Abstract
A recent flurry of genetic studies in mice has provided key insights into how the somatosensory system is organized at a cellular level to encode itch, pain, temperature, and touch. These studies are largely predicated on the idea that functional cell types can be identified by their unique developmental provenance and gene expression profile. However, the extent to which gene expression profiles can be correlated with functional cell types and circuit organization remains an open question. In this review we focus on recent progress in characterizing the sensory afferent and dorsal horn neuron cell types that process cutaneous somatosensory information and ongoing circuit studies that are beginning to bridge the divide between cell type and function.
Introduction
Sensory neurons in the peripheral nervous system (PNS) play a central role in monitoring the internal and external state of the body. Of particular importance are the cutaneous exteroceptive modalities that animals use for reflex actions that prevent tissue injury, for the control of movement, and to elicit affective behaviors necessary for socialization and well-being [1]. These cutaneous modalities are encoded by specialized sensory afferent cell types that innervate the skin [2–4] and relay a wide range of noxious and innocuous information to the spinal and medullary dorsal horn, a key waystation for processing cutaneous somatosensory inputs as well as visceral sensory information. The PNS neurons that innervate the dorsal horn are highly heterogeneous with respect to their anatomy, electrophysiological properties, gene expression profiles and function. This heterogeneity is mirrored in the dorsal horn interneurons (INs) that they innervate. How this cellular diversity relates to gene expression, physiology and connectivity is still not fully understood, nor is it clear how cell diversity contributes to sensory coding and to function.
Sensory afferent heterogeneity: does the labeled line theory hold true after all?
Most of what we know about the cellular makeup of PNS comes from the analysis of skin and muscle afferents. Historically, skin afferents were classified by a handful of markers (e.g. Calcitonin gene-related peptide (CGRP), Isolectin B4 (IB4), Neurofilament (NF)) and their nerve conduction properties (Aβ, Aδ, C). This classification system, which provided limited resolution of individual sensory cell types, began to change with the discovery of various Transient receptor potential (Trp) channels (e.g. TrpV1, TrpA2, TrpM8) and G-protein coupled receptors (e.g. Mas-related G-protein coupled receptor D (Mrgprd), Mrgpra3) that are expressed in subsets of sensory neurons. More recently, single-cell RNA sequencing (scRNA-seq) approaches have been employed to analyze either the entire population of dorsal root ganglia (DRG) [5••], or specific subsets like trigeminal ganglia (TG) neurons [6], neurons innervating the leg [7,8] or the colon [9••] (Figure 1). Strikingly, the comparison of TG and DRG neurons revealed that despite major differences in their innervation targets, the molecularly defined neuronal types were remarkably similar, arguing that neural crest derived TG and DRG sensory cell types share a common developmental program.
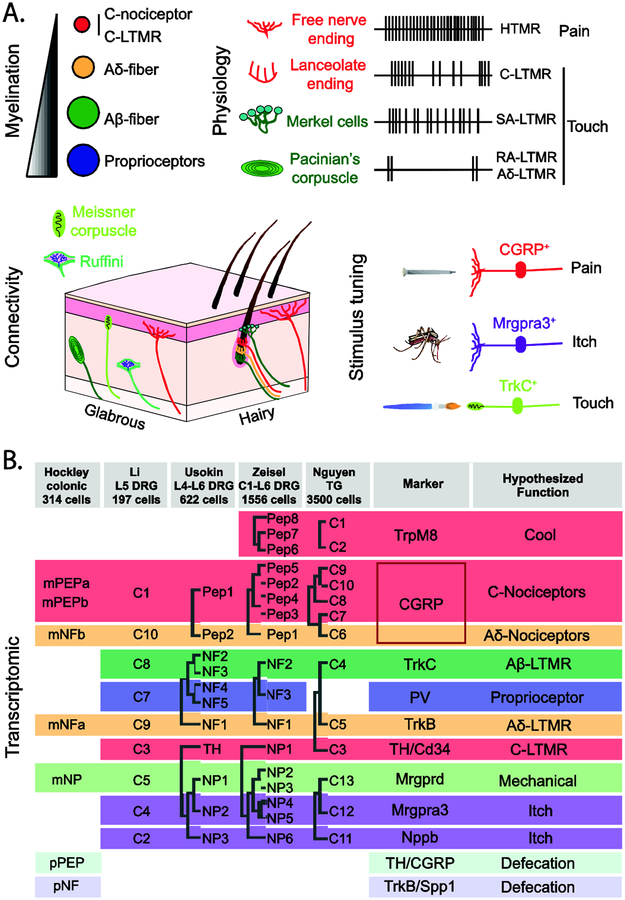
Figure 1. Transcriptomic analysis of sensory cell types.
A. Cutaneous sensory afferents are characterized by their myelination-conduction velocity profiles, firing patterns, connectivity and stimulus response.
B. Recent scRNA-seq studies [5••,6,7,8,9••] have identified distinct transcriptomic signatures for several types of sensory neurons, some of which are shared between cutaneous and visceral afferents.
ScRNA-seq has begun to resolve with greater precision the molecular composition of these sensory afferent populations. One example is the in-depth transcript analysis of nociceptive afferents that were traditionally subdivided into two broad classes based on their expression of peptidergic markers (e.g. CGRP) and IB4 binding. The TrpM8+ subset of sensory neurons expressing Tachykinin Precursor 1 (Tac1, also known as Substance P) but not CGRP segregate with a specific peptidergic population of primary afferents — the cool sensors [5••,6], whereas Ggta1, the alpha-galactosidase enzyme that confers IB4 binding, was found to be selectively expressed in two non-peptidergic neuron types characterized by the expression of Mrgprd and Mrgpra3 [5••]. Such analysis, and the accompanying online resources that allow access to the expression of ‘genes of interest’, promise further insights into sensory neuron diversity.
Transcriptome studies have revealed molecular signatures that are eithershared or unique to the exteroreceptive and interoreceptive sensory systems. For example, the colon is dually innervated by the pelvic and splanchnic nerves, with the majority of the colonic afferent subtypes represented in both nerves. Intriguingly, all of these so-called ‘mixed’ subtypes mapped onto defined transcriptomic populations previously found in the exteroceptive system, such as peptidergic afferents [9••]. Afferent populations that typically target the skin (itch afferents, CLTMRs and Aβ-LTMRs) were absent among colonic afferents, the notable exception being the Mrgprd+ subgroup of afferents that appear to be the cellular target of Htr4 antagonists, which are effective for the treatment of constipation [9••]. Colonic afferents also included a mysterious population that are transcriptomically related to an Aδ-LTMR population that innervates hair follicles. Since there are no hairs in the colon, the colonic counterparts must have a distinct function, perhaps acting as low-threshold mechanosensors to help coordinate motility and secretion. The profiling of colonic afferents also uncovered two populations that are exclusive to the pelvic nerve. These specialized afferents innervate the distal colon and may play specialized roles in urgency and defecation [9••].
The challenge is to correlate transcriptomic populations with well-established functional classes of sensory neurons. So far, the cell types defined by scRNA-seq paint a blurry picture, with transcriptomic profile and function aligned in a manner consistent with labeled-line transmission, while other cell types and modalities remain unmatched. For abundant afferents, such as peptidergic nociceptors, there are more molecular classes than currently described functional subtypes. These molecularly distinct populations may innervate different targets (e.g., vasculature, muscle, bone) or serve different functions in tissue homeostasis and repair. For rare afferents, functional diversity exceeds the number of distinct transcriptomic populations.
Spinal cord heterogeneity: insights from development
Rather less is known about cell type diversity in the spinal cord and how this relates to function, nor is it clear that sensory information is transmitted and gated within the spinal cord by “labeled lines”. So far, efforts to understand the cellular organization of sensory circuits in the spinal cord have centered on: a) developmentally regulated genes, including transcription factors that specify neuronal identity and function, or b) markers that are differentially expressed in the adult spinal cord, e.g. Protein Kinase C γ (PKCγ) and various neuropeptides. The neurons in the medulla and spinal dorsal horn are derived from progenitors that express the Lbx1 homeodomain transcription factor [10]. These Lbx1+ neurons can be further subdivided into inhibitory dI4/dILA and excitatory dI5/dILB neuron populations [11,12], the latter of which comprises a mix of local circuit INs and projection neurons [13] (Figure 2). The identification of key fate determinants: Tlx1/3, Lmx1b and Ascl1 (Mash1) for excitatory Lbx1+ neurons [14,15] and Ptf1α, Pax2, Lhx1/Lhx5 and Gbx½ for inhibitory Lbx1+ INs [16,17] prompted efforts to identify differentially expressed genes that molecularly parse these populations [14,18–20]. The genes identified in these screenings, which included the transcription factors Maf and nuclear orphan receptor ROR, and multiple neuropeptides, like Cholecystokinin (CCK), Somatostatin (Sst), Neuropeptide Y (NPY) and Dynorphin (Dyn), together with other identified developmental genes (e.g. Bhlhb5) revealed a complex yet incomplete picture of the molecular landscape of the dorsal horn [21–23•]. The scRNA-seq methodologies that can probe the molecular landscape with greater sensitivity are now providing a better measure of cell diversity in the dorsal horn [5••,24••,25••]. Nonetheless, there are still important issues that need to be addressed, including whether the transcriptomic signature is sufficient to identify bona fide functional cell types. Several complementary approaches are likely to be helpful in making determinations about cell type (Figure 2). The first is the use of scRNA-seq analysis to determine lineage relationship between cells in different clusters or within the same cluster to address whether they share a common developmental provenance. The second incorporates cellular features, such as morphology and electrophysiology. Understanding connectivity is also important, including the extent of common input and output within different clusters or within cells belonging to the same cluster. Finally, the functional characterization and interrogation of cell types is needed, which is now feasible using genetic models such as the mouse. Such studies are already underway and are beginning to reveal interesting relationships between molecular identity and function.
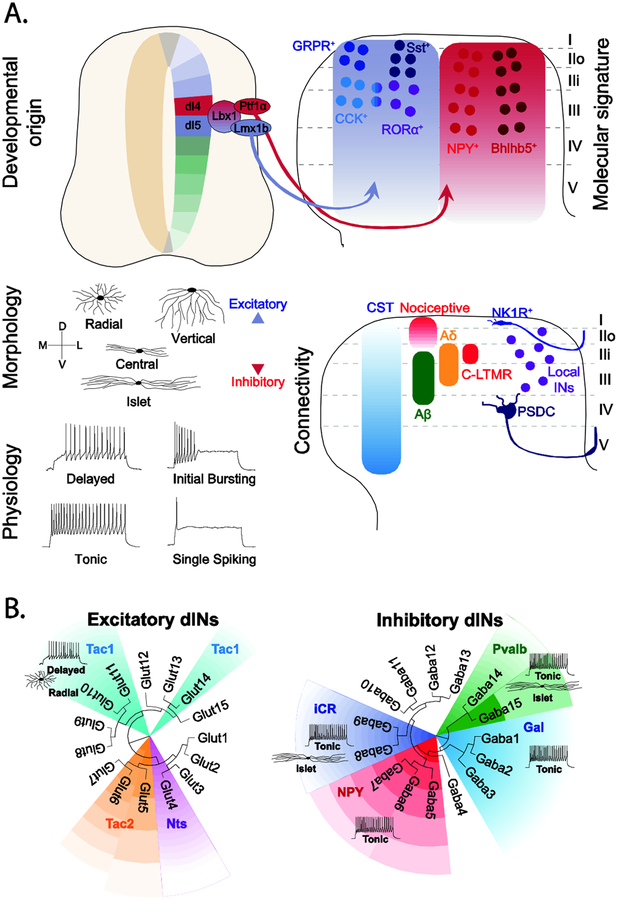
Figure 2. Dorsal horn neuron diversity.
A. Spinal cord cell types have been classified according to their developmental origin, expression of defined molecular markers, morphology, physiology and connectivity. Dorsal horn neurons that process and gate noxious and innocuous cutaneous sensory information arise from Lbx1+ dI4 and dI5 progenitors that are marked by the expression of Lbx1 and express several post-mitotic markers [10]. Dorsal horn neurons can also be classified according to their morphological and electrophysiological properties as exemplified by the classification of two neurochemically distinct neuron types: GRP+ and Tac1+ INs [48]. Neurons in the more superficial laminae, receive little corticospinal (CST) and strong noxious input, whereas neurons within the LTMR-RZ receive a unique mix of Aβ-, Aδ- and C-LTMR and CST input [35••]. Lamina position is also a determinant of identity, with NK1R+ projection neurons in lamina I contributing to the Spinothalamic Tract [52], and neurons within laminae III/IV being part of the Post-Synaptic Dorsal Column (PSDC) [35••].
B. ScRNA-seq analysis of dorsal horn neurons showing the transcriptomic clusters identified in Häring et al. [25••]) overlaid with known neurochemical markers, morphology and physiology [26•,28,30,48–51,53]. Tac1: Tachykinin 1, Tac2: Tachykinin 2, Nts: Neurotensin, iCR: inhibitory Calretinin, NPY: Neuropeptide Y, Pvab, Parvalbumin, Gal: Galanin.
Cellular diversity: form and function
Recent efforts to assess the contribution of molecularly defined cell types to somatosensation are indicative of functional specialization. This is perhaps best exemplified by the inhibitory IN cell types that gate itch, with the Bhlhb5+ and NPY∷Cre IN lineages inhibiting chemical and mechanical itch, respectively [21,26•]. Dyn-expressing Bhlhb5+ INs act to suppress chemical itch [23•,27•], whereas the NPY ∷ Cre INs specifically gate the light touch pathways necessary to drive mechanical itch [26•]. The story is however more complicated, with both IN populations also having roles in regulating pain [28,29•,30]. Mice in which the Dyn+ inhibitory INs have been ablated develop spontaneous mechanical allodynia [29•], which is consistent with these neurons gating Aβ inputs to dorsal horn neurons and inhibiting excitatory Sst+ neurons to transmit mechanical pain [29•,31]. There is also evidence that the NPY peptide is involved in gating pain, and by implication also the NPY+ INs [32,33]. This dual role in pain and itch suggest that both populations may comprise more specialized subsets of INs that have dedicated roles. The presence of such task-dependent specialization is best exemplified by RORβ+ inhibitory INs gating proprioceptive transmission during ongoing locomotion [34].
The excitatory neuron landscape is equally, if not more, complex, with descriptions of multiple molecular markers for different excitatory cell types [35••,36] and transcriptomic studies describing at least 10 excitatory dorsal IN clusters [5••,24••,25••]. Not surprisingly, multiple sensory functions can be attributed to broad populations of dorsal excitatory INs, with the conditional knockout of Tlx3 in Lbx1-derived dI5/dILB INs reducing sensitivity to itch, thermal sensitivity, static and dynamic touch [14]. However, this genetic manipulation results in the loss of several classes of excitatory INs including those that express Sst, Gastrin-Releasing Peptide Receptor (GRPR) and PKCγ, each of which might individually account for a subset of the observed somatosensory deficits [14]. Abraira et al. [35••] in comprehensively characterizing seven genetically defined excitatory IN populations in laminae IIi-IV that receive innocuous touch information, found a high degree of complexity with respect to their cellular properties, morphology and patterns of innervation. While some IN populations were relatively homogeneous, e.g. NeuroD4-derived INs, others such as the excitatory CCK+ INs displayed a range of morphologies and physiological properties. The molecular heterogeneity within the CCK+ IN population has further been confirmed by scRNA-seq studies [24••,25••]. Moreover, the CCK+ INs appear to contribute to multiple aspects of dynamic touch [35••,37•] raising the question as to whether the CCK+ IN population constitutes a single cell type with many functions, or comprises multiple cell types with more specialized functions.
Many aspects of the chemical itch pathway are consistent with labeled line transmission. Mrgpra3+ sensory neurons selectively transmit chloroquine-, BAM8–22- and SLIGRL-induced itch, whereas other chemical pruritogens, such as β-alanine and 5-HT are transmitted by Mrgprd and Nppb sensory neurons, respectively [5••]. Nonetheless, all three chemical itch pathways converge on dorsal horn GRPR+ INs that are essential and sufficient for chemical itch transmission [38,39]. However, another study has proposed a leaky gate model by which the intensity of GRP+ IN activation tips the balance for pain over itch [40•]. In contrast to chemical itch, pain or innocuous touch appeared to be encoded by a more distributed circuitry. Peptidergic and nonpeptidergic C-fiber afferents, as well as myelinated Aδ fibers, all contribute to the transduction of pain stimuli, with specific pain modalities often being transmitted by more than one fiber type. Moreover, Mrgprd+ polymodal nociceptive neurons innervate multiple excitatory cell types in lamina II, including radial, vertical and central cells [41]. Likewise multiple specialized LTMRs contribute to light touch sensitivity [2], which is not surprising given the somatosensory system’s capacity for a rich haptic representation of the external environment [1]. Indeed, LTMR inputs to excitatory INs in the LTMR-recipient zone (LTMR-RZ) are distributed across multiple genetically defined cell types [35••], with single excitatory INs receiving inputs from a plurality of LTMR types [35••,42•]. Ran et al. [43] in monitoring neuronal responses to heat and cold in the dorsal horn also observed widespread activation of neurons in response to noxious heat or noxious cold, with many INs responding to both stimuli. Taken together, these findings paint a more complex picture of sensory transmission in the dorsal horn, with multiple neuron types receiving and processing cutaneous inputs to the spinal cord. It is also consistent with the observation that five different excitatory spinal IN populations derived from dI5 INs – Vesicular Glutamate Transporter 3+ (VGluT3+), Sst+, CCK+, Calretinin+ (CR+) and PKCγ+ INs - contribute to the development of mechanical allodynia [29•,31,37•,44•,45•,46].
Is the genetic signature enough to define a functional cell type?
Ongoing functional studies in the mouse are beginning to yield insights into the relationship between genetically defined excitatory cell types and their roles in somatosensation. One such example is the analysis of the excitatory RORα+ INs in laminae IIi-III that are selectively innervated by LTMRs. The loss of dynamic touch sensitivity and the associated deficits in fine motor control following the ablation of RORα+ INs reflect their circuit connectivity in so far as they receive inputs from LTMRs and descending motor pathways, and they project onto spinal premotor INs and motor neurons [42•]. Interestingly, light touch sensitivity in these mice was not completely abolished, arguing other excitatory dorsal horn IN cell types contribute to light touch transmission. Candidates include the CCK+ INs that display impaired responses to cutaneous touch, either under physiological conditions or during allodynia [35••,37•]. However, given the 40% overlap in CCK and RORα expression [42•], the extent to which ablation of the CCK+ and RORα+ IN populations targets different neuron types is still an open question.
The diversity observed within the RORα+ and Sst+ INs demonstrates the limitations of defining cell types by the expression of a single gene. This complexity has ramifications for interpreting functional studies. The RORα+ INs are not homogeneous, both with respect to the markers they express (CCK, MafA and PKCγ) and their morphology (central, radial)[42•], raising the question of how this diversity relates to function and connectivity. Likewise, Sst expression encompasses multiple overlapping populations. Total RNA-seq analysis of the Sst+ INs indicates more than 13000 transcripts are uniquely expressed in this population [47]. These Sst+ INs are also phenotypically diverse, with ‘superficial’ Sst+ INs in lamina IIo being innervated by nociceptors, while ‘deep’ Sst+ INs in lamina IIi receive inputs from Aβ-, Aδ-, and C-LTMRs [29•]. Loss-of-function studies have shown the Sst+ INs are required for both acute mechanical pain and mechanical allodynia [29•], and it is tempting to speculate that these two functions are encoded by the superficial and deep populations, respectively. Untangling this heterogeneity will require more sophisticated intersectional approaches, with two or more molecular markers likely being necessary to label homogeneous and coherent subset of INs.
A further complication comes from how a “functional” cell type is defined, with neurons in many instances contributing to multiple circuits. Moreover, relating one cell type to a specific function has proved rather difficult due to the experimental manipulations and assays being used to assess cell type function. For example, optogenetic activation of the Sst+ INs increases histamine-induced scratching [31], which is mediated by the release of Sst and hyperpolarization of the Dyn-expressing Bhlbh5+ INs [27•]. This highlights the complication of interpreting functional changes when a neuropeptide is released in response to one stimulus and affects other modalities, as illustrated by the additional roles of Sst in itch and NPY in pain [27•,33]. Dynamic gene expression during development is another important nuance, with recombinase-dependent mouse reporter lines capturing the developmental history of gene expression and viral reporters that are introduced postnatally sometimes targeting a subset of these INs, resulting in different functional outcomes.
With regard to pain, there is also growing evidence that multiple IN cell types contribute to allodynia. One potential circuit might start with the activation of VGlut3+ INs in lamina III that in turn excite the more dorsal PKCγ+, CR+ and Sst+ IN populations [29•,44•,45•,46]. CCK+ INs that are recruited by descending corticospinal (CST) axons are also involved in generating mechanical allodynia [37•]. The extent to which these populations overlap has not been addressed nor is it known whether the proposed circuit for allodynia reflects multiple allodynic pathways or is the mere result of an oversampling of the same population by using different markers. Loss of Sst+ INs leads to the development of both static and dynamic allodynia, whereas loss of VGluT3+ INs, spares the static allodynia pathway, suggesting VGlut3+ INs might be a subset of Sst+ INs (these is indeed a 28% overlap between the two populations) [45•]. There are at least 3 types of Sst+ INs [29•], this diversity might reflect subpopulations that show overlap with CR, PKCγ, and VGlut3.
Outlook
ScRNA-seq has provided a new perspective on neuronal heterogeneity in the dorsal horn, and it has highlighted the insufficiency of a single gene to capture a specialized cell type, with many of the classical neuropeptide markers and calcium binding proteins being expressed in multiple cell types, as noted in the other neural structures including the hippocampus and cortex (CON this issue). Attempts to date to ascribe specific functions to the cell types identified by the scRNA-seq approach using activity dependent markers have shown promiscuous patterns of recruitment during different behaviors [24••,25••]. Thus, the combination of transcriptomic signature, morphological and physiological properties, connectivity, and laminar position is key to unravel the functional heterogeneity of these INs. There are now ongoing efforts to validate cell types using a combination of neurochemical markers, morphology and physiology [48–51,53] (Figure 2B). This, together with an understanding of their developmental origin constitutes the more holistic approach needed to bridge the gap between cell type and function.
A further challenge is to define the criteria for a consistent bioinformatic analysis of such extensive expression datasets. While transcriptomic analyses of peripheral sensory neurons reveals a high degree of similarity across studies [5••,6,7,8,9••], the picture for the spinal cord is more murky [5••,24••,25••]. Given that a consensus of spinal IN subtypes has not yet been reached and that many of the current genetic tools target somewhat heterogeneous populations, care is warranted in ascribing function to genetically defined IN classes.
Finally, in light of the complex integration of somatosensory input that occurs within the nervous system, there is a tremendous need for more sophisticated behavioral assays. For instance, many behavioral assays currently being used to evaluate cutaneous sensory responses have a binary endpoint (yes/no), with little attention to kinematics or motor sequence. In addition, most of these assays measure evoked responses rather than monitoring ongoing affective states. These limitations hightlight the need for new assays that will enable a more fine-grained interogation of the cell types that underlie somatosensation, both in health and in the pathological states of chronic pain and itch.
Highlights.
Single cell sequencing is elucidating neuronal heterogeneity within sensory circuits
Transcriptome, morphology, physiology and connectome underlie functional cell types
Distinct primary afferents types are tuned for distinct modalities of somatosensation
Understanding sensory integration in the spinal cord and beyond remains a major gap
Improved behavioral assays will facilitate the analysis of cell function and circuitry
Acknowledgements
Our work is supported by grants NIH grants NS0850586 and NS086372 to MG, and AR063772 and NS 096705 to Sarah Ross. Graziana Gatto was supported by an EMBO postdoctoralfellowship (ALTF 13–2015) and Salk Women in Science funding.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
Nothing to declare.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
References
- 1.McGlone F, Reilly D The cutaneous sensory system. Neurosci Biobehav Rev 34 (2010), pp. 148–159. [DOI] [PubMed] [Google Scholar]
- 2.Abraira VE, Ginty DD The Sensory Neurons of Touch. Neuron 79 (2013), pp. 618–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basbaum AI, Bautista DM, Scherrer G, Julius D Cellular and molecular mechanisms of pain. Cell 139 (2009), pp. 267–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong X, Dong X Peripheral and Central Mechanisms of Itch. Neuron 98 (2018), pp. 482–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5••.Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, Häring M, Braun E, Borm LE, La Manno G, et al. Molecular Architecture of the Mouse Nervous System. Cell 174 (2018), pp. 999–1014.e22.This massive sequencing effort encompasses cells throughout the nervous system including primary afferents and dorsal horn neurons, providing an updated view of diversity: http://mousebrain.org
- 6.Nguyen MQ, Wu Y, Bonilla LS, von Buchholtz LJ, Ryba NJP Diversity amongst trigeminal neurons revealed by high throughput single cell sequencing. PLoS One 12 (2017), pp. e0185543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li CL, Li KC, Wu D, Chen Y, Luo H, Zhao JR, Wang SS, Sun MM, Lu YJ, Zhong YQ, et al. Somatosensory neuron types identified by high-coverage single-cell RNA-sequencing and functional heterogeneity. Cell Res 26 (2016), pp. 83–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8•.Usoskin D, Furlan A, Islam S, Abdo H, Lönnerberg P, Lou D, Hjerling-Leffler J, Haeggström J, Kharchenko O, Kharchenko PV, et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci 18 (2015), pp. 145–153.This was the first scRNA-seq study of primary afferents, which is accompanied by online tools that allow researchers to investigate genes of interest https://linnarssonlab.org/dorsalhorn/
- 9••.Hockley JRF, Taylor TS, Callejo G, Wilbrey AL, Gutteridge A, Bach K, Winchester WJ, Bulmer DC, McMurray G, Smith ESJ Single-cell RNAseq reveals seven classes of colonic sensory neuron. Gut (2018), 10.1136/gutjnl-2017-315631.Gene expression profiles across colonic populations are available through an accompanying online resource http://hockley.shinyapps.io/ColonicRNAseq
- 10.Lai HC, Seal RP, Johnson JE Making sense out of spinal cord somatosensory development. Development 143 (2016), pp. 3434–3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Müller T, Brohmann H, Pierani A, Heppenstall PA, Lewin GR, Jessell TM, Birchmeier C The homeodomain factor lbx1 distinguishes two major programs of neuronal differentiation in the dorsal spinal cord. Neuron 34 (2002), pp. 551–562. [DOI] [PubMed] [Google Scholar]
- 12.Gross MK, Dottori M, Goulding M Lbx1 specifies somatosensory association interneurons in the dorsal spinal cord. Neuron 34 (2002), pp. 535–549. [DOI] [PubMed] [Google Scholar]
- 13.Szabo NE, da Silva RV, Sotocinal SG, Zeilhofer HU, Mogil JS, Kania A Hoxb8 Intersection Defines a Role for Lmx1b in Excitatory Dorsal Horn Neuron Development, Spinofugal Connectivity, and Nociception. J Neurosci 35 (2015), pp. 5233–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Y, Lopes C, Wende H, Guo Z, Cheng L, Birchmeier C, Ma Q Ontogeny of excitatory spinal neurons processing distinct somatic sensory modalities. J Neurosci 33 (2013), pp. 14738–14748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mizuguchi R, Kriks S, Cordes R, Gossler A, Ma Q, Goulding M Ascl1 and Gsh½ control inhibitory and excitatory cell fate in spinal sensory interneurons. Nat Neurosci 9 (2006), pp. 770–778. [DOI] [PubMed] [Google Scholar]
- 16.Huang M, Huang T, Xiang Y, Xie Z, Chen Y, Yan R, Xu J, Cheng L Ptf1a, Lbx1 and Pax2 coordinate glycinergic and peptidergic transmitter phenotypes in dorsal spinal inhibitory neurons. Dev Biol 322 (2008), pp. 394–405. [DOI] [PubMed] [Google Scholar]
- 17.Glasgow SM, Henke RM, Macdonald RJ, Wright CVE, Johnson JE Ptf1a determines GABAergic over glutamatergic neuronal cell fate in the spinal cord dorsal horn. Development 132 (2005), pp. 5461–5469. [DOI] [PubMed] [Google Scholar]
- 18.Bröhl D, Strehle M, Wende H, Hori K, Bormuth I, Nave KA, Müller T, Birchmeier C A transcriptional network coordinately determines transmitter and peptidergic fate in the dorsal spinal cord. Dev Biol 322 (2008), pp. 381–393. [DOI] [PubMed] [Google Scholar]
- 19.Hu J, Huang T, Li T, Guo Z, Cheng L c-Maf Is Required for the Development of Dorsal Horn Laminae III/IV Neurons and Mechanoreceptive DRG Axon Projections. J Neurosci 32 (2012), pp. 5362–5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wildner H, Das Gupta R, Brohl D, Heppenstall PA, Zeilhofer HU, Birchmeier C Genome-Wide Expression Analysis of Ptf1a- and Ascl1-Deficient Mice Reveals New Markers for Distinct Dorsal Horn Interneuron Populations Contributing to Nociceptive Reflex Plasticity. J Neurosci 33 (2013), pp. 7299–7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ross SE, Mardinly AR, McCord AE, Zurawski J, Cohen S, Jung C, Hu L, Mok SI, Shah A, Savner EM, et al. Loss of inhibitory interneurons in the dorsal spinal cord and elevated itch in Bhlhb5 mutant mice. Neuron 65 (2010), pp. 886–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ross SE, McCord AE, Jung C, Atan D, Mok SI, Hemberg M, Kim TK, Salogiannis J, Hu LL, Cohen S, et al. Bhlhb5 and Prdm8 form a repressor complex involved in neuronal circuit assembly. Neuron 73 (2012), pp. 292–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23•.Kardon AP, Polgár E, Hachisuka J, Snyder LM, Cameron D, Savage S, Cai X, Karnup S, Fan CR, Hemenway GM, et al. Dynorphin acts as a neuromodulator to inhibit itch in the dorsal horn of the spinal cord. Neuron 82 (2014), pp. 573–586.In this study, the authors identify dynorphin as a key neuromodulator of itch.
- 24••.Sathyamurthy A, Johnson KR, Matson KJE, Dobrott CI, Li L, Ryba AR, Bergman TB, Kelly MC, Kelley MW, Levine AJ Massively Parallel Single Nucleus Transcriptional Profiling Defines Spinal Cord Neurons and Their Activity during Behavior. Cell Rep 22 (2018), pp. 2216–2225.Comprehensive spinal neurons scRNA-seq studies characterizing the heterogeneity of dorsal and ventral IN populations. Supplemental Information include downloadable RNAseq datasets.
- 25••.Häring M, Zeisel A, Hochgerner H, Rinwa P, Jakobsson JET, Lönnerberg P, La Manno G, Sharma N, Borgius L, Kiehn O, et al. Neuronal atlas of the dorsal horn defines its architecture and links sensory input to transcriptional cell types. Nat Neurosci 21 (2018), pp. 869–880.This single-cell profiling study of dorsal horn neurons is accompanied by online tools that allow users to investigate genes of interest through a user-friendly interface https://linnarssonlab.org/dorsalhorn/
- 26•.Bourane S, Duan B, Koch SC, Dalet A, Britz O, Garcia-Campmany L, Kim E, Cheng L, Ghosh A, Ma Q, et al. Gate control of mechanical itch by a subpopulation of spinal cord interneurons. Science 350 (2015), pp. 550–554.Here the authors show that NPY+ INs gate mechanical but not chemical itch.
- 27•.Huang J, Polgár E, Solinski HJ, Mishra SK, Tseng PY, Iwagaki N, Boyle KA, Dickie AC, Kriegbaum MC, Wildner H, et al. Circuit dissection of the role of somatostatin in itch and pain. Nat Neurosci 21 (2018), pp. 707–716.This study proposes a differential role of Sst+ primary afferents and Sst+ dorsal INs in itch and pain.
- 28.Iwagaki N, Ganley RP, Dickie AC, Polgár E, Hughes DI, Del Rio P, Revina Y, Watanabe M, Todd AJ, Riddell JS A combined electrophysiological and morphological study of neuropeptide Y–expressing inhibitory interneurons in the spinal dorsal horn of the mouse. Pain 157 (2016), pp. 598–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29•.Duan B, Cheng L, Bourane S, Britz O, Padilla C, Garcia-Campmany L, Krashes M, Knowlton W, Velasquez T, Ren X, et al. Identification of Spinal Circuits Transmitting and Gating Mechanical Pain. Cell 159 (2014), pp. 1417–1432.In this comprehensive study the authors use selective ablation of several dorsal INs to identify key components of a circuit responsible for mechanical pain.
- 30.Chiang MC, Hachisuka J, Todd AJ, Ross Insight into SE B5-I spinal interneurons and their role in the inhibition of itch and pain. Pain 157 (2016), pp.:544–545. [DOI] [PubMed] [Google Scholar]
- 31.Christensen AJ, Iyer SM, François A, Vyas S, Ramakrishnan C, Vesuna S, Deisseroth K, Scherrer G, Delp SL In Vivo Interrogation of Spinal Mechanosensory Circuits. Cell Rep 17 (2016), pp. 1699–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Solway B, Bose SC, Corder G, Donahue RR, Taylor BK Tonic inhibition of chronic pain by neuropeptide Y. Proc Natl Acad Sci U S A 108 (2011), pp. 7224–7229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diaz-del Castillo M, Woldbye DPD, Heegaard AM Neuropeptide Y and its Involvement in Chronic Pain. Neuroscience 387 (2018), pp. 162–169. [DOI] [PubMed] [Google Scholar]
- 34.Koch SC, Del Barrio MG, Dalet A, Gatto G, Günther T, Zhang J, Seidler B, Saur D, Schüle R, Goulding M RORβ Spinal Interneurons Gate Sensory Transmission during Locomotion to Secure a Fluid Walking Gait. Neuron 96 (2017), pp. 1419–1431.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35••.Abraira VE, Kuehn ED, Chirila AM, Springel MW, Toliver AA, Zimmerman AL, Orefice LL, Boyle KA, Bai L, Song BJ, et al. The Cellular and Synaptic Architecture of the Mechanosensory Dorsal Horn. Cell 168 (2017), pp. 295–310.e19.In this study, the authors generate and characterize a broad array of genetic tools targeting populations of dorsal INs that are involved in the integration of tactile stimuli.
- 36.Del Barrio MG, Bourane S, Grossmann K, Schule R, Britsch S, O’Leary DD, Goulding M A transcription factor code defines nine sensory interneuron subtypes in the mechanosensory area of the spinal cord. PLoS One 8 (2013), pp. e77928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37•.Liu Y, Latremoliere A, Li X, Zhang Z, Chen M, Wang X, Fang C, Zhu J, Alexandre C, Gao Z, et al. Touch and tactile neuropathic pain sensitivity are set by corticospinal projections. Nature 561 (2018), pp. 547–550.In this study, authors nicely characterized how cortical input modulates the activity of spinal excitatory INs during physiological and allodynia conditions.
- 38.Sun YG, Chen ZF A gastrin-releasing peptide receptor mediates the itch sensation in the spinal cord. Nature 448 (2007), pp. 700–703. [DOI] [PubMed] [Google Scholar]
- 39.Mishra SK, Hoon MA The cells and circuitry for itch responses in mice. Science 340 (2013), pp. 968–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40•.Sun S, Xu Q, Guo C, Guan Y, Liu Q, Dong X Leaky Gate Model: Intensity-Dependent Coding of Pain and Itch in the Spinal Cord. Neuron 93 (2017), pp. 840–853.e5.Here the authors propose an intensity dependent coding of itch and pain through Grp+ INs.
- 41.Wang HH, Zylka MJ Mrgprd-Expressing Polymodal Nociceptive Neurons Innervate Most Known Classes of Substantia Gelatinosa Neurons. J Neurosci 29 (2009), pp. 13202–13209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42•.Bourane S, Grossmann KS, Britz O, Dalet A, Del Barrio MG, Stam FJ, Garcia-Campmany L, Koch SC, Goulding M Identification of a spinal circuit for light touch and fine motor control. Cell 160 (2015), pp. 503–515.In this study, the authors identify a selective population of INs which encodes tactile input key to elicit reflexive and corrective motor responses.
- 43.Ran C, Hoon MA, Chen X The coding of cutaneous temperature in the spinal cord. Nat Neurosci 19 (2016), pp. 1201–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44•.Peirs C, Williams SPG, Zhao X, Walsh CE, Gedeon JY, Cagle NE, Goldring AC, Hioki H, Liu Z, Marell PS, et al. Dorsal Horn Circuits for Persistent Mechanical Pain. Neuron 87 (2015), pp. 797–812.This study reveals that inflammatory and neuropathic injuries cause allodynia via distinct spinal circuits.
- 45•.Cheng L, Duan B, Huang T, Zhang Y, Chen Y, Britz O, Garcia-Campmany L, Ren X, Vong L, Lowell BB, et al. Identification of spinal circuits involved in touch-evoked dynamic mechanical pain. Nat Neurosci 20 (2017), pp. 804–814.In this study the authors show that a population of excitatory INs (VT3Lbx1) are required for touch-evoked dynamic allodynia.
- 46.Petitjean H, Pawlowski SA, Fraine SL, Sharif B, Hamad D, Fatima T, Berg J, Brown CM, Jan LY, Ribeiro-da-Silva A, et al. Dorsal Horn Parvalbumin Neurons Are Gate-Keepers of Touch-Evoked Pain after Nerve Injury. Cell Rep 13 (2015), pp. 1246–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chamessian A, Young M, Qadri Y, Berta T, Ji RR, Van de Ven T Transcriptional Profiling of Somatostatin Interneurons in the Spinal Dorsal Horn. Sci Rep 8 (2018), pp. 6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dickie AC, Bell AM, Iwagaki N, Polgár E, Gutierrez-Mecinas M, Kelly R, Lyon H, Turnbull K, West SJ, Etlin A, et al. Morphological and functional properties distinguish the substance P and gastrin-releasing peptide subsets of excitatory interneuron in the spinal cord dorsal horn. Pain (2018), 10.1097/j.pain.0000000000001406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gutierrez-Mecinas M, Furuta T, Watanabe M, Todd AJ A quantitative study of neurochemically defined excitatory interneuron populations in laminae I-III of the mouse spinal cord. Mol Pain 12 (2016), pp. 174480691662906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith KM, Boyle KA, Madden JF, Dickinson SA, Jobling P, Callister RJ, Hughes DI, Graham BA Functional heterogeneity of calretinin-expressing neurons in the mouse superficial dorsal horn: implications for spinal pain processing. J Physiol 593 (2015), pp. 4319–4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boyle KA, Gutierrez-Mecinas M, Polgár E, Mooney N, O’Connor E, Furuta T, Watanabe M, Todd AJ A quantitative study of neurochemically defined populations of inhibitory interneurons in the superficial dorsal horn of the mouse spinal cord. Neuroscience 363 (2017), pp. 120–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Todd AJ Neuronal circuitry for pain processing in the dorsal horn. Nat Rev Neurosci 11 (2010), pp. 823–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hughes DI, Sikander S, Kinnon CM, Boyle KA, Watanabe M, Callister RJ, Graham BA Morphological, neurochemical and electrophysiological features of parvalbumin-expressing cells: a likely source of axo-axonic inputs in the mouse spinal dorsal horn. J Physiol 590 (2012), pp. 3927–3951. [DOI] [PMC free article] [PubMed] [Google Scholar]