Abstract
Recent advances in cell-free gene expression (CFE) systems have enabled their use for a host of synthetic biology applications, particularly for rapid prototyping of genetic circuits and biosensors. Despite the proliferation of cell-free protein synthesis platforms, the large number of currently existing protocols for making CFE extracts muddles the collective understanding of how the extract preparation method affects its functionality. A key aspect of extract performance relevant to many applications is the activity of the native host transcriptional machinery that can mediate protein synthesis. However, protein yields from genes transcribed in vitro by the native Escherichia coli RNA polymerase are variable for different extract preparation techniques, and specifically low in some conventional crude extracts originally optimized for expression by the bacteriophage transcriptional machinery. Here, we show that cell-free expression of genes under bacterial σ70 promoters is constrained by the rate of transcription in crude extracts, and that processing the extract with a ribosomal runoff reaction and subsequent dialysis alleviates this constraint. Surprisingly, these processing steps only enhance protein synthesis in genes under native regulation, indicating that the translation rate is unaffected. We further investigate the role of other common extract preparation process variants on extract performance and demonstrate that bacterial transcription is inhibited by including glucose in the growth culture but is unaffected by flash-freezing the cell pellet prior to lysis. Our final streamlined and detailed protocol for preparing extract by sonication generates extract that facilitates expression from a diverse set of sensing modalities including protein and RNA regulators. We anticipate that this work will clarify the methodology for generating CFE extracts that are active for biosensing using native transcriptional machinery and will encourage the further proliferation of cell-free gene expression technology for new applications.
Keywords: cell-free synthetic biology, endogenous transcription, TX-TL, CFPS, CFE, cell extract, genetic circuitry, in vitro protein synthesis
Graphical Abstract

Cell-free systems are emerging as a prominent platform for use in synthetic biology.1,2 By mixing clarified cellular extracts, exogenously supplied energy sources and cofactors, and DNA-encoded genetic instructions, cell-free protein synthesis supports the basic processes of gene expression and metabolism in a convenient and engineerable in vitro reaction environment. Because the open environment enables flexibility for optimizing extract and reaction conditions and is amenable to high-throughput automation,3 cell-free gene expression (CFE) technology has found great utility in a wide range of contexts. Since their first application in deciphering the genetic code,4,5 cell-free systems have been successfully applied for the bulk production of model6–9 and therapeutic proteins.10–15 Beyond protein synthesis, CFE technologies have evolved more generally to enable complex and diverse functions, including prototyping cellular metabolism16–18 and glycosylation,19–21 expressing minimal synthetic cells, virus-like particles, and bacteriophages,7,22–27 portable on-demand manufacturing of pharmaceuticals,28,29 incorporation of noncanonical amino acids within proteins,30–34 prototyping of genetic circuitry,35–39 and sensing nucleic acids and small molecules through rapid, low-cost, and field-deployable molecular diagnostics.40–45 Most progress has occurred in CFE systems generated from Escherichia coli strains engineered for protein production, largely due to the bacterium’s well-characterized genetics and metabolism.1 However, there has been recent progress in adapting CFE protocols to make lysates from eukaryotic and nonmodel organisms, including yeast,46,47 Gram-positive bacteria,48,49 plants,50,51 and mammalian cells.52–54 CFE technology is therefore at the point of expanding beyond specialist laboratories and becoming a major toolbox throughout synthetic biology research, application, and education.35,5,56
A cell-free gene expression reaction is composed of three to four components that enable in vitro gene expression and metabolism: the clarified cellular lysate (or “extract”) that contains the requisite cellular machinery for protein synthesis; a buffered mixture of phosphorylated energy substrates, nucleoside triphosphates (NTPs), amino acids, salts, and other required cellular cofactors; the DNA templates that define the genetic program to be executed in the reaction; and any other optional exogenous cofactors, substrates, or inducers required for the reactions. Of these, the extract is the most labor-intensive component to prepare, requiring precise control over cell culture growth, lysis, and postlysis separation of unwanted cellular debris from the transcriptional and translational machinery that must remain behind in the final extract. Recent work has focused on optimizing performance of and expanding access to CFE technology by simplifying extract preparation protocols, including replacing lysis by homogenization with cheaper methods like sonication,57,58 bead-beating,57,59 enzymatic lysis,60 or flash-freezing,61 as well as reducing centrifugation intensity to speeds accessible on conventional benchtop instruments.62
While there have been many recent efforts to develop optimized methods for preparing highly active E. coli extracts, there has yet to be a targeted effort to deconstruct the effects of different protocol variations on extract performance that would ultimately provide simpler routes to obtain the same performance. Similarly, little is known about how specific variations in the protocol used to prepare extract impact its utility for different applications (e.g., high protein expression titers versus active genetic circuitry). Moreover, as a result of protocol and performance inconsistencies between research groups, many laboratories instead opt to use chemically defined, bottom-up reconstituted cell-free gene expression systems such as the “purified recombinant elements” (PURE) system.40,63,64 Although reconstituted in vitro protein synthesis platforms are powerful, their cost can be prohibitive, and they also lack the flexibility for strain engineering and cofactor and energy regeneration afforded by cellular extracts. A better understanding of how differences in extract preparation yield differences in functionality for CFE could therefore be important for broadening the adoption of cell-free technology for a range of applications.
In this work, we set out to characterize one such performance inconsistency: the functional inactivity of simple genetic programs using native bacterial regulators in an extract that had been previously simplified and optimized for bulk protein production. Specifically, we discovered that extracts optimized to yield high protein titers above 1000 ng/μL using T7 bacteriophage promoters to drive model reporter protein expression had 15-fold lower protein expression under the control of native E. coli σ70 promoters. Because transcriptional regulation of the native E. coli polymerase is crucial for many applications of CFE systems, we aimed to uncover which aspect of the extract preparation process caused this discrepancy, toward the goal of generating an improved CFE platform that supports gene expression from native E. coli regulators.
Here, we demonstrate that the transcriptional limitations from E. coli regulatory elements are removed by the addition of specific postlysis processing steps in the preparation of crude extracts for CFE. Specifically, we find that ribosomal runoff and dialysis steps are critical for recovering transcriptional activity from E. coli σ10 promoters. We also investigate the effects of other common extract preparation steps, such as flash freezing, and evaluate their impact on final extract performance. Combining these features, we create a modified cell-free extract preparation framework that can be carried out in a single 12-hour day or split across two shorter days and demonstrate its batch-to-batch reproducibility. Overall, this system imp roves the cell-free gene expression yield from bacterial σ70 promoters by 5-fold, speeds up the signal response time by 3-fold, and can be used to activate a wide array of synthetic genetic regulators and circuits, including RNA transcriptional activators and repressors, protein transcription factors, CRISPR-Cas9 cleavage, toehold translational switches, and translational riboswitches. This work serves to illuminate the “black box” of CFE and make cell-free technology more accessible, particularly for the numerous applications that require cell-free expression from bacterial transcriptional machinery. To this end, we have listed detailed protocol instructions for preparing active extract and running the CFE reaction in the Supplemental Note. We anticipate that this work will have impacts in cell-free synthetic biology and molecular diagnostics.
RESULTS AND DISCUSSION
Cell-Free Gene Expression from Bacterial Promoters in Crude Extract Is Limited.
A rapidly growing application area for bacterial cell-free systems is to use them for prototyping genetic elements (e.g., promoters, terminators) and circuits for function in living hosts.35 We therefore set out to assess the ability to activate native transcriptional machinery in the sonication-based CFE platform developed and optimized for protein production by Kwon and Jewett.58 We prepared cell free extracts from the Rosetta2 (DE3) pLysS strain of Escherichia coli, a derivative of E. coli BL21 supplemented with a plasmid encoding rare bacterial tRNAs to facilitate enhanced translation of recombinant proteins, following our published sonication-based protocol (Figure 1A, “No processing”, gray line).58 We then carried out cell-free gene expression of the model reporter superfolder green fluorescent protein (sfGFP). The sfGFP expression cassette contained a T7 bacteriophage promoter followed by the sfGFP coding sequence and T7 transcriptional terminator. A batch-mode transcription-translation reaction was carried out by adding the expression cassette template plasmid DNA to a mixture containing T7 RNA polymerase (RNAP), cell extract, and essential substrates (e.g., amino acids, nucleotides, energy substrates, cofactors, and salts) to a final volume of 15 μL and incubating this mixture at 30 °C for 15 h. sfGFP expression was quantified at the end of the reaction on a plate reader, which was then compared to a standard curve to derive protein concentrations. Consistent with previous work, our extract yielded greater than 1000 ng/μL of sfGFP, confirming that it was active for bulk protein production.
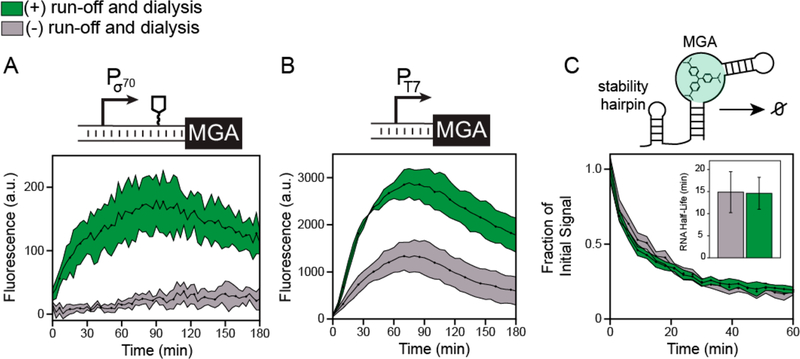
Figure 1.
The impact of different extract preparation procedures on cell-free gene expression (CFE) from different promoter systems. (A) Schematic for extract preparation. Combined transcription and translation activity differs based on extract preparation procedure and the use of endogenous or exogenous (bacteriophage) promoter systems. (B) Extract preparations that did not include postlysis processing steps yielded protein titers of ~ 1250 ng/μL when the sfGFP reporter is expressed from a T7 promoter (PT7) using exogenously added T7 RNA polymerase, independent of extract preparation steps. (C) Extracts prepared without postlysis processing showed poor yield (~50 ng/μL) when under the control of the consensus E. coli σ70 promoter (Pσ70) and E. coli RNA polymerase supplied from the lysate. The addition of processing steps to the lysate outlined in green in (A) improved protein expression yields from the bacterial σ70 promoter by 5× without impacting yields from the T7 promoter system. Protein yields are from overnight (15-h) in 15 μL reactions carried out in 2.0 mL tubes. Reporter constructs using the consensus E. coli σ70 promoter also encoded an RNA stability hairpin before the RBS and downstream coding sequence (Figure S1). Reactions were supplemented with 5 nM of a plasmid-based sfGFP expression cassette in 2.0 mL tubes, and protein yields were background-subtracted (i.e., no plasmid control) and measured by correlation to a known fluorescent standard. Error bars represent the standard deviation of the mean from three technical replicates drawn from a single batch of extract.
After validating gene expression under control of T7 RNAP, we next sought to investigate production of sfGFP under control of a bacterial σ70 promoter and the E. coli RNAP already present in the extract. Previous work has shown that bacterial cell-free gene expression systems can support expression from an array of native prokaryotic transcriptional components.7,65 To verify this in our system, we constructed an sfGFP expression cassette containing the consensus E. coli σ70 promoter followed by a predicted strong ribosome binding site (RBS), the sfGFP coding sequence, and the TrrnB transcriptional terminator from the E. coli rrnB ribosomal operon. To our surprise, we observed no expression of the reporter protein above background when this DNA construct was incubated with extract and transcription-translation reagents (Figure S1). Drawing inspiration from previous work that reported substantial improvements to cell-free expression productivity when strong secondary structure is added to the 5’ untranslated region of the reporter mRNA,65 we modified our expression construct to include the PHP14 mRNA stability hairpin66 immediately upstream of the ribosomebinding site on our reporter (refer to the Supporting Plasmid Files for more detailed information on plasmid architecture). With this construct, we observed 64 ± 14 ng/μL of constitutive protein production in the same reaction conditions, about 20-fold lower than that observed from the T7 promoter constructs (Figure 1B and 1C, gray bar). Since such low yields from cellfree expression would likely be insufficient for prototyping genetic circuits with naturally low ON states, we sought to optimize our extract preparation protocol to increase cell-free productivity using the endogenous bacterial transcriptional machinery.
Postlysis Processing Enhances CFE Yield from Bacterial Promoters.
We next sought to uncover which, if any, steps in the extract preparation protocol were responsible for poor expression from bacterial promoters. CFE extract preparation consists of five key steps: preculture, growth culture, cell harvest, cell lysis, and postlysis processing (Figure 1). Historically, a number of protocols have been developed for making productive extract, principally varying in the lysis and postlysis steps (Table 1).4,5,58,59,62 Due to its potential for scalability, equipment accessibility, and relatively low cost, we considered sonication as the preferable lysis method compared to physical lysis techniques such as homogenization or bead-beating. We therefore aimed to fix sonication as a design constraint while optimizing our extract preparation methods to enhance expression from the native transcription machinery.
Table 1.
Outline of Protocol Differences between Extract Preparation Methodsa
| extract protocol | Zubay, 19734 | Sun et al., 201359 | Kwon and Jewett, 201558 | this work |
|---|---|---|---|---|
| starter culture | one 16-h culture | two 8-h cultures | one 16-h culture | one 16-h culture |
| culture media composition | 2× YT | 2× YT + phosphate | 2× YT + phosphate + glucose | 2× YT + phosphate |
| lysis method | French press | bead-beating | sonication | sonication |
| postlysis centrifugation | 30 000g for 30 min | 12 000g for 10 min | 12 000g for 10 min | 12 000g for 10 min |
| run-off reaction | 80min | 80 min | strain-dependent; none for BL21 | 80 min |
| dialysis | 18 h | 3 h | none | 3 h |
| flash freeze | none | none | before lysis | optional |
Italicized: variables examined in this study.
The other major discriminant between common extract preparation protocols is in the postlysis processing, consisting of the runoff reaction, dialysis, and centrifugation steps. Both the runoff and dialysis steps originate from the pioneering work of Nirenberg and Matthaei5 and Zubay.4 During the runoff reaction, the extract is incubated at 37 °C to allow ribosomes to “run off’ their native transcripts, freeing them to translate recombinant transcripts.5,67 The clarified extract is then dialyzed against buffer in a step hypothesized to remove unwanted metabolic byproducts that accumulate during the runoff reaction such as inorganic phosphate.7,59,62 Previous work showed that protein synthesis from a T7 expression cassette is not improved in BL21-sourced extracts by including the runoff reaction or dialysis.58,68 However, we hypothesized that these steps might impact protein synthesis under control of native bacterial promoter systems. We thus aimed to systematically study the impact of these postlysis processing steps on protein expression from the native E. coli RNAP.
As an initial test, we directly adapted our clarification protocol to incorporate the classic runoff and dialysis steps (Figure 1A, “Postlysis processing”, green line). Consistent with previous work, in the same reaction conditions, we observed no difference in T7-driven protein synthesis from the additional clarification steps (1220 ± 85 ng/μL postlysis processed vs 1240 ± 89 ng/μL nonprocessed). However, we observed a 5-fold improvement in yield from the E. coli σ70 reporter containing the RNA stability hairpin to greater than 300 ng/μL (Figure 1B and C green bar). These yields are comparable to the reported titers for in vitro protein synthesis driven by a promoter of bacterial origin65 and approach, but are lower than the titers achievable from a synthetic σ70-regulated lambda phage promoter. We hypothesize that remaining differences in protein synthesis yield are mainly attributable to variation in plasmid architecture and our use of a phosphorylated energy substrate, which has been shown to enhance the rate of protein synthesis in the first few hours of the reaction at the expense of the reaction’s lifetime.69–71
Overall, these results supported our goal to design a CFE platform that permits robust expression from the native E. coli RNAP machinery. However, these experiments did not reveal why CFE expression under control of bacterial but not T7 promoters is sensitive to the runoff and dialysis steps.
Yield Improvements to Cell-Free Gene Expression Are Independent of Regulatory Element Strength.
We next sought to understand the improvement in cell-free expression yields from the postlysis processing steps. Specifically, we hypothesized that because the runoff reaction and dialysis steps had minimal effect on T7-driven expression, these steps selectively improve transcription by the native bacterial polymerase and do not affect the translational machinery. We therefore predicted that the postlysis processing steps would improve protein synthesis for constructs that had a range of promoter and ribosome binding site strengths.
To test this hypothesis, we measured cell-free expression protein yield from reporter constructs that contained synthetic E. coli σ70 promoters and ribosome binding sites that were predicted to have a range of strengths. In each case, we maintained the mRNA stability hairpin in the 5’ untranslated region. Protein yield was then quantified using 8 h kinetic CFE experiments at the 10 μL scale to enable continuous monitoring of sfGFP production, compared against a known fluorescein isothiocyanate (FITC) standard.
For all σ70 reporter designs tested, comparison of reactions using these extracts revealed at least a 3-fold improvement in final protein yield from the postlysis processed extract relative to the nonprocessed extract (Figures 2A and B). The improvement was most dramatic for the weakest predicted RBS and promoter designs, for which no expression of sfGFP could be observed over background in the nonprocessed condition. This result is important for two reasons. First, it shows that the enhanced protein yield from the E. coli transcriptional machinery due to postlysis processing is general regardless of regulatory element strength, suggesting that the omission of these steps causes transcriptional inhibition. Second, these data underscore the importance of postlysis processing for cell-free expression from reporter constructs that have weak native expression—for example, gene circuits or biosensors that are designed with a low ON state to prevent leak.
Figure 2.
Postlysis processing steps in extract preparation enhance cell-free gene expression yields from E. coli σ70 promoters. Extracts prepared with runoff and dialysis (green) increase the yield of an sfGFP reporter across constructs containing a range of synthetic (A) ribosome binding site and (B) E. coli σ70 promoter strengths, compared to extracts prepared without these steps (gray). (C) Titration of reporter construct DNA that contains a strong promoter and RBS in both extracts suggests that protein synthesis saturates above 10 nM of reporter template DNA in the processed extract but continues to increase in the nonprocessed extract. (D) Kinetics at the 20 nM reporter template DNA concentration show that despite having equal endpoint sfGFP production levels, protein production is 3× faster for the processed extract. Endpoint data in (A–C) are from 8 h experiments incubated and measured in a plate reader at 30 °C. Error bars represent the standard deviation of the mean across three technical replicates drawn from a single batch of extract.
On the basis of these observations, we hypothesized that if transcription by the bacterial polymerase is rate-limiting for gene expression in cell-free conditions, then protein production should scale with the DNA template concentration until either the available polymerase is saturated or transcription is no longer limiting. To test this, we performed kinetic CFE reactions as above on the strongest promoter and RBS construct over a range of template DNA concentrations. As predicted, the processed extract showed marked improvement over the nonprocessed extract at low DNA concentrations, and then both reactions saturated at the same final protein levels when 20 nM DNA template was used (Figure 2C). This result demonstrated that adding more template DNA can relieve transcriptional limitations up to a point, as we had anticipated. Interestingly, however, the two different extracts varied greatly in the kinetics of protein synthesis (Figure 2D), with the processed extract reaching its endpoint value about three times faster than the nonprocessed extract. Indeed, the quicker onset of sfGFP production in the processed extract was observed across all template concentrations (Figure S2).
We observed a much weaker dependence of CFE yield on DNA template concentration when the sfGFP reporter gene was transcribed by the T7 RNAP. Specifically, increasing the template concentration from 1 nM to 5 nM improved expression from the bacteriophage promoter by 17% in the nonprocessed and 49% in the processed extract (Figure S3). By contrast, when expression was driven by the endogenous bacterial promoter, final protein yield increased about 10-fold in both processed and nonprocessed extracts (Figure 2C).
Yield Improvements to Cell-Free Gene Expression Arise from Enhanced Transcriptional Activity.
Taken together, the above results suggest that transcription by the E. coli RNA polymerase can be the rate-determining step of a CFE reaction, and that although transcriptional constraints can be partially relaxed by adding more DNA template, the transcriptional activity in vitro can be linked to specific steps of the extract preparation. To validate our hypothesis that transcription limits cell-free gene expression, we next aimed to remove the confounding effect of reporter protein translation and quantify the transcription rate in each extract by monitoring gene expression at the RNA level. To do this, we replaced the sfGFP coding sequence in our strongest σ70 and T7 reporter template plasmids with the malachite green aptamer (MGA) sequence. Once transcribed, this RNA aptamer binds to malachite green and activates the dye’s fluorescence. The MGA reporter system enables a convenient fluorescence-based quantification of transcript levels and has been previously used in vitro.72–74 As expected, under the consensus E. coli σ70 promoter, we observed substantially higher expression of the MGA reporter in the processed extract over the course of a 3 h experiment (Figure 3A).
Figure 3.
Postlysis processing enhances the transcription rate but does not affect RNA degradation rates in CFE systems. (A) In vitro transcription of the malachite green RNA aptamer (MGA) from a strong E. coli σ70 promoter in a cell-free gene expression reaction containing the malachite green dye using either processed (green) or nonprocessed (gray) extracts. The construct encoded a 5’ stability hairpin before the MGA DNA sequence to enhance fluorescence observation. (B) In vitro transcription of the MGA construct from a T7 promoter and no 5’ stability hairpin. Kinetic data in (A) and (B) represent the average of three technical replicates, with shaded region representing plus-minus one standard deviation of the mean. (C) Characterization of RNA degradation rates. The malachite green RNA aptamer was purified with the 5’ stability hairpin, mixed with the malachite green dye and 10% extract by volume, and the decay in fluorescence over time was measured to quantify RNA degradation rates (kdegradation)· Inset: the RNA half-life (ln(2)/kdegradation) was estimated by fitting an exponential decay function to the fluorescence kinetics over the first 30 min to fit kdegradation. Errors in half-lives are propagated from three independent measurements and reported as standard deviation.
Notably, we observed almost no fluorescence from the MGA reporter in the nonprocessed condition, suggesting that RNA degradation keeps pace or outstrips weaker transcription rates in those extracts. The hypothesis that ribonuclease (RNase)-mediated degradation controls transcript abundance is supported by the observation that there is a peak in MGA expression at ~80 min, likely caused by continued degradation after transcriptional slow-down from the depletion of NTPs in the reaction.69
Interestingly, when transcribed from the T7 promoter, MGA production is also about 2-fold higher in the processed extract (Figure 3B). This result contradicts our earlier finding that the postlysis processing steps do not appreciably improve expression from the phage polymerase (Figure 1B). However, these results can be explained together by the presence of translational bottlenecks in CFE that limit protein but not mRNA production from the bacteriophage promoter—only by removing the translational bottlenecks can we directly observe differences. Because native RNases in the extract may be saturated at the high RNA levels generated by T7 transcription, it is difficult to estimate quantitatively how much the processing steps actually improve the bacteriophage transcription rate. However, the transcription-only assays do demonstrate that the extract processing steps are crucial for maintaining gene expression in transcriptionally limiting conditions.
Rather than activating transcription, an alternative mechanism for how the postlysis processing steps improve extract productivity would be the stabilization of transcripts by removing or inhibiting RNases present in the extract. In fact, batch-to-batch variation in extract productivity has previously been observed to be partly due to variability in RNA degradation rates between batches.36 To test for this in our extract protocol variations, we measured RNA degradation by spiking purified MGA and the malachite green dye into both processed and nonprocessed extracts and observed the decay of fluorescence over time (Figure 3C). Interestingly, we observed nearly identical half-lives for the RNA in each extract, indicating that the postlysis processing steps do not alter the observed rate of RNA degradation. Our measurements are consistent with previous estimates of MGA half-life in extract.36 It is possible that the measured half-lives are overestimated by this assay because the RNase machinery is saturated by MGA at high RNA concentrations. However, we believe this effect is representative of saturation conditions present in cell-free transcription-translation reactions, since the concentration of mRNA produced in such reactions is of a similar magnitude.3,75 We could not quantify the RNA degradation rate at lower concentrations of MGA due to its relatively small extinction coefficient and the low fluorescent signal over background. Identical measurements performed with MGA RNAs lacking a 5’ stability hairpin showed that the hairpin increases the RNA half-life by about 2-fold, underscoring the importance of RNA stability in transcriptionally limited systems (Figure S4).
Overall, these experiments demonstrate that the improvements in CFE yield from postlysis processing are due to improvements in transcriptional output of the reactions. Importantly, this effect is exacerbated in reactions where mRNA levels are limiting and DNA template loading impacts that arise from resource constraints can be neglected.75
Protocol Sensitivity Can Be Assessed in Transcriptionally Limiting Conditions.
Having demonstrated that transcriptional limitations in CFE reactions can be lifted by postlysis processing through a runoff reaction and dialysis, we next aimed to measure the sensitivity of extract productivity to other extract preparation protocol variations. Specifically, we sought to measure the impact that three common protocol variations would have on CFE reactions operating under transcriptional limitations. These include media conditions used to grow cells prior to lysis, flash freeze steps used as a convenient stopping point between growth and lysis, and the individual impacts of runoff and dialysis (Figure 4). From a practical standpoint, this analysis would facilitate the adoption of protocols to laboratories with less familiarity with the nuances of preparing extract. Moreover, the sensitivities could reveal useful insight into how each extract preparation protocol step impacts the overall productivity of the extract.
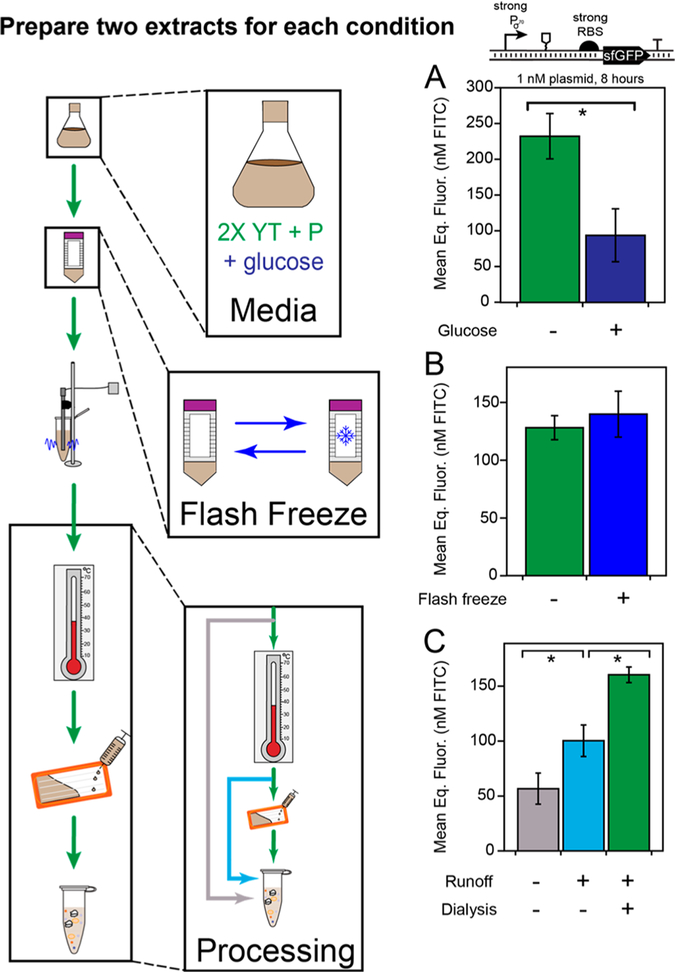
Figure 4.
Extract activity is sensitive to some protocol variants. (A) Addition of glucose (navy) to the 2× YT + P culture media reduces CFE yields from bacterial promoters under transcriptionally limiting conditions (p = 0.035). (B) Flash freezing the cell pellet before lysis does not significantly impact the extract’s productivity (p = 0.30). (C) Extract clarification by the Kwon et al.58 protocol for BL21 (gray), with runoff reaction (light blue), and with runoff reaction and dialysis (green) under transcriptionally limiting conditions shows that both the runoff and dialysis steps contribute to the observed increase in transcriptional activity (p = 0.039 for green vs gray and p = 0.031 for green vs light blue). All bars represent the average of six independent cell-free reactions, drawn from three technical replicates of two independently generated extracts made on different days. Error bars represent the standard deviations of the six measurements. In each case, unpaired one-tailed Student’s t tests were performed for N = 2 extracts using Welch’s correction for heteroscedasticity.
Before starting this analysis, we first had to assess the impact of batch–batch variability on our ability to discriminate meaningful differences between extract preparation protocol variations. To estimate batch-to-batch variability in our base protocol, we prepared three identical extracts on three different days, aiming to replicate all experimental conditions in the preparation. We then performed identical transcriptionally limited sfGFP synthesis reactions on all three extracts on three different days, with the aim of determining whether observed variation was due to extract–extract variation, experiment–experiment variation, or both. Our results (Figure S5A) showed that both extract and experimental variation are statistically significant (p = 0.030 for extract variability and 0.026 for experimental variability from a two-factor ANOVA). Despite these differences, the rankings for both extract and experimental productivity showed high correlation across all 3 days. Moreover, only the combination of the two highest-performing conditions (Experiment Day 2, Extract Batch 2) was found to be statistically significantly different than any other experiment–extract combination in pairwise t tests (Figure S5B).
Compared against previous studies, we observed lower batch–batch variability between individual extracts; using the endpoint fluorescence averaged across all 3 days, the coefficient of variation (CV) between the three extracts was 11%. This finding contrasts with previous work which has shown that yields from bacterial promoters in transcription-translation reactions can vary by more than 50% between batches intended to be identical.35 We hypothesize that one reason for the consistency in our approach was the use of sonication for cellular lysis rather than less precise techniques like bead-beating since sonication allows the amount of energy delivered to the sample to be precisely controlled. Nevertheless, achieving statistical reproducibility between extract preparations remains difficult. Despite preparing all media and buffers in tandem and inoculating the starter cultures from a clonal population, we still observed differences in growth conditions such that the time required to reach the target harvest optical density varied from 4 to 5 h. This is one possible explanation for the observed batch–batch variability, though we confirmed that in 2× YTP media, the Rosetta2 (DE3) pLysS strain is still growing in exponential phase at the harvest optical density (OD600) of 3.0 (Figure S6).
The use of cell-free extracts as a platform for molecular diagnostics requires that batches are consistent in the dynamic rate of protein production as well as their endpoint yields. We quantified this by the fluorescence in batch reactions after 90 min, a time at which the reactions are rapidly producing protein, and the relevant endpoint for previously published in vitro diagnostics.40 We observed only slightly greater variability between the extracts in the rate of protein synthesis (CV = 17%) (Figure S5B).
We also observed a small amount of experiment–experiment variability (CV = 11%) equivalent to the extract–extract variability. We attribute this variability to small variations in pipetting plasmid DNA from a concentrated stock into the reaction master mix, given the previous observation that protein synthesis yields under transcriptional limitations are exquisitely sensitive to template DNA concentration (Figure 2C). Because this amount of experimental variability will be the same for every reaction measured in a given experiment, we felt confident in our ability to discriminate extract productivities with statistically significant differences in final protein titers.
In this work, we compare only against extracts prepared in house, and observe a relatively small amount of batch variability compared to previous work.36 Further independent and crowdsourced method validation by external laboratories, as has been done previously for cellular gene expression characterization,76,77 would be a useful resource toward the proliferation of cell-free technology.
Having established our ability to discriminate extract batch variability from meaningful protocol differences in extract preparation, we next aimed to measure our protocol’s sensitivity against specific variations. We tested variation at every step except the sonication, which was kept constant as a design constraint, since previous work has shown that the sonication energy has a strong, strain-dependent impact on extract productivity.58 Specifically, the three variables that we tested were media composition for cell growth, flash freezing prior to lysis, and the postlysis processing steps individually rather than as a group, paying close attention to major protocol variations that exist in the literature with the aim of creating a robust protocol. We did not measure the impact of cell density at the point of harvest, as this has been recently studied in independent work.78 In each case, we performed the same sfGFP synthesis reactions under limiting bacterial transcription for 8 h, but also prepared two independent batches of extract following each protocol variant to remove any unknown bias present in single batches. Under these conditions, we expected to see the maximum sensitivity between protocol variants.
First, we tested different media formulations. Two common formulations used in the literature include 2× ΎT + P59 and 2× YT + P + G,58 where P stands for buffered potassium phosphate and G is for glucose. Although our earlier extracts were prepared with 2× YT + P media, we sought to determine if glucose could be supplemented to the media and bacterial transcription activity retained. We tested the impact of growing lysate chassis cells in media with and without glucose on the extract’s performance. Surprisingly, supplementation of glucose significantly diminished our observed yields under bacterial transcription control (Figure 4A, p = 0.035). While glucose was initially implemented by Kim and Choi79 to lower phosphatase activity in the lysate (î.e., hexose phosphatase), and maintained in the protocols developed by Jewett and Swartz to activate natural energy metabolism,16,17,80 its presence in the media is deleterious to endogenous transcription in the lysate. Thus, growing the cells on 2× YT + P was selected as a core design parameter moving forward. This result corroborates protocols developed by Noireaux and colleagues that similarly avoid the use of glucose in the media of their systems that activate natural transcription.59
Next, we aimed to test the impact of flash-freezing cells in liquid nitrogen prior to lysis. In the literature, flash-freezing has been an optional step implemented in some but not all protocols and has a practical advantage as a convenient stopping point if included. We therefore wondered if flash freezing would diminish the quality of the extract for expression from bacterial promoters. We observed that flash-freezing our cell pellet immediately before lysis has no appreciable impact on the final cell-free yield (Figure 4B). Since the additional clarification steps contribute 5 h of labor to the extract protocol, the ability to split the procedure across more than 1 day broadens its appeal, particularly for laboratories acclimating to the methods for extract preparation.
Finally, we sought to examine the impact of each postlysis clarification step individually, particularly the result of performing a runoff reaction’s incubation but not dialysis. We observed 50% recovery in the yield from the bacterial promoter from the runoff reaction with no dialysis, suggesting that the two steps have specific yet different effects on the physiochemical makeup of the extract (Figure 4C).
A Wide Array of Transcriptional Genetic Circuits Are Functional in a Modified Cell-Free Extract Preparation Framework.
Having observed the relaxation of transcriptional constraints in the constitutive expression of a reporter protein from a bacterial promoter, we next aimed to demonstrate the activation of a number of synthetic gene expression circuits driven by bacterial genetic machinery in our modified CFE system. We assembled a library of gene expression activators and repressors, functioning at both the RNA and protein levels, with a diversity of expression levels and tested them in both processed and nonprocessed extracts (Figure 5). The six circuits conditions that we assayed were as follows: small transcription-activating RNAs (STARs);81 RNA transcriptional repressors;82 the inducible protein LuxR transcription factor;83 CRISPR-Cas9-mediated DNA cleavage;84,85 toehold switch translation activating RNAs;86 and a translational theophylline riboswitch.87 The individual reaction conditions for each circuit are presented in Table S1. In each case, the reaction’s ON state was first maximized by adjusting magnesium concentration, a crucial determinant of CFE functionality, and then adjusting the concentration ratios for each plasmid supplied to the reaction. Additionally, each functional construct contained either the PHP stability hairpin or, in the case of the RNA regulators, other structural features in the 5’ UTR to limit RNase activity.
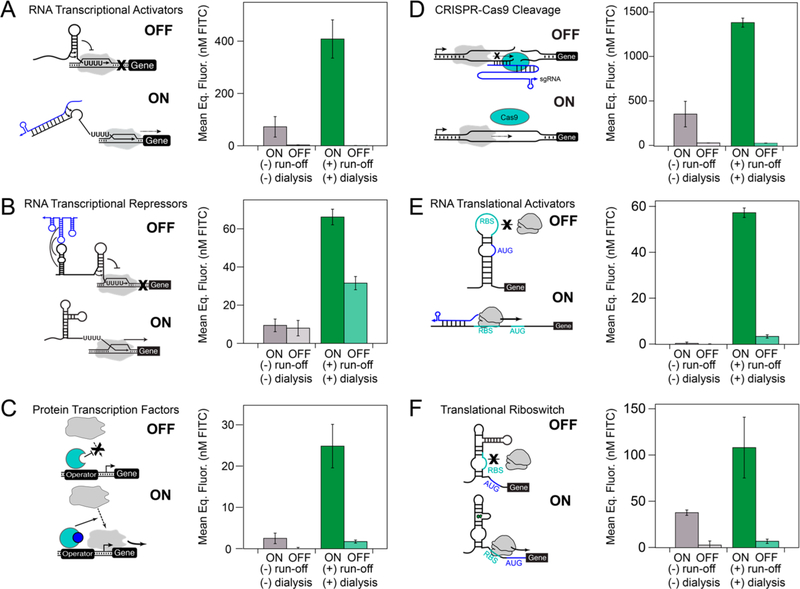
Figure 5.
Processed extracts enable cell-free expression of a wide array of genetic circuitry. Postlysis processing enables significantly higher ON states, including activation in some cases, for (A) small RNA transcriptional activators (STARs), (B) RNA transcriptional repressors, (C) LuxR, the ;Y-acyl homoserine lactone (AHL)-inducible σ70 transcription factor, (D) CRISPR-Cas9 cleavage, (E) toehold switch RNA translation activators, and (F) translational riboswitches. Each experiment was done in technical triplicate in either a postlysis processed (green) or nonprocessed (gray) extract for 8 h at 30 °C in a plate reader monitoring sfGFP production. The total plasmid DNA concentration was held constant in each reaction between the ON and OFF states of each circuit to remove variation caused by unwanted cell-free transcription and translation of the antibiotic resistance genes encoded on each plasmid. Complete reaction conditions including plasmid and inducer concentrations and optimal magnesium levels are included in Table S1. Kinetics of gene expression trajectories for each circuit are presented in Figure S7. Error bars represent the standard deviation of the mean for three technical replicates drawn from a single batch of each extract.
In all six circuits that we tested, the ON state for the processed extract was at least three times higher than the ON state for the nonclarified extract. In two circuits (transcription factors, toeholds), we could not observe signal above background in the nonprocessed extract. For the transcriptional repressors, the ON state for the nonprocessed extract was nearly indistinguishable from the OFF state (15% repression vs 52% repression in the processed extract). Although the circuits designed to have greater ON activity were at least functional in the nonprocessed extract, they were much slower: the STARs, CRISPR-Cas9, and riboswitch systems all responded around three times faster in the optimized extract (refer to Figure S7 for all kinetics). Since speed of response is a major advantage for cell-free biosensors over more conventional diagnostic methods, our extract system could to play an important role in the proliferation of this technology. Taken together, these data show that postlysis processing is critical to generate an extract system that enables function of diverse genetic circuits.
SUMMARY
In this work, we performed in-depth investigation into a modified method for production of cell-free extracts for in vitro synthetic biology. With this modified procedure, we demonstrated ~3–5-fold increases in cell-free production of a model reporter placed under control of a bacterial promoter. Equally important, we identified transcription from the σ70 promoter to be a key limiting step in CFE operating at nonsaturating plasmid DNA concentrations. This result opposes a common prevailing notion, mainly borne from experiments with bacteriophage polymerases, that energy regeneration for translation strictly determines final protein titer in in vitro transcription-translation reactions.88–90 With our optimized extracts, we were able to activate a diverse array of genetic circuitry in CFE, including RNA transcriptional activators, RNA translational activators, protein transcription factors, CRISPR-Cas9 nuclease activity, RNA translational repressors, and a translational riboswitch.
We observed that the key process improvements for activating cell-free gene expression from bacterial promoters were the inclusion of an 80 min runoff reaction and subsequent dialysis. Our observed increase in transcriptional output from including a runoff reaction challenges the common belief that the postlysis incubation is necessary to free the ribosomes from their actively translating templates. If true, that hypothesis would have suggested that the benefit from the runoff reaction is purely translational. As previous studies have demonstrated both strain-and organism-specific variance on the need for a runoff reaction in just bacteria, the mechanism for transcriptional rate enhancement remains unclear.58,91,92 We hypothesize that dialysis removes a small molecule global inhibitor of transcription that accumulates in the crude lysate during the runoff reaction. However, due to the complex and largely unknown metabolome of the extract, we were unable to identify the origin of this inhibition. Future studies may do well to further elucidate the mechanism, with an ultimate aim toward engineering a better source strain.
Previous work has shown the ability to activate native transcriptional machinery from E. coli sigma factors and regulatory circuits in CFE systems.7 However, it was unclear why their approach worked and others in the literature did not.58,62 This lack of understanding, compounded by disparities in reaction conditions and reagent compositions between published methods, has made it challenging for new laboratories to adopt cell-free gene expression technologies. To help alleviate this, we have provided a detailed extract preparation protocol that will aid new laboratories in adopting the approaches described here (Supporting Note). Additionally, as CFE systems gain popularity for prototyping genetic regulators, especially in nonmodel organisms, the link between extract preparation and resulting transcriptional activity will grow in importance. By illiiminating the importance of extract processing steps and highlighting their relevance to CFE, we anticipate that our work will expand the utility of cell-free systems throughout synthetic biology.
MATERIALS AND METHODS
Plasmid Construction and Purification.
A table of all plasmids and the DNA concentration used in each experiment of this study can be found in Table S1, and plasmid architectures can be found in Supporting Data File S1. Plasmids were assembled using either Gibson assembly or inverse PCR and purified using a Qiagen QIAfilter Midiprep Kit. pJBL002 and pJBL004 are from the literature93 and pJBL7022 is based on pJBLOOó from the literature.93 pJBL3859, 4860, 5816, and 5817 are from the literature.94 pJBL7008 is based on the theophylline riboswitch RS.E from the literature.87 pJBL2812 and 2814 are from the literature.42 pJBL7023 is from the literature.58 pJBL623 and 632 were a gift from Dr. Lei Qi (Stanford University).
Extract Preparation.
Refer to the detailed extract preparation protocol included in the Supplemental Note. Briefly, cell-free extract was prepared based on the protocol in the literature.58 Rosetta 2 cells were streaked onto chloramphenicol agar plates overnight, then inoculated into 30 mL LB supplemented with antibiotic and grown in a 250 mL baffled flask at 37 °C. After 16 h, 20 mL of the stationary culture was used to inoculate one liter of autoclaved growth media 2× YT + P (16 g/Ltryptone, 10 g/L yeast extract, 5 g/L sodium chloride, 7 g/L potassium phosphate dibasic, 3 g/L potassium phosphate monobasic) in a 2.5 L baffled Tunair flask. A separate bottle containing 7.2% glucose solution was autoclaved and added at the time of inoculation for 2× YTP + G experiments to a final glucose concentration of 1.8%.
Cultures were grown to the exponential phase optical density (OD6OO) 3.0 ± 0.2 for approximately 4 h at 37 °C and 200 rpm. The culture was divided in two and centrifuged for 15 min at 5000g at 4 °C to pellet the cells. The cell pellets were washed three times in 25 mL wash buffer (50 mM Tris, 14 mM Mg-glutamate, 60 mM K-glutamate, 2 mM DTT, brought to pH 7.7 with acetic acid) and recentrifuged between each wash at 5000g at 4 °C for 10 min. After the fourth and final centrifugation at 7000g at 4 °C for 10 min, pellets were resuspended in 1 mL wash buffer/g pellet and transferred to 1.6 mL microcentrifuge tubes. For the flash freeze experiments, cell pellets were submersed in liquid nitrogen before this step and thawed the next day.
The cell suspensions were then lysed by sonication on a QSonica Q125 sonicator with a 3.175 mm diameter probe at a frequency of 20 kHz and 50% amplitude by 10 s ON/OFF pulses for a total of 60 s (delivering ~350 J). The lysate was centrifuged for 10 min at 4 °C and 12 OOOg, and the supernatant was removed. For preparations including a runoff reaction, the tubes of crude lysate were covered in aluminum foil and were incubated shaking with lids on for 80 min at 37 °C and 200 rpm. The extract was recentrifuged for 10 min at 4 °C and 12 000g and the supernatant was removed. For preparations including a dialysis, the extract was injected into a 10K MWCO Slide-a-Lyzer dialysis cassette (ThermoFisher, 66380) and dialyzed against 600 mL of dialysis buffer (5 mM Tris, 14 mM Mg-glutamate, 60 mM K-glutamate, 1 mM DTT, pH 8.2) at 4 °C for 3 h. The dialyzed extract was removed from the cassette and centrifuged once more for 10 min at 4 °C and 12 00Og. The supernatant was removed, aliquoted, and snap-frozen in liquid nitrogen.
CFE Experiment.
Refer to the detailed description for the preparation of the cell-free expression experiment included in the Supplemental Note and the Supplemental Experimental Design Spreadsheet. The final CFE reaction mixture is composed of the following reagents: 10–20 mM magnesium glutamate; 10 mM ammonium glutamate; 130 mM potassium glutamate; 1.2 mM ATP; 0.850 mM each of GTP, UTP, and CTP; 0.034 mg/mL folinic acid; 0.171 mg/mL yeast tRNA; 2 mM amino acids; 30 mM PEP; 0.33 mM NAD; 0.27 mM CoA; 4 mM oxalic acid; 1 mM putrescine; 1.5 mM spermidine; 57 mM HEPES; 30% CFE extract by volume; plasmid DNA to the desired concentration (refer to Table S1 for plasmid DNA concentrations used in this study); and water. For reactions involving T7 RNAP expression, in-house purified T7 RNAP was doped into the reaction at 0.10 mg/mL. The optimal magnesium concentration was determined for each reporter construct and was found to be 16 mM magnesium glutamate in nearly all cases (for exceptions to this, as well as the inducer concentrations in Figure 5, refer to Table S1).
All kinetic CFE reactions were prepared on ice in triplicate at the 10 μL scale. 33 μL of a mixture containing the desired reaction components was prepared and then 10 μL was pipetted into three wells of a 384-well plate (Corning, 3712), taking care to avoid bubbles. Plates were sealed (Thermo Scientific, 232701) and sfGFP fluorescence (emission/excitation: 485/520 nm) was monitored every 5 min on a BioTek Synergy Him plate reader for 8 h at 30 °C. For the bulk endpoint experiments in Figure 1, reactions were prepared to the 15 μL scale in triplicate, pipetted into the bottom of a 2.0 mL microcentrifuge tube, and incubated without shaking at 30 °C overnight for 15 h. Final protein titers were calculated from a previously developed plate reader correlation to either sfGFP (high-yield overnight experiments) or FITC (low-yield kinetic experiments) (see below). For the malachite green experiments in Figure 3, fluorescence (emission/excitation: 615/650 nm) was measured every 3 min. For all experiments, a no-DNA negative control was prepared in triplicate for every extract being tested. All reported fluorescence values have been baseline-subtracted by the no-DNA condition, and all error bars include the propagated error from the no-DNA condition.
RNA Purification and RNA Degradation Experiments.
The malachite green RNA aptamer with and without the 5’ stability hairpin was purified from a runoff in vitro transcription of pJBL7004 or pJBL7005 using in-house prepared T7 RNAP. The RNA was purified by denaturing 8% urea-polyacrylamide gel electrophoresis, followed by elution into nuclease-free water overnight and ethanol precipitation.
For RNA degradation experiments, 2.2 μM of the purified RNA was doped into a 11 μL mixture containing 10% malachite green dye and 10% extract by volume, with the balance nuclease free water and quickly pipetted into a 384-well plate. Malachite green fluorescence (emission/excitation: 615/650 nm) was monitored every 3 min for 2 h. Half-lives were then calculated by fitting the fluorescence trajectories over the first half hour to the equation F = F0e−kt where t1/2 = In(2)/k.
Fluorescent Standard Calibration.
For bulk production of sfGFP in overnight reactions (Figures 1 and S3), we measured the reaction yield with a standard curve previously developed by incorporating radioactive C 14-leucine into the reporter protein and relating counts to endpoint fluorescence. For Figures 2, 4, 5, S1, S2, S5, and S7, in which far less sfGFP was deliberately produced per reaction, we instead developed a linear correlation based on a FITC standard diluted to low micromolar concentrations.
Data Availability.
All source data for main and SI figures was deposited open access in Northwestern’s arch database (https://arch.library.northwestern.edu/). Data can be accessed via https://doi.org/10.21985/N2318T.
Supplementary Material
ACKNOWLEDGMENTS
We extend a special thanks to our collaborators as well as William Poole and Dr. Richard Murray for thoughtful insight into and editing of this manuscript. This work was supported by the Air Force Research Laboratory Center of Excellence for Advanced Bioprogrammable Nanomaterials (C-ABN) Grant FA8650-15-2-5518 (to M.C.J.), the David and Lucile Packard Foundation (to M.C.J.), an NSF CAREER Award (1452441 to J.B.L.), the Camille Dreyfus Teacher-Scholar Program (to M.C.J. and J.B.L.), and Searle Funds at the Chicago Community Trust (to J.B.L.). A.D.S. was supported in part by the National Institutes of Health Training Grant (T32GM008449) through Northwestern University’s Biotechnology Training Program. The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies or endorsements, either expressed or implied, of the Air Force Research Laboratory, Air Force Office of Scientific Research, or US Government.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acssynbio.8b00430.
Supporting figures and tables, descriptions of plasmid architecture, and detailed protocol notes for extract preparation and reaction execution (PDF)
Supporting Data S1 (XLSX)
Experimental design spreadsheet (XLSX)
The authors declare no competing financial interest.
REFERENCES
- (1).Carlson ED, et al. (2012) Cell-Free Protein Synthesis: Applications Come of Age. Biotechnol. Adv 30 (5), 1185–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Hodgman CE, and Jewett MC (2012) Cell-Free Synthetic Biology: Thinking Outside the Cell. Metab. Eng 14 (3), 261–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Moore SJ, et al. (2018) Rapid acquisition and model-based analysis of cell-free transcription–translation reactions from nonmodel bacteria. Proc. Natl Acad. Sci. U. S. A 115, E4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Zubay G (1973) In Vitro Synthesis of Protein in Microbial Systems. Annu. Rev. Genet 7 (1), 267–287. [DOI] [PubMed] [Google Scholar]
- (5).Nirenberg MW, and Matthaei JH (1961) The Dependence of Cell-Free Protein Synthesis in E. Coli Upon Naturally Occurring or Synthetic Polyribonucleotides. Proc. Natl. Acad. Sci. U. S. A 47 (10), 1588–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Caschera F, and Noireaux V (2014) Synthesis of 2.3 mg/mL of protein with an all Escherichia coli cell-free transcription–translation system. Biochimie 99, 162–168. [DOI] [PubMed] [Google Scholar]
- (7).Garamella J, et al. (2016) The All E. coli TX-TL Toolbox 2.0: A Platform for Cell-Free Synthetic Biology. ACS Synth. Biol 5 (4), 344–355. [DOI] [PubMed] [Google Scholar]
- (8).Ezure T, et al. (2006) Cell-Free Protein Synthesis System Prepared from Insect Cells by Freeze-Thawing. Biotechnol. Prog 22 (6), 1570–1577. [DOI] [PubMed] [Google Scholar]
- (9).Goerke AR, et al. (2008) Cell-free metabolic engineering promotes high-level production of bioactive Gaussia princeps luciferase. Metab. Eng 10 (3), 187–200. [DOI] [PubMed] [Google Scholar]
- (10).Zawada JF, et al. (2011) Microscale to Manufacturing Scale-up of Cell-Free Cytokine Production–A New Approach for Shortening Protein Production Development Timelines. Biotechnol. Bioeng 108 (7), 1570–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Thoring L, et al. (2017) High-yield production of “difficult-to-express” proteins in a continuous exchange cell-free system based on CHO cell lysates. Sci. Rep 7 (1), 11710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Tran K, et al. (2018) Cell-free production of a therapeutic protein: Expression, purification, and characterization of recombinant streptokinase using a CHO lysate. Biotechnol. Bioeng 115 (1), 92–102. [DOI] [PubMed] [Google Scholar]
- (13).Yang J, et al. (2005) Rapid expression of vaccine proteins for B-cell lymphoma in a cell-free system. Biotechnol. Bioeng 89 (5), 503–511. [DOI] [PubMed] [Google Scholar]
- (14).Kanter G, et al. (2007) Cell-free production of scFv fusion proteins: an efficient approach for personalized lymphoma vaccines. Blood 109 (8), 3393–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Salehi A S. M, et al. (2016) Cell-free protein synthesis of a cytotoxic cancer therapeutic: Onconase production and a just-add-water cell-free system. Biotechnol. J 11 (2), 274–281. [DOI] [PubMed] [Google Scholar]
- (16).Karim AS, and Jewett MC (2016) A cell-free framework for rapid biosynthetic pathway prototyping and enzyme discovery. Metab. Eng 36, 116–126. [DOI] [PubMed] [Google Scholar]
- (17).Dudley QM, Anderson KC, and Jewett MC (2016) Cell-Free Mixing of Escherichia coli Crude Extracts to Prototype and Rationally Engineer High-Titer Mevalonate Synthesis. ACS Synth. Biol 5 (12), 1578–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Karim AS, et al. (2018) Controlling cell-free metabolism through physiochemical perturbations. Metab. Eng 45, 86–94. [DOI] [PubMed] [Google Scholar]
- (19).Schoborg JA, et al. (2018) A cell-free platform for rapid synthesis and testing of active oligosaccharyltransferases. Biotechnol. Bioeng 115 (3), 739–750. [DOI] [PubMed] [Google Scholar]
- (20).Kightlinger W, et al. (2018) Design of glycosylation sites by rapid synthesis and analysis of glycosyltransferases. Nat. Chem. Biol 14 (6), 627–635. [DOI] [PubMed] [Google Scholar]
- (21).Jaroentomeechai T, et al. (2018) Single-pot glycoprotein biosynthesis using a cell-free transcription-translation system enriched with glycosylation machinery. Nat. Commun 9 (1), 2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Shin J, Jardine P, and Noireaux V (2012) Genome Replication, Synthesis, and Assembly of the Bacteriophage T7 in a Single Cell-Free Reaction. ACS Synth. BioL 1 (9), 408–413. [DOI] [PubMed] [Google Scholar]
- (23).Rustad M, et al. (2018) Cell-free TXTL synthesis of infectious bacteriophage T4 in a single test tube reaction. Synth. Biol 3 (1), 002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Noireaux V, and Libchaber A (2004) A vesicle bioreactor as a step toward an artificial cell assembly. Proc. Natl. Acad. Sci. U. S. A 101 (51), 17669–17674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Bundy BC, Franciszkowicz MJ, and Swartz JR (2008) Escherichia coli-based cell-free synthesis of virus-like particles. Biotechnol. Bioeng 100 (1), 28–37. [DOI] [PubMed] [Google Scholar]
- (26).Adamala KP, et al. (2016) Engineering genetic circuit interactions within and between synthetic minimal cells. Nat. Chem 9, 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Shin J, and Noireaux V (2012) An E. coli Cell-Free Expression Toolbox: Application to Synthetic Gene Circuits and Artificial Cells. ACS Synth. Biol 1 (1), 29–41. [DOI] [PubMed] [Google Scholar]
- (28).Pardee K, et al. (2016). Portable, On-Demand Biomolecular Manufacturing. Cell 167 (1), 248–259. [DOI] [PubMed] [Google Scholar]
- (29).Sullivan CJ, et al. (2016) A cell-free expression and purification process for rapid production of protein biologies. Biotechnol. J 11 (2), 238–248. [DOI] [PubMed] [Google Scholar]
- (30).Schinn SM, et al. (2017) Rapid in vitro screening for the location-dependent effects of unnatural amino acids on protein expression and activity. Biotechnol. Bioeng 114 (10), 2412–2417. [DOI] [PubMed] [Google Scholar]
- (31).Shrestha P, Smith MT, and Bundy BC (2014) Cell-free unnatural amino acid incorporation with alternative energy systems and linear expression templates. New Biotechnol 31 (1), 28–34. [DOI] [PubMed] [Google Scholar]
- (32).Martin RW, et al. (2018) Cell-free protein synthesis from genomically recoded bacteria enables multisite incorporation of noncanonical amino acids. Nat. Commun 9 (1), 1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Hong SH, et al. (2014) Cell-free Protein Synthesis from a Release Factor 1 Deficient Escherichia coli Activates Efficient and Multiple Site-specific Nonstandard Amino Acid Incorporation. ACS Synth. Biol 3 (6), 398–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Hoon HS, et al. (2015) Improving Cell-Free Protein Synthesis through Genome Engineering of Escherichia coli Lacking Release Factor 1. ChemBioChem 16 (5), 844–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Takahashi MK, et al. (2015) Characterizing and prototyping genetic networks with cell-free transcription-translation reactions. Methods 86, 60–72. [DOI] [PubMed] [Google Scholar]
- (36).Takahashi MK, et al. (2015) Rapidly Characterizing the Fast Dynamics of RNA Genetic Circuitry with Cell-Free Transcription–Translation (TX-TL) Systems. ACS Synth. Biol 4 (5), 503–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Niederholtmeyer H, et al. (2015) Rapid cell-free forward engineering of novel genetic ring oscillators. eLife 4, No. e09771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Yim SS, et al. (2018) Multiplex transcriptional characterizations across diverse and hybrid bacterial cell-free expression systems. bioRxiv, DOI: 10.1101/42755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Watters KE (2018) Systematic discovery of natural CRISPR-Cas 12a inhibitors. Science 362, 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Pardee K (2016) Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell 165 (5), 1255–1266. [DOI] [PubMed] [Google Scholar]
- (41).Pardee K (2014) Paper-Based Synthetic Gene Networks. Cell 159 (4), 940–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Wen KY, et al. (2017) A Cell-Free Biosensor for Detecting Quorum Sensing Molecules in P. aeruginosa-infected Respiratory Samples. ACS Synth. Biol 6 (12), 2293–2301. [DOI] [PubMed] [Google Scholar]
- (43).Salehi ASM, et al. (2017) Cell-Free Protein Synthesis Approach to Biosensing hTRβ-Specific Endocrine Disruptors. Anal Chem 89 (6), 3395–3401. [DOI] [PubMed] [Google Scholar]
- (44).de los Santos ELC, et al. (2016) Engineering Transcriptional Regulator Effector Specificity Using Computational Design and In Vitro Rapid Prototyping: Developing a Vanillin Sensor. ACS Synth. Biol 5 (4), 287–295. [DOI] [PubMed] [Google Scholar]
- (45).Takahashi MK, et al. (2018) Alow-cost paper-based synthetic biology platform for analyzing gut microbiota and host biomarkers. Nat. Commun 9 (1), 3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Hodgman CE, and Jewett MC (2013) Optimized extract preparation methods and reaction conditions for improved yeast cell-free protein synthesis. Biotechnol Bioeng 110 (10), 2643–2654. [DOI] [PubMed] [Google Scholar]
- (47).Iizuka N, and Sarnow P (1997) Translation-Competent Extracts from Saccharomyces cerevisiae:Effects of L-A RNA, 5’ Cap, and 3’ Poly(A) Tail on Translational Efficiency of mRNAs. Methods 11 (4), 353–360. [DOI] [PubMed] [Google Scholar]
- (48).Li J, et al. (2017) Establishing a high yielding streptomyces-based cell-free protein synthesis system. Biotechnol Bioeng 114 (6), 1343–1353. [DOI] [PubMed] [Google Scholar]
- (49).Kelwick R, et al. (2016) Development of a Bacillus subtilis cell-free transcription-translation system for prototyping regulatory elements. Metab. Eng 38, 370–381. [DOI] [PubMed] [Google Scholar]
- (50).Harbers M (2014) Wheat germ systems for cell-free protein expression. FEBS Lett 588 (17), 2762–2773. [DOI] [PubMed] [Google Scholar]
- (51).Madin K, et al. (2000) A highly efficient and robust cell-free protein synthesis system prepared from wheat embryos: Plants apparently contain a suicide system directed at ribosomes. Proc. Natl. Acad. Sci. U. S. A 97 (2), 559–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Stech M, et al. (2017) Cell-free synliiesis of functional antibodies using a coupled in vitro transcription-translation system based on CHO cell lysates. Sci. Rep 7 (1), 12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Brödel AK, Sonnabend A, and Kubick S (2014) Cell-free protein expression based on extracts from CHO cells. Biotechnol. Bioeng 111 (1), 25–36. [DOI] [PubMed] [Google Scholar]
- (54).Martin RW, et al. (2017) Development of a CHO-Based Cell-Free Platform for Synthesis of Active Monoclonal Antibodies. ACS Synth. Biol 6 (7), 1370–1379. [DOI] [PubMed] [Google Scholar]
- (55).Stark JC (2018) BioBits Bright: Afluorescent synthetic biology education kit. Science Advances 4 (8), eaat5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Huang A (2018) BioBits Explorer: A modular synthetic biology education kit. Science Advances 4 (8), eaat5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Shrestha P, Holland TM, and Bundy BC (2012) Streamlined extract preparation for Escherichia coli-based cell-free protein synthesis by sonication or bead vortex mixing. Biotechniques 53 (3), 163–74. [DOI] [PubMed] [Google Scholar]
- (58).Kwon Y-C, and Jewett MC (2015) High-throughput preparation methods of crude extract for robust cell-free protein synthesis. Sci. Rep 5, 8663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Sun ZZ (2013) Protocols for Implementing an Escherichia coli Based TX-TL Cell-Free Expression System for Synthetic Biology. J. Visualized Exp No. 79, e50762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Fujiwara K, and Doi N (2016) Biochemical Preparation of Cell Extract for Cell-Free Protein Synthesis without Physical Disruption. PLoS One 11 (4), No. e0154614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Didovyk A, et al. (2017) Rapid and Scalable Preparation of Bacterial Lysates for Cell-Free Gene Expression. ACS Synth. Biol 6 (12), 2198–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Kim T-W, et al. (2006) Simple procedures for the construction of a robust and cost-effective cell-free protein synthesis system. J. Biotechnol 126 (4), 554–561. [DOI] [PubMed] [Google Scholar]
- (63).Shimizu Y, and Ueda T (2010) PURE Technology, In Cell-Free Protein Production: Methods and Protocols (Endo Y, Takai K, and Ueda T, Eds.) pp 11–21, Humana Press, Totowa, NJ. [Google Scholar]
- (64).Shimizu Y, et al. (2001) Cell-free translation reconstituted with purified components. Nat. Biotechnol 19, 751. [DOI] [PubMed] [Google Scholar]
- (65).Shin J, and Noireaux V (2010) Efficient cell-free expression with the endogenous E. Coli RNA polymerase and sigma factor 70. J. Biol. Eng 4, 8–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Carrier TA, and KeasHng JD (1999) Library of Synthetic 5’ Secondary Structures To Manipulate mRNA Stability in Escherichia coli. Biotechnol. Prog 15 (1), 58–64. [DOI] [PubMed] [Google Scholar]
- (67).Jermutus L, Ryabova LA, and Plückthun A (1998) Recent advances in producing and selecting functional proteins by using cell-free translation. Curr. Opin. Biotechnol 9 (5), 534–548. [DOI] [PubMed] [Google Scholar]
- (68).Liu DV, Zawada JF, and Swartz JR (2005) Streamlining Escherichia Coli S30 Extract Preparation for Economical Cell-Free Protein Synthesis. Biotechnol Prog 21 (2), 460–465. [DOI] [PubMed] [Google Scholar]
- (69).Jewett MC, and Swartz JR (2004) Substrate replenishment extends protein synthesis with an in vitro translation system designed to mimic the cytoplasm. Biotechnol. Bioeng 87 (4), 465–471. [DOI] [PubMed] [Google Scholar]
- (70).Jewett MC, and Swartz JR (2004) Mimicking the Escherichia coli cytoplasmic environment activates long-lived and efficient cell-free protein synthesis. Biotechnol. Bioeng 86 (1), 19–26. [DOI] [PubMed] [Google Scholar]
- (71).Kim D-M, and Swartz JR (2001) Regeneration of adenosine triphosphate from glycolytic intermediates for cell-free protein synthesis. Biotechnol. Bioeng 74 (4), 309–316. [PubMed] [Google Scholar]
- (72).Baugh C, Grate D, and Wilson C (2000) 2.8 Å crystal structure of the malachite green aptamer.J. Mol. Biol 301 (1), 117–128. [DOI] [PubMed] [Google Scholar]
- (73).Kolpashchikov DM (2005) Binary Malachite Green Aptamer for Fluorescent Detection of Nucleic Acids. J. Am. Chem. Soc 127 (36), 12442–12443. [DOI] [PubMed] [Google Scholar]
- (74).Yerramilli VS, and Kim KH (2018) Labeling RNAs in Live Cells Using Malachite Green Aptamer Scaffolds as Fluorescent Probes. ACS Synth. Biol 7 (3), 758–766. [DOI] [PubMed] [Google Scholar]
- (75).Siegal-Gaskins D, et al. (2014) Gene Circuit Performance Characterization and Resource Usage in a Cell-Free “Breadboard. ACS Synth. Biol 3 (6), 416–425. [DOI] [PubMed] [Google Scholar]
- (76).Canelas AB, et al. (2010) Integrated multilaboratory systems biology reveals differences in protein metabolism between two reference yeast strains. Nat. Commun 1, 145. [DOI] [PubMed] [Google Scholar]
- (77).Kelly JR, et al. (2009) Measuring the activity of BioBrick promoters using an in vivo reference standard. J. Biol Eng 3 (1), 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Failmezger J, et al. (2017) Cell-free protein synthesis from non-growing, stressed Escherichia coli. Sci. Rep 7 (1), 16524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Kim RG, and Choi CY (2000) Expression-independent consumption of substrates in cell-free expression system from Escherichia coli. J. Biotechnol 84 (1), 27–32. [DOI] [PubMed] [Google Scholar]
- (80).Jewett MC, and Swartz JR (2004) Mimicking the Escherichia coli cytoplasmic environment activates long-lived and efficient cell-free protein synthesis. Biotechnol. Bioeng 86 (1), 19–26. [DOI] [PubMed] [Google Scholar]
- (81).Chappell J, Takahashi MK, and Lucks JB (2015) Creating small transcription activating RNAs. Nat. Chem. Biol 11 (3), 214–220. [DOI] [PubMed] [Google Scholar]
- (82).Takahashi MK, and Lucks JB (2013) A modular strategy for engineering orthogonal chimeric RNA transcription regulators. Nucleic Acids Res 41 (15), 7577–7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Kuo A, Blough NV, and Dunlap PV (1994) Multiple N-acyl-L-homoserine lactone autoinducers of luminescence in the marine symbiotic bacterium Vibrio fìscheri. J Bacterial 176 (24), 7558–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Jinek M, et al. (2012) A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 337 (6096), 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Marshall R, et al. (2018) Rapid and Scalable Characterization of CRISPR Technologies Using an E. coli Cell-Free Transcription-Translation System. Mol. Cell 69 (1), 146–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Green AA (2014) Toehold Switches: De-Novo-Designed Regulators of Gene Expression. Cell 159 (4), 925–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Topp S (2010) Synthetic riboswitches that induce gene expression in diverse bacterial species. Appl. Environ. Microbiol 76, 7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Horvath N, et al. (2017) Toward a Genome Scale Sequence Specific Dynamic Model of Cell-Free Protein Synthesis in Escherichia coli. bioRxiv, DOI: 10.1101/215012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Kim H-C, and Kim D-M (2009) Methods for energizing cell-free protein synthesis.J. Biosci. Bioeng 108 (1), 1–4. [DOI] [PubMed] [Google Scholar]
- (90).Dong-Myung K, and Swartz JR (1999) Prolonging cell-free protein synthesis with a novel ATP regeneration system. Biotechnol Bioeng 66 (3), 180–188. [PubMed] [Google Scholar]
- (91).Des Soye BJ, et al. (2018) Establishing a High-Yielding Cell-Free Protein Synthesis Platform Derived from Vibrio natriegens. ACS Synth. Biol 7 (9), 2245–2255. [DOI] [PubMed] [Google Scholar]
- (92).Wiegand DJ (2018) Establishing a Cell-Free Vibrio natriegens Expression System. ACS Synth. Biol 7, 2475. [DOI] [PubMed] [Google Scholar]
- (93).Qi L, et al. (2012) Engineering naturally occurring trans-acting non-coding RNAs to sense molecular signals. Nucleic Acids Res 40 (12), 5775–5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Chappell J, et al. (2017) Computational design of small transcription activating RNAs for versatile and dynamic gene regulation. Nat. Commun 8 (1), 1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All source data for main and SI figures was deposited open access in Northwestern’s arch database (https://arch.library.northwestern.edu/). Data can be accessed via https://doi.org/10.21985/N2318T.