Abstract
The first, general glycosylation pathway in bacteria, the N-linked glycosylation system of Campylobacter jejuni, was discovered two decades ago. Since then, many diverse prokaryotic glycosylation systems have been characterized, including O-linked glycosylation systems that have no homologous counterparts in eukaryotic organisms. Shortly after these discoveries, glycosylation pathways were recombinantly introduced into E. coli creating the field of bacterial glycoengineering. Bacterial glycoengineering is an emerging biotechnological tool that harnesses prokaryotic glycosylation systems for the generation of recombinantly glycosylated proteins using E. coli as a host. Over the last decade, as our understanding of prokaryotic glycosylation systems has advanced, so too has the glycoengineering toolbox. Currently, glycoengineering utilizes two broad approaches to recombinantly glycosylate proteins, both of which can generate N- or O-linkages: oligosaccharyltransferase (OTase)-dependent and OTase-independent. This review discusses the applications of these bacterial glycoengineering techniques as they relate to the development of glycoconjugate vaccines, therapeutic proteins, and diagnostics.
Keywords: bioconjugate, E. coli, glycoengineering, glycosyltransferase, oligosaccharyltransferase
Introduction
Protein glycosylation, or the covalent attachment of carbohydrates to proteins, is a ubiquitous posttranslational modification. For the most part, protein glycosylation is characterized as either N-linked with glycans attached to asparagine residues, or as O-linked with glycans attached to serine or threonine residues. While the importance of eukaryotic glycosylation has been and continues to be a source of intensive research, prokaryotic glycosylation has only recently grabbed the attention of the scientific community with the discovery of a general N-linked protein glycosylation system in the ε-proteobacterium Campylobacter jejuni (Szymanski et al. 1999). Since the initial C. jejuni discovery, prokaryotic glycosylation systems have been described across a plethora of Gram-negative and Gram-positive bacteria and been shown to contribute towards normal bacterial physiology as well as pathogenesis (Nothaft, and Szymanski 2010; Iwashkiw et al. 2013; Schaffer, and Messner 2017). Given the straightforward nature of prokaryotic genetics, it was only a matter of time before protein glycosylation systems were engineered and exploited for the production of designer glycoproteins in a process termed “bacterial glycoengineering”.
Bacterial glycoengineering is a rapidly advancing field utilized to recombinantly glycosylate proteins mainly using E. coli as a model organism; however, other bacteria like Shigella and Salmonella have also been used. Because E. coli does not naturally encode for protein glycosylation machineries, it serves as an appropriate scaffold for bottom up glycoengineering applications for different types of glycoproteins. Moreover, recombineering-based strategies to remove major non-essential polysaccharide gene clusters, like the enterobacterial common antigen and host O antigen, from E. coli allow for even greater glycoengineering control (Yates et al. 2019). For bioconjugation purposes, bacterial glycosylation is classified as either oligosaccharyltransferase (OTase)-dependent or OTase-independent. OTase-dependent glycosylation occurs in the periplasmic space and is the en block transfer of glycans from pre-assembled lipid-linked precursors to acceptor proteins. Much like eukaryotic glycosylation, bacteria have evolved an N-linked OTase pathway, but also employ O-linked OTase systems that are unique to prokaryotic organisms. OTase-independent glycosylation occurs in the cytoplasm and relies on glycosyltransferases to transfer monosaccharides from nucleotide activated precursors for the sequential assembly of glycoproteins. Both OTase-dependent and -independent pathways are exploited for bioconjugating carbohydrates to proteins. In this review we outline recent advances in bacterial glycoengineering collectively used to develop glycoconjugate vaccines, protein drugs and diagnostics. In addition, we will introduce and discuss a relatively new field of bacterial glycoengineering revolving around outer membrane vesicles and their use as alternative vaccine delivery platform.
An introduction to glycoconjugate vaccines
Bacterial surface polysaccharides are some of the first, and most abundant, microbial components encountered by the immune system during infection (Comstock, and Kasper 2006). These polysaccharides, usually in the form of capsule or O antigen attached to lipid A, serve a multitude of purposes, including protecting microbial organisms from external threats and immune clearance. Given their abundance on invading organisms as well as their biochemical distinctness from eukaryotic carbohydrates, some microbial surface polysaccharides have been used as antigens for vaccine development. However, when polysaccharides are used alone in vaccine formulations, they usually act as T-cell independent antigens and therefore do not stimulate immunoglobulin class switching and long-term B cell memory. Moreover, polysaccharide vaccines alone do not elicit protection in vulnerable groups like infants and children under two years of age. This poor immune response can be overcome by covalently attaching a polysaccharide to a protein carrier in a process known as conjugation (De Gregorio, and Rappuoli 2014).
Traditionally, glycoconjugate vaccines are synthesized using a semi-synthetic approach where the polysaccharide is extracted from the target bacterium, purified, chemically modified and covalently linked to a carrier protein. This approach has resulted in the commercial licensure of multiple glycoconjugate vaccines to prevent colonization and infection by Haemophilus influenzae type B, and multiple serotypes of Streptococcus pneumoniae and Neisseria meningiditis. For detailed reviews on semi-synthetic or synthetic glycoconjugate vaccine production please refer to the following excellent review article (Berti, and Adamo 2018). Although conjugate vaccines produced chemically have seen immense commercial success (the glycoconjugate vaccine Prevnar 13 has been Pfizer’s best-selling product from 2015 to 2018 with over 24 billion USD in sales), their manufacturing processes are not without drawbacks; including, batch to batch variation, heterogenous product formation, large scale production of pathogenic organisms, and high manufacturing costs (Frasch 2009).
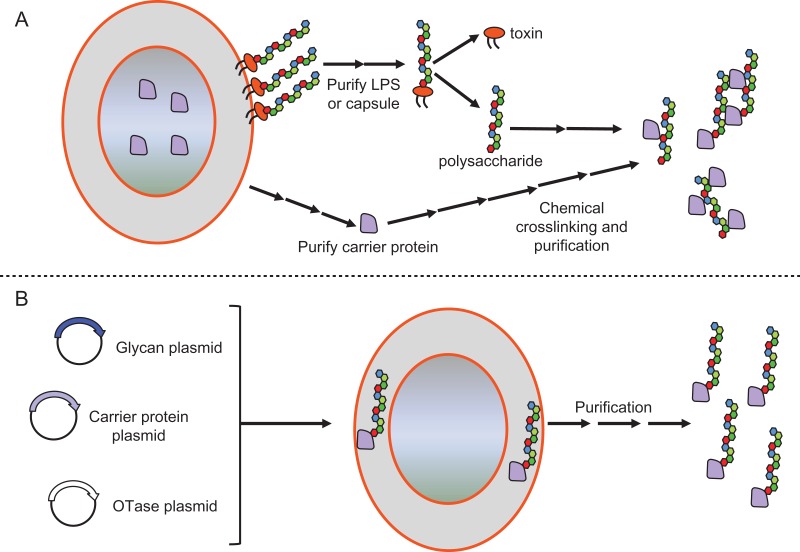
Over the last two decades, alternative strategies for producing glycoconjugate vaccines have emerged. These techniques are broad in their approach with some yielding vaccines closer to commercial licensure than others. Specifically, the advent of in vivo bacterial conjugations for manufacturing glycoconjugate vaccines have produced some of the most clinically advanced products to date. Commonly referred to as bioconjugation or protein glycan coupling technology (PGCT), the in vivo conjugation of polysaccharides to proteins for glycoconjugate vaccine production relies on OTases (Terra et al. 2012). It is generally considered that bioconjugation represents a simplification of the production and manufacturing process of glycoconjugate vaccines (Rappuoli et al. 2019) and is graphically represented in Figure 1.
Fig. 1.
Glycoconjugate vaccine production: Chemical conjugation compared to bioconjugation. (A) Glycoconjugate vaccines produced chemically require multiple processes for purification and detoxification of both the protein carrier as well as the polysaccharide. In addition, another set of processes are required to chemically crosslink the polysaccharides to proteins. (B) Bioconjugation produces glycoconjugate vaccines using E. coli as a host. The glycoengineered strain of E. coli simultaneously produces the lipid-linked polysaccharide, the protein carrier and an OTase, which will transfer the polysaccharide to the protein generating a bioconjugate.
Principles for glycoengineering bioconjugate vaccines
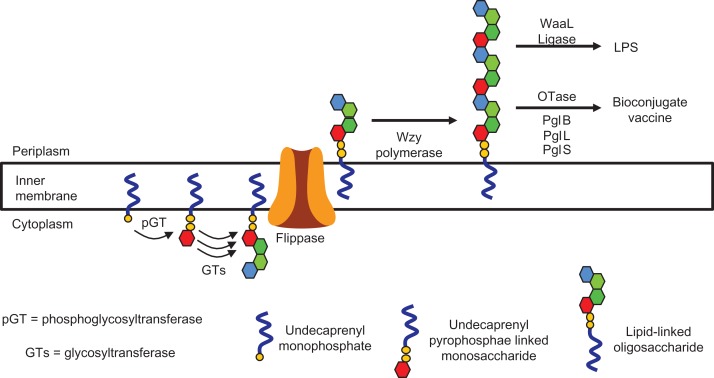
Prior to discussing the details of OTase-dependent bioconjugate vaccine production, it is helpful to introduce the steps required for assembly of lipid-linked oligosaccharides (LLOs). This process is summarized in Figure 2 for a pathway dependent on the generic polysaccharide polymerase Wzy. LPS synthesis and glycoprotein production share many homologies, which are exploited for bioconjugate vaccine production (Hug, and Feldman 2011). While both require undecaprenyl-pyrophosphate-linked carbohydrate precursors, LPS is dependent on the action of the WaaL O antigen ligase to transfer a polysaccharide to the outer core saccharide (Raetz, and Whitfield 2002); whereas, bacterial glycoproteins (in the context of OTase dependent glycosylation) depend on a specific OTase to transfer the glycan to an amino acid. The synthesis of the LLO is initiated by the transfer of a phosphosugar by an initiating phosphoglycosyltransferase, generating an undecaprenyl-pyrophosphate linked monosaccharide. Glycosyltransferases sequentially add monosaccharides from nucleotide activated precursors to the undecaprenyl-pyrophosphate linked monosaccharide (Valvano 2003). The LLO is subsequently flipped to the periplasm where it can be further polymerized into a heteropolymer consisting of many repeat units by Wzy (Raetz, and Whitfield 2002). Alternatively, a polysaccharide can be Wzy-independently generated by the addition of sugars to the undecaprenyl-pyrophosphate linked monosaccharide by different glycosyltransferases, and transported to the periplasm by an ABC transporter (Cuthbertson et al. 2010). A plethora of polysaccharides are synthesized as lipid-linked precursors including many capsular polysaccharides, teichoic acids, as well as the O antigen of lipopolysaccharide (Raetz, and Whitfield 2002; Whitfield 2006; Cuthbertson et al. 2010). OTases exclusively utilize undecaprenyl-pyrophosphate linked glycans as substrates (Wacker et al. 2006), and therefore glycans produced on other lipid carriers cannot be bioconjugated to proteins by the actions of OTases. While both ABC transporter and Wzy-dependent LLOs can be used by OTases, the majority of bioconjugate vaccines developed exploit polysaccharides synthesized in a Wzy-dependent manner.
Fig. 2.
Summary of lipid-linked oligosaccharide synthesis in a Wzy-dependent manner. Lipid-linked oligosaccharides are first assembled at the cytoplasmic leaflet of the inner-membrane, flipped to the periplasm, and polymerized by a Wzy polymerase where they then can be transferred by WaaL ligases or OTases to the outer core saccharide of LPS or proteins, respectively.
Because the WaaL ligase and OTases both use lipid-linked carbohydrates as substrates, it is well accepted that bacteria that possess WaaL do not carry genes encoding for OTases to diminish cross talks between these two pathways. Indeed, when developing strains of E. coli for glycoengineering purposes, one of the first mutations introduced is the deletion of the waaL gene to eliminate non-specific transfer of lipid-linked polysaccharides to the outer core saccharide of LPS (Feldman et al. 2005). While this is not essential for glycoengineering, it greatly increases the pool of LLOs that can be polymerized and transferred by the OTase to the target protein. In addition, endogenous, non-essential polysaccharide synthesis gene clusters, like the enterobacterial common antigen, can be removed to eliminate production of undesirable LLOs that could interfere with assembly of the target polysaccharide or that they themselves could be transferred by the OTase to a protein (Yates et al. 2019).
Both N-linking and O-linking OTases have been employed for biologically conjugating polysaccharides to carrier proteins for glycoconjugate vaccine production. Regardless of which OTase is employed, biological conjugations in any Gram-negative bacterium rely on three components (Figure 2): a genetic locus or loci that encode(s) for the polysaccharide biosynthesis proteins, a carrier protein to be glycosylated, and an OTase to transfer the desired carbohydrate to the carrier protein. While these three components are required, they do not necessarily need to be on three separate plasmids as outlined in Figure 2. In the following sections, we will introduce and discuss the N-linking OTase PglB and the O-linking OTases PilO, PglL, and PglS and discuss their roles’ in the production of glycoconjugate vaccines.
The N-linking oligosaccharyltransferase PglB and its use in glycoconjugate vaccine production
The N-linking OTase, PglB, which is a functional ortholog of the eukaryotic STT3 OTase (Wacker et al. 2002), was first discovered in the human gut pathogen Campylobacter jejuni. PglB glycosylates more than 80 proteins and plays an essential role in the pathobiology of C. jejuni (Szymanski et al. 2002; Scott et al. 2011, 2014); however, its biotechnological potential began to take shape in 2002 when the C. jejuni protein glycosylation system was functionally transferred into E. coli. At the time, the E. coli-PglB system was used to glycosylate native C. jejuni proteins with the native C. jejuni heptasaccharide or a permutation containing N-acetylglucosamine (GlcNAc), an artifact of heterologous expression in E. coli, instead of the natural 2,4-diacetamido-2,4,6-trideoxyglucose (diNAcBac) at the reducing end (Wacker et al. 2002). These glycoproteins generated in E. coli had limited biotechnological value, but, the stage had been set for producing recombinant glycoproteins with seemingly endless combinations. Three years later, the first OTase-dependent designer glycoprotein in E. coli was generated when the genetic locus encoding for the C. jejuni heptasaccharide was replaced with the locus encoding for the E. coli O7 polysaccharide, thus, enabling its transfer to one of the natural acceptor proteins, AcrA, by the OTase PglB (Feldman et al. 2005). This platform has since become the model for bioconjugate vaccine production and is also employed for developing some protein drugs and diagnostics discussed below.
The ability of PglB to transfer fully polymerized O7 polysaccharides was remarkable given that PglB naturally transfers only a single subunit of the C. jejuni heptasaccharide in its native host environment. In addition to transferring O7 polysaccharides, PglB was also able to transfer the E. coli O16 polysaccharide as well as the Pseudomonas aeruginosa O11 polysaccharide to AcrA simply by swapping out the biosynthetic machinery for these glycans in E. coli (Feldman et al. 2005). This feature is one of the many highlights of bioconjugate vaccine production, specifically, novel designer glycoconjugates can be rapidly engineered by exchanging the plasmid that encodes for the glycan biosynthetic machinery. Although groundbreaking, the first iteration of bioconjugate vaccines had obvious differences from those produced semi-synthetically; namely, the choice of carrier protein, which conventionally is an inactivated toxin (Berti, and Adamo 2018). Nevertheless, the relative ease at which these first generation bioconjugate vaccines were produced highlights the versatility and utility of in vivo conjugations for rapidly producing glycoconjugate vaccines against a plethora of bacteria.
The PglB N-glycosylation site consensus sequence (sequon) was determined to be D/E-X1-N-X2-S/T, where X1 and X2 can be any amino acid other than proline (Kowarik et al. 2006). Although the PglB sequon needed to be solvent exposed, it could be engineered onto many proteins, including a more conventional vaccine carrier, the Cholera toxin B subunit. With the advent of a transferable N-linking glycotag, other vaccine carriers could now be recombinantly engineered for use in bioconjugation systems. Second generation bioconjugate vaccines, consisting of polysaccharides attached to conventional vaccine carriers, were introduced not long after when Shigella O antigens were bioconjugated to a variant of the exotoxin A protein from Pseudomonas aeruginosa (EPA) (Ihssen et al. 2010). While PglB has been show to transfer long chain polysaccharides (>20 repeat units), an ideal property for glycoconjugate vaccine synthesis, it cannot transfer all polysaccharides. Specifically, PglB requires that the polysaccharide substrate contain an acetamido group in the C2 position (HexNAc) of the sugar linked directly to the lipid carrier, also known as the reducing end sugar (Wacker et al. 2006). As such, PglB has been mainly employed to develop bioconjugate vaccines for Shigella species, Staphylococcus aureus, and extraintestinal pathogenic E. coli (ExPEc), all pathogens with polysaccharides that contain HexNAc sugars at their reducing ends. However, after the crystal structure of PglB from C. lari was published, structure guided engineering was performed to alter the substrate specificity of PglB, which successfully tuned the enzyme to transfer polysaccharides with galactose at the reducing end (Ihssen et al. 2015). Polysaccharides with glucose at the reducing end are inefficiently transferred by PglB.
While the PglB generated bioconjugate vaccines against Shigella flexneri 2a (Riddle et al. 2016) and ExPEc (Huttner et al. 2017) are the most clinically advanced products, other bioconjugate vaccines against a variety of bacteria have been or are currently being explored by many academic groups (Table I). Specifically, bioconjugate vaccines against Burkholderia pseudomallei (Garcia-Quintanilla et al. 2014), E. coli O157 (Ma et al. 2014), Francisella tularensis (Cuccui et al. 2013; Marshall et al. 2018), Staphylococcus aureus (Wacker et al. 2014), and Streptococcus pneumoniae (Herbert et al. 2018; Reglinski et al. 2018) have been generated using the PglB system.
Table I.
Characteristics of bacterial oligosaccharyltransferases.
| OTase | PglB | PilO | PglL | PglS |
|---|---|---|---|---|
| Linkage Formed | N-linked | O-linked (serine) | O-linked (serine) | O-linked (serine) |
| Sequon or Glycotag | D/E-X1-N-X2-S/T | -TAWKPNYAPANAPKS | WPAAASAP* | Not determined; must contain serine 82 |
| Sequon/Glycotag Location on Fusion Proteins | Solvent exposed | C-terminal | N- or C-terminal | not determined |
| Sugar at the Reducing End of Substrate Transferred | Acetamido group at the C2 | Acetamido group at the C2 | Acetamido group at the C2 and Galactose | Acetamido group at the C2 /Galactose/Glucose |
| # of Repeat Subunits Transferred | Many (>15) | Up to 2 | Many (>15) | Many (>15) |
| Bacteria of Origin (experimentally tested) | Campylobacter, Helicobacter, Desulfovibrio | Pseudomonas, Acinetobacter, Dichelobacter | Neisseria, Vibrio, Acinetobacter, Burkholderia, Francisella | Acinetobacter |
| Bioconjugate Vaccines in Development | Shigella flexneri, Staphylococcus aureus, E. coli, Francisella tularensis, Streptococcus pneumoniae, Burkholderia pseudomallei | none | Shigella flexneri, Salmonella enterica serovar Paratyphi A | Streptococcus pneumoniae |
| OTase used for Diagnostic Development | Brucella and Enterohemorrhagic E. coli | none | none | none |
O-linking OTases and their roles in glycoconjugate vaccine production
Despite the fact that the lion’s share of academic and biotechnological development has focused on the use of PglB for bioconjugate vaccine production; the very first bacterial OTase characterized was actually an O-linking enzyme (Castric 1995). Unlike the single N-linking PglB system characterized, multiple O-linking OTases have been discovered. These enzymes do not have counterparts in eukaryotic organisms and are quite diverse, affording distinct biotechnological applications, particularly, when discussing the substrate specificity of each OTase. O-linking OTases display significant variation from their N-linking counterparts, and not just in their primary amino acid sequences, but also with respect to the apparent lack of a general glycosylation sequon. To date, three O-linking OTase systems have been characterized and are introduced based on their chronological discovery: The PilO OTase (also known as TfpO), the PglL/PglO OTase, and the most recently characterized PglS OTase, previously characterized as PglL by (Schulz et al. 2013) and as PglLComP by (Harding et al. 2015).
PilO—a type IV pilin specific OTase and its applications in glycoengineering
PilO orthologs, first characterized in Pseudomonas aeruginosa, preferentially transfer a single repeating subunit of their host strain’s O antigen to the carboxy-terminal serine residue of the type IV pilin, PilA, as a mechanism to defend against phage infection (Comer et al. 2002; DiGiandomenico et al. 2002; Harvey et al. 2018). In P. aeruginosa, PilO will promiscuously transfer a diverse set of O antigen repeat units to PilA, including the P. aeruginosa O5, the O7, and even the E. coli O157:H7 O antigen (Horzempa et al. 2006). Translational fusions containing the last 15 amino acids of the P. aeruginosa 1244 PilA sequence fused to the C-terminus of PhoA from E. coli are sufficient for PilO dependent glycosylation (Qutyan et al. 2010). PilO’s broad substrate specificity and the identification of a 15 amino acid glycosylation tag (glycotag) would seem to indicate a potential use for this OTase system in glycoconjugate vaccine production; however, for unknown reasons, PilO will not transfer long chain polysaccharides; and because of this, PilO has limited potential for glycoconjugate vaccine production. Recent advances in our understanding on the mechanisms of adaptive immunity towards glycoconjugate vaccines though has shown that some carbohydrate-protein antigens are processed to a single monosaccharide subunit on a peptide resulting in dominant CD4+ T-cell responses (Sun et al. 2019). As such, a role for PilO in future bioconjugate vaccine design may reemerge.
PglL—a general O-linking OTase employed for bioconjugate vaccine production
The second class of O-linking OTase termed PglL was first identified in Neisseria meningitidis and found to, again, glycosylate a major type IV pilin, PilE (Power et al. 2006). In Neisseria, PglL acts as a general OTase glycosylating at least 12 membrane-associated proteins with a carbohydrate that displays microheterogeneity between species and even strains (Vik et al. 2009; Borud et al. 2014). The carbohydrate can be a disaccharide containing the 2,4 diacetamido-2,4,6 trideoxyhexose (DADTH) or a glyceramido acetamido trideoxyhexose (GATDH) derivative as the reducing sugar linked to GlcNAc or a trisaccharide containing DADTH or GATDH as the reducing sugar with two galactose residues linked (Chamot-Rooke et al. 2007). Proteomics approaches have identified at least 12 proteins that are glycosylated by PglL; however, a clear O-linked PglL glycosylation sequon has been not identified. Instead, it was shown that PglL glycosylates its acceptor proteins in regions of low complexity (LCR) rich in alanine, proline and glycine (Vik et al. 2009). While many proteins are naturally glycosylated by PglL, the bulk of research has focused on PilE, the major type IV pilin in Neisseria, which is glycosylated at serine 63 (Marceau, and Nassif 1999).
When functionally transferred into E. coli, PglL will glycosylate PilE with most polysaccharides as PglL has very broad substrate specificity and naturally transfers glycans with a galactose at the reducing end as well as sugars containing an acetamido group at the C2 (Faridmoayer et al. 2007, 2008). Examples of these are the Salmonella O antigens. Unlike PilO, PglL will transfer long chain lipid-linked polysaccharides to proteins, which when combined with its extreme substrate specificity demonstrates a clear use for PglL in bioconjugate vaccine production. In spite of its naturally broader substrate specificity and ability to transfer long chain polysaccharides, PglL does not transfer lipid-linked carbohydrates that contain glucose at the reducing end.
Recently, a PglL specific glycotag (a transferable O-linked glycosylation motif) was derived from the acceptor protein PilE. The PilE glycotag, comprising 29 amino acids of the PilE protein from amino acids 45–73 (serine 63 is the site of glycosylation), was shown to be efficiently glycosylated by PglL when added to the N- or C- terminus of conventional carrier proteins like the exotoxin A of Pseudomonas aeruginosa, the tetanus toxin C fragment, or the cholera toxin B subunit (Pan et al. 2016). Further, the PglL glyctotag was reductively optimized to an 8 amino acid tag dubbed the MOOR (minimum optimum O-linked recognition) motif. Using different PglL glycotags translationally fused to the cholera toxin B subunit, bioconjugate vaccines against Shigella flexneri 2a and Salmonella enterica serovar Paratyphi A have been recombinantly produced, both of which were immunogenic and protective (Sun et al. 2018).
PglS—a recently characterized type IV pilin specific OTase encoded by Acinetobacter species
Recently, a third class of O-linking OTase was employed for bioconjugate vaccine production (Harding et al. 2019). Much like PilO and PglL, this third class of OTase, termed PglS, naturally glycosylates a pilin like protein, ComP (Porstendorfer et al. 2000; Schulz et al. 2013). A follow up study demonstrated that PglS was indeed a pilin specific OTase, likely, only glycosylating ComP as no other glycoproteins were identified using a comprehensive glycoprotein screening approach (Harding et al. 2015). Originally characterized as a PglL ortholog from the environmental bacterium Acinetobacter baylyi strain ADP1, PglS is in fact phylogenetically distinct from PglL proteins. Strains of Acinetobacter that encode for a PglS protein also encode for a PglL protein, which has been shown to act as the general OTase glycosylating at least seven membrane-associated proteins in a manner similar to Neisseria species (Iwashkiw, Seper et al. 2012). In addition, some strains of Acinetobacter also encode for PilO OTases, making Acinetobacter the only know genera of bacteria carrying genes for all three O-OTase families (PilO, PglL and PglS) (Iwashkiw, Seper et al. 2012; Harding et al. 2015).
Aside from phylogenetic differences, PglS glycosylates its cognate pilin at a unique serine site that is not conserved when compared to the site of glycosylation for PilE (the pilin target of PglL) or PilA (the pilin target for PilO), and is not contained within an LCR (Harding et al. 2019). However, the most notable difference lies in the polysaccharide substrates PglS transfers. PglS is the only known OTase, both N- or O-linking, capable of transferring polysaccharides with glucose at the reducing end. Many pathogens, like Streptococcus pneumoniae (Geno et al. 2015), Group B Streptococcus (Carboni et al. 2017), and Klebsiella pneumoniae (Pan et al. 2015), produce capsules that contain polysaccharides with glucose at the reducing and are thus potential targets for PglS dependent bioconjugate vaccine development. Indeed, PglS was used to generate a polyvalent pneumococcal bioconjugate vaccine against serotypes 8, 9 V, and 14 (all contain glucose at the reducing end) using the natural acceptor, ComP, as a carrier protein. In addition, a fragment of ComP lacking its first 28 amino acids was also able to serve as a glycotag when translationally fused to the C-terminus of exotoxin A of P. aeruginosa paving the way for incorporation of more conventional vaccine carriers in the PglS bioconjugation system (Harding et al. 2019).
Glycoengineered outer membrane vesicles as alternatives to glycoconjugate vaccines
Many Gram-negative bacterial species produce outer membrane vesicles (OMVs), which consist predominantly of outer membrane lipids, proteins and polysaccharides (Schwechheimer, and Kuehn 2015). Although their mechanisms of biogenesis are contentious, their inherent immunogenicity, due to the presence of the endotoxic lipid-A portion of lipopolysaccharide (LPS), is undeniable. LPS is one of the most prominent outer membrane components of Gram-negative bacteria and thus OMVs, making them attractive vaccine delivery vehicles with inherent adjuvant-like properties (Tan et al. 2018). While vaccines have been prepared using OMVs sourced directly from the bacterium for which the vaccine is targeted against, e.g., Bexsero—an OMV based vaccine produced from and targeting N. meningiditis (Gorringe, and Pajon 2012), we will focus our discussion on E. coli OMV glycoengineering strategies.
As a main component of OMVs, LPS has been the target of mutational engineering to not only reduce its toxicity but also introduce novel O antigen polysaccharides (OPS), which is dependent on the WaaL O antigen ligase (Raetz, and Whitfield 2002). Similarly to O-linking OTases, the WaaL O antigen ligase is an inner membrane protein with multiple transmembrane domains. In fact, both WaaL ligases and O-OTases contain protein domains from the Wzy_C superfamily and require lipid-linked carbohydrates as substrates (Power et al. 2006). Because the WaaL ligases display a highly relaxed substrate specificity towards both the lipid and carbohydrate moiety (Han et al. 2012) as well as the fact that both OPS and many CPS precursors are lipid-linked, they can interchangeably be engineered for incorporation into LPS molecules and subsequent surface exposure on OMVs. This approach has been utilized to generate diverse glycoengineered OMVs (geOMVs) in E. coli simply by introducing biosynthetic loci for different OPSs or CPSs (Valguarnera, and Feldman 2017).
Using iterations of this approach, many geOMVs have been developed as alternatives to traditional glycoconjugate vaccines and, importantly, have proven to be both immunogenic and protective. Specifically, chickens vaccinated with geOMVs expressing the C. jejuni heptasaccharide were found to have decreased C. jejuni gastrointestinal colonization and sera from mice vaccinated with geOMVs containing the serotype 14 CPS of S. pneumoniae demonstrated bactericidal killing in an opsonophagocytosis assay (Price et al. 2016), the gold standard for determining pneumococcal conjugate vaccine efficacy. Moreover, mice vaccinated with geOMVs expressing the OPS of Francisella tularensis subspecies tularensis (type A) strain Schu S4 demonstrated prolonged survival post challenge with the same strain as well as complete protection when challenged with F. tularensis subspecies holarctica (type B) live vaccine strains (Chen et al. 2016).
Additionally, non-WaaL ligase dependent mechanism for glycoengineering OMVs have been described. In one example, E. coli OMVs were engineered to display polysialic acid attached to the core saccharide of LPS, which when used as a vaccine, was able to elicit glycan specific antibodies against the polysialic acid capsule of Neisseria meningiditis serogroup B (MenB) (Valentine et al. 2016). The anti-polysialic acid antibodies were also able to enhance killing of MenB in a serum bactericidal assay, one of the best correlates of immunity for determining protective efficacy of vaccines against MenB. Moreover, incorporating polysaccharides directly onto the surface of OMVs without LPS as a substrate vessel for glycosylation has also been achieved. Specifically, OMVs decorated with poly-N-acetylglucosamine (PNAG) have been shown to induce broadly antimicrobial antibodies against both Staphylococcus aureus as well as F. tularensis subspecies holarctica (Stevenson et al. 2018). While glycoengineered OMVs hold much promise, the drawback of possible toxicity due to the presence of the LPS will need thorough evaluation.
Strategies for glycoengineered protein drugs produced using E. coli as a host
While it is inherently obvious that glycoconjugate vaccines contain both polysaccharide and protein moieties, it may be less well known that the majority of human protein drugs in preclinical development or currently approved for use are in fact glycoproteins too (Sethuraman, and Stadheim 2006). Many cellular platforms are being developed for precise glycosylation as current mammalian expression systems like Chinese hamster ovary (CHO) cells produce very heterogenous glycosylation patterns, while systems like Saccharomyces cerevisiae produce glycans that are highly mannosylated and thus differ from human glycans (Lalonde, and Durocher 2017). Further bacterial polysaccharides are quite distinct from eukaryotic glycans and usually immunogenic (Comstock, and Kasper 2006), which hinders the use of therapeutic proteins containing endogenous prokaryotic glycosylation profiles. While this is a valued trait for bioconjugate vaccine development, it does increase the complexity of glycoengineering human like glycans in a prokaryotic system; however, significant progress has been made using a variety of glycoengineering approaches. For the most part, two approaches are utilized for glycosylating therapeutic proteins in E. coli—OTase-independent (cytoplasmic) or OTase-dependent (periplasmic) glycosylation. In addition, chemoenzymatic remodeling of bacterial derived glycoproteins is a tactic for generating very precise glycosylation profiles, but this process relies on secondary in vitro methods for transglycosylation and will not be discussed in detail here.
OTase-dependent glycoprotein drug design and advances
Much like bioconjugate vaccine production, PglB is the best characterized and most utilized OTase for glycosylating therapeutic proteins in an OTase-dependent manner. A bacterial glycoprotein was decorated in vivo by PglB with a tetrasaccharide derived from Haemophilus influenzae, and fucosylated in vitro with a Helicobacter pylori fucosyltransferase to generate a LewisX glycan-containing glycoprotein (Hug et al. 2011). However, the yield was too low to be applied for commercial production. While glycosylation with bacterial glycans on therapeutic proteins may not be ideal, addition of two N-glycans on the single-chain fragment of the anti-His antibody 3D5 significantly increased solubility and resistance towards proteolytic degradation, indicating that even addition of simple N-glycans can introduce favorable biological favorable traits (Lizak et al. 2011). Subsequently, a mutated C. jejuni biosynthetic glycan cluster was engineered to express a linear hexasaccharide ((GalNAc)5-GlcNAc-), which could be transferred by PglB to a DQNAT sequon on carrier proteins. After PglB dependent transfer, the glycoprotein was purified and enzymatically trimmed to a single GlcNAc residue with an exo-alpha-N-acetylgalactosaminidase, enabling further chemoenzymatic extension with human glycans (Schwarz et al. 2010).
While trimming and chemoenzymatic steps can be used to extend glycosylation from an asparagine linked hexosamine, a more direct method for human-like glycosylation has been developed. In a first of its kind, the bottom-up synthesis of a bona fide human lipid-linked glycan (mannose3-N-acetylglucosamine2) was successfully engineered by introducing four eukaryotic glycosyltransferases (Alg13, Alg14, Alg1, and Alg2) into E. coli (Valderrama-Rincon et al. 2012). While the yields were low, the trimannosyl chitobiose glycan was transferred by PglB to variety of eukaryotic proteins containing engineered PglB sequons (DQNAT). Follow up studies using an innovative screening approach for rapid identification of PglB variants with greatly relaxed acceptor-site specificity identified PglB mutants that were able to glycosylate eukaryotic N-X-S/T sites of bovine pancreatic RNase A (Ollis et al. 2014). If fully optimized, PglB mutants combined with bottom-up glycan synthetic approaches could allow for the generation of therapeutic glycoproteins with very well defined and homogenous human glycosylation profiles.
In an alternative approach to cell-based OTase systems, cell-free systems have also been utilized to synthesize glycoproteins (Schoborg et al. 2018). While seemingly identical to OTase cell-based glycosylation in theory, this approach has the added advantage that the target protein no longer has to be targeted to the periplasm. Targeting non-periplasmic proteins (eukaryotic or prokaryotic) to the periplasm is partially responsible of the low efficiency of some examples, whereas, cell-free extracts bypass this step of in vivo OTase glycosylation. Specifically, cell-free extracts enriched in the desired lipid-linked glycans and the OTase (so far only PglB has been utilized, but the strategy could be employed for O-linking enzymes link PglL or PglS) are combined with a plasmid encoding for the acceptor protein. The acceptor protein, containing natural or engineered glycosylation sites, is translated in vitro and subsequently glycosylated in a one-pot reaction. Both prokaryotic and eukaryotic acceptor proteins, like erythropoietin, have been successfully glycosylated using the cell free system at yields comparable to other cell free protein synthesis systems (Jaroentomeechai et al. 2018).
OTase-Independent advances for glycoengineered protein drugs
OTase-independent mechanisms are reliant on dedicated cytoplasmic glycosyltransferases to sequentially add monosaccharides from nucleotide activated donors to a target protein. Two main approaches for OTase-independent glycosylation have been employed when using E. coli as a platform for therapeutic glycoprotein design: [1] the use of a novel family of prokaryotic glycosyltransferases first identified in Haemophilus influenzae (Grass et al. 2003; Gross et al. 2008; Choi et al. 2010) termed the N-glycosyltransferase (known as HMW1C in H. influenzae) or [2] the direct use of eukaryotic glycosyltransferases in E. coli.
The cytoplasmic N-glycosyltransferase naturally transfers a hexose from a nucleotide activated precursor to an asparagine residue located in an N-X-(S/T) consensus (Grass et al. 2010; Schwarz et al. 2011), which is similar to the eukaryotic acceptor site (recall that the PglB site is D/E-X1-N-X2-S/T is more stringent). NGT enzymes are inverting glycosyltransferases found in a restricted group of proteobacteria with the bulk of research focusing on HMW1C from H. influenzae and its homolog in Actinobacillus pleuropneumoniae, designated ApNGT. For glycoengineering purposes, the ApNGT system has been functionally transferred into E. coli and exploited for designer glycoprotein synthesis (Naegeli, Neupert et al. 2014). Given the similarities between the ApNGT and eukaryotic acceptor sites, ApNGT will glycosylate all three natural N-glycosylation sites (Asn51, Asn65, and Asn110) of human erythropoietin (hEPO) with a single glucose residue at each site. In addition, the constant region (Fc) of a human immunoglobulin (IgG1) antibody was also shown to be homogenously glycosylated with ApNGT at the natural occurring site or optimized site variants further demonstrating the potential utility of ApNGT for glycoengineering (Kightlinger et al. 2018). The glucosylated hEPO and Fc, while not the preferred final glycosylated versions, could serve as templates for downstream chemoenzymatic extension to make human like glycoproteins, however, this process relies on costly sugar donor substrates. While the preferred substrate for ApNGT is UDP-Glc in vivo, many nucleotide-activated hexoses or a pentose (xylose) can be transferred in vitro (Naegeli, Michaud et al. 2014); however, substituted hexoses (GlcNAc) are not compatible with the WT isoform. An engineered NGT Q469A variant, however, will transfer glucosamine (GlcN) to peptides in vitro (Song et al. 2017), which when paired with an N-acetyltransferase (GlmA) produces a single GlcNAc residue on the target Asp residue (Xu et al. 2017).
Alternatively, the NGT transferred glucose residue can serve as a priming point for polysialylation (Keys et al. 2017). In this system, the polysialyltransferase of N. meningiditis serogroup B was used to extend a di-sialyllactose, generated by combining the enzymatic activities of the glycosyltransferases ApNGT, LgtB and CstII. Using this system, a Designed Ankyrin Repeat Protein (DARPin) was polysialylated with either short, medium or long chain α2,8-linked sialic acid residues, which greatly increased the effective molecular size of the glycoconjugate and theoretically could serve to improve efficacy through reduced kidney diafiltration or physical shielding to slow immune recognition.
While advances for glycoengineering therapeutic N-linked glycoproteins in E. coli has thoroughly been progressing, O-linked glycoengineering is less developed. As described above, O-linking OTases like PglL and PglS have been employed for bioconjugate vaccine development, but they have not been used for therapeutic O-glycoprotein production. This is mainly due to fact that the recognition sequons for O-linking OTases are much larger when compared to those for PglB and NGT. While the eight amino acid MOOR glycotag of PglL may hold some promise, it is still large when compared to the N-X-S/T sequon recognized by NGTs and eukaryotic machinery. As such, engineering an eight amino acid tag into therapeutic proteins may present new obstacles that interfere with protein production and/or function. Nevertheless, O-linked glycoprotein production in the cytoplasm of E. coli is possible.
To this end, directly expressing eukaryotic glycosyltransferases in the cytoplasm of E. coli has shown the most promise. Early examples include glycoengineering mucin like O-glycosylation into E. coli by co-expressing the human GalNAc-T2 transferase, a UDP-GlcNAc to UDP-GalNAc epimerase (gne orthologs), and O-glycosylation acceptor proteins like Muc1 or putative O-glycosylation peptide tags fused to antibody Fab fragments (Henderson et al. 2011). Proteins modified with the Tn-Antigen (GalNAcα) were then subsequently used as primers for PEGylation as polyethylene glycol modification is an industry standard to improve serum half-life. However, alternative strategies to increase serum half-life are sought as PEGylated proteins may be immunogenic. Further, non-metabolized PEG may accumulate in tissues. Recently, a glycoengineered strain of E. coli was developed for the production and transfer of an authentic core-1 O-glycan structure, the T-antigen (Gal-β1,3-GalNAcα), to two human therapeutic proteins modified with GB1 tags, human interferon α-2b and human growth hormone (Du et al. 2018). Impressively, 74% of the modified IFNa2b was glycosylated with the T-Ag. As is the case for N-glycan engineering, the T-Ag modified proteins could serve as templates for chemoenzymatic extension; however, more elaborately engineered E. coli strains could encode sialyltransferases for the generations of polysialylated T-Ag containing glycoproteins produced completely within the cytoplasm of E. coli.
Glycoengineering glycoproteins for rapid diagnostics of infectious diseases
Early diagnosis of bacterial infectious diseases is crucial for their treatment. Current diagnostic methods include bacterial culture, direct immunofluorescence, immunoenzymatic assays, PCR-based methods, histology and immunohistochemistry (Caliendo et al. 2013). Although valuable, these methods often require specialized equipment, are slow, and may produce false positives. For bacterial agents that mount an adaptive immune response, these limitations can be circumvented by employing engineered glycoproteins for the detection of antibodies directly in the fluids of infected individuals (Iwashkiw, Fentabil et al. 2012). The first examples for employing glycoengineered proteins for diagnostic purposes has been characterized for detecting hemolytic uremic syndrome (HUS) and brucellosis.
Brucellosis is the most common bacterial zoonosis with over half a million new cases annually. Three Brucella species, B. abortus, B. melitensis, and B. suis are the common species that cause human brucellosis (Pappas et al. 2005). They can also infect domestic livestock, causing miscarriages and sterility leading to significant economic loss. To rapidly detect animals previously or actively infected with Brucella species, the C. jejuni protein AcrA was glycoengineered with the Yersinia enterocolitica O3 antigen, which is identical to the O antigen of B. abortus employing the N-OTase PglB. The recombinant glycoprotein was shown to efficiently diagnose human, bovine and porcine brucellosis (Iwashkiw, Fentabil et al. 2012; Ciocchini et al. 2014; Cortina et al. 2016). HUS is a systemic illness clinically defined by hemolytic anemia, thrombocytopenia and acute renal failure. HUS is caused by Shiga toxin-producing Escherichia coli (STEC) bacteria. Most STEC strains causing HUS belong to the O157 and the “big six” (O26, O11, O103, O121, O45 and O145) serotypes (Melli et al. 2015). By employing PglB as the OTase, Melli et al. again engineered the Campylobacter protein AcrA with the O157, O145 and O121 O antigens. Together, these serotypes account for approximately 80% of the E. coli strains causing HUS. The neo-glycoproteins are easily purified, and because they do not contain the lipid A-core portion of the LPS common to all the E. coli strains, reduce the rate of false positives. Using an enzyme-linked immunosorbent assay (ELISA), immunoglobulins (IgM) reactive to the polysaccharides were readily detected 1 day after the start of diarrhea. Although both the glycoengineered diagnostics for brucellosis and HUS are based on ELISA, current efforts are directed to adapting these tests for lateral flow immunoassays, which may enable rapid, point-of-care or field diagnosis of these bacterial infections.
Conclusions and Future Perspectives
Bacterial glycoengineering, while steadily advancing, still has many hurdles to cross before it equals or surpasses conventional approaches used for glycoprotein production like chemical conjugation for glycoconjugate vaccine manufacturing or the use of CHO cells for glycosylated protein drug synthesis. One of the major technical hurdles to overcome will be novel methods for increasing the yields of the fully glycosylated variants of the targeted therapeutic (vaccine or protein drug), such that these processes are in line with traditional low-cost features that make E. coli an attractive model organism. Nevertheless, optimism is high that the field of bacterial glycoengineering will continue to progress given the relative ease at which E. coli can be both genetically manipulated and the previously established processes implemented for large volumetric production of bacterially fermented products. As such, recruiting more academic and industrial partners/resources could greatly accelerate the development of bacterial glycoengineering as a more viable pharmaceutical platform given that this field is quite new when compared to those traditional platforms employed for decades to produce recombinant glycoproteins.
Funding
This work was supported by a National Institute of Allergy and Infectious Disease Small Business Technology Transfer grant (R41AI136633-01).
Conflict of interest statement
CH and MF have a financial interest in VaxNewMo, which is developing bioconjugate vaccines employing PglS.
References
- Berti F, Adamo R. 2018. Antimicrobial glycoconjugate vaccines: an overview of classic and modern approaches for protein modification. Chem Soc Rev. 47:9015–9025. [DOI] [PubMed] [Google Scholar]
- Borud B, Anonsen JH, Viburiene R, Cohen EH, Samuelsen AB, Koomey M. 2014. Extended glycan diversity in a bacterial protein glycosylation system linked to allelic polymorphisms and minimal genetic alterations in a glycosyltransferase gene. Mol Microbiol. 94:688–699. [DOI] [PubMed] [Google Scholar]
- Caliendo AM, Gilbert DN, Ginocchio CC, Hanson KE, May L, Quinn TC, Tenover FC, Alland D, Blaschke AJ, Bonomo RA et al. 2013. Better tests, better care: improved diagnostics for infectious diseases. Clin Infect Dis. 57(Suppl 3):S139–S170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carboni F, Adamo R, Fabbrini M, De Ricco R, Cattaneo V, Brogioni B, Veggi D, Pinto V, Passalacqua I, Oldrini D et al. 2017. Structure of a protective epitope of group B Streptococcus type III capsular polysaccharide. Proc Natl Acad Sci USA. 114:5017–5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castric P. 1995. pilO, a gene required for glycosylation of Pseudomonas aeruginosa 1244 pilin. Microbiology. 141(Pt 5):1247–1254. [DOI] [PubMed] [Google Scholar]
- Chamot-Rooke J, Rousseau B, Lanternier F, Mikaty G, Mairey E, Malosse C, Bouchoux G, Pelicic V, Camoin L, Nassif X et al. 2007. Alternative Neisseria spp. type IV pilin glycosylation with a glyceramido acetamido trideoxyhexose residue. Proc Natl Acad Sci USA. 104:14783–14788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Valentine JL, Huang CJ, Endicott CE, Moeller TD, Rasmussen JA, Fletcher JR, Boll JM, Rosenthal JA, Dobruchowska J et al. 2016. Outer membrane vesicles displaying engineered glycotopes elicit protective antibodies. Proc Natl Acad Sci USA. 113:E3609–E3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KJ, Grass S, Paek S, St Geme JW 3rd, Yeo HJ. 2010. The Actinobacillus pleuropneumoniae HMW1C-like glycosyltransferase mediates N-linked glycosylation of the Haemophilus influenzae HMW1 adhesin. PLoS One. 5:e15888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciocchini AE, Serantes DA, Melli LJ, Guidolin LS, Iwashkiw JA, Elena S, Franco C, Nicola AM, Feldman MF, Comerci DJ et al. 2014. A bacterial engineered glycoprotein as a novel antigen for diagnosis of bovine brucellosis. Vet Microbiol. 172:455–465. [DOI] [PubMed] [Google Scholar]
- Comer JE, Marshall MA, Blanch VJ, Deal CD, Castric P. 2002. Identification of the Pseudomonas aeruginosa 1244 pilin glycosylation site. Infect Immun. 70:2837–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comstock LE, Kasper DL. 2006. Bacterial glycans: key mediators of diverse host immune responses. Cell. 126:847–850. [DOI] [PubMed] [Google Scholar]
- Cortina ME, Balzano RE, Rey Serantes DA, Caillava AJ, Elena S, Ferreira AC, Nicola AM, Ugalde JE, Comerci DJ, Ciocchini AE. 2016. A Bacterial Glycoengineered Antigen for Improved Serodiagnosis of Porcine Brucellosis. J Clin Microbiol. 54:1448–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuccui J, Thomas RM, Moule MG, D’Elia RV, Laws TR, Mills DC, Williamson D, Atkins TP, Prior JL, Wren BW. 2013. Exploitation of bacterial N-linked glycosylation to develop a novel recombinant glycoconjugate vaccine against Francisella tularensis. Open Biol. 3:130002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthbertson L, Kos V, Whitfield C. 2010. ABC transporters involved in export of cell surface glycoconjugates. Microbiol Mol Biol Rev. 74:341–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gregorio E, Rappuoli R. 2014. From empiricism to rational design: a personal perspective of the evolution of vaccine development. Nat Rev Immunol. 14:505–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiandomenico A, Matewish MJ, Bisaillon A, Stehle JR, Lam JS, Castric P. 2002. Glycosylation of Pseudomonas aeruginosa 1244 pilin: glycan substrate specificity. Mol Microbiol. 46:519–530. [DOI] [PubMed] [Google Scholar]
- Du T, Buenbrazo N, Kell L, Rahmani S, Sim L, Withers SG, DeFrees S, Wakarchuk W. 2018. A Bacterial Expression Platform for Production of Therapeutic Proteins Containing Human-like O-Linked Glycans. Cell Chem Biol. 26(2):203–212.e5. [DOI] [PubMed] [Google Scholar]
- Faridmoayer A, Fentabil MA, Haurat MF, Yi W, Woodward R, Wang PG, Feldman MF. 2008. Extreme substrate promiscuity of the Neisseria oligosaccharyl transferase involved in protein O-glycosylation. J Biol Chem. 283:34596–34604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faridmoayer A, Fentabil MA, Mills DC, Klassen JS, Feldman MF. 2007. Functional characterization of bacterial oligosaccharyltransferases involved in O-linked protein glycosylation. J Bacteriol. 189:8088–8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman MF, Wacker M, Hernandez M, Hitchen PG, Marolda CL, Kowarik M, Morris HR, Dell A, Valvano MA, Aebi M. 2005. Engineering N-linked protein glycosylation with diverse O antigen lipopolysaccharide structures in Escherichia coli. Proc Natl Acad Sci USA. 102:3016–3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch CE. 2009. Preparation of bacterial polysaccharide-protein conjugates: analytical and manufacturing challenges. Vaccine. 27:6468–6470. [DOI] [PubMed] [Google Scholar]
- Garcia-Quintanilla F, Iwashkiw JA, Price NL, Stratilo C, Feldman MF. 2014. Production of a recombinant vaccine candidate against Burkholderia pseudomallei exploiting the bacterial N-glycosylation machinery. Front Microbiol. 5:381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geno KA, Gilbert GL, Song JY, Skovsted IC, Klugman KP, Jones C, Konradsen HB, Nahm MH. 2015. Pneumococcal Capsules and Their Types: Past, Present, and Future. Clin Microbiol Rev. 28:871–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorringe AR, Pajon R. 2012. Bexsero: a multicomponent vaccine for prevention of meningococcal disease. Hum Vaccin Immunother. 8:174–183. [DOI] [PubMed] [Google Scholar]
- Grass S, Buscher AZ, Swords WE, Apicella MA, Barenkamp SJ, Ozchlewski N, St Geme JW 3rd. 2003. The Haemophilus influenzae HMW1 adhesin is glycosylated in a process that requires HMW1C and phosphoglucomutase, an enzyme involved in lipooligosaccharide biosynthesis. Mol Microbiol. 48:737–751. [DOI] [PubMed] [Google Scholar]
- Grass S, Lichti CF, Townsend RR, Gross J, St Geme JW 3rd. 2010. The Haemophilus influenzae HMW1C protein is a glycosyltransferase that transfers hexose residues to asparagine sites in the HMW1 adhesin. PLoS Pathog. 6:e1000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J, Grass S, Davis AE, Gilmore-Erdmann P, Townsend RR, St Geme JW 3rd. 2008. The Haemophilus influenzae HMW1 adhesin is a glycoprotein with an unusual N-linked carbohydrate modification. J Biol Chem. 283:26010–26015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W, Wu B, Li L, Zhao G, Woodward R, Pettit N, Cai L, Thon V, Wang PG. 2012. Defining function of lipopolysaccharide O-antigen ligase WaaL using chemoenzymatically synthesized substrates. J Biol Chem. 287:5357–5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding CM, Nasr MA, Kinsella RL, Scott NE, Foster LJ, Weber BS, Fiester SE, Actis LA, Tracy EN, Munson RS Jr. et al. 2015. Acinetobacter strains carry two functional oligosaccharyltransferases, one devoted exclusively to type IV pilin, and the other one dedicated to O-glycosylation of multiple proteins. Mol Microbiol. 96:1023–1041. [DOI] [PubMed] [Google Scholar]
- Harding CM, Nasr MA, Scott NE, Goyette-Desjardins G, Nothaft H, Mayer AE, Chavez SM, Huynh JP, Kinsella RL, Szymanski CM et al. 2019. A platform for glycoengineering a polyvalent pneumococcal bioconjugate vaccine using E. coli as a host. Nat Commun. 10:891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey H, Bondy-Denomy J, Marquis H, Sztanko KM, Davidson AR, Burrows LL. 2018. Pseudomonas aeruginosa defends against phages through type IV pilus glycosylation. Nat Microbiol. 3:47–52. [DOI] [PubMed] [Google Scholar]
- Henderson GE, Isett KD, Gerngross TU. 2011. Site-specific modification of recombinant proteins: a novel platform for modifying glycoproteins expressed in E. coli. Bioconjug Chem. 22:903–912. [DOI] [PubMed] [Google Scholar]
- Herbert JA, Kay EJ, Faustini SE, Richter A, Abouelhadid S, Cuccui J, Wren B, Mitchell TJ. 2018. Production and efficacy of a low-cost recombinant pneumococcal protein polysaccharide conjugate vaccine. Vaccine. 36:3809–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horzempa J, Dean CR, Goldberg JB, Castric P. 2006. Pseudomonas aeruginosa 1244 pilin glycosylation: glycan substrate recognition. J Bacteriol. 188:4244–4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug I, Feldman MF. 2011. Analogies and homologies in lipopolysaccharide and glycoprotein biosynthesis in bacteria. Glycobiology. 21:138–151. [DOI] [PubMed] [Google Scholar]
- Hug I, Zheng B, Reiz B, Whittal RM, Fentabil MA, Klassen JS, Feldman MF. 2011. Exploiting bacterial glycosylation machineries for the synthesis of a Lewis antigen-containing glycoprotein. J Biol Chem. 286:37887–37894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttner A, Hatz C, van den Dobbelsteen G, Abbanat D, Hornacek A, Frolich R, Dreyer AM, Martin P, Davies T, Fae K et al. 2017. Safety, immunogenicity, and preliminary clinical efficacy of a vaccine against extraintestinal pathogenic Escherichia coli in women with a history of recurrent urinary tract infection: a randomised, single-blind, placebo-controlled phase 1b trial. Lancet Infect Dis. 17:528–537. [DOI] [PubMed] [Google Scholar]
- Ihssen J, Haas J, Kowarik M, Wiesli L, Wacker M, Schwede T, Thony-Meyer L. 2015. Increased efficiency of Campylobacter jejuni N-oligosaccharyltransferase PglB by structure-guided engineering. Open Biol. 5:140227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihssen J, Kowarik M, Dilettoso S, Tanner C, Wacker M, Thony-Meyer L. 2010. Production of glycoprotein vaccines in Escherichia coli. Microb Cell Fact. 9:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwashkiw JA, Fentabil MA, Faridmoayer A, Mills DC, Peppler M, Czibener C, Ciocchini AE, Comerci DJ, Ugalde JE, Feldman MF. 2012. Exploiting the Campylobacter jejuni protein glycosylation system for glycoengineering vaccines and diagnostic tools directed against brucellosis. Microb Cell Fact. 11:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwashkiw JA, Seper A, Weber BS, Scott NE, Vinogradov E, Stratilo C, Reiz B, Cordwell SJ, Whittal R, Schild S et al. 2012. Identification of a general O-linked protein glycosylation system in Acinetobacter baumannii and its role in virulence and biofilm formation. PLoS Pathog. 8:e1002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwashkiw JA, Vozza NF, Kinsella RL, Feldman MF. 2013. Pour some sugar on it: the expanding world of bacterial protein O-linked glycosylation. Mol Microbiol. 89:14–28. [DOI] [PubMed] [Google Scholar]
- Jaroentomeechai T, Stark JC, Natarajan A, Glasscock CJ, Yates LE, Hsu KJ, Mrksich M, Jewett MC, DeLisa MP. 2018. Single-pot glycoprotein biosynthesis using a cell-free transcription-translation system enriched with glycosylation machinery. Nat Commun. 9:2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keys TG, Wetter M, Hang I, Rutschmann C, Russo S, Mally M, Steffen M, Zuppiger M, Muller F, Schneider J et al. 2017. A biosynthetic route for polysialylating proteins in Escherichia coli. Metab Eng. 44:293–301. [DOI] [PubMed] [Google Scholar]
- Kightlinger W, Lin L, Rosztoczy M, Li W, DeLisa MP, Mrksich M, Jewett MC. 2018. Design of glycosylation sites by rapid synthesis and analysis of glycosyltransferases. Nat Chem Biol. 14:627–635. [DOI] [PubMed] [Google Scholar]
- Kowarik M, Young NM, Numao S, Schulz BL, Hug I, Callewaert N, Mills DC, Watson DC, Hernandez M, Kelly JF et al. 2006. Definition of the bacterial N-glycosylation site consensus sequence. EMBO J. 25:1957–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde ME, Durocher Y. 2017. Therapeutic glycoprotein production in mammalian cells. J Biotechnol. 251:128–140. [DOI] [PubMed] [Google Scholar]
- Lizak C, Fan YY, Weber TC, Aebi M. 2011. N-Linked glycosylation of antibody fragments in Escherichia coli. Bioconjug Chem. 22:488–496. [DOI] [PubMed] [Google Scholar]
- Ma Z, Zhang H, Shang W, Zhu F, Han W, Zhao X, Han D, Wang PG, Chen M. 2014. Glycoconjugate vaccine containing Escherichia coli O157:H7 O-antigen linked with maltose-binding protein elicits humoral and cellular responses. PLoS One. 9:e105215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marceau M, Nassif X. 1999. Role of glycosylation at Ser63 in production of soluble pilin in pathogenic Neisseria. J Bacteriol. 181:656–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall LE, Nelson M, Davies CH, Whelan AO, Jenner DC, Moule MG, Denman C, Cuccui J, Atkins TP, Wren BW et al. 2018. An O-Antigen Glycoconjugate Vaccine Produced Using Protein Glycan Coupling Technology Is Protective in an Inhalational Rat Model of Tularemia. J Immunol Res. 2018:8087916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melli LJ, Ciocchini AE, Caillava AJ, Vozza N, Chinen I, Rivas M, Feldman MF, Ugalde JE, Comerci DJ. 2015. Serogroup-specific bacterial engineered glycoproteins as novel antigenic targets for diagnosis of shiga toxin-producing-escherichia coli-associated hemolytic-uremic syndrome. J Clin Microbiol. 53:528–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naegeli A, Michaud G, Schubert M, Lin CW, Lizak C, Darbre T, Reymond JL, Aebi M. 2014. Substrate specificity of cytoplasmic N-glycosyltransferase. J Biol Chem. 289:24521–24532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naegeli A, Neupert C, Fan YY, Lin CW, Poljak K, Papini AM, Schwarz F, Aebi M. 2014. Molecular analysis of an alternative N-glycosylation machinery by functional transfer from Actinobacillus pleuropneumoniae to Escherichia coli. J Biol Chem. 289:2170–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothaft H, Szymanski CM. 2010. Protein glycosylation in bacteria: sweeter than ever. Nat Rev Microbiol. 8:765–778. [DOI] [PubMed] [Google Scholar]
- Ollis AA, Zhang S, Fisher AC, DeLisa MP. 2014. Engineered oligosaccharyltransferases with greatly relaxed acceptor-site specificity. Nat Chem Biol. 10:816–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan YJ, Lin TL, Chen CT, Chen YY, Hsieh PF, Hsu CR, Wu MC, Wang JT. 2015. Genetic analysis of capsular polysaccharide synthesis gene clusters in 79 capsular types of Klebsiella spp. Sci Rep. 5:15573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C, Sun P, Liu B, Liang H, Peng Z, Dong Y, Wang D, Liu X, Wang B, Zeng M et al. 2016. Biosynthesis of Conjugate Vaccines Using an O-Linked Glycosylation System. MBio. 7:e00443-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas G, Akritidis N, Bosilkovski M, Tsianos E. 2005. Brucellosis. N Engl J Med. 352:2325–2336. [DOI] [PubMed] [Google Scholar]
- Porstendorfer D, Gohl O, Mayer F, Averhoff B. 2000. ComP, a pilin-like protein essential for natural competence in Acinetobacter sp. Strain BD413: regulation, modification, and cellular localization. J Bacteriol. 182:3673–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power PM, Seib KL, Jennings MP. 2006. Pilin glycosylation in Neisseria meningitidis occurs by a similar pathway to wzy-dependent O-antigen biosynthesis in Escherichia coli. Biochem Biophys Res Commun. 347:904–908. [DOI] [PubMed] [Google Scholar]
- Price NL, Goyette-Desjardins G, Nothaft H, Valguarnera E, Szymanski CM, Segura M, Feldman MF. 2016. Glycoengineered Outer Membrane Vesicles: A Novel Platform for Bacterial Vaccines. Sci Rep. 6:24931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qutyan M, Henkel M, Horzempa J, Quinn M, Castric P. 2010. Glycosylation of pilin and nonpilin protein constructs by Pseudomonas aeruginosa 1244. J Bacteriol. 192:5972–5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz CR, Whitfield C. 2002. Lipopolysaccharide endotoxins. Annu Rev Biochem. 71:635–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappuoli R, De Gregorio E, Costantino P. 2019. On the mechanisms of conjugate vaccines. Proc Natl Acad Sci USA. 116:14–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reglinski M, Ercoli G, Plumptre C, Kay E, Petersen FC, Paton JC, Wren BW, Brown JS. 2018. A recombinant conjugated pneumococcal vaccine that protects against murine infections with a similar efficacy to Prevnar-13. NPJ Vaccines. 3:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle MS, Kaminski RW, Paolo CD, Porter CK, Gutierrez RL, Clarkson KA, Weerts HE, Duplessis C, Castellano A, Alaimo C et al. 2016. Safety and Immunogenicity of a Candidate Bioconjugate Vaccine against Shigella flexneri 2a Administered to Healthy Adults: a Single-Blind, Randomized Phase I Study. Clin Vaccine Immunol. 23:908–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer C, Messner P. 2017. Emerging facets of prokaryotic glycosylation. FEMS Microbiol Rev. 41:49–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoborg JA, Hershewe JM, Stark JC, Kightlinger W, Kath JE, Jaroentomeechai T, Natarajan A, DeLisa MP, Jewett MC. 2018. A cell-free platform for rapid synthesis and testing of active oligosaccharyltransferases. Biotechnol Bioeng. 115:739–750. [DOI] [PubMed] [Google Scholar]
- Schulz BL, Jen FE, Power PM, Jones CE, Fox KL, Ku SC, Blanchfield JT, Jennings MP. 2013. Identification of bacterial protein O-oligosaccharyltransferases and their glycoprotein substrates. PLoS One. 8:e62768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz F, Fan YY, Schubert M, Aebi M. 2011. Cytoplasmic N-glycosyltransferase of Actinobacillus pleuropneumoniae is an inverting enzyme and recognizes the NX(S/T) consensus sequence. J Biol Chem. 286:35267–35274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz F, Huang W, Li C, Schulz BL, Lizak C, Palumbo A, Numao S, Neri D, Aebi M, Wang LX. 2010. A combined method for producing homogeneous glycoproteins with eukaryotic N-glycosylation. Nat Chem Biol. 6:264–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C, Kuehn MJ. 2015. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol. 13:605–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott NE, Marzook NB, Cain JA, Solis N, Thaysen-Andersen M, Djordjevic SP, Packer NH, Larsen MR, Cordwell SJ. 2014. Comparative proteomics and glycoproteomics reveal increased N-linked glycosylation and relaxed sequon specificity in Campylobacter jejuni NCTC11168 O. J Proteome Res. 13:5136–5150. [DOI] [PubMed] [Google Scholar]
- Scott NE, Parker BL, Connolly AM, Paulech J, Edwards AV, Crossett B, Falconer L, Kolarich D, Djordjevic SP, Hojrup P et al. 2011. Simultaneous glycan-peptide characterization using hydrophilic interaction chromatography and parallel fragmentation by CID, higher energy collisional dissociation, and electron transfer dissociation MS applied to the N-linked glycoproteome of Campylobacter jejuni. Mol Cell Proteomics. 10:M000031–MCP000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethuraman N, Stadheim TA. 2006. Challenges in therapeutic glycoprotein production. Curr Opin Biotechnol. 17:341–346. [DOI] [PubMed] [Google Scholar]
- Song Q, Wu Z, Fan Y, Song W, Zhang P, Wang L, Wang F, Xu Y, Wang PG, Cheng J. 2017. Production of homogeneous glycoprotein with multisite modifications by an engineered N-glycosyltransferase mutant. J Biol Chem. 292:8856–8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson TC, Cywes-Bentley C, Moeller TD, Weyant KB, Putnam D, Chang YF, Jones BD, Pier GB, DeLisa MP. 2018. Immunization with outer membrane vesicles displaying conserved surface polysaccharide antigen elicits broadly antimicrobial antibodies. Proc Natl Acad Sci USA. 115:E3106–E3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P, Pan C, Zeng M, Liu B, Liang H, Wang D, Liu X, Wang B, Lyu Y, Wu J et al. 2018. Design and production of conjugate vaccines against S. Paratyphi A using an O-linked glycosylation system in vivo. NPJ Vaccines. 3:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Stefanetti G, Berti F, Kasper DL. 2019. Polysaccharide structure dictates mechanism of adaptive immune response to glycoconjugate vaccines. Proc Natl Acad Sci USA. 116:193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski CM, Burr DH, Guerry P. 2002. Campylobacter protein glycosylation affects host cell interactions. Infect Immun. 70:2242–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski CM, Yao R, Ewing CP, Trust TJ, Guerry P. 1999. Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol Microbiol. 32:1022–1030. [DOI] [PubMed] [Google Scholar]
- Tan K, Li R, Huang X, Liu Q. 2018. Outer Membrane Vesicles: Current Status and Future Direction of These Novel Vaccine Adjuvants. Front Microbiol. 9:783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terra VS, Mills DC, Yates LE, Abouelhadid S, Cuccui J, Wren BW. 2012. Recent developments in bacterial protein glycan coupling technology and glycoconjugate vaccine design. J Med Microbiol. 61:919–926. [DOI] [PubMed] [Google Scholar]
- Valderrama-Rincon JD, Fisher AC, Merritt JH, Fan YY, Reading CA, Chhiba K, Heiss C, Azadi P, Aebi M, DeLisa MP. 2012. An engineered eukaryotic protein glycosylation pathway in Escherichia coli. Nat Chem Biol. 8:434–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine JL, Chen L, Perregaux EC, Weyant KB, Rosenthal JA, Heiss C, Azadi P, Fisher AC, Putnam D, Moe GR et al. 2016. Immunization with Outer Membrane Vesicles Displaying Designer Glycotopes Yields Class-Switched, Glycan-Specific Antibodies. Cell Chem Biol. 23:655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valguarnera E, Feldman MF. 2017. Glycoengineered Outer Membrane Vesicles as a Platform for Vaccine Development. Methods Enzymol. 597:285–310. [DOI] [PubMed] [Google Scholar]
- Valvano MA. 2003. Export of O-specific lipopolysaccharide. Front Biosci. 8:s452–s471. [DOI] [PubMed] [Google Scholar]
- Vik A, Aas FE, Anonsen JH, Bilsborough S, Schneider A, Egge-Jacobsen W, Koomey M. 2009. Broad spectrum O-linked protein glycosylation in the human pathogen Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 106:4447–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker M, Feldman MF, Callewaert N, Kowarik M, Clarke BR, Pohl NL, Hernandez M, Vines ED, Valvano MA, Whitfield C et al. 2006. Substrate specificity of bacterial oligosaccharyltransferase suggests a common transfer mechanism for the bacterial and eukaryotic systems. Proc Natl Acad Sci USA. 103:7088–7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker M, Linton D, Hitchen PG, Nita-Lazar M, Haslam SM, North SJ, Panico M, Morris HR, Dell A, Wren BW et al. 2002. N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science. 298:1790–1793. [DOI] [PubMed] [Google Scholar]
- Wacker M, Wang L, Kowarik M, Dowd M, Lipowsky G, Faridmoayer A, Shields K, Park S, Alaimo C, Kelley KA et al. 2014. Prevention of Staphylococcus aureus infections by glycoprotein vaccines synthesized in Escherichia coli. J Infect Dis. 209:1551–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield C. 2006. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem. 75:39–68. [DOI] [PubMed] [Google Scholar]
- Xu Y, Wu Z, Zhang P, Zhu H, Zhu H, Song Q, Wang L, Wang F, Wang PG, Cheng J. 2017. A novel enzymatic method for synthesis of glycopeptides carrying natural eukaryotic N-glycans. Chem Commun (Camb). 53:9075–9077. [DOI] [PubMed] [Google Scholar]
- Yates LE, Natarajan A, Li M, Hale ME, Mills DC, DeLisa MP. 2019. Glyco-recoded Escherichia coli: Recombineering-based genome editing of native polysaccharide biosynthesis gene clusters. Metab Eng. 53:59–68. [DOI] [PubMed] [Google Scholar]