Summary
Ubiquitylation is a post-translational modification (PTM) that controls various cellular signaling pathways. It is orchestrated by a three-step enzymatic cascade know as the ubiquitin proteasome system (UPS). E3 ligases dictate the specificity to the substrates, primarily leading to proteasome-dependent degradation. Deregulation of the UPS components by various mechanisms contributes to the pathogenesis of cancer. This review focuses on E3 ligase-substrates pairings that are implicated in B-cell malignancies. Understanding the molecular mechanism of specific E3 ubiquitin ligases will present potential opportunities for the development of targeted therapeutic approaches.
The Ubiquitin Proteasome System
The ubiquitin proteasome system (UPS) plays a significant role in the regulation of cell growth and survival, in addition to maintaining cellular homeostasis. By means of the UPS, cells can precisely and temporally degrade approximately 80% of the entire proteome. However, failure to do so results in numerous diseases, including hematological malignancies and cancer1-3. Protein ubiquitylation is catalyzed by a three-step enzymatic cascade in which the ubiquitin is first activated by an E1 enzyme (ubiquitin-activating enzyme) and subsequently transferred to an E2 enzyme (ubiquitin-conjugating enzyme). Finally, ubiquitin is attached to a specific substrate that is selected by an E3 ubiquitin ligase that governs substrate specificity. One of the principal outputs of protein ubiquitylation is degradation via the proteasome complex. The proteasome comprises of a regulatory 19S cap complex that unfolds the substrates in an ATP-dependent manner and a catalytic 20S core complex that has proteolytic activities1. Proteins that are tagged by the ubiquitin chains are recognized, deubiquitylated, and unfolded by the 19S complex and subsequently fed through the inner channel of the 20S proteasome chamber, which cleaves proteins into peptides4.
Ubiquitin contains total eight attachment sites (seven lysine residues and the amino N-terminus) for the formation of polymeric chains5. Substrates can be modified at multiple lysine residues with a single ubiquitin molecule (multimono-ubiquitylation), or a single ubiquitin molecule can build a chain using ubiquitin as substrate6. Moreover, ubiquitin chains can be homotypic conjugates where they are elongated through the same lysine as in Lys11-, Lys48-, Lys63-linked chains or methionine (M-linked) residue, as in linear chains6. Lys-11-linked chains and Lys48-linked chains target proteins for the proteosomal degradation7, 8. On the other hand, Lys-63-linked chains regulate DNA repair, endocytic trafficking, NF-κB activation, and assembling a signaling complex for mRNA translation9-12. M-linked chains or linear ubiquitin chains play an important role in immune, inflammatory and NF-κB signaling13-15. The significance and the roles of Lys6-, Lys27-, Lys29-, Lys33- linked chains are still poorly understood although, recently, they have been implicated in DNA repair, trans-Golgi trafficking, and mitochondria damage16,17.
Ubiquitin ligases
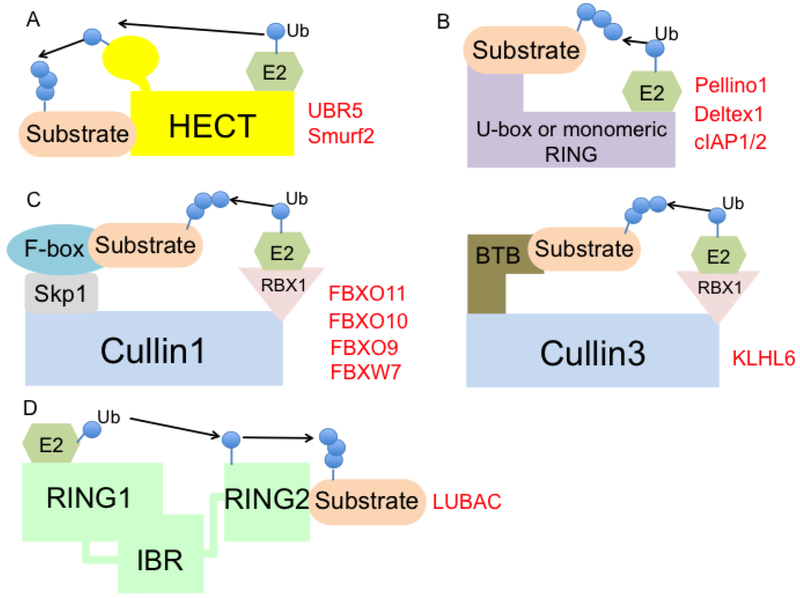
Ubiquitin ligases are categorized into different classes based on their specific structural configuration and the composition of subunits—HECT (homologous to E6-AP1 (E6-associated protein 1) carboxy-terminus)-type, RING (really interesting new gene)-finger-type, U-box-type, or RING-in-between RING (RBR)-type (Fig. 1).
Fig.1. Different types of E3 ubiquitin ligases.
Ubiquitin ligases are classified into different groups—HECT-type, RING-type or U-box type, Multi-subunit RING-type, and RING-Between-RING (RBR) type.
(A) The HECT-type E3 ligase directly accepts ubiquitin molecules from the E2 enzyme and transfers it to the target substrate. This includes Ubr5 and Smurf2.
(B) The RING-type ligase uses the RING-domain binding the E2 enzyme and the other end interacting with the substrate, which bridges in close proximity to transfer the ubiquitin from the E2 to the target. This includes Pellino1, Deltex1, cIAP1/2.
(C) Multi-subunit RING-type E3 ligase utilizes different CULLIN scaffolds, a substrate receptor, an adaptor, and a RING-domain protein for E2 enzyme recruitment. CULLIN1 uses SKP1 (adaptor), different F-box proteins (substrate receptor), and RBX1 (binding E2). This includes FBXO11, FBXO10, FBXO9, FBXW7. In contrast, CULLIN3 uses BTB protein (adaptor and substrate receptor) and RBX1. This includes KLHL6.
(D) RBR-type E3 ligases use combination mechanisms of the HECT-type and RING-type. This includes LUBAC.
The HECT-type E3 ligases are the only ones that demonstrate intrinsic catalytic activity, as they receive the ubiquitin from an E2 enzyme and transfer it to the substrate18. HECT-type E3 ligases are the first family of E3 ligases that have been described and consist of ~30 HECT domain E3 ligases in mammals. They play important roles in several biologic areas, including protein trafficking, cell growth and survival, immune regulation, and many others19. The N-terminus of the HECT-type E3 ligases mediates substrate targeting, while the C-terminus contains the conserved HECT domain, which interacts with the E2 and contains an active cysteine that accepts the ubiquitin-moiety. The topology of the HECT-type E3s with the E2s depends on the status of the ubiquitin transfer of E2 and the non-covalent interaction with all N-terminus, C-terminus, E2 and ubiquitin20, 21.
The RING-finger and the U-box- type E3 ligases act as a scaffold protein to bridge an E2 enzyme and a substrate proximally for ubiquitin conjugation (Fig. 1). The RING-finger-type E3 ligases are generally thought to be the biggest family of ligases and contain a Zn2+-coordinating domain with spaced cysteine and histidine residues, facilitating E2-dependent ubiquitylation22. The RING finger ubiquitin ligase family functions either as a monomer, dimer, or a multi-subunit complex. Homodimerization and/or heterodimerization usually occurs through the RING finger domain23.
Multi-subunit RING-type E3 ligases are exemplified by the CULLIN-RING-ligase (CRL) and the anaphase-promoting complex/cyclosome (APC/C)19. CRLs constitute the biggest family of other multi-component E3 ligases. These consist of a cullin scaffold protein (CULLIN 1, 2, 3, 4A, 4B, 5, 7), a substrate receptor, an adaptor, and a RING domain protein for E2 enzyme recruitment. A large body of evidence suggests that CRLs share a similar molecular architecture, where substrates are recruited at the N-terminal regions of the cullins, which comprise of an adaptor protein and a substrate receptor24, 25. For the well-characterized example of CULLIN1, the adaptor protein is SKP1, which recruits numerous substrate receptors (i.e., the F-box proteins). SKP1 can interact with both CULLIN1 and CULLIN7, while CULLIN2 and CULLIN5 utilize the elongin B/C adaptor proteins to recruit the substrate receptors; a family of proteins, which is named suppressor of cytokine signaling/elongin BC (SOCS/BC)-box-protein. CULLIN3 is unique; it utilizes a BTB domain-containing protein that can function both as an adaptor and a substrate receptor. CULLIN4A uses the adaptor protein DNA damage-binding protein-1 (DDB1), which in turns binds to substrate receptor proteins such as the DDB1 and CUL4 associated factors (DCAFs). The biological function of the cullins is involved in various cellular processes; these include cell cycle, signaling transduction, cell proliferation and survival, and DNA damage response24.
A third class of E3 ligases is that of the RBR E3 ligases, which consists of a RING1, an intermediate RING (IBR), and a RING2 domain (Fig. 1). These ligases use a unique mode of catalyzing the ubiquitin transfer by combining both the RING-type and HECT-type E3 ligase mechanisms. Similar to classical RING-type ligases, these recognize the E2 conjugated ubiquitin by the RING1 domain, and form a HECT-like intermediate by accepting the ubiquitin to the cysteine of the RING2 domain. The ubiquitin is finally transferred to the substrate by the RING226.
Ubiquitin ligases in B-cell lymphoid malignancies
Increasing evidence supports that the B-cell lymphoid malignancies develop from various stages of B-cells, hijacking the mechanisms that drive B-cell differentiation and activation27. For instance, mantle cell lymphoma (MCL) and chronic lymphocytic leukemia (CLL) originate from pre-germinal center (GC) mature B-cells, while most of the non-Hodgkin’s lymphomas and multiple myelomas (MMs) originate from the GC B-cells or B-cells that have gone through a germinal center reaction. Each of these B-cell lymphoid malignancies features different genomic alterations, including chromosomal translocations, amplifications, frameshift deletions, and mutations, to ultimately activate the oncogenic signaling programs that promote growth and survival28. Sequencing efforts have identified many E3 ubiquitin ligases that are mutated, amplified, or deleted in B-cell lymphoid malignancies, functioning either as a tumor suppressor or oncogene29. Importantly, the success of a proteasome inhibitor (bortezomib), in both MM and MCL, has further inspired the investigation of the biological significance of protein ubiquitylation and degradation in B-cell cancers30-32. Thus, the understanding of how the ubiquitylation of protein is achieved, and of the downstream molecular events, is critical for the development of therapeutic approaches. Targeting specific E3 ubiquitin ligases has garnered attention as it confers a selective advantage over a proteasome inhibitor by preventing any unwanted toxic effects on intra-cellular proteins.
In this review, we will discuss a wide spectrum of E3 ligases that have been implicated in the pathogenesis of B-cell malignancies (Fig.1 and Table.1).
Table. 1.
Summary of E3 ligases implicated in B-cell malignancies
| E3 ligases | Targets | Pathway associated in B- cell malignancies |
B-cell malignancies associated |
|---|---|---|---|
| KLHL6 | Roquin2 | BCR signaling, NF-κB pathway | ABC-DLBCL, CLL, FL, MM, MZL |
| LUBAC | NEMO/IKKγ | BCR signaling, NF-κB pathway | ABC-DLBCL |
| Pellino1 | BCL6 | BCR signaling, BCL6 stabilization | GCB-DLBCL, BL, PL |
| UBR5 | Katanin, E6AP, CTNNB1, hPXR, RNF168, CDK9, PEPCK1, TOPBP1, PAIP2, TRIP12 | DNA damage, cell cycle, chromosome regulation | MCL |
| Deltex1 | MEKK1, PKCθ, PLC-γ1, HIF-1α, c-FLIP | Notch activation | CLL, DLBCL |
| cIAP1/2 | RIP1, NIK, BCL10 | BCR signaling, NF-κB pathway | ABC-DLBCL |
| Smurf2 | Smads, Runx2, Id1, Smurf1, RNF20, YY1 | Transactivation of c-Myc | GCB-DLBCL, BL, FL |
| FBXO11 | CDT2, BCL6, BLMP-1, SNAIL | GC expansion, BCL6 up-regulation | GCB-DLBCL, BL |
| FBXO10 | BCL2, RAGE | Overexpression of BCL2 | GCB-DLBCL, MCL |
| FBXO9 | TEL2, TTI1, PPARγ | PI3K/TORC2/AKT pathway | MM |
| FBXW7 | NOTCH1, NOTCH4, cyclin E, c-Myc, c-Jun, Aurora B, MCL1, STAT3, p-STAT3 (Y705) | c-Myc, NOTCH, STAT3 stabilization, alternative NF-κB pathway | ABC-DLBCL, BL, MM, CML |
KLHL6
Kelch-like protein 6 (KLHL6) is a CULLIN3-based E3 ligase. KLHL6 utilizes the BTB domain to interact with CULLIN3 and the Kelch domain to recruit substrates33. KLHL6 is expressed at all stages of B-cell development and is up-regulated in sheep Peyer’s patch, human tonsil B cells, and germinal center (GC) B-cells34, 35. Klhl6−/− mice are viable, but they display a smaller spleen, reduced GC formation upon antigen-stimulation, and defects in mature B-cell populations34. Recently, Bertocci et al. have shown that the reduction of mature B cells is due to the inability of the transitional type 1 B cells to survive and segregate to the next stage36. Furthermore, type 1 B cells of Klhl6−/− mice display an overexpression of the cell cycle and proliferation genes, whose expression is down-regulated upon BAFF stimulation. Overall, KLHL6 seems to control BAFF-induced B cell differentiation by regulating the cell cycle pathway genes.
It is important to note that point mutations in the KLHL6 locus occur in B-cell malignancies. Most of the mutations hit diffuse large B-cell lymphoma (DLBCL), and more rarely, the chronic lymphocytic leukaemia (CLL), follicular lymphoma (FL), multiple myeloma (MM) and marginal zone lymphoma (MZL)37-44. Most KLHL6 mutations are missense, and localize in the BTB domain, resulting in a protein that lacks the ability to interact with CULLIN333. Although these cancer-associated mutations are equally stratified between the germinal center (GC)- and the activated B cell (ABC)-DLBCL subtypes, they correlate specifically with poorer survival rate in ABC-DLBCL patients. Whether these mutations would have the same loss-of-function phenotype in other B-cell malignancies still remains to be determined.
Roquin2 is the first bona-fide substrate of KLHL6 that was recently identified using proteomic approaches. It is an RNA-binding protein that promotes mRNA decay of tumor suppressors and NF-κB inhibitor genes, many of which are dependent on B-cell receptor (BCR) signaling. Thus, the loss of Roquin2 degradation in DLBCLs with KLHL6 mutations results in the hyperactivation of the NF-κB pathway, promoting cancer cell growth and proliferation33.
Further studies will be required to identify other substrates and to fully understand the contribution of KLHL6 to the pathogenesis of B-cell malignancies.
LUBAC
The linear ubiquitin chain assembly complex (LUBAC) is the only known E3 ligase responsible for the linear polyubiquitin chain formation13. LUBAC consists of heme-oxidized iron-responsive element-binding protein 2 (IRP2) ubiquitin ligase-1 [HOIL-1; also known as RanBP-type and C3HC4-type zinc finger-containing protein 1 (RBCK1)], the HOIL-1-interacting protein (HOIP; also known as RNF31) and the SH3 and multiple ankyrin repeat domains protein (SHANK)-associated RBCK1 homology (RH)-domain-interacting protein (SHARPIN). Both HOIL-1 and HOIP are RING-between-RING (RBR) E3 ligases, but HOIP is considered to be the key component for LUBAC catalytic activity. While HOIP alone is not sufficient to induce linear ubiquitin chains, any combination of the LUBAC components with HOIP can stimulate a catalysis of linear ubiquitin chains14,15. LUBAC function has been implicated in various signaling pathways such as TNF, Toll-like receptors (TLRs), CD40/CD40L, RIG-I, IL-1β, and the nucleotide-binding oligomerization domain 2 (NOD2)45.
Sharpin−/− mice develop chronic proliferative dermatitis with splenomegaly and systemic inflammation, while Hoip1−/− mice are embryonic lethal due to the vascular defects in the yolk sac46-48. Remarkably, while Hoil-1 −/− mice are viable without any abnormal phenotype49, human patients with a loss of function mutations in HOIL-1 develop chronic autoimmune symptoms with impaired TNF- and IL-1β-induced NF-κB activation50.
LUBAC can be recruited to the CARD11-BCL10- MALT1 (CBM) complex by binding K63-linked chains through its subunit RNF31. The RNF31 domain is also responsible for promoting NEMO (also known as IKKγ) poly-linear ubiquitylation, which controls the NF-κB pathway51,52. The ubiquitinated NEMO, in turn, brings additional IKK complexes that induce the auto-phosphorylation and activation of the IKKβ and increase NF-κB activation.
A subtype of DLBCL patients, called activated B-Cell (ABC)-DLBCL, carry missense mutations of the LUBAC member RNF31 at the positions Q584H and Q622L53. These mutations increase the ubiquitin ligase activity of LUBAC by enhancing the interaction between HOIL-1 and HOIP, which in turn promotes the NF-κB activity. In addition to NEMO, LUBAC sustains BCR signaling in the ABC-DLBCL cell lines by promoting MALT1 activity; a member of the CBM complex, which activates the IKK complex. Consistently, the depletion of components of LUBAC is toxic to the ABC-DLBCL cells53,54. Given that LUBAC controls various pathways that engage with NF-κB, it is conceivable that a small molecule inhibitor of LUBAC could be a potential therapeutic target in the ABC-DLBCLs.
PELI1
The Pellino protein family consists of three members: Pellino1 (also known as PRISM), Pellino2, and Pellino355. Pellinos contain a putative RING-like motif with a conserved pattern of cysteine and histidine residues, suggesting that it belongs to the RING class E3 ubiquitin ligase56. All three Pellino members have E3 ubiquitin ligase activity in vitro, and utilize various E2 enzymes to promote poly-ubiquitination via K11, K48, and K63-linked chains. The first evidence of Pellino proteins having ubiquitin ligase activity came from the identification of IRAK1 as a bona-fide substrate57,58. Importantly, RING-like motif mutants of Pellino are impaired in the IRAK1 ubiquitylation. The Pellino members also interact with IRAK4 kinase, Toll-like receptors (TLRs), and interleukin-1 (IL-1R) signaling molecules like TRAF6 and TAK1, regulating innate and adaptive immune responses57,59,60.
Pellino1−/− mice are viable without obvious phenotypes. However, they show a diminished response to TLR3 and TLR4 signaling, thus displaying resistance to septic shock caused by TLR engagement61. Consistently, the mice showed reduced activation of the NF-κB and decreased expression of pro-inflammatory genes like TNF and IL-6. In contrast, Pellino1 transgenic mice exhibit B-cell lymphoma formation with constitutive activation of B-cell receptor signaling62. In this context, Pellino1 has been shown to interact and promote K63-linked polyubiquitylation of BCL6, resulting in its stabilization and in the development of DLBCL, BL, and plasmablastic lymphoma (PL). Pellino1 is often highly expressed in high-grade B-cell cancers, correlating with the expression of oncoproteins such as BCL2, BCL6, c-MYC, and MUM163. High Pellino1 levels show poor prognosis and survival rates in DLBCL patients, suggesting Pellino1 as an oncogene and a potential therapeutic target in B-cell lymphoma. Although other two Pellino members play key roles in immune signaling, their contribution to the pathogenesis of B-cell malignancies remains currently unknown.
UBR5
UBR5 is a HECT-type E3 ligase, also known as EDD (E3 identified by differential display)64. The HECT domain in UBR5 has a unique feature, where its C-lobe does not have a surface for the binding of ubiquitin65. Instead, the UBA domain is used for ubiquitin binding and the MLLE/PABC domain is used for substrate interaction66, 67. UBR5 plays a key role in regulating the cell cycle, DNA damage response, and transcriptional and translational control by targeting a variety of substrates; such as KATANIN, E6AP, CTNNB1, hPXR, RNF168, CDK9, PEPCK1, TOPBP1, PAIP2, and TRIP1268-76. Furthermore, UBR5 is required for CHK2 phosphorylation and effective p53 activation77.
UBR5 is an essential gene as Ubr5−/− mice exhibit embryonic lethality due to defects in the vascular and yolk sac development, suggesting its role in vessel formation78. Despite this observation, non-synonymous mutations of UBR5 are detectable at high frequency (~18%) in MCL tumors and display a mutually exclusive distribution pattern with alterations of the major driver of MCL, cyclin D179,80. Most of the UBR5 mutations, including frameshift, affect the cysteine residue of the HECT domain or tend to occur in the HECT and PABC domains, resulting in a loss of E3 ligase activity. Gene ontology analysis reveals DNA damage, cell cycle, and chromosome biology as the predominant factors affected by such UBR5 mutations. Overall, these evidences suggest that UBR5 is a tumor suppressor in MCL, although the relevant substrate(s) responsible for the pathogenesis of the disease remains in question.
Deltex1
In mammals, the Deltex protein family consists of Deltex 1, 2, 3, and 4. These proteins contain two WWE domains, a proline-rich motif, and a RING-finger domain81,82. Deltex1 is a negative regulator of the Notch signaling pathway, controlling ubiquitination and the endosomal recycling of Notch81,83,84. Deltex is a transcriptional target of the nuclear factor of activated T cells (NFAT) and is well known for its involvement in T-cell tolerance. Mice expressing a Deltex1 deleted of its RING-finger domain, or those lacking the first WWE domain, show normal lymphocyte development and function, suggesting that these domains and the E3 ligase activity may be dispensable in lymphocyte differentiation85. Although, it is likely that other Deltex members are functionally redundant and compensate for the loss of Deltex1 function, Deltex1−/− mice exhibit a profound increase in T-cell activation, which manifests as cell proliferation and a resistance to anergy86. Deltex1−/− mice are more susceptible to autoimmune syndromes characterized by inflammation and to the production of auto-antibodies.
Mechanically, Deltex has been shown to stimulate the degradation of many proteins such as MAP kinase kinase kinase 1 (MEKK1), protein kinase Cθ, phospholipase C-γ1, HIF-1α, and c-FLIP87-90. Deltex1 has also been shown to promote the expression of ILT3, the immunoglobulin-like transcript 3, in CLL. ILT3 is a receptor expressed by myeloid cells, which inhibits the activation of AKT following BCR-stimulation through the recruitment of active SHIP1 (Phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase 1)91. Though ILT3 up-regulation is a marker of CLL cells, how Deltex1 synergistically acts with other specific CLL factors in the regulatory network remains to be understood.
Furthermore, DLBCL patients with non-synonymous mutations in the WWE1-domain of Deltex1 exhibit significantly worse survival than those with a wild-type Deltex192. The Deltex1 deleterious mutations release their negative regulation on Notch, thus promoting Notch activation in DLBCL. Interestingly, DLBCL tumors with Deltex1 mutations also show significantly lower Deltex1 expression. Pathway enrichment analysis studies demonstrate that the expression signatures IFN-γ, JAK-STAT, and the ubiquitin-proteasome signaling pathways inversely correlate with the Deltex1 expression. Alternately, the genes involved in BCR signaling, MAPK signaling, and chromatin remodeling are positively correlated with the Deltex1 expression. Although the Deltex1 seems to be a tumor-suppressing protein, the exact molecular mechanism is yet to be determined.
cIAP1/2
The cellular inhibitor of apoptosis 1 and 2 (cIAP1 and 2) proteins belong to the inhibitor of the apoptosis protein (IAP) family of proteins that share three baculovirus IAP-repeat (BIR) domains22. In addition to the IAP domains that recruit caspases and other proteins, the cIAP1/2 proteins contain a RING-finger domain suggesting that they might bind an E2 enzyme, and thus have E3 ligase activity. cIAP1/2 are involved in various signaling pathways including TNF-α mediated NF-κB activation93. These proteins directly interact with TRAF2 and the TNF receptors to promote a poly-ubiquitination of RIP1, utilizing K63-linked ubiquitin chains. This molecular event stimulates interaction with the IKK, LUBAC, and TAK1-TAB2/3 complexes52,94-96. cIAP1/2 have also been described to promote the K48-linked polyubiquitination and the proteasomal-degradation of NIK; the main kinase upstream of the alternative NF-κB pathway97,98.
ciap1−/− or ciap2−/− mice have defects in capase-1 activation and inflammasome assembly, causing resistance to peritonitis. Deletion of both the cIAP1/2 genes is not compatible with life as embryos die at E12. 5. Conditional ablation in B-cells leads to an increase of the B-cell numbers, characterized by hyperactivation of the alternative NF-κB pathway and defects in the germinal center immune response99. In keeping with the function of being the positive regulator of NF-κB, the cIAP1/2 copy number gains have been detected in ABC-DLBCL tumors95. In addition to RIP1, cIAP1/2 further contribute to the NF-κB activity by poly-ubiquitylating BCL10 with K63-linked chains, which in turn promotes IKK recruitment to the CBM complex.
Chemical inhibition (SMAC mimetics) and the genetic knockdown of cIAP1/2 are toxic to BCR-signaling dependent ABC-DLBCL cell lines, but not to BCR-signaling independent ones. SMAC is a small mitochondrial protein that binds and suppresses the IAP proteins during apoptosis. Mechanistically, SMAC mimetics promote auto-ubiquitylation and degradation of cIAP1/2, suppressing NF-κB pathway. Whether these small molecules have clinical impacts for treating ABC-DLBCL patients, as a single agent or with the combination of other BCR pathway inhibitors, still needs further investigation.
Smurf2
Smurf2 is a HECT-type E3 ubiquitin ligase and was initially considered a negative regulator of TGF-β signaling100,101. Later studies have shown that Smurf2 plays a role in various signaling pathways by targeting Smads, Runx2, Id1, Smurf1, TGF-beta receptor, and RING finger protein (RNF20) for ubiquitylation and degradation100,102-105. Moreover, Smurf2 is also an important regulator of senescence as it can activate p53 and pRB pathways106.
Smurf2−/− mice develop a wide spectrum of tumors in the liver, blood, lungs, and pituitary glands, but most notably in B-cell lymphoma107. Smurf deficient cells can bypass senescence and increase splenic B-cell proliferation leading to malignant lymphoma. In the context of lymphoma, a key substrate of Smurf2 is YY1, which is ubiquitylated and degraded. YY1 is a master regulator of the GC transcription program, and Smurf2 deficiency induces YY1-dependent transactivation of c-MYC, which contributes to lymphomagenesis. Human DLBCL, BL, and FL tumors show a decrease in the Smurf2 expression with elevated YY1 expression, which correlates with poorer survival rates only in DLBCL and BL108. This Smurf2-YY1-MYC axis represents the novel mechanism for lymphomagenesis in GC or post-GC B cells. Recently, Smurf2 has been shown to regulate genomic instability by affecting monoubiquitylation of histone H2B and function as a tumor suppressor102. Smurf2, thus, has multiple tumor suppressive roles by regulating genomic stability, cell proliferation, senescence, and apoptosis. However, the exact mechanism of how Smurf2 contributes to tumorigenesis in different cancer types remains an open question.
FBXO11
FBXO11 is a member of the F-box proteins family, which includes three different groups based on the substrate recognition domains—the FBXW (subclass with WD40 repeat domains), the FBXL (subclass with leucine-rich repeat domains), and the FBXO (subclass with various uncharacterized domains). In most cases, F-box proteins recognize their substrates through short and specific motifs, which are often called degrons5. Often, phosphorylation at the degron is the trigger for the efficient recruitment of the F-box protein and the consequent ubiquitylation and proteasomal degradation.
FBXO11 utilizes the C-terminal carbohydrate-binding proteins and sugar hydrolases (CASH) repeat domain as the substrate-binding domain. FBXO11 controls the cell cycle via the degradation of its substrate CDT2 (also known as DTL)109. CDT2 is a CRL4 complex which promotes a cell cycle exit, DNA replication, and cell migration through p21 and SEDT8 degradation110. FBXO11 also controls epithelial-mesenchymal transition (EMT), and cancer metastasis by degrading the substrates such as BLMP-1 and SNAIL111,112.
Fbxo11−/− mice exhibit perinatal lethality with facial clefting, while Fbxo11 haploinsufficient mice display reduced weight and a severe infection of the middle ear, as observed in Jeff (deaf mouse mutant) mice, suggesting a role in the inflammation113-115.
Significantly, the FBXO11 monoallelic deletions and inactivating mutations are observed in DLBCL, Burkitt’s lymphoma (BL) cell lines, and primary DLBCL patient samples. The mutations target the CASH domain, leading to a loss of the substrate interaction. In the context of B-cell malignancies, a key FBXO11 substrate is BCL6. BCL6 is a critical transcriptional factor that regulates B-cell development and differentiation and is often overexpressed in DLBCL by chromosomal translocation, hypermutation of its promoter, or deregulated proteolysis. This supports the notion that FBXO11 functions as a haploinsufficient tumor suppressor gene in DLBCL by mediating the degradation of the oncogenic BCL6 protein116. Accordingly, a re-expression of FBXO11 in FBOX11-null DLBCL cells has an antitumor effect by inhibiting cancer cell growth. Mice carrying Fbxo11 inactivation, specifically in GC B-cells, exhibits abnormal GC expansion, up-regulation of BCL6 protein levels, and B-cell lymphoproliferations117. Further studies will assess whether the additional substrates in germinal center B-cells contribute to the developments of any B-cell malignancies.
FBXO10
FBXO10 is another FBXO protein with the CASH domain43. The primary target of FBXO10 is the anti-apoptotic protein BCL2, although the receptor for advanced glycation end products (RAGE) has recently been identified as another substrate118,119. The BCL2 protein is localized to the outer membrane of the mitochondria and inhibits the function of pro-apoptotic proteins such as Bax and Bak. Overexpression of BCL2 induces cellular transformation, in concomitance with the alteration of oncogenes like c-Myc, causing aggressive B-cell lymphoid malignancies120.
BCL2 chromosomal translocation that places its promoter next to the immunoglobulin heavy chain locus leads to abnormally high transcription levels of BCL2 in follicular lymphoma121. Thus, it is conceivable that the deregulation of BCL2 proteolysis could be another mechanism for BCL2 overexpression and lymphoma development. In DLBCL, the FBXO10 mRNA level is down-regulated and frameshift, missense mutations, or deletions are also detected118. Specifically, the R44H mutation in the F-box domain abolishes FBXO10 interaction to SKP1, inhibiting the formation of the SCF complex. Furthermore, the V762L and R825W mutations in the CASH domain presumably lead to the disruption of substrate interaction. All FBXO10 mutants exhibit an impaired ability to degrade BCL2 and are much less toxic than the wild-type FBXO10 when overexpressed in lymphoma cell lines. Interestingly enough, the FBXO10 mutations detected in DLBCLs are heterozygous; thus, FBXO10 functions as a haploinsufficient tumor suppressor. Similarly, like in DLBCLs, high levels of BCL2 expression and low levels of FBXO10 expression are also observed in MCL122.
The BCL2 inhibitor, ABT-199, BTK inhibitor, and ibrutinib, have synergistic anti-tumor effects in both the BCR-dependent and BCR-independent MCL tumor growth. It will be important to investigate whether this inverse correlation between FBXO10 and BCL2 is a general regulation mechanism, which applies to other B-cell malignancies.
FBXO9
FBXO9 has been reported to control the proliferation of MM. Specifically, its mRNA level is elevated in MM cell lines and MM patients, correlating with copy number gains. Expression of FBXO9 correlates with a higher progression-free survival (PFS) and better response rates in MM patients treated with the proteasome inhibitor bortezomib123,124. FBXO9 promotes ubiquitylation and degradation of the mTORC1-bound telomere length regulation protein 2 (TEL2) and TELO2-interacting protein 1 (TTI1), upon growth factor withdrawal. This leads to the attenuation of mTORC1 signaling to restrain cell growth, but activates the PI3K/TORC2/AKT pathway, disengaging the inhibitory feedback and sustaining cell survival. Correspondingly, MM cells with FBXO9 overexpression exhibit low levels of S6K1 phosphorylation and high levels of AKT phosphorylation. Furthermore, it has been shown that the CK2 kinase, one that is frequently overexpressed in many different cancers, primes the FBXO9-dependent degradation of TEL2 and TTI1125. Thus, pharmacological inhibition of CK2 in MM cell lines could potentially be of a therapeutic advantage by promoting the stabilization of TEL2 and TTI1. Recently, the FBXO9 has been shown to promote proteasomal degradation of peroxisome proliferator-activated receptor gamma (PPARγ), playing a role in adipocyte differentiation and adipogenesis126. Further investigation to understand other cellular pathways regulated by FBXO9 as well as the assessment of the therapeutic windows might be required to target the FBXO9-TEL2 pathway.
FBXW7
FBXW7 is a member of the FBXW class, featuring eight WD40 repeats that assemble in a β-propeller binding pocket, interacting with phosphorylated substrates. Fbxw7−/− mice exhibit embryonic lethality due to defects in their vascular development, probably because of the stabilization of its substrates—NOTCH1 and NOTCH4127. In the bone marrow, conditional FBWX7 deletion promotes the p53 dependent-cell cycle entry and cellular apoptosis, leading to a premature decrease in hematopoietic stem cells128,129.
FBXW7 functions as a tumor suppressor by targeting multiple oncogenic proteins such as cyclin E, c-MYC, c-Jun, Aurora B, and MCL1130-134. Recently, FBXW7 has been shown to target STAT3 and pSTAT3 (Y705) regulating cellular apoptosis. The low expression of FBXW7 correlates with the high expression of STAT3, resulting in a poor prognosis in ABC-DLBCL patients135. Correspondingly, loss of function mutations of FBXW7 with frequent deletions and inactivating mutations are identified in many cancers136. Most missense mutations are heterozygous and occur within the WD40 domain; these mutations directly interfere with the recruitment of phosphorylated substrates, its localization, or translation. Some FBXW7 substrates such as c-MYC and NOTCH show degron mutations in cancer. For instance, in Burkitt’s lymphoma, mutations were identified in Threonine 58 of c-MYC that undergoes phosphorylation by GSK3β , causing c-MYC stabilization137.
However, the alteration of FBXW7 is infrequent in some B-cell malignancies, and evidences that FBXW7 serves as a pro-survival factor in MM124 and CML138 have been shown. In MM, FBXW7 is responsible for the degradation of p100, an inhibitor of the non-canonical NF-κB signaling pathway. MM cell lines are addicted to NF-κB activation. Thus, constitutive degradation of p100 is a requirement for MM growth and survival. As nuclear p100 is phosphorylated by glycogen synthase kinase (GSK3), it is conceivable that GSK3 inhibitors can be a potential therapeutic approach for MM patients. Although FBXW7 mostly functions as a tumor suppressor by degrading oncogenic proteins, it can also exert a pro-survival effect, supporting its role as both context- and substrate-dependent.
Concluding remarks
It has become apparent that the E3 ubiquitin ligases play a critical role in many biological processes through the regulation of substrate ubiquitylation. The precisely controlled and context-specific turnover of proteins in a timely manner is critical for normal cellular homeostasis. It is also evident that the deregulation of E3 ligases or their substrate degradation contributes to the pathogenesis of human diseases, including lymphoid malignancies.
The biology of different types of lymphoid cancers depends on the stages of B-cell differentiation from which they are derived. Moreover, different lymphoid malignancies are driven by various oncogenic signaling pathways, which are regulated by protein ubiquitin system at multiple levels.
One of key events in lymphoma arising from a post-GC B cells is the constitutive activation of NF-κB pathway that can promote cancer cell survival and proliferation. In this review, KLHL6, LUBAC, and cIAP1/2 are the prime examples of E3 ligases that contribute to stimulation of NF-κB transcriptional activity. All three ligases engage chronic active BCR signaling to promote NF-κB signaling, which is central to the pathogenesis of ABC-DLBCLs. Although they generate different polyubiquitin-linked chains (Lys-48, Lys-63, and linear) of distinct structures, they are directly affected by malignant lesions and deregulated by multiple mechanisms. Both aberrant proteolytic and non-proteolyic protein ubiquitination ultimately activate the survival NF-κB pathway.
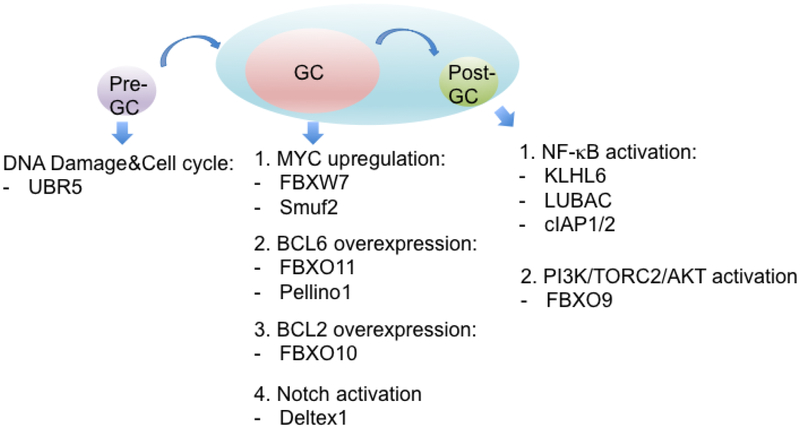
BCL6, BCL2, and c-MYC are dominant oncogenes deregulated by chromosomal rearrangement and missense mutations in lymphoma arising from germinal center B-cells such as BL, FL, or GCB-DLBCL. Protein ubiquitylation adds another layer to the biology of GC lymphoma as discussed in this review. Specifically, BCL6 is overexpressed and stabilized by inactivation of FBXO11 and Pellino1-induced Lys63-polyubiquitylation. MYC is up-regulated by Smurf2-mediated activation of YY1 or by mutations of its FBXW7 degron. In addition, the substrate interaction domain of FBXO10 is disrupted to impair the proteosomal degradation of BCL2. Thus, different lymphoma subtypes are characterized by inappropriate turnover of key cellular players for that particular stage of B-cell differentiation (Fig.2).
Fig.2. Different E3 ubiquitin ligases altered at particular stages of B-cell differentiation.
The relevant ubiquitin ligases that are genetically altered through overexpression, deletion, and mutation along with the subsequent pathways or substrates that are deregulated are shown at a pre-GC, GC, or post-GC stage.
Although our knowledge of the biochemical and biological functions of E3 ubiquitin ligases has increased in B-cell malignancies, a deeper characterization of cell-context dependent substrate regulation and biological relevance is needed. The recent approval of a general proteasome inhibitor, VELCADE or bortezomib, has demonstrated a great efficacy for the treatment of MM and MCL30-32. However, toxicity and the relevant side effects, which include anemia, neuropathy, and thrombocytopenia, in addition to bortezomib resistance, make this drug imperfect139-141. Recent discoveries have shown that thalidomide-based drugs target Cereblon (CRBN)142, a member of the CULLIN4 E3 ubiquitin ligase complex, to promote proteasomal degradation of specific targets143,144. This promising area of research has shown the achievement of higher drug potency, resulting in the rapid destabilization of targets via ubiquitin-dependent proteasomal degradation.
Highlights.
The Proteasome ubiquitylation System is deregulated in a variety of B-cell cancers
E3 ligase and substrate pairings in malignant B-cell cancer contribute to the initiation and maintenance of tumorigenesis
Understanding the mechanism of specific E3 ligases will help the development of potential therapeutic targets
Acknowledgements
This work was supported in part by grant R00-CA166181-04, R01-CA207513-01 from the National Cancer Institute and Gilead Sciences Research Scholars Program in Hematology/Oncology to L.B
Footnotes
Disclosure of potential conflicts of interest
The authors certify that they have no affiliations or involvement in any organization or entity with financial or non-financial interests with respect to the subject discussed in this manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hershko A & Ciechanover A The ubiquitin system. Annu Rev Biochem 67, 425–479 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Hershko A & Ciechanover A The ubiquitin system for protein degradation. Annu Rev Biochem 61, 761–807 (1992). [DOI] [PubMed] [Google Scholar]
- 3.Silverman JS, Skaar JR & Pagano M SCF ubiquitin ligases in the maintenance of genome stability. Trends Biochem Sci 37, 66–73 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun BC et al. The base of the proteasome regulatory particle exhibits chaperone-like activity. Nat Cell Biol 1, 221–226 (1999). [DOI] [PubMed] [Google Scholar]
- 5.Skaar JR, Pagan JK & Pagano M Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol 14, 369–381 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komander D & Rape M The ubiquitin code. Annu Rev Biochem 81, 203–229 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Chau V et al. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243, 1576–1583 (1989). [DOI] [PubMed] [Google Scholar]
- 8.Jin L, Williamson A, Banerjee S, Philipp I & Rape M Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell 133, 653–665 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng L et al. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103, 351–361 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Sun L & Chen ZJ The novel functions of ubiquitination in signaling. Curr Opin Cell Biol 16, 119–126 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Xu M, Skaug B, Zeng W & Chen ZJ A ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFalpha and IL-1beta. Mol Cell 36, 302–314 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spence J, Sadis S, Haas AL & Finley D A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol Cell Biol 15, 1265–1273 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirisako T et al. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J 25, 4877–4887 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerlach B et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature 471, 591–596 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Ikeda F et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-kappaB activity and apoptosis. Nature 471, 637–641 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gatti M et al. RNF168 promotes noncanonical K27 ubiquitination to signal DNA damage. Cell Rep 10, 226–238 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Yuan WC et al. K33-Linked Polyubiquitination of Coronin 7 by Cul3-KLHL20 Ubiquitin E3 Ligase Regulates Protein Trafficking. Mol Cell 54, 586–600 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Ciechanover A The ubiquitin-proteasome proteolytic pathway. Cell 79, 13–21 (1994). [DOI] [PubMed] [Google Scholar]
- 19.Rotin D & Kumar S Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol 10, 398–409 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Huang L et al. Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade. Science 286, 1321–1326 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Verdecia MA et al. Conformational flexibility underlies ubiquitin ligation mediated by the WWP1 HECT domain E3 ligase. Mol Cell 11, 249–259 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Lorick KL et al. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci U S A 96, 11364–11369 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lipkowitz S & Weissman AM RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat Rev Cancer 11, 629–643 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petroski MD & Deshaies RJ Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 6, 9–20 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Cardozo T & Pagano M The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol 5, 739–751 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Walden H & Rittinger K RBR ligase-mediated ubiquitin transfer: a tale with many twists and turns. Nat Struct Mol Biol (2018). [DOI] [PubMed] [Google Scholar]
- 27.Staudt LM & Dave S The biology of human lymphoid malignancies revealed by gene expression profiling. Adv Immunol 87, 163–208 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Staudt LM Oncogenic activation of NF-kappaB. Cold Spring Harb Perspect Biol 2, a000109 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sahasrabuddhe AA & Elenitoba-Johnson KS Role of the ubiquitin proteasome system in hematologic malignancies. Immunol Rev 263, 224–239 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Jagannath S et al. A phase 2 study of two doses of bortezomib in relapsed or refractory myeloma. Br J Haematol 127, 165–172 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Richardson PG et al. A phase 2 study of bortezomib in relapsed, refractory myeloma. N Engl J Med 348, 2609–2617 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Hambley B, Caimi PF & William BM Bortezomib for the treatment of mantle cell lymphoma: an update. Ther Adv Hematol 7, 196–208 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi J et al. Loss of KLHL6 promotes diffuse large B-cell lymphoma growth and survival by stabilizing the mRNA decay factor roquin2. Nat Cell Biol 20, 586–596 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kroll J et al. The BTB-kelch protein KLHL6 is involved in B-lymphocyte antigen receptor signaling and germinal center formation. Mol Cell Biol 25, 8531–8540 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupta-Rossi N et al. Specific over-expression of deltex and a new Kelch-like protein in human germinal center B cells. Mol Immunol 39, 791–799 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Bertocci B et al. Klhl6 Deficiency Impairs Transitional B Cell Survival and Differentiation. J Immunol 199, 2408–2420 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Lohr JG et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A 109, 3879–3884 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morin RD et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 476, 298–303 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garcia-Ramirez I et al. Crebbp loss cooperates with Bcl2 overexpression to promote lymphoma in mice. Blood 129, 2645–2656 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reddy A et al. Genetic and Functional Drivers of Diffuse Large B Cell Lymphoma. Cell 171, 481–494 e415 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Puente XS et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 475, 101–105 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lohr JG et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell 25, 91–101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weigert O et al. Molecular ontogeny of donor-derived follicular lymphomas occurring after hematopoietic cell transplantation. Cancer Discov 2, 47–55 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ganapathi KA et al. The genetic landscape of dural marginal zone lymphomas. Oncotarget 7, 43052–43061 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimizu Y, Taraborrelli L & Walczak H Linear ubiquitination in immunity. Immunol Rev 266, 190–207 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seymour RE et al. Spontaneous mutations in the mouse Sharpin gene result in multiorgan inflammation, immune system dysregulation and dermatitis. Genes Immun 8, 416–421 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Potter CS et al. Chronic proliferative dermatitis in Sharpin null mice: development of an autoinflammatory disease in the absence of B and T lymphocytes and IL4/IL13 signaling. PLoS One 9, e85666 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liang Y & Sundberg JP SHARPIN regulates mitochondria-dependent apoptosis in keratinocytes. J Dermatol Sci 63, 148–153 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peltzer N et al. HOIP deficiency causes embryonic lethality by aberrant TNFR1-mediated endothelial cell death. Cell Rep 9, 153–165 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Boisson B et al. Immunodeficiency, autoinflammation and amylopectinosis in humans with inherited HOIL-1 and LUBAC deficiency. Nat Immunol 13, 1178–1186 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tokunaga F et al. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat Cell Biol 11, 123–132 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Haas TL et al. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol Cell 36, 831–844 (2009). [DOI] [PubMed] [Google Scholar]
- 53.Yang Y et al. Essential role of the linear ubiquitin chain assembly complex in lymphoma revealed by rare germline polymorphisms. Cancer Discov 4, 480–493 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dubois SM et al. A catalytic-independent role for the LUBAC in NF-kappaB activation upon antigen receptor engagement and in lymphoma cells. Blood 123, 2199–2203 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Moynagh PN The roles of Pellino E3 ubiquitin ligases in immunity. Nat Rev Immunol 14, 122–131 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Rich T, Allen RL, Lucas AM, Stewart A & Trowsdale J Pellino-related sequences from Caenorhabditis elegans and Homo sapiens. Immunogenetics 52, 145–149 (2000). [DOI] [PubMed] [Google Scholar]
- 57.Schauvliege R, Janssens S & Beyaert R Pellino proteins are more than scaffold proteins in TLR/IL-1R signalling: a role as novel RING E3-ubiquitin-ligases. FEBS Lett 580, 4697–4702 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Butler MP, Hanly JA & Moynagh PN Kinase-active interleukin-1 receptor-associated kinases promote polyubiquitination and degradation of the Pellino family: direct evidence for PELLINO proteins being ubiquitinprotein isopeptide ligases. J Biol Chem 282, 29729–29737 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Strelow A, Kollewe C & Wesche H Characterization of Pellino2, a substrate of IRAK1 and IRAK4. FEBS Lett 547, 157–161 (2003). [DOI] [PubMed] [Google Scholar]
- 60.Smith H et al. The role of TBK1 and IKKepsilon in the expression and activation of Pellino 1. Biochem J 434, 537–548 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chang M, Jin W & Sun SC Peli1 facilitates TRIF-dependent Toll-like receptor signaling and proinflammatory cytokine production. Nat Immunol 10, 1089–1095 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park HY et al. Pellino 1 promotes lymphomagenesis by deregulating BCL6 polyubiquitination. J Clin Invest 124, 4976–4988 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Choe JY et al. PELI1 expression is correlated with MYC and BCL6 expression and associated with poor prognosis in diffuse large B-cell lymphoma. Mod Pathol 29, 1313–1323 (2016). [DOI] [PubMed] [Google Scholar]
- 64.Callaghan MJ et al. Identification of a human HECT family protein with homology to the Drosophila tumor suppressor gene hyperplastic discs. Oncogene 17, 3479–3491 (1998). [DOI] [PubMed] [Google Scholar]
- 65.Matta-Camacho E, Kozlov G, Menade M & Gehring K Structure of the HECT C-lobe of the UBR5 E3 ubiquitin ligase. Acta Crystallogr Sect F Struct Biol Cryst Commun 68, 1158–1163 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kozlov G et al. Structural basis of ubiquitin recognition by the ubiquitin-associated (UBA) domain of the ubiquitin ligase EDD. J Biol Chem 282, 35787–35795 (2007). [DOI] [PubMed] [Google Scholar]
- 67.Kozlov G et al. Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase. EMBO J 23, 272–281 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maddika S & Chen J Protein kinase DYRK2 is a scaffold that facilitates assembly of an E3 ligase. Nat Cell Biol 11, 409–419 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tomaic V et al. Regulation of the human papillomavirus type 18 E6/E6AP ubiquitin ligase complex by the HECT domain-containing protein EDD. J Virol 85, 3120–3127 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hay-Koren A, Caspi M, Zilberberg A & Rosin-Arbesfeld R The EDD E3 ubiquitin ligase ubiquitinates and up-regulates beta-catenin. Mol Biol Cell 22, 399–411 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ong SS et al. Stability of the human pregnane X receptor is regulated by E3 ligase UBR5 and serine/threonine kinase DYRK2. Biochem J 459, 193–203 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gudjonsson T et al. TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell 150, 697–709 (2012). [DOI] [PubMed] [Google Scholar]
- 73.Cojocaru M et al. Transcription factor IIS cooperates with the E3 ligase UBR5 to ubiquitinate the CDK9 subunit of the positive transcription elongation factor B. J Biol Chem 286, 5012–5022 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jiang W et al. Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol Cell 43, 33–44 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yoshida M et al. Poly(A) binding protein (PABP) homeostasis is mediated by the stability of its inhibitor, Paip2. EMBO J 25, 1934–1944 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Honda Y et al. Cooperation of HECT-domain ubiquitin ligase hHYD and DNA topoisomerase II-binding protein for DNA damage response. J Biol Chem 277, 3599–3605 (2002). [DOI] [PubMed] [Google Scholar]
- 77.Smits VA EDD induces cell cycle arrest by increasing p53 levels. Cell Cycle 11, 715–720 (2012). [DOI] [PubMed] [Google Scholar]
- 78.Saunders DN et al. Edd, the murine hyperplastic disc gene, is essential for yolk sac vascularization and chorioallantoic fusion. Mol Cell Biol 24, 7225–7234 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Meissner B et al. The E3 ubiquitin ligase UBR5 is recurrently mutated in mantle cell lymphoma. Blood 121, 3161–3164 (2013). [DOI] [PubMed] [Google Scholar]
- 80.Saba N & Wiestner A Do mantle cell lymphomas have an 'Achilles heel'? Curr Opin Hematol 21, 350–357 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kishi N et al. Murine homologs of deltex define a novel gene family involved in vertebrate Notch signaling and neurogenesis. Int J Dev Neurosci 19, 21–35 (2001). [DOI] [PubMed] [Google Scholar]
- 82.Storck S et al. Normal immune system development in mice lacking the Deltex-1 RING finger domain. Mol Cell Biol 25, 1437–1445 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Matsuno K, Diederich RJ, Go MJ, Blaumueller CM & Artavanis-Tsakonas S Deltex acts as a positive regulator of Notch signaling through interactions with the Notch ankyrin repeats. Development 121, 2633–2644 (1995). [DOI] [PubMed] [Google Scholar]
- 84.Hori K et al. Drosophila deltex mediates suppressor of Hairless-independent and late-endosomal activation of Notch signaling. Development 131, 5527–5537 (2004). [DOI] [PubMed] [Google Scholar]
- 85.Lehar SM & Bevan MJ T cells develop normally in the absence of both Deltex1 and Deltex2. Mol Cell Biol 26, 7358–7371 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hsiao HW et al. Deltex1 is a target of the transcription factor NFAT that promotes T cell anergy. Immunity 31, 72–83 (2009). [DOI] [PubMed] [Google Scholar]
- 87.Liu WH & Lai MZ Deltex regulates T-cell activation by targeted degradation of active MEKK1. Mol Cell Biol 25, 1367–1378 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hsu TS, Hsiao HW, Wu PJ, Liu WH & Lai MZ Deltex1 promotes protein kinase Ctheta degradation and sustains Casitas B-lineage lymphoma expression. J Immunol 193, 1672–1680 (2014). [DOI] [PubMed] [Google Scholar]
- 89.Hsiao HW et al. Deltex1 antagonizes HIF-1alpha and sustains the stability of regulatory T cells in vivo. Nat Commun 6, 6353 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hsu TS, Mo ST, Hsu PN & Lai MZ c-FLIP is a target of the E3 ligase deltex1 in gastric cancer. Cell Death Dis 9, 135 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zurli V et al. Ectopic ILT3 controls BCR-dependent activation of Akt in B-cell chronic lymphocytic leukemia. Blood 130, 2006–2017 (2017). [DOI] [PubMed] [Google Scholar]
- 92.Meriranta L et al. Deltex-1 mutations predict poor survival in diffuse large B-cell lymphoma. Haematologica 102, e195–e198 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mahoney DJ et al. Both cIAP1 and cIAP2 regulate TNFalpha-mediated NF-kappaB activation. Proc Natl Acad Sci U S A 105, 11778–11783 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bertrand MJ et al. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell 30, 689–700 (2008). [DOI] [PubMed] [Google Scholar]
- 95.Yang Y et al. Targeting Non-proteolytic Protein Ubiquitination for the Treatment of Diffuse Large B Cell Lymphoma. Cancer Cell 29, 494–507 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Varfolomeev E et al. IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell 131, 669–681 (2007). [DOI] [PubMed] [Google Scholar]
- 97.Annunziata CM et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell 12, 115–130 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vince JE et al. IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell 131, 682–693 (2007). [DOI] [PubMed] [Google Scholar]
- 99.Heard KN, Bertrand MJ & Barker PA cIAP2 supports viability of mice lacking cIAP1 and XIAP. EMBO J 34, 2393–2395 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kavsak P et al. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol Cell 6, 1365–1375 (2000). [DOI] [PubMed] [Google Scholar]
- 101.Lin X, Liang M & Feng XH Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-beta signaling. J Biol Chem 275, 36818–36822 (2000). [DOI] [PubMed] [Google Scholar]
- 102.Blank M et al. A tumor suppressor function of Smurf2 associated with controlling chromatin landscape and genome stability through RNF20. Nat Med 18, 227–234 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kaneki H et al. Tumor necrosis factor promotes Runx2 degradation through up-regulation of Smurf1 and Smurf2 in osteoblasts. J Biol Chem 281, 4326–4333 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kong Y, Cui H & Zhang H Smurf2-mediated ubiquitination and degradation of Id1 regulates p16 expression during senescence. Aging Cell 10, 1038–1046 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fukunaga E et al. Smurf2 induces ubiquitin-dependent degradation of Smurf1 to prevent migration of breast cancer cells. J Biol Chem 283, 35660–35667 (2008). [DOI] [PubMed] [Google Scholar]
- 106.Zhang H & Cohen SN Smurf2 up-regulation activates telomere-dependent senescence. Genes Dev 18, 3028–3040 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ramkumar C et al. Smurf2 regulates the senescence response and suppresses tumorigenesis in mice. Cancer Res 72, 2714–2719 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ramkumar C et al. Smurf2 suppresses B-cell proliferation and lymphomagenesis by mediating ubiquitination and degradation of YY1. Nat Commun 4, 2598 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Abbas T et al. CRL1-FBXO11 promotes Cdt2 ubiquitylation and degradation and regulates Pr-Set7/Set8-mediated cellular migration. Mol Cell 49, 1147–1158 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rossi M et al. Regulation of the CRL4(Cdt2) ubiquitin ligase and cellcycle exit by the SCF(Fbxo11) ubiquitin ligase. Mol Cell 49, 1159–1166 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Horn M et al. DRE-1/FBXO11-dependent degradation of BLMP-1/BLIMP-1 governs C. elegans developmental timing and maturation. Dev Cell 28, 697–710 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zheng H et al. PKD1 phosphorylation-dependent degradation of SNAIL by SCF-FBXO11 regulates epithelial-mesenchymal transition and metastasis. Cancer Cell 26, 358–373 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hardisty RE et al. The deaf mouse mutant Jeff (Jf) is a single gene model of otitis media. J Assoc Res Otolaryngol 4, 130–138 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hardisty-Hughes RE et al. A mutation in the F-box gene, Fbxo11, causes otitis media in the Jeff mouse. Hum Mol Genet 15, 3273–3279 (2006). [DOI] [PubMed] [Google Scholar]
- 115.Segade F et al. Association of the FBXO11 gene with chronic otitis media with effusion and recurrent otitis media: the Minnesota COME/ROM Family Study. Arch Otolaryngol Head Neck Surg 132, 729–733 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Duan S et al. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature 481, 90–93 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schneider C et al. FBXO11 inactivation leads to abnormal germinal-center formation and lymphoproliferative disease. Blood 128, 660–666 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chiorazzi M et al. Related F-box proteins control cell death in Caenorhabditis elegans and human lymphoma. Proc Natl Acad Sci U S A 110, 3943–3948 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Evankovich J et al. Receptor for advanced glycation end products is targeted by FBXO10 for ubiquitination and degradation. FASEB J 31, 3894–3903 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Korac P, Dotlic S, Matulic M, Zajc Petranovic M & Dominis M Role of MYC in B Cell Lymphomagenesis. Genes (Basel) 8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Leich E et al. Follicular lymphomas with and without translocation t(14;18) differ in gene expression profiles and genetic alterations. Blood 114, 826–834 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li Y et al. FBXO10 deficiency and BTK activation upregulate BCL2 expression in mantle cell lymphoma. Oncogene 35, 6223–6234 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fernandez-Saiz V et al. SCFFbxo9 and CK2 direct the cellular response to growth factor withdrawal via Tel2/Tti1 degradation and promote survival in multiple myeloma. Nat Cell Biol 15, 72–81 (2013). [DOI] [PubMed] [Google Scholar]
- 124.Busino L et al. Fbxw7alpha- and GSK3-mediated degradation of p100 is a pro-survival mechanism in multiple myeloma. Nat Cell Biol 14, 375–385 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Trembley JH, Wang G, Unger G, Slaton J & Ahmed K Protein kinase CK2 in health and disease: CK2: a key player in cancer biology. Cell Mol Life Sci 66, 1858–1867 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lee KW et al. F-box only protein 9 is an E3 ubiquitin ligase of PPARgamma. Exp Mol Med 48, e234 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tetzlaff MT et al. Defective cardiovascular development and elevated cyclin E and Notch proteins in mice lacking the Fbw7 F-box protein. Proc Natl Acad Sci U S A 101, 3338–3345 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Perry JM & Li L Self-renewal versus transformation: Fbxw7 deletion leads to stem cell activation and leukemogenesis. Genes Dev 22, 1107–1109 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Crusio KM, King B, Reavie LB & Aifantis I The ubiquitous nature of cancer: the role of the SCF(Fbw7) complex in development and transformation. Oncogene 29, 4865–4873 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhang W & Koepp DM Fbw7 isoform interaction contributes to cyclin E proteolysis. Mol Cancer Res 4, 935–943 (2006). [DOI] [PubMed] [Google Scholar]
- 131.King B et al. The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell 153, 1552–1566 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hoeck JD et al. Fbw7 controls neural stem cell differentiation and progenitor apoptosis via Notch and c-Jun. Nat Neurosci 13, 1365–1372 (2010). [DOI] [PubMed] [Google Scholar]
- 133.Teng CL et al. FBXW7 is involved in Aurora B degradation. Cell Cycle 11, 4059–4068 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Inuzuka H et al. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature 471, 104–109 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yao S et al. Fbw7 regulates apoptosis in activated B-cell like diffuse large B-cell lymphoma by targeting Stat3 for ubiquitylation and degradation. J Exp Clin Cancer Res 36, 10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wang Z, Liu P, Inuzuka H & Wei W Roles of F-box proteins in cancer. Nat Rev Cancer 14, 233–247 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bahram F, von der Lehr N, Cetinkaya C & Larsson LG c-Myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome-mediated turnover. Blood 95, 2104–2110 (2000). [PubMed] [Google Scholar]
- 138.Reavie L et al. Regulation of c-Myc ubiquitination controls chronic myelogenous leukemia initiation and progression. Cancer Cell 23, 362–375 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Stessman HA et al. Profiling bortezomib resistance identifies secondary therapies in a mouse myeloma model. Mol Cancer Ther 12, 1140–1150 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kouroukis TC et al. Bortezomib in multiple myeloma: systematic review and clinical considerations. Curr Oncol 21, e573–603 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zaal EA et al. Bortezomib resistance in multiple myeloma is associated with increased serine synthesis. Cancer Metab 5, 7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Fischer ES et al. Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide. Nature 512, 49–53 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lu J et al. Hijacking the E3 Ubiquitin Ligase Cereblon to Efficiently Target BRD4. Chem Biol 22, 755–763 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Winter GE et al. DRUG DEVELOPMENT. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 348, 1376–1381 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]