Abstract
Neuroblastoma (NB) is a sympathetic nervous system cancer for children, occupying approximately 15% of pediatric oncology deaths. BARD1, a tumor suppressor, is essential for genome stability by interaction with BRCA1. Here, we performed a systematic investigation for the association between SNPs in BARD1 and the risk of NB in Chinese population. After SNP screening in BARD1 gene, we performed case-control study of eleven selected SNPs in BARD1 with 339 NB patients and 778 cancer-free controls. The OR and 95% CI of these candidate SNPs were computed by logistic regression. After adjusted gender and age, seven out of eleven SNPs in BARD1 were significant associated with the risk of NB, including one SNP in 5'-UTR (rs17489363 G > A), two SNPs in exon (rs2229571 G > C and rs3738888 C > T), and four SNPs in intron (rs3768716 A > G, rs6435862 T > G, rs3768707 C > T and rs17487792 C > T). When stratified by the INPC, primary tumor site and the INSS, these seven SNPs were significant associated with GNB/NB, stage III/IV and adrenal origin of NB. Dual-luciferase reporter assay showed rs17489363 A allele-containing haplotypes (TAC, CAC, TAG and CAG), composed with rs34732883 T > C, and rs1129804 C > G, dramatically reduced the transcriptional activity of reporter gene. The major of our study showed that seven SNPs of BARD1 associated with increased NB risk in Chinese population, and four haplotypes could reduce transcription activity of BARD1.
Keywords: Neuroblastoma, Single nucleotide polymorphism, BARD1, Case-control study
Introduction
Neuroblastoma (NB) is the most common type of pediatric extracranial cancer, accounting for approximately 15% of all cancer deaths in children 1, 2. It could arise in any part of sympathetic nervous system, especially in adrenal medulla and abdomen 3. The clinical presentation is highly variable, ranging from local invasion and distant metastasis 4. Although advanced immunotherapy and multiple cytotoxic therapies are applied, the survival rate of high-risk NB remains below 50% 4, 5. However, the complex pathogenesis of NB is still not well understood. Therefore, it is of great significance to explore the molecular mechanism of NB in order to accelerate the development of novel drugs and therapy methods.
Although only about 1% of NB patients are heredofamilial, accumulating evidences indicate the complicated genomic abnormalities play significant roles in NB heterogeneity 6, 7. To recognize tumor genetic variations, genome-wide association studies (GWASs) are considered as a powerful and hypothesis-free method 8. Based on GWAS results, multiple genetic variations associated with NB risk have been identified, such as ALK, FLJ22536, FLJ44180, BARD1, LMO1, HACE1 and LIN28B9-15. The earliest GWAS on NB was performed by Maris and colleagues in 2008, including 1032 cases and 2043 controls in European American, which validated that chromosome 6p22 locus was associated with clinically aggressive NB10. In high-risk NB, they further found six SNPs were associated with NB susceptibility in the BRCA1 associated RING domain 1 (BARD1) gene 11. Moreover, these BARD1 SNPs has been confirmed in both Italians 16 and African-Americans 17. In Chinese population, three BARD1 SNPs have been studied in NB, but only stratified analysis demonstrated that rs3768716 and rs6435862 were risk factors for aggressive NB 18. As a result, further systematic research and functional assessment of SNPs in BARD1 gene need to be performed. In addition, identifying SNPs in Chinese population is informative and it can explore potential ethnic differences and strengthen the role of BARD1 as locus of NB susceptibility.
Herein, we performed a systematic screening of the SNPs in BARD1 and investigated the function of SNPs located in functional regions such as 5'-UTR and exon. We selected eleven candidate SNPs in BARD1 gene and carried out a case-control study in a total of 339 NB patients and 778 cancer-free controls. Association analysis indicated that seven SNPs contributed to NB risk. Functional evaluation of SNPs in 5'-UTR and exon region revealed that rs17489363 G > A might influence BARD1 promoter activity in four haplotypes. The major aim of our study was to discover the association between BARD1 SNPs and NB susceptibility, and further explore the precise molecular mechanism of the functional SNPs.
Methods and Materials
Ethics statement
This study was approved by the Ethics Committee of the Beijing Children's Hospital, Capital Medical University (Beijing, China). The written informed consents have been obtained from participants and/or their legal guardians involved in this study prior to inclusion in the study.
Subjects and clinical information
Volunteers were enrolled from January 2004 to December 2016 at Beijing Children's Hospital, which is the National Center for Children's Health (NCCH). A total of 339 NB patients and 778 cancer-free controls were recruited. Even the subjects were from all over the country, more than half of them were from Northern of China. To avoid minority differences of genetic background, all the volunteers in our research were Han Chinese children. According to the International Neuroblastoma Pathology Classification (INPC) 19, we stratified cases into three pathological groups, namely Neuroblastoma (NB), Ganglioneuroma (GN) and Ganglioneuroblastoma (GNB). In addition, the tumor stage was classified based on the International Neuroblastoma Staging System (INSS) by two professional pathologists. All controls were health children, admitted by physical examination center of Beijing Children's Hospital. All recruited volunteers were collected EDTA-anticoagulated blood samples for this research, and stored at -80oC.
SNPs screening
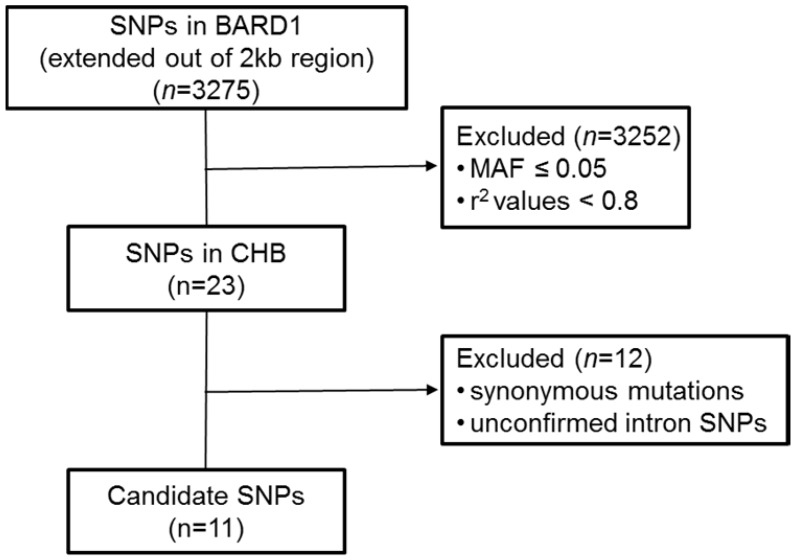
We used computer-facilitated strategy to select candidate SNPs in BARD1 gene. The following steps were operated (Figure 1). Firstly, candidate SNPs were filtered by using the Haploview (http://www.broad.mit.edu/mpg/haploview/) with selection criteria of minor allele frequency (MAF) > 0.05 and r2 values > 0.8 from Han Chinese in Beijing (CHB) in the 1000 Genomes database (https://www.ncbi.nlm.nih.gov/variation/tools/1000genomes/). The candidate SNPs screening region extended to a 2 kb (1kb upstream and 1kb downstream the gene). Secondly, to identify functional SNPs in BARD1, we excluded synonymous mutations and unconfirmed intron SNPs.
Figure 1.
The schematic for the strategy of SNP selection.
Sample preparation and genotyping
DNA Blood Mini Kit (Qiagen, Hilden, Germany) was used to extract Genomic DNA from 300μL peripheral blood of all samples, and then the DNAs were stored at -20oC until use. By using the SequenomMassArray platform, SNP genotyping was performed following the manufacturer's protocol (Sequenom, San Diego, USA) 13, 20.
Cell culture
Human embryonic kidney cell line (293T) and human neuroblastoma cell line (SH-SY5Y) were obtained from Cell Resource Center, Chinese Academy of Medical Sciences (CAMS, Beijing, China). The cells were cultured in DMEM supplemented with 10% fetal bovine serum, in a humidified, 5% CO2 incubator at 37°C.
Plasmid construction
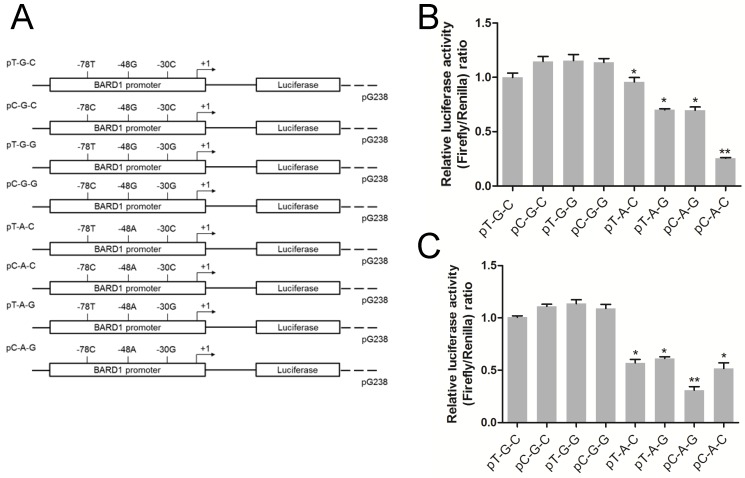
Three SNPs (rs34732883 T > C, rs17489363 G > A and rs1129804 C > G) located in 5'-UTR of BARD1. For these SNPs, a GV238 reporter (Genechem, Shanghai, China) plasmid encompassing 5'-UTR and truncated promoter region of BARD1 was constructed, designated as pT-G-C according to sequence analysis. Subsequently, this pT-G-C was used as a template, 5'-UTR site-specific mutagenesis in rs34732883 T > C, rs17489363 G > A and rs1129804 C > G were performed to generate other constructs containing all of the possible haplotypes, namely pC-G-C, pT-G-G, pC-G-G, pT-A-C, pC-A-C, pT-A-G and pC-A-G (Figure 2A).
Figure 2.
Dual-luciferase reporter assay with constructs containing BARD1 promoter. (A) Schematic of reporter gene constructs having the BARD1 promoter, with different haplotypes containing three SNPs in BARD1 5'-UTR (rs34732883, rs17489363 and 1129804). 293T cells (B) and SH-SY5Y (C) were co-transfected with 50 ng of the reporter plasmid and 10 ng of Renilla luciferase reporter plasmid for standardization of the transfection efficiencies. The luciferase activity was expressed as Mean ± S.E. of the ratio of firefly expression of BARD1 promoters. (*) indicate P < 0.05 with respect to the general (without stimulation). Abbreviation: NC, negative control.
The exon SNP rs1048108 located near zinc finger ring region, which played a key role in the interaction of BARD1 and Breast cancer 1 (BRCA1). To explore the function of rs1048108, BARD1 ORF expression plasmid (pCMV3-Myc-BARD1) was obtained from Sino Biological lnc (Beijing, China). This plasmid with rs1048108 C allele was named as BARD1-Myc-CC. Then, site-specific mutagenesis was performed to creating single nucleotide mutation in rs1048108 C > T, named as BARD1-Myc-TT accordingly. All constructs were sequenced to confirm their authenticity.
Daul-luciferase reporter assay
Total of 1×105 cells were plated in 24-well plates (Corning, NY, USA) and grew to 80-90% confluence for transient transfection with Lipofectamine 2000 Reagent (Life Technologies, Inc., Rockville, USA). Cells were then transfected with 50 ng/well of reporter plasmid. Identically, Renilla luciferase reporter plasmid (10 ng/well) was co-transfected for standardization of the transfection efficiencies. Luciferase activity was measured by the Dual-Luciferase Reporter Assay system (Promega, Madison, USA) on a microplate reader (CLARIOstar, BMG labtech, Germany) according to the manufacturer's protocol. Results were normalized by Renilla activity, and labeled as relative luciferase activity. Three independent transfection experiments were performed, and each was done in triplicate.
Co-immunoprecipitation (Co-IP) and Western blotting
To study the interaction between BARD1 and BRCA1, we obtained the human BRCA1 ORF expression plasmid (pcDNA3-HA-BRCA1) from Sino Biological lnc (Beijing, China). Co-IP was performed as follows. Briefly, 293T cells were co-transfected with plasmid containing BRCA1 gene (pcDNA3-HA-BRCA1) and plasmid containing different genotype BARD1 gene (BARD1-Myc-CC or BARD1-Myc-TT) for 48 h. Proteins were then extracted with RIPA cell lysis buffer (Dingguo, Beijing). Total cell protein extracts were incubated with anti-HA antibody (Abcam, UK) overnight at 4oC followed by immunoprecipitation with protein A agarose beads (Roche, Switzerland). The beads were washed, collected and resolved in prechilled RIPA lysis buffer for six times. Finally, the protein-agarose complexes were boiled for SDS-PAGE. Immunoblots were carried out according to the standard operating procedure. The relative densities of the desired protein bands were quantified by Image Lab software (Bio-Rad, USA).
Statistical Analysis
A two-sided χ2 test was performed to describe the distribution of the age and gender between cases and controls, as well as the deviation of genotypes. All samples were detected for Hardy-Weinberg equilibrium by Goodness-of-fit χ2 test. In order to get an exact result, we analyzed the association between the polymorphisms and NB susceptibility by using the multivariate logistic regression analysis to assess Odds ratios (ORs) and 95% Confidence intervals (95%CIs) with age and gander as covariates. The P value below 0.05 from either model was considered statistically significant. Bonferroni test was carried out to adjust for multiple comparisons in the single SNP analysis (P < 0.05/n, n=11). All statistical analyses described above were computed using SAS software (version 9.1; SAS Institute, Cary, NC). Haploview 4.221 was used to measure the LD and the haplotype.
Results
Clinical characteristics
To analyze the association between NB risk and BARD1 SNPs, 339 cases and 778 controls were recruited in Beijing Children's Hospital from January 2004 to December 2016 (Table 1). Two groups were matched with gender and age (P > 0.05). Pathological types classified by INPC were categorized into NB (240, 70.80%), GN (33, 9.73%), and GNB (59, 17.40%). According to INSS, the case group was divided into stage I (45, 13.27%), II (59, 17.40%), III (99, 29.20%), IV (109, 32.15%) and IVs (24, 7.08%). Besides, among these 339 cases, 152 (44.84%) of the primary tumors located in adrenal gland and 171 (51.33%) developed from non-adrenal regions including abdominal/retroperitoneal region (137, 40.41%), pelvic cavity (20, 5.90%), and other sites (14, 4.13%). The remaining 13 (3.83 %) cases were not classified into a specific region.
Table 1.
Distribution of Subject Characteristics
| Case (N=339) | Controls (N=778) | P* | ||||
|---|---|---|---|---|---|---|
| No. | (%) | No. | (%) | |||
| Sex | ||||||
| Male | 194 | 57.23 | 453 | 58.23 | 0.76 | |
| Female | 144 | 42.48 | 325 | 41.77 | ||
| Unknown | 1 | 0.29 | 0 | 0.00 | ||
| Age | ||||||
| ≤12months | 64 | 18.87 | 144 | 18.51 | 0.91 | |
| >12months | 275 | 81.13 | 633 | 81.36 | ||
| Unknown | 0 | 0.00 | 1 | 0.13 | ||
| Tumor stagea | ||||||
| I | 45 | 13.27 | ||||
| II | 59 | 17.40 | ||||
| III | 99 | 29.20 | ||||
| IV | 109 | 32.15 | ||||
| IVs | 24 | 7.08 | ||||
| Unknown | 3 | 0.88 | ||||
| Categoriesb | ||||||
| GN | 33 | 9.73 | ||||
| GNB | 59 | 17.4 | ||||
| NB | 240 | 70.80 | ||||
| Unknown | 7 | 2.06 | ||||
| Primary tumor sites | ||||||
| Adrenal | 152 | 44.84 | ||||
| Abdominal | 137 | 40.41 | ||||
| Pelvic | 20 | 5.90 | ||||
| Others | 14 | 4.13 | ||||
| Unknown | 13 | 3.83 | ||||
*Two-sided χ2 test. a According to INSS. b According to INPC.
Identification of SNPs
Based on the screening strategy (Figure 1), eleven candidate SNPs were selected for further study, including three SNPs in 5'-UTR (rs34732883, rs17489363 and rs1129804), three SNPs in exon (rs2229571, rs3738888 and rs1048108), and five SNPs in intron (rs3768716, rs6435862, rs3768707, rs17487792 and rs7587476) (Table 2).
Table 2.
Basic Information of Potential BARD1 SNPs
| # | Identity | Position(GRCh37) | Position | Base change | MAFa |
|---|---|---|---|---|---|
| 1 | rs17489363 | Ch2:214809617 | 5'-UTR | G>A | 0.18 |
| 2 | rs34732883 | Ch2:214809647 | 5'-UTR | T>C | 0.18 |
| 3 | rs1129804 | Ch2:214809599 | 5'-UTR | C>G | 0.18 |
| 4 | rs2229571 | Ch2:214780740 | exon | G>C | 0.34 |
| 5 | rs3738888 | Ch2:214730440 | exon | C>T | 0.04 |
| 6 | rs1048108 | Ch2:214809500 | exon | C>T | 0.37 |
| 7 | rs3768716 | Ch2:214771070 | intron | A>G | 0.15 |
| 8 | rs6435862 | Ch2:214807822 | intron | T>G | 0.09 |
| 9 | rs3768707 | Ch2:214780411 | intron | C>T | 0.23 |
| 10 | rs17487792 | Ch2:214778776 | intron | C>T | 0.14 |
| 11 | rs7587476 | Ch2:214789163 | intron | C>T | 0.23 |
Abbreviation: MAF, minor allele frequency.
a Data from CHB population in 1000 Genomes project
Since linkage disequilibrium (LD) represented correlating genotypes between SNPs at different loci 22, the pair-wise LD between these eleven SNPs were measured by Haploview software. The disequilibrium coefficient D' and r2 were represented of the proportion or the maximum possible disequilibrium. The results showed that three SNPs in 5'-UTR were in strong LD in our study population, with a D' of 0.99 (r2 = 0.97) for rs34732883 and rs1129804 (P < 0.001), a D' of 1.00 (r2 = 0.98) for rs34732883 and rs17489363 (P < 0.001), and a D' of 1.00 (r2 = 0.99) for rs17489363 and rs1129804 (P < 0.001). In exon, rs2229571 and rs1048108 also showed strong LD (D' = 0.88, r2 = 0.7, P < 0.001) (Supplement Figure 1).
Effect of BARD1 SNPs on NB Risk
To explore the effect of eleven candidate SNPs on NB risk, the genotype differences were compared between 339 NB cases and 778 controls. By multivariate logistic regression analysis, we found that seven SNPs were significantly associated with NB risk (Table 3), with one in 5'-UTR (rs17489363 G > A), two in exon (rs2229571 G > C and rs3738888 C > T), and four in intron (rs3768716 A > G, rs6435862 T > G, rs3768707 C > T and rs17487792 C > T). However, the relationship between different haplotypes and NB risk were not detected (data not shown).
Table 3.
Associations between BARD1 SNPs and NB Risk
| Cases(N=339) | Controls(N=778) | P | Adjusted ORa (95%CI) | ||||
|---|---|---|---|---|---|---|---|
| No. | (%) | No. | (%) | ||||
| rs17489363 | |||||||
| GG | 206 | 61.0 | 529 | 68.9 | 1.00 | ||
| GA | 114 | 33.7 | 217 | 28.3 | 0.03 | 1.26 (0.95-1.68) | |
| AA | 18 | 5.3 | 22 | 2.9 | 0.035 | 2.04 (1.06-3.92) | |
| rs34732883 | |||||||
| TT | 197 | 61.2 | 504 | 68.3 | 1.00 | ||
| CT | 109 | 33.9 | 212 | 28.7 | 0.082 | 1.24(0.93-1.66) | |
| CC | 16 | 5.0 | 22 | 3.0 | 0.088 | 1.81(0.93-3.56) | |
| rs1129804 | |||||||
| CC | 135 | 39.8 | 302 | 39.8 | 1.00 | ||
| CG | 163 | 48.1 | 254 | 46.6 | 0.84 | 1.08 (0.81-1.43) | |
| GG | 42 | 12.1 | 103 | 13.6 | 0.78 | 0.98(0.64-1.51) | |
| rs2229571 | |||||||
| CC | 67 | 20.0 | 232 | 30.1 | 1.00 | ||
| CG | 164 | 49.0 | 370 | 48 | <0.0001 | 1.74 (1.24-2.44) | |
| GG | 104 | 31.0 | 169 | 21.9 | <0.0001 | 1.84 (1.37-2.49) | |
| rs3738888 | |||||||
| CC | 323 | 95.3 | 749 | 97.7 | 1.00 | ||
| CT | 16 | 4.7 | 18 | 2.4 | 0.041 | 2.17 (1.04-4.50) | |
| TT | - | - | - | - | - | - | - |
| rs1048108 | |||||||
| CC | 147 | 43.6 | 307 | 39.8 | 1.00 | ||
| CT | 154 | 45.7 | 369 | 47.9 | 0.60 | 0.93(0.70-1.23) | |
| TT | 36 | 10.7 | 95 | 12.3 | 0.39 | 0.83(0.55-1.27) | |
| rs3768716 | |||||||
| AA | 211 | 63.0 | 555 | 71.8 | 1.00 | ||
| AG | 111 | 33.1 | 200 | 25.9 | 0.014 | 1.49 (1.12-1.99) | |
| GG | 13 | 3.9 | 18 | 2.3 | 0.26 | 1.53 (0.73-3.21) | |
| rs6435862 | |||||||
| TT | 232 | 68.8 | 594 | 76.9 | 1.00 | ||
| GT | 96 | 28.5 | 168 | 21.8 | 0.029 | 1.40(1.04-1.90) | |
| GG | 9 | 2.7 | 10 | 1.3 | 0.13 | 2.05(0.81-5.18) | |
| rs3768707 | |||||||
| CC | 183 | 54.6 | 499 | 64.4 | 1.00 | ||
| CT | 132 | 39.4 | 242 | 31.4 | 0.013 | 1.45(1.10-1.92) | |
| TT | 20 | 6.0 | 31 | 4.0 | 0.18 | 1.73(0.95-3.15) | |
| rs17487792 | |||||||
| CC | 209 | 64.7 | 545 | 72.4 | 1.00 | ||
| CT | 104 | 32.2 | 192 | 25.5 | 0.04 | 1.45(1.08-1.95) | |
| TT | 10 | 3.1 | 16 | 2.1 | 0.52 | 1.31(0.58-2.95) | |
| rs7587476 | |||||||
| CC | 179 | 54.9 | 343 | 59.5 | 1.00 | ||
| CT | 126 | 38.6 | 206 | 35.8 | 0.59 | 1.04(0.77-1.41) | |
| TT | 21 | 6.4 | 27 | 4.7 | 0.32 | 1.37(0.74-2.56) | |
aData were calculated by logistic regression analysis with adjustment for sex and age as covariate
Stratification analysis of identified SNPs
For further estimating the association between seven identified SNPs and NB, we stratified the cases according to tumor categories, tumor stages and primary tumor sites. As shown in Supplementary Table 1, six SNPs were associated with malignant type GNB/NB rather than mature type GN. With regards to tumor stages, NB in stage III was related to rs3768707 C > T and rs3768716 A > G, while NB in stage IV was related to rs2229571 C > G, rs6435862 T > G, rs17487792 C > T and rs3768716 A > G. Based on tumor location, all the SNPs showed significant association with primary NB located in adrenal. These results indicated that the seven SNPs in BARD1 might contribute to NB aggressiveness.
Effect of identified SNPs on BARD1 transcriptional activity
We have confirmed that rs17489363 G > A in 5'-UTR was a risk SNP and it had strong LD with rs34732883 T > C and rs1129804 C > G, therefore the potential haplotypes should be considered in BARD1 transcriptional activity. Thus, the Dual-luciferase reporter assay was performed. Compared to reporter gene expressions driven by the rs17489363 G allele BARD1 promoters, all of rs17489363 A allele-containing counterparts showed 1.2- to 4- fold lower transcriptional activity both in 293T and SH-SY5Y cells (P < 0.05, Figure 2B & 2C). The results suggested that rs17489363 G > A may play a major function on BARD1 promoter activity in these haplotypes.
The function of SNPs in BARD1 exon
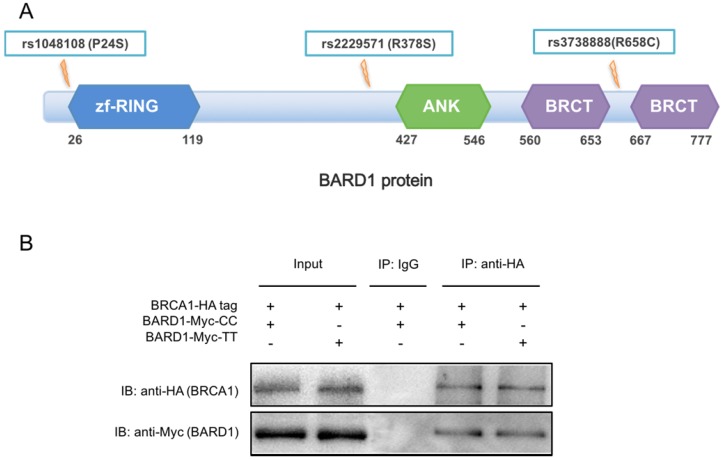
To explore the function of SNPs in BARD1 exon, we first overviewed the protein structure of BARD1. According to UniProt database (http://www.uniprot.org/), BARD1 had a BRCA1-interaction region (M26-E119), three ANK repeats (R427-P546) and two BRCT1 domains (M560-I653 and L667 -S777) (Figure 3A). The main function of BARD1 was to interact with BRCA1 via the zinc finger ring region and rs1048108 C > T (P24S) located near the domain. Moreover, rs1048108 had prominent LD with rs2229571 (D'=0.9, r2=0.7), suggesting potential correlating between these two SNPs (Supplementary Figure 1). Even rs2229571 (R378S) and rs3738888 (R658C) were risk SNPs, their sites were away from the functional domain. Thus, we chose rs1048108 C > T as the representative SNP for functional study of BARD1 protein. Therefore, we performed Co-IP to assess if rs1048108 could directly affect BARD1-BRCA1 interaction. As shown in Figure 3B, BARD1 protein containing rs1048108 T allele did not show any difference in BRCA1 binding capacity compared to the protein containing the rs1048108 C allele. This Co-IP result suggested that rs1048108 C > T (P24S) has no detectable influence on the BARD1/BRCA1 heterodimer.
Figure 3.
(A) The BARD1 structure. The amino acid mutation caused by rs1048108, rs2229571 and rs3738888 in BARD1 were indicated. Rs1048108 (P24S) located near zinc finger ring region, which played a key role in the interaction of BARD1 and BRCA1. (B) Effect of the rs1048108 C or T allele on BARD1-BRCA1 interaction. Co-IP result showed no difference in BARD1-BRCA1 interaction between wild type and mutant type of rs1048108 C > T (P24S).
Discussion
Neuroblastoma is the most common pediatric extracranial solid tumor and causes increasing concern on pediatrics due to its malignancy. Several studies have revealed that genetic factors could raise NB risk 9-15. BARD1, as an important tumor suppressor gene 23, has been verified significantly associated with NB susceptibility in different ethnicities 11, 16, 17, 24. However, the significance of BARD1 SNPs in Chinese population is far from clear. Thus, the present study was performed to screen significant SNPs in the whole length of BARD1 in Chinese population. Herein, seven risk SNPs for NB were identified and four rs17489363 A allele-containing haplotypes (composed with rs34732883 T > C and rs1129804 C > G) were demonstrated to affect transcriptional activity of BARD1 gene.
The seven identified SNPs included one in 5'-UTR (rs17489363 G > A), two in exon (rs2229571 G > C and rs3738888 C > T), and four in intron (rs3768716 A > G, rs6435862 T > G, rs3768707 C > T and rs17487792 C > T). Previous studies have demonstrated that rs17489363, rs3768716 and rs6435862 contributed to NB risk 11, 16, 17, 24. The study in South Chinese population reported that subjects carrying risk alleles of rs3768716 or rs6435862 had a significant trend on NB developing in clinical stage III/IV patients 18. However, the significant association was failed to be found in total subjects. In our study, significant association appeared in both total subjects and stratification analysis, the larger sample size might contribute to the difference (339 cases and 778 controls in our study vs 201 cases and 501 controls in the study on South Chinese population). Consistent with our results, Capasso et al. also elicited that rs3768716 and rs6435862 polymorphisms were the most significant variants that associated with NB risk in European ancestry 11. What's important is that a novel SNP rs3738888 was identified associated with NB risk in our study, which was not reported before in European, American African, Italian or Chinese 11, 16, 17, 24.
Functional studies of identified SNPs were further analyzed according to their locations. Among the seven identified SNPs, SNP rs17489363 G > A was located in 5' -UTR of BARD1. By using in silico approach, previous study found that rs17489363 might alter the transcriptional binding site of BARD1 25, and the latest study clarified this perspective in vitro 24. In the present study, we further confirmed the influence of rs17489363 on BARD1 transcriptional activity. Due to rs17489363 was in strong LD with rs34732883 and rs1129804 and LD represented correlating genotypes between these SNPs, the functions of different genotypes should be concerned. Compared to rs17489363 G allele-containing counterparts, the four rs17489363 A allele-containing haplotypes (composed with rs34732883 T > C and rs1129804 C > G) were shown to reduce the transcriptional activity of BARD1 gene. As a result, haplotypes containing rs17489363 A might play a key role in BARD1 promoter activity.
The BARD1's function is also related to protein structure. BARD1 interacts with BRCA1 via the zinc finger ring region 26-28 and to enhance the stability of BRCA1 29. According to UniProt database, rs1048108 C > T (P24S) located near the domain. Even rs2229571 (R378S) and rs3738888 (R658C) were found associated with NB susceptibility, their sites were away from the interaction region. Moreover, rs1048108 C > T is in prominent LD with rs2229571 G > C, which suggested potential correlating between these two SNPs. However, our result suggested that rs1048108 C > T (P24S) has no detectable influence on the BARD1/BRCA1 heterodimer.
For SNPs located in intron region, four SNPs (rs3768716 A > G, rs6435862 T > G, rs3768707 C > T and rs17487792 C > T) were significantly associated with NB susceptibility. The same results were also obtained in European ancestry 11. Moreover, previous study has reported that NB cells with the rs6435862 G risk allele could significantly increase the expression of one of BARD1 splice variant, BARD1β 30. Since the splice variant BARD1β was an oncogenic driver of high-risk NB tumorigenesis, the G risk allele may participant in the development of NB risk through increasing BARD1β expression 30.
Several limitations of this study should be mentioned. Firstly, all of the participants were recruited from Beijing Children's Hospital, including 339 cases and 778 controls. However, most of our patients were lived in the North of China. Although we have adjusted age and gender as the concomitant variable, we couldn't ignore the existent bias of participants' born places. Secondly, as there were eight haplotypes composed with rs34732883, rs17489363 and 1129804, the sample size of per group was small. Thus, the statistical power maybe limited. Therefore, we observed strong effects on BARD1 transcriptional activity in vitro, but found weak relationship between haplotypes and NB risk in SNP analysis. Thirdly, we did not complete 3-years' follow-up for monitoring far-reaching influence. In our future researches, both biological and clinical approaches would be considered and the sample size would be enlarged.
Conclusion
In conclusion, our study revealed the relationship between identified SNPs in BARD1 and NB susceptibility by systematically whole genetic scanning in Chinese population. Seven identified SNPs in BARD1 significantly contributed to NB risk. In addition, four rs17489363 A allele-containing haplotypes in 5'-UTR could reduce the promoter activity of BARD1 in vitro. These findings have provided novel clues to investigate the genetic mechanism of NB.
Supplementary Material
Supplementary figures and tables.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 31401067, 81472369, 81502144), Beijing Health System Top Level Technical Personnel Training Plan (NO. 20153079).
References
- 1.Maris J M, Hogarty M D, Bagatell R. et al. Neuroblastoma. Lancet. 2007;369:2106–20. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 2.London W B, Castleberry R P, Matthay K K. et al. Evidence for an age cutoff greater than 365 days for neuroblastoma risk group stratification in the Children's Oncology Group. J Clin Oncol. 2005;23:6459–65. doi: 10.1200/JCO.2005.05.571. [DOI] [PubMed] [Google Scholar]
- 3.Deyell R J, Attiyeh E F. Advances in the understanding of constitutional and somatic genomic alterations in neuroblastoma. Cancer Genet. 2011;204:113–21. doi: 10.1016/j.cancergen.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Maris J M. Recent advances in neuroblastoma. N Engl J Med. 2010;362:2202–11. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu A L, Gilman A L, Ozkaynak M F. et al. Anti-GD2 antibody with GM-CSF, interleukin-2, and isotretinoin for neuroblastoma. N Engl J Med. 2010;363:1324–34. doi: 10.1056/NEJMoa0911123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matthay K K, Maris J M, Schleiermacher G. et al. Neuroblastoma. Nat Rev Dis Primers. 2016;2:16078. doi: 10.1038/nrdp.2016.78. [DOI] [PubMed] [Google Scholar]
- 7.Capasso M, Diskin S J. Genetics and genomics of neuroblastoma. Cancer Treat Res. 2010;155:65–84. doi: 10.1007/978-1-4419-6033-7_4. [DOI] [PubMed] [Google Scholar]
- 8.Pandey J P. Genomewide association studies and assessment of risk of disease. N Engl J Med. 2010;363:2076–7. doi: 10.1056/NEJMc1010310. author reply 2077. [DOI] [PubMed] [Google Scholar]
- 9.Mosse Y P, Laudenslager M, Longo L. et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–5. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maris J M, Mosse Y P, Bradfield J P. et al. Chromosome 6p22 locus associated with clinically aggressive neuroblastoma. N Engl J Med. 2008;358:2585–93. doi: 10.1056/NEJMoa0708698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Capasso M, Devoto M, Hou C. et al. Common variations in BARD1 influence susceptibility to high-risk neuroblastoma. Nat Genet. 2009;41:718–23. doi: 10.1038/ng.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen le B, Diskin S J, Capasso M. et al. Phenotype restricted genome-wide association study using a gene-centric approach identifies three low-risk neuroblastoma susceptibility Loci. PLoS Genet. 2011;7(e):1002026. doi: 10.1371/journal.pgen.1002026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang K, Diskin S J, Zhang H. et al. Integrative genomics identifies LMO1 as a neuroblastoma oncogene. Nature. 2011;469:216–20. doi: 10.1038/nature09609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diskin S J, Capasso M, Schnepp R W. et al. Common variation at 6q16 within HACE1 and LIN28B influences susceptibility to neuroblastoma. Nat Genet. 2012;44:1126–30. doi: 10.1038/ng.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Capasso M, Diskin S, Cimmino F. et al. Common genetic variants in NEFL influence gene expression and neuroblastoma risk. Cancer Res. 2014;74:6913–24. doi: 10.1158/0008-5472.CAN-14-0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Capasso M, Diskin S J, Totaro F. et al. Replication of GWAS-identified neuroblastoma risk loci strengthens the role of BARD1 and affirms the cumulative effect of genetic variations on disease susceptibility. Carcinogenesis. 2013;34:605–11. doi: 10.1093/carcin/bgs380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Latorre V, Diskin S J, Diamond M A. et al. Replication of neuroblastoma SNP association at the BARD1 locus in African-Americans. Cancer Epidemiol Biomarkers Prev. 2012;21:658–63. doi: 10.1158/1055-9965.EPI-11-0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang R, Zou Y, Zhu J. et al. The Association between GWAS-identified BARD1 Gene SNPs and Neuroblastoma Susceptibility in a Southern Chinese Population. Int J Med Sci. 2016;13:133–8. doi: 10.7150/ijms.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimada H, Ambros I M, Dehner L P. et al. Terminology and morphologic criteria of neuroblastic tumors: recommendations by the International Neuroblastoma Pathology Committee. Cancer. 1999;86:349–63. [PubMed] [Google Scholar]
- 20.Gabriel S, Ziaugra L, Tabbaa D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr Protoc Hum Genet; 2009. Chapter 2:Unit 2.12. [DOI] [PubMed] [Google Scholar]
- 21.Barrett J C, Fry B, Maller J. et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 22.Al-Chalabi A. Genome-wide association studies. Cold Spring Harb Protoc. 2009;2009:pdb.top66. doi: 10.1101/pdb.top66. [DOI] [PubMed] [Google Scholar]
- 23.Cimmino F, Formicola D, Capasso M. Dualistic Role of BARD1 in Cancer. Genes (Basel); 2017. p. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.M A, SJ D, Z V, Fine mapping of 2q35 high-risk neuroblastoma locus reveals independent functional risk variants and suggests full-length BARD1 as tumor-suppressor.%A Cimmino F. International journal of cancer; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alshatwi A A, Hasan T N, Syed N A. et al. Identification of functional SNPs in BARD1 gene and in silico analysis of damaging SNPs: based on data procured from dbSNP database. PLoS One. 2012;7(e):43939. doi: 10.1371/journal.pone.0043939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu L C, Wang Z W, Tsan J T. et al. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat Genet. 1996;14:430–40. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- 27.Thai T H, Du F, Tsan J T. et al. Mutations in the BRCA1-associated RING domain (BARD1) gene in primary breast, ovarian and uterine cancers. Hum Mol Genet. 1998;7:195–202. doi: 10.1093/hmg/7.2.195. [DOI] [PubMed] [Google Scholar]
- 28.Fabbro M, Savage K, Hobson K. et al. BRCA1-BARD1 complexes are required for p53Ser-15 phosphorylation and a G1/S arrest following ionizing radiation-induced DNA damage. J Biol Chem. 2004;279:31251–8. doi: 10.1074/jbc.M405372200. [DOI] [PubMed] [Google Scholar]
- 29.Fox D 3rd, Le Trong I, Rajagopal P. et al. Crystal structure of the BARD1 ankyrin repeat domain and its functional consequences. J Biol Chem. 2008;283:21179–86. doi: 10.1074/jbc.M802333200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bosse K R, Diskin S J, Cole K A. et al. Common variation at BARD1 results in the expression of an oncogenic isoform that influences neuroblastoma susceptibility and oncogenicity. Cancer Res. 2012;72:2068–78. doi: 10.1158/0008-5472.CAN-11-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables.