Key Points
Question
Can genetic polymorphisms serve as biomarkers of prognosis and direct therapy outcomes in patients with pancreatic cancer amenable to resection?
Findings
This study used 2 independent cohorts of patients with pancreatic ductal adenocarcinoma who underwent resection of their tumors to perform a genome-wide screening for functional single-nucleotide polymorphisms that affect pancreatic cancer survival. Two single-nucleotide polymorphisms in known cancer-associated genes that were associated with tumor-associated survival after pancreatic resection were identified and validated.
Meaning
These common polymorphisms may be used as a noninvasive biomarker signature in a clinical setting that is readily available at the time of diagnosis to identify patients with a very low probability of survival, guide personalized treatment decisions, and direct patient stratification in clinical trials.
This study assesses 2 datasets on cohorts of patients with pancreatic ductal adenocarcinoma to identify and validate functional genetic variants that can serve as biomarkers for prospective treatment response and survival.
Abstract
Importance
Surgery currently offers the only chance for a cure in pancreatic ductal adenocarcinoma (PDAC), but it carries a significant morbidity and mortality risk and results in varying oncologic outcomes. At present, to our knowledge, there are no tests available before surgical resection to identify tumors with an aggressive biological phenotype that could guide personalized treatment strategies.
Objective
Identification of noninvasive genetic biomarkers that could direct therapy in patients whose cases are amenable to pancreatic cancer resection.
Design, Setting, and Participants
This multicenter study combined a prospective European cohort of patients with PDAC who underwent pancreatic resection (from University Hospital of Zurich, Zurich, Switzerland; Cantonal Hospital of Winterthur, Winterthur, Switzerland; and University Clinic of Ulm, Ulm, Germany) with data from the Cancer Genome Atlas database in the United States, which includes prospectively registered patients with PDAC. A genome-wide screening for functional single-nucleotide polymorphisms (SNPs) that affect PDAC survival was conducted using the European cohort for identification and the Cancer Genome Atlas cohort for validation. We used Cox proportional hazards models to screen for high-frequency polymorphic variants that are associated with allelic differences in tumor-associated survival and either result in an altered protein structure and function or reside in known regulatory noncoding genomic regions. The false-discovery rate method was applied for multiple hypothesis-testing corrections. Data analysis occurred from November 2017 to May 2018.
Exposures
Pancreatic resection.
Main Outcomes and Measures
Tumor-associated survival.
Results
A total of 195 patients in the European cohort were included, as well as 136 patients in the Cancer Genome Atlas cohort (overall median [range] age, 66 [19-87] years; 156 [47.1%] were women, and 175 [52.9%] were men). Two SNPs in noncoding, functional regions of genes that regulate cancer progression, invasion, and metastasis were identified (CHI3L2 SNP rs684559 and CD44 SNP rs353630). These were associated with survival after PDAC resection; patients who carry the risk alleles at 1 of both SNP loci had a 2.63-fold increased risk for tumor-associated death compared with those with protective genotypes (hazard ratio for survival, 0.38 [95% CI, 0.27-0.53]; P = 1.0 × 10−8).
Conclusions and Relevance
The identified polymorphisms may serve as a noninvasive biomarker signature of prospective survival after pancreatic resection that is readily available at the time of PDAC diagnosis. This signature can be used to identify a subset of high-risk patients with PDAC with very low survival probability who might be eligible for inclusion in clinical trials of new therapeutic strategies, including neoadjuvant chemotherapy protocols. In addition, the biological knowledge about these SNPs could help guide the development of individualized genomic strategies for PDAC therapies.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) accounts for 85% of all types of pancreatic cancer and is one of the most lethal malignant conditions, owing to its aggressive biological phenotype with an early local invasion, as well as a high metastatic potential.1,2 Despite this aggressive phenotype, PDAC displays a wide range of biology.3,4,5 As a result, early recurrence and metastases are observed in some individuals after pancreatic resection (about 15% to 20% will develop clinically evident metastatic disease, even within the recovery period) and long-term disease-free survival in others.3,4,5,6 The prognostication of such individual oncologic courses and responses to personalized treatment decisions could be made possible by the understanding of human germline genetic variation.7,8 This type of genetic information about the patient is present in the germline DNA and can be easily obtained by a technically simple blood or saliva test at the time of diagnosis. Indeed, for other cancer types, such as acute lymphoblastic leukemia, the US Food and Drug Administration already recommends genotyping of selected single-nucleotide polymorphisms (SNPs) in clinical practice.9
In PDAC, little is known about genetic polymorphisms that could influence the progression of an already-formed malignant tumor.10,11,12,13,14 A rare exception is a germline variant in intron 1 of the CD44 gene (SNP rs187115) that has been suggested to help select patients who are likely to benefit from pancreatic resection.15,16 However, the usefulness of SNP rs187115 to guide the development of genomic strategies for PDAC therapies is limited to date, since the underlying functional mechanisms of this polymorphism have yet to be inferred, and only about 10% of individuals carry the high-risk genotype. Moreover, genome-wide association studies designed to investigate the association of polymorphic variants with pancreatic cancer survival have largely failed to identify loci with consistent genome-wide significance levels.11,12,13,14 This is owing to the known inherent limitations of this agnostic study approach, such as a high rate of false-negative results likely, in part, owing to the required stringent control for multiple testing and biases associated with the selection of study participants.10,11,12,13,14
To overcome these limitations, we perform a genome-wide screening for high-frequency inherited variants that affect survival of patients with PDAC, based on existing biological knowledge about the regions where the polymorphisms reside. These common polymorphisms could direct the design of clinical trials, be used as biomarkers in surgical patients, and owing to their inferred functional mechanisms, guide the development of genomic strategies for PDAC therapies.
Methods
European Patient Cohort
This study includes patients with pancreatic cancer who were diagnosed in 2001 to 2016 at the University Hospital Zurich, Cantonal Hospital Winterthur (Winterthur, Switzerland), and University Hospital Ulm (Ulm, Germany) and included in 2 prospective biobank databases (at University Hospital Zurich and University of Ulm). All patients with PDAC who underwent pancreatic resection for whom high-quality DNA was extracted from pathologically confirmed tumor-free tissue or blood and from whom a full clinical data set was available were included. Approval was obtained from the Cantonal Ethics Committee Zurich and the Ethics Committee of the University of Ulm. The data were deidentified, and therefore individual patient consent was not required.
The Cancer Genome Atlas Patient Cohort
The Cancer Genome Atlas (TCGA) database includes patients with pancreatic cancer whose years of initial pathological diagnosis were 2007 to 2013. All prospectively registered patients with pathologically confirmed PDAC who underwent pancreatic resection and for whom germline genotyping results and a full clinical data set were available were included in this study. Approval from the NIH Data Access Committee was obtained. The data were deidentified, and therefore individual patient consent was not required.
Laboratory Analyses
The DNA extraction, quality-control, processing, genotyping, imputation, and histopathological protocols are detailed in the eMethods in the Supplement.17,18,19,20,21,22 The effects of nonsynonymous SNPs and the resulting amino acid substitutions on the structure and function of human proteins were assessed with the software tool PolyPhen-2 (Polymorphism Phenotyping version 2; http://genetics.bwh.harvard.edu/pph2/)23 with the default settings for structural query options.
The Ensembl Variant-Effect-Predictor was used to find overlaps of SNP loci with known regulatory regions as determined in the Ensembl Regulatory Build.24,25 The University of California, Santa Cruz Human Genome Browser (http://genome.ucsc.edu/cgi-bin/hgGateway) was used to access the regulatory features of the identified SNPs based on the integrated experimental data from the Encyclopedia of DNA Elements (ENCODE) project.26
Statistical Analysis
We used Cox proportional hazards models to estimate the impact of SNP genotype on tumor-associated death. The SNP genotypes were modeled as categorical variables. We tested the following models: (1) AA vs AB vs BB (with the homozygous genotypes being used as baseline), (2) AA/AB vs BB, and (3) AA vs AB/BB. We also accounted for other prognostic factors that are relevant for PDAC, namely American Joint Committee on Cancer (AJCC) stage (ie, I, II, III, or IV) and surgical margin status (ie, R0, R1, or R2); for these factors, the baseline levels were set to stage II and status R0, respectively. In addition, we calculated estimates for survival using a Kaplan-Meier analysis and the log-rank test. Missing data were handled on a complete-case analysis basis. Correction for multiple hypothesis testing was done using the false-discovery rate method.27 Values for q less than 0.05 were considered significant. All analyses were performed using the Survival Package of the R Statistical Language version 3.1 (R Foundation for Statistical Computing). Data analysis occurred from November 2017 to May 2018.
Sample Size
Based on previous studies,15,16 we conducted a power analysis to assess the required sample size to detect a 2-fold increase in risk (hazard ratio [HR], 2.00) for common variants (SNPs with a minimal allele frequency ≥0.20) with an overall probability of an event as in the discovery set of 0.70, a power of 0.80, and a type I error rate (α) of less than .05. This resulted in a minimum sample size of 148 patients. Therefore, the number of patients included in our discovery set (n = 195), which is based on the availability of tissue material for DNA extraction and analysis and the histopathological or clinical follow-up data of all patients included in the database who underwent pancreatic resection, meets the minimum requirements of the power analysis. If the number of patients was less than 148 for a specific variant, the SNP was removed.
Results
A total of 195 of the 479 individuals in the European hospital databases were included. In addition, the 136 patients with PDAC of the 185 total patients in the TCGA were included. Across both cohorts, the median (range) age was 66 (19-87) years; 156 (47.1%) of the patients were women, and 175 (52.9%) were men (Table 1).
Table 1. Clinical and Histopathological Data.
| Characteristic | Patients, No. (%) | ||
|---|---|---|---|
| European Cohort | TCGA Cohort | Both Cohorts | |
| Median age,a y | 66 (19-87) | 65 (35-85) | 66 (19-87) |
| Median observation time,a mo | 17.5 (1-135) | 14.3(0-67) | 16 (0-135) |
| Frequency of cases assessed for inclusion | 479 | 185 | 664 |
| Frequency of included cases | |||
| Total | 195 (100) | 136 (100) | 331 (100) |
| Female | 94 (48.2) | 62 (45.6) | 156 (47.1) |
| Male | 101 (51.8) | 74 (54.4) | 175 (52.9) |
| Frequency of excluded cases | |||
| Total excluded | 284 (100) | 49 (100) | 333 (100) |
| Missing follow-up data | 2 (0.7) | 5 (10.2) | 7 (2.1) |
| No normal tissue and/or blood sampels | 16 (5.6) | 8 (16.3) | 24 (7.2) |
| Missing histopathologic data | 8 (2.8) | 9 (18.4) | 17 (5.1) |
| Histology other than PDAC | 142 (50.0) | 27 (55.1) | 169 (50.8) |
| No pancreatic resection | 29 (10.2) | 0 | 29 (8.7) |
| Failed DNA quality controlb | 87 (30.6) | NA | 87 (26.1) |
| Status at last follow-up | |||
| Died of tumor-associated causes | 135 (69.2) | 65 (47.8) | 200 (60.4) |
| Alive | 42 (21.5) | 56 (41.2) | 98 (29.6) |
| Died of causes not associated with tumors | 8 (4.1) | 10 (7.4) | 18 (5.4) |
| In-hospital death | 10 (5.1) | 5 (3.7) | 15 (4.5) |
| Stage | |||
| I | 7 (3.6) | 12 (8.8) | 19 (5.7) |
| II | 166 (85.1) | 116 (85.3) | 282 (85.2) |
| III | 8 (4.1) | 4 (2.9) | 12 (3.6) |
| IV | 14 (7.1) | 4 (2.9) | 18 (5.4) |
| Tumor resectionc | |||
| R0 | 121 (62.1) | 86 (63.2) | 207 (62.5) |
| R1 | 64 (32.8) | 45 (33.1) | 109 (32.9) |
| R2 | 10 (5.1) | 5 (3.7) | 15 (4.5) |
Abbreviations: NA, not applicable; PDAC, pancreatic ductal adenocarcinoma; TCGA, the Cancer Genome Atlas.
Included cases only.
Failed Illumina DNA quality control testing or achieved SNP call rate less than 90%.
Tumor resections labeled R0 are radical; those labeled R1 and R2 are not radical.
Identification of Functional Noncoding DNA Elements and Nonsynonymous, Coding Functional Polymorphic Variants
The genotypes of 2 338 671 SNPs were determined with the high-density Human Omni 2.5-8 (Illumina) SNP array in the European discovery set, which included 195 patients with PDAC who had undergone pancreatic resection (Table 1; eFigure 1 in the Supplement). This array provides a comprehensive genome-wide coverage with a tag SNP content from the 1000 Genomes Project pilot data.18 To begin to search for polymorphisms that had frequencies that would allow for biomarkers to be used in a clinical setting, we restricted the set of SNPs to include only common polymorphic variants with a minimal allele frequency of 0.20 or more.28 Only SNPs with high-confidence genotype calls that also did not show extreme departures from Hardy-Weinberg equilibrium (P value ≥1.00 × 10−4) were included in the analysis, to ensure a high quality of the genotypes.29 This resulted in a total number of 358 895 SNPs.
Since most of the genotyped variants reside in noncoding genomic regions to which no regulatory features have been ascribed, we reasoned that functional polymorphisms are more likely to be located within regulatory DNA elements of the genome, such as promoters and their flanking regions, intergenic and intragenic enhancers, and untranslated regions. Indeed, many SNPs identified in cancer risk genome-wide association studies are considerably enriched in noncoding functional DNA elements, as defined by the ENCODE project.26,30 To begin searching for such variants, we used the Ensembl Variant-Effect-Predictor to find overlaps of SNP loci with known regulatory regions.24,25 These regions are inferred from a collection of publicly available experimental genome-wide assays, including ones from the ENCODE project, in which the annotated features encompass promoters, promoter-flanking regions, enhancers, CCCTC-binding factor (CTCF) binding sites, transcription factor binding sites, 3′ untranslated regions, and open-chromatin regions. Importantly, we found that the Variant-Effect-Predictor tool could identify both genomic regions in which 2 molecularly well-characterized, known functional variants that affect cancer risk reside, namely mouse double minute 2 homologue (MDM2) SNP309 (rs2279744) and KIT ligand p53 response element SNP (rs4590952).30,31,32,33 In total, 37 977 of the 358 895 SNPs that had passed the initial filtering step were found to reside in such noncoding functional DNA elements.
Next, we were interested in identifying common coding SNPs that change the protein sequence and are also expected to affect protein function. In close similarity to the search for functional noncoding DNA elements, we hypothesized that such polymorphisms are more likely to represent true functional and potentially actionable variants. To this end, the effect of nonsynonymous SNPs and the resulting amino acid substitutions on the structure and function of human proteins was prognosticated using the software tool PolyPhen-2.23 In brief, these assessments of the effect of nonsynonymous SNPs on the biological function of a protein are based on a number of features, including sequence and phylogenetic and structural information characterizing the substitution.23 Of the 358 895 SNPs from the first filtering step, 2571 polymorphisms were located in exons and found to affect the protein sequence of the genes in which they reside. Of these, 369 SNPs were assessed by PolyPhen-2 as having a deleterious functional outcome on the protein product. These variants were added to the final set of polymorphisms, resulting in a total of 38 346 SNPs that were included in the survival analysis.
Allelic Differences in Tumor-Associated Survival and Prognosticative Value of SNPs
Without applying any further restrictions to this data set at this point, we first performed a multivariate Cox regression analysis (adjusted to the known prognostic factors of AJCC stage and resection status) in the European data set to search for SNPs that show allelic differences in tumor-associated death, using models that directly compare the 3 (biallelic) genotypes (AA vs AB vs BB) as well as both dominant models (AX vs BB and AA vs BX) and report the top 73 SNP hits with a P value threshold of .001 (eTable 1 in the Supplement). In an attempt to validate these observations, we were interested in exploring whether any of those 73 candidate SNPs show a similar association with PDAC survival in an independent data set. To this end, we used the publicly available TCGA pancreatic cancer database, which is supervised by the National Cancer Institute and at present is made up of data on 136 patients with PDAC who underwent surgical resection for their tumors and for whom normal tissue as well as a full clinical data set is available.34 Of the 73 nominally most significant polymorphisms in the European data set, a total of 45 SNPs was also represented directly or by a proxy variant in high-linkage disequilibrium on the Affymetrix array used by TCGA (eTable 1 in the Supplement). Cox regression models were calculated for each of those 45 top-hit SNPs in the TCGA cohort. After correction for multiple hypothesis testing using the false-discovery rate method, we noted that 2 of the 45 variants also showed considerable allelic differences in their ability to prognosticate tumor-associated survival in the TCGA cohort specifically (SNP rs684559 [q = 0.0257] and SNP rs353630 [q = 0.0452]; Table 2; eTable 1 in the Supplement).
Table 2. Cox Multivariate Regression Analysis in the European and Cancer Genome Atlas Cohorts for Single-Nucleotide Polymorphisms rs353630 and rs684559a.
| Cohort | European Cohort | Cancer Genome Atlas Cohort | Merged Cohort | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Single-Nucleotide Polymorphism | No. | Hazard Ratio (95% CI) | P Value | No. | Hazard Ratio (95% CI) | P Value | Q Value | No. | Hazard Ratio (95% CI) | P Value |
| rs353630 (CD44)a | ||||||||||
| T/T | 20 | 1 [Reference] | NA | 6 | 1 [Reference] | NA | NA | 26 | 1 [Reference] | NA |
| C/T | 67 | 0.35 (0.19-0.65) | 8.47 × 10−4 | 56 | 0.23 (0.07-0.70) | 9.81 × 10−3 | 0.083 | 123 | 0.40 (0.23-0.66) | 3.30 × 10−4 |
| C/C | 105 | 0.40 (0.22-0.72) | 2.25 × 10−3 | 74 | 0.17 (0.06-0.51) | 1.72 × 10−3 | NA | 179 | 0.39 (0.24-0.66) | 1.30 × 10−4 |
| C allele | 172 | 0.38 (0.22-0.67) | 8.76 × 10−4 | 130 | 0.19 (0.07-0.56) | 2.74 × 10−3 | 0.045 | 302 | 0.40 (0.24-0.65) | 1.20 × 10−5 |
| rs684559 (CHI3L2)b | ||||||||||
| A/A | 30 | 1 [Reference] | NA | 18 | 1 [Reference] | NA | NA | 48 | 1 [Reference] | NA |
| A/G | 84 | 0.40 (0.24-0.66) | 2.89 × 10−4 | 70 | 0.28 (0.12-0.61) | 1.59 × 10−3 | 0.027 | 154 | 0.42 (0.28-0.63) | 2.90 × 10−4 |
| G/G | 79 | 0.430 (0.26-0.72) | 1.25 × 10−3 | 48 | 0.27 (0.12-0.61) | 1.59 × 10−3 | NA | 127 | 0.45 (0.30-0.68) | 1.25 × 10−3 |
| G allele | 163 | 0.41 (0.26-0.65) | 1.75 × 10−4 | 118 | 0.27 (0.13-0.58) | 7.80 × 10−4 | 0.026 | 281 | 0.44 (0.30-0.63) | 2.00 × 10−4 |
| Combined biomarker signaturec | ||||||||||
| Risk-indicating | 47 | 1 [Reference] | NA | 23 | 1 [Reference] | NA | NA | 70 | 1 [Reference] | NA |
| Protective | 148 | 0.37 (0.24-0.55) | 1.30 × 10−6 | 113 | 0.20 (0.10-0.39) | 4.20 × 10−6 | NA | 261 | 0.38 (0.27-0.53) | 1.00 × 10−8 |
Abbreviation: NA, not applicable.
All calculations were performed independently for each single-nucleotide polymorphism and adjusted for tumor stage (American Joint Committee on Cancer stages I through IV) and R status (ie, R0, R1, and R2).
The rs353630 and rs684559 genotypes of 3 and 2 patients, respectively, were not determined succesfully in the European cohort.
The risk-indicating genotypes were ones with the rs353630 T/T and/or rs684559 A/A genotypes; protective is defined by the presence of the rs353630 C allele and rs684559 G allele.
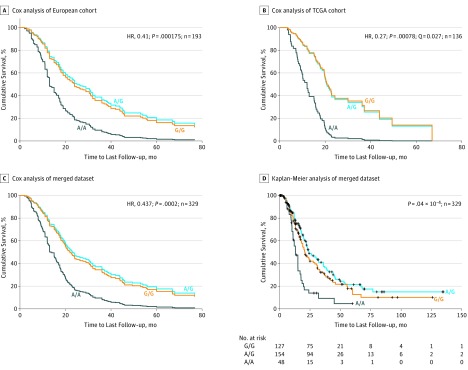
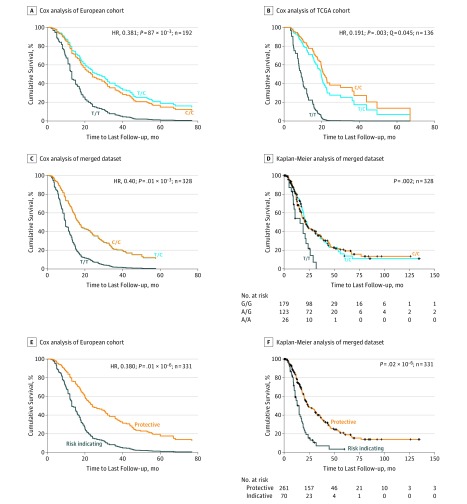
The most significant outcomes were seen for SNP rs684559, which is located in a 5′ regulatory region of the CHI3L2 gene. Specifically, the 30 patients in the European data set who were homozygous for the minor A allele of SNP rs684559 had a 2.43-fold increased risk for tumor-associated death compared with the 163 patients who carry 1 or 2 G alleles (HR, 0.41 [95% CI, 0.26-0.65]; P = 1.75 × 10−4; Table 2, Figure 1A). Accordingly, a Kaplan-Meier estimate demonstrated the shortest survival time for patients with an A/A genotype compared with those with a G allele (mean survival time, 18.1 vs 41.7 months; P = 7.0 × 10−4; eTable 2 and eFigure 2 in the Supplement). In line with this analysis, 18 individuals from the TCGA cohort with the A/A genotype had a 3.70-fold increase in risk and shortest estimated survival time when compared with 118 patients who carry the G allele (HR = 0.27 [95% CI, 0.13-0.58]; P = 7.80 × 10−4, by Cox multivariate analysis; P = .001 by log-rank test; Table 2 Figure 1B; eTable 2 and eFigure 2 in the Supplement). Next, we performed the survival analysis after merging both cohorts, which resulted in a data set that consisted of 331 patients who underwent pancreatic resection (including 329 individuals with successfully determined rs684559 genotypes) with 200 tumor-associated deaths. In this merged data set, we found that the risk for tumor-associated death was increased and survival time shortened for patients carrying the A/A genotype compared with individuals with the G allele (HR, 0.44 [95% CI, 0.30-0.63]; P = 2.00 × 10−4 by Cox multivariate analysis; P = 4.00 × 10−6 by log-rank test; Tables 2 and 3; Figure 1C and D). Similar results were observed for the second identified SNP that remained significant after multiple hypothesis testing correction. Specifically, individuals homozygous for the minor T allele of rs353630, which is located in an enhancer in intron 1 of the CD44 gene, had a significantly increased risk for tumor-associated death compared with patients with at least 1 C allele (European cohort: HR, 0.38 [95% CI, 0.22-0.67]; P = 8.76 × 10−4; TCGA cohort: HR, 0.19 [95% CI, 0.07-0.56]; P = 2.74 × 10−3; merged cohorts: HR, 0.40 [95% CI, 0.24-0.65]; P = 1.20 × 10−5). These individuals also had shorter survival times relative to those with 1 or 2 C alleles (mean survival time: T/T, 16.6 [95% CI, 12.7-20.4] months; C/T, 37.7 [95% CI, 28.3-47.1] months; C/C, 38.6 [95% CI, 31.4-45.8]; P = .007) (Tables 2 and 3; Figure 2; eTable 2 and eFigure 2 in the Supplement).
Figure 1. Survival Analysis of Individuals With CHI3L2 Single-Nucleotide Polymorphism rs684559.
A genome-wide screen for polymorphic variants that affect pancreatic ductal adenocarcinoma (PDAC) survival identified a potentially functional single-nucleotide polymorphism (SNP) in an active regulatory genomic region of the CHI3L2 gene. The A/A genotype of the identified SNP (rs684559) correlates significantly with poorer tumor-specific survival after resection of PDAC. A, Results of the Cox analysis in 193 patients with PDAC from the European cohort who underwent a surgical resection of their primary pancreatic tumors (adjusted to the known independent prognostic factors tumor stage and resection status). Patients with an A/A genotype were associated with the shortest survival time compared with those with 1 or 2 G alleles. B, Results of the Cox analysis in 136 patients with PDAC from the publicly available Cancer Genome Atlas database who underwent a surgical resection of their tumors. Patients with an A/A genotype had the shortest survival time compared with those with a G allele. The results were significant after correction for multiple testing (Q = 0.027). C and D, The graphs display the survival curves of 329 patients in the merged data set in the Cox multivariate analysis and the Kaplan-Meier survival curves, respectively. The P values in the Kaplan-Meier analysis (D) were determined with the log-rank test. HR indicates hazard ratio; TCGA, the Cancer Genome Atlas.
Table 3. Survival Time Analysis in the Merged Data Set That Includes Both Pancreatic Ductal Adenocarcinoma Cohorts Based on Kaplan-Meier Estimates.
| Group | Patients, No. | Tumor-Associated Deaths, No. | Mean Value, Estimate (SE) [95% CI], mo | Median Value, Estimate (SE) [95% CI], mo | Log-rank P Value |
|---|---|---|---|---|---|
| rs353630 (CD44) | |||||
| T/T | 26 | 19 | 16.6 (2.0) [12.7-20.4] | 16.0 (4.2) [7.8-24.2] | .007 |
| C/T | 123 | 68 | 37.7 (4.8) [28.3-47.1] | 22.7 (1.9) [19.0-26.5] | |
| C/C | 179 | 110 | 38.6 (3.7) [31.4-45.8] | 20.4 (2.2) [15.9-24.8] | |
| C-allele | 302 | 178 | 39.1 (3.1) [33.1-45.2] | 21.4 (1.3) [19.0-23.9] | .002 |
| rs684559 (CHI3L2) | |||||
| A/A | 48 | 35 | 17.2 (2.2) [12.9-21.5] | 13.0 (1.3) [10.5-15.5] | 1.00 × 10−5 |
| A/G | 154 | 85 | 43.1 (4.6) [34.2-52.1] | 24.0 (3.4) [17.4-30.6] | |
| G/G | 127 | 78 | 35.1 (4.1) [27.1-43.1] | 20.8 (2.3) [16.4-25.3] | |
| G-allele | 281 | 163 | 40.2 (3.2) [33.9-46.5] | 23.0 (1.9) [19.3-26.7] | 4.00 × 10−6 |
| AJCC | |||||
| 1 | 19 | 7 | 46.0 (9.2) [28.0-64.0] | 21.9 (NA) | .005 |
| 2 | 282 | 169 | 38.3 (3.1) [32.2-44.3] | 22.5 (1.3) [19.9-25.1] | |
| 3 | 12 | 8 | 14.5 (2.0) [10.6-18.3] | 17.0 (2.0) [13.0-21.0] | |
| 4 | 18 | 16 | 18.0 (3.4) [11.3-24.7] | 15.0 (3.2) [8.8-21.2] | |
| R status | |||||
| 0 | 207 | 123 | 42.1 (3.7) [34.70-49.4] | 23.6 (3.0) [17.8-29.4] | 2.00 × 10−4 |
| 1 | 109 | 65 | 29.0 (4.3) [20.6-37.5] | 18.5 (1.7) [15.2-21.8] | |
| 2 | 15 | 12 | 13.9 (2.3) [9.4-18.4] | 15.0 (5.0) [5.3-24.7] | |
| Signature | |||||
| Risk-indicatinga | 70 | 51 | 17.0 (1.7) [13.6-20.4] | 15.0 (1.5) [12.1-17.9] | 2.00 × 10−8 |
| Protectiveb | 261 | 149 | 41.9 (3.4) [35.3-48.6] | 24.0 (3.0) [18.1-29.9] |
Abbreviations: AJCC, American Joint Commission on Cancer; NA, not applicable.
The risk-indicating genotypes are the rs353630 T/T and/or rs684559 A/A genotypes.
The protective genotypes are those that include an rs353630 C allele and/or rs684559 G allele.
Figure 2. Survival Analysis of Individuals With CD44 Single-Nucleotide Polymorphism rs353630 and Biomarker Signature .
Analysis of tumor-specific survival after resection of pancreatic ductal adenocarcinoma (PDAC). A, Results of the Cox analysis in 192 patients with PDAC from 3 European centers who underwent a surgical resection of their pancreatic tumors (adjusted to the known independent prognostic factors tumor stage and resection status). Patients with a single-nucleotide polymorphism (SNP) rs353630 T/T genotype were associated with the shortest survival time compared with those with 1 or 2 C alleles. B, Results of the Cox analysis in 136 patients with PDAC from the publicly available Cancer Genome Atlas database who underwent a surgical resection of their tumors. Patients with a SNP rs353630T/T genotype had the shortest survival time compared with those with a C allele. The results were significant after correction for multiple testing (Q = 0.0451). C, and D, The graphs displays the survival curves for SNP rs353630 of 328 patients in the merged data set in the Cox multivariate analysis (C) and the Kaplan-Meier survival curves (D). E, Cox analysis of tumor-associated survival with a 2-dimensional biomarker signature that combines both SNP loci (rs684559 and rs353630) in the merged data set that comprises of 331 PDAC patients (adjusted to the known independent prognostic factors tumor stage and resection status). The risk-indicating signature includes the genotypes of each individual SNP that has individually been shown to associate with an increased risk of death, precisely rs684559 A/A or rs353630 T/T. The protective signature consists of the remaining genotype combinations: at least 1 C allele of rs353630 and 1 G allele of rs684559. F, Biomarker signature: the graphs display the Kaplan-Meier survival curves in the merged data set. The P values in D and F were determined with the log-rank test. HR indicates hazard ratio; TCGA, the Cancer Genome Atlas.
Next, we directly compared the predictive power of each of the identified SNPs with the clinical factors R status (ie, R0, R1, and R2) and AJCC stage. To do this in a way that would not be limited in statistical power, we performed a survival analysis after merging both cohorts.11,15,35,36,37 When we conducted a Kaplan-Meier analysis using SNPs rs684559, rs353630, R status, and AJCC status as prognosticative factors independently, we determined that all 4 factors were statistically significant (rs353630, mean survival time: patients with 1 or 2 C alleles: 39.1 months; patients with T/T genotype: 16.6 months; P = .002; rs684559, mean survival time: patients with 1 or 2 G alleles: 40.2 months; patients with A/A genotype: 17.2 months; P = 4.0 × 10−6; AJCC stage, mean survival times: stage 1, 46.0 months; stage 2, 38.3 months; stage 3, 14.5 months; stage 4, 18.0 months; P = .005; R status, mean survival times: R0, 42.1 months; R1, 29.0 months; R2, 13.9 months; P = 2.00 × 10−4; Table 3; Figure 1D and Figure 2D; and eFigure 3 in the Supplement). To further assess the significance and the prognosticative power of SNPs rs684559 and rs353630, we used a likelihood ratio test between the models that includes either 1 of both SNPs and a reduced model that considers only R status and AJCC stage (eTable 3 in the Supplement). We observed that the 2 models, which include either rs684559 or rs353630, fit significantly better than the reduced one (rs684559: HR, 0.40 [95% CI, 0.24-0.65]; P = 2.48 × 10−4; rs353630: HR, 0.44 [95% CI, 0.30-0.61]; P = .004). This comparison shows that the inclusion of the genotypes at this locus together with the clinical factors will have greater predictive power than the clinical information alone.
A Novel Biomarker Signature for PDAC Survival
Finally, we were interested whether the genetic information of both SNPs combined can further increase prognosticative power. To this end, we merged the genotypes associated with a lower rate of survival in each individual SNP by grouping patients with a T/T genotype of rs353630 or the A/A genotype of rs684559 into a joint risk–indicating biomarker signature and compared this with the grouped set of the remaining alleles that individually confer a survival advantage (at least 1 C allele of rs353630 and 1 G allele of rs684559). After stratification, we found that 47 individuals from the European cohort who were homozygous for either the minor T allele of SNP rs353630 or the minor A allele of SNP rs684559 had a 2.74-fold increased risk for tumor-associated death compared with 148 patients with at least 1 C allele of rs353630 and at least 1 G allele of rs684559 (HR, 0.37 [95% CI, 0.24-0.55]; P = 1.30 × 10−6 per Cox multivariate analysis; Table 2; eFigure 4 in the Supplement). In the TCGA data set, 23 individuals with the risk-indicating biomarker signature had a 5.08-fold increase in risk compared with the remaining 113 patients with PDAC (HR, 0.20 [95% CI, 0.10-0.39]; P = 4.20 × 10−6; Table 2; eFigure 4 in the Supplement). Accordingly, a Kaplan-Meier survival estimate demonstrated the shortest mean survival time for patients with the risk-indicating signature vs the protective signature in each cohort (European, 18.1 [95% CI, 14.1-22.1] months vs 43.7 [95% CI, 35.7-51.6] months; P = 3.00 × 10−5; TCGA, 13.0 [95% CI, 9.8-16.2] months vs 30.3 [95% CI, 24.5-36.0] months; P = 1.00 × 10−5; eTable 2 and eFigure 4 in the Supplement). Lastly, we performed the survival analysis after merging both data sets. The fold-increase of risk for tumor-associated death for those with the risk-indicating biomarker signature was 2.63 (HR, 0.38 [95% CI, 0.27-0.53]; P = 1.00 × 10−8; Table 2; Figure 2E), whereby 70 patients with a T/T genotype of rs353630 or the A/A genotype of rs684559 were estimated to survive a median of 15.0 (95% CI, 12.1-17.9) months after resection of their tumors, compared with 24.0 (95% CI, 18.1-29.9) months for patients with a C allele of rs353630 and a G allele of rs684559 (P = 2.00 × 10−8; Table 3; Figure 2F). When we used a likelihood ratio test between the model that includes the 2-dimensional biomarker signature and a reduced model that considers only SNP rs684559, R status, and AJCC stage, we found that the former model fits significantly better than the reduced one (HR, 0.38 [95% CI, 0.27-0.53]; P = 6.31 × 10−4).
Discussion
In this report, we expand previous methodology to uncover functional inherited genetic variants that affect the clinical course of cancer.15,16 Using this screening approach, we identified 2 common polymorphisms that are located in regulatory regions of the CHI3L2 and the CD44 genes and associated with allelic differences in survival after PDAC resection. This observation was subsequently validated in an independent study cohort, the publicly available TCGA pancreatic cancer database.
The genotype of the identified SNPs can be determined before the initiation of any treatment by a technically straightforward blood test, which is readily available at the time of diagnosis and is used to identify patients who are at higher risk for faster tumor progression. Other prognostic factors currently in clinical use (completeness of resection and tumor stage) or those suggested to be useful in a clinical setting (postoperative carbohydrate antigen 19-9 serum levels)38 can only be accurately determined after surgical resection and therefore have limited relevance to treatment decisions. In contrast with those prognostic factors, the proposed SNP-based biomarker signature may be used to identify individuals with an aggressive tumor biology who may be considered for alternative systemic treatment strategies, such as neoadjuvant chemotherapy, in keeping with the definition and criteria of borderline resectable pancreatic ductal adenocarcinoma recently proposed in Sendai, Japan.39 Indeed, the adoption of neoadjuvant systemic protocols, as opposed to upfront surgery, is thought to provide several advantages, including the treatment of micrometastatic disease in a subset of patients with an aggressive cancer phenotype who are most likely to benefit from early administration of systemic therapy.6,40 In addition, this biomarker signature also has a potential to affect adjuvant treatment protocols.
Limitations
However, 1 limitation of this study is the sample size that did not allow for an adjustment of the survival analysis to the type of administered systemic treatment, and future studies are necessary to elucidate the association of the SNPs with individual therapeutic protocol outcomes. Indeed, upcoming clinical trials of new systemic therapeutic strategies in patients with resectable PDAC should consider a stratification of patients by the identified SNP genotypes.
Conclusions
The protein CD44 is a transmembrane glycoprotein involved in a vast range of cellular processes, such as regulation of growth, survival, differentiation, and motility.41 In pancreatic cancer, CD44 has been shown to affect the invasiveness, progression, and metastatic phenotype of this tumor type.42,43,44 In contrast, CHI3L2 is 1 of 3 human members of the chitinase-like protein family (CLPs), which are lectins combining properties of cytokines and growth factors.45 Human chitinase-like proteins are secreted by cancer cells, macrophages, neutrophils, and other cells and regulate cancer cell proliferation, tumor microenvironment, and metastatic spread.45,46 Experimental data on DNA-protein interactions from the ENCODE project47 (specifically functional DNA markers, including DNase I–hypersensitivity sites; H3K4me1, H3K4me3, and H3K27Ac marks; and chromatin interactions and chromatin state segmentation), demonstrated that the identified SNPs (rs684559 and rs353630) reside in highly active regulatory genomic regions of the CHI3L2 and CD44 genes (data not shown). These observations offer grounds for further experimental investigation into the molecular and cellular mechanisms underlying the allelic differences in survival reported in this article. Importantly, this type of molecular knowledge is also key to providing a basis for the development of individualized drug therapies. It is tempting to speculate that the identified SNPs could be used in the future to select groups of patients who are more likely to respond to CHI3L2 and CD44 targeted therapies, which are currently being investigated in both preclinical and clinical trials45,48,49,50 (ClinicalTrials.gov: NCT03009214).
eMethods. Online Methods.
eTable 1. Cox survival analysis of top SNPs with a cut-off P value of P < .001 in the European discovery and the TCGA validation PDAC cohorts (adjusted for AJCC stage and R-status)
eTable 2. Survival time analysis in the European and TCGA data sets based on Kaplan-Meier estimates
eTable 3. Hazard Ratios
eFigure 1. Workflow diagram of the survival screen.
eFigure 2. Survival analysis of tumor-specific survival after resection of PDAC. (A) and (B) Kaplan-Meier survival curves for SNP rs353630 in the European and TCGA cohort, respectively. (C) and (D) Kaplan-Meier survival curves for SNP rs684559 in the European and TCGA cohort, respectively.
eFigure 3. Kaplan-Meier survival curves for the two independent prognostic factors AJCC stage (A) and resection margin status (B) in the merged data set.
eFigure 4. Analysis of tumor-related survival for the biomarker signature that combines both SNP loci (rs684559 and rs353630). (A) and (B) Cox multivariate regression survival curves for the combination signature in the European and TCGA cohort, respectively (adjusted to the known independent prognostic factors tumor stage and resection status). (C) and (D) Kaplan-Meier survival curves for the combination signature in the European and TCGA cohort, respectively.
References
- 1.Lennon AM, Wolfgang CL, Canto MI, et al. The early detection of pancreatic cancer: what will it take to diagnose and treat curable pancreatic neoplasia? Cancer Res. 2014;74(13):-. doi: 10.1158/0008-5472.CAN-14-0734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371(11):1039-1049. doi: 10.1056/NEJMra1404198 [DOI] [PubMed] [Google Scholar]
- 3.Heestand GM, Murphy JD, Lowy AM. Approach to patients with pancreatic cancer without detectable metastases. J Clin Oncol. 2015;33(16):1770-1778. doi: 10.1200/JCO.2014.59.7930 [DOI] [PubMed] [Google Scholar]
- 4.Yekebas EF, Bogoevski D, Cataldegirmen G, et al. En bloc vascular resection for locally advanced pancreatic malignancies infiltrating major blood vessels: perioperative outcome and long-term survival in 136 patients. Ann Surg. 2008;247(2):300-309. doi: 10.1097/SLA.0b013e31815aab22 [DOI] [PubMed] [Google Scholar]
- 5.Mueller S, Engleitner T, Maresch R, et al. Evolutionary routes and KRAS dosage define pancreatic cancer phenotypes. Nature. 2018;554(7690):62-68. doi: 10.1038/nature25459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolff RA. Adjuvant or neoadjuvant therapy in the treatment in pancreatic malignancies: where are we? Surg Clin North Am. 2018;98(1):95-111. doi: 10.1016/j.suc.2017.09.009 [DOI] [PubMed] [Google Scholar]
- 7.Vazquez A, Bond EE, Levine AJ, Bond GL. The genetics of the p53 pathway, apoptosis and cancer therapy. Nat Rev Drug Discov. 2008;7(12):979-987. doi: 10.1038/nrd2656 [DOI] [PubMed] [Google Scholar]
- 8.Grochola LF, Zeron-Medina J, Mériaux S, Bond GL. Single-nucleotide polymorphisms in the p53 signaling pathway. Cold Spring Harb Perspect Biol. 2010;2(5):a001032. doi: 10.1101/cshperspect.a001032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wheeler HE, Maitland ML, Dolan ME, Cox NJ, Ratain MJ. Cancer pharmacogenomics: strategies and challenges. Nat Rev Genet. 2013;14(1):23-34. doi: 10.1038/nrg3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang H, Wei P, Chang P, et al. Genetic polymorphisms associated with pancreatic cancer survival: a genome-wide association study. Int J Cancer. 2017;141(4):678-686. doi: 10.1002/ijc.30762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu C, Kraft P, Stolzenberg-Solomon R, et al. Genome-wide association study of survival in patients with pancreatic adenocarcinoma. Gut. 2014;63(1):152-160. doi: 10.1136/gutjnl-2012-303477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willis JA, Olson SH, Orlow I, et al. A replication study and genome-wide scan of single-nucleotide polymorphisms associated with pancreatic cancer risk and overall survival. Clin Cancer Res. 2012;18(14):3942-3951. doi: 10.1158/1078-0432.CCR-11-2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Innocenti F, Owzar K, Cox NL, et al. A genome-wide association study of overall survival in pancreatic cancer patients treated with gemcitabine in CALGB 80303. Clin Cancer Res. 2012;18(2):577-584. doi: 10.1158/1078-0432.CCR-11-1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rizzato C, Campa D, Talar-Wojnarowska R, et al. Association of genetic polymorphisms with survival of pancreatic ductal adenocarcinoma patients. Carcinogenesis. 2016;37(10):957-964. doi: 10.1093/carcin/bgw080 [DOI] [PubMed] [Google Scholar]
- 15.Stracquadanio G, Vrugt B, Flury R, et al. CD44 SNP rs187115: A novel biomarker signature that predicts survival in resectable pancreatic ductal adenocarcinoma. Clin Cancer Res. 2016;22(24):6069-6077. doi: 10.1158/1078-0432.CCR-16-0058 [DOI] [PubMed] [Google Scholar]
- 16.Vazquez A, Grochola LF, Bond EE, et al. Chemosensitivity profiles identify polymorphisms in the p53 network genes 14-3-3τ and CD44 that affect sarcoma incidence and survival. Cancer Res. 2010;70(1):172-180. doi: 10.1158/0008-5472.CAN-09-2218 [DOI] [PubMed] [Google Scholar]
- 17.Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31(21):3555-3557. doi: 10.1093/bioinformatics/btv402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Auton A, Brooks LD, Durbin RM, et al. ; 1000 Genomes Project Consortium . A global reference for human genetic variation. Nature. 2015;526(7571):68-74. doi: 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang DK, Johns AL, Merrett ND, et al. Margin clearance and outcome in resected pancreatic cancer. J Clin Oncol. 2009;27(17):2855-2862. doi: 10.1200/JCO.2008.20.5104 [DOI] [PubMed] [Google Scholar]
- 20.Esposito I, Kleeff J, Bergmann F, et al. Most pancreatic cancer resections are R1 resections. Ann Surg Oncol. 2008;15(6):1651-1660. doi: 10.1245/s10434-008-9839-8 [DOI] [PubMed] [Google Scholar]
- 21.Wittekind C, Compton C, Quirke P, et al. A uniform residual tumor (R) classification: integration of the R classification and the circumferential margin status. Cancer. 2009;115(15):3483-3488. doi: 10.1002/cncr.24320 [DOI] [PubMed] [Google Scholar]
- 22.Munding J, Uhl W, Tannapfel A. [R classification and pancreatic ductal adenocarcinoma—R 0 is R 0] [article in German]. Z Gastroenterol. 2011;49(10):1423-1427. doi: 10.1055/s-0031-1281750 [DOI] [PubMed] [Google Scholar]
- 23.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248-249. doi: 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLaren W, Gil L, Hunt SE, et al. The Ensembl variant effect predictor. Genome Biol. 2016;17(1):122. doi: 10.1186/s13059-016-0974-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zerbino DR, Wilder SP, Johnson N, Juettemann T, Flicek PR. The ensembl regulatory build. Genome Biol. 2015;16(1):56. doi: 10.1186/s13059-015-0621-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M; ENCODE Project Consortium . An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57-74. doi: 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57(1):289-300. doi: 10.2307/2346101 [DOI] [Google Scholar]
- 28.Park J-H, Gail MH, Weinberg CR, et al. Distribution of allele frequencies and effect sizes and their interrelationships for common genetic susceptibility variants. Proc Natl Acad Sci U S A. 2011;108(44):18026-18031. doi: 10.1073/pnas.1114759108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76(5):887-893. doi: 10.1086/429864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeron-Medina J, Wang X, Repapi E, et al. A polymorphic p53 response element in KIT ligand influences cancer risk and has undergone natural selection. Cell. 2013;155(2):410-422. doi: 10.1016/j.cell.2013.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bond GL, Hu W, Bond EE, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119(5):591-602. doi: 10.1016/j.cell.2004.11.022 [DOI] [PubMed] [Google Scholar]
- 32.Post SM, Quintás-Cardama A, Pant V, et al. A high-frequency regulatory polymorphism in the p53 pathway accelerates tumor development. Cancer Cell. 2010;18(3):220-230. doi: 10.1016/j.ccr.2010.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grochola LF, Müller TH, Bond GL, Taubert H, Udelnow A, Würl P. MDM2 SNP309 associates with accelerated pancreatic adenocarcinoma formation. Pancreas. 2010;39(1):76-80. doi: 10.1097/MPA.0b013e3181b9f105 [DOI] [PubMed] [Google Scholar]
- 34.Aguirre AJ, Hruban RH, Raphael BJ; Cancer Genome Atlas Research Network. Electronic address: andrew_aguirre@dfci.harvard.edu; Cancer Genome Atlas Research Network . Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell. 2017;32(2):185-203.e13. doi: 10.1016/j.ccell.2017.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson DC, Weinhold N, Mitchell JS, et al. Genome-wide association study identifies variation at 6q25.1 associated with survival in multiple myeloma. Nat Commun. 2016;7:10290. doi: 10.1038/ncomms10290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo Q, Schmidt MK, Kraft P, et al. ; kConFab Investigators . Identification of novel genetic markers of breast cancer survival. J Natl Cancer Inst. 2015;107(5):djv081. doi: 10.1093/jnci/djv081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stadler ZK, Thom P, Robson ME, et al. Genome-wide association studies of cancer. J Clin Oncol. 2010;28(27):4255-4267. doi: 10.1200/JCO.2009.25.7816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berger AC, Garcia M Jr, Hoffman JP, et al. Postresection CA 19-9 predicts overall survival in patients with pancreatic cancer treated with adjuvant chemoradiation: a prospective validation by RTOG 9704. J Clin Oncol. 2008;26(36):5918-5922. doi: 10.1200/JCO.2008.18.6288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Isaji S, Mizuno S, Windsor JA, et al. International consensus on definition and criteria of borderline resectable pancreatic ductal adenocarcinoma 2017. Pancreatology. 2018;18(1):2-11. doi: 10.1016/j.pan.2017.11.011 [DOI] [PubMed] [Google Scholar]
- 40.Jang J-Y, Han Y, Lee H, et al. Oncological benefits of neoadjuvant chemoradiation with gemcitabine versus upfront surgery in patients with borderline resectable pancreatic cancer: a prospective, randomized, open-label, multicenter phase 2/3 trial. Ann Surg. 2018;268(2):215-222. doi: 10.1097/SLA.0000000000002705 [DOI] [PubMed] [Google Scholar]
- 41.Ponta H, Sherman L, Herrlich PA. CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol. 2003;4(1):33-45. doi: 10.1038/nrm1004 [DOI] [PubMed] [Google Scholar]
- 42.Zöller M. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer. 2011;11(4):254-267. doi: 10.1038/nrc3023 [DOI] [PubMed] [Google Scholar]
- 43.Li C, Heidt DG, Dalerba P, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67(3):1030-1037. doi: 10.1158/0008-5472.CAN-06-2030 [DOI] [PubMed] [Google Scholar]
- 44.Jiang W, Zhang Y, Kane KT, et al. CD44 regulates pancreatic cancer invasion through MT1-MMP. Mol Cancer Res. 2015;13(1):9-15. doi: 10.1158/1541-7786.MCR-14-0076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kzhyshkowska J, Yin S, Liu T, Riabov V, Mitrofanova I. Role of chitinase-like proteins in cancer. Biol Chem. 2016;397(3):231-247. doi: 10.1515/hsz-2015-0269 [DOI] [PubMed] [Google Scholar]
- 46.Shao R, Hamel K, Petersen L, et al. YKL-40, a secreted glycoprotein, promotes tumor angiogenesis. Oncogene. 2009;28(50):4456-4468. doi: 10.1038/onc.2009.292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kundaje A, Meuleman W, Ernst J, et al. ; Roadmap Epigenomics Consortium . Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317-330. doi: 10.1038/nature14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Faibish M, Francescone R, Bentley B, Yan W, Shao R. A YKL-40-neutralizing antibody blocks tumor angiogenesis and progression: a potential therapeutic agent in cancers. Mol Cancer Ther. 2011;10(5):742-751. doi: 10.1158/1535-7163.MCT-10-0868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wood NJ. Pancreatic cancer: pancreatic tumour formation and recurrence after radiotherapy are blocked by targeting CD44. Nat Rev Gastroenterol Hepatol. 2014;11(2):73. doi: 10.1038/nrgastro.2014.1 [DOI] [PubMed] [Google Scholar]
- 50.Li L, Hao X, Qin J, et al. Antibody against CD44s inhibits pancreatic tumor initiation and postradiation recurrence in mice. Gastroenterology. 2014;146(4):1108-1118. doi: 10.1053/j.gastro.2013.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods. Online Methods.
eTable 1. Cox survival analysis of top SNPs with a cut-off P value of P < .001 in the European discovery and the TCGA validation PDAC cohorts (adjusted for AJCC stage and R-status)
eTable 2. Survival time analysis in the European and TCGA data sets based on Kaplan-Meier estimates
eTable 3. Hazard Ratios
eFigure 1. Workflow diagram of the survival screen.
eFigure 2. Survival analysis of tumor-specific survival after resection of PDAC. (A) and (B) Kaplan-Meier survival curves for SNP rs353630 in the European and TCGA cohort, respectively. (C) and (D) Kaplan-Meier survival curves for SNP rs684559 in the European and TCGA cohort, respectively.
eFigure 3. Kaplan-Meier survival curves for the two independent prognostic factors AJCC stage (A) and resection margin status (B) in the merged data set.
eFigure 4. Analysis of tumor-related survival for the biomarker signature that combines both SNP loci (rs684559 and rs353630). (A) and (B) Cox multivariate regression survival curves for the combination signature in the European and TCGA cohort, respectively (adjusted to the known independent prognostic factors tumor stage and resection status). (C) and (D) Kaplan-Meier survival curves for the combination signature in the European and TCGA cohort, respectively.