Abstract
The derivation of induced pluripotent stem cells (iPSCs) over a decade ago sparked widespread enthusiasm for the development of new models of human disease, enhanced platforms for drug discovery, and more widespread use of autologous cell-based therapy. Early studies using directed differentiation of iPSCs frequently uncovered cell-level phenotypes in monogenic diseases, but translation to tissue-level and organ-level diseases has required development of more complex, 3D, multicellular systems. Organoids and human–rodent chimaeras more accurately mirror the diverse cellular ecosystems of complex tissues and are being applied to iPSC disease models to recapitulate the pathobiology of a broad spectrum of human maladies, including infectious diseases, genetic disorders and cancer.
Introduction
More than 10 years ago, the discovery that mature human somatic cells from the skin or blood could be reprogrammed to a pluripotent state and then differentiated along alternative cell lineages offered the theoretical opportunity for personalized cell-based autologous therapies in a wide array of diseases1–4. Enthusiasm for patient-specific therapies based on induced pluripotent stem cells (iPSCs) rose with the rapid advances in precise DNA editing that followed. However, for the development of optimized disease models as platforms to uncover new drugs, human patient-derived iPSCs must effectively differentiate to cell states that faithfully recapitulate hallmarks of diseased cells and tissues. Therefore, the clinical utility of iPSC-derived products relies heavily on the state of the art of directed differentiation, cell state conversion and tissue engineering. Now, a decade later, where do we stand in attaining these goals (Fig. 1)?
Figure 1. Progress in iPSC-based therapies.
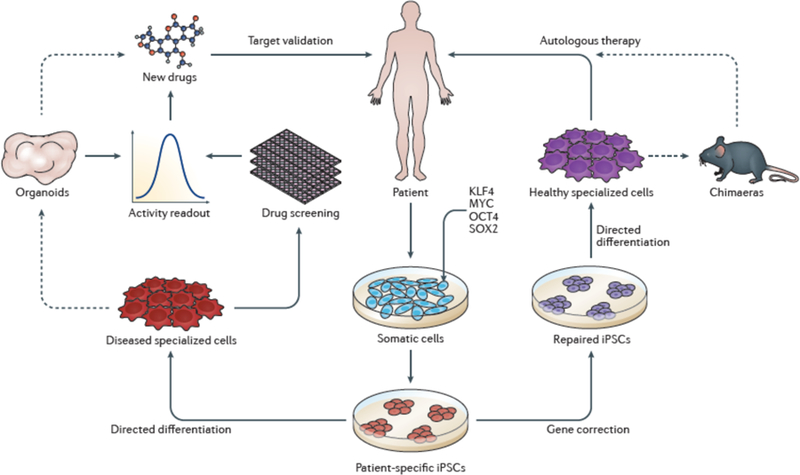
Upon the discovery of iPSCs, approaches to cell-based therapy and drug discovery were proposed22. Here, we illustrate the progress made toward these goals in the past decade. Iterative improvements in reprogramming methodologies have increased the efficiency of iPSC derivation and the quality of iPSC lines in use. As a stride toward autologous iPSC-based therapy, gene editing using CRISPR–Cas9 technology has been widely applied to iPSCs to enable repair of disease-causing genetic lesions. As discussed in this review, remarkable advances have been made approaches to in directed differentiation that can be used to derive gene-corrected terminally differentiated cells. However, with rare exceptions150, widespread autologous iPSC-based cell therapy remains out of reach. Optimization of transplantation and directed differentiation in chimaeric models as well as improvement in the efficiency and scalability of generating clinical-grade cells will continue to advance toward the goal of autologous therapy in varied organ systems. Non-corrected cells differentiated from patient-derived iPSCs have been used in drug screening and validation studies largely using cell-level readouts23. Currently, a handful of candidate drugs identified in iPSC-based systems are under study in human trials23. We anticipate that increased sophistication of iPSC-based disease models using organoids will integrate into these approaches to drug discovery by improving preclinical drug screening, design and validation using highly disease-relevant readouts to accelerate candidate therapies into clinical trials. Figure adapted with permission from Ref. 22.
Since the first isolation of human embryonic stem cells (ESCs) in 1998, much effort has focused on the differentiation of ESCs to mature tissues with the long-term objective of allogeneic cell therapy5. Protocols were developed to differentiate ESCs to neural cells6,7, cardiomyocytes8, pancreatic β-cells9 and haematopoietic cells10,11, which were leveraged into early ESC-based disease models12–14. Given that the isolation of primary diseased ESC lines was limited by the requirement for preimplantation genetic diagnosis in the context of in vitro fertilization12, genetic manipulation of normal ESC lines was used to introduce disease-relevant molecular defects using contemporary technologies such as RNA interference or homologous recombination13,14. These seminal studies sustained optimism for modelling diseases using ESCs; after all, these human systems lack the limitations of evolutionary divergence inherent to the interpretation of disease studies in model organisms.
The emergence of reprogramming technology enabled the use of disease-specific iPSCs in established directed differentiation protocols, circumventing many limitations of disease modelling in ESCs, such as the need for gene editing15–18 (Fig. 1). Cell-level phenotypes were identified in cells differentiated from patient-specific iPSCs across a panoply of diseases, including impaired motor neuron differentiation in iPSCs from patients with spinal muscular atrophy15; decreased neuronal connectivity, neurite formation and synaptic protein expression in neurons derived from iPSCs obtained from patients diagnosed as having schizophrenia19; prolonged action potential in cardiomyocytes derived from patients with familial long-QT syndrome16; defects in neural differentiation in iPSCs from patients with familial dysautonomia20; or diminished colony formation in haematopoietic progenitors derived from patients with Fanconi anaemia21. These phenotypes seem robust, reproducible and disease-relevant, are measurable in cell culture-based systems, and reflect diseases intrinsic to the cellular level.
Although these systems have markedly advanced the understanding of mechanisms underlying the modelled disorders, human disease occurs within the context of complex multicellular ecosystems wherein interactions occur between cells, extracellular matrices, tissues, organ systems and pathogens. Consequently, more complex iPSC-based differentiation systems are currently being developed to faithfully recapitulate human tissue-level and organ-level dysfunction, which have recently been incorporated into disease modelling, drug screening and the study of host–pathogen interactions. Furthermore, advances in xenotransplantation have facilitated the development of ever more complex iPSC-derived human tissues for the study of disease. However, widely pervasive barriers to further advancement of disease modelling in iPSCs exist, including the persistence of embryonic or fetal identity in derived tissues and background genetic variability of iPSC lines.
Here, we take stock of the progress made in modelling human disease using iPSCs, discuss recent studies that define the extent of the current frontiers of disease modelling using iPSCs and review the challenges impeding further progress. Improvements in differentiation and re-specification of iPSCs toward more complex 3D tissues as well as engraftment of iPSC-derived cells to form chimeric animals are accelerating the field toward highly faithful multicellular disease models. Furthermore, as the leading edge of iPSC research has advanced, it has also broadened such that inroads are now being made in using sophisticated iPSC-based models to interrogate host–pathogen interactions. We will discuss these recent advances that we believe have driven iPSC research ever closer to long-awaited therapeutic breakthroughs. In synthesizing the implications of these innovations, we present a case to support continued optimism in iPSCs as imminently yielding faithful disease models, validated drugs and cell-based therapies.
Organoids: complex tissues in a dish
Following the isolation of human ESCs, differentiation protocols typically modulated morphogen exposure in 2D tissue culture or within embryoid bodies with the aim of recapitulating the patterning events of early embryogenesis to first specify a desired germ layer followed by a specific target lineage6–11. Upon the development of patient-specific iPSCs, these techniques were applied to disease modelling to derive an affected cell lineage17. A flurry of studies over a period of about 6 years starting in 2008 described cellular-level phenotypes in iPSC-based models across a wide spectrum of diseases22. Although drug screening efforts using these systems identified candidate small-molecule therapies16,20,23–25, unfortunately, to date, translation from cell-based systems to human trials has not yet satisfied early hopes and expectations.
A significant advance in disease modelling with iPSCs occurred with the development of iPSC-derived organoids26,27. Organoids are 3D multicellular aggregates derived from stem cells that differentiate and self-organize to recapitulate the structural features and cell–cell interactions of mature tissues28. The soluble and biophysical cues employed to guide organoid differentiation from PSCs have been incrementally refined to generate increasingly complex “tissues in a dish” 26,27. Furthermore, investigators have achieved specification of particular regions of organs such as the brain and gastrointestinal tract29,30. Here, we will review key studies that have advanced human iPSC organoid technology (Table 1, Table 2) and discuss where these advances fit in the broader scheme of iPSC disease modelling and drug discovery (Fig. 2).
Table 1.
Phenotypes modelled in 2D and 3D iPSC-based systems
| Tissue | 2D Phenotype | 3D Phenotype | References |
|---|---|---|---|
| Blood | Oligopotent differentiation | Multipotent differentiation, engraftment | 10,11,108 |
| Neural | Neural differentiation, gene expression, neurite formation | Cortical organization, regional specification, cell-cell interactions, neuronal migration | 15,19,20,31,32,40,42 |
| Cardiac | Action potential, contractility | Self-organization, integration of biophysical cues | 16,64 |
| Gastrointestinal | Differentiation | Bile secretion, motility, cell-cell interactions | 46–49 |
Table 2.
Key studies of disease modelling using organoids derived from human iPSCs.
| Organ system | Site | Cell types analysed | Diseases modelled | Gene, locus or intervention | Disease phenotype | Ref. |
|---|---|---|---|---|---|---|
| Neural | Cerebrum | Neurons, NPCs, RG, retina, choroid plexus, meninges | Microcephaly | CDK5RAP2 | Premature neuronal differentiation | 31 |
| Neural | Cerebrum | Neurons, RG | Seckel syndrome microcephaly | CENPJ | Smaller organoids, diminished RG, premature neuronal differentiation | 39 |
| Neural | Cerebrum | Neurons, RG, NPCs | Autism | N/A | Increased GABAergic neuron fate | 40 |
| Neural | Cerebrum | Neurons, NPCs | Rett syndrome | MECP2 | Increased ventricular area, impaired neurogenesis | 41 |
| Neural | Cerebrum | NESCs | Miller–Dieker lissencephaly | 17p13.3 deletion | Smaller organoids, poor neurite growth, apoptosis | 42 |
| Neural | Cerebral | NPCs | Glioblastoma | MYC and others | Organoid overgrowth | 43 |
| Gastrointestinal | Gastric | Epithelial, neuroendocrine | Helicobacter pylori | Infection | Increased epithelial proliferation | 30 |
| Gastrointestinal | Intestinal | Epithelial, enteric nervous system | Hirschsprung disease | PHOX2B | Impaired enteric nervous system organization | 46 |
| Gastrointestinal | Liver | Cholangiocytes | Alagille syndrome | Pharmacologic notch inhibition | Impaired organoid formation | 49 |
| Gastrointestinal | Liver | Cholangiocytes | Cystic fibrosis | CFTR | Dysfunctional epithelial transport | 48 |
| Gastrointestinal | Colon | Colonocytes | Colon cancer | APC | Epithelial proliferation | 50 |
| Kidney | Glomeruli, proximal tubules | Podocytes, epithelial cells, endothelial cells | Polycystic kidney disease | PKD1, PKD2 | Cyst formation | 54 |
| Lung | Airway | Epithelial cells | Cystic fibrosis | CFTR | Impaired forskolin-induced swelling | 58 |
| Lung | Alveolus | Type 2 epithelial cells | Surfactant deficiency | SFTPB | Diminished surfactant production | 59 |
| Heart | Myocardium | Cardiomyocytes | Regeneration | Injury | Fetal-like cardiomyocyte proliferation | 60 |
NESCs, neural epithelial stem cells; NPC, neural progenitor cell; RG, radial glia.
Figure 2. Application of iPSC-derived organoids to disease modelling and drug discovery.
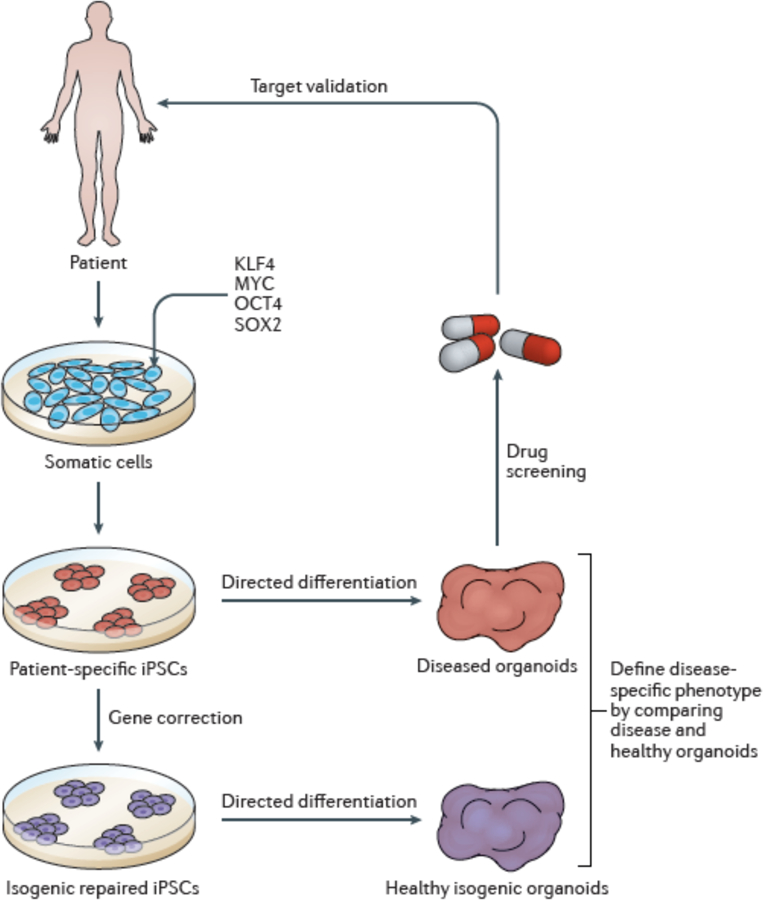
Remarkable progress has been made in the differentiation of increasingly complex multicellular and diverse organoid systems across many tissues. We propose that parallel differentiation of organoids from patient-derived iPSCs as well as genetically corrected, isogenic control iPSCs will allow attribution of an organoid-level disease phenotype to a specific molecular lesion. Once a clear organoid-level readout is established, diseased organoids can be used in drug screening and validation studies.
Neural organoids.
Neural differentiation of PSCs has been intensely investigated, as neurectoderm seems to be a default cell fate induced upon removal of PSCs from culture conditions that maintain pluripotency6,7. Early studies in ESCs and iPSCs utilized neural differentiation methodologies to model neurologic disorders15,19,20. As a great stride forward, the development of 3D cerebral organoids derived from human PSCs31 built upon these foundational neural differentiation protocols but additionally provided instructive morphogenetic cues followed by growth in a suspension bioreactor for up to 70 days31,32. These organoids contained functional neurons with cortical self-organization, specified forebrain and hindbrain regions and, remarkably, even included differentiation of immature retina and choroid plexus31,32. Refinement of this methodology has led to the specification of further specific regions of the brain such as hippocampus and cerebellum, the derivation of functional photoreceptors and astrocytes, and induction of cortical folding29,33–38. Along with this innovation, such meticulously patterned organoids likely bear less variability in structure compared to their predecessors.
The remarkable complexity of neural organoids has been harnessed to model a variety of monogenic and polygenic neurologic, developmental and psychiatric disorders that have collectively provided deep insight into the pathobiology of neurologic diseases. Human iPSC-derived neural organoids were initially leveraged to model microcephaly due to compound heterozygous mutations in CDK5RAP2, which encodes a centrosomal protein that localizes to the spindle poles during mitosis, finding that patient-specific neural organoids underwent premature neuroepithelial differentiation, showed aberrant radial glial orientation and smaller areas of differentiated neural tissue31. Similar phenotypes were observed in neural organoids derived from iPSCs from patients with Seckel syndrome with microcephaly caused by mutations in CENPJ39, another centrosomal protein that function in the regulation of microtubule assembly and nucleation. Neural organoids derived from iPSCs from patients with autism showed normal early neuronal differentiation but a relative increase in inhibitory GABAergic neuron fate over glutamatergic fate due to overproduction of the transcriptional repressor FOXG1, suggesting this differentiation imbalance as a mechanism underlying autism pathogenesis40. Using iPSCs bearing MECP2 mutations, derived from patients with Rett syndrome, neural organoids showed impaired neurogenesis and reduced neuronal migration41. Cortical organoids from iPSCs from patients with lissencephaly due to Miller–Dieker syndrome, which is caused by a heterozygous deletion at 17p13.3, showed poor neurite growth and apoptosis in the ventricular zone, and impaired neuronal migration42.
Recently, neural organoids derived from human PSCs have been used as a platform for the study of brain tumours43. Cerebral organoids containing neural stem and progenitor cells were transduced with vectors to introduce mutations commonly found in brain tumours, resulting in overgrowth of transduced cells43. Thus, compared to 2D systems, neural organoids have enhanced our understanding of the pathobiology of neurologic diseases.
Gastrointestinal and liver organoids.
Gastrointestinal organoids have been intensely studied for the past 10 years28. Intestinal organoids can be derived from adult intestinal stem cells44 as well as PSCs45. By using a strict schedule of morphogen exposure to specify endoderm commitment, foregut and hindgut fate, organoids can be generated from human PSCs containing functional enterocytes as well as goblet cells, Paneth cells and neuroendocrine cells45. Subsequently, by modulating retinoic acid signalling to define posterior foregut specification, gastric antral organoids could be generated with a complex antral epithelial structure, which were used for modelling infection with Helicobacter pylori30. Another layer of complexity was added when PSC-derived neural precursor cells were incorporated into intestinal organoids, forming a functional enteric nervous system capable of mediating contractile waves46. Defects in neuronal migration into the primitive intestine underlie Hirschsprung disease, which frequently requires surgical resection of large tracts of aganglionic intestine due to impaired motility. Human iPSC-derived intestinal organoids combined with neuronal precursor cells deficient in the PHOX2B gene — which when mutated in humans causes a complete loss of intestinal innervation — showed impaired organization of an enteric nervous system46. Together, these studies have ushered in advanced regional specification and complexity of iPSC-derived gastrointestinal organoids and resulted in marked improvements in disease modelling.
2D differentiation protocols have proven effective in generating hepatocytes from iPSCs for a variety of liver disorders47. More recently, cholangiocytes derived from iPSCs with hepatobiliary disease have been derived in 3D organoid systems48,49. Two similar differentiation protocols have been described, both beginning with initial directed differentiation to definitive endoderm and hepatoblasts followed by specification to cholangiocyte-like cells in 3D culture48,49. These cholangiocyte organoids could efflux bile acids and possessed functional secretory action48,49, enabling the modelling of Alagille syndrome, a condition of impaired bile duct formation caused by disrupted Notch signalling49. In cholangiocyte organoids differentiated from a patient with polycystic liver disease, the synthetic somatostatin analogue octreotide decreased the size of organoids, consistent with the action of this drug in patients with polycystic liver disease49. Cholangiocyte organoids derived from iPSCs from patients with cystic fibrosis carrying the F508del CFTR mutation showed impaired chloride transport and diminished CFTR-dependent, forskolin-induced swelling that was rescued with the CFTR corrector drug VX-80948,49. In each of these cases, modelling of a complex disease phenotype was uniquely enabled by development of a 3D organoid.
Gastrointestinal organoids derived from PSCs can be used to model intestinal neoplasia. Precise morphogen-directed specification of hindgut endoderm can yield colonic organoids with morphologic crypts containing goblet cells, epithelial cells and neuroendocrine cells50. iPSCs from patients with familial adenomatous polyposis with germline mutations in the APC gene generated colonic organoids with increased nuclear localization of β-catenin and proliferation compared to control organoids50.
Kidney organoids.
The discovery of human iPSCs raised enthusiasm for the potential differentiation of functional renal tissue for autologous transplantation; thus, renal differentiation of iPSCs and modelling of kidney disorders has received much attention51,52. Derivation of human kidney organoids from iPSCs was first reported in 2015, as complex 3D structures containing nephron components (glomeruli and proximal and distal tubules) as well as stroma and vasculature53. Subsequently, it was shown that iPSCs can be differentiated to ureteric bud progenitor-like cells that, upon culture in 3D, develop into primitive ureteric buds that can be applied to modelling polycystic kidney disease (PKD)52. Another method first differentiates human iPSCs to epiblast spheroids in 3D and then to kidney organoids by inhibiting GSK3β signalling54. Genetic ablation of the PKD1 or PKD2 genes in human iPSCs in this system results in the formation of aberrant cysts54. Methods for generating kidney organoids from human iPSCs continue to advance in complexity, such that it is reasonable to anticipate further refinement of renal disease models55,56. As an alternative system, models of functional vascularized iPSC-derived glomeruli dependent on laminar flow provide a platform for modelling functional glomerular disease in the future57.
Lung organoids.
Two recent studies have highlighted the power of human iPSC-derived lung organoids in disease modelling58,59, by directed differentiation of human iPSCs to an Nkx2.1+ airway progenitor capable of subsequent patterning into either proximal or distal airway cells58. Culture of Nkx2.1+ progenitors in 3D with low WNT activation resulted in the reproducible formation of proximal airway organoids containing cells of secretory, goblet and basal lineages58. iPSCs from patients with cystic fibrosis homozygous for the F508del mutation efficiently formed proximal airway organoids but showed impaired forskolin-induced swelling compared to organoids from a control human iPSC line58. Attributing this phenotype to CFTR dysfunction, genetic correction of F508del in iPSCs rescued forskolin-induced swelling58, thereby revealing the power of gene editing to confirm genotype–phenotype relationships in iPSC disease models.
Human iPSC-derived Nkx2.1+ lung progenitor cells can also be differentiated to distal alveolar organoids in 3D culture59. These alveolar organoids contain functional type 2 alveolar epithelial cells that contain lamellar bodies and secrete surfactant proteins59. Using iPSCs from a patient deficient in surfactant protein B, the investigators found that differentiated type 2 alveolar epithelial cells did not contain lamellar bodies or synthesize SFTPB, a phenotype reversible by Cas9-mediated correction of the SFTPB mutations59.
[H2] Cardiac organoids.
Recently, considerable advances have been made in the development of self-organizing cardiac organoids. iPSC-derived cardiac organoids show features of fetal-like differentiation and have been used to model cardiomyocyte regeneration following injury60. This finding reinforces the notion that cardiomyocytes and other tissues derived from iPSCs often show fetal-like differentiation, which can hamper modelling of adult disease61–63. Progressive refinement of differentiation protocols has recently generated complex cardiomyocyte organoids with a central void chamber64. Furthermore, iPSCs have been used to engineer 3D cardiac muscle tissue to evaluate the effect of mechanical forces, metabolism and the extracellular matrix on cardiomyocyte maturation65,66. Theses studies incorporate aspects of tissue engineering using iPSCs. Interestingly, iPSCs have also been used to model cardiomyopathy using 3D organ-on-a-chip technology, a concept gaining traction in many other disease systems that provides the added dimension of modelling vascular perfusion26,57,67,68.
We have described a handful of the innovative advances that have been made in the derivation of increasingly complex human iPSC-derived organoids in efforts to advance toward the goal of autologous iPSC-based therapies (Table 2, Figure 2). As this frontier has advanced, these organoid systems have yielded invaluable insight into disease pathogenesis, building on the foundational studies modelling disease with iPSCs in 2D systems. In many of these studies, gene correction in iPSCs can reverse disease phenotypes at the organoid level, providing proof-of-principle that this approach could be applied to autologous iPSC-based therapy of solid organ diseases in the future. It should be noted that trade-offs exist in the adoption of organoid-based systems relative to 2D culture systems. Novel organoid culture technologies designed to address the inherently decreased throughput of these models should be widely applied for possible future use in genomic or drug screens33,66. Presumed batch effects arising from variation in in organoid organization and differentiation between experiments and iPSC cell lines must be considered, with appropriate quality controls incorporated into experimental designs. Limited oxygen penetration limits growth in culture, and future systems will build on early studies of perfused organoid xenografts69,70. Furthermore, human genetic variation that can impact iPSC differentiation may influence the comparison of organoids from patients with the modelled disease and healthy control individuals71,72. Future studies will balance self-organization and tissue engineering approaches to tailor organoids to specific organ systems and pathophysiologic processes31,65. We anticipate that future studies using iPSC-derived cells differentiated along specific lineages assembled into ever more complex and functional organoids and engineered tissues will be exploited to further understand disease pathogenesis at the tissue and organ levels26,46,56,73.
Modelling host–pathogen interactions using iPSCs
Although the earliest iPSC-based disease models focused on genetic diseases15,17, it has been shown that cells terminally differentiated from human iPSCs are susceptible to infection with human pathogens, opening a new frontier for probing host–pathogen interactions. Human iPSC-based systems overcome the limitation of species specificity of infectious pathogenicity and inflammatory responses, which has limited the translation of findings from model organisms to humans74,75. Moreover, iPSC-based systems have recently provided platforms upon which to screen for novel antimicrobials76.
[H2] Modelling infections at the cellular level using iPSCs.
Cells differentiated from human iPSCs can be targets of pathogenic viral infection. One early study used a model of HIV infection of T-cells to show that iPSC-derived natural killer (NK) cells could mediate potent anti-HIV activity through direct cytotoxicity of target cells, cytokine elaboration, and antibody-dependent cellular cytotoxicity77. HIV replication in target cells could be further enhanced by expression of a chimeric extracellular CD4 domain fused to the intracellular domain of CD3ζ in human iPSC-derived NK cells78. Using CRISPR–Cas9, the introduction of the CCR5del32 mutation into human iPSCs rendered differentiated monocytes resistant to HIV infection, providing proof-of-principle for iPSC-based therapy for HIV79. In a model of herpes simplex virus 1 (HSV-1) encephalitis, neurons differentiated from iPSCs from patients deficient in either TLR3 or UNC-93B showed increased susceptibility to HSV-1 infection, consistent with the sensitivity of such patients to HSV-1 encephalitis80. Human iPSC-derived macrophages have also been used to model Dengue virus infection81.
[H2] Modelling viral infection in 3D systems.
These studies of viral interaction with human iPSC-derived cells have laid the foundation for recent studies using organoids (Table 3), with several timely studies modelling Zika virus infection. Two landmark publications illustrated the value of iPSC-derived organoids by showing that Zika infection impairs growth and causes cell death within human iPSC-derived neurospheres82,83. Subsequently, it was reported that recent Zika isolates induced differentiation of neural precursor cells in the ventricular zone of human neurospheres due to disruption of the centrosome thereby promoting incorrect orientation of the mitotic division plane84. Recently, Zika virus was again shown to impair organoid growth, but infection also induced a lissencephaly-like phenotype in a model of cerebral folding37. It has been proposed that the AXL protein is important for Zika infection of neural progenitor cells85. However, ablation of AXL in cerebral organoids does not affect Zika infection86. These observations were extended by engineering a miniaturized spinning bioreactor system — a requirement for neural organoid derivation31 — to generate forebrain, midbrain, and hypothalamic organoids by modification of directed differentiation stimuli33. This forebrain differentiation system was used to model Zika virus infection at various developmental time points, finding that infection of neural progenitor cells in early organoids resulted in markedly decreased organoid size and decreased ventricular zone thickness and dilated ventricular structures33. This miniaturized spinning bioreactor system could serve as a platform for drug discovery using neural organoids in the future. In another study, human iPSC-derived neural progenitor cells (NPCs) were screened for compounds that block Zika infection and clear Zika virus from infected NPCs76; that is, human iPSC-derived forebrain organoids were used to validate the ability of hit compounds to inhibit Zika virus infection of neural tissue76.
Table 3.
Key studies modelling infectious disease using human iPSCs.
| Target | Pathogen | Phenotype | Ref. |
|---|---|---|---|
| T cells | HIV | Target cell killing by NK cells differentiated from iPSCs | 77,78 |
| Monocytes | HIV | Introduction of CCR5del32 mutation causes HIV resistance | 79 |
| Neurons | HSV-1 | Neurons derived from iPSCs from patients with TLR3 or UNC-93B mutations show increased susceptibility | 80 |
| Neural organoids | Zika | Impaired organoid growth, decreased ventricular zone thickness, increased ventricle size, decreased cortical folding | 31,33,37,76,82–86 |
| Gastric organoids | Helicobacter pylori | Increased epithelial cell proliferation | 30 |
| Intestinal organoids | Clostridium difficile | Impaired epithelial barrier function | 90 |
| Intestinal organoids | Salmonella | Epithelial cell invasion | 91 |
iPSCs, induced pluripotent stem cells; NK, natural killer.
Organoids derived from human PSCs have been used to model infection of epithelial tissue. Human ESC-derived intestinal organoids have been used to model rotavirus pathobiology87. A sophisticated system of branching human lung organoids derived from PSCs has been employed to model infection with respiratory syncytial virus, where infected epithelial cells swell and shed, as seen in human tissues88. These studies demonstrate the power of organoid systems in recapitulating human viral infection.
[H2] Bacterial infections.
Organoids have been used to model bacterial infection in the gastrointestinal system. Human gastric antrum organoids were microinjected with H. pylori, and the morphologic and biochemical response of epithelial cells was measured30. H. pylori closely associated with gastric epithelium, induced c-Met phosphorylation and epithelial proliferation, recapitulating aspects of H. pylori infectious pathobiology30,89. Intestinal organoids derived from human ESCs have been used to model infection with Clostridium difficile, a major cause of hospital-acquired colitis90,91. Toxigenic C. difficile associated with intestinal epithelial cells and impaired organoid barrier function90. Human iPSC-derived intestinal organoids were used to model Salmonella infection92. Salmonella injected into intestinal organoids associated with microvilli and invaded epithelial cells, forming Salmonella-containing vacuoles within epithelial cells92. Human ESC-derived enterospheres containing adult-like intestinal epithelial cells were used to model infection with the Escherichia coli strain O157:H793. E. coli induced a transcriptional inflammatory response in PSC-derived intestinal epithelial cells93.
Collectively, these studies show that human PSC-derived systems are amenable to modelling host–pathogen interactions across a variety of infectious pathogens and organ systems. We expect that further sophistication of organoid systems, particularly by the incorporation of immune and inflammatory cells, will lead to broadening of the repertoire of human PSC-based infectious disease models. We speculate that human iPSC models will play a particularly crucial role in the field of infectious disease in the future due to the species specificity of host–pathogen interactions that limits translation of findings from non-human models to clinically relevant therapies. Furthermore, scaling human iPSC organoid models for drug screening will provide a major advance in the identification and validation of novel antibiotics and antivirals33,76.
Human–animal chimaeras
Although increasing in complexity, maturation and cellular diversity, confinement of organoid-based disease models to the tissue culture setting limits analysis of interactions with the circulatory, nervous and immune systems, hormones and other metabolic mediators that are present in vivo26. Furthermore, organoids rely on self-organization in culture, missing many morphogenetic cues present in vivo28. In modelling blood diseases, in vitro differentiation systems rarely incorporate the niche factors that feed into specification of haematopoietic differentiation and bona fide hematopoietic stem cells94,95. Recently, these limitations have been addressed by advances in the xenotransplantation of cells and organoids differentiated from human PSCs. Given the recent explosion of cell-based therapies under investigation in the preclinical or clinical settings for many diseases96–100, iPSC-based chimaeric disease modelling by xenotransplantation has emerged as a reliable tool to fuel these advances by faithfully modelling human disease (Fig. 3, Box 1). Here, we will focus on key studies in several organ systems.
Figure 3. iPSC-based chimaeric models.
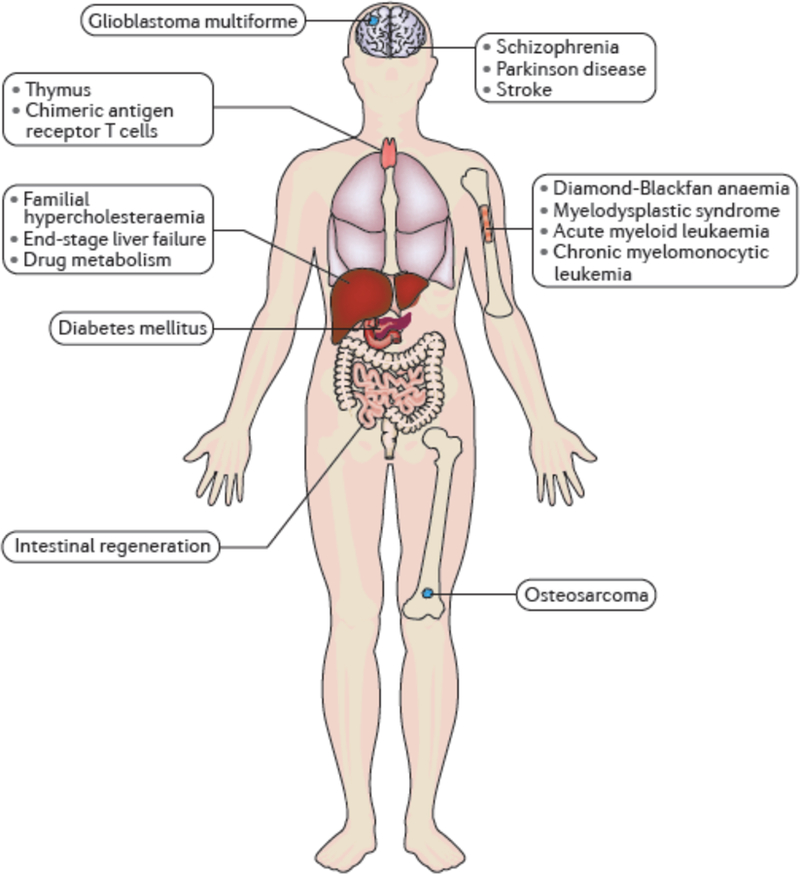
Summary of the current state of human iPSC chimaera models, illustrating the diversity of tissues and organs modelled as chimaeras and the classes of disease that have been modelled in chimaeras.
Box 1. Considerations in xenotransplantation of cells derived from iPSCs.
The efficiency of engraftment of human tissue into mice to generate chimaeric models relies on immunocompromized murine hosts that enable the production of ‘humanized’ mice. This technology has been driven by research in haematopoiesis, where models supportive of engraftment of human blood cells have greatly enhanced our understanding of human haematopoietic stem and progenitor cells (HSPCs)141. Since the first report of the scid mutation in 1983, which impaired B-cell and T-cell development in mice142, iterative addition of further mutations that compromise host immunity has enhanced the receptiveness of genetically engineered mice to human haematopoietic grafts141,143. The triple mutant NOD.Prkdcscid Il2rgtm1Wjl strain is receptive to engraftment by re-specified HSPCs derived from human iPSCs106–108, enabling formation of haematopoietic chimaeras from iPSCs following irradiation. However, besides ablating native haematopoietic cells to ‘clear space’ for an incoming exogenous graft, irradiation damages the supporting haematopoietic microenvironment and causes genetic toxicity144. Future studies of the function of HSPCs in chimaeric mice should make use of new models that do not require genotoxic conditioning such as antibody-mediated conditioning or NOD.Prkdcscid Il2rgtm1Wjl mice bearing the W41 mutation in c-kit that can be engrafted with human HSPCs in the absence of conditioning144,145.
Immunodeficient mouse models are now in widespread use to generate non-haematopoietic chimaeras using human iPSC-derived cells. Such strains have been used to generate neural122,126, liver70, solid tumor118,119 and pancreas chimaeras131. These studies illustrate the applicability of immunocompromized mice, not only to iPSC-based disease modelling but also to tissue engineering and development of approaches to transplantation.
Haematopoietic chimaeras.
Since the isolation of human ESCs, efforts have focused on using directed differentiation to generate haematopoietic stem and progenitor cells (HSPCs) with the potential for engraftment into recipient humanized mice as a first step to cellular therapy for blood disorders11,94,95. Initial efforts yielded haematopoietic progenitors with restricted myeloid and erythroid lineage potential, resembling the primitive progenitors arising within the yolk sac in early embryogenesis10,11. More recently, fine-tuning of the signalling axes that delineate primitive versus definitive haematopoietic fate (for example, activin–nodal and WNT) has led to the directed differentiation of definitive-like progenitors defined by in vitro lymphoid potential94,101,102. However, even the most sophisticated morphogen directed differentiation protocols designed to derive definitive HSPCs from PSCs do not yield bona fide haematopoietic stem cells (HSCs) with the capacity for high-level long-term engraftment into immunodeficient mice required to produce chimaeric disease models94,101. Indeed, derivation of engraftable HSCs from PSCs by directed differentiation in the absence of transgene induction in vitro remains elusive103.
Our group aimed to unlock the capacity for in vivo engraftment in HSPCs derived via directed differentiation from PSCs to enable improved disease modelling and to advance iPSC-based haematopoietic therapy. Predicated on the finding that primitive-like HSPCs derived from murine ESCs could be re-specified to a definitive phenotype with multi-lineage engraftment potential and self-renewal by enforced expression of the homeotic transcription factor HOXB4104,105, our group performed an in vivo screen for transcription factors that drive engraftment of primitive-like human HSPCs derived from PSCs106. Expression of HOXA9, ERG, RORA, SOX4 and MYB could expand primitive-like progenitors derived from human iPSCs in culture and confer short-term myeloid and erythroid engraftment in immunocompromized mice106. We used this system to model the congenital erythroid aplasia of Diamond-Blackfan anaemia (DBA) using iPSCs from patients carrying mutations in the ribosomal protein genes RPS19 or RPL5107. Engraftment of re-specified human DBA iPSC-derived progenitors to generate chimeric mice revealed markedly impaired erythroid differentiation, recapitulating the human DBA phenotype107. Re-specified, expandable progenitors were used to screen for small molecules that could rescue erythropoiesis in cells from patients with DBA, identifying the autophagy inducer SMER28 as a candidate therapeutic that was also capable of rescuing erythropoiesis in chimeric mice107. Recently, our group reported transcription factor-mediated re-specification of definitive haematopoietic progenitors from human iPSCs to engraftable, self-renewing, multi-lineage HSCs108. Given that human iPSC lines from many genetic blood disorders exist13,109–111, we anticipate that this system will support deeper modelling of blood disorders in the future. As in vitro HSPC production remains inefficient in this system, translation to transplantation therapy remains dependent on further advances and will likely interface with further development of human HSPC xenotransplantation (Box 1).
Chimaeras to model cancer.
Xenografting of tumors into immunodeficient mouse models has been a key technique in cancer research for several years112. The ability to form haematopoietic chimaeras has been exploited in human iPSC models of haematologic malignancies113,114. Acute myeloid leukemia (AML) blasts bearing translocations in the MLL locus can be efficiently reprogrammed to pluripotency with ablation of aberrant MLL-driven DNA methylation, with effective differentiation to non-haematopoietic lineages113. However, upon directed differentiation to haematopoietic progenitors, these cells show aberrant self-renewal in culture and engraft in vivo as AML113. This system was used to model the clonal architecture of human AML, identifying differential drug susceptibility of individual iPSC-derived subclones113. A contemporaneous study derived human iPSC lines from patients spanning the spectrum from normal haematopoiesis, nonmalignant clonal haematopoiesis, low-grade and high-grade myelodysplastic syndrome (MDS), to AML114. This study reported that only haematopoietic progenitors generated by directed differentiation from iPSCs reprogrammed from AML blasts could xenograft as leukaemia114. Interestingly, as opposed to AML, B-acute lymphoblastic leukaemia (B-ALL) blasts are resistant to reprogramming to pluripotency115. However, it was recently reported that transient expression of the MLL–AF4 fusion oncogene in haematopoietic progenitors from human iPSCs could induce multi-lineage engraftment, with the eventual development of B-ALL upon long-term engraftment, presenting a novel model of human B-ALL using human iPSCs116. Recently, iPSCs reprogrammed from a patient with chronic myelomonocytic leukemia were used to generate teratomas containing engraftable leukemic cells117.
Human iPSCs-based chimaeras have also been used to model solid tumours. Reprogramming of human glioblastoma multiforme (GBM) cells to iPSCs erased hypermethylation of promoters of genes with tumour-suppressive function118. Upon directed differentiation of iPSCs to NPCs, these cells retained malignant potential, forming aggressive tumours when xenotransplanted into mouse striatum118. Osteoblasts differentiated from iPSCs obtained from patients with Li-Fraumeni syndrome (LFS) bearing germline TP53 mutations, who are predisposed to developing a variety of solid and liquid malignancies, formed osteosarcoma-like tumours upon subcutaneous injection in immunodeficient mice119. Future studies using organoids derived from iPSCs and xenotransplanted into mice could provide new insight into solid tumour genesis and progression.
Neural chimaeras.
Xenotransplantation of human neural tissue has been employed for decades to study human disease120. The isolation and culture of human ESCs and development of iPSC technology, as well as the propensity of human PSCs to undergo neural differentiation and even engraft model organisms presented the potential for novel cell-based therapies6,121. Neurons differentiated from human iPSCs can integrate, form functional synapses and incorporate into neural circuitry when xenografted into the developing mouse brain122. NSCs differentiated from iPSCs will engraft in rat brains and differentiate into neurons and astrocytes and may enhance recovery in a model of ischemic stroke123,124. Dopaminergic neurons differentiated from either human or nonhuman primate iPSCs can engraft in the brains of rats and improve function in a model of Parkinson disease125. Dopaminergic neurons from nonhuman primate iPSCs could also be transplanted autologously, with long-term engraftment in vivo125. Such compelling pre-clinical evidence of functional recovery has provided a strong foundation for human clinical testing in Parkinson disease, which is imminent. Adoptive cellular therapy to repair neural connections within the hippocampus is under investigation for treatment of schizophrenia98. Interestingly, in a recent study, iPSCs derived from patients with childhood-onset schizophrenia were differentiated to glial precursor cells and xenotransplanted into the neonatal mouse brain where they showed aberrant migration and astrocyte differentiation126. Recently, engraftment of neural organoids derived from human PSCs was described, with perfusion via the host vasculature and electrophysiologic evidence of neuronal activity, providing a platform for modelling a panoply of neurologic disease69. Together, these models demonstrate the value of chimaeric models in modelling human neurologic disease with iPSCs as well as the feasibility of iPSC-based cell therapy in these diseases.
Solid organ chimaeras.
Human iPSC-derived organoids have been engrafted into mice to incorporate vasculature and interaction with normal host physiology70,127. Hepatic endoderm cells derived from human iPSCs can be combined with endothelial and mesenchymal stem cells in culture where they will organize into primitive hepatic buds70. Upon xenotransplantation, these buds formed functional connections with murine vasculature, liberated human albumin into mouse serum, and generated human-specific drug metabolites70. Transplantation of these liver buds could enhance survival in a mouse model of liver failure70. Hepatocytes differentiated from iPSCs from patients with familial hypercholesterolemia have been xenotransplanted to model this disease128. Human intestinal organoids derived from iPSCs mature into intestinal tissue when engrafted under the mouse kidney capsule and form a supportive mesenchyme127. In response to ileocaecal resection, engrafted intestinal tissue showed evidence of proliferation and crypt fission as part of the systemic response to intestinal removal127. Furthermore, human ESCs can be xenotransplanted following differentiation to thymic epithelium129,130. Recently, it was shown that human iPSC-derived β-cells could be engrafted into mouse pancreas with secretion of human insulin131.
Modelling adoptive immunity.
Human iPSCs have been used to model adoptive T-cell therapy in cancer and infectious disease, and potentially represent a source of unlimited, customized T-cells for immunotherapy. Chimeric antigen receptor (CAR) T-cells are approved for use in the treatment of B-ALL99. CD19-targeted CAR T-cells differentiated from human iPSCs are effective in targeting human CD19+B-ALL cells in a chimaeric model of leukaemia132. Production of iPSCs from human T-lymphocytes specific for the WT1 antigen, followed by differentiation to cytotoxic T-cells, yields and engraftment into chimeric mice bearing WT1+ human leukaemia prolonged recipient survival133. Human mucosal-associated invariant T-cells can be reprogrammed to iPSCs, differentiated to the T cell lineage, and xenotransplanted into mice where they are effective against Mycobacterium abscessus inoculation134.
NK cells derived from human iPSCs have been assayed in chimaeric models of disease. Adoptive transfer of NK cells differentiated from human iPSCs can inhibit replication of HIV in vivo78. NK cells derived from iPSCs bearing a CAR against Mesothelin target ovarian cancer cells in a chimeric model135. Together, these studies demonstrate the validity of preclinical models of adoptive immunity in identifying potential immune cell-based therapies.
Conclusions and perspectives
We have reviewed the progress in disease modelling using human iPSCs over the past decade, highlighting ever more sophisticated 3D organoid systems and chimaeric models that are being exploited for discovery of new therapeutic approaches. Although successful in differentiating patient-derived iPSCs to target cells with measurable phenotypic defects, has the field advanced to a point where these models are of sufficient accuracy to support identification of new therapies affecting the treatment of a wide range of diseases? As we await the impact of iPSC technology on human health, we posit that this broad objective might yet remain beyond the current state of the art of iPSC research. We caution that in many of systems, further refinement of the directed differentiation is required before the validity of candidate interventions can be confirmed. One reason for this is the propensity of human iPSCs to differentiate to cells with an immature embryonic or fetal identity, rather than a fully mature adult state. In our experience, this has been a significant barrier in modelling haematopoietic differentiation from iPSCs, as evidenced by the expression of predominantly embryonic and fetal globins in erythrocytes derived from iPSCs, which impedes the modelling of disorders of adult globins, such as thalassemia and sickle cell disease106,136. Similar effects have been reported in iPSC-derived intestinal organoids61, cardiomyocytes63 and pancreatic β-cells62. Engraftment of iPSC-derived tissue as chimaeras may enhance maturation owing to the integration of niche-derived signals61,70.
A second challenge is genetic heterogeneity of iPSC lines. Studies of iPSCs derived from large populations have demonstrated variability in differentiated cell phenotypes and gene expression, which can be attributable to background genetic variation71,72,137,138. Although cohorts of iPSC lines can be exploited to model human genetic heterogeneity71,72,137,138, this effect can confound disease modelling in the absence of appropriate controls (Box 2). Genetic correction of disease-causing mutations in patient-derived iPSCs should be used to generate otherwise isogenic control iPSCs for use as controls in disease models.
Box 2. Human genetic variability in iPSCs and its impact on disease modelling.
For the past 30 years, human genetic variation has been used as a tool in genomic research, enabling identification of disease-associated genes by positional cloning. Recent studies using large cohorts of iPSCs have highlighted the impact of genetic variation on iPSC function and differentiation. iPSCs derived from unrelated, independent donors without apparent disease phenotypes have been shown to differ in methylation profiles, gene expression and efficiency of directed differentiation, effects which occur independently of the method used for reprogramming146,147. These data underline the importance of rigorous controls when using iPSCs for disease modelling, to avoid attribution of a disease phenotype to background genetic variation rather than to a suspected disease-causing mutation in a monogenic disease gene (Fig. 2).
In a key study, Kilpinen and colleagues analysed over 700 iPSC lines from 301 healthy donors, enabling investigation of variation in genetic and epigenetic factors both between and within individual donors138. By analysing copy number variation, gene expression and proteomics across these cell lines, the authors estimated that 5–46% of phenotype variability, including differentiation efficiency, is attributable to genetic variation. A second key study performed whole-genome sequencing on 215 iPSC lines from different donors, identifying over 22 million single nucleotide variants and over 15,000 copy number variants71. By using RNA sequencing, the authors reported the existence of quantitative trait loci (QTL) affecting 4,622 protein-coding genes, some of which affected transcription factor binding to regulatory regions71.
Warren and colleagues exploited genetic variation between iPSC lines to functionally validate variants identified in a genome-wide association study72. In this study, the authors derived 68 iPSC lines from peripheral blood cells from 34 patients included in the Framingham Heart study, with the objective of experimentally validating the association of a specific single nucleotide polymorphism (SNP) with metabolic phenotypes by measuring cellular readouts relevant to cardiovascular disease in cells differentiated from iPSCs72. As expected, efficiencies of differentiation of iPSCs to hepatocytes and adipocytes were highly variable72. However, using metabolomics, the authors found that the candidate QTL did associate with effects on lipid metabolism in cells differentiated from these iPSC lines72.
Collectively, these studies highlight the impact of human background genetic variability on gene expression and differentiation phenotypes in iPSCs. Thus, in disease models, a specific disease-associated phenotype elicited in iPSCs should be interpreted with caution in the absence of appropriate control iPSC lines, such as those wherein a suspected disease-causing genetic defect has been repaired or complemented. Efforts should be made to use control iPSC lines as genetically similar as possible to disease lines in disease modelling experiments58,59,148,149.
Since the first description of diseased iPSCs, models have advanced from differentiated 2D cultures of single lineage cells reporting cellular-level outputs to increasingly complex 3D organoids and in vivo chimaeras to now incorporate perturbed cell–tissue and tissue–organ level interactions into disease models. The acceleration of this research in the past few years supports our continued optimism that iPSC-based models will continue to approach the complexity of human multisystem disease. We anticipate that further advances in xenotransplantation and organoid technologies will contribute to the overcoming of the final hurdles on the path to autologous cell-based therapies and the identification of new drugs in iPSC systems (Fig. 1). Translation of interspecies chimaera and blastocyst complementation technologies to generate chimaeric tissue specified by endogenous in vivo morphogen signals may lead to future advances in chimaeric disease models139,140. Furthermore, ongoing improvement in iPSC differentiation protocols, organoid systems, and chimeric models will broaden the current frontier of iPSC-based cell therapy with the potential to revolutionize diverse disciplines including infectious disease, oncology and tissue transplantation.
Acknowledgements
We apologize for the important studies not included here due to space constraints.
Glossary
- Batch effects
Variation between experimental replicates due to differences in cellular source or reagent lot.
- Blasts
Undifferentiated, immature hematopoietic cells. These can be either rare, normal hematopoietic progenitors within healthy hematopoietic organs or transformed leukemic cells arrested at an early state of differentiation.
- Cas9
A bacterial DNA endonuclease that uses RNAs to localize and cleave targeted sequences within a genome. This enzyme has been exploited as a technology for precise genomic editing to introduce or correct specific genetic mutations in IPSCs.
- Cell state conversion
Process by which a cell is converted from one identity to another, frequently via the expression of exogenous transcription factors.
- Chimeric antigen receptor T-cells
Genetically manipulated T-cells bearing a modified T-cell receptor against a specific target.
- Directed differentiation
The use of morphogens and environmental signals to direct the state of pluripotent cells toward a desired lineage.
- Immunotherapy
A therapeutic approach using modulation of the immune system.
- Induced pluripotent stem cells
Pluripotent cells derived from terminally differentiated somatic cells generated by reprogramming via introduction of a defined set of transcription factors.
- Isogenic
A term to describe two cells, tissues, or organisms that have the same genotypes.
- Morphogen
A growth factor or chemical signal that regulates cellular differentiation or tissue patterning.
- Organ-on-a-chip
An experimental system where tissue architecture and cellular composition are assembled on a fabricated synthetic matrix.
- Quantitative trait locus
A DNA sequence whose variation contributes to the manifested heterogeneity of a quantitative polygenic trait.
- Tissue engineering
Synthetic recapitulation of the cellular composition and matrix structure of a target tissue.
- Xenotransplantation
Transplantation of cells from a certain species into a different species.
Footnotes
Competing interests
G. Q. D. holds intellectual property relevant to development of cell and drug therapies based on IPSC technology.
References
- 1.Park IH et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 451, 141–146, 10.1038/nature06534 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Yu J et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920, 10.1126/science.1151526 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872, 10.1016/j.cell.2007.11.019 (2007).References 1–3 provide the first descriptions of induced pluripotent stem cells from human cells.
- 4.Takahashi K & Yamanaka S Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676, 10.1016/j.cell.2006.07.024 (2006).This is the first description of induced pluripotent stem cells.
- 5.Thomson JA et al. Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 (1998).This paper describes the isolation of human embryonic stem cells.
- 6.Zhang SC, Wernig M, Duncan ID, Brustle O & Thomson JA In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol 19, 1129–1133, 10.1038/nbt1201-1129 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Reubinoff BE et al. Neural progenitors from human embryonic stem cells. Nat Biotechnol 19, 1134–1140, 10.1038/nbt1201-1134 (2001). [DOI] [PubMed] [Google Scholar]
- 8.He JQ, Ma Y, Lee Y, Thomson JA & Kamp TJ Human embryonic stem cells develop into multiple types of cardiac myocytes: action potential characterization. Circ Res 93, 32–39, 10.1161/01.RES.0000080317.92718.99 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Assady S et al. Insulin production by human embryonic stem cells. Diabetes 50, 1691–1697 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Kaufman DS, Hanson ET, Lewis RL, Auerbach R & Thomson JA Hematopoietic colony-forming cells derived from human embryonic stem cells. Proc Natl Acad Sci U S A 98, 10716–10721, 10.1073/pnas.191362598 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chadwick K et al. Cytokines and BMP-4 promote hematopoietic differentiation of human embryonic stem cells. Blood 102, 906–915, 10.1182/blood-2003-03-0832 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Eiges R et al. Developmental study of fragile X syndrome using human embryonic stem cells derived from preimplantation genetically diagnosed embryos. Cell Stem Cell 1, 568–577, 10.1016/j.stem.2007.09.001 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Tulpule A et al. Knockdown of Fanconi anemia genes in human embryonic stem cells reveals early developmental defects in the hematopoietic lineage. Blood 115, 3453–3462, 10.1182/blood-2009-10-246694 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Urbach A, Schuldiner M & Benvenisty N Modeling for Lesch-Nyhan disease by gene targeting in human embryonic stem cells. Stem Cells 22, 635–641, 10.1634/stemcells.22-4-635 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Ebert AD et al. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature 457, 277–280, 10.1038/nature07677 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moretti A et al. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med 363, 1397–1409, 10.1056/NEJMoa0908679 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Park IH et al. Disease-specific induced pluripotent stem cells. Cell 134, 877–886, 10.1016/j.cell.2008.07.041 (2008).This is an early example of reprogramming of diseased somatic cells to generate induced pluripotent stem cells.
- 18.Maehr R et al. Generation of pluripotent stem cells from patients with type 1 diabetes. Proc Natl Acad Sci U S A 106, 15768–15773, 10.1073/pnas.0906894106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brennand KJ et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature 473, 221–225, 10.1038/nature09915 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee G et al. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature 461, 402–406, 10.1038/nature08320 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu GH et al. Modelling Fanconi anemia pathogenesis and therapeutics using integration-free patient-derived iPSCs. Nat Commun 5, 4330, 10.1038/ncomms5330 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinton DA & Daley GQ The promise of induced pluripotent stem cells in research and therapy. Nature 481, 295–305, 10.1038/nature10761 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi Y, Inoue H, Wu JC & Yamanaka S Induced pluripotent stem cell technology: a decade of progress. Nat Rev Drug Discov 16, 115–130, 10.1038/nrd.2016.245 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Itzhaki I et al. Modelling the long QT syndrome with induced pluripotent stem cells. Nature 471, 225–229, 10.1038/nature09747 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Lee G et al. Large-scale screening using familial dysautonomia induced pluripotent stem cells identifies compounds that rescue IKBKAP expression. Nat Biotechnol 30, 1244–1248, 10.1038/nbt.2435 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu C, Oikonomopoulos A, Sayed N & Wu JC Modeling human diseases with induced pluripotent stem cells: from 2D to 3D and beyond. Development 145, 10.1242/dev.156166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCauley HA & Wells JM Pluripotent stem cell-derived organoids: using principles of developmental biology to grow human tissues in a dish. Development 144, 958–962, 10.1242/dev.140731 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dutta D, Heo I & Clevers H Disease Modeling in Stem Cell-Derived 3D Organoid Systems. Trends Mol Med 23, 393–410, 10.1016/j.molmed.2017.02.007 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Quadrato G, Brown J & Arlotta P The promises and challenges of human brain organoids as models of neuropsychiatric disease. Nat Med 22, 1220–1228, 10.1038/nm.4214 (2016). [DOI] [PubMed] [Google Scholar]
- 30.McCracken KW et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 516, 400–404, 10.1038/nature13863 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lancaster MA et al. Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379, 10.1038/nature12517 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mariani J et al. Modeling human cortical development in vitro using induced pluripotent stem cells. Proc Natl Acad Sci U S A 109, 12770–12775, 10.1073/pnas.1202944109 (2012).References 31 and 32 describe early neural organoids derived from IPSCs.
- 33.Qian X et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell 165, 1238–1254, 10.1016/j.cell.2016.04.032 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muguruma K, Nishiyama A, Kawakami H, Hashimoto K & Sasai Y Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. Cell Rep 10, 537–550, 10.1016/j.celrep.2014.12.051 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Jo J et al. Midbrain-like Organoids from Human Pluripotent Stem Cells Contain Functional Dopaminergic and Neuromelanin-Producing Neurons. Cell Stem Cell 19, 248–257, 10.1016/j.stem.2016.07.005 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quadrato G et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature 545, 48–53, 10.1038/nature22047 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y et al. Induction of Expansion and Folding in Human Cerebral Organoids. Cell Stem Cell 20, 385–396 10.1016/j.stem.2016.11.017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sloan SA et al. Human Astrocyte Maturation Captured in 3D Cerebral Cortical Spheroids Derived from Pluripotent Stem Cells. Neuron 95, 779–790 10.1016/j.neuron.2017.07.035 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gabriel E et al. CPAP promotes timely cilium disassembly to maintain neural progenitor pool. EMBO J 35, 803–819, 10.15252/embj.201593679 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mariani J et al. FOXG1-Dependent Dysregulation of GABA/Glutamate Neuron Differentiation in Autism Spectrum Disorders. Cell 162, 375–390, 10.1016/j.cell.2015.06.034 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mellios N et al. MeCP2-regulated miRNAs control early human neurogenesis through differential effects on ERK and AKT signaling. Mol Psychiatry 23, 1051–1065, 10.1038/mp.2017.86 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bershteyn M et al. Human iPSC-Derived Cerebral Organoids Model Cellular Features of Lissencephaly and Reveal Prolonged Mitosis of Outer Radial Glia. Cell Stem Cell 20, 435–449 10.1016/j.stem.2016.12.007 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bian S et al. Genetically engineered cerebral organoids model brain tumor formation. Nat Methods 15, 631–639, 10.1038/s41592-018-0070-7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sato T et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265, 10.1038/nature07935 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Spence JR et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470, 105–109, 10.1038/nature09691 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Workman MJ et al. Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nat Med 23, 49–59, 10.1038/nm.4233 (2017).Combination of neural and intestinal cells from IPSCs to generate complex intestinal tissue.
- 47.Rashid ST et al. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J Clin Invest 120, 3127–3136, 10.1172/JCI43122 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ogawa M et al. Directed differentiation of cholangiocytes from human pluripotent stem cells. Nat Biotechnol 33, 853–861, 10.1038/nbt.3294 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Sampaziotis F et al. Cholangiocytes derived from human induced pluripotent stem cells for disease modeling and drug validation. Nat Biotechnol 33, 845–852, 10.1038/nbt.3275 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crespo M et al. Colonic organoids derived from human induced pluripotent stem cells for modeling colorectal cancer and drug testing. Nat Med 23, 878–884, 10.1038/nm.4355 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Freedman BS et al. Reduced ciliary polycystin-2 in induced pluripotent stem cells from polycystic kidney disease patients with PKD1 mutations. J Am Soc Nephrol 24, 1571–1586, 10.1681/ASN.2012111089 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xia Y et al. Directed differentiation of human pluripotent cells to ureteric bud kidney progenitor-like cells. Nat Cell Biol 15, 1507–1515, 10.1038/ncb2872 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Takasato M et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526, 564–568, 10.1038/nature15695 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Freedman BS et al. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat Commun 6, 8715, 10.1038/ncomms9715 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Z et al. 3D Culture Supports Long-Term Expansion of Mouse and Human Nephrogenic Progenitors. Cell Stem Cell 19, 516–529, 10.1016/j.stem.2016.07.016 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taguchi A & Nishinakamura R Higher-Order Kidney Organogenesis from Pluripotent Stem Cells. Cell Stem Cell 21, 730–746 10.1016/j.stem.2017.10.011 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Musah S et al. Mature induced-pluripotent-stem-cell-derived human podocytes reconstitute kidney glomerular-capillary-wall function on a chip. Nat Biomed Eng 1, 10.1038/s41551-017-0069 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCauley KB et al. Efficient Derivation of Functional Human Airway Epithelium from Pluripotent Stem Cells via Temporal Regulation of Wnt Signaling. Cell Stem Cell 20, 844–857 10.1016/j.stem.2017.03.001 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jacob A et al. Differentiation of Human Pluripotent Stem Cells into Functional Lung Alveolar Epithelial Cells. Cell Stem Cell 21, 472–488 10.1016/j.stem.2017.08.014 (2017).References 58 and 59 demonstrate the derivation of complex lung organoids and their application in disease modeling.
- 60.Voges HK et al. Development of a human cardiac organoid injury model reveals innate regenerative potential. Development 144, 1118–1127, 10.1242/dev.143966 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Finkbeiner SR et al. Transcriptome-wide Analysis Reveals Hallmarks of Human Intestine Development and Maturation In Vitro and In Vivo. Stem Cell Reports, 10.1016/j.stemcr.2015.04.010 (2015). [DOI] [PMC free article] [PubMed]
- 62.Pagliuca FW et al. Generation of functional human pancreatic beta cells in vitro. Cell 159, 428–439, 10.1016/j.cell.2014.09.040 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ronaldson-Bouchard K et al. Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature 556, 239–243, 10.1038/s41586-018-0016-3 (2018).Demonstration of the maturation of IPSC-derived cardiomyocytes from a fetal to adult state.
- 64.Hoang P, Wang J, Conklin BR, Healy KE & Ma Z Generation of spatial-patterned early-developing cardiac organoids using human pluripotent stem cells. Nat Protoc 13, 723–737, 10.1038/nprot.2018.006 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abilez OJ et al. Passive Stretch Induces Structural and Functional Maturation of Engineered Heart Muscle as Predicted by Computational Modeling. Stem Cells 36, 265–277, 10.1002/stem.2732 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mills RJ et al. Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc Natl Acad Sci U S A 114, E8372–E8381, 10.1073/pnas.1707316114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang G et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med 20, 616–623, 10.1038/nm.3545 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brown JA et al. Recreating blood-brain barrier physiology and structure on chip: A novel neurovascular microfluidic bioreactor. Biomicrofluidics 9, 054124, 10.1063/1.4934713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mansour AA et al. An in vivo model of functional and vascularized human brain organoids. Nat Biotechnol 36, 432–441, 10.1038/nbt.4127 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Takebe T et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 499, 481–484, 10.1038/nature12271 (2013). [DOI] [PubMed] [Google Scholar]
- 71.DeBoever C et al. Large-Scale Profiling Reveals the Influence of Genetic Variation on Gene Expression in Human Induced Pluripotent Stem Cells. Cell Stem Cell 20, 533–546 10.1016/j.stem.2017.03.009 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Warren CR et al. Induced Pluripotent Stem Cell Differentiation Enables Functional Validation of GWAS Variants in Metabolic Disease. Cell Stem Cell 20, 547–557 10.1016/j.stem.2017.01.010 (2017). [DOI] [PubMed] [Google Scholar]
- 73.Turco MY et al. Trophoblast organoids as a model for maternal-fetal interactions during human placentation. Nature, 10.1038/s41586-018-0753-3 (2018). [DOI] [PMC free article] [PubMed]
- 74.Seok J et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A 110, 3507–3512, 10.1073/pnas.1222878110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen P, Stanojcic M & Jeschke MG Differences between murine and human sepsis. Surg Clin North Am 94, 1135–1149, 10.1016/j.suc.2014.08.001 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou T et al. High-Content Screening in hPSC-Neural Progenitors Identifies Drug Candidates that Inhibit Zika Virus Infection in Fetal-like Organoids and Adult Brain. Cell Stem Cell 21, 274–283 10.1016/j.stem.2017.06.017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ni Z et al. Human pluripotent stem cells produce natural killer cells that mediate anti-HIV-1 activity by utilizing diverse cellular mechanisms. J Virol 85, 43–50, 10.1128/JVI.01774-10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ni Z, Knorr DA, Bendzick L, Allred J & Kaufman DS Expression of chimeric receptor CD4zeta by natural killer cells derived from human pluripotent stem cells improves in vitro activity but does not enhance suppression of HIV infection in vivo. Stem Cells 32, 1021–1031, 10.1002/stem.1611 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ye L et al. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Delta32 mutation confers resistance to HIV infection. Proc Natl Acad Sci U S A 111, 9591–9596, 10.1073/pnas.1407473111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lafaille FG et al. Impaired intrinsic immunity to HSV-1 in human iPSC-derived TLR3-deficient CNS cells. Nature 491, 769–773, 10.1038/nature11583 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lang J et al. An hPSC-Derived Tissue-Resident Macrophage Model Reveals Differential Responses of Macrophages to ZIKV and DENV Infection. Stem Cell Reports, 10.1016/j.stemcr.2018.06.006 (2018). [DOI] [PMC free article] [PubMed]
- 82.Garcez PP et al. Zika virus impairs growth in human neurospheres and brain organoids. Science 352, 816–818, 10.1126/science.aaf6116 (2016). [DOI] [PubMed] [Google Scholar]
- 83.Cugola FR et al. The Brazilian Zika virus strain causes birth defects in experimental models. Nature 534, 267–271, 10.1038/nature18296 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gabriel E et al. Recent Zika Virus Isolates Induce Premature Differentiation of Neural Progenitors in Human Brain Organoids. Cell Stem Cell 20, 397–406 10.1016/j.stem.2016.12.005 (2017). [DOI] [PubMed] [Google Scholar]
- 85.Nowakowski TJ et al. Expression Analysis Highlights AXL as a Candidate Zika Virus Entry Receptor in Neural Stem Cells. Cell Stem Cell 18, 591–596, 10.1016/j.stem.2016.03.012 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wells MF et al. Genetic Ablation of AXL Does Not Protect Human Neural Progenitor Cells and Cerebral Organoids from Zika Virus Infection. Cell Stem Cell 19, 703–708, 10.1016/j.stem.2016.11.011 (2016).References 82–86 apply neural organoids to modeling Zika virus pathogenesis.
- 87.Finkbeiner SR et al. Stem cell-derived human intestinal organoids as an infection model for rotaviruses. MBio 3, e00159–00112, 10.1128/mBio.00159-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen YW et al. A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat Cell Biol 19, 542–549, 10.1038/ncb3510 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Churin Y et al. Helicobacter pylori CagA protein targets the c-Met receptor and enhances the motogenic response. J Cell Biol 161, 249–255, 10.1083/jcb.200208039 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Leslie JL et al. Persistence and toxin production by Clostridium difficile within human intestinal organoids result in disruption of epithelial paracellular barrier function. Infect Immun 83, 138–145, 10.1128/IAI.02561-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Paredes-Sabja D, Shen A & Sorg JA Clostridium difficile spore biology: sporulation, germination, and spore structural proteins. Trends Microbiol 22, 406–416, 10.1016/j.tim.2014.04.003 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Forbester JL et al. Interaction of Salmonella enterica Serovar Typhimurium with Intestinal Organoids Derived from Human Induced Pluripotent Stem Cells. Infect Immun 83, 2926–2934, 10.1128/IAI.00161-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nadkarni RR et al. Functional Enterospheres Derived In Vitro from Human Pluripotent Stem Cells. Stem Cell Reports 9, 897–912, 10.1016/j.stemcr.2017.07.024 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sturgeon CM, Ditadi A, Awong G, Kennedy M & Keller G Wnt signaling controls the specification of definitive and primitive hematopoiesis from human pluripotent stem cells. Nat Biotechnol 32, 554–561, 10.1038/nbt.2915 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang L, Li L, Menendez P, Cerdan C & Bhatia M Human embryonic stem cells maintained in the absence of mouse embryonic fibroblasts or conditioned media are capable of hematopoietic development. Blood 105, 4598–4603, 10.1182/blood-2004-10-4065 (2005). [DOI] [PubMed] [Google Scholar]
- 96.Tzannou I et al. Off-the-Shelf Virus-Specific T Cells to Treat BK Virus, Human Herpesvirus 6, Cytomegalovirus, Epstein-Barr Virus, and Adenovirus Infections After Allogeneic Hematopoietic Stem-Cell Transplantation. J Clin Oncol 35, 3547–3557, 10.1200/JCO.2017.73.0655 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bollard CM & Heslop HE T cells for viral infections after allogeneic hematopoietic stem cell transplant. Blood 127, 3331–3340, 10.1182/blood-2016-01-628982 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Donegan JJ & Lodge DJ Cell-based therapies for the treatment of schizophrenia. Brain Res 1655, 262–269, 10.1016/j.brainres.2016.08.010 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Maude SL et al. Tisagenlecleucel in Children and Young Adults with B-Cell Lymphoblastic Leukemia. N Engl J Med 378, 439–448, 10.1056/NEJMoa1709866 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mandai M et al. Autologous Induced Stem-Cell-Derived Retinal Cells for Macular Degeneration. N Engl J Med 376, 1038–1046, 10.1056/NEJMoa1608368 (2017). [DOI] [PubMed] [Google Scholar]
- 101.Ditadi A et al. Human definitive haemogenic endothelium and arterial vascular endothelium represent distinct lineages. Nat Cell Biol 17, 580–591, 10.1038/ncb3161 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kennedy M et al. T lymphocyte potential marks the emergence of definitive hematopoietic progenitors in human pluripotent stem cell differentiation cultures. Cell Rep 2, 1722–1735, 10.1016/j.celrep.2012.11.003 (2012). [DOI] [PubMed] [Google Scholar]
- 103.Wahlster L & Daley GQ Progress towards generation of human haematopoietic stem cells. Nat Cell Biol 18, 1111–1117, 10.1038/ncb3419 (2016). [DOI] [PubMed] [Google Scholar]
- 104.Lu YF et al. Engineered Murine HSCs Reconstitute Multi-lineage Hematopoiesis and Adaptive Immunity. Cell Rep 17, 3178–3192, 10.1016/j.celrep.2016.11.077 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kyba M, Perlingeiro RC & Daley GQ HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell 109, 29–37 (2002). [DOI] [PubMed] [Google Scholar]
- 106.Doulatov S et al. Induction of multipotential hematopoietic progenitors from human pluripotent stem cells via respecification of lineage-restricted precursors. Cell Stem Cell 13, 459–470, 10.1016/j.stem.2013.09.002 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Doulatov S et al. Drug discovery for Diamond-Blackfan anemia using reprogrammed hematopoietic progenitors. Sci Transl Med 9, 10.1126/scitranslmed.aah5645 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sugimura R et al. Haematopoietic stem and progenitor cells from human pluripotent stem cells. Nature 545, 432–438, 10.1038/nature22370 (2017).Description of engraftable, functional, multilineage hematopoietic stem and progenitor cells derived from IPSCs.
- 109.Muller LU et al. Overcoming reprogramming resistance of Fanconi anemia cells. Blood 119, 5449–5457, 10.1182/blood-2012-02-408674 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Uchida N et al. Efficient Generation of beta-Globin-Expressing Erythroid Cells Using Stromal Cell-Derived Induced Pluripotent Stem Cells from Patients with Sickle Cell Disease. Stem Cells 35, 586–596, 10.1002/stem.2517 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Niu X et al. Combining Single Strand Oligodeoxynucleotides and CRISPR/Cas9 to Correct Gene Mutations in beta-Thalassemia-induced Pluripotent Stem Cells. J Biol Chem 291, 16576–16585, 10.1074/jbc.M116.719237 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Davies G, Duke D, Grant AG, Kelly SA & Hermon-Taylor J Growth of human digestive-tumour xenografts in athymic nude rats. Br J Cancer 43, 53–58 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chao MP et al. Human AML-iPSCs Reacquire Leukemic Properties after Differentiation and Model Clonal Variation of Disease. Cell Stem Cell 20, 329–344 10.1016/j.stem.2016.11.018 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kotini AG et al. Stage-Specific Human Induced Pluripotent Stem Cells Map the Progression of Myeloid Transformation to Transplantable Leukemia. Cell Stem Cell 20, 315–328 10.1016/j.stem.2017.01.009 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Munoz-Lopez A et al. Development Refractoriness of MLL-Rearranged Human B Cell Acute Leukemias to Reprogramming into Pluripotency. Stem Cell Reports 7, 602–618, 10.1016/j.stemcr.2016.08.013 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tan YT et al. Respecifying human iPSC-derived blood cells into highly engraftable hematopoietic stem and progenitor cells with a single factor. Proc Natl Acad Sci U S A 115, 2180–2185, 10.1073/pnas.1718446115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Taoka K et al. Using patient-derived iPSCs to develop humanized mouse models for chronic myelomonocytic leukemia and therapeutic drug identification, including liposomal clodronate. Sci Rep 8, 15855, 10.1038/s41598-018-34193-1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Stricker SH et al. Widespread resetting of DNA methylation in glioblastoma-initiating cells suppresses malignant cellular behavior in a lineage-dependent manner. Genes Dev 27, 654–669, 10.1101/gad.212662.112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lee DF et al. Modeling familial cancer with induced pluripotent stem cells. Cell 161, 240–254, 10.1016/j.cell.2015.02.045 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Aguayo AJ, Kasarjian J, Skamene E, Kongshavn P & Bray GM Myelination of mouse axons by Schwann cells transplanted from normal and abnormal human nerves. Nature 268, 753–755 (1977). [DOI] [PubMed] [Google Scholar]
- 121.Barker RA, Parmar M, Studer L & Takahashi J Human Trials of Stem Cell-Derived Dopamine Neurons for Parkinson’s Disease: Dawn of a New Era. Cell Stem Cell 21, 569–573, 10.1016/j.stem.2017.09.014 (2017). [DOI] [PubMed] [Google Scholar]
- 122.Espuny-Camacho I et al. Pyramidal neurons derived from human pluripotent stem cells integrate efficiently into mouse brain circuits in vivo. Neuron 77, 440–456, 10.1016/j.neuron.2012.12.011 (2013). [DOI] [PubMed] [Google Scholar]
- 123.Yuan T et al. Human induced pluripotent stem cell-derived neural stem cells survive, migrate, differentiate, and improve neurologic function in a rat model of middle cerebral artery occlusion. Stem Cell Res Ther 4, 73, 10.1186/scrt224 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jensen MB, Yan H, Krishnaney-Davison R, Al Sawaf A & Zhang SC Survival and differentiation of transplanted neural stem cells derived from human induced pluripotent stem cells in a rat stroke model. J Stroke Cerebrovasc Dis 22, 304–308, 10.1016/j.jstrokecerebrovasdis.2011.09.008 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sundberg M et al. Improved cell therapy protocols for Parkinson’s disease based on differentiation efficiency and safety of hESC-, hiPSC-, and non-human primate iPSC-derived dopaminergic neurons. Stem Cells 31, 1548–1562, 10.1002/stem.1415 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Windrem MS et al. Human iPSC Glial Mouse Chimeras Reveal Glial Contributions to Schizophrenia. Cell Stem Cell 21, 195–208 10.1016/j.stem.2017.06.012 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Watson CL et al. An in vivo model of human small intestine using pluripotent stem cells. Nat Med 20, 1310–1314, 10.1038/nm.3737 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yang J et al. Generation of Human Liver Chimeric Mice with Hepatocytes from Familial Hypercholesterolemia Induced Pluripotent Stem Cells. Stem Cell Reports 8, 605–618, 10.1016/j.stemcr.2017.01.027 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Parent AV et al. Generation of functional thymic epithelium from human embryonic stem cells that supports host T cell development. Cell Stem Cell 13, 219–229, 10.1016/j.stem.2013.04.004 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sun X et al. Directed differentiation of human embryonic stem cells into thymic epithelial progenitor-like cells reconstitutes the thymic microenvironment in vivo. Cell Stem Cell 13, 230–236, 10.1016/j.stem.2013.06.014 (2013). [DOI] [PubMed] [Google Scholar]
- 131.Ma H, Wert KJ, Shvartsman D, Melton DA & Jaenisch R Establishment of human pluripotent stem cell-derived pancreatic beta-like cells in the mouse pancreas. Proc Natl Acad Sci U S A 115, 3924–3929, 10.1073/pnas.1702059115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Themeli M et al. Generation of tumor-targeted human T lymphocytes from induced pluripotent stem cells for cancer therapy. Nat Biotechnol 31, 928–933, 10.1038/nbt.2678 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Maeda T et al. Regeneration of CD8alphabeta T Cells from T-cell-Derived iPSC Imparts Potent Tumor Antigen-Specific Cytotoxicity. Cancer Res 76, 6839–6850, 10.1158/0008-5472.CAN-16-1149 (2016). [DOI] [PubMed] [Google Scholar]
- 134.Wakao H et al. Expansion of functional human mucosal-associated invariant T cells via reprogramming to pluripotency and redifferentiation. Cell Stem Cell 12, 546–558, 10.1016/j.stem.2013.03.001 (2013). [DOI] [PubMed] [Google Scholar]
- 135.Li Y, Hermanson DL, Moriarity BS & Kaufman DS Human iPSC-Derived Natural Killer Cells Engineered with Chimeric Antigen Receptors Enhance Anti-tumor Activity. Cell Stem Cell 23, 181–192 e185, 10.1016/j.stem.2018.06.002 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Vanuytsel K et al. Induced pluripotent stem cell-based mapping of beta-globin expression throughout human erythropoietic development. Blood Adv 2, 1998–2011, 10.1182/bloodadvances.2018020560 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pashos EE et al. Large, Diverse Population Cohorts of hiPSCs and Derived Hepatocyte-like Cells Reveal Functional Genetic Variation at Blood Lipid-Associated Loci. Cell Stem Cell 20, 558–570 10.1016/j.stem.2017.03.017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kilpinen H et al. Common genetic variation drives molecular heterogeneity in human iPSCs. Nature 546, 370–375, 10.1038/nature22403 (2017).This study highlights the genetic variability between IPSC lines derived from different individuals that can impact differentiation.
- 139.Yamaguchi T et al. Interspecies organogenesis generates autologous functional islets. Nature 542, 191–196, 10.1038/nature21070 (2017). [DOI] [PubMed] [Google Scholar]
- 140.Yang Y et al. Derivation of Pluripotent Stem Cells with In Vivo Embryonic and Extraembryonic Potency. Cell 169, 243–257 10.1016/j.cell.2017.02.005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shultz LD et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J Immunol 174, 6477–6489 (2005). [DOI] [PubMed] [Google Scholar]
- 142.Bosma GC, Custer RP & Bosma MJ A severe combined immunodeficiency mutation in the mouse. Nature 301, 527–530 (1983). [DOI] [PubMed] [Google Scholar]
- 143.Greiner DL et al. Improved engraftment of human spleen cells in NOD/LtSz-scid/scid mice as compared with C.B-17-scid/scid mice. Am J Pathol 146, 888–902 (1995). [PMC free article] [PubMed] [Google Scholar]
- 144.Palchaudhuri R et al. Non-genotoxic conditioning for hematopoietic stem cell transplantation using a hematopoietic-cell-specific internalizing immunotoxin. Nat Biotechnol 34, 738–745, 10.1038/nbt.3584 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.McIntosh BE et al. Nonirradiated NOD,B6.SCID Il2rgamma−/− Kit(W41/W41) (NBSGW) mice support multilineage engraftment of human hematopoietic cells. Stem Cell Reports 4, 171–180, 10.1016/j.stemcr.2014.12.005 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kyttala A et al. Genetic Variability Overrides the Impact of Parental Cell Type and Determines iPSC Differentiation Potential. Stem Cell Reports 6, 200–212, 10.1016/j.stemcr.2015.12.009 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kajiwara M et al. Donor-dependent variations in hepatic differentiation from human-induced pluripotent stem cells. Proc Natl Acad Sci U S A 109, 12538–12543, 10.1073/pnas.1209979109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hockemeyer D & Jaenisch R Induced Pluripotent Stem Cells Meet Genome Editing. Cell Stem Cell 18, 573–586, 10.1016/j.stem.2016.04.013 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chen JR et al. Effects of genetic correction on the differentiation of hair cell-like cells from iPSCs with MYO15A mutation. Cell Death Differ 23, 1347–1357, 10.1038/cdd.2016.16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mandai M, Kurimoto Y & Takahashi M Autologous Induced Stem-Cell-Derived Retinal Cells for Macular Degeneration. N Engl J Med 377, 792–793, 10.1056/NEJMc1706274 (2017). [DOI] [PubMed] [Google Scholar]


