Abstract
The fMRI-based functional connectome was shown to be sufficiently unique to allow individual identification (fingerprinting). We aimed to test whether a fNIRS-based connectome could also be used to identify individuals. Forty-four participants performed experimental protocols that consisted of two periods of resting-state interleaved by a cognitive task period. Connectome identification was performed for all possible pairwise combinations of the three periods. The influence of hemodynamic global variation was tested using global signal regression and principal component analysis. High identification accuracies well-above chance level (2.3%) were observed overall, being particularly high (93%) to the oxyhemoglobin signal between resting conditions. Our results suggest that fNIRS is a suitable technique to assess connectome fingerprints.
1. Introduction
The scientific understanding of brain dynamics and organization has greatly advanced with the development of techniques based on the neuro-hemodynamic coupling, particularly BOLD fMRI. Recently, the observation of significant interindividual variabilities in human neuroanatomy and brain function has received increasing attention [1], and has been consistently associated with individual differences in cognitive and behavioral profiles [2]. In fMRI research, the functional connectome, defined as the whole-brain profile of functional connectivity, has been shown to vary significantly among participants [3–5]. An individual functional connectome is represented as a matrix composed by the functional connectivities between distinct brain regions. Remarkable mounting evidence supports the claim that this connectivity matrix is sufficiently unique to allow for individual identification or “connectome fingerprinting” [3–8]. Moreover, mono or dizygotic twins can also be identified based only on their functional connectome, suggesting that this functional architecture is partially heritable [9,10]. Applications of the connectome fingerprint approach have already been proposed for investigating neurodevelopment [6], cognition [4] and psychiatric conditions [4–6].
Findings suggest that connectome fingerprint is a potential marker of mental health in adolescents during neurodevelopment and can provide information about stability of functional brain in adults [6]. Disorders on the stability of functional brain indicates some psychiatric disturbs, as schizophrenia [5]. To date, the connectome fingerprinting approach has been restricted to fMRI studies. Another technic, also based on neurovascular coupling, is the functional near-infrared spectroscopy (fNIRS). Despite presenting a lower spatial resolution and only sparsely sampling the cortical surface in comparison with the fine whole-brain representation of fMRI, fNIRS might provides reliable cortical hemodynamic information with high temporal resolution, relative low cost and great portability [11–14]. Several studies have demonstrated that fNIRS is a suitable tool to evaluate brain functional connectivity [15–18]. Besides supporting the feasibility of fNIRS for the study of meaningful functional connectivity variations, these previous findings also suggest the possibility of assessing individual connetome profiles [17].
Here we aimed to investigate whether functional connectomes, determined from fNIRS data, contain sufficient information to uniquely identify an individual in a data set. This investigation might contribute for: (i) providing an independent evidence for the existence of individual functional connectomes or “fingerprints” in humans and (ii) allowing the expansion of such a remarkable fMRI finding to populations where performing an MRI scanning is impossible or inappropriate. These contributions might be of particular relevance considering the potential application of fNIRS connectome to the study of child neurodevelopment and specific clinical situations.
In the present study, fNIRS data from 44 participants was collected during resting-state and a range of cognitive tasks, following a rest-task-rest sequence. In order to evaluate fNIRS-based connectome identification, a functional connectivity matrix was constructed for each participant and acquisition period. Conventional preprocessing steps were performed and the influence of each of these steps on individual identification accuracies was determined. Moreover, to further investigate the potential influence of global hemodynamic variation on the identification accuracy, the data were analyzed both before and after removing global signal (using both Global Signal Regression, GSR; and Partial Component Analysis, PCA). Applying global signal regression to fNIRS signal at least partially removed physiological non-neuronal sources of noise [19].
2. Methods
2.1 Experimental protocol
A total of 44 undergraduate students (13 women, average (standard deviation) age = 22 (3.1) years old) were recruited. Exclusion criteria included self-reported current or past neurological or psychiatric conditions, recent use of psychoactive substances or of drugs with cardiovascular effects. Procedures were approved by the local Ethics Committee and all participants were thoroughly informed about the experimental procedure before signing the consent form.
The experimental design consisted of two 6-minute resting-state sessions interleaved with a task period (i.e., rest1-task-rest2). Participants were instructed to fixate their gaze at a white cross at the center of a monitor with a black background during the resting periods and on the baseline blocks of the mixed block event-related task. The tasks were the same applied in a seminal fMRI work on the mapping of functional networks engaged across tasks on different cognitive domains, namely the control network [20]. Based on this study, we analyzed all tasks together in order to represent a task-related cognitive state in contrast with the task-independent resting-state. Further details on the tasks design are presented on Table 1 and the detailed analysis of task-related activations will be object of an independent report. Three groups of participants performed different tasks: the first group performed an object naming task, the second a simple language task, and the third group performed both a simple visual and a complex language task, in that order.
Table 1. Task’s description.
| Group 1 | Group 2 | Group 3 | ||
|---|---|---|---|---|
| Task | Object Naming | Matching | All the same/ one different judgment | Abstract/concrete judgment |
| Stimuli | Images | Symbols/letters | 4 Gabor patches | Nouns |
| Input modality | Visual | Visual | Visual | Auditory |
| Output modality | Speech | Button | Button | Button |
| # Participants | 15 | 12 | 17* | 17* |
| Task period (s) | 105 | 125 | 130 | 105 |
| # Task periods | 2 | 2 | 3 | 3 |
| Baseline (s) | 51 | 40 | 37.5 | 37.5 |
| # Baseline periods | 3 | 3 | 4 | 4 |
| Stimulus duration (ms) | 200 | 500 | 300 | Varied |
the same participants performed the two tasks in the same order.
2.2 Data acquisition and preprocessing
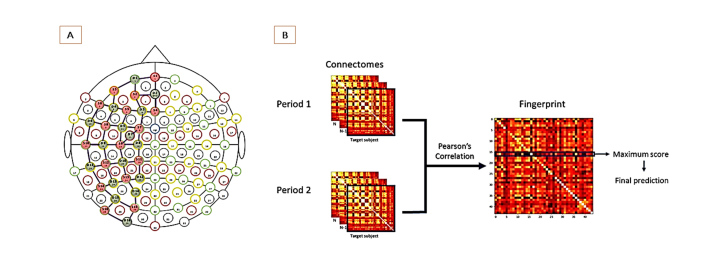
A continuous-wave NIRS system (NIRScout 16x16, NIRx Medical Technologies LLC, Berlin, Germany) was used for acquisition (wavelengths: 760nm and wl2-850nm). A total of 32 optodes (16 sources and 16 detectors) were used with to form 49 channels (3 cm of distance between source and detector). In order to maximize the covering of cortical surface, the 49 source-detector pairs (channels) were placed over the entire left hemisphere. The position of sources and detectors was established using the International 10/20 system for EEG recording (Fig. 1(a)). All pre-processing steps were applied using the Homer2 software: (1) the raw data was first converted from optical intensities to optical densities; (2) a bandpass filter (0.01-0.08Hz) was then applied to the data using the function “hmrBandpassFilt”; (3) lastly, optic density was converted to oxyhemoglobin and deoxyhemoglobin concentrations. The Partial Pathlength Factor (PPF) parameter was set at 6 for both wavelengths. A global signal was computed by averaging out all channels' time series for each participant. This signal was then regressed out from each channel signal (Global Signal Regression – GSR [19]). A Principal Component Analysis (PCA) was computed using the “hmrPCAFilter” function on Homer2.
Fig. 1.
(a) The configuration of the fNIRS channel assembly was based on the 10-20 system. Pairs of optodes were placed to maximize left hemisphere coverage. (b) To compute a “fingerprint matrix”, the Pearson correlation was calculated between each individual connectivity matrix from one period database (e.g rest 1) with all the other connectivity matrices from the database representing another period (e.g rest 2 or task). Thus, row represent the target connectome and column the predictor. In other words, each pixel (i,j) represent the correlation of a connectome i of one participant from one database (e.g., rest 1) with connectome j of any given participant from the other database (e.g., rest 2), and a correct individual identification corresponds to a maximal correlation coefficient at the diagonal. Accuracy can be determined by the total number of correct identifications divided by the number of participants.
2.3 Connectome fingerprinting analysis
Pearson correlation coefficients were used to calculate the channel-to-channel functional connectivity and to construct the individual connectome matrices. One 49x49 matrix was obtained for each individual, each chromophore and each acquisition period, i.e., for resting period 1, tasks period and resting period 2. The method described in the reference [4] was then applied for the three “databases” constructed by polling these matrices according to acquisition period. In this so-called fingerprinting procedure, the individual identification is determined by computing the Pearson’s correlation coefficient of each individual connectivity matrix from one database with all the other connectivity matrices from the other database. The procedure is illustrated in Fig. 1(b). Comparing all subjects in two databases hence produces a 44x44 matrices (here called the “fingerprinting matrices”) in which lines represent ordinate individual connectome matrix of one database (period) and columns represent the ordinate individual connectome matrix of another database. If the maximal correlation coefficient in the row of such matrix corresponded to the same individual on the column (i.e., if the maximum value is at the diagonal of the matrix), the identification was correct. Accuracies for each condition and chromophore were calculated by determining the number of correct identifications divided by the total number of iterations (e.g., individuals).
3. Results
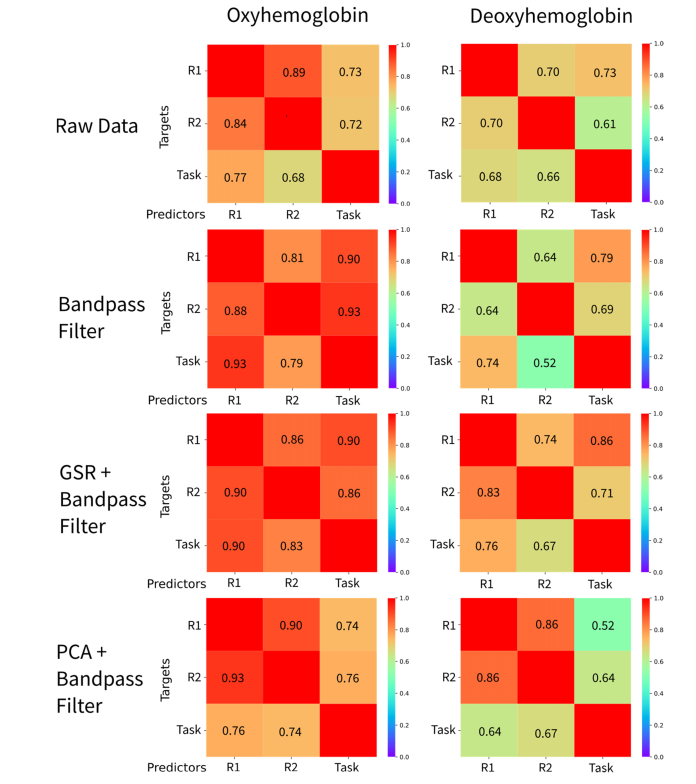
The accuracies of identification for all the possible combinations (rest 1 vs rest 2, rest 1 vs task, rest 2 vs task; for HbO and HbR concentrations) are described in Fig. 2. All accuracies were well-above the average chance level of 2.3% (see results of permutations in Fig. 4), ranging between 52% and 93%. The higher accuracy levels were obtained in the comparisons between resting states. Compelling high accuracies were obtained with filtered data but without removing global signal (with both GSR or PCA) of the oxyhemoglobin (HbO) signal. Besides presenting lower identification accuracies overall, connectome fingerprinting analysis based on deoxyhemoglobin (HbR) signal was roughly similar before (raw data) and after filtering the data or removing global signal.
Fig. 2.
Individual identification accuracies based on the fingerprint procedure for each possible comparison between targets and predictors of individual connectivity matrices, for oxy and deoxyhemoglobin signals.
Fig. 4.
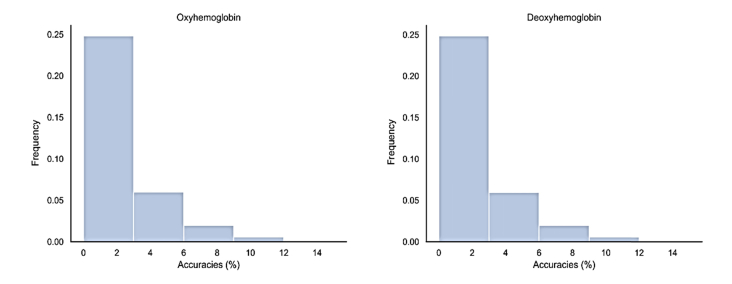
Histograms of the results of a hundred thousand permutations, simulating random individual identification.
In summary, identification based on oxyhemoglobin clearly outperformed identification based on deoxyhemoglobin. The results also suggest that accuracies obtained after removing global signal with PCA were generally similar to applying GSR.
In order to evaluate the possible identification of individuals based on the connectivity pattern of their global regressed fNIRS signal, we used the connectome matrix constructed from the signal of one chromophore to identify the matrix constructed from the other one. Hence, the HbO connectivity matrix of one individual was used to identify their respective HbR connectivity matrix (Fig. 3). For each comparison between acquisition periods (rest 1, task and rest 1), the two values of identification accuracies were calculated (identifying HbO from HbR-based connectomes and vice versa). When values differed, the lowest one was reported. For all comparisons, identification accuracies were well-above chance level, ranging from 41% to 73%. Identifications between HbO and HbR matrices using signals from the same acquisition period (represented on the diagonal of Fig. 3) outperformed accuracies of identification from signals of different periods. The greater performance (73%) was achieved for the comparison between the second resting state period.
Fig. 3.
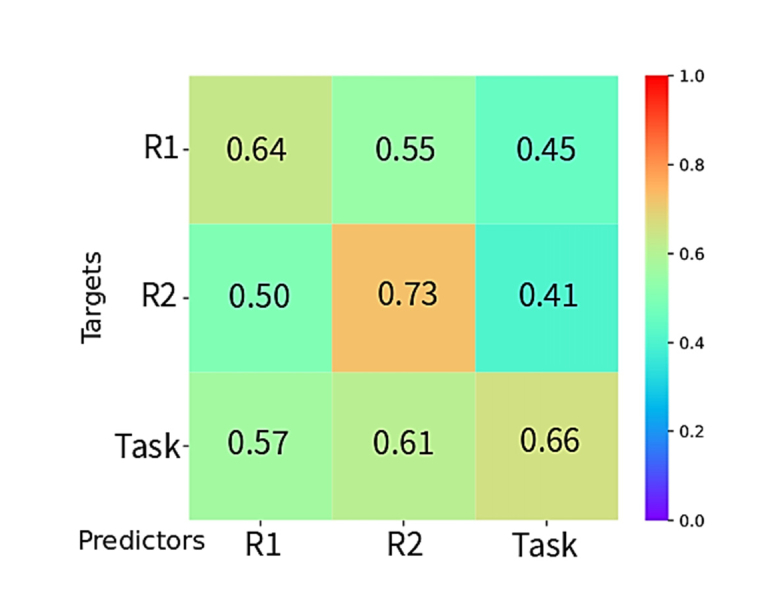
Accuracy values for the comparisons between oxy and deoxyhemoglobin data, for the conditions (rest 1, task, rest 2).
The average probability to identify an individual by chance in our data set was of 1/44 ~2.3%. To further evaluate the distribution of probability for hit random, a hundred thousand permutations were performed. Permutations results for both HbO and HbR matrices (Fig. 4) show that the chance to identify an individual at random peaks at 10%, with a greater density between 0 and 2%.
4. Discussion
The main objective of this study was to evaluate the feasibility of extending the fMRI connectome fingerprinting analysis to fNIRS data. The obtained results show identification accuracies peaking at 93% for the oxyhemoglobin and at 86% for the deoxyhemoglobin signal after filtering the data to a range corresponding to neural slow frequency fluctuations, but without removing global signal. Moreover, the maximum identification accuracy was observed for the oxyhemoglobin signal in the comparison between resting conditions. Our results thus suggest that fNIRS-based cortical connectome might capture the individual differences in connectivity profiles reported on fMRI studies, further supporting the existence of uniquely individual functional connectome architectures. However, whether these high identification accuracies are exclusively due to differences in brain functional architecture remains uncertain, since possible contributions of extra-cortical signals and differences in the representation of cortical areas by the optodes positioning cannot be completely ruled out. In favor of a major contribution of cortical signal, a significant improvement in identification accuracy was observed after filtering the signal for frequencies associated with the neural slow fluctuations underling intrinsic functional connectivity, particularly for the HbR signal.
All studies on connectome fingerprints, to the present date, were based on fMRI data sets. To the best of our knowledge, this is the first attempt to apply this approach to fNIRS data. Findings of individual connectome identification through the fingerprinting approach has been successfully replicated in various fMRI data sets [4–8,21–27]. As both fNIRS and fMRI signals relies on neurovascular coupling mechanisms, we hypothesized that fNIRS could also replicate such robust fMRI findings. Though the hemodynamic data acquired with fNIRS are restricted to superficial layers of the cerebral cortex, its signal appears to contain highly individual information. Our findings are indeed in agreement with previous fMRI fingerprinting results showing almost pinpoint identification accuracy for resting-state functional networks with extensive superficial nodes (e.g. fronto-parietal network) [3,4,8].
Similarly to the BOLD fMRI signal, fNIRS signals can provide indirect information about cortical neural activity through the variations of oxy and deoxyhemoglobin concentrations. However, despite providing information about some aspects of neural activity, fNIRS-derived functional connectivity also contains a non-neural component, particularly on its global signal (i.e., the average signal across all channels [19]). To evaluate the possibility that an individual's global neurovascular functional architecture contributes to their connectome-based identification, the fNIRS data was analysed either before and after removing global signal with GSR or PCA [28, 29].
Besides removing systemic global artifacts, both PCA and GSR might also remove global signals of neural origin [19].
However, some limitations of this study should be taken into account. Besides the relatively small sample, participants did not take off the cap between acquisitions. Hence, common individual sources of noise may have artificially contributed to increases identification accuracies Among these sources, different representations of cortical areas between individuals due to variation in optodes positioning is a major concern. We emphasize that our results is a first evidence for a fNIRS based connectome fingerprint, but further work is warranted to confirm these findings, extend it to determined the differential influence of large scale networks on the fingerprint and to determine the arrangements that maximize identification accuracy. Therefore, further studies should acquire fNIRS data of the same individuals longitudinally and with at least two independent and subsequent acquistions.
In short, here we provide the first evidence for assessing the unique architecture (or fingerprint) of the human functional connectome using fNIRS. Further studies are still desired for validating these findings in independent data sets and including intervals between acquisitions. Moreover, in our study the channels were positioned only over the left hemisphere, thus further investigations should explore the optimal optodes positions in order to maximize the efficiency of individual identification.
Characterizing individual variability in neural activity and connectivity might have great impact in many open questions in cognitive neuroscience. For example, recent fMRI have applied functional connectome fingerprinting to: (1) characterize functional networks of neurodevelopmental trajectories [6]; (2) understand patterns of affective relationship between parents and their children [30]; and (3) predict the expression of psychiatric symptoms or disorders [5–7]. The fNIRS-based cortical connectomes fingerprinting approach could provide information about the stabilization of functional networks before and after interventions in realistic settings [31]. Particularly, fingerprint cortical connectome analysis might help to elucidade the trajectories of neural functional development in children. In overall, here we provide evidence that fNIRS-based fingerprinting can be developed as an important new tool to improve neurodevelopmental and mental health research [4–8].
Acknowledgments
We would like to thank reviewers, for the comments and suggestions that contributed to improve several aspects of this study. Also, we would like to thank Gabriela Melo to help improve this paper writing.
Funding
São Paulo Research Fundation (FAPESP) (2018/21934-5); National Council for Scientific and Technological Development (CNPQ); Coordination for the Improvement of Higher Education Personnel (CAPES).
Disclosures
The authors declare that there are no conflicts of interest related to this article.
References
- 1.Mueller S., Wang D., Fox M. D., Yeo B. T. T., Sepulcre J., Sabuncu M. R., Shafee R., Lu J., Liu H., “Individual variability in functional connectivity architecture of the human brain,” Neuron 77(3), 586–595 (2013). 10.1016/j.neuron.2012.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanai R., Rees G., “The structural basis of inter-individual differences in human behaviour and cognition,” Nat. Rev. Neurosci. 12(4), 231–242 (2011). 10.1038/nrn3000 [DOI] [PubMed] [Google Scholar]
- 3.Miranda-Dominguez O., Mills B. D., Carpenter S. D., Grant K. A., Kroenke C. D., Nigg J. T., Fair D. A., “Connectotyping: model based fingerprinting of the functional connectome,” PLoS One 9(11), e111048 (2014). 10.1371/journal.pone.0111048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Finn E. S., Shen X., Scheinost D., Rosenberg M. D., Huang J., Chun M. M., Papademetris X., Constable R. T., “Functional connectome fingerprinting: identifying individuals using patterns of brain connectivity,” Nat. Neurosci. 18(11), 1664–1671 (2015). 10.1038/nn.4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaufmann T., Alnæs D., Brandt C. L., Bettella F., Djurovic S., Andreassen O. A., Westlye L. T., “Stability of the Brain Functional Connectome Fingerprint in Individuals With Schizophrenia,” JAMA Psychiatry 75(7), 749–751 (2018). 10.1001/jamapsychiatry.2018.0844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaufmann T., Alnæs D., Doan N. T., Brandt C. L., Andreassen O. A., Westlye L. T., “Delayed stabilization and individualization in connectome development are related to psychiatric disorders,” Nat. Neurosci. 20(4), 513–515 (2017). 10.1038/nn.4511 [DOI] [PubMed] [Google Scholar]
- 7.Biazoli C. E., Jr., Salum G. A., Pan P. M., Zugman A., Amaro E., Jr., Rohde L. A., Miguel E. C., Jackowski A. P., Bressan R. A., Sato J. R., “Commentary: Functional connectome fingerprint: identifying individuals using patterns of brain connectivity,” Front. Hum. Neurosci. 11, 47 (2017). 10.3389/fnhum.2017.00047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sato J. R., White T. P., Biazoli C. E., Jr., “Commentary: A test-retest dataset for assessing long-term reliability of brain morphology and resting-state brain activity,” Front. Neurosci. 11, 85 (2017). 10.3389/fnins.2017.00085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miranda-Dominguez O., Feczko E., Grayson D. S., Walum H., Nigg J. T., Fair D. A., “Heritability of the human connectome: A connectotyping study,” Netw Neurosci 2(2), 175–199 (2018). 10.1162/netn_a_00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ribeiro F. L., Pinaya W. H. L., Sato J. R., Junior C. E. B., “Unique individual factors shape brain hubs organization of the human functional connectome,” Bioarxiv https://www.biorxiv.org/content/10.1101/437335v1 (2018).
- 11.Balardin J. B., Zimeo Morais G. A., Furucho R. A., Trambaiolli L., Vanzella P., Biazoli C., Jr., Sato J. R., “Imaging Brain Function with Functional Near-Infrared Spectroscopy in Unconstrained Environments,” Front. Hum. Neurosci. 11, 258 (2017). 10.3389/fnhum.2017.00258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duan L., Zhang Y.-J., Zhu C.-Z., “Quantitative comparison of resting-state functional connectivity derived from fNIRS and fMRI: A simultaneous recording study,” Neuroimage 60(4), 2008–2018 (2012). 10.1016/j.neuroimage.2012.02.014 [DOI] [PubMed] [Google Scholar]
- 13.Cui X., Bray S., Bryant D. M., Glover G. H., Reiss A. L., “A quantitative comparison of NIRS and fMRI across multiple cognitive tasks,” Neuroimage 54(4), 2808–2821 (2011). 10.1016/j.neuroimage.2010.10.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.R. M. Forti, A. Alessio, and R. C. Mesquita, “Characterization of the NIRS Hemodynamic Response Function with Independent Component Analysis,” in Biomedical Optics 2014 (2014).
- 15.Sasai S., Homae F., Watanabe H., Taga G., “Frequency-specific functional connectivity in the brain during resting state revealed by NIRS,” Neuroimage 56(1), 252–257 (2011). 10.1016/j.neuroimage.2010.12.075 [DOI] [PubMed] [Google Scholar]
- 16.Mesquita R. C., Franceschini M. A., Boas D. A., “Resting state functional connectivity of the whole head with near-infrared spectroscopy,” Biomed. Opt. Express 1(1), 324–336 (2010). 10.1364/BOE.1.000324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dalmis M. U., Akin A., “Similarity analysis of functional connectivity with functional near-infrared spectroscopy,” J. Biomed. Opt. 20(8), 086012 (2015). 10.1117/1.JBO.20.8.086012 [DOI] [PubMed] [Google Scholar]
- 18.Lu C. M., Zhang Y. J., Biswal B. B., Zang Y. F., Peng D. L., Zhu C. Z., “Use of fNIRS to assess resting state functional connectivity,” J. Neurosci. Methods 186(2), 242–249 (2010). 10.1016/j.jneumeth.2009.11.010 [DOI] [PubMed] [Google Scholar]
- 19.Murphy K., Fox M. D., “Towards a consensus regarding global signal regression for resting state functional connectivity MRI,” Neuroimage 154, 169–173 (2017). 10.1016/j.neuroimage.2016.11.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dosenbach N. U. F., Visscher K. M., Palmer E. D., Miezin F. M., Wenger K. K., Kang H. C., Burgund E. D., Grimes A. L., Schlaggar B. L., Petersen S. E., “A core system for the implementation of task sets,” Neuron 50(5), 799–812 (2006). 10.1016/j.neuron.2006.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kocsis L., Herman P., Eke A., “The modified Beer-Lambert law revisited,” Phys. Med. Biol. 51(5), N91–N98 (2006). 10.1088/0031-9155/51/5/N02 [DOI] [PubMed] [Google Scholar]
- 22.Amico E., Goñi J., “Mapping hybrid functional-structural connectivity traits in the human connectome,” Netw Neurosci 2(3), 306–322 (2018). 10.1162/netn_a_00049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finn E. S., Scheinost D., Finn D. M., Shen X., Papademetris X., Constable R. T., “Can brain state be manipulated to emphasize individual differences in functional connectivity?” Neuroimage 160, 140–151 (2017). 10.1016/j.neuroimage.2017.03.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horien C., Noble S., Finn E. S., Shen X., Scheinost D., Constable R. T., “Considering factors affecting the connectome-based identification process: Comment on Waller et al,” Neuroimage 169, 172–175 (2018). 10.1016/j.neuroimage.2017.12.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vanderwal T., Eilbott J., Finn E. S., Craddock R. C., Turnbull A., Castellanos F. X., “Individual differences in functional connectivity during naturalistic viewing conditions,” Neuroimage 157, 521–530 (2017). 10.1016/j.neuroimage.2017.06.027 [DOI] [PubMed] [Google Scholar]
- 26.Waller L., Walter H., Kruschwitz J. D., Reuter L., Müller S., Erk S., Veer I. M., “Evaluating the replicability, specificity, and generalizability of connectome fingerprints,” Neuroimage 158, 371–377 (2017). 10.1016/j.neuroimage.2017.07.016 [DOI] [PubMed] [Google Scholar]
- 27.Lee T.-H., Miernicki M. E., Telzer E. H., “Families that fire together smile together: Resting state connectome similarity and daily emotional synchrony in parent-child dyads,” Neuroimage 152, 31–37 (2017). 10.1016/j.neuroimage.2017.02.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfeifer M. D., Scholkmann F., Labruyère R., “Signal Processing in Functional Near-Infrared Spectroscopy (fNIRS): Methodological Differences Lead to Different Statistical Results,” Front. Hum. Neurosci. 11, 641 (2018). 10.3389/fnhum.2017.00641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murphy K., Fox M. D., “Towards a consensus regarding global signal regression for resting state functional connectivity MRI,” Neuroimage 154, 169–173 (2017). 10.1016/j.neuroimage.2016.11.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gratton C., Laumann T. O., Nielsen A. N., Greene D. J., Gordon E. M., Gilmore A. W., Nelson S. M., Coalson R. S., Snyder A. Z., Schlaggar B. L., Dosenbach N. U. F., Petersen S. E., “Functional Brain Networks Are Dominated by Stable Group and Individual Factors, Not Cognitive or Daily Variation,” Neuron 98(2), 439–452 (2018). 10.1016/j.neuron.2018.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balardin J. B., Zimeo Morais G. A., Furucho R. A., Trambaiolli L., Vanzella P., Biazoli C., Jr., Sato J. R., “Imaging brain function with functional near-infrared spectroscopy in unconstrained environments,” Front. Hum. Neurosci. 11, 258 (2017). 10.3389/fnhum.2017.00258 [DOI] [PMC free article] [PubMed] [Google Scholar]