass Isotopologue Search Engine) that compares UHR-FTMS m/z data against our metabolite m/z library (calculated with mass accuracy to the 5th decimal point) to discern all known lipids and their 13C isotopologues, including hypothetical lipids, while simultaneously taking into account all of the major adducts (here H+, Na+, K+ and NH4+) [50, 51]. An in-house developed natural abundance (NA) correction algorithm [52, 53] was applied to simultaneously examine the distribution of naturally occurring 13C isotopologues of the unlabeled lipids to help verify the assigned molecular formulae, and to eliminate non-monoisotopic 13C isotopologues from further analysis. For statistical classification, we used only high accuracy monoisotopic m/z values that mapped to lipid molecular formulae, and multiple adducts of each were tracked throughout to avoid redundancy. Below, such m/z values are referred to as “lipid features”, and neither molecular formulae nor lipid names were directly used.
The number of assigned lipid features in each sample varied from 1 to 70. After combining all samples into a master file, the data set had a total of 430 such lipid features. Prior to multivariate statistical analyses, MS1 peaks arising from solvent blanks and known contaminants were removed from the lipid feature lists. As absolute intensities vary from sample to sample, the lipid features must be normalized. The intensities of the lipid features in each sample were thus normalized to the summed intensities of all mass peaks that were non-zero in 20%, 50%, 75%, 97%, 100% of all samples. This is equivalent to estimating the mole fraction of each lipid feature present, and therefore can be used for determining relative changes in composition. We found that normalization using the summed intensities of lipid features that were non-zero in 20% of all samples provided the best statistical outcome according to the ROC analysis.
2.3 Multivariate statistical analyses
2.3.1 Principal Component Analysis (PCA) and Orthogonal Partial Least Square Discriminant Analysis (OPLS-DA)
PCA and supervised OPLS-DA were performed using the SIMCA-P software package (Umetrics, Umea Sweden) to visualize the distributions of the high dimensional lipids profiles in different classes. OPLS-DA [54] is an supervised approach where the class labels are used to extract both group-predictive and group-unrelated variations in the high dimensional data. The explained variation (R2) of each component in PCA and OPLS-DA was reported.
2.3.2 Random Forest (RF)
Random Forest is a supervised classifier developed by Breiman [55] that assembles prediction results of a number of classification and regression trees (CART). Bootstrap sampling was used in the CARTs with random training sampling and replacement to fit each tree. The prediction results were calculated by averaging the results of all trained tree predictors. Bootstrap sampling and ensemble methods provided superior performance for RF analysis. Besides classification, RF provided the importance of the lipid features based on the Gini impurity reduction in every tree [55, 56] [57]. The proximity matrix was then embedded in a 2-D plot to visualize the separation among different classes and to detect outliers. The RF classification analysis was performed using scikit-learn (version 0.18rc2) library in Python (version 2.7.13). The proximity analysis was performed with the Random Forest package (version 4.6–12) in R (version 3.3.1).
2.3.3 LASSO
In parallel to RF analysis, we performed the LASSO regression analysis [58] on the same datasets. Specifically, a multinomial regression model was implemented to classify subjects into normal, early stage lung cancer, or late stage lung cancer groups, where a predicted probability for an individual belonging to each of the three groups was obtained from the model. A grouped lasso penalty was used for feature selection, which ensured that the multinomial coefficients for a variable were all in or out together in the model. The analysis was performed based on the glmnet package (version 2.0–5) in R (version 3.3.1).
2.3.4 Classification performance evaluation
For both methods, the classification performance was evaluated by 5-fold cross validation, where four fifths of the data were used for feature selection and model construction, and the area under the Receiver Operation Characteristic curve (AUROC), sensitivity and specificity of the model were evaluated based on the hold-off one fifth data. The exact m/z list with the highest importances were selected by the RF or by LASSO, representing the important lipid features based on all data. The number of lipid features selected were 16 in RF and automatically decided by the algorithm in LASSO. If the selected lipids features were of low signal-to-noise ratio, overlapped contaminants, or other artifact peaks, they were removed and the selection repeated. Classification tests were performed only after all top lipid features no longer overlapped with noise, contaminant, or other artifact peaks. For the final RF and LASSO classification test, the top 16 and 7 lipid features were selected, respectively. For RF, the 5-fold cross validation was replicated 500 times, and the average AUROC, sensitivity, specificity as well as their 95% confidence intervals were reported.
3. Results
3.1. Exploratory analysis with PCA and OPLS-DA
We first analyzed the normalized and blank-removed exosomal lipid data using classical unsupervised PCA and supervised OPLS-DA methods [54] to visualize data outliers. As shown in Figure S3, only a few outliers were evident in both types of analysis. We also noted that the PCA method did not yield a clear separation of normal from the early or late stage lung cancer subjects using the first two components (Fig. S3A). Although the separation with the OPLS-DA method was somewhat better (Fig. S3B), this supervised method tended to overfit models to data.
3.1 Classification performance of Random Forest
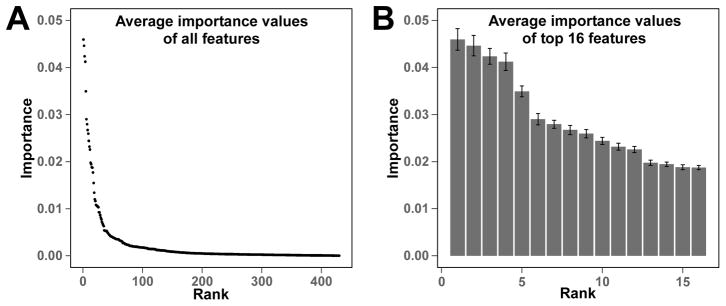
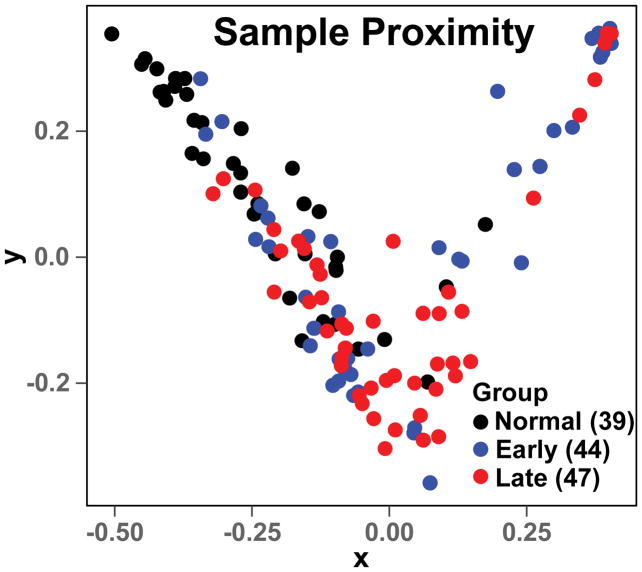
The Gini importance of a total 430 lipid features was calculated using the RF method. The number of decision trees was set to 500 based on the results of parameter tuning tests. The importance status is shown in Figure 1A. Based on the 500 decision tree test, about 2/3 of the 430 features had an importance value equal or close to 0. This showed that only 1/3 of the assigned lipid features had the capacity to discriminate different lung cancer stages. The 16 lipid features with highest Gini importance (Figure 1B) were selected for classification. The classification results of normal versus early or late lung cancer as well as early versus late lung cancer are shown in Table 1 and Figure S4. The calculated AUROCs for the normal versus cancer were ≥ 0.85 with low standard deviations, which shows the promise of using the exosomal lipid features for classifying lung cancer. In contrast, the AUROC of early versus late stage cancer was much lower (0.64), which suggests a lower potential for exosomal lipid features as classifiers of different stages of lung cancer. The AUROC results were consistent with the RF proximity plot (Figure 2), which showed good clustering of normal versus cancer with few outliers but not early versus late stage cancer.
Figure 1. Gini importance of lipid features discerned from the plasma exosomal MS data of normal and lung cancer subjects by Random Forest.
The Gini importance was calculated as described in the Methods.
(A) The Gini importance of all 430 assigned lipid features in the entire exosomal MS data set. (B) The Gini importance of the top 16 lipid features averaged over 500 tree predictors. Error bars represent standard deviations.
Table 1.
Exosomal lipid-based classification of normal versus early and late stage NSCLC using Random Forest and LASSO.
| Subjects | AUROC | Std | 95% CI | Sensitivity | Std | 95%CI | Specificity | Std | 95% CI | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Random Forest (with top 16 features) | ||||||||||||
|
| ||||||||||||
| Normal vs_Early | 0.85 | 0.09 | 0.62 | 0.99 | 0.77 | 0.16 | 0.43 | 1.00 | 0.72 | 0.17 | 0.38 | 1.00 |
| Normal vs Late | 0.88 | 0.08 | 0.69 | 1.00 | 0.84 | 0.12 | 0.57 | 1.00 | 0.72 | 0.16 | 0.42 | 1.00 |
| Early vs Late | 0.64 | 0.12 | 0.41 | 0.84 | 0.67 | 0.16 | 0.31 | 1.00 | 0.54 | 0.17 | 0.22 | 0.89 |
|
| ||||||||||||
| LASSO | ||||||||||||
|
| ||||||||||||
| Normal vs_Early | 0.79 | 0.04 | 0.71 | 0.85 | 0.65 | 0.09 | 0.46 | 0.78 | 0.77 | 0.05 | 0.66 | 0.85 |
| Normal vs Late | 0.77 | 0.04 | 0.68 | 0.83 | 0.54 | 0.08 | 0.37 | 0.70 | 0.82 | 0.04 | 0.73 | 0.89 |
| Early vs Late | 0.51 | 0.05 | 0.41 | 0.61 | 0.33 | 0.09 | 0.16 | 0.51 | 0.73 | 0.09 | 0.53 | 0.90 |
Figure 2. The Random Forest proximity plot of the lung cancer exosome mass data set.
The RF analysis was performed as described in Methods with mass ion intensities of each sample normalized to the summed intensities of lipid features that were non-zero in 20% of all samples. The top 16 lipid features from Fig. 1 were used for the proximity plot generation. The distance between two samples in the proximity plot represents their similarity during tree generating in RF.
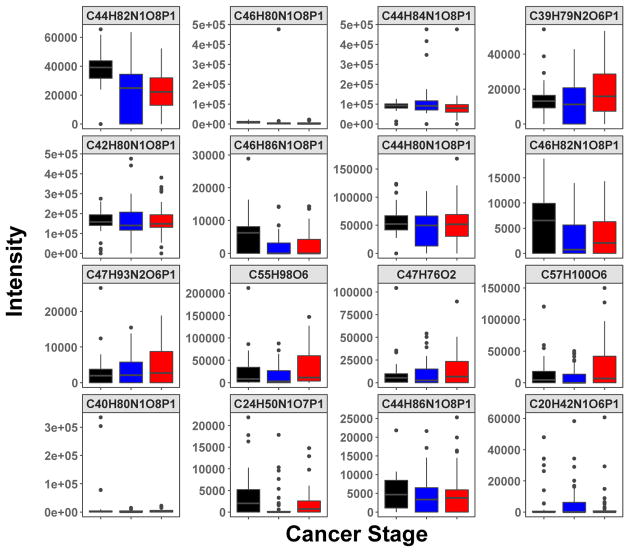
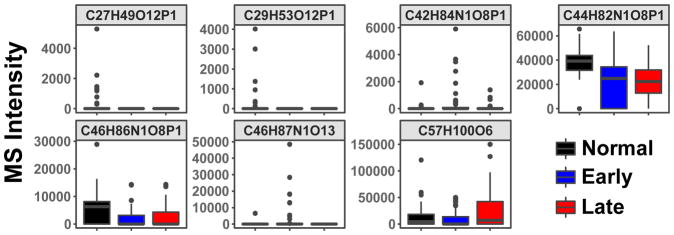
The distribution of the MS peak intensity of the top 16 lipid features (shown as molecular formulae) for classifying the three subject groups is shown as boxplots in Figure 3. They illustrated both positive and negative changes from normal to early and late stage lung cancer. These 16 lipid features were assigned based on both accurate mass and MS2 fragmentation patterns, as shown in Table 2. Many of the top lipid features mapped to phosphatidylcholines (PC) containing polyunsaturated fatty acyl (PUFA) chains, two were SM known to be enriched in exosomes, and two were lysophosphatidylcholines (LPC) shown to promote exosome biogenesis and lymphocyte chemotaxis [59]. However, it should be noted that these assignments are not fully validated.
Figure 3. The boxplots of intensities distribution of top RF-selected 16 lipid features for normal, early, and late-stage lung cancer subjects.
The top 16 lipid features were selected according to their Gini importance values (cf. Fig. 1). These features were assigned to molecular formulae (as shown) based on their accurate masses using PREMISE and validated as specific lipid species via their MS2 fragmentation patterns (cf. Table 2).
 Non cancer;
Non cancer;
 Early stage;
Early stage;
 Last stage
Last stage
Table 2.
Assignment of exosomal lipids that distinguish normal from early or late stage NSCLC using RF and LASSO.
| MF1 | adduct | accurate mass2 | lipid assignment3 | RF | LASSO |
|---|---|---|---|---|---|
|
| |||||
| C44H82N1O8P1 | [M+H]+ | 784.58508 | PC(18:1_18:2) | Yes | Yes |
| C44H84N1O8P1 | [M+H]+ | 786.60073 | PC(18:0_18:2) | Yes | No |
| C46H80N1O8P1 | [M+H]+ | 806.56943 | PC(16:0_22:6) | Yes | No |
| C42H80N1O8P1 | [M+H]+ | 758.56943 | PC(16:0_18:2) | Yes | No |
| C39H79N2O6P1 | [M+H]+ | 703.57485 | SM(d18:1_16:0) | Yes | No |
| C46H86N1O8P1 | [M+H]+ | 812.61638 | PC(18:0_20:3) | Yes | Yes |
| C44H80N1O8P1 | [M+H]+ | 782.56943 | PC(16:0_20:4) | Yes | No |
| C46H82N1O8P1 | [M+H]+ | 808.58508 | PC(16:0_22:5) | Yes | No |
| C47H76O2 | [M+Na]+ | 695.57375 | CE(20:4) | Yes | No |
| C55H98O6 | [M+Na]+ | 877.72556 | TAG(52:5) | Yes | No |
| C47H93N2O6P1 | [M+H]+ | 813.68440 | SM(d18:1_24:1) | Yes | No |
| C44H86N1O8P1 | [M+H]+ | 788.61638 | PC(18:0_18:1) | Yes | No |
| C40H80N1O8P1 | [M+H]+ | 734.56943 | PC(16:0_16:0) | Yes | No |
| C57H100O6 | [M+Na]+ | 903.74121 | TAG(54:6) | Yes | Yes |
| C24H50N1O7P1 | [M+H]+ | 496.33977 | LysoPC(16:0) | Yes | No |
| C20H42N1O6P1 | [M+H]+ | 424.28225 | LysoPC-pmg(12:0)4 | Yes | No |
Molecular formula
estimated mass error <±0.1 ppm. These lipid features (as defined in the text) were verified by further MS1 analysis as described in the Methods, except that the resolving power was set to 500,000 at m/z=200.
In this study lipid features (monoisotopic accurate m/z values) were used for classification. The molecular formulae and lipid name assignments are interpretations listed for the reader. The assignments were based on accurate mass and MS2 fragmentation patterns. CE, cholesterol esters; TAG, triacylglyceride; LysoPC-pmg, lysophosphatidylcholine-plasmalogen; PC phosphatidyl choline; SM sphingomyelin. Nomenclature according to [60,61].
This molecular formula and lipid assignment was the closest from our comprehensive lipids database. Among non-lipids there is the possibility of the non-phosphate 31P-containing compound C19H37N8O1P1, which is inconsistent with the phosphocholine fragment in the MS2 data (Fig. S2).
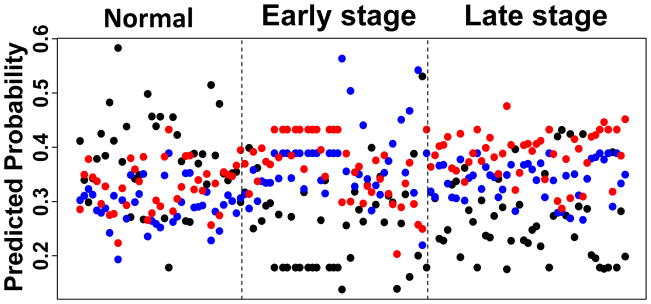
3.2 Classification performance of LASSO
The LASSO method selected 7 out of the 430 lipid features to construct a multinomial regression model. Figure 4 shows the model-predicted probabilities for each subject to be in each of the three disease status groups. For many patients, the predicted probability of belonging to the true disease group was the highest, indicating that the model was able to accurately classify a large fraction of the subjects. The MS intensity distributions of the 7 features in the three subject groups were plotted in Figure 5. We were able to confirm the lipid identity on 3 out of the 7 lipid features, which overlapped with those revealed by the RF method (Table 2). To more rigorously evaluate the performance of LASSO, a 5-fold cross validation was performed as described in the Methods section. The AUROCs for discriminating normal versus early and late stage lung cancer were 0.79 and 0.77, respectively (Table 1), which was somewhat lower than those for the RF method. However, the LASSO method gave higher specificity indices than the RF method (Table 1).
Figure 4. The LASSO predicted score of the lung cancer exosome mass data set.
The LASSO score was calculated as described in the Methods. For each sample, predicted probability scores of belonging to each of the normal, early stage, and late stage groups were calculated based on a 7-feature model obtained from LASSO. A higher score indicates a higher probability of belonging to the group. Black dots are from normal subjects, blue are from early (I, II) and red are from late stage (III, IV) lung cancer subjects.
Figure 5. Boxplots of MS intensities distribution of LASSO-selected 7 lipid features for normal, early, and late-stage lung cancer subjects.
Normalized intensities of the seven lipid species identified by LASSO as described in the text. The molecular formulae are derived from the accurate m/z and isotopologue distributions as described in the Methods. These features were assigned to molecular formulae (as shown) based on their accurate m/z values using PREMISE and three of which were cross-validated as specific lipid species via their MS2 fragmentation patterns (cf. Table 2).
4. Discussion and Conclusions
We have applied two new orthogonal multivariate statistical tools (RF and LASSO) to classify different stages of NSCLC versus healthy individuals based on UHR-FTMS analysis of lipid profiles of plasma exosomes from peripheral blood, a form of “liquid biopsy”. The data sets were large and highly sparse with many zero values and a high dynamic range, making accurate classification difficult by the classical methods (cf. Fig. S1). Using our in-house program PREMISE and NA correction to assign, verify by isotope distribution, and deisotoped m/z values to map to monoisotopic lipid structures, we assigned 430 lipid features by class, and only the sub-ppm accuracy m/z values were used for statistical analyses. Their importance to the classification was determined using RF and LASSO. For the RF method, these enabled the choice of 16 lipid features for the classification, which gave good AUROCs with reasonable sensitivity and specificity indices for discriminating normal subjects from early and late stage NSCLC patients (Table 1). In comparison, the LASSO method provided 7 features for classification, which gave somewhat lower AUROC values but higher specificity indices for the same types of classification. It is also interesting to note that three of the validated lipid features overlapped between the two methods (Table 2), which added confidence to their utility in classifying NSCLC.
The final data sets were scrutinized at multiple levels of quality control, both at the sample level, and subsequent data collection and multivariate statistical analyses. We emphasize the importance of removing contaminants/spectral artifacts and exploring normalization of the MS raw data for subsequent statistical analysis. Initial analysis by RF and LASSO without adequate correction for solvent impurities and Orbitrap spectral artifacts gave unreasonably high AUROC values of close to 1.0 for both methods. Some of the apparent classifiers turned out to be solvent impurities and spectral artifacts. After extensive investigations including manual curation of thousands of peaks and multiple iterations of artifact corrections and different normalization methods, we were able to validate the 15/16 lipid features of the RF method and 3 of the LASSO method, which should greatly improve the accuracy of the classification. Although the assignment for the 16th lipid feature with m/z=424.28225 (Table 2) was made based on the molecular formula and the tandem MS data (Fig. S2), the top candidate is not generally found in biological systems. Absolute identification will require additional information, which is beyond the scope of this study.
Since the RF and LASSO methods are independent approaches, and the congruence of these methods in terms of the overall accuracy gives us greater confidence of a meaningful result. We consider that this combined statistical approach with extensive tools for quality control represents a step forward in biomarker analysis of complex and sparse dataset. The total number of subjects used was moderate; we are in the second phase of validation to increase the overall study size with a complete blinded validation set to assess the overall accuracy for classification of NSCLC.
It should be noted that the majority of our lung cancer cohorts were smokers and many with some forms of inflammatory co-morbidities such as chronic obstructive pulmonary disease (COPD). COPD is considered to be a high risk factor for lung cancer development. Also noted is the moderate number of subjects used for this report. In a future study, we plan to increase the study size with a blinded validation set to assess the overall accuracy for NSCLC classification and to determine if exosomal lipid classifiers can be developed to discriminate COPD or other inflammatory lung diseases from early stage lung cancer.
In conclusion, both RF and LASSO-based multivariate statistical analyses of plasma exosomal lipid profiles were highly informative in discriminating normal from early and late stage lung cancer subjects with a moderate study size. The selected may not only be useful as lung cancer biomarkers but when fully validated (cf. Table 2) could also be related to important functions in exosome biogenesis and immune cell interactions.
Supplementary Material
Highlights.
The lipid profiles of peripheral blood plasma exosomes distinguish between early stage lung cancer and healthy individuals
A large number of lipids is resolved in direct infusion UHR-FTMS
A workflow for quality control and statistical analysis of the exosomal lipid data has been established
The multivariate techniques LASSO and Random Forest were both able to identify lipid features that successfully classify early stage lung cancer patient from healthy individuals
Acknowledgments
We thank Timothy Fahrenholz for collecting mass spectra in the early phase of this work.
Funding. This work was supported in part by NIH grants 1P01CA163223-01A1 (to ANL and TWMF), 1U24DK097215-01A1 (to RMH, TWMF, and ANL), 1R03CA222449-01 (to JL and RMH), the Redox Metabolism Shared Resource(s) of the University of Kentucky Markey Cancer Center (P30CA177558), and the Kentucky Lung Cancer Research Program KLCRP 3048112440 (to TWMF, ANL, SA).
Abbreviations
- AUROC
area under the receiver operator characteristic curve
- UHR-FTMS
ultra high-resolution Fourier transform mass spectrometry
- LASSO
Least Absolute Shrinkage and Selection Operator
- NSCLC
non-small cell lung cancer
- PCA
Principal Component Analysis
- OPLS-DA
Orthogonal partial least square discriminant analysis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hayat MJ, Howlader N, Reichman ME, Edwards BK. Cancer statistics, trends, and multiple primary cancer analyses from the surveillance, epidemiology, and end results (SEER) program. Oncologist. 2007;12:20–37. doi: 10.1634/theoncologist.12-1-20. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011. CA: a cancer journal for clinicians. 2011;61:212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA: a cancer journal for clinicians. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 4.Hopenhayn C, Jenkins TM, PJ The burden of lung cancer in Kentucky. J Ky Med Assoc. 2003;101:15–20. [PubMed] [Google Scholar]
- 5.Greenberg AK, Lee MS. Biomarkers for lung cancer: clinical uses. Curr Opin Pulm Med. 2007;13:249–255. doi: 10.1097/MCP.0b013e32819f8f06. [DOI] [PubMed] [Google Scholar]
- 6.Hayat MJ, Howlader N, Reichman ME, Edwards BK. Cancer statistics, trends, and multiple primary cancer analyses from the Surveillance, Epidemiology, and End Results (SEER) Program. Oncologist. 2007;12:20–37. doi: 10.1634/theoncologist.12-1-20. [DOI] [PubMed] [Google Scholar]
- 7.Unger M, Pause A. Progress, and Reassessment in Lung Cancer Screening. N Engl J Med. 2006;355:1822–1824. doi: 10.1056/NEJMe068207. [DOI] [PubMed] [Google Scholar]
- 8.Collins LG, Haines C, Perkel R, Enck RE. Lung cancer: Diagnosis and management. American Family Physician. 2007;75:56–63. [PubMed] [Google Scholar]
- 9.Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, Gareen IF, Gatsonis C, Marcus PM, Sicks JD. Reduced lung-cancer mortality with low-dose computed tomographic screening. The New England journal of medicine. 2011;365:395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, Shukla SA, Guo GW, Brooks AN, Murray BA, Imielinski M, Hu X, Ling SY, Akbani R, Rosenberg M, Cibulskis C, Ramachandran A, Collisson EA, Kwiatkowski DJ, Lawrence MS, Weinstein JN, Verhaak RGW, Wu CJ, Hammerman PS, Cherniack AD, Getz G, Artyomov MN, Schreiber R, Govindan R, Meyerson M N. Canc Genome Atlas Res. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nature Genetics. 2016;48:607-+. doi: 10.1038/ng.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collisson EA, Campbell JD, Brooks AN, Berger AH, Lee W, Chmielecki J, Beer DG, Cope L, Creighton CJ, Danilova L, Ding L, Getz G, Hammerman PS, Hayes DN, Hernandez B, Herman JG, Heymach JV, Jurisica I, Kucherlapati R, Kwiatkowski D, Ladanyi M, Robertson G, Schultz N, Shen RL, Sinha R, Sougnez C, Tsao MS, Travis WD, Weinstein JN, Wigle DA, Wilkerson MD, Chu AD, Cherniack AD, Hadjipanayis A, Rosenberg M, Weisenberger DJ, Laird PW, Radenbaugh A, Ma SG, Stuart JM, Byers LA, Baylin SB, Govindan R, Meyerson M, Rosenberg M, Gabriel SB, Cibulskis K, Sougnez C, Kim J, Stewart C, Lichtenstein L, Lander ES, Lawrence MS, Getz, Kandoth C, Fulton R, Fulton LL, McLellan MD, Wilson RK, Ye K, Fronick CC, Maher CA, Miller CA, Wendl MC, Cabanski C, Ding L, Mardis E, Govindan R, Creighton CJ, Wheeler D, Balasundaram M, Butterfield YSN, Carlsen R, Chu AD, Chuah E, Dhalla N, Guin R, Hirst C, Lee D, Li HYI, Mayo M, Moore RA, Mungall AJ, Schein JE, Sipahimalani P, Tam A, Varhol R, Robertson AG, Wye N, Thiessen N, Holt RA, Jones SJM, Marra MA, Campbell JD, Brooks AN, Chmielecki J, Imielinski M, Onofrio RC, Hodis E, Zack T, Sougnez C, Helman E, Pedamallu CS, Mesirov J, Cherniack AD, Saksena G, Schumacher SE, Carter SL, Hernandez B, Garraway L, Beroukhim R, Gabriel SB, Getz G, Meyerson M, Hadjipanayis A, Lee S, Mahadeshwar HS, Pantazi A, Protopopov A, Ren XJ, Seth S, Song XZ, Tang JB, Yang LX, Zhang JH, Chen PC, Parfenov M, Xu AW, Santoso N, Chin L, Park PJ, Kucherlapati R, Hoadley KA, Auman JT, Meng SW, Shi Y, Buda E, Waring S, Veluvolu U, Tan DH, Mieczkowski PA, Jones CD, Simons JV, Soloway MG, Bodenheimer T, Jefferys SR, Roach J, Hoyle AP, Wu JY, Balu S, Singh D, Prins JF, Marron JS, Parker JS, Hayes DN, Perou CM, Liu JZ, Cope L, Danilova L, Weisenberger DJ, Maglinte DT, Lai PH, Bootwalla MS, Van Den Berg DJ, Triche T, Baylin SB, Laird PW, Rosenberg M, Chin L, Zhang JH, Cho J, DiCara D, Heiman D, Lin P, Mallard W, Voet D, Zhang HL, Zou LH, Noble MS, Lawrence MS, Saksena G, Gehlenborg N, Thorvaldsdottir H, Mesirov J, Nazaire MD, Robinson J, Getz G, Lee W, Aksoy BA, Ciriello G, Taylor BS, Dresdner G, Gao JJ, Gross B, Seshan VE, Ladanyi M, Reva B, Sinha R, Sumer SO, Weinhold N, Schultz N, Shen RL, Sander C, Ng S, Ma S, Zhu JC, Radenbaugh A, Stuart JM, Benz CC, Yau C, Haussler D, Spellman PT, Wilkerson MD, Parker JS, Hoadley KA, Kimes PK, Hayes DN, Perou CM, Broom BM, Wang J, Lu YL, Ng PKS, Diao LX, Byers LA, Liu WB, Heymach JV, Amos CI, Weinstein JN, Akbani R, Mills GB, Curley E, Paulauskis J, Lau K, Morris S, Shelton T, Mallery D, Gardner J, Penny R, Saller C, Tarvin K, Richards WG, Cerfolio R, Bryant A, Raymond DP, Pennell NA, Farver C, Czerwinski C, Huelsenbeck-Dill L, Iacocca M, Petrelli N, Rabeno B, Brown J, Bauer T, Dolzhanskiy O, Potapova O, Rotin D, Voronina O, Nemirovich-Danchenko E, Fedosenko KV, Gal A, Behera M, Ramalingam SS, Sica G, Flieder D, Boyd J, Weaver J, Kohl B, Thinh DHQ, Sandusky G, Juhl H, Duhig E, Illei P, Gabrielson E, Shin J, Lee B, Rogers K, Trusty D, Brock MV, Williamson C, Burks E, Rieger-Christ K, Holway A, Sullivan T, Wigle DA, Asiedu MK, Kosari F, Travis WD, Rekhtman N, Zakowski M, Rusch VW, Zippile P, Suh J, Pass H, Goparaju C, Owusu-Sarpong Y, Bartlett JMS, Kodeeswaran S, Parfitt J, Sekhon H, Albert M, Eckman J, Myers JB, Cheney R, Morrison C, Gaudioso C, Borgia JA, Bonomi P, Pool M, Liptay MJ, Moiseenko F, Zaytseva I, Dienemann H, Meister M, Schnabel PA, Muley TR, Peifer M, Gomez-Fernandez C, Herbert L, Egea S, Huang M, Thorne LB, Boice L, Salazar AH, Funkhouser WK, Rathmell WK, Dhir R, Yousem SA, Dacic S, Schneider F, Siegfried JM, Hajek R, Watson MA, McDonald S, Meyers B, Clarke B, Yang IA, Fong KM, Hunter L, Windsor M, Bowman RV, Peters S, Letovanec I, Khan KZ, Jensen MA, Snyder EE, Srinivasan D, Kahn AB, Baboud J, Pot DA, Shaw KRM, Sheth M, Davidsen T, Demchok JA, Yang LM, Wang ZN, Tarnuzzer R, Zenklusen JC, Ozenberger BA, Sofia HJ, Travis WD, Cheney R, Clarke B, Sanja D, Duhig E, Funkhouser WK, Illei P, Farver C, Rekhtman N, Sica G, Suh J, Tsao MS N. Canc Genome Atlas Res. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, Sougnez C, Auclair D, Lawrence MS, Stojanov P, Cibulskis K, Choi K, de Waal L, Sharifnia T, Brooks A, Greulich H, Banerji S, Zander T, Seidel D, Leenders F, Ansen S, Ludwig C, Engel-Riedel W, Stoelben E, Wolf J, Goparju C, Thompson K, Winckler W, Kwiatkowski D, Johnson BE, Janne PA, Miller VA, Pao W, Travis WD, Pass HI, Gabriel SB, Lander ES, Thomas RK, Garraway LA, Getz G, Meyerson M. Mapping the Hallmarks of Lung Adenocarcinoma with Massively Parallel Sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammerman PS, Lawrence MS, Voet D, Jing R, Cibulskis K, Sivachenko A, Stojanov P, McKenna A, Lander ES, Gabriel S, Getz G, Sougnez C, Imielinski M, Helman E, Hernandez B, Pho NH, Meyerson M, Chu A, Chun HJE, Mungall AJ, Pleasance E, Robertson AG, Sipahimalani P, Stoll D, Balasundaram M, Birol I, Butterfield YSN, Chuah E, Coope RJN, Corbett R, Dhalla N, Guin R, Hirst AC, Hirst M, Holt RA, Lee D, Li HI, Mayo M, Moore RA, Mungall K, Nip KM, Olshen A, Schein JE, Slobodan JR, Tam A, Thiessen N, Varhol R, Zeng T, Zhao Y, Jones SJM, Marra MA, Saksena G, Cherniack AD, Schumacher SE, Tabak B, Carter SL, Pho NH, Nguyen H, Onofrio RC, Crenshaw A, Ardlie K, Beroukhim R, Winckler W, Hammerman PS, Getz G, Meyerson M, Protopopov A, Zhang JH, Hadjipanayis A, Lee S, Xi RB, Yang LX, Ren XJ, Zhang HL, Shukla S, Chen PC, Haseley P, Lee E, Chin L, Park PJ, Kucherlapati R, Socci ND, Liang YP, Schultz N, Borsu L, Lash AE, Viale A, Sander C, Ladanyi M, Auman JT, Hoadley KA, Wilkerson MD, Shi Y, Liquori C, Meng SW, Li L, Turman YJ, Topal MD, Tan DH, Waring S, Buda E, Walsh J, Jones CD, Mieczkowski PA, Singh D, Wu J, Gulabani A, Dolina P, Bodenheimer T, Hoyle AP, Simons JV, Soloway MG, Mose LE, Jefferys SR, Balu S, O’Connor BD, Prins JF, Liu J, Chiang DY, Hayes DN, Perou CM, Cope L, Danilova L, Weisenberger DJ, Maglinte DT, Pan F, den Berg DJ, Triche T, Herman JG, Baylin SB, Laird PW, Getz G, Noble M, Voet D, Saksena G, Gehlenborg N, DiCara D, Zhang JH, Zhang HL, Wu CJ, Liu SY, Lawrence MS, Zou LH, Sivachenko A, Lin P, Stojanov P, Jing R, Cho J, Nazaire MD, Robinson J, Thorvaldsdottir H, Mesirov J, Park PJ, Chin L, Schultz N, Sinha R, Ciriello G, Cerami E, Gross B, Jacobsen A, Gao J, Aksoy BA, Weinhold N, Ramirez R, Taylor BS, Antipin Y, Reva B, Shen RL, Mo Q, Seshan V, Paik PK, Ladanyi M, Sander C, Akbani R, Zhang NX, Broom BM, Casasent T, Unruh A, Wakefield C, Cason RC, Baggerly KA, Weinstein JN, Haussler D, Benz CC, Stuart JM, Zhu JC, Szeto C, Scott GK, Yau C, Ng S, Goldstein T, Waltman P, Sokolov A, Ellrott K, Collisson EA, Zerbino D, Wilks C, Ma S, Craft B, Wilkerson MD, Auman JT, Hoadley KA, Du Y, Cabanski C, Walter V, Singh D, Wu JY, Gulabani A, Bodenheimer T, Hoyle AP, Simons JV, Soloway MG, Mose LE, Jefferys SR, Balu S, Marron JS, Liu Y, Wang K, Liu J, Prins JF, Hayes DN, Perou CM, Creighton CJ, Zhang YQ, Travis WD, Rekhtman N, Yi J, Aubry MC, Cheney R, Dacic S, Flieder D, Funkhouser W, Illei P, Myers J, Tsao MS, Penny R, Mallery D, Shelton T, Hatfield M, Morris S, Yena P, Shelton C, Sherman M, Paulauskis J, Meyerson M, Baylin SB, Govindan R, Akbani R, Azodo I, Beer D, Bose R, Byers LA, Carbone D, Chang LW, Chiang D, Chu A, Chun E, Collisson E, Cope L, Creighton CJ, Danilova L, Ding L, Getz G, Hammerman PS, Hayes DN, Hernandez B, Herman JG, Heymach J, Ida C, Imielinski M, Johnson B, Jurisica I, Kaufman J, Kosari F, Kucherlapati R, Kwiatkowski D, Ladanyi M, Lawrence MS, Maher CA, Mungall A, Ng S, Pao W, Peifer M, Penny R, Robertson G, Rusch V, Sander C, Schultz N, Shen RL, Siegfried J, Sinha R, Sivachenko A, Sougnez C, Stoll D, Stuart J, Thomas RK, Tomaszek S, Tsao MS, Travis WD, Vaske C, Weinstein JN, Weisenberger D, Wheeler D, Wigle DA, Wilkerson MD, Wilks C, Yang P, Zhang JJ, Jensen MA, Sfeir R, Kahn AB, Chu AL, Kothiyal P, Wang Z, Snyder EE, Pontius J, Pihl TD, Ayala B, Backus M, Walton J, Baboud J, Berton DL, Nicholls MC, Srinivasan D, Raman R, Girshik S, Kigonya PA, Alonso S, Sanbhadti RN, Barletta SP, Greene JM, Pot DA, Tsao MS, Bandarchi-Chamkhaleh B, Boyd J, Weaver J, Wigle DA, Azodo IA, Tomaszek SC, Aubry MC, Ida CM, Yang P, Kosari F, Brock MV, Rogers K, Rutledge M, Brown T, Lee B, Shin J, Trusty D, Dhir R, Siegfried JM, Potapova O, Fedosenko KV, Nemirovich-Danchenko E, Rusch V, Zakowski M, Iacocca MV, Brown J, Rabeno B, Czerwinski C, Petrelli N, Fan Z, Todaro N, Eckman J, Myers J, Rathmell WK, Thorne LB, Huang M, Boice L, Hill A, Penny R, Mallery D, Curley E, Shelton C, Yena P, Morrison C, Gaudioso C, Bartlett JS, Kodeeswaran S, Zanke B, Sekhon H, David K, Juhl H, Van Le X, Kohl B, Thorp R, Tien NV, Van Bang N, Sussman H, Phu BD, Hajek R, PhiHung N, Khan KZ, Muley T, Shaw KRM, Sheth M, Yang L, Buetow K, Davidsen T, Demchok JA, Eley G, Ferguson M, Dillon LAL, Schaefer C, Guyer MS, Ozenberger BA, Palchik JD, Peterson J, Sofia HJ, Thomson E, Meyerson M N. Canc Genome Atlas Res. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skoulidis F, Byers LA, Diao LX, Papadimitrakopoulou VA, Tong P, Izzo J, Behrens C, Kadara H, Parra ER, Canales JR, Zhang JJ, Giri U, Gudikote J, Cortez MA, Yang C, Fan YH, Peyton M, Girard L, Coombes KR, Toniatti C, Heffernan TP, Choi M, Frampton GM, Miller V, Weinstein JN, Herbst RS, Wong KK, Zhang JH, Sharma P, Mills GB, Hong WK, Minna JD, Allison JP, Futreal A, Wang J, Wistuba, Heymach JV. Co-occurring Genomic Alterations Define Major Subsets of KRAS-Mutant Lung Adenocarcinoma with Distinct Biology, Immune Profiles, and Therapeutic Vulnerabilities. Cancer Discovery. 2015;5:860–877. doi: 10.1158/2159-8290.CD-14-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 16.Lane AN, Fan TW-M, Bousamra M, II, Higashi RM, Yan J, Miller DM. Stable Isotope-Resolved Metabolomics (SIRM) in Cancer Reseach with Clinical Applications of Non-Small Cell Lung Cancer. Omics. 2011;15:173–182. doi: 10.1089/omi.2010.0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zaugg K, Yao Y, Reilly PT, Kannan K, Kiarash R, Mason J, Huang P, Sawyer SK, Fuerth B, Faubert B, Kalliomaki T, Elia A, Luo X, Nadeem V, Bungard D, Yalavarthi S, Growney JD, Wakeham A, Moolani Y, Silvester J, Ten AY, Bakker W, Tsuchihara K, Berger SL, Hill RP, Jones RG, Tsao M, Robinson MO, Thompson CB, Pan G, Mak TW. Carnitine palmitoyltransferase 1C promotes cell survival and tumor growth under conditions of metabolic stress. Genes Dev. 2011;25:1041–1051. doi: 10.1101/gad.1987211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beloribi-Djefaflia S, Vasseur S, Guillaumond F. Lipid metabolic reprogramming in cancer cells. Oncogenesis. 2016;5:e189. doi: 10.1038/oncsis.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muralidharan-Chari V, Clancy JW, Sedgwick A, D’Souza-Schorey C. Microvesicles: mediators of extracellular communication during cancer progression. Journal of cell science. 2010;123:1603–1611. doi: 10.1242/jcs.064386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zech D, Rana S, Buchler MW, Zoller M. Tumor-exosomes and leukocyte activation: an ambivalent crosstalk. Cell Commun Signal. 2012;10:37. doi: 10.1186/1478-811X-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rak J. Microparticles in Cancer. Seminars in Thrombosis and Hemostasis. 2010;36:888–906. doi: 10.1055/s-0030-1267043. [DOI] [PubMed] [Google Scholar]
- 22.Liu C, Yu S, Zinn K, Wang J, Zhang L, Jia Y, Kappes JC, Barnes S, Kimberly RP, Grizzle WE, Zhang HG. Murine mammary carcinoma exosomes promote tumor growth by suppression of NK cell function. Journal of immunology. 2006;176:1375–1385. doi: 10.4049/jimmunol.176.3.1375. [DOI] [PubMed] [Google Scholar]
- 23.Couzin J. Cell biology: The ins and outs of exosomes. Science. 2005;308:1862–1863. doi: 10.1126/science.308.5730.1862. [DOI] [PubMed] [Google Scholar]
- 24.Wysoczynski M, Ratajczak MZ. Lung cancer secreted microvesicles: underappreciated modulators of microenvironment in expanding tumors. International journal of cancer Journal international du cancer. 2009;125:1595–1603. doi: 10.1002/ijc.24479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janowska-Wieczorek A, Wysoczynski M, Kijowski J, Marquez-Curtis L, Machalinski B, Ratajczak J, Ratajczak MZ. Microvesicles derived from activated platelets induce metastasis and angiogenesis in lung cancer. Int J Cancer. 2005;113:752–760. doi: 10.1002/ijc.20657. [DOI] [PubMed] [Google Scholar]
- 26.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arscott WT, Camphausen KA. EGFR isoforms in exosomes as a novel method for biomarker discovery in pancreatic cancer. Biomark Med. 2011;5:821. doi: 10.2217/bmm.11.80. [DOI] [PubMed] [Google Scholar]
- 28.Gesierich S, Berezovskiy I, Ryschich E, Zoller M. Systemic induction of the angiogenesis switch by the tetraspanin D6.1A/CO-029. Cancer Res. 2006;66:7083–7094. doi: 10.1158/0008-5472.CAN-06-0391. [DOI] [PubMed] [Google Scholar]
- 29.Costa-Silva B, Aiello NM, Ocean AJ, Singh S, Zhang H, Thakur BK, Becker A, Hoshino A, Mark MT, Molina H, Xiang J, Zhang T, Theilen TM, Garcia-Santos G, Williams C, Ararso Y, Huang Y, Rodrigues G, Shen TL, Labori KJ, Lothe IMB, Kure EH, Hernandez J, Doussot A, Ebbesen SH, Grandgenett PM, Hollingsworth MA, Jain M, Mallya K, Batra SK, Jarnagin WR, Schwartz RE, Matei I, Peinado H, Stanger BZ, Bromberg J, Lyden D. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 2015;17:816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, Singh S, Williams C, Soplop N, Uryu K, Pharmer L, King T, Bojmar L, Davies AE, Ararso Y, Zhang T, Zhang H, Hernandez J, Weiss JM, Dumont-Cole VD, Kramer K, Wexler LH, Narendran A, Schwartz GK, Healey JH, Sandstrom P, Labori KJ, Kure EH, Grandgenett PM, Hollingsworth MA, de Sousa M, Kaur S, Jain M, Mallya K, Batra SK, Jarnagin WR, Brady MS, Fodstad O, Muller V, Pantel K, Minn AJ, Bissell MJ, Garcia BA, Kang Y, Rajasekhar VK, Ghajar CM, Matei I, Peinado H, Bromberg J, Lyden D. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frydrychowicz M, Kolecka-Bednarczyk A, Madejczyk M, Yasar S, Dworacki G. Exosomes - structure, biogenesis and biological role in non-small-cell lung cancer. Scandinavian journal of immunology. 2015;81:2–10. doi: 10.1111/sji.12247. [DOI] [PubMed] [Google Scholar]
- 32.Safaei R, Larson BJ, Cheng TC, Gibson MA, Otani S, Naerdemann W, Howell SB. Abnormal lysosomal trafficking and enhanced exosomal export of cisplatin in drug-resistant human ovarian carcinoma cells. Mol Cancer Ther. 2005;4:1595–1604. doi: 10.1158/1535-7163.MCT-05-0102. [DOI] [PubMed] [Google Scholar]
- 33.Yu DD, Wu Y, Shen HY, Lv MM, Chen WX, Zhang XH, Zhong SL, Tang JH, Zhao JH. Exosomes in development, metastasis and drug resistance of breast cancer. Cancer science. 2015;106:959–964. doi: 10.1111/cas.12715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rahman MA, Barger JF, Lovat F, Gao M, Otterson GA, Nana-Sinkam P. Lung cancer exosomes as drivers of epithelial mesenchymal transition. Oncotarget. 2016 doi: 10.18632/oncotarget.10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao X, Yu S, Li S, Wu J, Ma R, Cao H, Zhu Y, Feng J. Exosomes: decreased sensitivity of lung cancer A549 cells to cisplatin. PLoS One. 2014;9:e89534. doi: 10.1371/journal.pone.0089534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beloribi S, Ristorcelli E, Breuzard G, Silvy F, Bertrand-Michel J, Beraud E, Verine A, Lombardo D. Exosomal lipids impact notch signaling and induce death of human pancreatic tumoral SOJ-6 cells. PLoS One. 2012;7:e47480. doi: 10.1371/journal.pone.0047480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beloribi-Djefaflia S, Siret C, Lombardo D. Exosomal lipids induce human pancreatic tumoral MiaPaCa-2 cells resistance through the CXCR4-SDF-1alpha signaling axis. Oncoscience. 2015;2:15–30. doi: 10.18632/oncoscience.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parolini I, Federici C, Raggi C, Lugini L, Palleschi S, De Milito A, Coscia C, Iessi E, Logozzi M, Molinari A, Colone M, Tatti M, Sargiacomo M, Fais S. Microenvironmental pH is a key factor for exosome traffic in tumor cells. The Journal of biological chemistry. 2009;284:34211–34222. doi: 10.1074/jbc.M109.041152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Plebanek MP, Mutharasan RK, Volpert O, Matov A, Gatlin JC, Thaxton CS. Nanoparticle Targeting and Cholesterol Flux Through Scavenger Receptor Type B-1 Inhibits Cellular Exosome Uptake. Scientific reports. 2015;5:15724. doi: 10.1038/srep15724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carracedo A, Gironella M, Lorente M, Garcia S, Guzman M, Velasco G, Iovanna JL. Cannabinoids induce apoptosis of pancreatic tumor cells via endoplasmic reticulum stress-related genes. Cancer Res. 2006;66:6748–6755. doi: 10.1158/0008-5472.CAN-06-0169. [DOI] [PubMed] [Google Scholar]
- 41.Madhavan B, Yue S, Galli U, Rana S, Gross W, Muller M, Giese NA, Kalthoff H, Becker T, Buchler MW, Zoller M. Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int J Cancer. 2015;136:2616–2627. doi: 10.1002/ijc.29324. [DOI] [PubMed] [Google Scholar]
- 42.Komatsu S, Ichikawa D, Takeshita H, Morimura R, Hirajima S, Tsujiura M, Kawaguchi T, Miyamae M, Nagata H, Konishi H, Shiozaki A, Otsuji E. Circulating miR-18a: a sensitive cancer screening biomarker in human cancer. In vivo (Athens, Greece) 2014;28:293–297. [PubMed] [Google Scholar]
- 43.Zoller M. Pancreatic cancer diagnosis by free and exosomal miRNA. World J Gastrointest Pathophysiol. 2013;4:74–90. doi: 10.4291/wjgp.v4.i4.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Que R, Ding G, Chen J, Cao L. Analysis of serum exosomal microRNAs and clinicopathologic features of patients with pancreatic adenocarcinoma. World J Surg Oncol. 2013;11:219. doi: 10.1186/1477-7819-11-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jakobsen KR, Paulsen BS, Bæk R, Varming K, Sorensen BS, Jørgensen MM. Exosomal proteins as potential diagnostic markers in advanced non-small cell lung carcinoma. Journal of Extracellular Vesicles. 2015;4 doi: 10.3402/jev.v3404.26659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rabinowits G, Gercel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10:42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 47.Baranyai T, Herczeg K, Onodi Z, Voszka I, Modos K, Marton N, Nagy G, Maeger I, Wood MJ, El Andaloussi S, Palinkas Z, Kumar V, Nagy P, Kittel A, Buzas EI, Ferdinandy P, Giricz Z. Isolation of Exosomes from Blood Plasma: Qualitative and Quantitative Comparison of Ultracentrifugation and Size Exclusion Chromatography Methods. Plos One. 2015;10 doi: 10.1371/journal.pone.0145686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koga K, Matsumoto K, Akiyoshi T, Kubo M, Yamanaka N, Tasaki A, Nakashima H, Nakamura M, Kurok S, Tanaka M, Katano M. Purification, characterization and biological significance of tumor-derived exosomes. Anticancer Research. 2005;25:3703–3707. [PubMed] [Google Scholar]
- 49.Fan TW-M. Sample Preparation for Metabolomics Investigation. In: Fan TW-M, Lane AN, Higashi RM, editors. The Handbook of Metabolomics: Pathway and Flux Analysis, Methods in Pharmacology and Toxicology. Springer Science; New York: 2012. pp. 7–27. [DOI] [Google Scholar]
- 50.Lane AN, Fan TWM, Xie X, Moseley HN, Higashi RM. Stable isotope analysis of lipid biosynthesis by high resolution mass spectrometry and NMR. Anal Chim Acta. 2009;651:201–208. doi: 10.1016/j.aca.2009.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lane AN, Fan TW, Higashi RM. Isotopomer-based metabolomic analysis by NMR and mass spectrometry. Methods Cell Biol. 2008;84:541–588. doi: 10.1016/S0091-679X(07)84018-0. [DOI] [PubMed] [Google Scholar]
- 52.Carreer WJ, Flight RM, Moseley HN. A Computational Framework for High-Throughput Isotopic Natural Abundance Correction of Omics-Level Ultra-High Resolution FT-MS Datasets. Metabolites. 2013;3 doi: 10.3390/metabo3040853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moseley HN. Correcting for the effects of natural abundance in stable isotope resolved metabolomics experiments involving ultra-high resolution mass spectrometry. BMC Bioinformatics. 2010;11:139. doi: 10.1186/1471-2105-11-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Worley B, Powers R. Multivariate Analysis in Metabolomics. Current Metabolomics. 2013;1:92–107. doi: 10.2174/2213235X11301010092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Breiman L. Random forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 56.Qi Y. Random Forest for Bioinformatics. In: Zhang CMY, editor. Ensemble Machine Learning. Springer; Boston: 2012. pp. 307–323. [Google Scholar]
- 57.Menze BH, Kelm BM, Masuch R, Himmelreich U, Bachert P, Petrich W, Hamprecht FA. A comparison of random forest and its Gini importance with standard chemometric methods for the feature selection and classification of spectral data. BMC Bioinformatics. 2009;10:213. doi: 10.1186/1471-2105-10-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tibshirani R. Regression shrinkage and selection via the lasso. J Royal Statistical Society, Series B (Methodological) 1996:267–288. [Google Scholar]
- 59.Subra C, Laulagnier K, Perret B, Record M. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie. 2007;89:205–212. doi: 10.1016/j.biochi.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 60.Fahy E, Subramaniam S, Murphy RC, Nishijima M, Raetz CRH, Shimizu T, Spener F, van Meer G, Wakelam MJO, Dennis EA. Update of the LIPID MAPS comprehensive classification system for lipids. Journal of Lipid Research. 2009;50:S9–S14. doi: 10.1194/jlr.R800095-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liebisch G, Vizcaíno JA, Köfeler H, Trötzmüller M, Griffiths WJ, Schmitz G, Spener F, Wakelam MJO. Shorthand notation for lipid structures derived from mass spectrometry. J Lipid Res. 2013;54:1523–1530. doi: 10.1194/jlr.M033506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.