This study investigates the utility of reinterpreting the genomic test results of pediatric patients who previously underwent genomic epilepsy testing.
Key Points
Question
How often do genomic test result interpretations change?
Findings
In this study of reported genetic variants from clinical genomic epilepsy tests, 67 of 185 pediatric patients (36.2%) had variants reclassified. A clinically significant change in the interpretation occurred in 19 of 61 patients (31.1%) with a genetic diagnosis during the 5-year study.
Meaning
The findings of this study suggest that pediatric patients with epilepsy and previous genomic test results should have their test results reinterpreted at least every 2 years and before further genetic testing.
Abstract
Importance
Clinical genomic tests that examine the DNA sequence of large numbers of genes are commonly used in the diagnosis and management of epilepsy in pediatric patients. The permanence of genomic test result interpretations is not known.
Objective
To investigate the value of reinterpreting previously reported genomic test results.
Design, Setting, and Participants
This study retrospectively reviewed and reinterpreted genomic test results from July 1, 2012, to August 31, 2015, for pediatric patients who previously underwent genomic epilepsy testing at a single tertiary care pediatric health care facility. Reinterpretation of previously reported variants was conducted in May 2017.
Main Outcomes and Measures
Patient reports from clinical genomic epilepsy tests were reviewed, and all reported genetic variants were reinterpreted using 2015 consensus standards and guidelines for interpreting hereditary genetic variants. Three classification tiers were used in the reinterpretation: pathogenic or likely pathogenic variant, variant of uncertain significance (VUS), or benign or likely benign variant.
Results
A total of 309 patients had genomic epilepsy tests performed (mean [SD] age, 5.6 [0.8] years; 163 [52.8%] male), and 185 patients had a genetic variant reported. The reported variants resulted in 61 patients with and 124 patients without a genetic diagnosis (VUS variants only). On reinterpretation of all reported variants, 67 of the 185 patients (36.2%) had a change in variant classification. Of the 67 patients with a genetic variant change in interpretation, 21 (31.3%) experienced a change in diagnosis. During the 5 years of the study, 19 of 61 patients (31.1%) with a genetic diagnosis and 48 of 124 patients (38.7%) with undiagnosed conditions (VUS only) had their results reclassified. Review of genomic reports issued during the final 2 years of the study identified reclassification of variants in 4 of 16 patients (25.0%) with a pathogenic or likely pathogenic variant and 11 of 41 patients (26.8%) with a VUS.
Conclusions and Relevance
The identified high rate of reinterpretation in this study suggests that interpretation of genomic test results has rapidly evolved during the past 5 years. These findings suggest that reinterpretation of genomic test results should be performed at least every 2 years.
Introduction
Genomic testing is used to evaluate many pediatric neurologic diseases, including intractable epilepsy and neuromuscular disorders. A recent study1 recommended broad genetic sequencing in the routine evaluation of early childhood epilepsies. Although it is technically feasible to analyze hundreds of genes in a single genomic test,2 the interpretation and clinical significance of genetic variants are evolving. Laboratory guidelines (College of American Pathologists and American College of Medical Genetics and Genomics [ACMG]) recommend periodic review of reported genetic variants.3,4 However, no clear recommendations guide the frequency of review of previously issued laboratory diagnoses.
Current clinical genomic tests include disease-specific panels, whole exome sequencing, and whole genome sequencing, which are powerful tools when used in combination with clinical knowledge.5 Recent reports6,7 have focused on the discovery and identification of new disease associations by reanalysis of genomic data. Only a few genomic studies8,9,10,11 have expanded the scope of reanalysis to include all gene variants previously reported. In a recent genomic reanalysis study8 of 494 patients with intellectual disability and/or developmental delay, the disease significance of a gene variant was upgraded in 23 patients and downgraded in 7 patients. Another previous study9 examining the reinterpretation of small panels of genes associated with hypertrophic cardiomyopathy found frequent changes in genetic interpretation. Similarly, periodic reinterpretation may apply new therapeutic associations to previously reported gene variants.12,13 Advances in publicly available databases and standardized interpretation criteria have improved the clinical interpretation of gene variants.4,14,15,16 In this study, we examined the frequency and significance of gene variant reclassification from previously interpreted epilepsy genomic test results.
Methods
This retrospective study included patients from a tertiary care pediatric health system (Children's Health Dallas, Dallas, Texas) with clinical genomic tests performed using next-generation sequencing epilepsy gene panels (July 1, 2012, to August 31, 2015). A single clinical laboratory (GeneDx) performed the tests and the initial interpretation of all the results. Initial genetic test results provided by the laboratory included interpretations equivalent to pathogenic variant, likely pathogenic variant, variant of uncertain significance, or negative test with no clinically significant variants. No benign or likely benign (B-LB) variants were initially reported. The laboratory in this study has a practice of routinely reclassifying variants and providing updated reports. In addition, this same laboratory routinely reinterprets variants and updates the significance of variants within the public database ClinVar and provides updated patient reports. This study was approved by the University of Texas Southwestern Medical Center Institutional Review Board with waiver of informed consent. Data were deidentified on collection and subsequently analyzed.
Reinterpretation of previously reported variants was conducted in May 2017 in 2 stages. First, variants were screened for new information in population frequency (Exome Aggregation Consortium, 1000 Genomes phase 315) (>1%) or clinical association (Human Gene Mutation Database, ClinVar16) databases. Second, identified variants were reinterpreted using the 2015 ACMG variant classification criteria4 and then compared with ClinVar updates from the original laboratory.
A clinically significant change of diagnosis was defined as P-LP variant downgraded to a VUS or B-LB variant or VUS upgraded to P-LP. Variant reinterpretation was performed by consensus among a clinical geneticist (G.G.), a clinical neurologist (D.M.T.), and a clinical genomics laboratory director (J.Y.P.).
Linear regression statistical analysis was performed using GraphPad Prism, version 7 (GraphPad Software). Statistical tests included R2, nonzero slope test, and comparison of the slopes (P < .05 was considered to be statistically significant).
Results
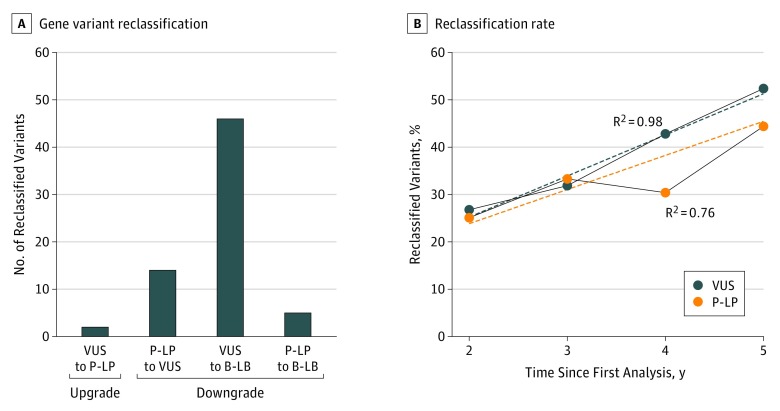
Genomic epilepsy tests were performed for 309 patients (mean [SD] age, 5.6 [0.8] years; 163 [52.8%] male), and 185 patients had a reported genetic variant. A laboratory diagnosis of at least 1 VUS (124 of 185 [67.0%]) and/or P-LP (61 of 185 [33.0%]) variant was initially reported. Thirty-five patients had both a VUS and a P-LP variant reported. After reanalyzing all cases with reported variants from the past 5 years, 67 of the 185 patients (36.2%) with a reported gene variant had a revised classification. A clinically significant change in diagnosis occurred in 21 of the 185 patients (11.4%) (Figure 1). Nineteen P-LP variants were downgraded in pathogenicity (14 to VUS and 5 to B-LB), and 2 VUSs were upgraded to P-LP (Table). Nine of these 21 reclassified variants were in concordance with the original laboratory data in upgraded or downgraded significance as reported in ClinVar and/or the patient's updated medical record.
Figure 1. Patients With Diagnostic Changes Based on Variant Classification.
A, Patients with reclassification of gene variants from each of the categories. There were patients with both a variant of uncertain significance (VUS) and a pathogenic or likely pathogenic (P-LP) variant reclassified; these patients are represented only once in this figure. B, Reclassification rate plotted as the fraction of reclassified variants for each year testing was performed. Solid lines indicate the fraction of patients with a reclassified variant; dotted lines, the extrapolated slopes for the change in VUS classification (9% per year) and P-LP variant classification (6% per year). The R2 and slope values were calculated using linear regression. B-LB indicates benign or likely benign.
Table. Reclassified Clinically Significant P-LP Variants and VUSsa.
| Gene | OMIM | Variant | Laboratory Initial Classification | Laboratory Revised Classification in ClinVar or Patient Medical Record | Other ClinVar Classification | Study Revised Classification | Clinical Significance |
|---|---|---|---|---|---|---|---|
| ARX | 300382 | c.426_458 dup33, p.Gly143_Ala153dup | P | None | P | VUS | Diagnostic |
| CLN5b | 608102 | c.2 T>C, p.Pro398Ser | LP | VUS | VUS | VUS | Carrier |
| CLN5 | 608102 | c.1192 C>T, p.Met1 | P | VUS | VUS, LB | VUS | Carrier |
| CTSD | 116840 | c.751 G>A, p.Asp251Asn | LP | None | VUS | VUS | Diagnostic |
| EFHC1 | 608815 | c.629 A>T, p.Asp210Val | LP | B | VUS, LB, B | B | Diagnostic |
| KCNQ2 | 602235 | c.1028 C>G, p.Ala343Gly | LP | None | None | VUS | Therapeutic |
| PCDH19 | 300460 | c.379 C>T, p.Pro127Ser | LP | None | None | VUS | Therapeutic |
| PNKP | 605610 | c.1029 + 2 T>C | P | LP | LP, VUS | LBc | Diagnostic |
| PNPO | 603287 | c.782 C>T, p.Pro261Leu | LP | VUS | VUS | VUS | Therapeutic |
| POLGb | 174763 | c.803 G>C, p. Gly268Ala | P | VUS | VUS, LB | LB | Carrier/diagnostic |
| PPT1 | 600722 | c.362 + 5 G>A | P | None | None | VUS | Diagnostic |
| SCN1A | 182389 | c.2681 C>G, p.Thr894Ser | LP | None | None | VUS | Therapeutic |
| SCN1A | 182389 | c.5168 C>T, p.Ser1723Phe | LP | None | None | VUS | Therapeutic |
| SCN1A | 182389 | c.5131 G>A, p.Gly1711Ser | LP | None | None | VUS | Therapeutic |
| SCN2A | 182390 | c.2695 G>A, p.Gly899Ser | LP | None | None | VUS | Therapeutic |
| SCN2A | 182390 | c.3997 G>A, p. Ala1333Thr | LP | P | None | VUS | Therapeutic |
| SCN8A | 600702 | c.5614 C>G, p.Arg1872Gly | VUS | LP | P, LP | P-LP | Therapeutic |
| TBC1D24 | 613577 | c.457 G>A, p.Glu153Lys | VUS | None | None | P-LP | Diagnostic |
| TSC2 | 191092 | c.599 + 3 A>G | LP | LB | VUS | LB | Cancer screening |
Abbreviations: B, benign; LB, likely benign; LP, likely pathogenic; P-LP, pathogenic or likely pathogenic; P, pathogenic; VUS, variant of uncertain significance.
Reclassifications that affected diagnosis, therapy, or carrier status are shown. Laboratory initial classification is the original laboratory interpretation. Laboratory revised classification in ClinVar or patient medical record is the original laboratory’s reinterpretation reported in ClinVar and/or in the patient's medical record. Other ClinVar classification is other laboratories’ interpretations. Study-revised classification is this study’s reinterpretation.
Two gene variants (CLN5 and POLG) were each seen in 2 separate probands.
For PNKP c.1029 + 2T>C, this study used the database Exome Aggregation Consortium, which has a frequency of 3.6% in a European (Finnish) subpopulation. This was used to interpret the variant as likely benign; however, a more recently available and larger database (Genome Aggregation Database) shows a European (Finnish) subpopulation frequency of 0.1%.
Next, we examined the likelihood of reclassification over time. The reclassification rates for patients with P-LP variants and VUSs were similar, with nonsignificant differences in slope (P-LP, 6% change per year; VUS, 9% change per year; P = .24) (Figure 1). In total, 19 of 61 patients (31.1%) had a change in genetic diagnosis. Examination of a subset of the most recent 2 years of data identified 4 of 16 P-LP variants (25.0%) and 11 of 41 VUSs (26.8%) as reclassified.
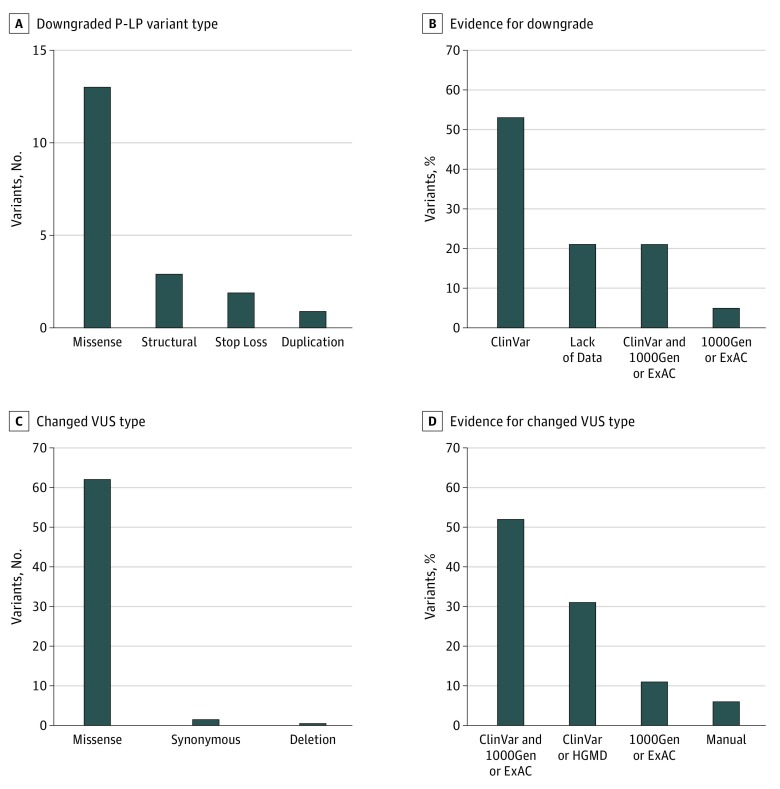
The most frequently downgraded P-LP variants were missense (n = 13) (Figure 2). In most cases, protein-truncating variants were downgraded because of new population database information that indicated a high frequency of loss-of-function variants in healthy individuals. Overall, P-LP variants were reclassified because of new information in clinical databases (10 of 19 [52.6%]), population databases (1 of 19 [5.3%]), or a combination of both (4 of 19 [21.1%]) or the absence of data in either (4 of 19 [21.1%]) (Figure 2). Many of the downgraded P-LP variants or VUSs were similarly downgraded by the original reporting laboratory as observed in the laboratory’s ClinVar and/or medical record updates.
Figure 2. Variants Reinterpreted in the Study.
Number of downgraded pathogenic or likely pathogenic (P-LP) variants (A) and evidence for downgrade (B) and number of changed variant of uncertain significance (VUS) types (C) and evidence for change (D). The most frequently downgraded P-LP variant type was missense (n = 13) followed by protein truncating variants (structural, n = 3; stop-loss, n = 2) and duplications (n = 1). For VUS types that changed, the most frequent type was missense (n = 62) followed by synonymous (n = 2) and deletions (n = 1). The dominant category of protein consequence was missense variants in the P-LP variant (68.4%) and VUS (95.4%). The evidence used to reclassify the variants was as follows: ClinVar, information posted to the ClinVar database; lack of data, no evidence in the ClinVar database or the Human Gene Mutation Database; HGMD, population frequency in the Human Gene Mutation Database; 1000Gen, population frequency in the 1000 Genomes database; ExAC, population frequency information in the Exome Aggregation Consortium database; and manual, determined when variant was synonymous or a protein-truncating variant.
Of the 124 patients with no definitive diagnosis (VUS only), 2 (1.6%) had a VUS upgraded to P-LP (Table); however, 46 of 124 patients (37.1%) had their variants downgraded to B-LB. Most reclassified VUSs were missense variants (n = 62) followed by synonymous or deletion variants (Figure 2). In reviewing VUSs, new information in clinical databases (20 of 65 [30.8%]), population databases (7 of 65 [10.8%]), or a combination of both (34 of 65 [52.3%]) provided the basis for revising the VUS classification (Figure 2).
Discussion
On the basis of our single institution experience, 67 of 185 patients (36.2%) had revised gene variant interpretations. A clinically significant change in diagnosis occurred for 21 patients (11.4%). During the 5-year study period, patients with P-LP variants had a yearly reclassification rate of 5.5% and patients with VUSs had a yearly reclassification rate of 8.7%. This change in reclassification affected 19 of 61 patients (31.1%) with a P-LP variant and 48 of 124 patients (38.7%) with a VUS. In the final 2 years of the study, 4 of 16 patients (25.0%) with a P-LP variant and 11 of 41 patients (26.8%) with a VUS were affected.
Compared with the 36.2% variant reclassification rate observed in this study, another recent study8 that examined changes to P-LP variants and VUSs found a lower rate of reclassification (6%; 23 patients with variants upgraded in significance and 6 patients with variants downgraded in significance). The difference in the rate of reclassification may be because of the longer period of review in our study (5 years) compared with the previous study8 (2 years). Of more importance, the previous study8 examined patients with intellectual disability and/or developmental delay, whereas our study was focused on pediatric patients with epilepsy. We anticipate that both the patient’s disease and the passage of time will influence the degree to which reinterpretation will occur.
We recommend that reinterpretation of genetic test results for pediatric patients with epilepsy be performed at a minimum frequency of every 2 years. The reinterpretation should not be limited to genomic tests but should include all previously reported genetic tests. Before the initiation of the study, we had anticipated that the variants requiring reclassification might decrease after the publication of the 2015 ACMG guidelines4 or the release of population databases in 2014.15 On the contrary, the pattern of reclassifications was unchanged.
Limitations
This study was limited by an absence of an analysis of B-LB variants that may be upgraded to P-LP variants or VUSs; these variants were unavailable for examination. In addition, we used a consensus interpretation approach and did not examine variations in interpretation among health care professionals; however, this was mitigated by our frequent positive correlation (9 of 21), with upgraded and downgraded interpretations posted by the original laboratory to ClinVar and/or patient’s medical record (Table). Also, it is possible that our reinterpretation of variants in this study may not have included all of the evidence available in the literature.
Conclusions
This study suggests that clinical laboratories should commit to reinterpretation of P-LP variants and VUSs, and corrected laboratory reports should be communicated to treating physicians. Furthermore, physicians should understand and communicate to patients that medical knowledge is dynamic and genetic test interpretations may change over time. In summary, patients with previous epilepsy genomic test results should have their test results reinterpreted at least every 2 years and before further genetic testing. Applying these recommendations to other genomic tests is worth considering and should be investigated in future studies.
References
- 1.Berg AT, Coryell J, Saneto RP, et al. Early-life epilepsies and the emerging role of genetic testing. JAMA Pediatr. 2017;171(9):-. doi: 10.1001/jamapediatrics.2017.1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Comprehensive Epilepsy Panel GeneDx. https://www.genedx.com/test-catalog/available-tests/comprehensive-epilepsy-panel/. Updated December 2016. Accessed May 2018.
- 3.College of American Pathologists Sequence variants—interpretation and reporting In: Molecular Pathology Checklist: CAP Accreditation Program. Northfield, IL: College of American Pathologists; July 28, 2015. [Google Scholar]
- 4.Richards S, Aziz N, Bale S, et al. ; ACMG Laboratory Quality Assurance Committee . Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405-424. doi: 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nambot S, Thevenon J, Kuentz P, et al. ; Orphanomix Physicians’ Group . Clinical whole-exome sequencing for the diagnosis of rare disorders with congenital anomalies and/or intellectual disability: substantial interest of prospective annual reanalysis. Genet Med. 2018;20(6):645-654. doi: 10.1038/gim.2017.162 [DOI] [PubMed] [Google Scholar]
- 6.Costain G, Jobling R, Walker S, et al. Periodic reanalysis of whole-genome sequencing data enhances the diagnostic advantage over standard clinical genetic testing. Eur J Hum Genet. 2018;26(5):740-744. doi: 10.1038/s41431-018-0114-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walsh R, Thomson KL, Ware JS, et al. ; Exome Aggregation Consortium . Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med. 2017;19(2):192-203. doi: 10.1038/gim.2016.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hiatt SM, Amaral MD, Bowling KM, et al. Systematic reanalysis of genomic data improves quality of variant interpretation. Clin Genet. 2018;94(1):174-178. doi: 10.1111/cge.13259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aronson SJ, Clark EH, Varugheese M, Baxter S, Babb LJ, Rehm HL. Communicating new knowledge on previously reported genetic variants. Genet Med. 2012;14(8):713-719. doi: 10.1038/gim.2012.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrison SM, Dolinsky JS, Knight Johnson AE, et al. Clinical laboratories collaborate to resolve differences in variant interpretations submitted to ClinVar. Genet Med. 2017;19(10):1096-1104. doi: 10.1038/gim.2017.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wright CF, McRae JF, Clayton S, et al. Making new genetic diagnoses with old data: iterative reanalysis and reporting from genome-wide data in 1,133 families with developmental disorders. [published online January 11, 2018]. Genet Med. 2018. doi: 10.1038/gim.2017.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mudigoudar B, Weatherspoon S, Wheless JW. Emerging antiepileptic drugs for severe pediatric epilepsies. Semin Pediatr Neurol. 2016;23(2):167-179. doi: 10.1016/j.spen.2016.06.003 [DOI] [PubMed] [Google Scholar]
- 13.Hani AJ, Mikati MA. Current and emerging therapies of severe epileptic encephalopathies. Semin Pediatr Neurol. 2016;23(2):180-186. doi: 10.1016/j.spen.2016.06.001 [DOI] [PubMed] [Google Scholar]
- 14.Lek M, Karczewski KJ, Minikel EV, et al. ; Exome Aggregation Consortium . Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285-291. doi: 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The 1000 Genomes Project Consortium; Auton A, Brooks LD, Durbin RM, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68-74. doi: 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.ClinVar Database https://www.ncbi.nlm.nih.gov/clinvar/. Accessed May 2018.