Abstract
Background: A subset of women who are co-infected with Human Immunodeficiency Virus type 1 (HIV) and Human papillomavirus (HPV), progress rapidly to invasive cervical cancer regardless of antiretroviral therapy (ART) or immune status. We posit that HIV/HPV co-infection along with specific host HLA II -DRB1 and -DQB1 alleles play a major role in cervical cancer development.
Methodology: We conducted a hospital-based genetic susceptibility case-control study in Cape Town, South Africa. We recruited 256 women of the same race, from which a total of 624 HLA-DRB1 and -DQB1 class II genotypes were studied. We characterized HLA II candidate genes using PCR based, Luminex intermediate resolution genotyping and confirmed significant associated genotypes at four-digit resolution by high resolution gel typing. We analyzed 160 alleles from cancer, 64 alleles from pre-cancer and 400 alleles from healthy control women. Whole blood was used for HIV antibody test and HLA II typing. Cervical tumor tissue biopsies were used for HPV genotyping. Tests were statistically significant if p<0.05.
Results: Women who were co-infected with HIV/HPV had advanced cervical disease compared to women who were HIV negative. HLA class II -DQB1*03:01 and -DQB1*06:02 alleles were associated with cervical cancer in HIV/HPV co-infected women (p=0.001 and p<0.0001, respectively) while HLA class II -DRB1*13:01 and -DQB1*03:19 were rare or absent in women with cervical disease when compared to the control population (p=0.012 and 0.011, respectively).
Conclusion: We describe associations between HLA class II genotypes with cervical cancer, or likely protection from cervical cancer disease in HIV/HPV co-infected South African women. Identifying mechanisms that give rise to this likely protective HLA association will provide insight into development of immune-based prevention measures.
Keywords: HIV/HPV co-infection, HLA II allele association, cervical cancer susceptibility
Introduction
Each year, cervical cancer accounts for 528,000 new cases and 266,000 deaths worldwide 1. Despite the recent implementation of human papillomavirus (HPV) vaccine programs, cervical cancer still remains the most common cause of cancer-related mortality in women in Sub-Saharan Africa 2. Approximately ninety-nine percent of all cervical cancer cases are associated with persistent infection with oncogenic HPV, which is a causative organism for cervical cancer 3. Furthermore, women with Human Immunodeficiency Virus type 1 (HIV) infection are more likely to have a concurrent persistence of HPV infection than women who are not HIV positive 3. However, only a small subset of HIV/HPV co-infected women will develop invasive cervical cancer 4. This is regardless of HIV suppression with antiretroviral therapy (ART) or with high CD4 cell count 5.
Although associations between HPV infection and Human Leukocyte Antigen (HLA) genotypes have previously been described 6, the relationship between HIV/HPV co-infection, class II HLA and cervical cancer development has not been reported 7. It is also unclear what the immune mechanism would be 8. It is well established that T cell recognition of virally infected cells works through class I restricted epitope recognition and down-regulation of the viral peptide-HLA complex on the infected cell may lead to protection from cytotoxic T cells 9. This would impair the ability of the individual to clear virus and thus allow HPV infection to persist 10. Moreover, apart from the frequent overexpression of HPV E6 and E7 viral oncoproteins in cervical cancer, HPV16 E5 protein has been shown to interfere with both HLA Class I and II antigen presentation 11. This suggests that the HPV may inhibit CD4+ helper T-cells recognition through down-regulation of HLA class II molecules and thereby evade host immunity. This effect might be exacerbated by the added effect of HIV co-infection 12. HIV Tat proteins can directly interact with the pRb/p130/p107/p53 tumor suppressor genes and induce increased cell proliferation which may promote the effect of HPV oncoproteins E6 and E7 in the rapid cervical carcinogenesis 13,14.
The etiology of cervical cancer has been related to certain high-risk HLA class II genes 15. Polymorphisms in HLA-DRB1 and -DQB1 genes are hypothesized to play a role in carcinogenesis of cervical cancer 6. Despite considerable scientific interest, findings of different published studies have been inconsistent 10,16,17. Several studies have reported the protective effects of HLA-DRB1*13:01-DQB1*06:03 haplotypes on cervical cancer development and a positive association between HLA -DQB1*03:02 genes and cervical disease progression. However, this has not consistently been found in different population groups 18,19,20.
This study seeks to determine whether host HLA-DRB1 and -DQB1 backgrounds in HIV/HPV co-infected South African women, influence cervical cancer disease development. Our results add new knowledge to the existing theories of rapid cervical cancer progression in HIV positive women and aim to further inform on individualized host-directed cervical cancer prevention.
Methods
Research ethics
Ethics approval was obtained from the Human Research Ethics Committee of the University of Cape Town (HREC903/2015) and the National Health Laboratory Service (NHLS). Study approval was also obtained from the Western Cape provincial Department of Health.
Sample size, study population and selection criteria
We recruited a total of 256 Black South African women in a hospital-based genetic susceptibility case-control study conducted at the Groote Schuur Hospital, Cape Town, in the Western Cape province of South Africa. The recruitment processes were conducted from June 2016 to March 2017. The race of cases and the controls were the same in order to reduce the bias due to the diversity of HLA II genes within South Africa 21. We categorized our cases and controls as 'Black African' in terms of South Africa's five official population categories similar to a previous study 22. We sought consent from women who attended the outpatient Gynaecological cancer assessment clinic, the colposcopy clinic or who were admitted to the Gynaecology emergency room, to participate in the study. Our control group consisted of age-matched healthy women from the same study population. The control group was compiled from historical data derived from 200 archived records at the NHLS, Laboratory for Tissue Immunology at the Groote Schuur Hospital. We compiled data from unrelated and related bone marrow, renal and other organ transplants age-matched female donors, which represented the background population for the HLA class II genes. These HLA typing results from the control group were counter checked by a second experienced person in the laboratory in order to confirm the allele genotypes, sex, age and race as “Black South African” women.
Specimen collection
We used written and signed consent forms in the language of the subject's choice in front of a witness, and after detailed discussion with patients regarding the aims and nature of the study. A trained registered nurse who was fluent in the local languages explained the details of the study and answered questions from the patients before their consent was requested. Peripheral blood (8ml) was collected using two EDTA tubes (BD Vacutainer®, South Africa) for genomic DNA isolation and HLA typing. After cervical visual inspection, Gynecologists collected punch biopsies of abnormal cervical lesions. A small part of the tissue biopsy was preserved in Digene® specimen transport medium (Qiagen, South Africa) and then stored at -800C until processed in order to preserve the genomic DNA for HPV genotyping. Another part of the same biopsy was stored in formalin and sent to the laboratory for histopathology analyses at the NHLS Anatomical Pathology Laboratory similar to a published study 23. All patients were recruited in this study before the initiation of the radiotherapy or chemotherapy cancer treatment in order to avoid DNA damage during cancer therapy 24.
HIV antibody test
According to the South African HIV-testing algorithm, 20 µl of the collected peripheral whole blood was used for rapid HIV antibody testing (Determine, Alere, Inc.) for all cases 25. Whereas, for the control group, a retrospective follow-up was done on the NHLS bone marrow registry and organ transplant database for HIV status. In this study population, all women in the control group were HIV negative.
HPV DNA genotyping and detection
Genomic DNA was extracted from cervical tumor tissue specimens using Qiagen ® QIAamp DNA Mini purification kit (Qiagen, South Africa) according to a protocol from the manufacturer. The concentration of the extracted DNA was quantified by a Nanodrop® spectrophotometer. Due to high genomic DNA concentration from the tissue biopsies, the DNA was diluted using nuclease- free water (Thermo Fisher, South Africa) to reach a recommended final concentration of 20ng/100 µl of the total DNA. Then the HPV genotyping and detection tests were performed by using Linear array® PCR based HPV genotyping kit (Roche, South Africa). The HPV status was only genotyped for cases according to availability of tissue specimens in this study.
HLA Typing
Whole blood collected from patients was used to extract genomic DNA by using protocol for the modified salting out blood DNA extraction method 26. A Nanodrop® spectrophotometer was used to measure the concentration and purity of the extracted DNA using the A260/280 ratio to be at least 1.8 to 1.9. The DNA was further diluted using nuclease free water (Thermo Fisher, South Africa) to reach a recommended final concentration of 30ng/100µl. We used intermediate resolution by commercial kits using the Luminex platform for reverse sequence-specific oligonucleotide primers (SSOP) HLA typing technology (Immucor, England) 27. The PCR was conducted by multiple thermal cyclers using both PCR-SSOP and PCR-sequence-specific primers (SSP) supplied by the manufacturer in the HLA II typing kit. Amplification reactions were done according to manufacturer instructions. The HLA II allele results were analyzed by HLA II data analysis software program Match It ® 28. We further confirmed the HLA II genotypes that gave significant associations with Olerup® SSP (Immucor Inc. South Africa) HLA II high resolution gel typing as described by manufacturer's protocol.
Statistical analyses
We analyzed a total of 624 *DRB1 *DQB1 HLA II genes. We characterized 160 alleles from cancer patients, 64 alleles from pre-cancer patients and 400 alleles from unrelated age-matched women in the healthy control group. Allele frequencies for HLA-DRB1and HLA-DQB1 were calculated by direct counting similar to a published study 29. The observed genotype frequencies in the controls were tested for Hardy-Weinberg equilibrium. The HLA allele frequencies for DRB1 and DQB1 alleles were compared between the cases and controls using the Fisher's exact test with 2 × 2 tables or, where appropriate, by the χ2 test with Mantel-Haenszel correction. The Bonferroni correction for multiple tests was not required due to the nature of our study 30. The odds ratios (ORs), 95% confidence intervals (95% CIs) and the p-values calculated for multiple comparisons were considered significant at p<0.05.
Results
Table 1 shows that, without consideration of HIV co-infection status, in terms of cervical cancer susceptibility, HLA-DQB1*03:01 (p=0.002, OR 4.66, 95% CI 1.85-11.72) and HLA-DQB1*06:02 (p=0.002, OR 2.68, 95% CI 1.45-4.96) were significantly associated with cervical cancer disease in cases compared to the healthy control group. Furthermore, HLA-DRB1*13:01 (p=0.012, OR 0.18, 95% CI 0.02-0.75) and HLA-DQB1*03:19 (p=0.011, OR 0.11, 95% CI 0.003-0.74) were found to be significantly low or absent for the relative risk of cervical disease development, suggestive of protective alleles.
Table 1.
Relative risk for cervical cancer development with specific DRB1and DQB1 allele exposure without consideration of HIV co-infection status.
| Relative Risk for pre-cancer | Relative Risk for cancer | ||||||
|---|---|---|---|---|---|---|---|
| HLA DRB1 | Cancer Allele frequency (2n=83) |
Pre-cancer Allele frequency (2n= 33) |
Healthy controls (Allele frequency) (2n=205) |
Odds ratio (95%CI) | P-value | Odds ratio (95%CI) | P-value |
| *01:01 | 0.024 (2) | 0.03 (1) | 0.024 (5) | 1.26(0.03-11.79) | 0.59 | 1(0.19-5.26) | >0.999 |
| *01:02 | 0.1 (8) | 0.121 (4) | 0.05 (11) | 2.45(0.5-9.0) | 0.135 | 1.91(0.6-5.4) | 0.193 |
| *03:01 | 0.072 (6) | 0.09 (3) | 0.073 (15) | 1.28(0.22-4.92) | 0.721 | 1(0.31-2.86) | >0.999 |
| *03:02 | 0.072 (6) | 0.15 (5) | 0.093 (19) | 1.8(0.47-5.43) | 0.345 | 0.77(0.24-2.12) | 0.817 |
| *04:01 | 0.036 (3) | 0 | 0.024 (5) | N/A | N/A | 1.52(0.23-8.01) | 0.693 |
| *04:04 | 0 | 0.09 (3) | 0.02 (4) | 5.07(0.7-31.3) | 0.057 | N/A | N/A |
| *04:05 | 0 | 0.03 (1) | 0.005 (1) | 6.42(0.08-506) | 0.257 | N/A | N/A |
| *07:01 | 0.084 (7) | 0.06 (2) | 0.07 (14) | 0.86(0.09-4.17) | >0.999 | 1.27(0.42-3.54) | 0.62 |
| *08:01 | 0.012 (1) | 0.03 (1) | 0.005 (1) | 6.4(0.1-506) | 0.257 | 2.5(0.03-198.7) | 0.491 |
| *08:04 | 0.012 (1) | 0.06 (2) | 0.04 (8) | 1.6(0.3-7.9) | 0.632 | 0.3(0.01-2.34) | 0.454 |
| *09:01 | 0.036 (3) | 0.03 (1) | 0.01 (2) | 3.2(0.1-62.6) | 0.361 | 3.86(0.43-46.75) | 0.143 |
| *10:01 | 0.06 (5) | 0 | 0.024 (5) | N/A | N/A | 2.6(0.58-11.6) | 0.155 |
| *11:01 | 0.1 (8) | 0.09 (3) | 0.112 (23) | 0.8(0.14-2.9) | >0.999 | 0.86(0.32-2.1) | 0.835 |
| *11:02 | 0.084 (7) | 0.03 (1) | 0.08 (16) | 0.37(0.01-2.6) | 0.48 | 1.1(0.37-2.98) | 0.813 |
| *11:04 | 0.024 (2) | 0 | 0.005 (1) | N/A | N/A | 5.1(0.26-302.5) | 0.197 |
| *12:01 | 0.036 (3) | 0.06 (2) | 0.024 (5) | 2.6(0.24-16.69) | 0.249 | 1.52(0.23-8.01) | 0.693 |
| *13:01 | 0.024 (2) | 0 | 0.122 (25) | N/A | N/A | 0.18(0.02-0.75) | 0.012 |
| *13:02 | 0.036 (3) | 0.09 (3) | 0.07 (14) | 1.4(0.2-5.3) | 0.713 | 0.5(0.1-1.9) | 0.411 |
| *13:03 | 0.072 (6) | 0 | 0.03 (7) | N/A | N/A | 2.2(0.6-7.9) | 0.208 |
| *14:01 | 0.024 (2) | 0 | 0.01 (2) | N/A | N/A | 2.5(0.18-35.45) | 0.323 |
| *15:01 | 0.05 (4) | 0.03 (1) | 0.024 (5) | 1.26(0.03-11.79) | 0.594 | 2.05(0.4-9.79) | 0.281 |
| *15:03 | 0.05 (4) | 0 | 0.083 (17) | N/A | N/A | 0.57(0.13-1.82) | 0.452 |
|
HLA DQB1 |
(2n=77) | (2n=31) | (2n=195) | ||||
| *02:01 | 0.078 (6) | 0.065 (2) | 0.092 (18) | 0.67(0.07-3.06) | >0.999 | 0.82(0.26-2.27) | 0.815 |
| *02:02 | 0.13 (10) | 0.032 (1) | 0.154 (30) | 0.18(0.03-1.19) | 0.09 | 0.81(0.33-1.82) | 0.706 |
| *03:01 | 0.169 (13) | 0.063 (2) | 0.04 (8) | 1.6(0.16-8.54) | 0.632 | 4.66(1.85-11.72) | 0.002 |
| *03:02 | 0.04 (3) | 0.063 (2) | 0.056 (11) | 1.1(0.12-5.42) | 0.696 | 0.67(0.12-2.6) | 0.763 |
| *03:19 | 0.013 (1) | 0 | 0.1 (20) | N/A | N/A | 0.11(0.003-0.74) | 0.011 |
| *04:02 | 0.1 (8) | 0.129 (4) | 0.067 (13) | 2.1(0.45-7.3) | 0.265 | 1.6(0.55-4.36) | 0.322 |
| *04:04 | 0 | 0 | 0.005 (1) | N/A | N/A | N/A | N/A |
| *05:01 | 0.065 (5) | 0.194 (6) | 0.113 (22) | 1.87(0.56-5.34) | 0.24 | 0.54(0.15-1.54) | 0.268 |
| *05:03 | 0.026 (2) | 0 | 0.05 (10) | N/A | N/A | 0.49(0.05-2.27) | 0.519 |
| *06:02 | 0.324 (25) | 0.29 (9) | 0.149 (29) | 2.3(0.85-5.81) | 0.07 | 2.68(1.45-4.96) | 0.002 |
| *06:03 | 0.013 (1) | 0.032 (1) | 0.072 (14) | 0.43(0.01-3.02) | 0.7 | 0.17(0.004-1.15) | 0.075 |
| *06:04 | 0.013 (1) | 0 | 0.077 (15) | N/A | N/A | 0.16(0.004-1.05) | *0.046 |
| *06:06 | 0 | 0.097 (3) | 0 | N/A | N/A | N/A | N/A |
| *06:09 | 0.026 (2) | 0.032 (1) | 0.02 (4) | 1.58(0.03-16.6) | 0.527 | 1.26(0.11-8.96) | >0.999 |
*The significant p-value was disregarded because the confidence interval crosses '1'
Table 2 shows consideration of HIV co-infection status, whereby in HIV/HPV co-infected women who had cervical cancer, HLA-DQB1*03:01 (p=0.001, OR 5.6, 95% CI 1.9-16.9) and -DQB1*06:02 (p<0.0001, OR 4.5, 95% CI 2.2-9.0) were significantly associated with the risk of disease. However, HLA DRB1*13:01, HLA-DQB1*03:19 showed likely protection against cervical cancer due to completely absence or rare in both HIV/HPV co-infected and HIV negative cervical cancer population.
Table 2.
Relative risk for developing cervical cancer in HIV/HPV co-infected women with specific *DRB1 *DQB1 gene exposure. The cancers may be HIV negative but they are not HPV negative.
| HLA DRB1 |
HIV/HPV Co-infected cancer | HIV Negative cancer | Healthy Controls Allele frequency (2n=205) |
Relative Risk for HIV Negative cancer | Relative Risk for HIV/ HPV Co-infected cancer |
|||
|---|---|---|---|---|---|---|---|---|
| Allele frequency (2n=60) |
Allele frequency (2n=23) |
ODDS ratio(95%CI) | P-value | ODDS ratio(95%CI) | P-value | |||
| *01:01 | 0.017 (1) | 0.043 (1) | 0.024 (5) | 1.9(0.04-17.7) | 0.469 | 0.7(0.01-6.3) | >0.99 | |
| *01:02 | 0.1 (6) | 0.087 (2) | 0.05 (11) | 1.7(0.2-8.7) | 0.623 | 1.9(0.57-6.17) | 0.227 | |
| *03:01 | 0.05 (3) | 0.13 (3) | 0.073 (15) | 1.95(0.33-7.8) | 0.399 | 0.7(0.12-2.5) | 0.771 | |
| *03:02 | 0.05 (3) | 0.13 (3) | 0.093 (19) | 1.5(0.26-5.8) | 0.464 | 0.5(0.1-1.9) | 0.425 | |
| *04:01 | 0.05 (3) | 0 | 0.024 (5) | N/A | N/A | 2.1(0.32-11.3) | 0.384 | |
| *04:04 | 0 | 0 | 0.02 (4) | N/A | N/A | N/A | N/A | |
| *04:05 | 0 | 0 | 0.005 (1) | N/A | N/A | N/A | N/A | |
| *07:01 | 0.083 (5) | 0.087 (2) | 0.068 (14) | 1.33(0.14-6.5) | 0.663 | 1.3(0.34-3.9) | 0.775 | |
| *08:01 | 0.017 (1) | 0 | 0.005 (1) | N/A | N/A | 3.5(0.04-275.3) | 0.399 | |
| *08:04 | 0.017 (1) | 0 | 0.04 (8) | N/A | N/A | 0.4(0.01-3.3) | 0.689 | |
| *09:01 | 0.05 (3) | 0 | 0.01 (2) | N/A | N/A | 5.4(0.6-65.6) | 0.077 | |
| *10:01 | 0.083 (5) | 0 | 0.024 (5) | N/A | N/A | 3.7(0.81-16.5) | *0.049 | |
| *11:01 | 0.083 (5) | 0.13 (3) | 0.112 (23) | 1.22(0.21-4.6) | 0.728 | 0.7(0.21-2.1) | 0.638 | |
| *11:02 | 0.067 (4) | 0.13 (3) | 0.08 (16) | 1.8(0.31-7.2) | N/A | 0.9(0.2-2.8) | >0.99 | |
| *11:04 | 0.033 (2) | 0 | 0.005 (1) | N/A | N/A | 7.1(0.36-421.3) | 0.128 | |
| *12:01 | 0.017 (1) | 0.087 (2) | 0.024 (5) | 3.9(0.35-25.54) | 0.145 | 0.7(0.01-6.3) | >0.999 | |
| *13:01 | 0 | 0.087 (2) | 0.122 (25) | 0.7(0.1-3.2) | >0.99 | N/A | N/A | |
| *13:02 | 0.033 (2) | 0.043 (1) | 0.068 (14) | 0.6(0.01-4.5) | >0.99 | 0.5(0.1-2.15) | 0.537 | |
| *13:03 | 0.1 (6) | 0 | 0.034 (7) | N/A | N/A | 3.1(0.83-11.4) | 0.081 | |
| *14:01 | 0.017 (1) | 0.043 (1) | 0.01 (2) | 4.7(0.08-93) | 0.27 | 1.74(0.03-33.8) | 0.536 | |
| *15:01 | 0.067 (4) | 0 | 0.024 (5) | N/A | N/A | 2.9(0.55-13.9) | 0.118 | |
| *15:03 | 0.067 (4) | 0 | 0.083 (17) | N/A | N/A | 0.8(0.19-2.6) | >0.99 | |
|
HLA DQB1 |
(2n=56) |
(2n=21) |
(2n=195) |
|||||
| *02:01 | 0.036 (2) | 0.19 (4) | 0.092 (18) | 2.2(0.5-7.9) | 0.247 | 0.36(0.08-1.6) | 0.263 | |
| *02:02 | 0.125 (7) | 0.143 (3) | 0.154 (30) | 0.9(0.16-3.3) | >0.99 | 0.78(0.27-1.9) | 0.674 | |
| *03:01 | 0.196 (11) | 0.1 (2) | 0.041 (8) | 2.4(0.23-13.2) | 0.259 | 5.6(1.9-16.9) | 0.001 | |
| *03:02 | 0.036 (2) | 0.048 (1) | 0.056 (11) | 0.8(0.02-6.2) | >0.99 | 0.61(0.06-2.9) | 0.739 | |
| *03:19 | 0 | 0.048 (1) | 0.103 (20) | 0.4(0.01-2.9) | 0.702 | N/A | N/A | |
| *04:02 | 0.054 (3) | 0.238 (5) | 0.067 (13) | 4.2(1.3-13.3) | 0.022 | 0.8(0.14-3) | >0.99 | |
| *04:04 | 0 | 0 | 0.005 (1) | N/A | N/A | N/A | N/A | |
| *05:01 | 0.054 (3) | 0.1 (2) | 0.113 (22) | 0.8(0.1-3.7) | *0.017 | 0.44(0.1-1.6) | 0.218 | |
| *05:03 | 0.018 (1) | 0.048 (1) | 0.051(10) | 0.9(0.02-6.9) | >0.99 | 0.33(0.01-2.4) | 0.465 | |
| *06:02 | 0.446 (25) | 0 | 0.149 (29) | N/A | N/A | 4.5(2.2-9.0) | <0.0001 | |
| *06:03 | 0 | 0.048 (1) | 0.072(14) | 0.63(0.01-4.6) | >0.999 | N/A | N/A | |
| *06:04 | 0.018 (1) | 0 | 0.077 (15) | N/A | N/A | 0.22(0.01-1.47) | 0.131 | |
| *06:09 | 0.018 (1) | 0.048 (1) | 0.02 (4) | 2.3(0.05-24.9) | 0.41 | 0.86(0.02-8.9) | >0.99 | |
*The significant p-value was disregarded because the confidence interval crosses '1'.
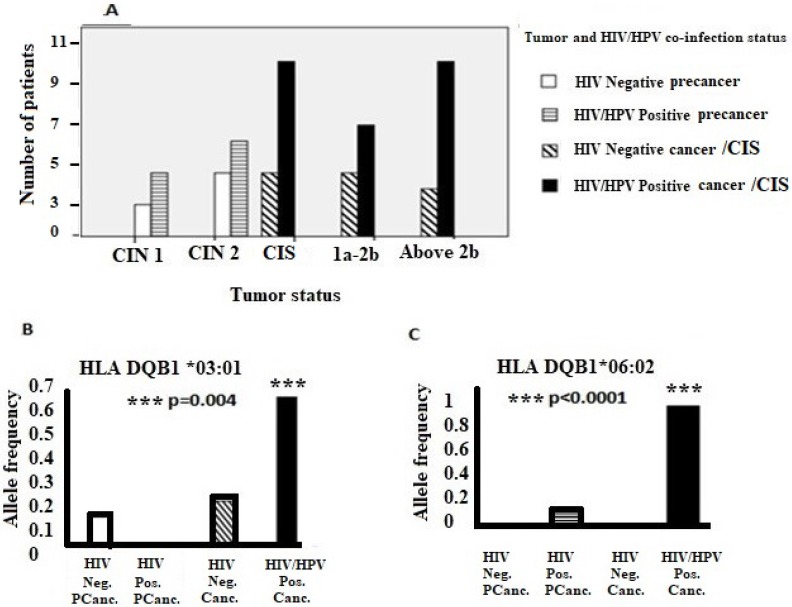
In addition, the majority of women who were co-infected with HIV/HPV had advanced cervical disease compared to women who were HIV negative. Moreover, alleles DQB1*03:01 and DQB1*06:02 were significantly associated with HIV/HPV-co-infected cervical cancer population. (Figure 1).
Figure 1.
Association of tumor status, specific HLA II alleles and HIV/HPV co-infection. (A) HIV/HPV co-infection, tumor status and the number of patients. (B) and (C) show associations of HLA-DQB1 *03:01 and HLA-DQB1 *06:02 alleles, respectively, in HIV/HPV co-infected cervical cancer patients. Where; FIGO = International Federation of Obstetrics and Gynaecology; 1a-2b = FIGO cervical cancer stage 1a up to stage 2b; Above 2b = FIGO cervical cancer stage above stage 2b; CIN = Cervical intraepithelial neoplasia; 1= Mild, 2= Moderate; CIS = Carcinoma In situ; Neg. Pcanc.= Negative precancer; Pos. Pcanc. = Positive precancer; Neg. Canc. = Negative cancer;Pos. Canc. = Positive cancer.
Table 3 shows comparisons between intermediate, SSP and Olerup high resolution HLA II typing only for significant alleles. We confirmed that the susceptible alleles for cervical cancer disease in HIV/HPV co-infected women were DQB1*03:01:01:01 and DQB1*06:02:01:01.
Table 3.
Association of HLA II alleles with cervical cancer susceptibility, likely protection and HIV/HPV co-infection after confirmation with high resolution gel typing.
| Cervical cancer and HLA II alleles | |||||||
|
Significant
alleles |
Luminex intermediate HLA typing |
SSP high
resolution HLA typing |
Olerup high
resolution HLA typing |
Cancer Patients
*(2n=83) |
Healthy
controls *(2n=205) |
Odds ratio (95% CI) | p-value |
| Susceptibility | DQB1*03:01 | DQB1*03:01 | DQB1*03:01:01:01 | 0.157 (13) | 0.039 (8) | 4.66(1.85-11.72) | 0.002 |
| DQB1*06:02 | DQB1*06:02 | DQB1*06:02:01:01 | 0.301(25) | 0.141(29) | 2.68(1.38-5.17) | 0.002 | |
| Likely protection | DRB1*13:01 | DRB1*13:01 | DRB1*13:01:01:01 | 0.024(2) | 0.122(25) | 0.18(0.02-0.75) | 0.012 |
| DQB1*03:19 | DQB1*03:19 | DQB1*03:19:01 | 0.012(1) | 0.098(20) | 0.11(0.003-0.74) | 0.011 | |
| Cervical cancer, HIV/HPV co-infection and HLA II alleles | |||||||
| Luminex intermediate HLA typing |
SSP high resolution
HLA typing |
Olerup high
resolution HLA typing |
Cancer
Patients * (2n=83) |
Healthy
controls * (2n=205) |
Odds ratio (95% CI) |
p-value | |
| Susceptibility | DQB1*03:01 | DQB1*03:01 | DQB1*03:01:01:01 | 0.108 (9) | 0.039 (8) | 4.4(1.4-13.78) | 0.001 |
| DQB1*06:02 | DQB1*06:02 | DQB1*06:02:01:01 | 0.301(25) | 0.141(29) | 4.5(2.2-9.0) | <0.0001 | |
*Total number of alleles (N) from cancer patients and healthy controls do not add up because we are showing only significant alleles in HIV/HPV co-infected women. Where; SSP = sequence specific primers.
Discussion
This is the first study to comprehensively investigate the association of HLA class II -DRB1 and -DQB1 alleles, at the molecular level, with cervical cancer disease in HIV/HPV co-infected women. Other studies have reported the presence of HLA II alleles -DQB1*03:01 31, and -DQB1*06:02 32, and cervical cancer association, but in populations with unknown HIV status.
In our study, in a different population, we report a unique finding on the same HLA II alleles -DQB1*03:01 and -DQB1*06:02 association with HIV/HPV co-infection in women with cervical cancer. As HLA-II -DRB1*13:01 and -DQB1*03:19 were significantly absent or rare from women with cervical cancer, we may infer that these are likely protective alleles for cervical cancer development in our study population. This may suggest that individuals with HLA-DRB1*13:01 and -DQB1*03:19 alleles background, may effectively present the oncogenic HPV antigens to the immune system to provide a level of protective immunity. These findings are novel and have never been reported in cervical cancer disease studies in any population.
Studies done amongst different racial or ethnic groups of patients with unknown HIV status have shown the possible correlation of HLA II molecules with cervical cancer development 19,33. For example, studies done in Bolivia 34, Brazil 35, China 29 , Costa Rica 36, Mexico 30, Senegal 37, Tunisia 10 and Venezuela 38, have reported cervical cancer disease associations with different HLA II alleles and HPV infection but none of them reported about HIV infected patients in their study populations. In another study, however, Hu et al. 39 reported that HLA II- DQB1*06:02 was protective against cervical cancer progression in Chinese patients with unknown HIV status. The inconsistency in the results of other studies to date amongst different populations, including ours, suggests that there are other, as yet unknown, factors that influence cervical cancer disease development.
The persistence of oncogenic HPV infection frequently leads to cervical cancer development 36,10,40. Our results, however, add new evidence that presence of HIV/HPV co-infection in addition with specific class II -DRB1 and -DQB1 alleles polymorphisms may aggravate or protect against cervical cancer disease development in some women. HIV as a co-infection may not be a direct risk factor for cervical cancer disease, although HIV positive status has been shown to influence the relative risk for cervical disease development. This means that HPV infection alone may influence cervical cancer development to a significant extent. When it is combined with a specific HLA II gene of risk for cervical cancer development and in the presence of HIV as a co-infection, the combination may increase the relative risk for cervical cancer disease development.
The persistence of the HPV in some women with specific HLA II alleles nearly always leads to cervical cancer. This effect can be further exacerbated by the pro-oncogenic effects of HIV co-infection, regardless of ART initiation, although the exactly mechanism is not yet clear 7. However, de Jong et al 41, suggested that absence of functional HPV16-specific CD4+ T-cell immune response found in women with cervical cancer, may explain further development of the disease despite immune reconstitution following ART initiation or in a competent CD4+ T-cell count. We acknowledge that studies on the association of HLA class II alleles and cervical cancer progression or susceptibility show inconsistent results worldwide 31, 42,43,44. However, our findings show that HLA class II DRB1*13:01, DQB1*03:01, DQB1*03:19 and DQB1*06:02 alleles can modulate the cervical carcinogenesis in HIV/HPV co-infected women.
Our study did not have enough sample size for the cases in order to assess specific HPV types with cervical cancer risk according to HLA II alleles and HIV status. Hospital-based studies are likely to have some selection bias because we studied only referred women with HIV and cervical disease so they don't really represent HIV/HPV co-infected women in the general population who have been diagnosed but not referred to the hospital yet. We did not study detailed information on immunocompromised status (CD4 + cell counts, CD4% of lymphocytes, CD45 white cell counts), HIV and HPV viral loads, initiation, regime and duration of ART, and the time of acquisition of HIV or HPV co-infection. Furthermore, we may have missed the effects of window period in some of the cases because we used HIV antibody test which takes at least three months for the antibodies to be detected in the blood since infection. Retrospective HLA data for the control group could add another bias to our study.
Conclusions
For the first time, our study provides evidence on the association of HLA class II -DRB1, -DQB1 alleles with cervical cancer development amongst HIV/HPV co-infected 'South African Black' women. This is a novel association from a new study population on the possible influence of variations of HLA class II alleles and HIV/HPV co-infection on cervical cancer development. In view of our findings we suggest, it is highly likely that specific HLA class II -DQB1*03:01 and -DQB1*06:02 alleles in the presence of HIV/HPV co-infection influence cervical cancer disease development in “Black South African” women. Furthermore, HLA class II -DRB1*13:01 and -DQB1*03:19 were rare or absent in women with cervical disease when compared with the control population. Therefore, identifying mechanisms that give rise to this likely protective association of HLA II alleles will provide insight into development of immune-based prevention measures. Additional research with whole genomic loci for allele association studies may reveal new potential targets for individualized cervical cancer molecular prevention in HIV/HPV co-infected women. Furthermore, analyses of other immune-related genes and chromosomes out of the HLA-II region may also enlighten our understanding regarding cervical cancer disease progression in HIV/HPV co-infected women.
Acknowledgments
The authors would like to thank:
Miss Lerato Majara, from the Division of Human Genetics, Department of Pathology, Faculty of Health Sciences, University of Cape Town, South Africa.
Professor Tim Quinlan, from the Health Economics and HIV/AIDS Research Division (HEARD) at the University of KwaZulu-Natal, Durban, South Africa for his assistance in discussing and reviewing early drafts of this article.
The staff members of the National Health Laboratory Service, Laboratory for Tissue Immunology, at Groote Schuur Hospital, Cape Town, South Africa for their assistance in HLA typing and further confirmation of the HLA typing results from the control group allele genotypes, sex, age and race as “Black South African” women.
This work is based on the research supported in part by;
Postgraduate Academic Mobility for African Physician-Scientists (PAMAPS), organized by the Faculty of Medicine, University of Ibadan, Nigeria and funded by the European Union.
South African Medical Research Council, Clinical Gynaecological Cancer Research Centre, University of Cape Town, South Africa.
National Research Foundation of South Africa (Grant Number: 64815)
National Health Laboratory Service of South Africa (Grant Number: 004_94678)
Opinions, findings and conclusions or recommendations expressed in this study generated by the listed organizations supported research are that of the authors alone, and that the organizations accept no liability whatsoever in this regard.
References
- 1.Chambuso RS, Kaambo E, Stephan S. Observed Age Difference and Clinical Characteristics of Invasive Cervical Cancer Patients in Tanzania; A Comparison between HIV-Positive and HIV-Negative Women. Journal of Neoplasm; 2017. p. 2. (3) [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. International journal of cancer Journal international du cancer. 2015;136(5):E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Williamson A-L. The Interaction between Human Immunodeficiency Virus and Human Papillomaviruses in Heterosexuals in Africa. Journal of Clinical Medicine. 2015;4(4):579. doi: 10.3390/jcm4040579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chambuso RS, Shadrack S, Lidenge SJ, Mwakibete N, Medeiros RM. Influence of HIV/AIDS on Cervical Cancer: A Retrospective Study From Tanzania. Journal of Global Oncology; 2017. JGO002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denny L, Adewole I, Anorlu R, Dreyer G, Moodley M, Smith T. et al. Human papillomavirus prevalence and type distribution in invasive cervical cancer in sub-Saharan Africa. International journal of cancer. 2014;134(6):1389–98. doi: 10.1002/ijc.28425. [DOI] [PubMed] [Google Scholar]
- 6.Meys R, Purdie KJ, de Koning MN, Quint KD, Little AM, Baker F. et al. HLA Immunogenotype Determines Persistent Human Papillomavirus Virus Infection in HIV-Infected Patients Receiving Antiretroviral Treatment. The Journal of infectious diseases. 2016;213(11):1717–24. doi: 10.1093/infdis/jiw038. [DOI] [PubMed] [Google Scholar]
- 7.Chambuso R, Gray CM, Kaambo E, Rebello G, Ramesar R. Impact of Host Molecular Genetic Variations and HIV/HPV Co-infection on Cervical Cancer Progression: A Systematic review. Oncomedicine. 2018;3:82–93. [Google Scholar]
- 8.Alves BM, Prellwitz IM, Siqueira JD, Meyrelles AR, Bergmann A, Seuanez HN. et al. The effect of human leukocyte antigen G alleles on human papillomavirus infection and persistence in a cohort of HIV-positive pregnant women from Brazil. Infect Genet Evol. 2015;34:339–43. doi: 10.1016/j.meegid.2015.06.027. [DOI] [PubMed] [Google Scholar]
- 9.Das Ghosh D, Mukhopadhyay I, Bhattacharya A, Roy Chowdhury R, Mandal NR, Roy S. et al. Impact of genetic variations and transcriptional alterations of HLA class I genes on cervical cancer pathogenesis. International Journal of Cancer. 2017;140(11):2498–508. doi: 10.1002/ijc.30681. [DOI] [PubMed] [Google Scholar]
- 10.Ben Othmane Y, Ghazouani E, Mezlini A, Lagha A, Rais M, Kochkar R. et al. HLA class II susceptibility to cervical cancer among Tunisian women. Bulletin du cancer. 2012;99(9):81–6. doi: 10.1684/bdc.2012.1623. [DOI] [PubMed] [Google Scholar]
- 11.Zhang B, Li P, Wang E, Brahmi Z, Dunn KW, Blum JS. et al. The E5 protein of human papillomavirus type 16 perturbs MHC class II antigen maturation in human foreskin keratinocytes treated with interferon-γ. Virology. 2003;310(1):100–8. doi: 10.1016/s0042-6822(03)00103-x. [DOI] [PubMed] [Google Scholar]
- 12.Tshabalala M, Morse GD, Zijenah LS. HLA genetic polymorphisms: role in HIV-1 susceptibility, disease progression and treatment outcomes. Retrovirology: Research and Treatment. 2013;5:1. [Google Scholar]
- 13.Nicol AF, Pires AR, de Souza SR, Nuovo GJ, Grinsztejn B, Tristao A. et al. Cell-cycle and suppressor proteins expression in uterine cervix in HIV/HPV co-infection: comparative study by tissue micro-array (TMA) BMC cancer. 2008;8:289. doi: 10.1186/1471-2407-8-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vernon SD, Hart CE, Reeves WC, Icenogle JP. The HIV-1 tat protein enhances E2-dependent human papillomavirus 16 transcription. Virus research. 1993;27(2):133–45. doi: 10.1016/0168-1702(93)90077-z. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X, Lv Z, Yu H, Wang F, Zhu J. The HLA-DQB1 gene polymorphisms associated with cervical cancer risk: A meta-analysis. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2015;73:58–64. doi: 10.1016/j.biopha.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 16.Mehta AM, Mooij M, Brankovic I, Ouburg S, Morre SA, Jordanova ES. Cervical Carcinogenesis and Immune Response Gene Polymorphisms: A Review. J Immunol Res. 2017;2017:8913860. doi: 10.1155/2017/8913860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivansson EL, Magnusson JJ, Magnusson PKE, Erlich HA, Gyllensten UB. MHC loci affecting cervical cancer risk: distinguishing the effects of HLA-DQB1 and non-HLA genes TNF, LTA, TAP1 and TAP2. Genes And Immunity. 2008;9:613. doi: 10.1038/gene.2008.58. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X, Zhang L, Tian C, Yang L, Wang Z. Genetic variants and risk of cervical cancer: epidemiological evidence, meta-analysis and research review. BJOG: an international journal of obstetrics and gynaecology. 2014;121(6):664–74. doi: 10.1111/1471-0528.12638. [DOI] [PubMed] [Google Scholar]
- 19.Leo PJ, Madeleine MM, Wang S, Schwartz SM, Newell F, Pettersson-Kymmer U. et al. Defining the genetic susceptibility to cervical neoplasia—A genome-wide association study. PLOS Genetics. 2017;13(8):e1006866. doi: 10.1371/journal.pgen.1006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei L-z, Wang H-l, Liu X, Lu Y-p, Xu F, Yuan J-q. et al. Meta-Analysis on the Relationship between HLA-DRBl Gene Polymorphism and Cervical Cancer in Chinese Population. PloS one. 2014;9(2):e88439. doi: 10.1371/journal.pone.0088439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paximadis M, Mathebula TY, Gentle NL, Vardas E, Colvin M, Gray CM. et al. Human leukocyte antigen class I (A, B, C) and II (DRB1) diversity in the black and Caucasian South African population. Human immunology. 2012;73(1):80–92. doi: 10.1016/j.humimm.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 22.Tshabalala M, Mellet J, Pepper MS. Human leukocyte antigen diversity: a southern African perspective. Journal of immunology research; 2015. p. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.USA Q. Digene-hc2-dna-collection-device-flyer. wwwtheHPVtestcom. www.qiagen.com/hpv. 2008.
- 24.López-González A, Ibeas Millán P, Provencio M. DNA damage and repair in cancer therapy. Journal of Cancer Science and Therapy; 2013. [Google Scholar]
- 25.Department of Health RoSA. NATIONAL HIV COUNSELLING AND TESTING POLICY GUIDELINES. In: Health Do, editor. www.health-e.org.za: Department of Health.
- 2015.p. 8-12.
- 26.Nasiri H, Forouzandeh M, Rasaee MJ, Rahbarizadeh F. Modified salting-out method: high-yield, high-quality genomic DNA extraction from whole blood using laundry detergent. J Clin Lab Anal. 2005;19(6):229–32. doi: 10.1002/jcla.20083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bunce M. PCR-Sequence-Specific Primer Typing of HLA Class I and Class II Alleles. In: Powis SH, Vaughan RW, editors. MHC Protocols. Totowa, NJ: Humana Press; 2003. p. 143-71. [DOI] [PubMed] [Google Scholar]
- 28.Mellet J, Gray CM, Pepper MS. HLA typing: Conventional techniques v. next-generation sequencing2015. [DOI] [PubMed]
- 29.Hu Y, Wu J-Z, Zhu H, Zhang S-H, Zhu Y-Y, Wu Y-Y. et al. Association of HLA-DRB1, HLA-DQB1 Polymorphisms with HPV 16 E6 Variants among Young Cervical Cancer Patients in China. Journal of Cancer. 2017;8(12):2401–9. doi: 10.7150/jca.19809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernal-Silva S, Granados J, Gorodezky C, Aláez C, Flores-Aguilar H, Cerda-Flores RM, HLA-DRB1 Class II antigen level alleles are associated with persistent HPV infection in Mexican women; a pilot study. Infectious agents and cancer; 2013. p. 31. (8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madeleine MM, Johnson LG, Smith AG, Hansen JA, Nisperos BB, Li S. et al. Comprehensive analysis of HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 loci and squamous cell cervical cancer risk. Cancer Res. 2008;68(9):3532–9. doi: 10.1158/0008-5472.CAN-07-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuping W, Yulong C, Longyu L, Yicheng C, Zehuan L, Benrong L. et al. Polymorphic amino acids at codons 9 and 37 of HLA-DQB1 alleles may confer susceptibility to cervical cancer among Chinese women. International Journal of Cancer. 2006;118(12):3006–11. doi: 10.1002/ijc.21746. [DOI] [PubMed] [Google Scholar]
- 33.Chattopadhyay K. A comprehensive review on host genetic susceptibility to human papillomavirus infection and progression to cervical cancer. Indian Journal of Human Genetics. 2011;17(3):132–44. doi: 10.4103/0971-6866.92087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cervantes J, Lema C, Valentina Hurtado L, Andrade R, Hurtado Gomez L, Torrico L. et al. HLA-DRB1*1602 allele is positively associated with HPV cervical infection in Bolivian Andean women. Human Immunology. 2003;64(9):890–5. doi: 10.1016/s0198-8859(03)00163-0. [DOI] [PubMed] [Google Scholar]
- 35.de Araujo Souza PS, Maciag PC, Ribeiro KB, Petzl-Erler ML, Franco EL, Villa LL. Interaction between polymorphisms of the Human Leukocyte Antigen and HPV-16 Variants on the risk of invasive cervical cancer. BMC cancer. 2008;8:246. doi: 10.1186/1471-2407-8-246. - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang SS, Bratti MC, Rodriguez AC, Herrero R, Burk RD, Porras C. et al. Common variants in immune and DNA repair genes and risk for human papillomavirus persistence and progression to cervical cancer. The Journal of infectious diseases. 2009;199(1):20–30. doi: 10.1086/595563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin P, Koutsky LA, Critchlow CW, Apple RJ, Hawes SE, Hughes JP, HLA Class II DR-DQ and Increased Risk of Cervical Cancer among Senegalese Women1. Cancer Epidemiology, Biomarkers & Prevention; 2001. Vol. 10:1037-45. [PubMed] [Google Scholar]
- 38.Dao DD, Sierra-Torres CH, Robazetti SC, de Gomez MN, Konig R, Lema C. et al. HLA-DQB1 and cervical cancer in Venezuelan women. Gynecologic oncology. 2005;96(2):349–54. doi: 10.1016/j.ygyno.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 39.Hu JM, Sun Q, Li L, Liu CX, Chen YZ, Zou H. et al. Human leukocyte antigen-DRB1*1501 and DQB1*0602 alleles are cervical cancer protective factors among Uighur and Han people in Xinjiang, China. International Journal of Clinical and Experimental Pathology. 2014;7(9):6165–71. [PMC free article] [PubMed] [Google Scholar]
- 40.Al-Daraji WI, Smith JHF. Infection and Cervical Neoplasia: Facts and Fiction. International Journal of Clinical and Experimental Pathology. 2009;2(1):48–64. [PMC free article] [PubMed] [Google Scholar]
- 41.de Jong A, van Poelgeest MIE, van der Hulst JM, Drijfhout JW, Fleuren GJ, Melief CJM. et al. Human Papillomavirus Type 16-Positive Cervical Cancer Is Associated with Impaired CD4+ T-Cell Immunity against Early Antigens E2 and E6. Cancer Research. 2004;64(15):5449–55. doi: 10.1158/0008-5472.CAN-04-0831. [DOI] [PubMed] [Google Scholar]
- 42.Madeleine MM, Brumback B, Cushing-Haugen KL, Schwartz SM, Daling JR, Smith AG. et al. Human leukocyte antigen class II and cervical cancer risk: a population-based study. The Journal of infectious diseases. 2002;186(11):1565–74. doi: 10.1086/345285. [DOI] [PubMed] [Google Scholar]
- 43.Krul EJT, Schipper RF, Schreuder GMT, Fleuren GJ, Kenter GG, Melief CJM. HLA and Susceptibility to Cervical Neoplasia. Human Immunology. 1999;60:337–42. doi: 10.1016/s0198-8859(98)00127-x. [DOI] [PubMed] [Google Scholar]
- 44.Cuzick J, Terry G, Ho L, Monaghan J, Lopes A, Clarkson P. et al. Association between high-risk HPV types, HLA DRB1* and DQB1* alleles and cervical cancer in British women. British journal of cancer. 2000;82(7):1348–52. doi: 10.1054/bjoc.1999.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]