This cohort study examines the prevalence of germline mutations in patients with known renal cell carcinoma predisposition genes and other cancer-associated genes and identifies clinical and pathologic factors associated with germline mutations.
Key Points
Question
What is the prevalence of germline mutations in known renal cell carcinoma predisposition genes and other cancer-associated genes and what are the clinicopathologic factors associated with mutations?
Findings
In this cohort study of 254 patients with advanced renal cell carcinoma unselected for inherited cancer risk factors, 5.5% had mutations in renal cell carcinoma–associated genes and 10.5% in other cancer-associated genes. Among patients with non–clear cell renal cell carcinoma, 20.0% had any germline mutation, and 9% had a mutation diagnostic of hereditary leiomyomatosis (eg, FH mutation).
Meaning
The results of this study suggest that germline mutations may be frequent in patients with advanced renal cell carcinoma; genetic testing should be considered, especially for patients with advanced non–clear cell renal cell carcinoma.
Abstract
Importance
Identification of patients with hereditary renal cell carcinoma (RCC) is important for cancer screening and, in patients with advanced disease, for guiding treatment. The prevalence of cancer-related germline mutations in patients with advanced RCC and the phenotypes associated with some rare mutations are unknown.
Objectives
To examine the prevalence of germline mutations in both known RCC predisposition genes and other cancer-associated genes and to identify clinical and pathologic factors associated with germline mutations.
Design, Setting, and Participants
In this cohort study conducted from October 1, 2015, to July 31, 2017, 254 of 267 patients with advanced (American Joint Committee on Cancer stage III or IV) RCC who were seen in medical oncology or urology clinics agreed to germline sequencing and disclosure of results under an institutional protocol of matched tumor-germline DNA sequencing.
Main Outcomes and Measures
Mutation prevalence and spectrum in patients with advanced RCC were determined. Clinical characteristics were assessed by mutation status.
Results
Of the 254 patients (median age [range], 56 [13-79] years; 179 [70.5%] male; 211 [83.1%] non-Hispanic white), germline mutations were identified in 41 (16.1%); 14 (5.5%) had mutations in syndromic RCC-associated genes (7 in FH, 3 in BAP1, and 1 each in VHL, MET, SDHA, and SDHB). The most frequent mutations were CHEK2 (n = 9) and FH (n = 7). Of genes not previously associated with RCC risk, CHEK2 was overrepresented in patients compared with the general population, with an odds ratio of RCC of 3.0 (95% CI, 1.3-5.8; P = .003). Patients with non–clear cell RCC were significantly more likely to have an RCC-associated gene mutation (9 [11.7%] of 74 vs 3 [1.7%] of 177; P = .001), and 8 (10.0%) had a mutation in a gene that could guide therapy. Of patients with mutations in RCC-associated genes, 5 (35.7%) failed to meet current clinical guidelines for genetic testing.
Conclusions and Relevance
Of patients with non–clear cell RCC, more than 20% had a germline mutation, of which half had the potential to direct systemic therapy. Current referral criteria for genetic testing did not identify a substantial portion of patients with mutations, supporting the role of a more inclusive sequencing approach.
Introduction
Renal cell carcinoma (RCC) is among the 10 most frequently diagnosed cancers in the United States, affecting approximately 64 000 patients per year.1 Approximately 30% of patients are initially diagnosed with locoregional (stage III) or metastatic disease.2,3 Renal cell carcinoma comprises a heterogenous group of cancers. Clear cell RCC (ccRCC), the most common subtype, is characterized by loss of function of the von Hippel–Lindau (VHL) protein.4 Additional histologic subtypes are collectively known as non–clear cell (nccRCC) and include papillary type I and II, chromophobe, microphthalmia transcription factor family translocation associated, collecting duct, medullary, and other rare subtypes.5 Tumors that do not meet criteria for any established subtypes are categorized as unclassified.6
Several autosomal dominant inherited cancer syndromes predispose patients to ccRCC and nccRCC.7,8 Among these are VHL, hereditary leiomyomatosis and RCC (HLRCC), hereditary papillary RCC, and Birt-Hogg-Dubé, caused by germline mutations in VHL (OMIM 608537), FH (OMIM 136850), MET (OMIM 164860), and FLCN (OMIM 607273), respectively.4 There is also an increased risk of RCC among patients with germline mutations in BAP1 (OMIM 603089), SDHB (OMIM 185470), SDHC (OMIM 602413), SDHD (OMIM 602690), TSC1 (OMIM 605284), TSC2 (OMIM 191092), and MITF (OMIM 156845).9,10,11,12,13
Inherited RCC syndromes are thought to account for 5% of all cases; however, these estimates were derived from mostly early-stage RCC, and no studies have specifically looked at advanced disease.14,15 Recent studies16,17,18 suggest that germline mutations may be more frequent in patients with advanced cancer compared with those with early-stage disease. Identifying patients with inherited mutations has become particularly relevant because mutations in certain genes, such as FH and MET, can guide systemic therapy or clinical trial eligibility.19,20,21,22 Furthermore, identification of germline mutations could provide useful information for patients and family members for cancer risk stratification and early detection. Despite the potential benefits of identifying germline carriers, there is still uncertainty of who should be referred for genetic testing.
We assessed the frequency of germline mutations in 76 cancer-associated genes in patients with advanced RCC unselected for inherited syndrome risk factors, such as age at onset, multifocal disease, or family history. Our aims were to examine the prevalence of germline mutations in known RCC predisposition genes and other cancer-associated genes and to identify clinical and pathologic factors associated with germline mutations.
Methods
Patient Selection
From October 1, 2015, through July 31, 2017, a total of 267 patients with advanced (AJCC stage III or IV) RCC seen in medical oncology or urology clinics at Memorial Sloan Kettering Cancer Center (MSKCC) were offered germline sequencing and disclosure of results under an institutional protocol of matched tumor-germline DNA sequencing. All patients with advanced nccRCC or advanced ccRCC who participated in clinical trials were approached. Patients were unselected for family history of cancer, age at onset, multifocal tumors, or personal history of multiple malignant tumors. All patients viewed a standard pretest education video and were offered pretest genetic counseling. If pathogenic or likely pathogenic variants were detected, patients discussed the results with a certified cancer genetic counselor (A.A.), who then coordinated family cascade testing, as appropriate. Clinical and family history data were obtained from medical records and self-administered questionnaires. Pathologic test results were reviewed by genitourinary pathologists (Y.-B.C.) at the institution. A total of 128 patients were previously described in a large series that covered multiple tumor types, including patients with localized disease who are not described here; clinical annotation, including RCC tumor subtypes, was also not reported.18 The study was approved by the MSKCC Institutional Review Board. All patients provided written informed consent. Data were deidentified except to the investigators of the study.
Sequencing and Interpretation of Variants
Tumor and blood samples from patients were sequenced using the Memorial Sloan Kettering–Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT), a next-generation sequencing assay that achieves hybridization capture with target-specific probe from exons of at least 341 cancer-associated genes in the first iteration and 468 in the most recent, as described previously (eTable 1 in the Supplement).23 Germline variants in 76 genes associated with cancer predisposition included in the panel were further analyzed.24 All variants with less than 1% frequency in the public database ExAC were interpreted. Only pathogenic or likely pathogenic variants (associated with disease causation and henceforth referred to as pathogenic variants) were considered to be deleterious and are included in this analysis. Pathogenicity was determined by a clinical molecular geneticist or molecular pathologist according to American College of Medical Genetics (ACMG) criteria and updated as of January 2018.25 Variants of unknown significance were reviewed but are not reported in this analysis. Mutations were classified as high (relative risk [RR], >4), intermediate (RR, 2-4) or low (RR, <2) penetrant, recessive, or of uncertain clinical actionability based on disease risks and prior modeling.26,27,28 The Fraction and Allele-Specific Copy Number Estimates From Tumor Sequencing (FACETS) algorithm was used to evaluate loss of heterozygosity (LOH) in the locus of the variants.29
Comparison of Guidelines-Based Personal and Family History–Based Testing vs Agnostic Testing
Family history, race/ethnicity, and clinical features were abstracted from self-administered questionnaires and medical records. The ACMG guidelines were used to determine indicated genetic tests (eTable 2 in the Supplement).27 For patients who met the guidelines, we assumed that a multigene panel test would be ordered and would include the following genes: VHL, FH, FLCN, MET, SDHB, SDHC, SDHD, BAP1, TSC1, TSC2, TP53 (OMIM 191170), and MITF. A pathogenic variant would be considered as incremental if it was detected by sequencing but not by testing based on application of published guidelines.
Statistical Analysis
Demographic and clinical characteristics of the RCC cohort are presented using descriptive statistics. The prevalence of pathogenic or likely pathogenic germline mutations and RCC-associated gene mutations are reported in the RCC cohort and among patients with ccRCC and nccRCC. Clinical characteristics of patients with an RCC-associated gene mutation were compared with those of patients with no RCC-associated gene mutations using the Fisher exact test. Allele frequencies in the cohort were compared with allele frequencies in a noncancer population obtained from the public database ExAC minus cases derived from The Cancer Genome Atlas. Additional details of ExAC are available at http://exac.broadinstitute.org/faq. Known pathogenic and likely pathogenic variants were collapsed by gene and burden tests performed among the RCC cohort vs the noncancer cohort from the ExAC database; odds ratios (ORs) were computed. Statistical analysis was performed using R, version 3.3.2 with RStudio, version 1.0.136 (R Foundation for Statistical Computing). A 2-sided Fisher exact test P < .05 was considered to be statistically significant.
Results
Patient Characteristics
During the study period, 267 individuals with advanced RCC consented to tumor-normal testing with MSK-IMPACT and were offered disclosure of germline results under a separate protocol. Of these, 254 (95.1%) (median [range] age, 56 [13-79] years; 179 [70.5%] male and 75 [29.5%] female; 211 [83.1%] non-Hispanic white) consented to receive the germline results. The primary reason for declining was potential cause of worry; other reasons are listed in eTable 3 in the Supplement. Demographic and clinical characteristics are given in eTable 4 in the Supplement. Among the 254 patients, 177 had ccRCC (69.7%), 74 had nccRCC (29.1%), and 3 (1.2%) had both. Overall, 33 patients (13.0%) had a history of a second malignant tumor, excluding nonmelanoma skin cancers. The most frequent secondary malignant tumors were prostate in 8 patients (3.1%), breast in 4 patients (1.6%), and melanoma in 3 (1.2%). Overall, 14 patients (5.5%) had bilateral or multifocal disease, and 24 (9.4%) reported a family history of RCC.
Frequency of Mutations and Comparison With the Population
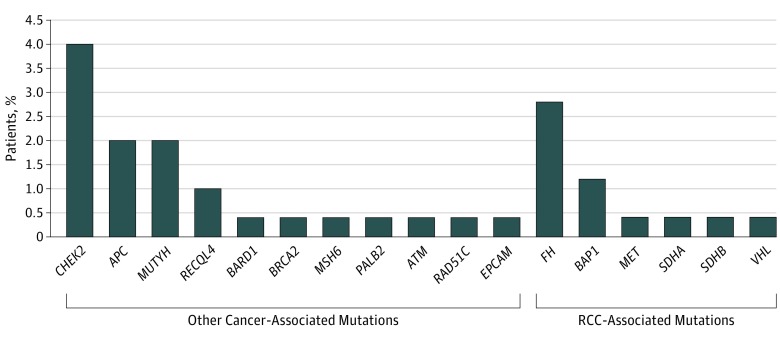
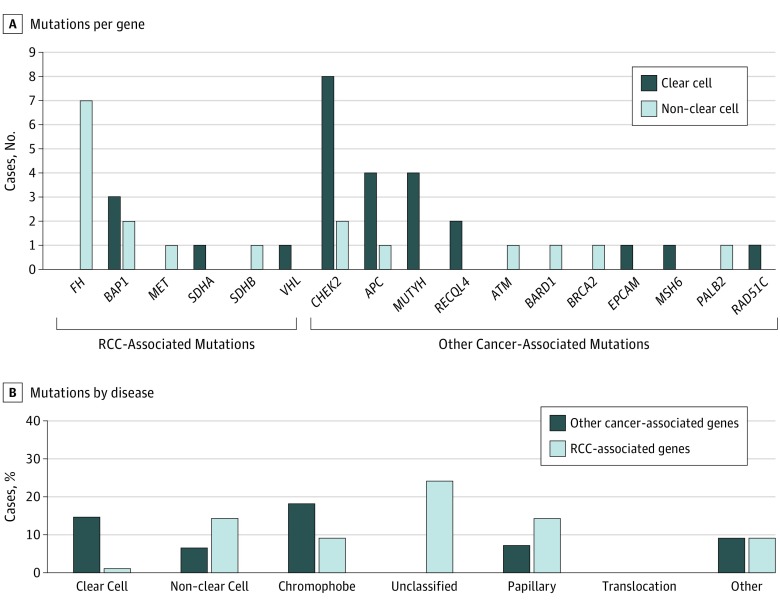
A total of 41 patients (16.1%) carried pathogenic or likely pathogenic germline variants in 17 different cancer-predisposition genes (Figure 1). No individuals had more than 1 germline mutation. Fourteen patients (5.5%) carried mutations in RCC-associated genes (7 in FH, 3 in BAP1, and 1 each in VHL, MET, SDHA, and SDHB). Twenty-seven patients (10.5%) carried mutations in genes not clearly associated with RCC, including 9 (3.5%) in CHEK2 (OMIM 604373); 2 of these were in the variant CHEK2 p.Ile157Thr, which is a low-penetrance breast cancer risk allele of uncertain actionability.30 Of the 41 patients, 17 patients (41.5%) carried mutations of high penetrance, 9 (22.0%) of moderate penetrance, 12 (29.3%) of low penetrance or uncertain clinical actionability, and 3 (7.3%) in genes linked to autosomal recessive syndromes (eTable 5 in the Supplement). Of the 177 patients with ccRCC, 25 (14.1%) had a germline mutation and 3 (1.7%) in an RCC-associated gene (Figure 2). Of the 74 patients with nccRCC, 13 (17.5%) had a germline mutation, with 9 (12.2%) in an RCC-associated gene. Of the 3 patients with both ccRCC and nccRCC, all 3 had germline mutations, 2 in BAP1 and 1 in CHEK2.
Figure 1. Frequency and Distribution of Pathogenic Germline Mutations.
Germline mutations were found in 41 patients (16.1%): RCC-associated mutations in 14 (5.5%) and other cancer-associated mutations in 27 (10.6%). Renal cell carcinoma (RCC)–associated germline mutations include mutations in BAP1, FH, MET, SDHA, SDHB, and VHL.
Figure 2. Pathogenic Mutations by Histologic Subtype.
Two patients with BAP1 mutation had both clear cell and non–clear cell renal cell carcinoma; 1 patient with CHEK2 mutation had clear cell and non–clear cell renal cell carcinoma.
For genes not traditionally associated with RCC, we compared mutation frequencies with those in the general population. The odds of mutations significantly exceeded the odds in the population only for CHEK2 (OR, 3.0; 95% CI, 1.3-5.8; P = .003) when all variants were considered (eTable 6 in the Supplement).
Factors Associated With Germline Mutations
We analyzed prevalence of mutations among patients thought to be at higher risk for inherited syndromes, including those with family history of RCC, early onset (≤46 years of age), and multifocal disease at diagnosis. Patients with nccRCC or multifocal RCC were significantly more likely to have an RCC-associated mutation (eTable 7 in the Supplement). Seven patients (9.5% of the nccRCC cohort) had germline FH mutations diagnostic of the hereditary syndrome HLRCC; all patients had tumors of unclassified histologic type or identified as FH deficient. All but one tumor, for which tissue was limited, were identified by genitourinary pathologists (Y.-B.C.) as suggestive of features of HLRCC. Median age at diagnosis for FH-positive patients was 49 years (range, 23-58 years); 4 presented with metastatic disease, and 3 later developed metastatic disease (range, 10 months to 10 years). None had multifocal RCC or family history of RCC. Of the other benign tumors associated with HLRCC, uterine fibroids were present in all women, but cutaneous leiomyomas were identified in only 1 patient after the HLRCC diagnosis was known.
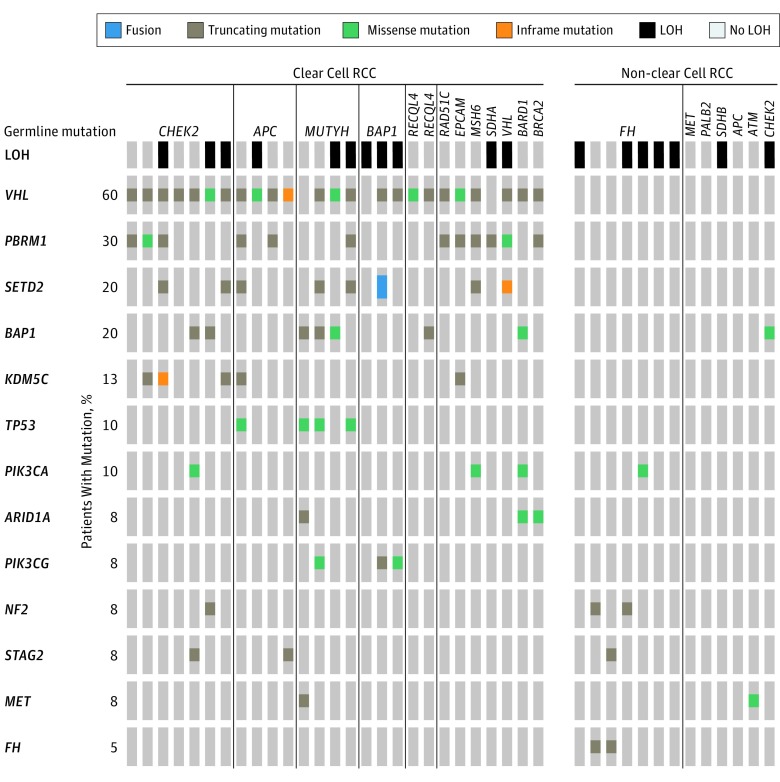
Three patients had BAP1 mutations: a woman in her 60s with ccRCC, a man in his 40s with ccRCC and a chromophobe tumor, and a man in his 80s with ccRCC, multiple papillary tumors, and colon cancer (pedigrees in the eFigure in the Supplement). Staining for BAP1 protein showed lack of expression in the ccRCC tumors of all 3 patients and the chromophobe tumor (eFigure in the Supplement). The papillary tumors were not available for immunohistochemical analysis.
Application of Clinical Genetic Counseling Referral Criteria
Overall, 99 patients (39.0%) would have met ACMG criteria for clinical genetics referral. Of 14 patients with RCC-associated mutations, 5 (35.7%) would not have met referral criteria, including 3 patients with FH, 1 with VHL, and 1 with SDHA. Of 12 patients with high or moderate penetrance mutations not related to RCC (ATM [OMIM 607585], BRCA2 [OMIM 600185], CHEK2 excluding variant I157T, MSH6 [OMIM 600678], PALB2 [OMIM 610355], RAD51C [OMIM 602774]), 7 patients would not have met referral criteria. At the time of analysis, at least 6 relatives of patients with mutations underwent germline testing through our clinic; several were also found to be carriers and referred for discussion of dedicated cancer screening.
Somatic Mutations in Patients With Germline Mutations
Tumor sequencing data were available for all but 1 patient. For 3 patients with both ccRCC and nccRCC, only the ccRCC was sequenced. In patients with germline mutations, we show the distribution of somatic variants and LOH in the tumor at the loci of interest (Figure 3). Of 9 patients with germline CHEK2 mutations, 4 (44.4%) had LOH. Of 7 patients with germline FH mutations, 5 (71.4%) had LOH and 2 (28.6%) had a somatic second hit.
Figure 3. Somatic Mutations and Loss of Heterozygosity (LOH) in Patients With Germline Mutations.
We investigated the clinical utility of tumor-only testing for guiding therapy using previously described methods.31 No patients had a level 1 or level 2A standard of care somatic biomarker predictive of response in RCC. A total of 36 (14.2%) had a level 2B or 3 somatic mutation that was a predictive biomarker in other cancers but not routinely used in RCC (eTable 8 in the Supplement).
Discussion
We report the prevalence of germline cancer-associated mutations in patients with advanced RCC unselected for suspicion of a hereditary syndrome. To date, approximately 5% of all RCC cases have been linked to high-penetrance cancer predisposition syndromes.4 Most studies14,15 on hereditary RCC, however, have been limited to selected, high-risk populations, and none to our knowledge have specifically studied patients with advanced disease. In this study, we found that among 254 patients with advanced RCC, 41 (16.1%) harbored pathogenic germline mutations, with 14 (5.5%) in RCC-associated genes and 27 (10.5%) in non–RCC-associated genes. The mutation rate was even higher among patients with uncommon RCC variants: more than 20% of patients with an nccRCC had germline mutations, of which half could help direct therapy according to previously published data from prospective clinical trials.19,20,32,33
Previous data16,17 suggest that the prevalence and spectrum of germline mutations differ between early- and late-stage disease in several cancers. In an analysis of 1040 patients with diverse cancers, a recent study18 found that the prevalence of germline, cancer-predisposing mutations was higher among patients with metastatic compared with localized disease (22.3% vs 8.3%; P < .001). In a separate study34 of 1235 patients with RCC with all stages of disease who were referred for multigene germline testing, 6% had mutations, but the spectrum of mutations differed from that in our cohort. The most frequent mutations were in FLCN (1.8% vs 0% in our cohort). Despite the enrichment in that series for suspected hereditary syndromes, FH mutations were less prevalent (1.3% vs 3% in our cohort) and only 2 BAP1 mutations were identified. FH and BAP1 mutations may be associated with more aggressive RCC, which could account for their overrepresentation in the current cohort of patients with advanced cancer.35,36
We found that 9% of all patients with advanced nccRCC had a germline FH mutation diagnostic of HLRCC, a higher percentage than previously reported.37,38 Hereditary leiomyomatosis and RCC is an autosomal dominant inherited syndrome associated with RCC and uterine and cutaneous leiomyomas. These patients were difficult to identify by clinical criteria. None had a family history of RCC, and typical cutaneous lesions were not identified by the treating oncologists. Although expert genitourinary pathologists identified histologic features suggestive of HLRCC, it is unclear whether nonspecialist pathologists would be able to draw the same conclusions.39,40 Relatives who are also found to carry FH mutations should be considered for RCC screening. Early detection may increase the likelihood of cure and survivorship.38,41 We also found that 2 of 3 individuals with BAP1 mutations had nccRCC tumors, which, to our knowledge, have never been reported in BAP1 carriers.42 The available chromophobe tumor had loss of expression of BAP1 on immunohistochemical analysis, which is rare in chromophobe tumors.43 The possible association of BAP1 mutations with nccRCC should be investigated in other cohorts.
Novel biomarkers predictive of therapy response are needed in RCC.31,44 In this cohort, 10% of patients with nccRCC had a predictive germline biomarker, none of which would have been identified with somatic-only sequencing. In RCC, several germline mutations are prognostic indicators of response to therapies. In a phase 2 biomarker study20 of a dual MET/VEGFR2 in patients with papillary RCC, the presence of a germline MET mutation was associated with response. Another phase 2 study33 of bevacizumab plus erlotinib for patients with sporadic or hereditary papillary RCC showed significant activity with the combination, particularly in those with HLRCC. On the basis of this study,33 the 2018 National Comprehensive Cancer Network guidelines added bevacizumab plus everolimus or erlotinib as options specifically for patients with germline FH mutations diagnostic of HLRCC.45 Because of the high positive germline mutation rate among patients with nccRCC, the difficulty using clinical criteria to identify them, and the potential for therapeutic actionability, patients with advanced nccRCC should be referred for genetic counseling and appropriate testing.
Compared with the general population, patients with RCC had a significantly increased frequency of germline CHEK2 mutations. Germline CHEK2 mutations are associated with increased susceptibility to several cancers. Few studies46,47 have found an increased risk of RCC among carriers of the 1100deC truncating mutation and Polish founder mutations, with ORs ranging from 2.1 to 3.6. We found an OR of 3.0 of RCC when all variants of CHEK2 were considered. A potential role of CHEK2 in the pathogenicity of RCC is also supported by the LOH in the tumor in 4 of 9 germline CHEK2 mutants. Although there are currently no RCC-specific screening recommendations for individuals with CHEK2 mutations, there may be incremental screening for other cancers, justifying including this gene on RCC panel tests.28
For patients with advanced RCC, current clinical guidelines for referral to genetic counseling miss patients who could benefit from genetic counseling. The ACMG has developed practice guidelines that consider age at diagnosis, tumor multifocality, family history, and histologic findings.27 We found that despite almost 40% of patients in this cohort meeting the broad guideline criteria, 36% of patients with high-penetrance RCC-associated mutations would have been missed. With potentially increasing numbers of mutations identified in patients and relatives, optimal cancer screening schedules need to be studied. For example, there are no consensus screening guidelines for several of the more rare RCC genetic syndromes, including BAP1-associated cancer syndrome, although some groups have suggested screening schedules.48
Limitations
There are limitations to this study. Many patients in this cohort also participated in clinical trials. Therefore, patients who contributed to this study most likely were younger and had fewer comorbid conditions than the general cancer population. For example, the median age at diagnosis in this study was 56 years compared with 64 years in the United States.1 Nevertheless, this limitation may underestimate the number with germline mutations because most trials exclude patients with a history of other malignant tumors. We acknowledge that some Ashkenazi Jewish founder mutations, such as APC (OMIM 611731) p.Ile1307Lys and CHEK2 p.Ser428Phe, may be overrepresented in our cohort. Although the association of APC and RCC risk is unclear, for CHEK2, we also performed a burden test excluding the p.Ser428Phe variant, and the association with RCC was still present (eTable 6 in Supplement). Finally, although we analyzed 254 patients, the sample size was still limited, and further studies should be conducted.
Conclusions
Our results suggest that germline mutations in cancer-associated genes in patients with advanced RCC may be prevalent, and many of these mutations can be used to guide therapy. Phenotype-directed or tumor-only testing would have failed to identify most patients with actionable mutations. A broader approach to tumor-normal sequencing of all patients with advanced RCC, especially those with nccRCC, might help identify individual patients for whom targeted therapies are indicated, as well as family members who may benefit from preventive interventions tailored to their increased cancer risk.
eTable 1. Genes Included in the Somatic and Germline MSK-IMPACT Assay
eTable 2. Current Referral Indications for Genetic Counseling Referral for Patients With Renal Cell Carcinoma
eTable 3. Reasons for Declining Genetic Testing
eTable 4. Demographic Characteristics
eTable 5. Detail on Pathogenic Mutations
eTable 6. CHEK2 Variants and Association with Renal Cell Carcinoma
eTable 7. Associations Between Clinicopathologic Characteristics and Presence of RCC-Associated Germline Mutation
eTable 8. OncoKB Levels of Evidence and Somatic Biomarkers in Cohort
eFigure. Families of BAP1 Germline Mutant Patients and Tumor Immunohistochemistry
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7-30. [DOI] [PubMed] [Google Scholar]
- 2.Stephenson AJ, Chetner MP, Rourke K, et al. . Guidelines for the surveillance of localized renal cell carcinoma based on the patterns of relapse after nephrectomy. J Urol. 2004;172(1):58-62. [DOI] [PubMed] [Google Scholar]
- 3.Dabestani S, Thorstenson A, Lindblad P, Harmenberg U, Ljungberg B, Lundstam S. Renal cell carcinoma recurrences and metastases in primary non-metastatic patients: a population-based study. World J Urol. 2016;34(8):1081-1086. [DOI] [PubMed] [Google Scholar]
- 4.Linehan WM. Genetic basis of kidney cancer: role of genomics for the development of disease-based therapeutics. Genome Res. 2012;22(11):2089-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sankin A, Hakimi AA, Hsieh JJ, Molina AM. Metastatic non-clear cell renal cell carcinoma: an evidence based review of current treatment strategies. Front Oncol. 2015;5:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kovacs G, Akhtar M, Beckwith BJ, et al. . The Heidelberg classification of renal cell tumours. J Pathol. 1997;183(2):131-133. [DOI] [PubMed] [Google Scholar]
- 7.Linehan WM, Lerman MI, Zbar B. Identification of the von Hippel-Lindau (VHL) gene. Its role in renal cancer. JAMA. 1995;273(7):564-570. [PubMed] [Google Scholar]
- 8.Zbar B, Glenn G, Merino M, et al. . Familial renal carcinoma: clinical evaluation, clinical subtypes and risk of renal carcinoma development. J Urol. 2007;177(2):461-465. [DOI] [PubMed] [Google Scholar]
- 9.Bertolotto C, Lesueur F, Giuliano S, et al. ; French Familial Melanoma Study Group . A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature. 2011;480(7375):94-98. [DOI] [PubMed] [Google Scholar]
- 10.Malinoc A, Sullivan M, Wiech T, et al. . Biallelic inactivation of the SDHC gene in renal carcinoma associated with paraganglioma syndrome type 3. Endocr Relat Cancer. 2012;19(3):283-290. [DOI] [PubMed] [Google Scholar]
- 11.Popova T, Hebert L, Jacquemin V, et al. . Germline BAP1 mutations predispose to renal cell carcinomas. Am J Hum Genet. 2013;92(6):974-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vanharanta S, Buchta M, McWhinney SR, et al. . Early-onset renal cell carcinoma as a novel extraparaganglial component of SDHB-associated heritable paraganglioma. Am J Hum Genet. 2004;74(1):153-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pasini B, McWhinney SR, Bei T, et al. . Clinical and molecular genetics of patients with the Carney-Stratakis syndrome and germline mutations of the genes coding for the succinate dehydrogenase subunits SDHB, SDHC, and SDHD. Eur J Hum Genet. 2008;16(1):79-88. [DOI] [PubMed] [Google Scholar]
- 14.Shuch B, Vourganti S, Ricketts CJ, et al. . Defining early-onset kidney cancer: implications for germline and somatic mutation testing and clinical management. J Clin Oncol. 2014;32(5):431-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gudbjartsson T, Jónasdóttir TJ, Thoroddsen A, et al. . A population-based familial aggregation analysis indicates genetic contribution in a majority of renal cell carcinomas. Int J Cancer. 2002;100(4):476-479. [DOI] [PubMed] [Google Scholar]
- 16.Pritchard CC, Mateo J, Walsh MF, et al. . Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med. 2016;375(5):443-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson DR, Wu YM, Lonigro RJ, et al. . Integrative clinical genomics of metastatic cancer. Nature. 2017;548(7667):297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandelker D, Zhang L, Kemel Y, et al. . Mutation detection in patients with advanced cancer by universal sequencing of cancer-related genes in tumor and normal DNA vs guideline-based germline testing. JAMA. 2017;318(9):825-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choueiri TK, Plimack E, Arkenau HT, et al. . Biomarker-based phase II trial of savolitinib in patients with advanced papillary renal cell cancer. J Clin Oncol. 2017;35(26):2993-3001. [DOI] [PubMed] [Google Scholar]
- 20.Choueiri TK, Vaishampayan U, Rosenberg JE, et al. . Phase II and biomarker study of the dual MET/VEGFR2 inhibitor foretinib in patients with papillary renal cell carcinoma. J Clin Oncol. 2013;31(2):181-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voss MH, Molina AM, Chen YB, et al. . Phase II trial and correlative genomic analysis of everolimus plus bevacizumab in advanced non-clear cell renal cell carcinoma. J Clin Oncol. 2016;34(32):3846-3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gordon MS, Hussey M, Nagle RB, et al. . Phase II study of erlotinib in patients with locally advanced or metastatic papillary histology renal cell cancer: SWOG S0317. J Clin Oncol. 2009;27(34):5788-5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng DT, Mitchell TN, Zehir A, et al. . Memorial Sloan Kettering–Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17(3):251-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng DT, Prasad M, Chekaluk Y, et al. . Comprehensive detection of germline variants by MSK-IMPACT, a clinical diagnostic platform for solid tumor molecular oncology and concurrent cancer predisposition testing. BMC Med Genomics. 2017;10(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richards S, Aziz N, Bale S, et al. ; ACMG Laboratory Quality Assurance Committee . Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt LS, Linehan WM. Genetic predisposition to kidney cancer. Semin Oncol. 2016;43(5):566-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hampel H, Bennett RL, Buchanan A, Pearlman R, Wiesner GL; Guideline Development Group, American College of Medical Genetics and Genomics Professional Practice and Guidelines Committee and National Society of Genetic Counselors Practice Guidelines Committee . A practice guideline from the American College of Medical Genetics and Genomics and the National Society of Genetic Counselors: referral indications for cancer predisposition assessment. Genet Med. 2015;17(1):70-87. [DOI] [PubMed] [Google Scholar]
- 28.Tung N, Domchek SM, Stadler Z, et al. . Counselling framework for moderate-penetrance cancer-susceptibility mutations. Nat Rev Clin Oncol. 2016;13(9):581-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 2016;44(16):e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu C, Wang Y, Wang QS, Wang YJ. The CHEK2 I157T variant and breast cancer susceptibility: a systematic review and meta-analysis. Asian Pac J Cancer Prev. 2012;13(4):1355-1360. [DOI] [PubMed] [Google Scholar]
- 31.Chakravarty D, Gao J, Phillips SM, et al. . OncoKB: a precision oncology knowledge base [published online May 16, 2017]. JCO Precis Oncol. doi: 10.1200/PO.17.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voss MH, Chen D, Marker M, et al. . Tumor genomic analysis for 128 renal cell carcinoma (RCC) patients receiving first-line everolimus: correlation between outcome and mutations status in MTOR, TSC1, and TSC2. J Clin Oncol. 2017;35(6)(suppl):484-484. [Google Scholar]
- 33.Srinivasan R, Su D, Stamatakis L, et al. . Mechanism based targeted therapy for hereditary leiomyomatosis and renal cell cancer (HLRCC) and sporadic papillary renal cell carcinoma: interim results from a phase 2 study of bevacizumab and erlotinib. Eur J Cancer. 2014;50(suppl 6):8. [Google Scholar]
- 34.Nguyen KA, Syed JS, Espenschied CR, et al. . Advances in the diagnosis of hereditary kidney cancer: initial results of a multigene panel test. Cancer. 2017;123(22):4363-4371. [DOI] [PubMed] [Google Scholar]
- 35.Patel VM, Handler MZ, Schwartz RA, Lambert WC. Hereditary leiomyomatosis and renal cell cancer syndrome: an update and review. J Am Acad Dermatol. 2017;77(1):149-158. [DOI] [PubMed] [Google Scholar]
- 36.Hakimi AA, Ostrovnaya I, Reva B, et al. ; ccRCC Cancer Genome Atlas (KIRC TCGA) Research Network investigators . Adverse outcomes in clear cell renal cell carcinoma with mutations of 3p21 epigenetic regulators BAP1 and SETD2: a report by MSKCC and the KIRC TCGA research network. Clin Cancer Res. 2013;19(12):3259-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pithukpakorn M, Toro JR. Hereditary leiomyomatosis and renal cell cancer In: Pagon RA, Adam MP, Ardinger HH, et al. , eds. GeneReviews(R). Seattle: University of Washington; 1993-2018. [Google Scholar]
- 38.Menko FH, Maher ER, Schmidt LS, et al. . Hereditary leiomyomatosis and renal cell cancer (HLRCC): renal cancer risk, surveillance and treatment. Fam Cancer. 2014;13(4):637-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toro JR, Nickerson ML, Wei MH, et al. . Mutations in the fumarate hydratase gene cause hereditary leiomyomatosis and renal cell cancer in families in North America. Am J Hum Genet. 2003;73(1):95-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alam NA, Rowan AJ, Wortham NC, et al. . Genetic and functional analyses of FH mutations in multiple cutaneous and uterine leiomyomatosis, hereditary leiomyomatosis and renal cancer, and fumarate hydratase deficiency. Hum Mol Genet. 2003;12(11):1241-1252. [DOI] [PubMed] [Google Scholar]
- 41.Chan MMY, Barnicoat A, Mumtaz F, et al. . Cascade fumarate hydratase mutation screening allows early detection of kidney tumour: a case report. BMC Med Genet. 2017;18(1):79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pilarski R, Rai K, Cebulla C, Abdel-Rahman M. BAP1 tumor predisposition syndrome In: Pagon RA, Adam MP, Ardinger HH, et al. , eds. GeneReviews(R). Seattle: University of Washington; 1993-2018. [PubMed] [Google Scholar]
- 43.Ho TH, Kapur P, Joseph RW, et al. . Loss of PBRM1 and BAP1 expression is less common in non-clear cell renal cell carcinoma than in clear cell renal cell carcinoma. Urol Oncol. 2015;33(1):23.e9-23.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zehir A, Benayed R, Shah RH, et al. . Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23(6):703-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.National Comprehensive Cancer Network Kidney Cancer (Version 1.20184) https://www.nccn.org/professionals/physician_gls/pdf/kidney.pdf. Accessed October 30, 2017.
- 46.Cybulski C, Górski B, Huzarski T, et al. . CHEK2 is a multiorgan cancer susceptibility gene. Am J Hum Genet. 2004;75(6):1131-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Näslund-Koch C, Nordestgaard BG, Bojesen SE. Increased risk for other cancers in addition to breast cancer for CHEK2*1100delC heterozygotes estimated from the Copenhagen General Population Study. J Clin Oncol. 2016;34(11):1208-1216. [DOI] [PubMed] [Google Scholar]
- 48.Star P, Goodwin A, Kapoor R, et al. . Germline BAP1-positive patients: the dilemmas of cancer surveillance and a proposed interdisciplinary consensus monitoring strategy. Eur J Cancer. 2018;92:48-53. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eTable 1. Genes Included in the Somatic and Germline MSK-IMPACT Assay
eTable 2. Current Referral Indications for Genetic Counseling Referral for Patients With Renal Cell Carcinoma
eTable 3. Reasons for Declining Genetic Testing
eTable 4. Demographic Characteristics
eTable 5. Detail on Pathogenic Mutations
eTable 6. CHEK2 Variants and Association with Renal Cell Carcinoma
eTable 7. Associations Between Clinicopathologic Characteristics and Presence of RCC-Associated Germline Mutation
eTable 8. OncoKB Levels of Evidence and Somatic Biomarkers in Cohort
eFigure. Families of BAP1 Germline Mutant Patients and Tumor Immunohistochemistry