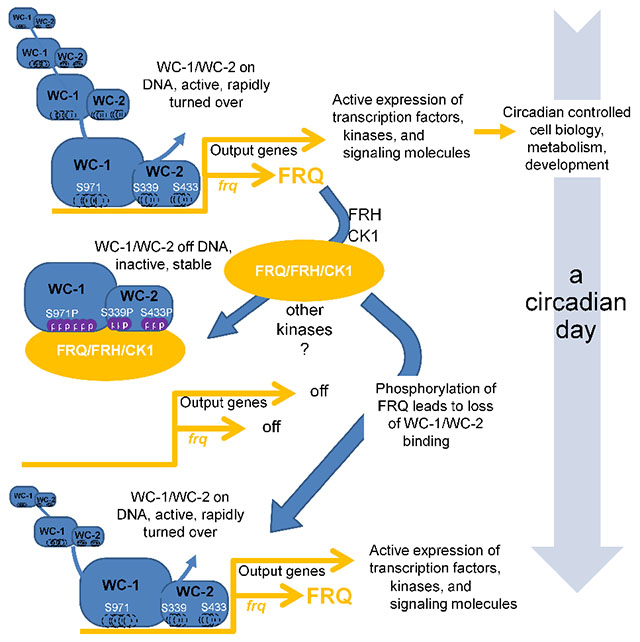
Summary
In the negative feedback loop driving fungal and animal circadian oscillators, negative elements (FRQ, PERs, CRYs) are understood to inhibit their own expression in part by promoting the phosphorylation of their heterodimeric transcriptional activators (e.g. WC-1/WC-2 (WCC), BMAL1/CLOCK). However, correlations between heterodimer activity and phosphorylation are weak, contradictions exist, and mechanistic details are almost wholly lacking. We report mapping of 80 phosphosites on WC-1 and 15 on WC-2, and elucidation of the time-of-day-specific code, requiring both a group of phosphoevents on WC-1 and two distinct clusters on WC-2, that governs circadian repression, leading to feedback loop closure. Combinatorial control via phosphorylation also governs rhythmic WCC binding to the promoters of clock-controlled genes mediating the essential first step in circadian output, a group encoding both transcription factors and signaling proteins. These data provide a basic mechanistic understanding for fundamental events underlying circadian negative feedback and output, key aspects of circadian biology.
Keywords: phosphorylation, WC-1, WC-2, FRQ, frq transcription, feedback loop, C-box, DNA binding, clock-controlled genes (ccgs)
Graphical Abstract
eTOC:
In fungal and animal circadian oscillators, phosphorylation of heterodimeric transcriptional activators (e.g. WC-1/WC-2, BMAL1/CLOCK) is associated with their cyclical inhibition, leading to rhythmicity. Wang et al. found and explored 95 phosphorylation sites on WC-1/WC-2, revealing that distinct clusters on both proteins must be modified to close the feedback loop. FRQ, whose expression is promoted by WC-1/WC-2, in turn promotes these essential phosphorylations.
Introduction
Circadian clocks regulate a wide variety of physiological and molecular events in eukaryotes and certain prokaryotes, and in fungi and animals, the core of the oscillator is a transcription/translation negative feedback loop. Circadian transcription activation complexes, such as BMAL1/CLOCK in mammals and WCC in Neurospora, are heterodimers in which the partners interact by PAS domains. WCC exemplifies this, consisting of White Collar-1 (WC-1) and White Collar-2 (WC-2). WC-1 contains a transactivation domain, a light-sensing LOV domain (dispensable for clock function), two classic Per-Arnt-Sim (PAS) domains, and both DBD and zinc finger (ZnF) domains required for DNA binding; WC-2 has PAS and ZnF DNA binding domains (Linden et al., 1999; Wang et al., 2015). WCC serves as the transcriptional activator for the pacemaker gene frequency (frq) by binding to either the Clock-box (C-box) mediating circadian function in the dark or the Proximal Light-Response Element (PLRE) mediating frq light-induction. The frq gene product, FRQ, acts with FRH (FRQ-Interacting RNA Helicase) and Casein kinase 1 (the FFC complex) leading to repression of WCC activity and thereby closing the positive arm of the circadian feedback loop (Cha et al., 2015; Dunlap, 1999).
Protein phosphorylation has been linked to regulation of DNA binding as well as to protein stability, protein-protein interaction, enzymatic activity, and sub-cellular localization, all of which have been invoked in the control of circadian systems (e.g. Lipton et al., 2015; Luciano et al., 2018; Narasimamurthy et al., 2018; Robles et al., 2017). In Neurospora, FRQ is tightly controlled by over 100 time-of-day specific phosphorylations (Baker et al., 2009; Garceau et al., 1997; Tang et al., 2009) that determine its activities and binding partners and eventually lead to its inactivation (Larrondo et al., 2015). The WCC also undergoes extensive phosphorylation (Linden et al., 1999), although conflicting observations have been made in relating WCC activity to its phosphorylation status. Work focusing on either WC-1 (He et al., 2005) or WC-2 (Schafmeier et al., 2005) showed that overall phosphorylation status negatively correlated with DNA binding and suggested a model in which phosphorylation might directly effect repression; however, in knockouts of the HAD-domain family phosphatase CSP-6, WCC is always hyperphosphorylated but strains still show sustained rhythms with normal circadian period lengths (Zhou et al., 2018). Some WC-1 and WC-2 phosphosite mutations affect rhythmicity (e.g. He et al., 2006; Sancar et al., 2009), but strains that cannot be phosphorylated at these residues still show clear overt rhythms (Wang et al., 2015). A mutant that impacts FRQ/FRH association with the WCC results in WCC hyperphosphorylation, high activity, and low stability (Shi et al., 2010), compatible with a “black-widow” model for transcriptional activators that links activity with hyperphosphorylation and degradation (Tansey, 2001). Comparable studies in mammalian clocks have similarly identified a variety of phosphorylated residues (see Discussion), but there is no unifying widely accepted mechanistic explanation for how these modifications effect closure of the feedback loop. Despite hints, the exact role of phosphorylation in establishing repression of circadian heterodimeric activators remains elusive.
To understand the circadian feedback loop at a mechanistic level, it is necessary to define the posttranslational modifications (PTMs) of a circadian heterodimeric activator such as the WCC, and to determine how they contribute to repression of the heterodimer’s transcriptional activity. To this end, we mapped phosphosites on the WCC and then functionally tested all PTMs on WC-1 and on WC-2 to describe the combinatorial code essential for rhythmicity. The WCC can be phosphorylated at >95 residues and, surprisingly, multiple phosphoevents on both WC-1 and WC-2, together, are needed to compromise the circadian DNA binding activity of the WCC and bring about repression. Furthermore, the mechanism used to effect repression is also applied by the cell to remove the WCC from other circadian-regulated genes in the first essential step of circadian output. These data provide a mechanistic basis for understanding how PTMs lead to repression of a circadian heterodimeric transcriptional activator, and also how regulation of this activator leads seamlessly into the first step of circadian output.
Results
Mapping In vivo WCC Phosphorylation Sites
To purify WCC, a tandem V5-10× His-3× FLAG (VHF tag) was placed via homologous recombination at the resident wc-1 locus C-terminal to the ORF, creating the wc-1VHF allele which exhibited normal WT circadian rhythms and light-dependent phosphorylation (e.g. Wang et al., 2014; Wang et al., 2015). Endogenously expressed WC-1VHF and its interactor, WC-2, were isolated from liquid cultures and purified following a three (FLAG, His and V5)-step protocol (Figure 1A). WC-1 and WC-2 protein bands were excised and subjected to protease digestion followed by MS analyses. Phosphosites were mapped on WCC isolated from four growth conditions: constant light (LL) when the WCC light-activity is high, FRQ is constantly made, and all FRQ phosphoisoforms coexist in the cell (Baker et al., 2009); following a 15 min light-pulse (LP15), when the WCC senses light and is hyperphosphorylated independent of the circadian clock (Linden et al., 1999); DD15 (~CT4; 15 hours into darkness) in the circadian subjective morning when WCC circadian activity peaks; DD24, (~CT14) in the circadian evening, when WCC activity is maximally repressed. Under all these conditions, 75 serine, threonine, and tyrosine (Ser/Thr/Tyr) phosphorylation events were unambiguously detected on WC-1 (Figure 1B), 66 of which are novel (Figure S1A), along with 5 additional ambiguous sites. 15 phosphosites were mapped on WC-2 (Figure 1C), 14 of which are novel (Figure S1B). All nine WCC phosphosites previously reported (He et al., 2005; Sancar et al., 2009) were common to both light-exposed and dark-only samples. Through the use of a combination of proteases during MS sample preparation, coverage of both proteins was close to 100% (Figure S1C).
Figure 1.
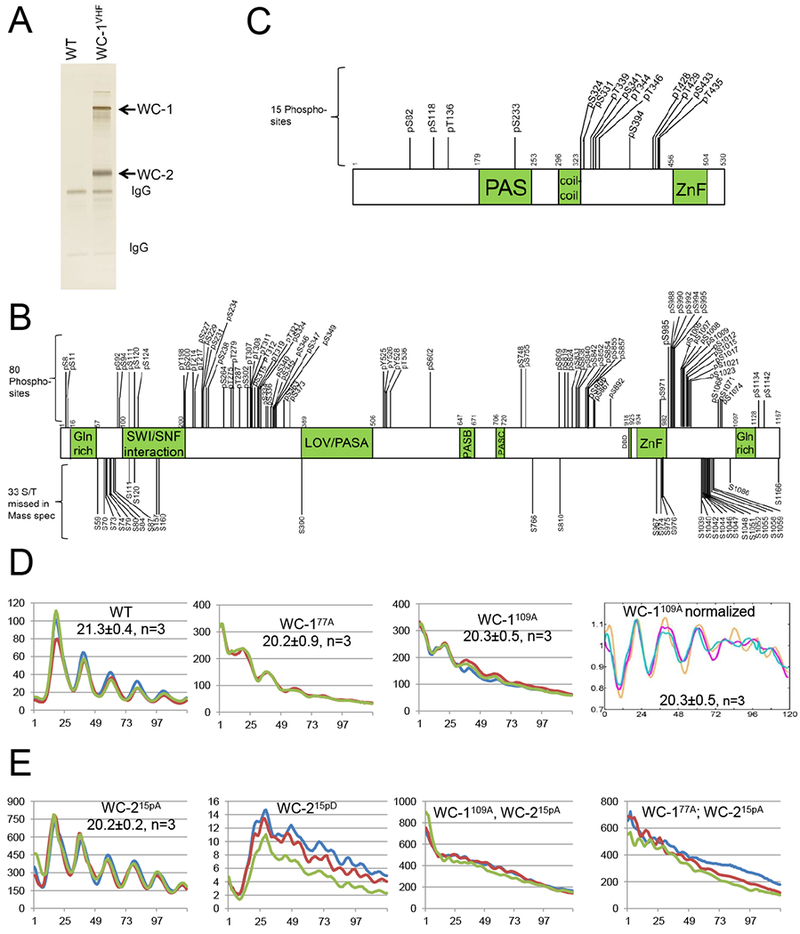
Identification of phosphorylation sites on WC-1 and WC-2. A A representative silver-stained gel showing WCC purified from a culture grown in the dark for 24 hr. B C. Schematic of WC-1 (B) and WC-2 (C) showing the position of phosphorylated residues identified by MS/MS. WC-1, 1,167 amino acids, contains a ZnF, zinc finger DNA binding domain; DBD, defective in DNA binding; LOV, light-, oxygen-, and voltage-sensing; and PAS, Per-Arnt-Sim domains. WC-2, 530 amino acids, contains PAS, coiled-coil, and ZnF domains. Vertical bars with numbers represent identified phosphorylation sites (above) and Ser/Thr phosphosites undetected by MS/MS (below). D Analysis of WCC activity by the frq C-box fused with firefly luciferase (frq C-box-luc transcriptional fusion). Middle two panels show circadian oscillator function with reduced amplitude and slightly shortened period (± SD) in WC-177A and in WC-1109A lacking nearly all WC-1 phosphosites. Time in hours is on the X-axis. Different colored lines represent three replicates. The panel on the right shows baseline subtracted and detrended data. E Oscillator function in wc-2 mutants in which all confirmed phosphosites are converted to Ala (left, WC-215pA; enhanced WCC activity as noted by scale bar) or Asp (mid left, WC-215pD; loss of rhythmicity and severely depressed WCC activity). Right two panels show enhanced WCC activity and arrhythmicity in strains lacking phosphorylatable sites on both WC-1 and WC-2.
Because the data (Figure 1B) report only presence or absence of WC-1 phosphorylation at a given site, we used phosphatase treatment and stable-isotope labeling (Wu et al., 2011) to better define the extent to which sites were phosphorylated at DD24, the time of maximum repression of WCC activity. Although not all WC-1 phosphosites were detected in this assay, phosphosites near to WC-1’s ZnF showed near stoichiometric phosphorylation (Figure S2), consistent with this region being involved in circadian repression.
Phosphorylation of Both WC-1 and WC-2 is Required for Circadian Repression
Knockin constructs (wc-1KI and wc-2KI) were used to replace Δwc-1 and Δwc-2 loci respectively with wc-1V5 and wc-2 sequences and rescue rhythmicity (Wang et al., 2014; Wang et al., 2015). Constructs in which all detected phosphorylated S/T/Y residues (Figures 1B and C) were deleted or mutated individually or in combination to Ala (Table S1) were also transformed into the Δwc-1 or Δwc-2 strains, rendered homokaryotic through backcrossing to ras-1bd, and analyzed for periodicity on race tubes. No single wc-1 and wc-2 phosphomutants, or clusters of mutants, were seen to be arrhythmic although minor period changes were noted in wc-1S971A, and wc-1S990A (both 2 hr shorter than WT; Table S1); mutation of the cluster of 5 phosphosites previously reported to be essential for overt rhythms (S988, S990, S992, S994, S995) (He et al., 2005) did not cause arrhythmicity (Table S1; Wang et al., 2015). 33 additional Ser/Thr to Ala mutations made in regions not identified as phosphorylated by MS also failed to disrupt rhythmicity (Table S2), strongly suggesting that multiple phosphoevents are required for feedback closure.
To confirm that multiple WCC phosphorylations must function cooperatively, all 80 WC-1 phosphosites (Figure 1B) together were mutated to Ala to create wc-180pA, along with the mutant bearing the 33 additional S/T to A changes, a total of 113 Ala mutations (wc-1113A). To facilitate screening a large variety of constructs, we switched assays for rhythmicity using, in place of race tubes, a reporter of the core circadian oscillator in which the C-box of the frq promoter is used to drive luciferase (e.g. Gooch et al., 2008; Larrondo et al., 2015). Using this assay both wc-180pA and wc-1113A displayed arrhythmic phenotypes (Figure S3A); however, this was traced to reduced WC-1 stability leading to low WC-1 amounts (Figures S3B and S3C). Regions near to the WC-1 LOV domain are known to be important for stability (Wang et al., 2015), and we identified four sites in WC-1(S363, S373, S390, Y525) that were rarely or never detected as phosphorylated (Figure S1A) but that when mutated to Ala resulted in the low WC-1 expression (Figure S3B). Restoration of these back to WT restored both WC-1 levels (wc-1109A, Figure S3B lower panel) and rhythmicity with a quasi-normal period length (Figure 1D). Similarly in the context of wc-180A, restoration of just S363, S373 and Y525 restored WC-1 levels and rhythmicity (wc-177A, Figure 1D and S3B). Interestingly, and despite the extremely low levels of WC-1, both WC-180pA and WC-1113A mediate normal light-induction of FRQ (Figure S3B) and they interact normally with WC-2, FRQ, and FRH (Figure S3D). Taken altogether, however, these data led to the surprising conclusion that the loss of no combination of real or potential phosphorylations on WC-1 alone, even 109 of them, is sufficient to eliminate the repression of the WCC needed to close the feedback loop of the core clock.
When the 15 WC-2 phosphosites (Figure 1C) were mutated individually to Ala, only T428A and T429A showed period effects (+3 hrs and +1 hr; Table S1), so as was done with WC-1, all 15 sites were collectively mutated to Ala to create wc-215pA. Surprisingly, wc-215pA has levels of protein comparable to WT (Figure S4A) and remains nicely rhythmic both by race tube and luciferase analyses (Figure 1E, S4B). We did not expect this: As with WC-1, loss of all detectable WC-2 phosphosites is not sufficient to interfere with closure of the circadian feedback loop. At this point, wondering if indeed any phosphorylation events were important, we generated wc-215pD in which all 15 WC-2 phosphosites were together mutated to Asp to mimic phosphorylation (accepting the caveat that Ser to Asp mutations may have other effects). In contrast to wc-215pA, wc-215pD had elevated WC-2 levels but extremely low FRQ expression under circadian conditions (Figure S4A) and very low, erratic WCC-driven luciferase expression under circadian conditions in the dark (Figure 1E; note scale bar). This suggested that phosphorylation does matter.
Taken altogether, these data suggested the unappealing possibility that phosphoevents on WCC might be sufficient to interfere with rhythmicity but, given the strong rhythms seen with wc-177pA and wc-215pA, not actually necessary for circadian repression - in short, a colossal unwanted diversion. However, the results did serve to sharpen the questions asked: To be relevant, phosphorylations ought to be FRQ-dependent and to influence rhythmicity while not unduly influencing WCC levels. In this context we knew (Wang et al., 2015) that circadian function of the WCC requires the ZnFs and nearby basic regions of both WC-1 and WC-2, suggesting that phosphorylation of both WC-1 and WC-2, each mediated by FRQ, might function together. In fact, both double mutant strains, wc-1109A; wc-215pA or wc-177A; wc-215pA, displayed very strong but arrhythmic/noncircadian WCC-driven luciferase signal under circadian conditions in the dark (Figure 1E). Although the relative contribution of WC-1 may be greater than WC-2 given the rhythms seen in wc-177A vs. wc-215pA (Figures 1D,E, S4A), these results are consistent with a model in which circadian repression of the WCC requires phosphorylation of both WC-1 and WC-2.
Isolation of the Key Phosphoevents on WCC Required for Closure of the Circadian Feedback Loop
To test this model and identify the phosphosites of WC-1 and of WC-2 required for circadian feedback loop closure, we adopted a strategy in which all phosphosites in large sections of each protein were mutated to Ala, and the mutants combined with the totally unphosphorylatable partner (wc-1109A or wc-215pA as appropriate) (Figure 2). For example, we generated a series of wc-1 mutants derived from wc-1109A (Figure 2A) in which all phosphorylatable residues between amino acids (aa) 603 and 1167 of WC-1 were changed to Ala (wc-1603-1167pA) or were combined with Ala mutations of Ser/Thr in regions not identified by MS (wc-1603-1167A), while keeping aa 1-602 WT and potentially phosphorylatable in both mutants; wc-11-602pA and wc-11-602A covered the other half of WC-1. Upon analysis, both wc-11-602A and the double mutant wc-11-602A; wc-215pA showed clear rhythms; wc-1603-1167A alone is perfectly rhythmic, but both wc-1603-1167A; wc-215pA and wc-1603-1167pA; wc-215pA are arrhythmic (Figure 2A, Figure S4C, Table S4). This indicates that the key phosphoresidues on WC-1 are located between aa 603-1167, and are among the phosphosites identified by MS. WC-1 phosphosites within aa 603-1167 were then further divided into two groups (aa 603-966 and aa 967-1167) and in each group phosphosites altogether were mutated to Ala and combined with wc-215pA: wc-1967-1167pA; wc-215pA and wc-1967-1167A; wc-215pA are arrhythmic indicating that within these regions are phosphosites essential for rhythmicity (Figure 2A). This strategy narrowed the search to seven phosphosites: S971, S985, S988, S990, S992, S994, and S995, of which all except S985 had >90% stoichiometries at DD24 when the WCC is maximally repressed (Figure S2). When these six sites were mutated to Ala (wc-1S971A,S988A, S990A, S992A, S994A, S995A) and combined with WC-215pA or WC-210pA, the double mutant showed elevated levels of C-box-driven luciferase but was profoundly arrhythmic (Figures 2C, S4C); phosphorylation of these six sites is crucial for repressing the WCC to close the feedback loop. To further test the importance of these phosphosites, all were mutated together to Asp to mimic the negative charge of phosphorylation. wc-1S971D, S988D, S990D, S992D, S994D, S995D; wc-215pD showed greatly reduced C-box-driven luciferase expression (~1% of the Ala-substitution strain) consistent with strong repression of WCC activity, and complete arrhythmicity (Figure 2C). In tests of each individual site among S971, S988, S990, S992, S994 and S995, the novel site, S971, plays a major role in repression: wc-1S988A S990A, S992A S994A S995A; wc-215pA still shows a rhythm at least for three days (Figure 2C); wc-1S971A has a 2 hr shorter period and a higher FRQ level at DD16; wc-1S971D loses rhythmicity quickly and has undetectable FRQ at DD16 or DD24 despite an increased level of WC-1 (Figure 2D), suggesting that phosphorylation of S971 elicits strong inhibition of WCC activity specifically in the dark (Figure 2D). Of the WC-1 phosphosites S988, S990, S992, S994 and S995, the first three that are closer to the ZnF play a somewhat more important role in WCC repression than S994 or S995 (Figure S4C).
Figure 2.
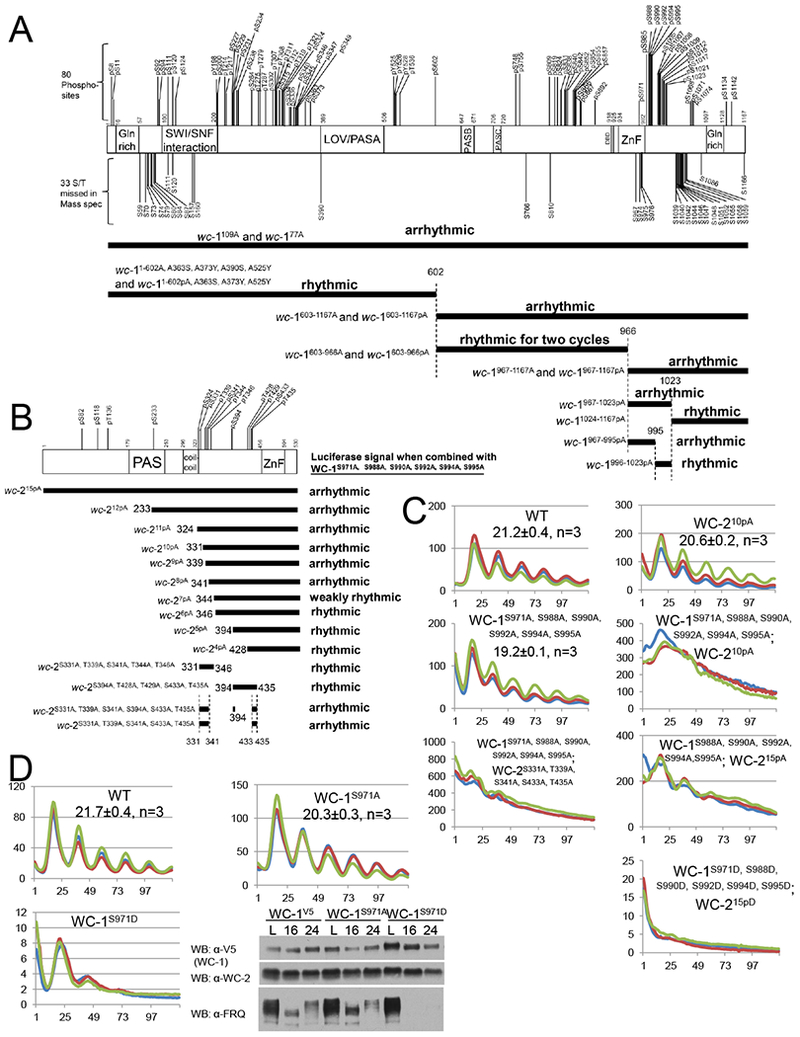
Identification of key phosphosites on WC-1 and WC-2 required for the circadian feedback loop closure. Schematic of WC-1 (A upper) and WC-2 (B upper) showing the position of phosphorylated residues as in Figure 1BC. Below these is the strategy used to identify the phosphorylation events on WC-1 (A lower) and WC-2 (B lower) essential for rhythmicity. Each horizontal bar represents a wc-1 (A) or wc-2 (B) mutant with phosphosites falling in the region of the bar mutated to Ala altogether. In the presence of wc-215pA (A) or wc-1S971A, S988A, S990A, S992A, S994A, S995A (B) respectively, the circadian rhythms of the wc-1 (A) or wc-2 (B) phosphomutants were measured by frq C-box-luc ; phenotypes (Table S4) were as noted. C WCC activity in WT and wcc mutants (y-axis) as measured by frq C-box-luc bioluminescence. D Phosphorylation of WC-1 S971 plays a key role in repressing the circadian WCC activity. WCC activity in strains of the noted genotypes was monitored by frq C-box-luc; note scale bars. Right panel: Western blot showing FRQ, WC-1, and WC-2 levels in WT (wc-1V5), wc-1S971A, and wc-1S971D in light and at 16 and 24 hrs in darkness.
Using a similar mutagenesis strategy, when a series of wc-2 phosphosite mutants (Figure 2B) were generated based on wc-215pA, backcrossed into wc-1S971A, S988A, S990A, S992A, S994A, S995A, and analyzed by the luciferase assay, arrhythmicity was observed from WC-212pAto WC-28pA; WC-27pA was weakly rhythmic and WC-26pA, WC-25pA, and WC-24pA showed clear rhythms (Figure 2B, 2C, S4C). These data indicate that multisite phosphorylation on WC-2, in combination with multisite phosphorylation on WC-1, is required for the repressive actions closing the feedback loop. Further narrowing WC-210pA into two groups in the background of wc-1S971A, S988A, S990A, S992A, S994A, S995A, both WC-2S394A, T428A, S429A, S433A, T435A and WC-2S331A, T339A, S341A, T344A, T346A display clear rhythms (Figure S4C), suggesting that two clusters of phosphosites on WC-2 co-determine WCC repression. These were further refined to wc-2S331A, T339A, S341A, S433A, T435A, in which S331A, T339A, and S341A are close to the WC-2 coiled-coil domain while S433A and T435A are near the ZnF (Figure 2B). The arrhythmicity of wc-1S971A, S988A, S990A, S992A, S994A, S995A, wc-2 S331A, T339A, S341A, S433A, T435A indicates that repression requires the two clusters of WC-2 phosphorylation events (Figure 2C). In addition to arrhythmia observed in wc-1/wc-2 phosphorylation mutants, additive effects of the circadian periods were also seen in wcc strains bearing multiple phosphorylation mutations (Figure S4D), indicating that WCC phosphorylation events function together.
WCC Phosphosite Mutants Bind to the frq Promoter at All Times
Repression of the WCC is believed to be initiated while it is, on balance, still DNA-bound, but to progress within hours to the state where the WCC no longer is bound to DNA. ChIP-PCR was used to assess the effect of WCC phosphosite mutations on binding to the C-box, which peaks at DD16 and becomes low at DD24 (Belden et al., 2007; He et al., 2006; Hurley et al., 2014). WCC is found constantly and strongly associated with the C-box in arrhythmic wc-1S971A, S988A, S990A, S992A, S994A, S995A; wc-210pA and in wc-1603-1167pA; wc-215pA whereas WC-215pD never binds (Figure 3). These data indicate that phosphorylation modulates the strength of WCC DNA binding and can lead to removal of the complex from DNA.
Figure 3.
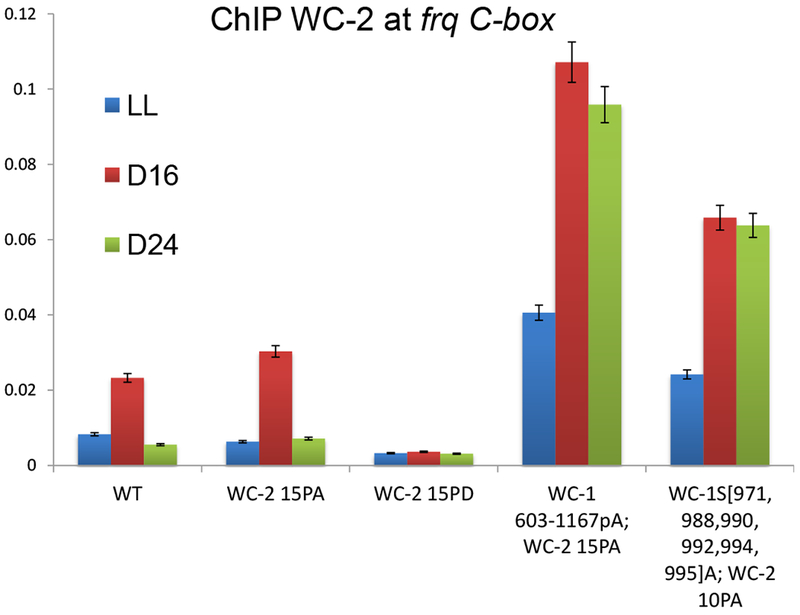
Mutation of phosphorylation sites on both WC-1 and WC-2 together leads to constant binding to the frq promoter C-box. Quantitative ChIP PCR experiments were carried out using WC-2 antibody with C-box- specific primer sets (Table S5) in indicated wc-1 phosphomutants. Average values are plotted as a percentage of the total; error bars show SEMs (n=3).
Much stronger luciferase signals were observed in arrhythmic strains such as wc-1S971A, S988A, S990A, S992A, S994A, S995A; wc-210pA or wc-1603-1167pA; wc-215pA than in WT (Figure 2C and S4B), consistent with stronger frq promoter binding (Figure 3). These data suggest that multisite phosphorylation, including sites required for the feedback loop closure, may also contribute to the robustness of frq transcription, that is, to circadian amplitude.
Detection of FRQ-Promoted Phosphorylation Events on WC-1 and WC-2
To bypass the need for MS in assessing the role of FRQ in promoting WCC phosphorylation, we adapted Phos-tag, a phosphate-binding chemical that specifically recognizes and binds phosphorylated Ser/Thr/Tyr (Kinoshita et al., 2006), to detect single phosphorylation events on WC-1 and WC-2 (See STAR methods). To validate the system, all Ser/Thr/Tyr in WC-1 and WC-2 were mutated to Ala to generate, respectively, wc-1STYtoA and wc-2STYtoA, and then a single Ser-to-Ala mutation was reversed in each protein to its WT residue. Thus, in wc-1STYtoA; A971S only aa 971 can be phosphorylated. WCC protein sequences were scanned using KinasePhos (Wong et al., 2007) to predict kinases responsible for specific phosphorylations (Table S3), and purified WCC was treated with specific kinases in vitro to confirm that single phosphorylations were identifiable (Figure S5). Single phosphorylations including WC-1 S94, S831, S971, or S1015 and WC-2 S118 and S324 were unambiguously detected (Figures S5A, S5B); WC-1 and WC-2 can be phosphorylated at these sites independent of other priming phosphorylation events. Looking in vivo, no phosphorylation was detected on WC-1113A and WC-1109A or WC-215pA (Figure 4A, S6A) suggesting that MS data covers all relevant phosphorylation sites. Phosphorylation of S990 and S1015 on WC-1 and S433 on WC-2 was detected as a single band shift in the gels although phosphorylation was not detected at some other sites (Figure S6B) suggesting that phosphorylation might be dependent on phosphorylation of nearby sites that were mutated in wc-1113A and wc-215pA.
Figure 4.
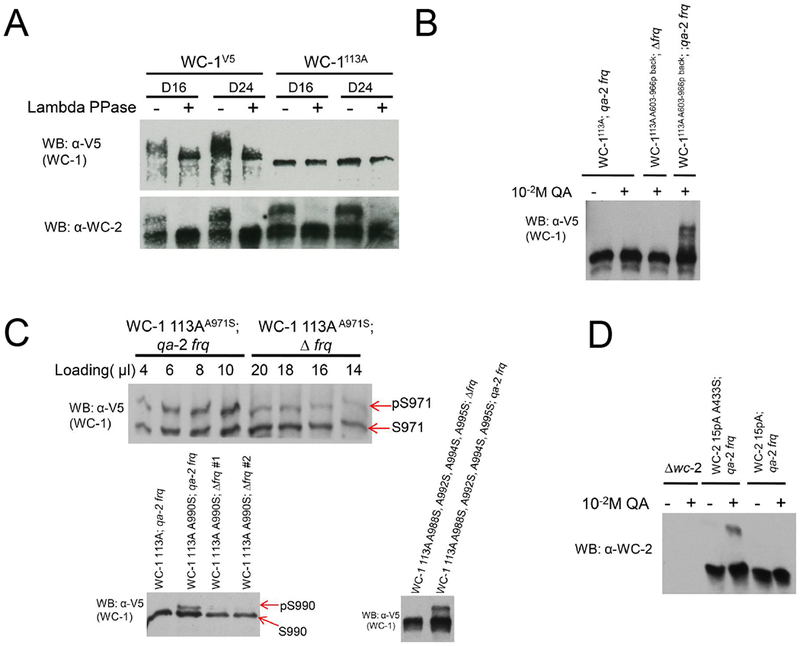
FRQ promotes phosphorylation of certain residues in WC-1 and WC-2. A Western blot showing complete loss of phosphorylation in wc-1113A. Western blots were performed using the 149:1 gel containing 20 μM Phos-tag (See Methods). B FRQ-induced phosphorylation. In WC-1113A A603-966p back, all Ser/Thr between aa 603 and aa 966 were returned to normal (phosphorylatable) sequence, whereas all other WC-1 Ser/Thr remained converted to Ala. These strains were either backcrossed to introduce Δfrq or transformed with qa-2 promoter-driven frq at the csr locus. To cultures in constant light, inducer (10−2 M QA) was added and the cultures immediately moved to dark for 16 hr. Phos-tag Western blots as in A are shown. C In the context of unphosphorylatable WC-1113A, Ser was restored at indicated sites; e.g. in wc-1113A, A971S the only Ser is at 971. Mutants were placed in a Δfrq or QA-inducible frq background and assayed for WC-1 phosphorylation as in B. Note differences in amount loaded in the top panel. D In the context of the unphosphorylatable wc-215pA, Ser was introduced to aa 433 (wc-215pA, A433S). This allele was placed in an inducible frq background in the presence or absence of inducer (10−2 M QA) and assayed for WC-2 phosphorylation as in B.
Using this assay we explored the role of FRQ in promoting the WCC phosphorylations that actuate circadian negative feedback. The FRQ-FRH complex associates with WC-1 through its DBD domain (Wang et al., 2015) that is very close to the WC-1 ZnF and to S971, S988, S990, S992, S994, and S995; this suggests that FRQ-promoted phosphorylation of WC-1 might occur at these key phosphosites required for feedback loop closure (Figure 2A,C). To assess this, discrete regions of normal sequence WC-1 were added back in the context of WC-1113A; because the FRQ level in wc-1113A is significantly reduced in the dark compared to WT (Figure S3B), in these strains FRQ was over-expressed under the control of the qa-2 promoter. In this assay FRQ elicits phosphorylations in the C-terminal half of WC-1 (Figures 4B, S7A). When Ala971 in WC-1113A was mutated back to Ser, a very low level of S971 phosphorylation was still observed even in the absence of FRQ, while over-expression of FRQ strongly promotes the phosphorylation of S971 (Figure 4C), suggesting that FRQ-induced repression of the WCC is partially through S971. FRQ also promotes phosphorylation of WC-1 S990, S1015, and site(s) among S988, S992, S994, and S995 (Figure 4C, S7A), and S433 on WC-2 (Figure 4D).
If FRQ-driven phosphorylation influenced the interaction of WC-1 with WC-2 it could affect DNA-binding and effect repression. However, control experiments discount this: WC-1 still strongly interacts with WC-2 even when FRQ is over-expressed (Figure S8A) and a fusion of WC-1 with WC-2 into a single fusion protein is able to support a normal clock (Figure S8B,C).
WCC Repression, the Black Widow Hypothesis, and A Biological Significance for Circadian Oscillator Amplitude
In the presence of unmutated WC-2, wc-1109A, wc-177A, wc-1603-1167A, and wc-1603-1167pA all supported a rhythmic core circadian oscillator as reported by frq C-box-luc (Figure 1D, S4C, and Table S4) but are completely arrhythmic on race tubes (Figure 5A), an inconsistency in the core clock vs. circadian output that suggests an importance in output for phosphorylations beyond those required for circadian repression. A similar disconnect between oscillator rhythmicity and output was seen in mutations of the SWI/SNF complex required for circadian activation via the WCC at the frq C-box (Wang et al., 2014), and in mutants of the phosphatase CSP-6 that acts on WC-1(Zhou et al., 2018). To emphasize this point, race tubes were compared for two strains constructed in the background of WC-1109A: WC-1 1-970A, 996-1167A has normal levels of WC-1 but lacks all phosphorylations except those required for rhythmicity, whereas WC-1S971A,S988A,S990A,S992A,S994A,S995A lacks only those phosphorylations required for rhythmicity. Oscillators in both strains cycle for >5 days (Figures 2C,S4C) but overt rhythmicity is worse in WC-11-970A, 996-1167A (Figure 5A, S7B); comparable data for WC-1603-1167pA (Figure 5A) confirm this trend, suggesting a role for phosphorylations outside of the oscillator-essential region in maintaining circadian output.
Figure 5.
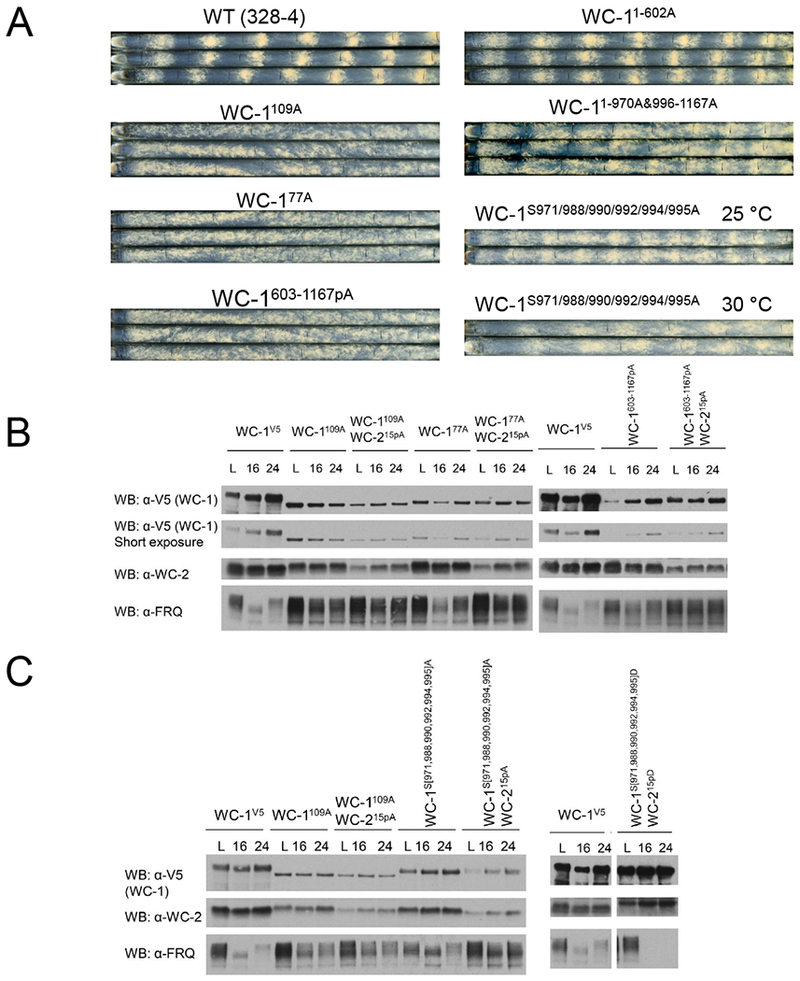
Loss of overt rhythmicity and reduction of oscillator amplitude in strains lacking phosphorylation around the WC-1 ZnF. A Race tube assays of WT (328-4) and wc-1 phosphomutants as indicated; WC-11-602A retained S363, S373, S390, and Y525 to maintain WC-1 at a level sufficient for core oscillator rhythmicity. B Effects of loss of WCC phosphorylation on levels of WC-1, WC-2, and FRQ. Left panel: Western blot showing reduced levels of WC-1 and WC-2 with increased FRQ in constant light (L), DD16 (~CT4, subjective morning), and DD24 (~CT14, subjective evening). Right panel: Same as left, but examining more restricted regions of WC-1. C Mutation of WCC residues essential for feedback repression has dramatic effects on WCC and FRQ levels; the level of FRQ is inversely proportional to the amount of WCC. Experimental conditions as in B.
Unlike circadian period, phase or output amplitude, core oscillator amplitude is a phenomenon that is measured but for which biological significance remains hard to ascribe. To better understand the relationship between oscillator amplitude and output we tracked levels of the WCC and FRQ in these mutants with and without WC-215pA (Figure 5B). In these strains, levels of WC-1 are greatly reduced, WC-2 modestly reduced, and FRQ greatly increased such that peak and trough levels are virtually the same in some cases. Similar data - low levels of a circadian transcriptional activator with high levels of repressor - have been reported before in circadian systems (He et al., 2006; Kondratov et al., 2003; Kwon et al., 2006; Schafmeier et al., 2005; Shi et al., 2010; Stratmann et al., 2012) and used to support the “Black Widow” hypothesis for transcriptional activators (Tansey, 2001) in which activators such as WCC or BMAL1/CLOCK are marked for turnover as a requisite part of their cycle in activating transcription. This hypothesis predicts that WC-1 phosphosite mutants that still run a core oscillator, even without overt output (e.g. wc-177A or wc-1109A) should have higher levels of WC-1 than truly uninhibitable strains (e.g. wc-1109A;WC-215pA or WC-1S971A, S988A, S990A, s992A, s994A, s995A;WC-215pA), and that phosphosite mutants that are never active (e.g. WC-1S971D, S988D, S990D, S992D, S994D, S995D;WC-215pD) should have very high levels of WC-1 and very low levels of FRQ. Data confirming these predictions (Figures 5 B,C) can be interpreted in terms of the output defects seen in Figure 5A. The function of the Black Widow (of activity-associated turnover) in circadian output is to eliminate the activator rather than only partially repress it. This creates robust oscillator amplitude (peak vs. trough) reflected in high cycles of FRQ expression, sufficient not only to drive the next oscillator cycle but also to drive robust cycles in the direct WCC targets that provide the first step of circadian output. When oscillator amplitude drops, with only partial repression as reflected in higher activity troughs and lower peaks in C-box-luc cycles, lower FRQ expression, and incomplete WC-1 turnover, the result is a dampened core oscillation and a loss of overt circadian output. These data, in which C-box-luc directly reports WCC transactivation (Figure 1D) and race tubes report the end result of the transcriptional network governing circadian output (Figure 5A), may provide a window on the significance of circadian oscillator molecular amplitude. This intimate view of the role of the WCC in the oscillator and output also makes predictions about WCC binding to target genes encoding transcription factors in the first tier of circadian output.
Rhythmic Binding of the WCC to Its Targets Genome-wide Depends on the Combination of Phospho-events on WC-1 and WC-2
The WCC not only drives transcription of frq, but also rhythmically binds to about 30 other promoters of clock-controlled genes (ccgs) leading to their rhythmic expression (Hurley et al., 2014; Smith et al., 2010); this is the first essential step in output. However, the number of WCC targets is somewhat plastic given the necessity of arbitrary cutoffs set in all ChIP studies; genes weakly bound by the WCC might be missed. When ChIP-seq was used to identify WCC targets, all known WCC targets were seen along with two additional genes, ncu06593 and ncu07024 (Figures 6A and S9A).
Figure 6.
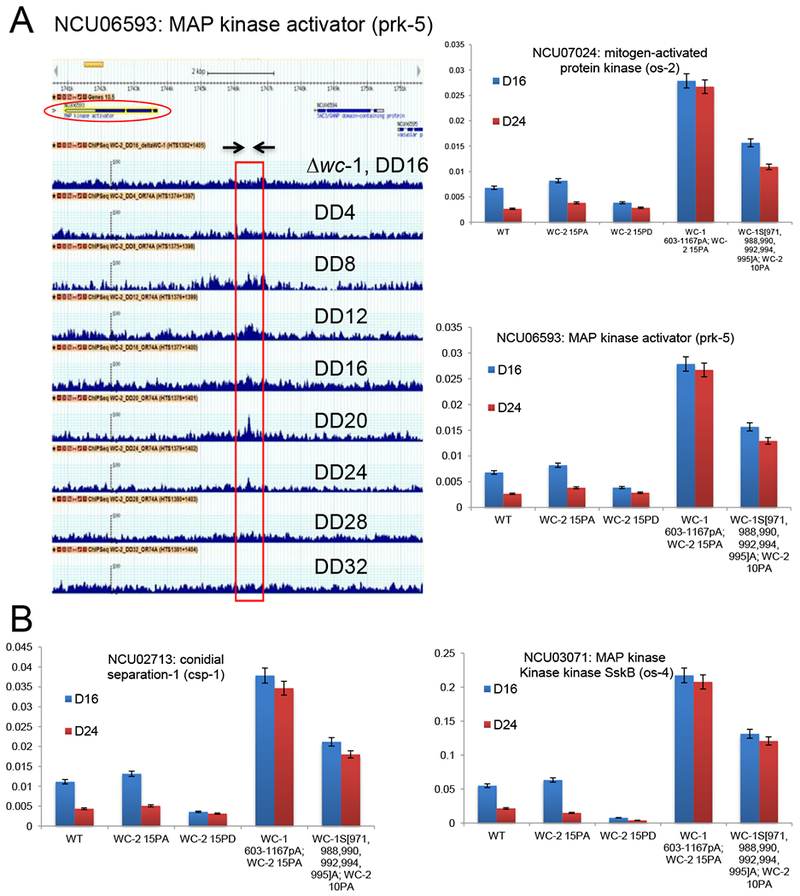
Mutations of phosphosites on the WCC that impact repression within the core oscillator also have direct effects on rhythmic binding to DNA targets in the first step of circadian output. A Identification of novel direct targets of the WCC. G-browse data for ncu06593 (encoding MAP kinase activator prk-5) from a series of ChIP-seq experiments using anti-WC-2. Top; gene map with the transcription unit circled. Below this are Δwc-1 (negative control for WCC binding) and WC-2 binding assessed at 4 hr intervals from DD4 to DD32. The peak of binding is marked by the box; arrows indicate the primer set (Table S5) used to detect the region. Right; ChIPqPCR data assessing WC-2 binding to this site and to the promoter of ncu07024, in WT and four mutants as shown, revealing that mutant strains that have lost circadian repression within the feedback loop are also altered for binding related to circadian output. Binding was assessed at DD16 (~CT4, subjective morning) and DD24 (~CT14, subjective evening). Error bars are SEMs (n=3). B Binding of WC-2 to two other direct targets of the WCC within the first step of circadian output in WT and mutants as shown. See Figure S9 for other examples of output genes.
Utilizing these novel as well as previously identified WCC targets, the importance of WCC phosphorylation to output gene binding was examined. We speculated that the same set of WCC phosphorylation events required for rhythmic transcription of frq (Figure 2C) would also be essential for rhythmic WCC binding to other targets genome-wide in the first step of circadian output. To test this, 13 genes whose promoters display a clear pattern of rhythmic WCC binding were examined by ChIP-PCR at two times using the wcc phosphomutants as indicated (Figure 6B, S9B): at D16 when the WCC is active and strongly associates with DNA and at D24 when the complex is inactive and mostly off DNA. Consistent with C-box-luc assays (Figure 2), WCC binds less strongly to all its targets at D24 than D16 in rhythmic strains, but binds all 13 targets very strongly and to the same degree at both times in the arrhythmic wc-1603-1167pA;wc-215A and wc-1S971A, S988A, S990A, S992A, S994A, S995A;wc-210pA backgrounds (Figure 6B, S9B). The same set of WCC phosphorylations used to repress frq expression in closing the feedback loop also represses expression of ccgs driving output.
Re-evaluation of Kinases and Phosphatases Required for the Circadian Clock
PKA and PP2A they have been suggested as key factors in the core oscillator based in part on analysis of overt rhythms. Given the disconnect between oscillator and output rhythmicity seen here and elsewhere (Wang et al., 2014; Zhou et al., 2018), we re-examined strains bearing Δpkac-1, Δpkac-2, (Huang et al., 2007), or rgb-1RIP, the RIP-inactivated allele of the rgb-1 regulatory subunit of PP2A required for action on WCC (Schafmeier et al., 2005; Yang et al., 2004). All three strains displayed functional oscillators with at most minor period effects (Figure 7A). While other kinases may have roles (Table S3), at this point the only kinase known to be essential for core circadian oscillator function is CK1, with CK2 having an essential role in compensation (Mehra et al., 2009).
Figure 7.
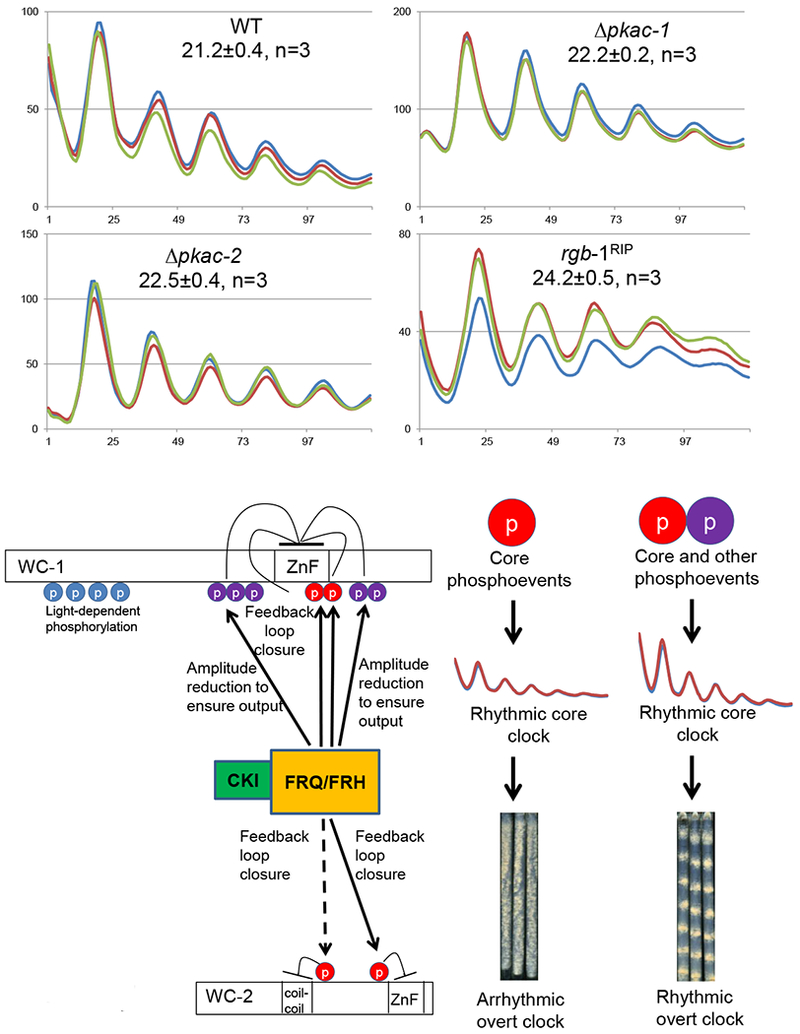
Re-assessment of kinases and a phosphatase previously implicated in regulation of WCC activity, and a summary model for the role of phosphorylation in regulating WCC activity. A PKA and PP2A are not required for WCC circadian activity. WCC activity was monitored by frq C-box-luc in the Δpkac-1, Δpkac-2, and in the rgb-1RIP strain that is defective in PP2A activity on the WCC. B Working model for the role of phosphorylation in regulating WCC activity. In a circadian cycle, FRQ/FRH/CK1 promotes phosphorylation of key residues (red circles) on both WC-1 and WC-2 to repress WCC activity in the feedback loop of the core clock, and also induces phosphorylation near the ZnF region of WC-1 (purple circles) that acts to depress the circadian amplitude as an aid to enhance robustness of circadian outputs.
Discussion
Circadian clocks are finely tuned auto-regulatory cellular feedback loops of significant importance to human health. How the feedback loop is closed is a question of central important to Chronobiology. Phosphorylation events impacting WCC repression were dissected via proteomics, genomics, genetics, and biochemistry, revealing that WC-1 and WC-2 are subject to multisite phosphorylation, promoted by FRQ, that determines circadian repression. Prior studies of WCC phosphorylation chiefly focused on its overall modification status and led to conflicting observations on the role of each partner and the relationship between modification status and WCC activity. By identifying all phosphosites and focusing on roles of individual sites, the picture has simplified with the surprising answer that FRQ-promoted phosphorylation of key sites in both proteins, in WC-1 near the DNA-binding ZnF and in WC-2 at two distinct locations, near to the coiled-coil and the ZnF, is required for repression. We note that this result allows the strong prediction that phosphorylation of most of these sites, in particular WC-1S971, should be rhythmic. No single phosphosite mutants have dramatic phenotypes (Tables S1 and S2) suggesting that repression in vivo is the result of gradual FRQ-promoted phosphorylation, but it is unknown whether FRQ directly recruits kinases other than CK1 to the WCC, or if the physical interaction between the WC-1 DBD and FRQ makes WC-1 available for kinases not physically recruited by FRQ (Wang et al., 2015). These data may inform understanding of the mammalian clock where BMAL1 and CLOCK also heterodimerize, play a role like that of WC-1 and WC-2, and like them are co-dependently hyperphosphorylated (e.g.Kondratov et al., 2003). BMAL1-dependent CLOCK phosphorylation is inhibited by CRY and promoted by PERs (Matsumura et al., 2014), and phosphorylation at various residues negatively correlates with nuclear accumulation and DNA binding (Yoshitane et al., 2009) or degradation (Spengler et al., 2009). In Drosophila, a balance of CK1 phosphorylation with phosphatases regulates dCLK modification (Kim and Edery, 2006) at more than a dozen dCLK phosphosites whose mutation results in period and entrainment defects (Lee et al., 2014). BMAL1 is also phosphorylated (Robles et al., 2010), and phosphorylation at S90 promotes CLOCK/BMAL1 heterodimerization and nuclear localization (Tamaru et al., 2009). In general, as in Neurospora, hyperphosphorylation is associated with elevated activity as well as instability, consistent with a “black-widow” model wherein activated heterodimers are degraded (Kondratov et al., 2003; Kwon et al., 2006; Stratmann et al., 2012); however, there are many loose ends such that a unifying mechanistic model for the role of phosphorylation has not emerged.
Considering repression more generally, in mammals, a repressive complex containing only PERs, CRYs and CK1 is assembled in the cytoplasm, enters the nucleus, and quantitatively binds CLOCK/BMAL1 while phosphorylating CLOCK and possibly other proteins of the complex in a manner that reduces its affinity for DNA (Aryal et al., 2017). That this happens in vitro suggests that no other proteins are needed. This complex is reminiscent of the FRQ/FRH/CK1 complex found associated with the WCC coincident with repression (Baker et al., 2009). In addition to phosphorylation, a large body of data suggest inhibitory roles for chromatin-modifying and RNA-binding proteins recruited to DNA-bound CLOCK-BMAL1 (e.g. Kim et al., 2014; Koike et al., 2012; Padmanabhan et al., 2012; Xu et al., 2015), mutations of which lead to changes in period or rhythm amplitude. Although inhibition may occur hours before the heterodimer dissociates from DNA, it remains hard to determine what events or interactions initiate repression versus contribute to it or consolidate it. Phosphorylation can influence interactions as well as DNA binding, and the data of Aryal et al. (2017) are not inconsistent with a view in which early phosphorylations within the complex could lead to downstream events on and off chromatin. While data from Neurospora also suggest roles for chromatin modifications in repression (e.g. Belden et al., 2007; Wang et al., 2014), the discrete nature of the phosphorylation events associated with repression, their correlation with changes in DNA binding, and their apparent necessity and sufficiency for repression, all suggest that initiation of repression may begin with these phosphorylations, while other events support it. Thus, phosphorylations but not necessarily loss of DNA binding may be key to repression.
What kinases and phosphatases are responsible for the key WCC phosphorylations? Kinase motif analyses show that key sites might be targets of multiple kinases (Table S3), and consistent with this, no individual nonessential kinase knockouts are arrhythmic. Similarly, although both PP2A (regulated by RGB-1) and CSP-6 dephosphorylate WC-1 (Yang et al., 2004; Zhou et al., 2018), neither rgb-1RIP nor Δcsp-6 is arrhythmic (Figure 7B; Zhou et al., 2018). There may be redundancy among kinases and phosphatases acting in the clock.
What happens to WCC that has been hyperphosphorylated through FRQ-promoted phosphorylation? Late in the circadian day when WCC is hyperphosphorylated and repressed, FRQ is also hyperphosphorylated and the two complexes interact very little (Baker et al., 2009; Hong et al., 2008). WC-1 is always found in the nucleus, even at DD24 (CT14) (Hong et al., 2008) when the WCC is maximally repressed, so phosphorylation-mediated nuclear export of the WCC could contribute to but not be the sole source of repression. Consistent with this, WC-1ΔDBD that fails to bind the frq promoter in the dark still resides in the nucleus (Wang et al., 2015), so the failure of WCC to bind DNA does not result in its nuclear export. Bulk WC-1 has a shorter half-life when active at CT4 than when inactive at CT14, but nuclear WC-1 always appears less stable than total WC-1 (Hong et al., 2008). Taken together, when the WCC is hyperphosphorylated and repressed, it can still be found in the nucleus and may be degraded and resynthesized or dephosphorylated to initiate a new circadian cycle. In Neurospora, expression of ~ 40% of the ~10,000 coding genes are clock-controlled (Hurley et al., 2014) but only ~30 genes are direct targets of the WCC (Hurley et al., 2014; Smith et al., 2010; this study). Data here suggest that a specific combination of phosphosignals on the WCC determines its rhythmic DNA binding to a few ccgs genome-wide (Figure 6 and Figure S9) including transcription factors tcf-21 (ncu02724), sub-1 (ncu01154), and csp-1 (ncu02713) and kinases casein kinase 1b (ncu04005), MAP kinase OS-2 (ncu07024), and MAP kinase kinase kinase SskB (ncu03071), suggesting that the WCC-FFC oscillator directly impacts signaling pathways as well as transcription programs.
Multisite phosphorylation is a widespread phenomenon seen in most organisms, although it is still poorly understood due to its complexity. The strategy used here for dissecting multisite phosphorylation on WC-1 and WC-2 may be prove feasible for discovering key phosphoevents involved in other biological processes. Our data suggest multiple roles for FRQ-promoted phosphorylation of the WCC (Figure 7C). Phosphorylation of a small number of clock-essential sites on both WC-1 and WC-2 is required to close the feedback loop, but phosphorylation of other sites more distal from these serves to modulate circadian oscillator amplitude, thereby ensuring robust circadian output.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents should be directed to and will be fulfilled by the corresponding author, Jay C. Dunlap (jay.c.dunlap@dartmouth.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Strains and growth conditions
328-4 (ras-1bd A) was used as a clock-WT strain in the race tube analyses. WT in the luciferase assays was 661-4a (ras-1bd A) that contains the frq C-box fused to codon-optimized firefly luciferase gene (transcriptional fusion) at the his-3 locus. Neurospora transformation was performed as previously reported (Colot et al., 2006). Race tube medium contains 1×Vogel’s salts, 0.17% arginine, 1.5% bacto-agar, and 50 ng/mL biotin with glucose at 0.1%, and liquid culture medium (LCM) contains 1×Vogel’s, 0.5% arginine, 50 ng/mL biotin and 2% glucose. Quinic acid (QA) was added into race tube medium at the concentration of 10−2 M. Unless otherwise specified, race tubes were cultured in constant light for 16–24 hr at 25 °C an d then transferred to the dark at 25 C. Images of replicate race tubes when displayed show vertical lines where the growth front was marked every 24 h. WC-1 tagged with V5-10×His-3×FLAG was generated previously (Wang et al., 2014). wc-1 and wc-2 mutants were made as previously described (Wang et al., 2015). All wc-1 and wc-2 constructs bearing mutations were targeted to their native loci respectively. All wc-1 and wc-2 mutant strains were in the ras-1bd genetic background, and all wc-1 mutants have a V5 tag at their C termini.
METHOD DETAILS
Protein lysate and Western Blot
Procedures for preparation of protein lysates and Western blots (WB) were followed as described. For WB, 15 μgs of whole-cell protein lysate were loaded per lane in a SDS gel. Antibodies against WC-1, WC-2, FRQ, and FRH have been described previously. V5 antibody (Thermo Pierce) was diluted 1:5000 for use as the primary antibody.
Identification of the WCC phosphorylation sites
WCC purification and the preparation of mass spectrometry samples were carried out as previously described (Wang et al., 2014).
Phos-tag gel
The Phos-tag reagent was purchased from ApexBio. To analyze the phosphorylation profiles of WC-1 and WC-2, 20 μM Phos-tag was added to the 6.5% SDS-PAGE gels containing a ratio of 149:1 acrylamide/bisacrylamide.
Quantification of phosphosites on WC-1
Phosphorylation quantification of WC-1 was carried out as described in Wu et al., 2011. Briefly, WC-1 was purified from a culture grown for 24 hr in the dark in three separate experiments. For each replicate, WC-1 was subject to digestion with proteinase K. Two identical peptide aliquots were subjected to either 0.5 μL of Alkaline Phosphatase, Calf Intestinal (CIP) (NEB) treatment at 37 °C for 1 hr or a mock reaction. After the treatment, peptides in the phosphatase-treated sample were chemically labeled by reductive methylation using deuteroformaldehyde to dimethylate free amines, while the untreated sample was chemically labeled with formaldehyde. Subsequently, the two aliquots were mixed, resulting in a 1:1 ratio for all peptides uninfluenced by the phosphatase treatment. The mixture was then analyzed by mass spectrometry to identify and quantify peptide species.
Immunoprecipitation (IP) assay
IP was performed as previously described (Wang et al., 2015). Briefly, 2 mgs of total protein were incubated with 20 μL of V5 agarose (Sigma-Aldrich) rotating at 4 °C fo r 2 hrs. The agarose beads were washed with the protein extraction buffer (50 mM HEPES [pH 7.4], 137 mM NaCl, 10% glycerol, 0.4% NP-40) twice and eluted with 50 μL of 5× SDS sample buffer at 99 °C for 5 min.
Kinase and lambda-phosphatase assay
WCC immunoprecipitated by V5 antibody-conjugated beads from 2 mg lysate was subjected to a kinase assay in the presence of 500 μM ATP. The reaction mixtures were incubated for 60 min at 30Ό, stopped with 5× SDS s ample buffer, heated to 99° and subjected to SDS-PAGE analysis in the presence of 20 μM Phos tag. Recombinant CDK-1, CKI, CKII, and MAPK were purchased from New England Biolabs. Lambda-phosphatase treatment was performed in a 50 μL reaction containing immunoprecipitated WCC from 2 mg lysate and 2 μL λ-phosphatase (New England Biolabs) in the phosphatase buffer supplied by the manufacturer. After incubating at 30°C for 60 min, 50 μl of 5× SDS sample buffer was added to the mixture, heated at 99 °C for 5 min, and subjected to electrophoresis.
Other techniques
Luciferase assays were performed as previously described (Larrondo et al., 2015). Briefly, cultures bearing the frq C-box luciferase transcriptional reporter inserted into the his-3 locus were grown in 96 well cultures in race tube medium containing luciferin in constant light for 24 hrs at 25°C and then transferred to darkness at 25°C to initiate circadian cycling. Light production was monitored by CCD camera every hour with a 10 min exposure for 5 days, data were extracted using NIH Image J with a custom macro and period lengths were calculated using custom Matlab software.
Chromatin immunoprecipitation (ChIP) experiments were done as previously described using fresh tissues (Belden et al., 2007; Wang et al., 2014). All primer pairs used for ChIP-PCRs were listed in Table S5.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-FRQ (rabbit) | Jay Dunlap (Garceau et al., 1997) | N/A |
| Anti-WC-1 (rabbit) | Jay Dunlap (Froehlich et al., 2002) | N/A |
| Anti-WC-2 (rabbit) | Jay Dunlap (Denault et al., 2001) | N/A |
| Anti-V5 (mouse) | Thermo Pierce | N/A |
| Anti-HA (rabbit) | Abcam | Ab9110 |
| Goat Anti-Rabbit IgG (H + L)-HRP Conjugate | BIO-RAD | #1706515 |
| Goat Anti-Mouse IgG (H + L)-HRP Conjugate | BIO-RAD | #1706516 |
| Bacterial and Virus Strains | ||
| E.coli, DH5alpha, Electro-competent | PROTEIN EXPRESS | 961-008 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Protease Inhibitor Cocktail | Roche | 11836170001 |
| Phos-tag reagent | ApexBio | F4002 |
| SuperSignal West Pico ECL (Pierce) | Thermo Fisher | 34078 |
| Critical Commercial Assays | ||
| CDK1-cyclin B | New England Biolabs | P6020 |
| Casein Kinase I (CK1) | New England Biolabs | P6030 |
| Casein Kinase II (CK2) | New England Biolabs | P6010S |
| p42 MAP Kinase (MAPK) | New England Biolabs | P6080 |
| Lambda Protein Phosphatase (Lambda PP) | New England Biolabs | P0753 |
| Alkaline Phosphatase, Calf Intestinal (CIP) | New England Biolabs | M0290 |
| Anti-V5 Agarose Affinity Gel antibody produced in mouse | Sigma-Aldrich | A7345 |
| SilverQuest Staining Kit | Invitrogen | LC6070 |
| Experimental Models: Organisms/Strains | ||
| S. cerevisiae: FY834 | Fungal Genetics Stock Center (Winston et al., 1995) | FGSC #9721 |
| N. crassa: WT: 328-4 | Fungal Genetics Stock Center | FGSC #1858 |
| N. crassa: WT: 661-4a (Pfrq(c-box)@his-3) A | Gooch et al., 2008 | N/A |
| N. crassa: −1-6: wc-1V5::bar+; ras-1bd | Wang et al., 2014 | −1-6 |
| N. crassa: ras-1bd; Δwc-1::hph+; Δmus-52::hph+ a | Wang et al., 2014 | 21-9 |
| N. crassa: ras-1bd; Δwc-2::hph+; Δmus-52::hph+ a | Wang et al., 2014 | 173-1 |
| N. crassa: Δpkac-1::hph+ | Fungal Genetics Stock Center (Colot et al., 2006) | FGSC#11513 |
| N. crassa: Δpkac-2::hph+ | Fungal Genetics Stock Center (Colot et al., 2006) | FGSC#11433 |
| N. crassa: rgb-1RIP | Fungal Genetics Stock Center | FGSC#8380 |
| Recombinant DNA | ||
| Plasmid: pRS426: Yeast episomal vector with URA3 marker, and Ampicillin resistance | Fungal Genetics Stock Center (Colot et al., 2006) | pRS426 |
| Software and Algorithms | ||
| NIH ImageJ with a custom macro and period lengths were calculated using custom MATLAB software | Larrondo et al., 2015 | N/A |
| ChronOSX 2.1 |
Highlights:
95 phosphosites on WC-1 and WC-2 were found and their roles in circadian feedback defined
Circadian negative feedback requires specific phosphorylations on both WC-1 and WC-2
A complex including FRQ, FRH, CK1 promotes the essential phosphorylations on WC-1/WC-2
Phosphorylations abolish WC-1/WC-2 DNA binding, impacting both Core Oscillator and Output
Acknowledgments
We thank the Fungal Genetics Stock Center for Neurospora strains and Dr. Oded Yarden for help with transforming the rgb-1RIP strain. This work was supported by grants from the NIH to J.C.D. (R35GM118021) and J.J.L. (R35GM118022) and A.N.K (R35GM119455). The Orbitrap Fusion Tribrid mass spectrometer was acquired with support from NIH (S10-OD016212). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests: The authors have declared that no competing interests exist.
References
- Aryal RP, Kwak PB, Tamayo AG, Gebert M, Chiu PL, Walz T, and Weitz CJ (2017). Macromolecular Assemblies of the Mammalian Circadian Clock. Mol Cell 67, 770–782 e776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CL, Kettenbach AN, Loros JJ, Gerber SA, and Dunlap JC (2009). Quantitative proteomics reveals a dynamic interactome and phase-specific phosphorylation in the Neurospora circadian clock. Mol Cell 34, 354–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belden WJ, Loros JJ, and Dunlap JC (2007). Execution of the circadian negative feedback loop in Neurospora requires the ATP-dependent chromatin-remodeling enzyme CLOCKSWITCH. Molec Cell 25, 587–600. [DOI] [PubMed] [Google Scholar]
- Cha J, Zhou M, and Liu Y (2015). Mechanism of the Neurospora circadian clock, a FREQUENCY-centric view. Biochemistry 54, 150–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, Weiss RL, Borkovich KA, and Dunlap JC (2006). A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci U S A 103, 10352–10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap JC (1999). Molecular bases for circadian clocks. Cell 96, 271–290. [DOI] [PubMed] [Google Scholar]
- Garceau NY, Liu Y, Loros JJ, and Dunlap JC (1997). Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell 89, 469–476. [DOI] [PubMed] [Google Scholar]
- Gooch VD, Mehra A, Larrondo LF, Fox J, Touroutoutoudis M, Loros JJ, and Dunlap JC (2008). Fully codon-optimized luciferase uncovers novel temperature characteristics of the Neurospora clock. Eukaryot Cell 7, 28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Cha J, He Q, Lee H, Yang Y, and Liu Y (2006). CKI and CKII mediate the FREQUENCY-dependent phosphorylation of the WHITE COLLAR complex to close the Neurospora circadian negative feedback loop. Genes and Dev 20, 2552–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Shu H, Cheng P, Chen S, Wang L, and Liu Y (2005). Light-independent phosphorylation of WHITE COLLAR-1 regulates its function in the Neurospora circadian negative feedback loop. J Biol Chem 280, 17526–17532. [DOI] [PubMed] [Google Scholar]
- Hong CI, Ruoff P, Loros JJ, and Dunlap JC (2008). Closing the circadian negative feedback loop: FRQ-dependent clearance of WC-1 from the nucleus. Genes Dev 22, 3196–3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Chen S, Li S, Cha J, Long C, Li L, He Q, and Liu Y (2007). Protein kinase A and casein kinases mediate sequential phosphorylation events in the circadian negative feedback loop. Genes Dev 21, 3283–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JM, Dasgupta A, Emerson JM, Zhou X, Ringelberg CS, Knabe N, Lipzen AM, Lindquist EA, Daum CG, Barry KW, et al. (2014). Analysis of clock-regulated genes in Neurospora reveals widespread posttranscriptional control of metabolic potential. Proc Natl Acad Sci U S A 111, 16995–17002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EY, and Edery I (2006). Balance between DBT/CKIepsilon kinase and protein phosphatase activities regulate phosphorylation and stability of Drosophila CLOCK protein. Proc Natl Acad Sci U S A 103, 6178–6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Kwak PB, and Weitz CJ (2014). Specificity in circadian clock feedback from targeted reconstitution of the NuRD corepressor. Mol Cell 56, 738–748. [DOI] [PubMed] [Google Scholar]
- Kinoshita E, Kinoshita-Kikuta E, Takiyama K, and Koike T (2006). Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics 5, 749–757. [DOI] [PubMed] [Google Scholar]
- Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, and Takahashi JS (2012). Transcriptional Architecture and Chromatin Landscape of the Core Circadian Clock in Mammals. Science 338, 349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov RV, Chernov MV, Kondratova AA, Gorbacheva VY, Gudkov AV, and Antoch MP (2003). BMAL1-dependent circadian oscillation of nuclear CLOCK: posttranslational events induced by dimerization of transcriptional activators of the mammalian clock system. Genes Dev 17, 1921–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I, Lee J, Chang SH, Jung NC, Lee BJ, Son GH, Kim K, and Lee KH (2006). BMAL1 shuttling controls transactivation and degradation of the CLOCK/BMAL1 heterodimer. Mol Cell Biol 26, 7318–7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrondo LF, Olivares-Yanez C, Baker CL, Loros JJ, and Dunlap JC (2015). Circadian rhythms. Decoupling circadian clock protein turnover from circadian period determination. Science 347, 1257277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Jeong EH, Jeong HJ, Yildirim E, Vanselow JT, Ng F, Liu YX, Mahesh G, Kramer A, Hardin PE, et al. (2014). Phosphorylation of a Central Clock Transcription Factor Is Required for Thermal but Not Photic Entrainment. Plos Genetics 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden H, Ballario P, Arpaia G, and Macino G (1999). Seeing the light: News in Neurospora blue light signal transduction. Adv Genet 41, 35–54. [DOI] [PubMed] [Google Scholar]
- Lipton JO, Yuan ED, Boyle LM, Ebrahimi-Fakhari D, Kwiatkowski E, Nathan A, Guttler T, Davis F, Asara JM, and Sahin M (2015). The Circadian Protein BMAL1 Regulates Translation in Response to S6K1-Mediated Phosphorylation. Cell 161, 1138–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano AK, Zhou W, Santana JM, Kyriakides C, Velazquez H, and Sessa WC (2018). CLOCK phosphorylation by AKT regulates its nuclear accumulation and circadian gene expression in peripheral tissues. J Biol Chem 293, 9126–9136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura R, Tsuchiya Y, Tokuda I, Matsuo T, Sato M, Node K, Nishida E, and Akashi M (2014). The mammalian circadian clock protein period counteracts cryptochrome in phosphorylation dynamics of circadian locomotor output cycles kaput (CLOCK). J Biol Chem 269, 32064–32072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra A, Shi M, Baker CL, Colot HV, Loros JJ, and Dunlap JC (2009). A role for casein kinase 2 in the mechanism underlying circadian temperature compensation. Cell 137, 749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimamurthy R, Hunt SR, Lu Y, Fustin JM, Okamura H, Partch CL, Forger DB, Kim JK, and Virshup DM (2018). CK1delta/epsilon protein kinase primes the PER2 circadian phosphoswitch. Proc Natl Acad Sci U S A 115, 5986–5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan K, Robles MS, Westerling T, and Weitz CJ (2012). Feedback regulation of transcriptional termination by the mammalian circadian clock PERIOD complex. Science 337, 599–602. [DOI] [PubMed] [Google Scholar]
- Robles MS, Boyault C, Knutti D, Padmanabhan K, and Weitz CJ (2010). Identification of RACK1 and Protein Kinase C alpha as Integral Components of the Mammalian Circadian Clock. Science 327, 463–466. [DOI] [PubMed] [Google Scholar]
- Robles MS, Humphrey SJ, and Mann M (2017). Phosphorylation Is a Central Mechanism for Circadian Control of Metabolism and Physiology. Cell Metab 25, 118–127. [DOI] [PubMed] [Google Scholar]
- Sancar G, Sancar C, Brunner M, and Schafmeier T (2009). Activity of the circadian transcription factor White Collar Complex is modulated by phosphorylation of SP-motifs. FEBS Lett 583, 1833–1840. [DOI] [PubMed] [Google Scholar]
- Schafmeier T, Haase A, Kaldi K, Scholz J, Fuchs M, and Brunner M (2005). Transcriptional feedback of Neurospora circadian clock gene by phosphorylation-dependent inactivation of its transcription factor. Cell 122, 235–246. [DOI] [PubMed] [Google Scholar]
- Shi M, Collett M, Loros JJ, and Dunlap JC (2010). FRQ-interacting RNA helicase mediates negative and positive feedback in the Neurospora circadian clock. Genetics 184, 351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KM, Sancar G, Dekhang R, Sullivan CM, Li S, Tag AG, Sancar C, Bredeweg EL, Priest HD, McCormick RF, et al. (2010). Transcription factors in light and circadian clock signaling networks revealed by genomewide mapping of direct targets for neurospora white collar complex. Eukaryot Cell 9, 1549–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spengler ML, Kuropatwinski KK, Schumer M, and Antoch MP (2009). A serine cluster mediates BMAL1-dependent CLOCK phosphorylation and degradation. Cell Cycle 8, 4138–4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratmann M, Suter DM, Molina N, Naef F, and Schibler U (2012). Circadian Dbp transcription relies on highly dynamic BMAL1-CLOCK interaction with E boxes and requires the proteasome. Mol Cell 48, 277–287. [DOI] [PubMed] [Google Scholar]
- Tamaru T, Hirayama J, Isojima Y, Nagai K, Norioka S, Takamatsu K, and Sassone-Corsi P (2009). CK2alpha phosphorylates BMAL1 to regulate the mammalian clock. Nat Struct Mol Biol 16, 446–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang CT, Li S, Long C, Cha J, Huang G, Li L, Chen S, and Liu Y (2009). Setting the pace of the Neurospora circadian clock by multiple independent FRQ phosphorylation events. Proc Natl Acad Sci U S A 106, 10722–10727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tansey WP (2001). Transcriptional activation: risky business. Genes Dev 15, 1045–1050. [DOI] [PubMed] [Google Scholar]
- Wang B, Kettenbach AN, Gerber SA, Loros JJ, and Dunlap JC (2014). Neurospora WC-1 recruits SWI/SNF to remodel frequency and initiate a circadian cycle. PLoS Genet 10, e1004599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Zhou X, Loros JJ, and Dunlap JC (2015). Alternative use of DNA binding domains by the Neurospora White Collar Complex dictates circadian regulation and light responses. Molecular and Cellular Biology 36, 781–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F, Dollard C, and Ricuperohovasse SL (1995). Construction of a Set of Convenient Saccharomyces-Cerevisiae Strains That Are Isogenic to S288c. Yeast 11, 53–55. [DOI] [PubMed] [Google Scholar]
- Wong YH, Lee TY, Liang HK, Huang CM, Wang TY, Yang YH, Chu CH, Huang HD, Ko MT, and Hwang JK (2007). KinasePhos 2.0: a web server for identifying protein kinase-specific phosphorylation sites based on sequences and coupling patterns. Nucleic Acids Res 35, W588–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Haas W, Dephoure N, Huttlin EL, Zhai B, Sowa ME, and Gygi SP (2011). A large-scale method to measure absolute protein phosphorylation stoichiometries. Nat Methods 8, 677–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Gustafson CL, Sammons PJ, Khan SK, Parsley NC, Ramanathan C, Lee HW, Liu AC, and Partch CL (2015). Cryptochrome 1 regulates the circadian clock through dynamic interactions with the BMAL1 C terminus. Nature structural & molecular biology 22, 476–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, He Q, Cheng P, Wrage P, Yarden O, and Liu Y (2004). Distinct roles for PP1 and PP2A in the Neurospora circadian clock. Genes Dev 18, 255–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshitane H, Takao T, Satomi Y, Du NH, Okano T, and Fukada Y (2009). Roles of CLOCK phosphorylation in suppression of E-box-dependent transcription. Mol Cell Biol 29, 3675–3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Wang B, Emerson JM, Ringelberg CS, Gerber SA, Loros JJ, and Dunlap JC (2018). A HAD family phosphatase CSP-6 regulates the circadian output pathway in Neurospora crassa . PLoS Genet 14, e1007192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


