Abstract
Multiple genome-wide studies have identified associations between outcome of HIV infection and polymorphisms in/around the gene encoding the HIV co-receptor CCR5, but the functional basis for the strongest of these associations, rs1015164 A/G, is unknown. We found that rs1015164, located 34KB downstream of CCR5, marks variation in an ATF1 binding site that controls expression of the antisense long non-coding RNA CCR5AS. Knockdown or enhancement of CCR5AS expression resulted in a corresponding change in CCR5 expression on CD4+ T cells. CCR5AS interfered with interactions between the RNA-binding protein Raly and the CCR5 3’ untranslated region, protecting CCR5 mRNA from Raly-mediated degradation. Reduction in CCR5 expression through inhibition of CCR5AS diminished infection of CD4+ T cells with CCR5-tropic HIV in vitro. These data represent a rare determination of the functional importance of a genome-wide disease association where expression of a long non-coding RNA affects level of HIV infection and disease progression.
Introduction
The non-coding RNAs are classified either as small (<200 bp) or long (>200 bp) transcripts without protein coding potential. Long non-coding RNA (lncRNA) genes are numerous, exceeding the number of protein-coding genes, and are emerging as epigenetic regulators with a prominent role in health and disease. LncRNAs regulate gene expression, chromatin organization, cellular trafficking, RNA decay, and translation. Notably, the localization, function, decay, and turnover of proteins are also influenced by lncRNAs (reviewed in1). In several viral infections, cellular lncRNAs have been shown to regulate viral replication2, or cellular metabolic3 and immune responses4.
A large number of intergenic single nucleotide polymorphisms (SNPs) associate with human disease outcomes (GWAS catalog; http://www.ebi.ac.uk/gwas), many of which are in or near lncRNA genes and may mark differential expression or function of the lncRNAs5. Recently, the influence of some of these SNPs on expression or function of lncRNA genes, their protein coding targets, and/or disease outcomes has been demonstrated in cancer6, the chronic immune-mediated disorder celiac disease7, and pneumococcal bacteremia8, but any potential role of lncRNA polymorphisms in viral infections remains uncharacterized.
Human chromosome 3p21–22 harbors a cluster of chemokine receptor genes, several of which can serve as co-receptors for HIV-19. CCR5 is the major co-receptor of HIV and variation in its expression influences the outcome of HIV-1 infection. Complete loss of CCR5 surface expression due to homozygosity for a 32-base pair (bp) deletion in CCR5 (CCR5-Δ32) results in resistance to HIV infection10, 11. There is a decrease in CCR5 surface expression on T cells among CCR5-Δ32 heterozygous subjects compared to those carrying two functional copies of the gene12, 13. Additional SNPs and haplotypes have been shown to associate with CCR5 transcription, surface expression levels, and HIV-1 susceptibility14, 15, 16, 17, 18. CCR5 surface expression levels influence multiple aspects of HIV pathogenesis and disease outcomes, such as HIV entry12, 13, viral load19, progression to AIDS19, neutralizing activity of HIV-1-specific antibodies20, immune reconstitution during highly active antiretroviral therapy19, 21, and the treatment efficacy of CCR5 blockers and entry inhibitors22, where in each instance, low CCR5 surface expression is protective. Genetic associations of CCR5 and CCR2 gene polymorphisms with HIV-1 pathogenesis are well established16, 23, 24, 25, including an intergenic SNP (rs1015164 A/G) downstream of the CCRL2 gene, which showed genome-wide significant association with HIV infection outcomes in meta-analyses that collectively examined genotyping data from 6,315 HIV-1-infected patients26. The rs1015164 SNP was found to have a genome-wide effect independent of other SNPs in the region, including CCR5-Δ32, after correction for ethnicity, gender and cohort (p = 1.5×10−19). Here we show that this SNP is in close genomic proximity to RP-11–24F11.2, an anti-sense non-coding RNA gene that overlaps with CCR5, and marks expression of the transcript encoded by the gene. We termed this novel transcript CCR5AS. Our data reveal that higher expression levels of CCR5AS, as a consequence of a variant in an ATF1 transcription factor binding site that rs1015164A marks, enhance CCR5 mRNA stability thereby increasing CCR5 mRNA and cell surface expression, and providing the functional basis for the association between rs1015164A and lack of HIV control.
Results
The rs1015164 SNP associates with HIV outcome after infection
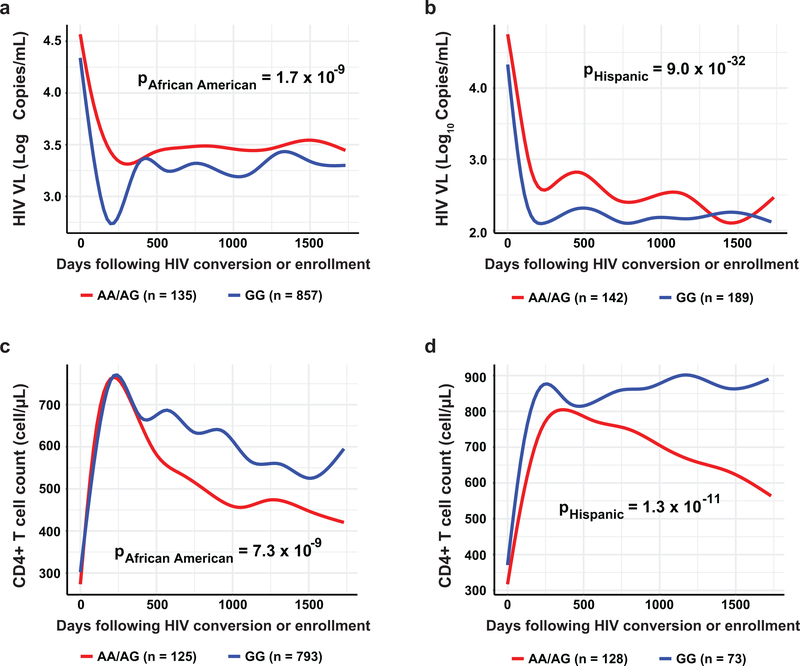
The CCR5Δ32 mutation is present almost exclusively in people of European descent and confers nearly complete protection from HIV infection in homozygotes and slower progression to AIDS in heterozygotes for the mutation10, 11. Other CCR5 variants that associate with outcome to HIV infection, including rs1015164, however, are present across many populations, and some of these affect CCR5 expression13, 27, 28, 29. Among 2,745 quantitative trait loci in a monocyte transcriptome-wide scan, rs1015164 was identified as a marker of CCR5 mRNA expression30. We tested for an effect of rs1015164 on viral load after HIV-1 infection in three ethnic groups: African Americans, Hispanics, and Japanese. Although the rs1015164A allele was less frequent in the African American and Hispanic cohorts, and homozygous individuals were rare, patients carrying at least one rs1015164A allele (AA+AG) had significantly higher viral load (increase of 0.24 log10 copies/ml for AA/AG, PAfrican American = 1.7×10−9; increase of 0.58 log10 copies/ml for AA/AG, PHispanic = 9.0×10−32; Fig. 1a, b) and decreased CD4+ T cell counts (−67.1 cells/μl for AA/AG, PAfrican American = 7.3×10−9; −121.7 cells/μl for AA/AG, PHispanic = 1.3×10−11; Fig. 1c, d) over time. These results extend the effect of rs1015164 beyond people of European descent as reported previously26, to Hispanics and African Americans. The rs1015164A allele was also less frequent in a sampling of Japanese patients compared to Europeans (Supplementary Fig. 1a); nevertheless, the AA genotype was significantly associated with higher viral loads in these patients (Supplementary Fig. 1b), pointing to a uniform deleterious effect of rs1015164A in HIV-1 infection across distinct populations. The consistency of the rs1015164 effect across the populations tested speak to a single functional mechanism explaining these associations.
Fig. 1. rs1015164 A/G variation associates with HIV-1 viral load and CD4 T-cell counts across distinct populations.
HIV-infected subjects followed prospectively were grouped according to rs1015164 genotype (GG and GA+AA). VL and CD4+ T cell counts are plotted against time following seroconversion or date of enrollment (censored at ~ 5 years). HIV longitudinal viral load is shown for a, African American (n = 992, AA+AG = 135, GG = 857); and b, Hispanics (n = 331, AA+AG = 142, GG = 189). Longitudinal CD4+ T cell counts are shown for c, African American (n = 918, AA+AG = 125, GG = 793); and d, Hispanics (n = 301, AA+AG = 128, GG = 173). The lines are best fit (LOWESS lines) to unadjusted VL or CD4 counts. Analysis of the log10 transformed HIV VL and CD4+ T-cell count at each timepoint was performed using the lmer function in R. We allowed for random effects due to the time post enrolment. Likelihood ratio p-values using the ANOVA function in R were calculated to compare nested models fit under a maximum-likelihood scenario.
rs1015164 marks expression of a novel lncRNA, CCR5AS
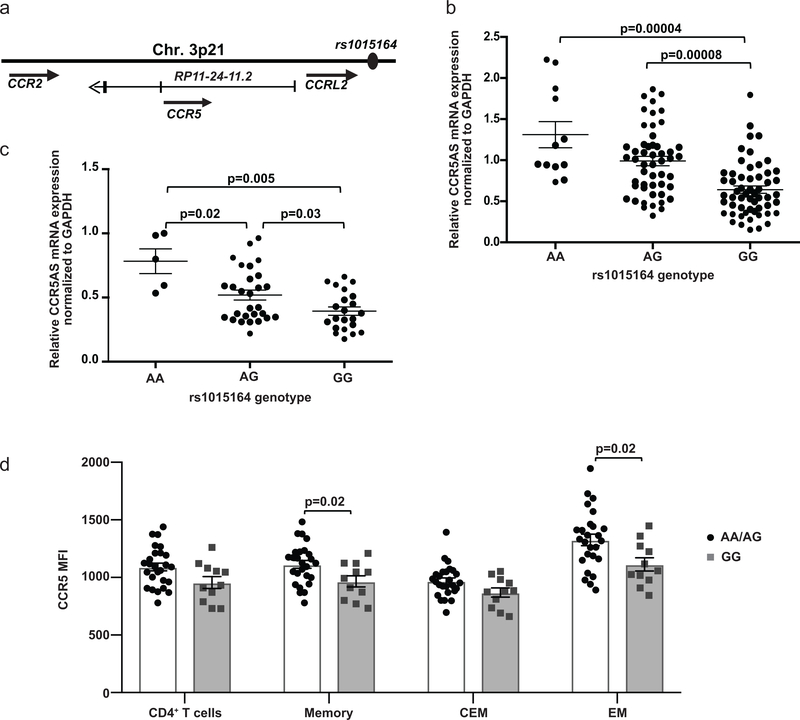
The rs1015164 SNP maps to the 5’ upstream region of a non-coding RNA gene RP-11–24F11.2 (Fig. 2a), so we tested whether this gene was transcribed. We detected and quantified a lncRNA transcript in total RNA from peripheral blood lymphocytes (PBLs) by qPCR using RP-11–24F11.2-specific primers as confirmed by sequencing of the amplicons. Because the non-coding RNA is transcribed from the anti-sense strand of the CCR5 gene, the lncRNA transcript was termed CCR5AS. The rs1015164A allele, which we found (Fig. 1) and was previously found to associate with higher viral loads26, associated with higher expression levels of CCR5AS in PBLs (Fig. 2b). As the primary cellular targets for HIV are CD4+ T cells, we tested and observed an association of rs1015164 genotype with CCR5AS expression levels in this cell type specifically, as well (Fig. 2c). In addition, rs1015164 genotype showed a significant correlation with CCR5 cell surface expression in bulk memory CD4+ T cells (p = 0.02, Fig. 2d, Supplementary Fig. 2a), and effector memory CD4+ T cells (p = 0.02, Fig. 2d, Supplementary Fig. 2a). The rs1015164 SNP also marks expression of CCR5AS (ENSG00000223552.1) in whole blood, colon, visceral adipose, heart, brain, subcutaneous adipose and lung tissue (https://www.gtexportal.org/; Supplementary Fig. 2b).
Fig. 2. rs1015164A/G variation marks differential expression of CCR5AS and CCR5.
a, Schematic representation of the RP11–24-11.2 gene that encodes CCR5AS and the position of rs1015164. b, Endogenous CCR5AS expression levels were estimated in total RNA from peripheral blood lymphocytes (PBL) (n = 119, rs1015164 AA=12, AG=51, GG= 56) by qPCR assay. The mean ±SE are depicted as horizontal and vertical bars for each group, respectively. Non-parametric Wilcoxon-Mann-Whitney tests were used for statistical comparisons and two tailed p values are indicated. c, CCR5AS levels in total RNA from CD4+ T cells were estimated from healthy donors (n=55; AA=5, AG=28, GG= 22). Statistics are as described in (b). d, PBMCs isolated from whole blood of donors without CCR5Δ32 (n=37; AA=10, AG=16, GG=11) were stained for CD3, CD4, CCR5, CCR7, CD45RA, and CD45RO, and the geometric mean fluorescence intensity (MFI) was measured for CCR5+ cells within bulk CD4+ T cells, bulk memory CD4+ T cells (CD45RO+CD45RA-), central effector memory (CEM; CCR7+) CD4+ T cells, and effector memory CD4+ T cells (EM; CCR7-). The mean ±SE are depicted as horizontal and vertical bars for each group, respectively. Statistical significance was determined by Mann-Whitney tests.
Amplification of the 3’ and 5’ ends of the CCR5AS transcript uncovered the presence of two different isoforms, which are identical at the 5’ end. The short isoform lacks exon 2 and has a truncated exon 4 (Supplementary Fig. 2c). Both isoforms are more abundant in the cytoplasmic, relative to nuclear, RNA fractions from CD4+ T cells and the Hut-78 human T cell lymphoma cell line, both of which express CCR5 and CCR5AS (Supplementary Fig. 2d). While the transcript did not contain any open reading frames longer than 20 codons, studies have suggested that several lncRNAs may in fact encode short peptides31. To test this possibility, we analyzed the CCR5AS transcript sequence using phyloCSF (phylogenetic codon substitution frequency)32, a conservation-based method that estimates whether a multispecies nucleotide sequence alignment in a specific locus is more likely to represent a protein coding than a non-coding transcript. The low phyloCSF score (−98.7) of CCR5AS is in line with that of other known non-coding RNAs than it is with that of known protein coding transcripts. No coding potential is predicted for the transcript by other methods such as RNAcode, coding potential assessment tool (CPAT), small ORF riboseq coding (http://fantom.gsc.riken.jp/cat/ )33.
CCR5AS enhances expression of the HIV co-receptor CCR5
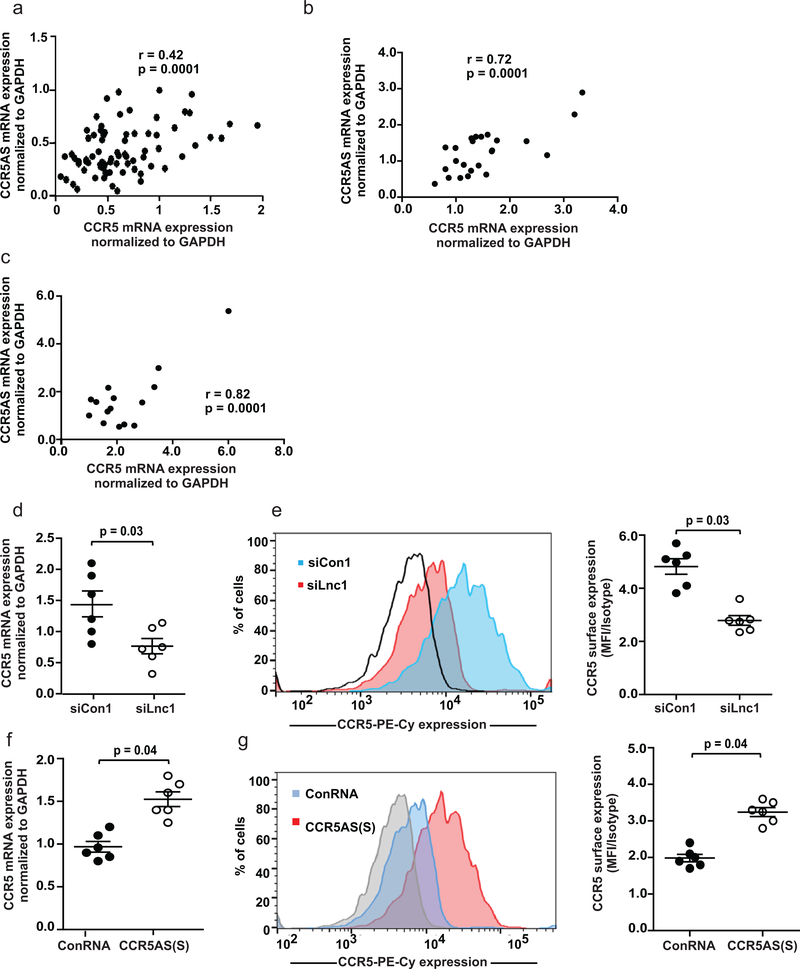
LncRNAs can affect the expression of nearby genes (reviewed in34). Given that the CCR5 gene is completely encompassed within the locus encoding CCR5AS (intron 2 specifically; Fig. 2a), we tested whether CCR5AS may be involved in differential expression of CCR5. Only a proportion of peripheral blood CD4+ T cells express CCR535. Nevertheless, CCR5AS expression levels in CD4+ T cells showed a significant, though moderate, positive correlation with levels of CCR5 mRNA expression (r=0.42, p=0.0001; Fig. 3a). Stronger correlations between CCR5AS and CCR5 mRNA levels were observed in peripheral memory CD4+ T cells and monocytes (r=0.72, p=0.0001 and r=0.82, p=0.0001, respectively; Fig. 3b, c), a larger proportion of which are known to express CCR535. Two distinct short interfering RNAs (siRNA) were used to silence CCR5AS expression (Supplementary Fig. 3a, b). The siRNA-mediated knockdown of CCR5AS resulted in reduced mRNA and surface expression of CCR5 (Fig. 3d, e; Supplementary Fig. 3c, d), whereas overexpression of the CCR5AS short form increased mRNA and surface expression of CCR5 in primary CD4+ T cells (Fig. 3f, g), as did overexpression of the CCR5AS long form (Supplementary Fig. 3e, f). Taken together, these data indicate that CCR5AS might function as a regulator of CCR5 expression levels. Since siRNA-mediated downregulation of CCR5AS also reduced mRNA and cell surface expression of CCR5 in the Hut-78 T cell line (Supplementary Fig. 4a, b), Hut-78 cells were used to further probe the molecular mechanism of CCR5AS-mediated regulation of CCR5.
Fig. 3. CCR5AS enhances CCR5 mRNA and cell surface expression.
The Pearson’s correlation coefficient was used to determine correlation between expression levels of CCR5AS and CCR5 mRNA levels in a, CD4+ T cells isolated from PBLs of normal donors (n = 55; r=0.42, p=0.0001); b, memory CD4+ T cells isolated from PBLs of normal donors (n = 24; r=0.72, p=0.0001); c, monocytes isolated from PBLs of normal donors (n = 16; r=0.82, p=0.0001). d, Peripheral blood CD4+ T cells from healthy donors (n = 6) were transfected with 300nm siRNA targeting CCR5AS (siLnc1) or a control siRNA (siCon1) and CCR5 mRNA was measured 24 hours post-transfection. The mean ±SE are depicted as horizontal and vertical bars for each group, respectively. Paired t test was used for statistical comparisons and two tailed p value is indicated. e, Cell surface expression of CCR5 was measured 24 hours after siRNA transfection (n = 6). A representative histogram for one of 6 donors tested is shown. Red and blue curves depict CCR5 cell surface expression of siLnc1 and siCon1 transfected cells, respectively. The open black curve depicts isotype control. Fold change in expression levels was calculated as the ratio of MFI of CCR5 vs isotype control. The mean ±SE (n = 6) are depicted as horizontal and vertical bars for each group, respectively. Paired t test was used for statistical comparisons and two tailed p value is indicated. f, CD4+ T cells from healthy donors (n = 6) were transfected with in vitro transcribed CCR5AS short form [CCR5AS(S)] or scrambled RNA control (ConRNA). CCR5 mRNA was measured 24 hours post-transfection. The mean ±SE are depicted as horizontal and vertical bars for each group, respectively. Statistics are as described in (e). g, Cell surface expression of CCR5 was measured 24 hours after CCR5AS lncRNA transfection. A histogram of one of 6 comparable experiments performed is shown. Red and blue curves depict CCR5 cell surface expression on CCR5AS(S) and ConRNA transfected cells, respectively. The gray curve depicts isotype controls. Fold change in expression levels was calculated as the ratio of MFI of CCR5 vs isotype control. The mean ±SE (n = 6) are depicted as horizontal and vertical bars for each group, respectively. Statistics are as described in (e).
Binding of the CCR5AS transcript to Raly protects CCR5 mRNA from degradation
Cytoplasmic lncRNAs have been shown to affect expression of their targets post-transcriptionally (reviewed in36). We assessed the effect of CCR5AS knockdown on cellular CCR5 mRNA by estimating the decay rates of metabolically labeled CCR5 mRNA in Hut-78 cells treated with control siRNA (siCon1) or CCR5AS-targeting siRNA (siLnc1). The decay of CCR5 mRNA was significantly faster in siLnc1 treated cells (Supplementary Fig. 4c), indicating that silencing of CCR5AS results in degradation of CCR5 mRNA.
Cytoplasmic lncRNAs can function as sponges of microRNAs (miRNAs) and rescue expression of the miRNA targets. CCR5AS and CCR5 transcripts share binding sites for two miRNA (miR-197 and miR-1224), but in vitro transcribed CCR5AS lncRNA with mutations in the binding sites of either or both of these miRNAs still enhanced CCR5 mRNA expression (Supplementary Fig. 5), indicating that CCR5AS-mediated regulation of CCR5 expression is independent of the shared miRNA binding sites.
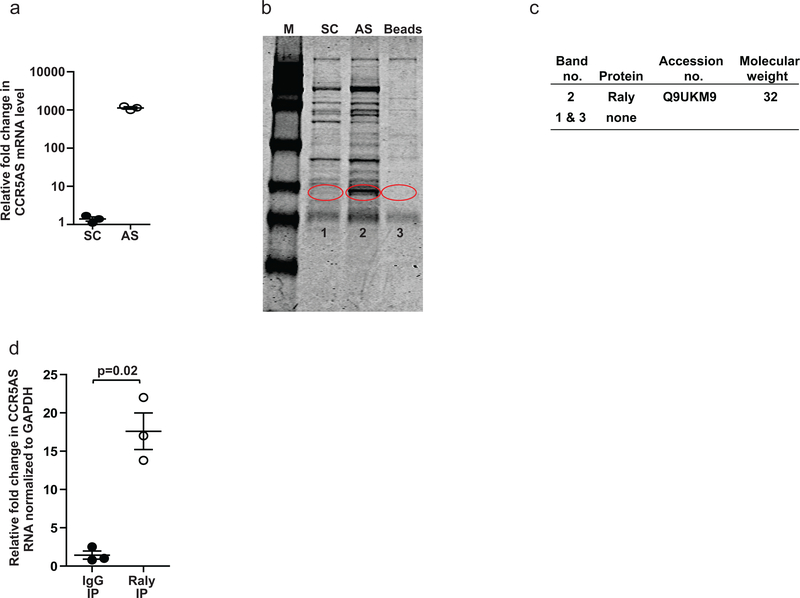
There are several examples of lncRNAs that alter gene expression via the recruitment or sequestration of proteins involved in post-transcriptional regulation of translation37 or mRNA stability38, 39, 40. In order to determine the protein binding partners of CCR5AS lncRNA, we carried out RNA antisense purification (RAP) using a CCR5AS-specific probe that enriched CCR5AS transcript 1000 fold (Fig. 4a). The proteins pulled down were separated by polyacrylamide gel electrophoresis (PAGE), and prominent bands with differential intensity were analyzed by mass-spectrometry (MS) to identify the proteins that bound specifically to CCR5AS (Fig. 4b). The Raly RNA binding protein was identified specifically in the CCR5AS-specific (AS) pulldown, but not the scrambled control (SC) pulldown (Fig. 4c), a finding that was further confirmed in an antisense pull-down using immunoblotting (Supplementary Fig. 6a). In silico analysis also showed strong prediction of Raly binding to both isoforms of CCR5AS (Supplementary Fig. 6b). To confirm the endogenous CCR5AS interaction with Raly, we performed RNA immunoprecipitation (RIP), in which a version of Raly protein tagged with Myc at the C-terminal end of the protein coding sequence was overexpressed. Myc-tagged proteins were immunoprecipitated using anti-Myc antibody-coated magnetic beads, and immunoprecipitation of Raly protein was confirmed by Western blot (Supplementary Fig. 6c). Protein-bound RNA was analyzed by qPCR, showing that endogenous CCR5AS was enriched significantly in the Raly Myc-pulldown (Raly-IP) as compared to control IgG coated beads (IgG-IP; Fig. 4d). These results indicate that CCR5AS RNA interacts with Raly.
Fig. 4. CCR5AS binds Raly.
a, RNA antisense pulldown of endogenous CCR5AS was carried out using a biotinylated antisense probe for specific pulldown of CCR5AS (AS) or the scrambled control sequence (SC). The mean ±SE (n = 6) are depicted as horizontal and vertical bars for each group, respectively (n=3), Student’s t test was used for statistical analysis and two tailed p value is indicated. b, Coomassie blue stained PAGE gel separating the RBPs that were pulled down with SC, AS, or Avidin beads in the absence of any labeled RNA (beads). A distinct protein band, which appeared only in the AS lane (Band 2) and proteins of corresponding molecular weight in the SC lane (Band 1) and the beads only lane (Band 3) were excised and analyzed by MS. The blot is derived from a single experiment. c, Raly was identified by LC-MS/MS in the RBPs pulled down using the AS probe (Band 2), and no RBPs were detected in Band 1 from the SC lane or Band 3 from the beads only control. d, A Myc-tagged Raly protein sequence was transfected into Hut-78 cells. The c-Myc tagged protein was immunoprecipitated using anti-Myc antibody coated magnetic beads. The protein bound RNA was analyzed by qPCR. Endogenous CCR5AS was enriched significantly in the Raly Myc-pulldown (Myc-IP) as compared to IgG coated beads (IgG-IP). The mean ±SE (n = 3) are depicted as horizontal and vertical bars for each group, respectively. Student’s t test was used for statistical comparison and two tailed p value is indicated.
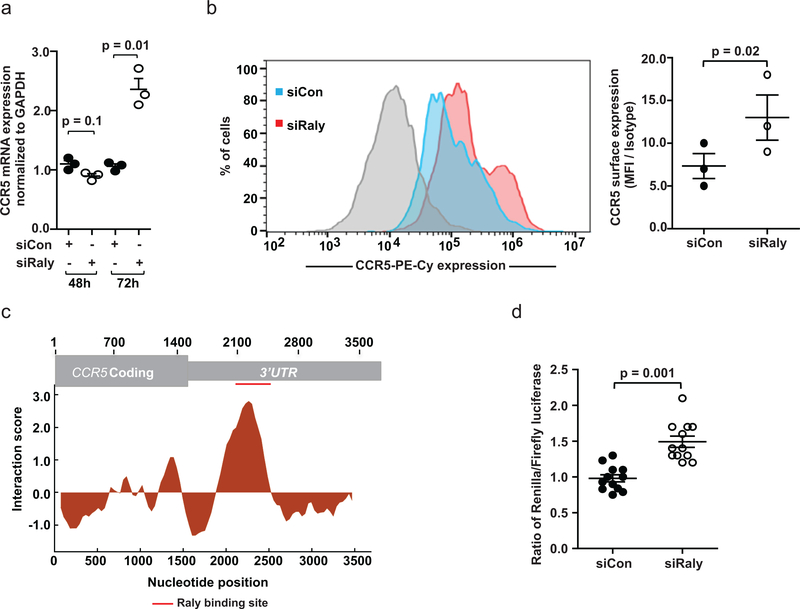
Given the interaction between Raly and CCR5AS, we tested for an effect of Raly on CCR5 expression level. Knockdown of Raly using a Raly-targeting siRNA (siRaly) was confirmed by immunoblot (Supplementary Fig. 6d). The suppressive effect of Raly on CCR5 expression was evident by enhanced mRNA expression and surface expression of CCR5 72 hours after siRNA mediated knockdown of Raly protein (Fig. 5a,b), a finding that was concordant with in silico analysis that strongly predicted binding of Raly to CCR5 3’UTR (Fig. 5c). These findings were further supported by cloning the CCR5 3’UTR region downstream of Renilla luciferase in a dual luciferase vector psicheck2 (Supplementary Fig. 6e), transfecting the construct into Raly-inhibited cells, and testing the effect of Raly inhibition on luciferase reporter activity. The siRNA mediated knockdown of Raly protein increased the luciferase activity of the CCR5 3’UTR construct as compared to that in the control siRNA transfected cells (Fig. 5d). Thus, depletion of Raly enhances CCR5 expression.
Fig. 5. Raly regulates CCR5 expression.
a, The Hut-78 cell line was transfected with either control siRNA (siCon) or Raly siRNA (siRaly). Cells were harvested at 48h and 72h post-transfection. CCR5 mRNA expression was upregulated after 72h in the siRaly-treated cell lines. The mean ±SE (n = 3) are depicted as horizontal and vertical bars for each group, respectively. Non-parametric t test was used for statistical comparisons and a two tailed p value is indicated. b, CCR5 surface expression was upregulated after 72h in the siRaly treated cells (red curve) as compared to the siCon treated cells (blue curve). The gray curve indicates isotype control. The histogram is representative of one of the three experiments. The mean ±SE (n=3) are depicted as horizontal and vertical bars for each group, respectively. Paired t test was used for statistical comparisons and two tailed p value is indicated. c, In silico prediction of the interaction between the CCR5 transcript and the Raly protein was done using a freely available algorithm (http://service.tartaglialab.com/newsubmission/globalscore), which integrates properties of protein and RNA structures into overall binding propensity. The interaction score predicts global and local interactions between RBP and lncRNA. A high interaction score (≈3) predicts a strong interaction between Raly protein and CCR5 transcript in the 3’UTR. d, CCR5 3’UTR was cloned downstream of Renilla luciferase into a dual luciferase reporter psicheck2 vector. Hut-78 cells were transfected with either the non-targeting siCon or siRaly. Seventy-two hours after siRNA transfection, the CCR5 3’UTR luciferase constructs were transfected into siCon or siRaly transfected Hut-78 cells. Twenty four hours after transfection of the luciferase constructs, the cells were lysed and luciferase activity was measured. Renilla and firefly luciferase activity were estimated by dual luciferase assays and presented as the normalized ratio of Renilla vs Firefly luciferase activity. The data represent six replicates in each group from two independent experiments. The mean ±SE are depicted as horizontal and vertical bars for each group, respectively. Non-parametric Wilcoxon-Mann-Whitney tests were used for statistical comparisons and two tailed p value is indicated.
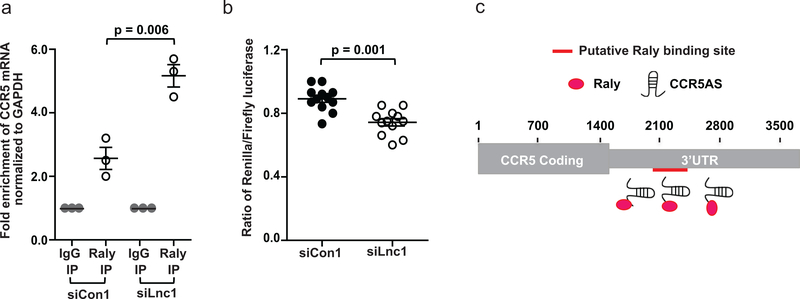
We hypothesized that CCR5AS may enhance stability of CCR5 mRNA by sequestering Raly, thereby keeping it from binding to the CCR5 3’UTR and degrading CCR5 transcripts. We tested directly whether the interaction between CCR5AS and Raly interferes with binding of Raly to the CCR5 3’UTR using Hut-78 cells transfected with Myc-tagged Raly plus either siLnc1 (to knockdown CCR5AS mRNA) or the control siCon1. Pulldown of Myc-tagged Raly protein from cellular lysates (input) by immunoprecipitation with anti-Myc antibody (Raly-IP), but not control IgG (IgG-IP), was confirmed by Western blot (Supplementary Fig. 6f). Changes in the association between Raly and CCR5 3’UTR with or without silencing of CCR5AS were then measured by RIP. Enrichment of CCR5AS and CCR5 3’UTR in the Raly-bound RNA was estimated by qPCR. CCR5AS was found to be highly enriched in Raly pulldown from the siCon1 treated cells, but absent in Raly pulldown from the siLnc1 transfected cells in which CCR5AS was silenced (Supplementary Fig. 6g). As predicted by in silico analysis (Fig. 5c), CCR5 3’UTR was also found to be enriched in the Raly pulldown in siCon1 treated cells; however, association between the CCR5 3’UTR and Raly was significantly enhanced upon CCR5AS knockdown (Fig. 6a). Further, transfection of Hut-78 cells with vector containing the CCR5 3’UTR region downstream of Renilla luciferase (Supplementary Fig. 6e), followed by CCR5AS knockdown, resulted in diminished luciferase activity of the CCR5 3’UTR construct as compared to the control cells (Fig. 6b). We assessed the effect of Raly knockdown on cellular CCR5 mRNA stability by estimating the decay rates of metabolically labeled CCR5 mRNA in cells treated with control siRNA (siCon) or Raly-targeting siRNA (siRaly). The decay of CCR5 mRNA was significantly slower in siRaly treated cells (Supplementary Fig. 6h), indicating that silencing of Raly results in increased stability of CCR5 mRNA. Overall, the data indicate that CCR5AS interferes with binding of Raly to the CCR5 3’UTR (Fig. 6c), thereby enhancing CCR5 mRNA stability.
Fig. 6. CCR5AS inhibits binding of Raly to CCR5 3’UTR.
a, Hut-78 cells were transfected with a vector encoding Myc-tagged Raly protein along with either Control siRNA (siCon1) or LncRNA siRNA (siLnc1). Cells were lysed and RNA immunoprecipitation was carried out using anti-Myc antibody or control IgG. Fold enrichment of CCR5 3’UTR in the pulldown, as determined using qPCR, was higher in the absence of CCR5AS (siLnc1 treated cells) as compared to the control (siCon1). Mean ± SE (n=3) are presented as vertical columns with error bars. Statistical significance was determined using non-parametric t test and two tailed p values are indicated between the comparison groups. b, Hut-78 cells were transfected with either a non-targeting siCon1 or siLnc1, along with the CCR5 3’UTR luciferase construct. Twenty-four hours after transfection, the cells were lysed and luciferase activity was measured. Silencing of CCR5AS reduces the expression of the CCR5 3’UTR luciferase construct. Renilla and firefly luciferase activity were estimated by dual luciferase assays and presented as the normalized ratio of Renilla vs Firefly luciferase activity. The data represent six replicates in each group from two independent experiments. The mean ±SE are depicted as horizontal and vertical bars for each group, respectively. Non-parametric Wilcoxon-Mann-Whitney tests were used for statistical comparisons and two tailed p values are indicated. c, Schematic representation of a model proposing CCR5AS interference with binding of Raly to CCR5 3’UTR.
Knockdown of CCR5AS reduces the susceptibility of CD4+ T cells to HIV-1 infection
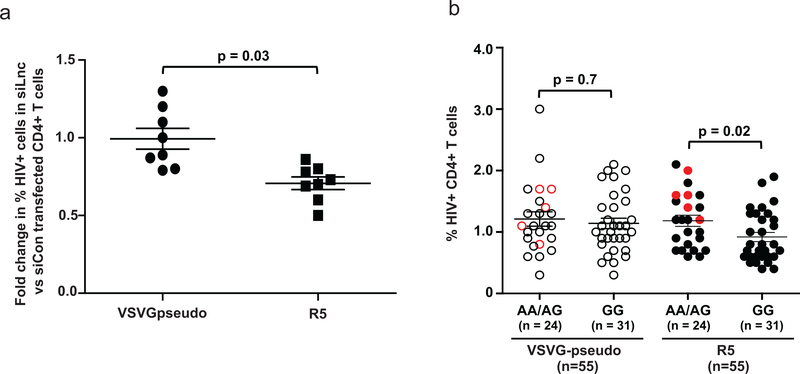
The potential impact of CCR5AS-mediated regulation of CCR5 expression levels on HIV infection of CD4+ T cells was tested using an in vitro infection assay in which purified CD4+ T cells were transfected with siRNA to downregulate CCR5AS (siLnc1) or with control siRNA (siCon1). The CCR5AS siRNA transfected cells were infected with R5-tropic (CCR5 dependent) or VSV-G pseudotyped (CCR5 independent) HIV-1 vectors encoding a green fluorescent protein (GFP). The proportions of GFP+ cells in the siLnc1 treated cells were compared to that of the siCon1 treated cells. The siRNA-mediated silencing of CCR5AS led to 25–50% reduction in the proportion of GFP+ T cells in the cultures infected with the R5-tropic virus, whereas downregulation of CCR5AS did not affect the proportion of GFP+ cells in the cultures infected with the CCR5-independent VSV-G pseudotyped vector (Fig. 7a; Supplementary Fig. 7a-f). Similarly, primary CD4+ T cells from rs1015164AA/AG donors showed a significant increase in infection with R5-tropic virus as compared to CD4+ T cells from donors with the rs1015164GG genotype (p=0.02; Fig. 7b; Supplementary Fig. 7b-d), but there was no difference in infection with the VSV-G pseudotyped vector in donors with these rs1015164 genotypes. These data indicate that silencing of CCR5AS reduces infection of T cells with a CCR5-dependent virus.
Fig. 7. CCR5AS expression levels associate with susceptibility to CCR5-dependent HIV infection.
a, Peripheral blood CD4+ T cells from 8 unrelated HIV negative donors, all with the rs1015164AG genotype, were transfected with control siRNA (siCon1) or CCR5AS siRNA (siLnc1). Twenty-four hours post transfection, the cells were infected with either VSV-G pseudotyped HIV virus encoding GFP or a CCR5-tropic HIV virus encoding GFP, and percent GFP expressing cells was measured after 96 hours. Fold change in % HIV+ cells was calculated as a ratio of HIV+ cells in siLnc1 transfected cells vs HIV+ cells in siCon1 transfected cells. The siRNA mediated silencing of CCR5AS led to 25–50% reduction in the proportion of GFP+ T cells in the cultures infected with the CCR5-tropic virus, but the downregulation of CCR5AS did not affect the proportion of GFP+ cells in the cultures infected with VSV-G pseudotyped vector. The mean ±SE are depicted as horizontal and vertical bars for each group. Statistical significance was determined using non-parametric t test and two tailed p value is indicated. Each dot represents one donor (n = 8 for each viral infection). b, Peripheral blood CD4+ T cells from 55 unrelated HIV negative donors grouped according to rs1015164 genotype (AA=5, AG=19, GG=31) were infected with either VSV-G pseudotyped HIV virus encoding GFP or a CCR5-tropic HIV virus encoding GFP, and percent GFP expressing cells was measured after 96 hours. The mean ±SE are depicted as horizontal and vertical bars for each group. Statistical significance was determined using a non-parametric t test and two tailed p value is indicated. Each dot represents one donor (n = 55 for each viral infection) and red dots represent donors with rs1015164AA genotype.
Our results suggest that the rs1015164A allele marks high CCR5AS expression. High CCR5AS enhances CCR5 expression post-transcriptionally by sequestering the RNA binding protein (RBP) Raly from the CCR5 3’UTR, and thereby protecting CCR5 mRNA from Raly-mediated decay, resulting in higher surface expression of CCR5, enhanced HIV infection, and higher HIV-1 viral loads (Supplementary Fig. 8).
rs2027820 is in perfect linkage disequilibrium with rs1015164, and influences the binding of activating transcription factor 1 (ATF1)
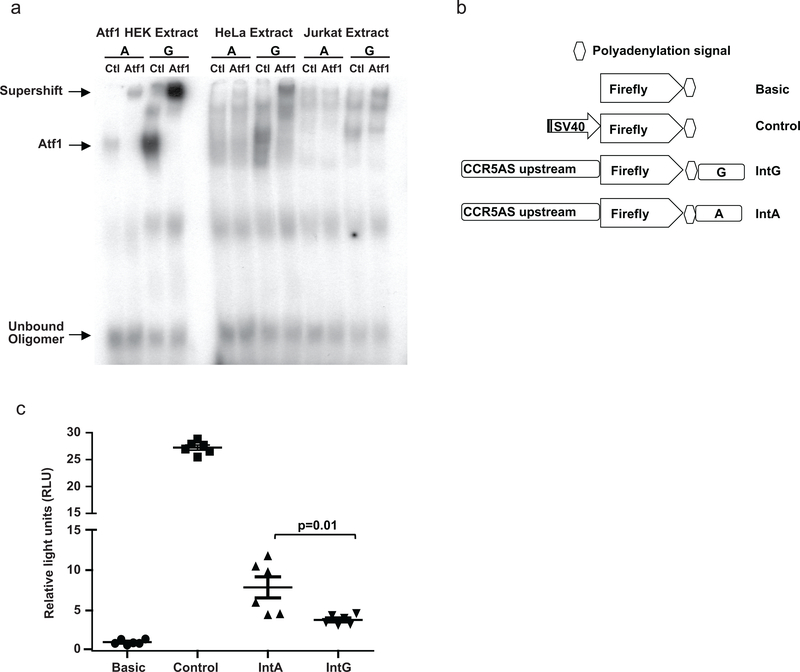
Bioinformatic analysis indicated that the rs1015164 SNP does not appear to be in a transcription factor binding site. There are, however, additional SNPs located upstream of CCR5AS (rs35937775) and in the intronic regions of CCR5AS (rs6441975, rs2027820, rs2373232) that are in perfect/near perfect linkage disequilibrium (LD) (r2=0.9–1) with rs1015164 in Europeans, Japanese and Africans (http://www.internationalgenome.org). Only one of these four SNPs, rs2027820, which is located in the first intron of CCR5AS, was predicted to lie within a transcription factor binding site and alter binding potential. The minor allele rs2027820G (in LD with rs1015164A, which associates with higher CCR5AS expression levels; r2=1 in Europeans and Africans, r2=0.94 in Japanese) maintains an intact binding site for ATF141, and is therefore predicted to enhance CCR5AS expression, whereas the major allele, rs2027820A (in LD with rs1015164G), disrupts the binding site. The presence of an ATF1 binding site within a sequence containing the rs2027820G variant was confirmed by electrophoretic mobility shift assay (EMSA) using rs2027820G vs. A oligonucleotides (Fig. 8a). As predicted, the A variant had greatly diminished binding compared to the G variant using Jurkat and HeLa nuclear extracts, as well as nuclear extracts from HEK 293 cells transfected with ATF1. We further tested the enhancer effect of the rs2027820 alleles using a luciferase reporter construct. A 1000 base pair fragment immediately upstream of the transcription start site of CCR5AS and a 500 base pair intronic fragment containing the rs2027820A/G polymorphic site were cloned upstream and downstream of the firefly luciferase gene, respectively (Fig. 8b). Relative luciferase activity of the construct containing rs2027820G (intact ATF1 binding site) was significantly higher as compared to the construct containing rs2027820A (disrupted ATF1 binding site) (Fig. 8c). These data support a model in which the CCR5AS intronic rs2027820 variant, which is in virtually perfect LD with rs1015164, regulates expression of CCR5AS through differential binding of ATF1.
Fig. 8. ATF1 binds rs2027820G more strongly than rs2027820A.
a, An electrophoretic mobility shift assay (EMSA) was performed with 32P-labelled oligonucleotides containing the core binding sequence for ATF1 containing the A or G variant of rs2027820. Nuclear extracts of recombinant human ATF1-transfected HEK 293, or untransfected HeLa or Jurkat cells were incubated with each of the oligonucleotides (indicated as A or G) in the presence of a control antibody (Ctl for ATF1-transfected HEK 293 extracts was specific for Sp1; Ctl for HeLa and Jurkat extracts was nonspecific IgG) or ATF1-specific antibody (ATF1). The presence of a shift (marked Atf1) indicates that the oligonucleotide binds a protein contained in the corresponding nuclear extract. A supershift is observed when an anti-ATF1 antibody is added, but not when Sp1 antibody or control IgG is added. A single experiment was performed. b, A fragment of 1000 base pairs immediately upstream of the transcription start site of CCR5AS, and a 500 base pair intronic fragment containing the rs2027820A/G polymorphic site were cloned upstream and downstream of the firefly luciferase gene, respectively, in a PGL3-basic vector. The firefly luciferase constructs with basic vector (no promoter), control vector (SV40 promoter), IntA (rs2027820A), and IntG (rs rs2027820G) were transfected along with Renilla luciferase encoding vector in a 293T derivative cell line (Lenti-X). c, Luciferase activity was measured 24 hours post-transfection and relative activity of Firefly luciferase normalized to Renilla was determined as relative light units (RLU). The IntG vector showed significantly higher luciferase activity as compared to the IntA vector. The data represent three replicates in each group from two independent experiments experiments. The mean ±SE are depicted as horizontal and vertical bars for each group, respectively. Statistical significance was determined using non-parametric t test and two tailed p value is indicated.
Discussion
The chemokine receptor gene cluster on human chromosome 3 encodes co-receptors for HIV-1, including the major co-receptor CCR5. Multiple independent candidate gene approaches and subsequent genome-wide studies have identified clear associations between polymorphisms within/near to CCR5, most prominently the CCR5Δ32 mutation, and outcomes of HIV infection. Reduction or absence of CCR5 expression due to heterozygosity or homozygosity for CCR5Δ32, respectively, also associates with HBV recovery42, whereas increased risk of fatal West Nile Virus infection associates with homozygosity for this mutation43, indicating the general importance of CCR5 expression levels in the immune response to viral infection. The work presented here demonstrates a novel mechanism of differential CCR5 regulation through the expression level of a lncRNA, CCR5AS. Our results reveal that a variant located upstream of CCR5AS, rs1015164A/G, associates with CCR5AS expression. High expression of CCR5AS (marked by rs1015164A) enhances CCR5 expression post-transcriptionally by sequestering the RBP Raly, inhibiting the binding of Raly to the CCR5 3’UTR, and thereby protecting CCR5 mRNA from Raly-mediated decay. Interestingly, the CCR5AS lncRNA sequence, as well as its genomic position with respect to the CCR5 gene, are conserved in non-human primates, raising the possibility that its regulatory function may also be conserved.
The rs1015164A/G variant has been shown to have a genome-wide significant effect on HIV infection outcomes in people of European ancestry, and its effect is independent of CCR5-Δ32 and other polymorphisms in the region26. We extend these studies showing a deleterious effect of rs1015164A in untreated HIV-1 infected African Americans, Hispanics and Japanese, in whom the CCR5-Δ32 mutation is virtually absent. Thus, differential CCR5AS expression may be the primary means for regulating CCR5 expression levels worldwide. Although CCR5 is not a receptor for HBV, CCR5-Δ32 has been shown to associate with spontaneous recovery of HBV42, apparently through immunological mechanisms44, 45. The rs1015164 genotype may also associate with outcome after HBV infection in a manner consistent with lower CCR5 expression (i.e. rs1015164G) conferring protection, as we observed a trend towards enhanced HBV recovery with increasing copy number of rs1015164G (p for trend = 0.04; data not shown). These data suggest a general importance of CCR5 expression levels and the mechanisms responsible for this differential regulation in diseases where CCR5 contributes to viral entry and/or the immune response.
The rs1015164 SNP is intergenic and is not predicted to be located in a transcription factor binding site; therefore, it is unlikely to be directly responsible for differential CCR5AS expression levels. However, a variant located in the first intron of CCR5AS, rs2027820, may directly affect expression levels of this gene, as it is located within a binding site for the transcription factor ATF1 and it is in virtually perfect LD with rs1015164. ATF1 belongs to the cAMP-regulated enhancer-binding (CREB) family of transcription factors and is expressed in a wide variety of cell types41. We show that rs2027820G, which is in LD with rs1015164A, allows binding of ATF1 and enhances protein expression in a luciferase assay relative to rs2027820A, indicating that the rs2027820G variant enhances CCR5AS expression levels, whereas the A variant confers poor binding of ATF1 and relatively poor expression of CCR5AS.
The majority of lncRNAs are in low abundance and localize to the nucleus, where they participate in transcriptional regulation through diverse mechanisms (reviewed in46). However, some localize to the cytoplasm, which we have observed for CCR5AS. While cytoplasmic lncRNAs can modulate mRNA stability or translation (reviewed in36, 47) by directly binding to their mRNA targets, others harbor miRNA binding sites that are in common with those present in the protein coding mRNA targets of the miRNAs. This competitive sequestration or “sponging” of the miRNAs protects the protein coding mRNA targets from miRNA-mediated repression. Similarly, lncRNAs can serve as ‘decoys’ of RBPs, sequestering RBPs from mRNA, and thereby regulating mRNA splicing, decay or translation. LncRNA mediated modulation of the molecular and spatial availability of RBPs has been shown to affect cell cycle, transformation, differentiation and the immune response. We showed that CCR5AS behaves as a decoy for the RBP Raly, preventing its binding to and repression of CCR5 such that the expression levels of these two RNA species are positively correlated.
Surveys of published GWAS show that most disease or trait-associated polymorphisms occur in non-coding regions48 with an enrichment in regulatory elements or expression quantitative trait loci (eQTL)49, suggesting that the basis for these disease associations are due in part to regulation of gene expression. Only 3% of the entire transcriptome is protein-coding50, signifying the vast predominance of non-coding RNAs in the human transcriptome. Thus, it is possible that a sizable portion of previously identified intergenic GWAS SNPs could be marking variation in function or expression of non-coding RNAs.
The functional explanations for the vast majority of disease-associated SNPs identified by GWAS remain unknown. Here we provide the underlying biological basis for the genome-wide association between the rs1015164A/G variant and outcome after HIV infection, establishing a foundation for potential therapeutic interventions.
Methods
Samples
Healthy European American (EA) donors recruited at the National Cancer Institute, Frederick, MD were used for determination of rs1015164 genotypes and the characterization of CCR5AS expression and transcript sequence. Data from a total of 1323 HIV-1-infected individuals (African American = 992, Hispanics = 331), were used in this study. The subjects in this study were enrolled into one of six cohorts: the AIDS linked to the Intravenous Experience (ALIVE) 51, the Massachusetts General Hospital controller cohort (MGHCC) (http://www.hivcontrollers.org/hivcontrollers), the Study on the Consequences of Protease Inhibitor Era (SCOPE) 52, the AIDS Clinical Trial Group cohort (ACTG) (https://actgnetwork.org), the U.S. military HIV Natural History Study (USMNHS) (www.idcrp.org), and the Multicenter AIDS Cohort Study (MACS). Five hundred and four chronically HIV-1 subtype B-infected, ART-naïve Japanese individuals (464 men and 40 women) were recruited in the National Center for Global Health and Medicine during 2000–201053. The respective institutional review boards approved the study, and all subjects gave written informed consent.
rs1015164 genotyping
DNA samples were amplified from genomic DNA by PCR using specific primers for the 200bp flanking rs1015164. The amplicons were sequenced in both directions with the same primers using an ABI-31730XL DNA analyzer (Applied Biosystems).
Statistical Analyses
Analysis of the log10 transformed HIV VL and CD4+ T-cell count at each timepoint was performed with the lmer function in R and presence of CCR5Δ32 was used as a covariate. We allowed for random effects due to the time post enrolment. Likelihood ratio p-values using the ANOVA function in R were calculated to compare nested models fit under a maximum-likelihood scenario. The p values for the effect of rs1015164 genotype on HIV-1 viral load in the Japanese cohort were generated by the Mann-Whitney U test. SAS 9.1 (SAS Institute) was used for data management and statistical analyses of the HBV cohort. PROC FREQ was used to compute frequencies on individual variables. Statistical significance refers to two-sided p values of <0.05. Odds ratios and p values were calculated by using conditional logistic regression with PROC LOGISTIC adjusting for HIV-1 status and ethnicity.
Rapid amplification of cDNA ends (RACE) and quantitative PCR (qPCR)
Peripheral blood was obtained from healthy donors and lymphocytes were separated using lymphocyte separation medium as per manufacturer’s instructions (Lonza). Total RNA was extracted from peripheral blood lymphocytes (PBLs; RNeasy Universal kit, Qiagen). Each sample was treated with gDNA eliminator to remove genomic DNA. The RNAs were quantified using the HT RNA Lab Chip (Caliper, Life Sciences), and all samples had an RNA quality score of >8. In order to determine the transcript boundaries, 3’RACE and 5’ RACE were performed using 3’ and 5’ Rapid amplification systems (Invitrogen) according to the manufacturer’s instructions. One microgram of RNA was used to initiate first strand cDNA synthesis at the poly(A) tail of the mRNA. The 3’ and 5’ ends of CCR5AS were amplified using gene specific primers (GSP) and a universal primer (UAP) (sequences provided in the list of primers). Amplicons were electrophoresed on 2% agarose gels to determine their length, and then they were cloned and sequenced to confirm the specificity of amplification.
The mRNA expression level was measured by qPCR. Briefly, reverse transcription was performed with 900ng of total RNA using the high capacity RNA to cDNA kit (Applied Bioscience) in a volume of 10μl. CCR5 coding region, CCR5 3’UTR, CCR5AS and GAPDH transcripts were amplified by SYBR green qPCR using the threshold cycle (CT) method54 in either a Viia7 machine (Applied Bioscience) or a Quantstudio 5 machine (Applied Biosystems). Each qPCR reaction included 6μl of power SYBR green PCR mastermix (Applied Biosystems), 200nM primers that specifically amplified the gene of interest or the housekeeping gene (GAPDH), and 2 μl of cDNA (1:20 dilution) in a total volume of 12 μl. The genes were amplified using the following conditions: 50°C for 2 minutes, 95°C for 2 minutes followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. The specificity of primers was tested by melt curve analysis using a dissociation step following the qPCR protocol and confirmed by sequencing the amplicons. The primers were found to amplify the targeted transcripts specifically and did not cross react with any other locus. The average expression levels of the genes were normalized to that of GAPDH RNA using the 2-ΔΔCt method54. Oligonucleotide primer sequences are provided in the list of primers and probes.
Construction of CCR5 3’UTR and CCR5AS luciferase reporters
The complete 3’UTR fragment of the CCR5 gene was amplified from genomic DNA and inserted downstream of the Renilla luciferase gene in the psicheck2 vector.
A 1000 base pair fragment immediately upstream of the transcription start site of CCR5AS and the intronic fragment containing the rs2027820A/G polymorphic site were cloned upstream and downstream of the firefly luciferase gene, respectively, in a PGL3-basic vector (Promega). The vectors were termed intA (rs2027820A) and intG (rs2027820G) as per the allele at the polymorphic site. The sequences of the primers used for sequencing are provided in the list of primers and probes.
Cell culture, cell transfection and luciferase reporter assays
CCR5 3’UTR
The Hut-78 cell line was maintained in RPMI 1640 (Gibco) medium with 10% heat inactivated fetal bovine serum (FBS; Atlanta Biologicals). Hut-78 or primary CD4+ T cells from donors with rs1015164AG genotype were plated at a density of 0.5×106 cells/well in a 12 well plate and transfected with 100nM final concentration of either Control siRNA or gene-targeting (CCR5AS or Raly) siRNA optimized using TransIT-X2® (Mirus Bio LLC) protocol. The cells were incubated for 24 hours in a 37°C CO2 incubator before determining mRNA expression or the cell-surface expression of CCR5. Hut-78 cells were plated at a density of 0.5×106 cells/well and transfected with 1μg/well of the CCR5 3’UTR reporter constructs in the psichek2 vectors using the optimized TransIT-X2® (Mirus Bio LLC) protocol. Transfected Hut-78 cells were incubated for 24 hours at 37°C in a CO2 incubator. The cells were lysed, and Firefly and Renilla luciferase activity were measured using the Dual Luciferase Reporter Assay System (Promega). Renilla luciferase activity was normalized relative to Firefly luciferase activity for each transfection. All experiments were performed with six replicates in two independent experiments.
CCR5AS
100ng of PGL3 vectors IntA or IntG were transfected into a Lenti-X cell line (derivative of 293T cells; Clontech) along with 6ng of Renilla vector. The cells were incubated for 24 hours and Firefly and Renilla luciferase activity were measured as described above. Firefly luciferase activity was normalized relative to Renilla luciferase activity for each transfection. Two independent experiments were performed with 3 replicates each.
Antibodies and flow cytometry
Bulk PBMCs isolated from whole blood via Ficoll-Paque PLUS (GE Healthcare) were stained for expression of CCR5 with the following antibodies (Biolegend): CD3-FITC clone UCHT1, CD4-PerCp-Cy5.5 clone OKT4, CCR5-PE-Cy7 clone J418F1, CCR7-APC clone G043H7, CD45RA-BV605 clone HI100, and CD45RO-BV785 clone UCHL1. Flow cytometry was performed on a BD Fortessa flow cytometer (BD Biosciences) and data were analyzed with FlowJo version 10.1.
CCR5 expression on the surface of Hut-78 cell line or CD4+ T cells was analyzed by staining with a PE-Cy7 conjugated antibody (Biolegend, clone J418F1), and expression was measured using an LSRII flow cytometer (BD Biosciences) or an AccuriC6 flow cytometer (BD Biosciences). The histograms were plotted using FlowJo software version 10.
RNA antisense pull down (RAP), polyacrylamide gel electrophoresis (PAGE) and Western blot analyses
An RNA antisense pull down protocol was adapted from a previously published method55. Hut-78 cells were UV-crosslinked (at 254nm) to capture RNA bound proteins. The cross-linked cells were lysed in the presence of protease and RNAse inhibitors. 50bp long biotinylated antisense oligonucleotide probes tiled across the entire CCR5AS transcript or biotinylated scrambled probe were synthesized. Probes were mixed with the lysate and hybridized at 37°C for 2–3 hours. RNA-protein complexes were purified with streptavidin coated magnetic beads and the bead bound complexes were eluted. Enrichment of CCR5AS RNA was analyzed using qPCR. Proteins captured on the beads were separated with Tris-glycine 4–12% gradient protein gel (Invitrogen) electrophoresis and detected by Coomassie-blue stain. Western blot was carried out using anti-Raly antibody (Invitrogen) and an anti-rabitt horse radish peroxidase (HRP) conjugated secondary antibody.
Identification of proteins by nano LC-tandem Mass Spectrometry (nLC-MS/MS)
The gel bands were excised with gel cutting racked tips and digested with trypsin as described previously56. Tryptic digests of proteins extracted from the gels (“in-gel” digests), were separated with a reversed-phase column using a linear gradient. Eluted peptides were subjected to reverse-phase micro-capillary nLC-MS/MS analysis using an Eksigent HPLC system (Eksigent) directly interfaced with an Orbitrap LTQ XL mass spectrometer (Thermo Fisher). The eluted ions were analyzed by full precursor MS scans acquired with the FT Orbitrap analyzer operated at a resolving power of 30,000 (400–2,000 m/z). MS spectrum was followed by eight MS/MS spectra, where the eight most abundant multiply charged ions were selected for MS/MS sequencing. Raw data were analyzed with Proteome Discoverer 1.4 (Thermo Fisher; https://www.thermofisher.com/order/catalog/product/IQLAAEGABSFAKJMAUH) software and searched against the SwissProt database restricted to human entries by using the MASCOT http://www.matrixscience.com/help/seq_db_setup_sprot.html) search engine. The precursor-ion tolerance was 10 ppm and the fragment-ion tolerance was 0.8 Da. Enzymatic digestion was specified as trypsin, with up to 2 missed cleavages allowed.
RNA immuno-precipitation (RIP)
Hut-78 were transfected with a plasmid encoding Raly cDNA with a Myc-tag (Clone RC210723, Origene) along with either siCon1 or siLnc1. For RNA immunoprecipitation of ribonucleoprotein (RNP) complexes from whole-cell extracts, the transfected cells were lysed in 20 mM Tris-HCl at pH 7.5, 100 mM KCl, 5mM MgCl2 and 0.5% NP-40 for 10 min on ice and centrifuged at 10,000 RPM for 15 min at 4°C. The supernatants were incubated with magnetic beads coated either with IgG or anti-Myc antibodies overnight at 4°C. Afterwards, the beads were washed with TBS-T buffer, the complexes were incubated with 20 units of RNase-free DNase I (15 min at 37°C) and further incubated with 0.1% SDS/0.5 mg/ml Proteinase K (15min at 55°C) to remove DNA and proteins, respectively. The RNPs isolated from the RIP were further assessed by immunoblot blot using an anti-Myc antibody (Clone 9E10) and an anti-mouse HRP conjugated secondary antibody for detection of Myc tagged proteins and by qPCR for detection of CCR5AS and CCR5 3’UTR.
CCR5 mRNA decay
CCR5 mRNA RNA decay was determined by 5-ethnyluridine (EU) pulse-labeling of RNA using the Click-iT Nascent RNA Capture Kit (Invitrogen, Thermo Scientific). Hut-78 cells were transfected with siCon1 or siLnc1. Four hours after transfection, the cells were pulsed with EU (0.2 mM/1×106/ml). Eighteen hours post-pulsing, the cells were spun down, washed with plain RPMI-1640 medium, supplemented with fresh growth medium and harvested at one hour intervals for 4 hours. Total RNA was isolated from cells and quantitated. The EU-labeled RNA was biotinylated, precipitated, and captured using the streptavidin coated magnetic beads as per the manufacturer’s protocol. RNA captured on beads was used for cDNA synthesis and qPCR analysis (primer sequences provided in the list of primers and probes).
Hut-78 cells were transfected with siCon or siRaly. Sixty hours after transfection, the cells were pulsed with EU (0.2 mM/1×106/ml). Eighteen hours post-pulsing, the cells were spun down, washed with plain RPMI-1640 medium, supplemented with fresh growth medium, and harvested at one hour intervals for 4 hours. Total RNA was isolated from cells and quantitated. The EU-labeled RNA was biotinylated, precipitated, and captured using the streptavidin coated magnetic beads as per the manufacturer’s protocol. RNA captured on beads was used for cDNA synthesis and qPCR analysis.
HIV-1 constructs and virus production
A Δenv HIV-1 plasmid encoding GFP and GFP-encoding R5-tropic HIV-1 vector derived from the NL4–3 molecular clone 57,58,59 was provided by N. Manel and D. Littman (New York University, New York, New York, USA). The env gene was derived from BaL HIV-1 and it is encoded within a vector containing all of the HIV genes60. Viral particles were produced by transfecting 293T cells with the respective HIV-1 plasmids and, if applicable, with pCG-VSV-G, using TransIT-293 (Mirus) in OptiMEM per the manufacturer’s instructions. Supernatants containing infectious viral particles were harvested 48 hours after transfection, centrifuged, treated with DNase I (20 U/ml) at room temperature for 1 hour, and stored at –80°C.
In vitro infection assays
CD4+ T cells were negatively isolated from frozen peripheral blood lymphocytes with purity greater than 95% of all viable lymphocytes. The CD4+ T cells from healthy donors with rs1015164AG genotype were transfected with 300nM siRNAs using Mirus transfection reagent. Eight hours after transfection, the cells were resuspended in culture medium supplemented with 20% fetal calf serum, and human recombinant IL-2 (hrIL-2; 50U/ml). Twenty four hours after transfection, CCR5AS expression was assessed by qPCR and cells were infected with R5 HIV-1 at 0.01 multiplicity of infection (MOI) for 4 hours at 37°C or with VSV-G psuedotyped HIV-1 virus at a tissue culture infective dose (TCID50) of 5000 to maintain similar levels of infection between the VSV and R5 viruses as per the previously published protocol60. After 2 washes, cells were resuspended in growth medium and plated at 5 × 105 cells/well in a 24-well plate. At regular intervals, the cultures were fed by removing and replacing one-half of the culture supernatant with fresh growth medium supplemented with hrIL-2 and were analyzed after 96 hours by flow cytometric assessment of GFP expression.
CD4+ T cells were negatively isolated from frozen peripheral blood lymphocytes were resuspended in culture medium supplemented with 20% fetal calf serum, and human recombinant IL-2 (hrIL-2; 50U/ml) for 24 hours. CD4+ T cells from healthy donors with rs1015164 AA, AG, or GG genotype (N=55) were infected with R5 HIV-1 at 0.01 MOI or with VSV-G psuedotyped HIV-1 at TCID50 of 5000 for 4 hours at 37°C. After 2 washes, cells were resuspended in growth medium and plated at 5 × 105 cells/well in a 24-well plate. At regular intervals, the cultures were fed by removing and replacing one-half of the culture supernatant with fresh growth medium supplemented with hrIL-2 and were analyzed after 96 hours by flow cytometric assessment of GFP expression.
Electrophoretic Mobility Shift Assay
Nuclear extracts prepared from HeLa and Jurkat cell lines via the CellLyticNuCLEAR extraction kit (Sigma-Aldrich) were stored at −80°C until use. Lysate from HEK 293 transfected with recombinant human Atf1 (Creative BioMart) was used as a positive control. Double-stranded DNA oligonucleotide probes incorporating the predicted ATF-1 binding sequence (Fwd: 5’- GTGAAAGCTTGTTACGTAGGTAACCTTTTGT-3’; Rev: 5’- GGACAAAAGGTTACCTACGTAACAAGCTTT-3’) or the mutant sequence (Fwd: 5’- GTGAAAGCTTGTTACATAGGTAACCTTTTGT-3’; Rev: 5’- GGACAAAAGGTTACCTATGTAACAAGCTTT-3’) were synthesized and labelled using Klenow fragment of DNA polymerase I (Invitrogen) with [α−32P]deoxycytidine triphosphate (3000 Ci/mmol; PerkinElmer Life and Analytical Sciences). The core binding site for ATF1 is underlined and the rs2027820 variant is bolded. 32P-labeled double stranded oligonucleotides were purified using the mini Quick Spin Oligo Columns (Roche). The DNA-protein binding reactions were performed in a 20 μl volume containing 10 μg nuclear extract, 1 μg poly(dI-DC) (Sigma-Aldrich), 1 μl 32P-labelled oligonucleotide probe (10,000 cpm) and 1 μL either control nonspecific IgG antibody (mouse IgG, sc-2025, Santa Cruz Biotechnology) or Sp1-specific antibody (Clone E-3, Santa Cruz Biotechnology) or ATF1-specific antibody (clone C41–5.1, Santa Cruz Biotechnology) as described previously61. The reactions were then loaded on a 5% polyacrylamide gel and electrophoresed in 0.5x TBE for 2.5 hours at 120 V. The gel was visualized with autoradiography.
Bioinformatic Analysis of CCR5 and CCR5AS Interaction Sites with Raly protein
In silico prediction of the interaction between the CCR5AS transcript as well as CCR5 mRNA and the Raly protein was done using a freely available algorithm (http://service.tartaglialab.com/new_submission/globalscore), which integrates properties of protein and RNA structures into overall binding propensity. Global score predicts global and local interactions between RBP and lncRNA.
List of primers and probes
qPCR primers
CCR5ASFw TCCTGGTCCCCGTATTGAAT
CCR5ASRev AGGAAGGTATGTGGTGACCA
CCR5Fw GTCCCTTCTGGGCTCACTAT
CCR5Rev CCCTGTCAAGAGTTGACACATTGTA
CCR53’UTRFw GCCTGCCCAGTGCACACAAG
CCR53’UTRRev CCCCCACCCCCATTCAGTCT
GAPDHFw GGTGAAGGTCGGAGTCAACG
GAPDHRev GTTGAGGTCAATGAAGGGGTC
siRNA target sequences
siCon1 TAAGGCTATGAAGAGATACTC
siLnc1 CCCACCCGCUGAUUCAAUATT
siCon2 TCTCGTCGGCTCACGACCTGAAGTA
siLnc2 TCTAATGCGGCCTCTCACCAGTGTT
RACE amplification and sequencing primers
3’RACE GSP1 CTCACCAGTGTTCGCAGAAA
UAP GGCCACGCGTCGACTAGTACTTTTTTTTTTTTTTTT
5’RACE GSP1 TCCGGGGACTTCGGCACCAAATG
5’RACE GSP2 TTTCTGCGAACACTGGTGAGAGG
Biotinylated probes for RNA antisense pulldown
AS1 (BTN)TATTGAATCAGCGGGTGGGTTTCTGCGAACACTGGTGAGAGGCCGCATTA
AS2 (BTN)ACTGTCATGGCAGGTTCCTGGTCCCCGTATTGAATCAGCGGGTGGGTTTC
AS3 (BTN)AGTGAGATGCCTTCTGAATATGTGCCCACAAGAAGTTGTGTCTAAGTCTGGTTCTC
SC (BTN)CCTAAAGGTTAAGTCGCCCTCGCTCG TTTCATAGGGTCGAAGCACTGCAAGC
PCR amplification and sequencing for rs1015164A/G
F1 GTCCCACTTCCTTTCTGTGG
R1 GGCCTGGGTAGGTTTTTAGC
Cloning of CCR5AS upstream region and intron in luciferase vector
LNC11.2UPMLUIF GATAAACGCGTAAATGGGAGATGATTTTCCCC
LNC11.2UPXHOIF TTATCCTCGAGTCAGAGTGTCACATGCCACAAG
LNC11.2INTBAMHI GATAAGGATCCAATGACAGATTAGATAAAGAAAGTGTGG
LNC11.2INTSALI TTATCGTCGACGGGACAGATTAAATTATCATCAGTC
Reporting Summary
Further information on research design is available in the Life Sciences Reporting Summary linked to this areticle linked to this article.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgements
The project was supported by NIAID (AI120900; AI140956; S. Kulkarni), HU-CFAR (S. Kulkarni), Cowles fellowship (S. Singh), Institutional funds from the Texas Biomedical Research Institute and the Ragon Institute of MGH, MIT and Harvard. This project has been funded in whole or in part with federal funds from the Frederick National Laboratory, Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This Research was supported in part by the Intramural Research Program of the NIH, Frederick National Lab, and Center for Cancer Research. See extended acknowledgements in Supplementary Information for full details.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Kopp F & Mendell JT Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 172, 393–407 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qiu L et al. Long Non-coding RNAs: Regulators of Viral Infection and the Interferon Antiviral Response. Frontiers in microbiology 9, 1621 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang P, Xu J, Wang Y & Cao X An interferon-independent lncRNA promotes viral replication by modulating cellular metabolism. Science 358, 1051–1055 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Chen YG, Satpathy AT & Chang HY Gene regulation in the immune system by long noncoding RNAs. Nat. Immunol 18, 962–972 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar V et al. Human disease-associated genetic variation impacts large intergenic non-coding RNA expression. PLoS Genet 9, e1003201 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ling H et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 23, 1446–1461 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castellanos-Rubio A et al. A long noncoding RNA associated with susceptibility to celiac disease. Science 352, 91–95 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kenyan Bacteraemia Study G et al. Polymorphism in a lincRNA Associates with a Doubled Risk of Pneumococcal Bacteremia in Kenyan Children. Am. J. Hum. Genet 98, 1092–1100 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berger EA, Murphy PM & Farber JM Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol 17, 657–700 (1999). [DOI] [PubMed] [Google Scholar]
- 10.McLaren PJ & Carrington M The impact of host genetic variation on infection with HIV-1. Nat. Immunol 16, 577–583 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naranbhai V & Carrington M Host genetic variation and HIV disease: from mapping to mechanism. Immunogenetics 69, 489–498 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paxton WA et al. Reduced HIV-1 infectability of CD4+ lymphocytes from exposed-uninfected individuals: association with low expression of CCR5 and high production of beta-chemokines. Virology 244, 66–73 (1998). [DOI] [PubMed] [Google Scholar]
- 13.Wu L et al. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J. Exp. Med 185, 1681–1691 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez E et al. Race-specific HIV-1 disease-modifying effects associated with CCR5 haplotypes. Proc. Natl. Acad. Sci. U. S. A. 96, 12004–12009 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawamura T et al. R5 HIV productively infects Langerhans cells, and infection levels are regulated by compound CCR5 polymorphisms. Proc. Natl. Acad. Sci. U. S. A 100, 8401–8406 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin MP et al. Genetic acceleration of AIDS progression by a promoter variant of CCR5. Science 282, 1907–1911 (1998). [DOI] [PubMed] [Google Scholar]
- 17.Mummidi S et al. Evolution of human and non-human primate CC chemokine receptor 5 gene and mRNA. Potential roles for haplotype and mRNA diversity, differential haplotype-specific transcriptional activity, and altered transcription factor binding to polymorphic nucleotides in the pathogenesis of HIV-1 and simian immunodeficiency virus. J. Biol. Chem 275, 18946–18961 (2000). [DOI] [PubMed] [Google Scholar]
- 18.Thomas SM et al. CCR5 expression and duration of high risk sexual activity among HIV-seronegative men who have sex with men. AIDS 20, 1879–1883 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gervaix A et al. Response to treatment and disease progression linked to CD4+ T cell surface CC chemokine receptor 5 density in human immunodeficiency virus type 1 vertical infection. J. Infect. Dis 185, 1055–1061 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Choudhry V et al. Increased efficacy of HIV-1 neutralization by antibodies at low CCR5 surface concentration. Biochem. Biophys. Res. Commun 348, 1107–1115 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reynes J, Baillat V, Portales P, Clot J & Corbeau P Relationship between CCR5 density and viral load after discontinuation of antiretroviral therapy. JAMA 291, 46 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Heredia A et al. CCR5 density levels on primary CD4 T cells impact the replication and Enfuvirtide susceptibility of R5 HIV-1. AIDS 21, 1317–1322 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Dean M et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science 273, 1856–1862 (1996). [DOI] [PubMed] [Google Scholar]
- 24.Mummidi S et al. Genealogy of the CCR5 locus and chemokine system gene variants associated with altered rates of HIV-1 disease progression. Nat. Med 4, 786–793 (1998). [DOI] [PubMed] [Google Scholar]
- 25.Smith MW et al. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC), ALIVE Study. Science 277, 959–965 (1997). [DOI] [PubMed] [Google Scholar]
- 26.McLaren PJ et al. Polymorphisms of large effect explain the majority of the host genetic contribution to variation of HIV-1 virus load. Proceedings of the National Academy of Sciences of the United States of America (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sciaranghella G et al. CCR5 Expression Levels in HIV-Uninfected Women Receiving Hormonal Contraception. J. Infect. Dis 212, 1397–1401 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joshi A et al. CCR5 promoter activity correlates with HIV disease progression by regulating CCR5 cell surface expression and CD4 T cell apoptosis. Sci Rep 7, 232 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Picton AC, Shalekoff S, Paximadis M & Tiemessen CT Marked differences in CCR5 expression and activation levels in two South African populations. Immunology 136, 397–407 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeller T et al. Genetics and beyond--the transcriptome of human monocytes and disease susceptibility. PLoS ONE 5, e10693 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ingolia NT, Lareau LF & Weissman JS Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin MF, Jungreis I & Kellis M PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 27, i275–282 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hon CC et al. An atlas of human long non-coding RNAs with accurate 5’ ends. Nature 543, 199–204 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guil S & Esteller M Cis-acting noncoding RNAs: friends and foes. Nat Struct Mol Biol 19, 1068–1075 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Lee B, Sharron M, Montaner LJ, Weissman D & Doms RW Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc. Natl. Acad. Sci. U. S. A 96, 5215–5220 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rashid F, Shah A & Shan G Long Non-coding RNAs in the Cytoplasm. Genomics Proteomics Bioinformatics 14, 73–80 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoon JH et al. LincRNA-p21 suppresses target mRNA translation. Mol. Cell 47, 648–655 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gong C & Maquat LE lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3’ UTRs via Alu elements. Nature 470, 284–288 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kretz M et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 493, 231–235 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao ZD et al. Energy stress-induced lncRNA FILNC1 represses c-Myc-mediated energy metabolism and inhibits renal tumor development. Nat Commun 8, 783 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rehfuss RP, Walton KM, Loriaux MM & Goodman RH The cAMP-regulated enhancer-binding protein ATF-1 activates transcription in response to cAMP-dependent protein kinase A. J. Biol. Chem 266, 18431–18434 (1991). [PubMed] [Google Scholar]
- 42.Thio CL et al. Genetic protection against hepatitis B virus conferred by CCR5Delta32: Evidence that CCR5 contributes to viral persistence. J. Virol 81, 441–445 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Glass WG et al. CCR5 deficiency increases risk of symptomatic West Nile virus infection. J. Exp. Med 203, 35–40 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kakimi K, Guidotti LG, Koezuka Y & Chisari FV Natural killer T cell activation inhibits hepatitis B virus replication in vivo. J. Exp. Med 192, 921–930 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moreno C et al. CCR5 deficiency exacerbates T-cell-mediated hepatitis in mice. Hepatology 42, 854–862 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Sun Q, Hao Q & Prasanth KV Nuclear Long Noncoding RNAs: Key Regulators of Gene Expression. Trends Genet. 34, 142–157 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Noh JH, Kim KM, McClusky WG, Abdelmohsen K & Gorospe M Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip Rev RNA 9, e1471 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maurano MT et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Degner JF et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature 482, 390–394 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.