Abstract
Oxylipins are metabolites with a variety of biological functions. However, the biosynthetic pathway is widely unknown. It is considered that the first step is the oxygenation of polyunsaturated fatty acids like linoleic acid. Therefore, a lipoxygenase (LOX) from the edible basidiomycete Agrocybe aegerita was investigated. The AaeLOX4 was heterologously expressed in E. coli and purified via affinity chromatography and gel filtration. Biochemical properties and kinetic parameters of the purified AaeLOX4 were determined with linoleic acid and linolenic acid as substrates. The obtained Km, vmax and kcat values for linoleic acid were 295.5 μM, 16.5 μM · min-1 · mg-1 and 103.9 s-1, respectively. For linolenic acid Km, vmax and kcat values of 634.2 μM, 19.5 μM · min-1 · mg-1 and 18.3 s-1 were calculated. Maximum activities were observed at pH 7.5 and 25 °C. The main product of linoleic acid conversion was identified with normal-phase HPLC. This analysis revealed an explicit production of 13-hydroperoxy-9,11-octadecadienoic acid (13-HPOD). The experimental regio specificity is underpinned by the amino acid residues W384, F450, R594 and V635 considered relevant for regio specificity in LOX. In conclusion, HPLC-analysis and alignments revealed that AaeLOX4 is a 13-LOX.
Introduction
Oxylipins are lipoxygenase-derived metabolites with a variety of different structures and biological functions. These metabolites can be found in plants, mammals, bacteria, algae, mosses and fungi [1]. In mammals, they are involved in the regulation of immune response. In recent years, human lipid mediators like e.g. hepoxilins and trioxilins, which play an important role in tissue healing, organ protection, pain reduction and host defense received a lot of attention [2, 3].
In plants, they serve as signaling molecules, are involved in mediate stress response or in defense against pathogens, whereas in fungi they are associated with sporulation and the sexual as well as asexual life cycle [4, 5]. Lipoxygenases (LOX) are non-heme iron dependent dioxygenases which catalyze the insertion of molecular oxygen at a (1Z,4Z)-pentadiene motif, which occurs e.g. in polyunsaturated fatty acids (PUFAs), in a regio- and stereospecific manner [2, 3, 6, 7]. Additionally, fungal LOX with manganese instead of iron in the active site were also described [8, 9]. The oxidation of PUFAs leads to a reactive hydroperoxide and is considered to be the first step in the oxylipin-pathway. In higher fungi of the phyla Basidiomycota and Ascomycota, C18-PUFAs are predominantly present, whereas C20-PUFAs can be found only in very minor amounts [1]. This indicates that the oxygenation of linoleic-, linolenic- or arachidonic acid is the initial step in oxylipin-biosynthesis. Subsequent steps lead to a variety of oxylipins such as C8 volatile compounds which contribute to the significant flavor of mushroom. Among them, the most important and well known is oct-1-en-3-ol derived from linoleic acid [10]. The pathways, leading to oct-1-en-3-ol and other C8 volatile compounds are unknown. Several studies showed that fungi can convert linoleic acid into different oxylipins. It was found that the button mushroom Agaricus bisporus is able to produce oct-1-en-3-ol in several steps from linoleic acid. These steps include amongst others an oxygenation of linoleic acid to 10-hydroperoxyoctadecadienoic acid (10-HPOD). It was proposed that LOX generate the needed 10-HPOD, but up to now the corresponding enzyme yet has to be found [11, 12, 13]. Another study with A. bisporus revealed the conversion of linoleic acid into 8(R)-hydroxy-9Z,12Z-octadecadienoic acid and 8(R)-11(S)dihydroxy-9Z,12Z-octadecadienoic acid [14]. It also has been reported, that a mycelial pellet homogenate of an oyster mushroom Pleurotus pulmonaris produced oct-1-en-3-ol and the 10-oxo-acid from linoleic acid [15]. In general, studies on basidiomycetous LOX are scarce with only two LOX described so far [6, 10]. This shortcoming is addressed in this paper and a new LOX from the black poplar mushroom Agrocybe aegerita, also known to produce C8 volatile compounds [16, 17], is expressed, purified and characterized.
Materials and methods
Cloning and protein expression of AaeLOX4
The codon optimized AaeLOX4 gene (Aaelox4), whose original cDNA was deposited under the accession number MK451709, was commercially purchased and cloned into the plasmid pET28a (BioCat GmbH, Heidelberg, Germany). For protein expression, pET28a/AaeLOX4 plasmid was transformed into E. coli BL21(DE3) Gold. Recombinant E. coli cells were cultivated in Terrific Broth medium (TB) containing 12 g tryptone, 24 g yeast extract and 5 g glycerol, supplemented with 50 mg·L-1 kanamycin as selection marker, at 37 °C until an OD600 of 0.5 was reached. Expression was induced by adding isopropyl-β-d-thiogalactopyranoside to a final concentration of 0.5 mM. Expression temperature was reduced to 24 °C and cultivated for another 24 h. Cells were harvested by centrifugation (4,000 g, 30 min, 4 °C) and stored at -20 °C until further use.
Protein purification
The cell pellet was thawed on ice and resuspended in lysis-buffer (50 mM phosphate, 300 mM NaCl, pH 7.5). Disruption of cells was carried out by sonification (3 cycles for 60 s on, 60 s rest) on ice using a sonifier (Bandelin Sonopuls, Berlin, Germany). After complete disruption, cell debris was removed by centrifugation (14,000 g, 30 min, 4 °C). The resulting supernatant was loaded twice on a 5 mL HisTrap column (GE-Healthcare), washed with 5 column volumes lysis-buffer containing 25 mM imidazole and eluted with 5 column volumes with the same buffer containing 500 mM imidazole. Gel filtration was performed on a Superdex 200 16/60 (GE Healthcare) using a buffer containing 50 mM Tris(hydroxymethyl)amino-methane (Tris/HCl), 300 mM NaCl, 10% (v/v) glycerol, pH 7.5 and a flow rate of 0.2 mL·min-1. Elution was collected in 2 mL fractions and analyzed via SDS-PAGE. Fractions with purified Aae -LOX4 were used for further analysis.
Lipoxygenase assay
The substrate solution for the LOX assay was prepared as follows. 10 μL linoleic acid or linolenic acid was added to 915 μL ddH2O containing 15 μL Tween 20 and 60 μL 1M NaOH. Aliquots were stored at -20 °C and thawed before use. LOX activity was determined by recording the formation of the conjugated double bond at 234 nm (ε = 25,000 M-1·cm-1) on a Nanophotometer (Implen, Munich, Germany). Typical reaction mixture contained 0.5 mM linoleic acid, 20 μL enzyme solution and 50 mM phosphate buffer, pH 7.5 to a final volume of 1 mL.
Determination of pH- and temperature-optimum
For the determination of the pH-optimum, three different buffers were used: 50 mM acetate buffer, pH 4.5–6; 50 mM phosphate buffer, pH 6.5–8 and 50 mM borate buffer, pH 8.5–10. Effects of the temperature were determined by incubating the reaction mixture at different temperatures, ranging from 15 °C– 65 °C.
Kinetic parameters
Michaelis-Menten kinetics were calculated by incubating purified AaeLOX4 with different linoleic acid or linolenic acid concentrations ranging from 0.025 mM– 2 mM. All measurements were carried out in 50 mM phosphate buffer at pH 7.5 and 25 °C. Initial formation rates of the conjugated double bond were recorded at 234 nm in triplicates and consulted for calculating Km, vmax and kcat.
Product analysis
Product specificity of AaeLOX4 was determined by incubating 1 mM linoleic acid, 20 μL AaeLOX4 and 50 mM phosphate buffer, pH 7.5 in a final volume of 1 mL. Reaction progress was followed by recording the formation of the conjugated double bond at 234 nm. Incubations were stopped when no further increase in extinction was detectable. The resulting hydroperoxy linoleic acids were extracted by adding 1 mL n-hexane. For HPLC-analysis, the extracts were dried under a continuous N2 stream and dissolved in n-hexane/2-propanol/acetic acid (100/5/1, v/v/v). Normal-phase-HPLC was carried out on a Nucleodur SiOH 100–5 column (Macherey-Nagel; 4.6x250 mm, 5 μm particle size) with an isocratic eluent system of n-hexane/2-propanol/acetic acid (100/5/1, v/v/v) at a flow rate of 1 mL·min-1. Elution was followed at 234 nm for hydroperoxy fatty acids and 210 nm for unsaturated fatty acids. Product specificity was evaluated by comparison with 13-HPOD and 9-HPOD standards prepared with commercial soybean LOX-1 [10]. Control experiments were carried out like described above without adding enzyme to the reaction mixture.
Results
Lipoxygenases of the black poplar mushroom Agrocybe aegerita
In the recently sequenced genome of Agrocybe aegerita (also known as Cyclocybe aegerita and Pholiota aegerita), five genes coding for putative LOX have been annotated and labelled as LOX1 (AAE3_00896), LOX2 (AAE3_01552), LOX3 (AAE3_09652), LOX4 (AAE3_04864) and LOX5 (AAE3_07753) ([18], www.thines-lab.senckenberg.de/agrocybe_genome). Deduced amino acid sequences of A. aegerita LOX genes and already characterized fungal LOX from Ascomycota (Fusarium oxysporum and Gaeumannomyces graminis) and Basidiomycota (Pleurotus sapidus and Pleurotus ostreatus) were used for phylogenetic analysis (http://www.phylogeny.fr/ using default parameters). LOX separated into two distinct sections with the characterized LOX from G. graminis [19] on one side and all basidiomycetous LOX as well as the characterized LOX from F. oxysporum [20] on the other side (S1 Fig). The latter group split into three parts: i) a large cluster containing both characterized LOXs from the Pleurotus species and four putative LOXs from A. aegerita (LOX1, LOX2, LOX4, LOX5), ii) LOX3 from A. aegerita and iii) the LOX from F. oxysporum.
Heterologous expression and purification
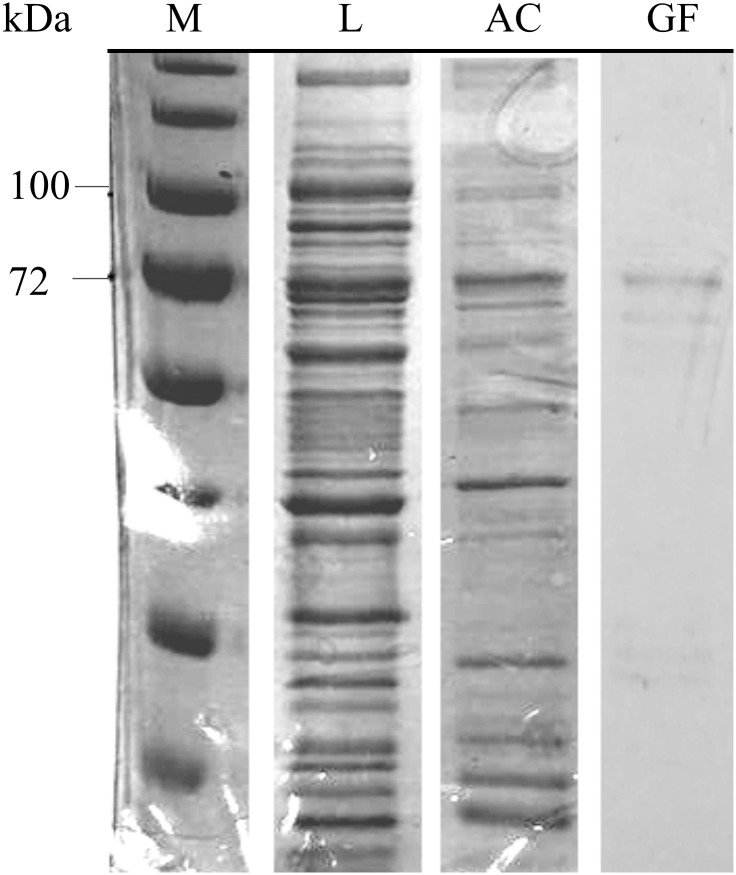
Active and soluble AaeLOX4 was obtained from cultures of E. coli BL21(DE3) Gold carrying the pET28a/AaeLOX4 plasmid. SDS-PAGE analysis of cell lysate indicated that AaeLOX4 is present in the soluble lysate fraction (Fig 1). Thus, the cell lysate was directly applied onto an affinity chromatographic column, which resulted in the removal of crude impurities. Subsequent gel filtration led to pure AaeLOX4 (Fig 1). The main band had an estimated molecular weight of around 80 kDa confirming the predicted molecular weight of the amino acid sequence of 79 kDa. Overall, a 1,351-fold purification yield was obtained (Table 1).
Fig 1. SDS-PAGE analysis of AaeLOX4 expression and purification.
M = marker, L = cleared lysate, AC = Purification after affinity chromatography, GF = Purified enzyme after gel filtration.
Table 1. Summary of the purification steps of AaeLOX4 in E. coli.
| Step | Total protein (mg) |
Specific activity (U · mg-1) |
Purification (fold) |
|---|---|---|---|
| Crude extract | 146.97 | 0.038 | 1.0 |
| Affinity chromatography | 37.16 | 0.329 | 8.6 |
| Gel filtration | 0.09 | 51.34 | 1351.1 |
LOX biochemical properties and characteristics
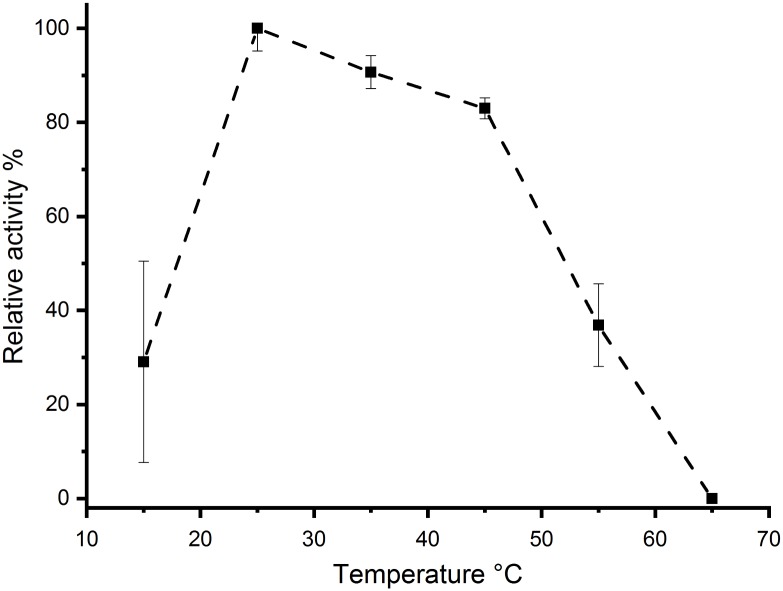
The activity of AaeLOX4 was determined in a temperature range between 15 °C and 65 °C. While the relative activity from 25 °C to 45 °C resulted in a steady loss of about one-fifth of activity, a further temperature increase to 55 °C caused a dramatic loss in activity and no activity could be detected at 65 °C. At the lowest analyzed temperature of 15 °C, one-third of the maximal LOX activity remained (Fig 2).
Fig 2. Effects of the temperature on AaeLOX4 activity.
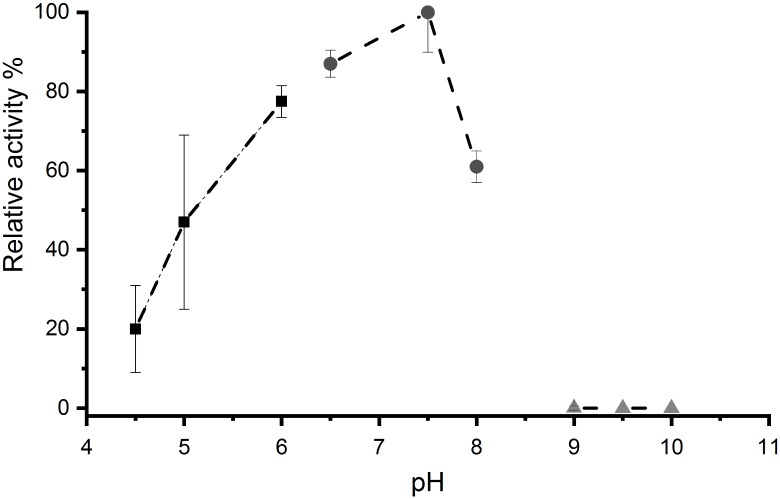
With increasing pH of the buffer system used for LOX activity assays, a steady activity enhancement is obtained reaching its maximum at pH 7.5. Further increase of the pH showed a drastic activity loss with no detectable activity from pH 9 and above. In the acidic environment, AaeLOX4 showed more tolerable activity properties. Decreasing the pH from 7.5 to 6, resulted in a loss of about 20% activity. Nevertheless, at pH 5.0 the relative activity decreased to 50%, while 20% relative activity was left at pH 4.5 (Fig 3).
Fig 3. Effects of pH on AaeLOX4 activity.
50 mM acetate buffer (squares), 50 mM phosphate buffer (circles), 50 mM borate buffer (triangles).
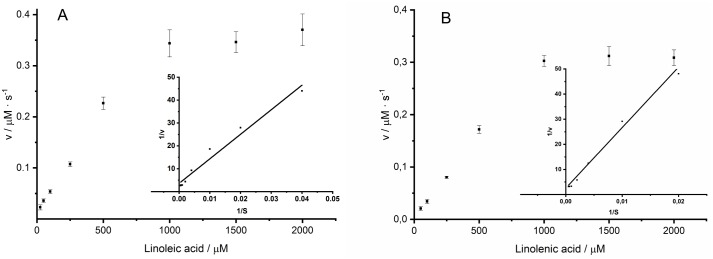
Michaelis-Menten parameters were deduced on basis of the Lineweaver-Burk plot depicting AaeLOX4 activity with linoleic acid or linolenic acid concentrations between 0.025–2 mM (Fig 4). The obtained Km, vmax and kcat values for linoleic acid were 295.5 μM, 16.5 μM min-1 mg-1 and 103.9 s-1, respectively. For linolenic acid as substrate Km, vmax and kcat values of 634.2 μM, 19.5 μM · min-1 · mg-1 and 18.3 s-1 were calculated (Table 2). Substrate specificity of AaeLOX4 was estimated by comparing the relative activities towards different PUFAs (Table 3). The enzyme showed the highest specificity for linoleic acid and a high relative activity for linolenic acid (88.8%). However, arachidonic acid showed the lowest relative activity with 7.3%.
Fig 4. Velocity of AaeLOX4 dependent on linoleic acid (A) and linolenic acid (B) concentrations.
Values represent the means of triplicates and their standard deviation. Inset represents Lineweaver-Burk plot.
Table 2. Kinetic parameters of AaeLOX4.
| Substrate | Km / μM | vmax / μM · min-1 · mg-1 | kcat / s-1 |
|---|---|---|---|
| linoleic acid | 295.5 | 16.5 | 103.9 |
| linolenic acid | 634.2 | 19.5 | 18.3 |
Table 3. Substrate specificity of AaeLOX4.
| Substrate | Relative activity / % |
|---|---|
| linoleic acid | 100 |
| linolenic acid | 88.8 |
| arachidonic acid | 7.3 |
HPLC-analysis of products
Specificity of AaeLOX4 was analyzed by normal-phase HPLC. The elution profile of AaeLOX4 shows two major products which elute at 3.12 min and 3.81 min (Fig 5). 13-Z,E-HPOD and 13-E,E-HPOD as well as 9-Z,E-HPOD and 9-E,E-HPOD have been prepared with soybean LOX-1 according to Kuribayashi et al. [10]. 13-Z,E-HPOD and 13-E,E-HPOD elute at 3.33 min and 3.79 min, respectively, whereas the isomeric 9-HPOD mixture elute as one peak at 5.79 min. The negative control showed no peaks at these retention times. The comparison between AaeLOX4 products and soybean LOX-1 products revealed that AaeLOX4 produces highly specific 13-HPOD. A production of 9-HPOD by AaeLOX4 was not detectable and ruled out by comparing the UV-spectra of 9-HPOD eluting at 5.79 min from soybean LOX-1 with peaks in this range in the elution profile of AaeLOX4 products.
Fig 5. NP-HPLC analysis of the oxygenated products of linoleic acid.
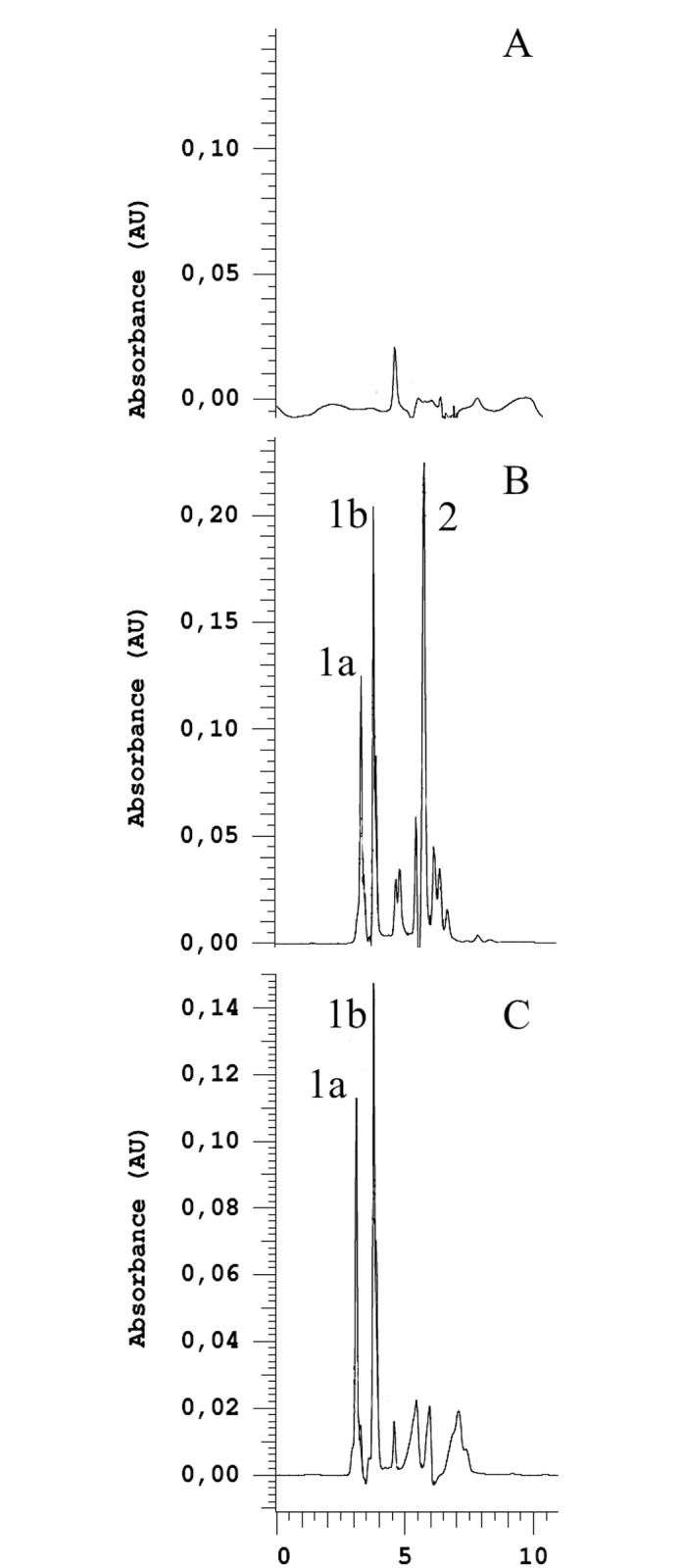
Chromatogram was recorded at 234 nm. (A) negative control. Extract of the reaction mixture lacking enzyme. (B) Extract of the reaction products from soybean LOX-1. (C) Extract of the reaction products from AaeLOX4. (1a) 13-Z,E-HPOD, (1b) 13-E,E-HPOD, (2) 9-Z,E-HPOD + 9-E,E-HPOD.
Discussion
In this study, we were able to express a soluble and active AaeLOX4 from A. aegerita in E. coli. As far as we know, this is the second basidiomycetous LOX recombinantly expressed in E. coli. While Plagemann et al. [5] used an approach with co-expression of chaperones to accomplish a soluble LOX expression from cDNA of P. sapidus, we reached the soluble expression with a codon optimized gene. Affinity chromatography with following gel filtration led to a highly purified protein in sufficient concentrations for further characterization experiments. The observed influence of temperature and pH on AaeLOX4 with its maximum activity at 25 °C and pH 7.5 is in accordance with LOXs from other fungi, such as a wildtype LOX from P. ostreatus, having its maximum activity at 25 °C and pH values between 7.5 and 8.0 [10]. However, the recombinant LOX from P. sapidus showed a temperature range with highest activities ranging from 25 °C to 35 °C. The optimal pH value of P. sapidus LOX was 7.0 and in contrast to AaeLOX4 showed activity up to a pH of 9.5 [5]. Interestingly, AaeLOX4 showed differences in the kinetic parameters for linoleic acid compared to other LOXs from Basidiomycota. The Km values for the LOXs from P. sapidus and from P. ostreatus are 40.3 μM and 130 μM, which are both lower than our calculated Km of 295.5 μM. On the other hand, the obtained vmax of 16.5 μM min-1 mg-1 is comparable with the LOX from P. ostreatus (23.4 μM min-1 mg-1) and the kcat value of 103.9 s-1 is comparable with the LOX from P. sapidus (157 s-1) [5, 10]. Analogous Km and vmax values were also found for a LOX in oriental melon (230.3 μM and 15.25 μM min-1 mg-1) [21]. Interestingly, a LOX originating the bacteria Myxococcus xanthus shows comparable Km values for linoleic acid (380 μM) but drastically lower Km with linolenic acid and other C20-PUFAs like arachidonic acid or eicosapentaenoic acid [6]. Soybean LOX-1 shows a lower Km of 150 μM but shares a comparable vmax of 13.5 μM min-1 mg-1 [22]. The catalytic efficiency of AaeLOX4 was higher with linoleic acid than with linolenic acid. This finding reflects the estimated relative activities towards different PUFAs which revealed a preference for linoleic acid. High activity was also found for linolenic acid, whereas arachidonic acid showed poor activity. These results are comparable with the LOXs from P. ostreatus and F. oxysporum [10, 20]. The analysis of product specificity revealed that 13-HPOD (13-Z,E-HPOD and 13-E,E-HPOD) was the only detectable product and thus, AaeLOX4 can be considered as a 13-LOX. Equally, LOXs from P. sapidus, P. ostreatus, F. oxysporum and G. graminis showed a specific conversion of linoleic acid to 13-HPOD [5, 10, 19, 20]. Nevertheless in contrast to AaeLOX4, LOXs from P. sapidus, P. ostreatus and F. oxysporum produced minor amounts of 9-HPOD. Whereas the LOX from G. graminis showed no significant synthesis of 9-HPOD [19]. This finding was confirmed with amino acid sequence alignments which showed that various determinants responsible for the regio- and stereo-specificity can also be found in AaeLOX4 (S2 Fig). Three positions, which are responsible for the depth of substrate penetration and thus are responsible for regio-specificity, have been described for mammalian LOX [23, 24]. While space-filling amino acids at these positions lead to oxygenation near the methyl end, enlargement of the active site favors oxygenations towards the carboxy end [24]. Applying this concept on AaeLOX4, the bulky amino acids at the crucial positions (W384, F450 and V635) are present in the amino acid sequence of AaeLOX4 and result in the transformation of linoleic acid to 13-HPOD (Fig 5). Also the LOXs from P. ostreatus, P. sapidus and F. oxysporum led to a specific production of 13-HPOD [5, 10, 20]. Consistently, all these LOXs share the same bulky amino acids at the crucial positions [23, 24]. According to Andreou et al. [25, 26] and Hornung et al. [27], regio-specificity is mainly determined by the orientation of the substrate. Based on their model, substrate orientation with the methyl end first leads to an oxygenation at C-13. The invers orientation of the substrate therefore is proposed to oxygenate C-9. Additionally, reduced space in the active site is also considered to favor a substrate orientation with its methyl end towards the active site. This model involves the importance of a positive charged amino acid at the end of the active site with no interference from another amino acid masking the positive charge to stabilize the carboxy end of the substrate. On the one hand the LOXs from P. ostreatus, P. sapidus and F. oxysporum share a lysine, considered to stabilize the negative charged carboxy end of the substrate and therefore differ in their amino acid sequence compared to AaeLOX4 where this lysine is replaced by an arginine which could of course fulfil the same purpose. On the other hand, the alignment shows that the LOXs from P. ostreatus, P. sapidus, F. oxysporum and AaeLOX4 share a phenylalanine near the positive charged amino acids lysine or arginine which masks the charge and therefore leads to a substrate orientation with the methyl end penetrating the active site leading to a region-specific oxygenation at C-13 [25, 26, 27]. Phylogenetic analysis shows that the fungal LOXs cluster in different groups. Whether LOXs of the same group harbor similar enzymatic properties is an important question, which can help to depict distinct enzyme function. This assumption is reasonable by comparing the Km values for linoleic acid LOXs in the phylogenetic tree. FoxLOX and GgrLOX, show low Km values of 4.4 and 6.9 μM while the subgroup of PosLOX and PsaLOX show distinctly higher Km values of 130 and 40.3 μM, respectively. The characterized AaeLOX4, which does not cluster with other subgroups of characterized LOX showed an even higher Km value of 295.5.
Supporting information
Aae—Agrocybe aegerita, Fox—Fusarium oxysporum, Ggr—Gaeumannomyces graminis, Pos—Pleurotus ostreatus, Psa—Pleurotus sapidus; AaeLOX1, AaeLOX2, AaeLOX3, AaeLOX4 (MK451709), AaeLOX5, FoxLOX (KNB01601), GgrLOX (AAK81882), PsaLOX (CCV01580), PosLOX (CCV01578).
(TIF)
Agrocybe aegerita AaeLOX1, AaeLOX2, AaeLOX3, AaeLOX4, AaeLOX5, Pleurotus sapidus PsaLOX, Pleurotus ostreatus PosLOX, Fusarium oxysporum FoxLOX, Gaeumannomyces graminis GgrLOX. Alignment was carried out by using Clustal Omega with default parameters. The highlighted amino acid residues involved in ligand binding are indicated as “L”. Stereo specificity related amino acids are indicated as “B” [24], “H” [27] and “S” [23].
(TIF)
Acknowledgments
We would like to thank the anonymous reviewer for providing valuable feedback that helped to improve the paper.
Data Availability
Gene sequence for AaeLOX4 is available from the GenBank database under the accession number MK451709.
Funding Statement
The grant RU 2137/1 from the Deutsche Forschungsgemeinschaft (http://www.dfg.de/en/) was awarded to MR. DK and MR used facilities of the LOEWE Center for Insect Biotechnology & Bioresources financed by the Hessen State Ministry of Higher Education, Research and the Arts (http://www.proloewe.de/en/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Brodhun F, Feussner I. Oxylipins in fungi. FEBS J. 2011;278(7):1047–1063. 10.1111/j.1742-4658.2011.08027.x [DOI] [PubMed] [Google Scholar]
- 2.An JU, Song YS, Kim KR, Ki YJ, Yoon DY, Oh DK. Biotransformation of polyunsaturated fatty acids to bioactive hepoxilins and trioxilins by microbial enzymes. Nat Commun. 2018;9(1):128 10.1038/s41467-017-02543-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee IG, An JU, Ko YJ, Park JB, Oh DK. Enzymatic synthesis of new hepoxilins and trioxilins from polyunsaturated fatty acids. Green Chem. 2019. 10.1039/c9gc01031a [DOI] [Google Scholar]
- 4.Kock JLF, Venter P, Linke D, Schewe T, Nigam S. Biological dynamics and distribution of 3-hydroxy fatty acids in the yeast Dipodascopsis uninucleata as investigated by immunofluorescence microscopy. Evidence for a putative regulatory role in the sexual reproductive cycle 1. FEBS Lett. 1998;427(3):345–348. [DOI] [PubMed] [Google Scholar]
- 5.Plagemann I, Zelena K, Arendt P, Ringel PD, Krings U, Berger R. LOXPsa1, the first recombinant lipoxygenase from a basidiomycete fungus. J Mol Catal B: Enzym. 2013;87(1):99–104. [Google Scholar]
- 6.An JU, Hong SH, Oh DK, Regiospecificity of a novel bacterial lipoxygenase from Myxococcus xanthus for polyunsaturated fatty acids. Biochim Biophys Acta, Mol Cell Biol Lipids. 2018;8(8):823–833. [DOI] [PubMed] [Google Scholar]
- 7.Liavonchanka A, Feussner I. Lipoxygenases: occurrence, functions and catalysis. J Plant Physiol. 2006;163(3):348–357. 10.1016/j.jplph.2005.11.006 [DOI] [PubMed] [Google Scholar]
- 8.Hamberg M, Su C, Oliw E. Manganese Lipoxygenase. Discovery of a bis-allylic hydroperoxide as product and intermediate in a lipoxygenase reaction. J Biol Chem. 1998;273(21):13080–13088. 10.1074/jbc.273.21.13080 [DOI] [PubMed] [Google Scholar]
- 9.Oliw EH, Jernerén F, Hoffmann I, Sahlin M, Garscha U. Manganese lipoxygenase oxidizes bis-allylic hydroperoxides and octadecenoic acids by different mechanisms. Biochim Biophys Acta. 2011;1811(3):138–147. 10.1016/j.bbalip.2010.12.002 [DOI] [PubMed] [Google Scholar]
- 10.Kuribayashi T, Kaise H, Uno C, Hara T, Hayakawa T, Joh T. Purification and Characterization of Lipoxygenase from Pleurotus ostreatus. J Agric Food Chem. 2002;50(5):1247–1253. [DOI] [PubMed] [Google Scholar]
- 11.Wurzenberger M, Grosch W. The enzymic oxidative breakdown of linoleic acid in mushrooms (Psalliota bispora). Z Lebensm Unters Forsch. 1982;175(3):186–190. [Google Scholar]
- 12.Wurzenberger M, Grosch W. Origin of the oxygen in the products of the enzymatic cleavage reaction of linoleic acid to 1-octen-3-ol and 10-oxo-trans-8-decenoic acid in mushrooms (Psalliota bispora). Biochim Biophys Acta. 1984;794(1):18–24. [Google Scholar]
- 13.Wurzenberger M, Grosch W. Stereochemistry of the cleavage of the 10-hydroperoxide isomer of linoleic acid to 1-octen-3-ol by a hydroperoxide lyase from mushrooms (Psalliota bispora). Biochim Biophys Acta. 1984;795(1):163–165. [Google Scholar]
- 14.Wadman M, van Zadelhoff G, Hamberg M, Visser T, Veldink G, Vliegenthart JG. Conversion of linoleic acid into novel oxylipins by the mushroom Agaricus bisporus. Lipids. 2005;40(11):1163–1170. [DOI] [PubMed] [Google Scholar]
- 15.Assaf S, Hadar Y, Dosoretz C. Biosynthesis of 13-hydroperoxylinoleate, 10-oxo-8-decenoic acid and 1-octen-3-ol from linoleic acid by a mycelial-pellet homogenate of Pleurotus pulmonarius. J Agric Food Chem. 1995;43(8):2173–2178. [Google Scholar]
- 16.Rapior S, Breheret S, Talou T, Pélissier Y, Milhau M, Bessière JM. Volatile Components of Fresh Agrocybe aegerita and Tricholoma sulfureum. Cryptogam Mycol. 1998;19(1):15–23. [Google Scholar]
- 17.Kleofas V, Sommer L, Fraatz MA, Zorn H, Rühl M . Fruiting body production and aroma profile analysis of Agrocybe aegerita cultivated on different substrates. Nat Resour. 2014;5(6):233–240. [Google Scholar]
- 18.Gupta DK, Rühl M, Mishra B, Kleofas V, Hofrichter M, Herzog R, et al. The genome sequence of the commercially cultivated mushroom Agrocybe aegerita reveals a conserved repertoire of fruiting-related genes and a versatile suite of biopolymer-degrading enzymes. BMC Genomics. 2018;19(1):48 10.1186/s12864-017-4430-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su C, Oliw E. Manganese Lipoxygenase. Purification and Characterization. J Biol Chem. 1998;273(21):13072–13079. 10.1074/jbc.273.21.13072 [DOI] [PubMed] [Google Scholar]
- 20.Brodhun F, Cristobal-Sarramian A, Zabel S, Newie J, Hamberg M, Feussner I. An 13S-Lipoxygenase with an α-Linolenic Acid Specific Hydroperoxidase Activity from Fusarium oxysporum. PLoS ONE. 2013;8(5):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao S, Chen H, Zhang C, Tang Y, Liu J, Qi H. Heterologous Expression and Biochemical Characterization of Two Lipoxygenases in Oriental Melon, Cucumis melo var. makuwa Makino. PLoS ONE. 2016;11(4):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinto M, García-Barrado J, Macías P. Oxidation of resveratrol catalyzed by soybean lipoxygenase. J Agric Food Chem. 2003;51(6):1653–1657. 10.1021/jf025818d [DOI] [PubMed] [Google Scholar]
- 23.Sloane D, Leung R, Craik C, Sigal E. A primary determinant for lipoxygenase positional specificity. Nature. 1991;354(6349):149–152. 10.1038/354149a0 [DOI] [PubMed] [Google Scholar]
- 24.Borngräber S, Browner M, Gillmor S, Gerth C, Anton M, Fletterick R, et al. Shape and Specificity in Mammalian 15-Lipoxygenase Active Site. J Biol Chem. 1999;274(52):37345–37350. 10.1074/jbc.274.52.37345 [DOI] [PubMed] [Google Scholar]
- 25.Andreou A, Feussner I. Lipoxygenases—Structure and reaction mechanism. Phytochemistry. 2009;70(13–14):1504–1510. 10.1016/j.phytochem.2009.05.008 [DOI] [PubMed] [Google Scholar]
- 26.Andreou A, Hornung E, Kunze S, Rosahl S, Feussner I. On the substrate binding of linoleate 9-lipoxygenases. Lipids. 2009;44(3):207–215. 10.1007/s11745-008-3264-4 [DOI] [PubMed] [Google Scholar]
- 27.Hornung E, Kunze S, Liavonchanka A, Zimmermann G, Kühn D, Fritsche K. Identification of an amino acid determinant of pH regiospecificity in a seed lipoxygenase from Momordica charantia. Phytochemistry. 2008;69(16):2774–2780. 10.1016/j.phytochem.2008.09.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Aae—Agrocybe aegerita, Fox—Fusarium oxysporum, Ggr—Gaeumannomyces graminis, Pos—Pleurotus ostreatus, Psa—Pleurotus sapidus; AaeLOX1, AaeLOX2, AaeLOX3, AaeLOX4 (MK451709), AaeLOX5, FoxLOX (KNB01601), GgrLOX (AAK81882), PsaLOX (CCV01580), PosLOX (CCV01578).
(TIF)
Agrocybe aegerita AaeLOX1, AaeLOX2, AaeLOX3, AaeLOX4, AaeLOX5, Pleurotus sapidus PsaLOX, Pleurotus ostreatus PosLOX, Fusarium oxysporum FoxLOX, Gaeumannomyces graminis GgrLOX. Alignment was carried out by using Clustal Omega with default parameters. The highlighted amino acid residues involved in ligand binding are indicated as “L”. Stereo specificity related amino acids are indicated as “B” [24], “H” [27] and “S” [23].
(TIF)
Data Availability Statement
Gene sequence for AaeLOX4 is available from the GenBank database under the accession number MK451709.