Abstract
Myxoid liposarcoma is a malignant lipogenic tumor that develops in deep soft tissues. While local control rates are good, current chemotherapy options remain ineffective against metastatic disease. Myxoid liposarcoma is characterized by the FUS-DDIT3 fusion oncoprotein that is proposed to function as an aberrant transcription factor, but its exact mechanism of action has remained unclear. To identify the key functional interacting partners of FUS-DDIT3, this study utilized immunoprecipitation-mass spectrometry (IP-MS) to identify the FUS-DDIT3 interactome in whole cell lysates of myxoid liposarcoma cells, and results showed an enrichment of RNA processing proteins. Further quantitative MS analyses of FUS-DDIT3 complexes isolated from nuclear lysates showed that members of several chromatin regulatory complexes were present in the FUS-DDIT3 interactome, including NuRD, SWI/SNF, PRC1, PRC2, and MLL1 COMPASS-like complexes. Co-immunoprecipitation validated the associations of FUS-DDIT3 with BRG1/SMARCA4, BAF155/SMARCC1, BAF57/SMARCE1, and KDM1A. Data from this study provides candidates for functional validation as potential therapeutic targets, particularly for emerging epigenetic drugs.
Abbreviations: C/EBP, CCAAT/enhancer-binding protein; ChIP, chromatin immunoprecipitation; CV, coefficient of variation; DDIT3, DNA damage inducible transcript 3; FUS, fused in sarcoma; IP-MS, immunoprecipitation-mass spectrometry; KDM1A, lysine demethylase 1A; MAP4K4, mitogen-activated protein kinase kinase kinase kinase 4; NONO, non-POU domain containing octamer binding; NuRD, nucleosome remodeling and deacetylation; PES, protein enrichment score; PRC, polycomb repressive complex; PSPC1, paraspeckle component 1; SFPQ, splicing factor proline and glutamine rich; SP3, single-pot solid-phase-enhanced sample preparation; SPAG5, sperm associated antigen 5; SWI/SNF, SWItch/Sucrose non-fermentable; TMT, tandem mass tags
Introduction
Myxoid liposarcoma is a translocation-associated malignancy with peak incidence in younger adults between 30 and 50 years of age. The great majority of cases of myxoid liposarcoma are characterized by a t(12;16)(q13;p11) reciprocal translocation which results in the in-frame fusion of fused in sarcoma (FUS) to DNA damage inducible transcript 3 (DDIT3) [1]. About 3% of cases have an alternative t(12;22)(q13;q12) translocation that creates an EWSR1-DDIT3 fusion [2], [3].
FUS is a member of a highly conserved and ubiquitously expressed group of proteins termed the FET family, which also includes the proteins Ewing sarcoma breakpoint region 1 (EWSR1) and TATA binding protein-associated factor 15 (TAF15); all three share a similar domain structure [4]. FUS is a multifunctional protein involved in several cellular pathways, including transcriptional regulation, DNA repair and splicing regulation [5]. Unlike the near-ubiquitously expressed FUS, DDIT3 (also known as CHOP: C/EBP homologous protein), a basic leucine zipper transcription factor, has limited and tightly regulated expression. DDIT3 functions as a cellular stress sensor that is expressed at a very low level in normal physiology, but can be rapidly induced in response to endoplasmic reticulum stress, nutrient deprivation, DNA damage, cellular growth arrest or hypoxia [6]. DDIT3 is also a member of the CCAAT/enhancer-binding protein (C/EBP) family, and has been implicated in the negative regulation of cellular differentiation [6].
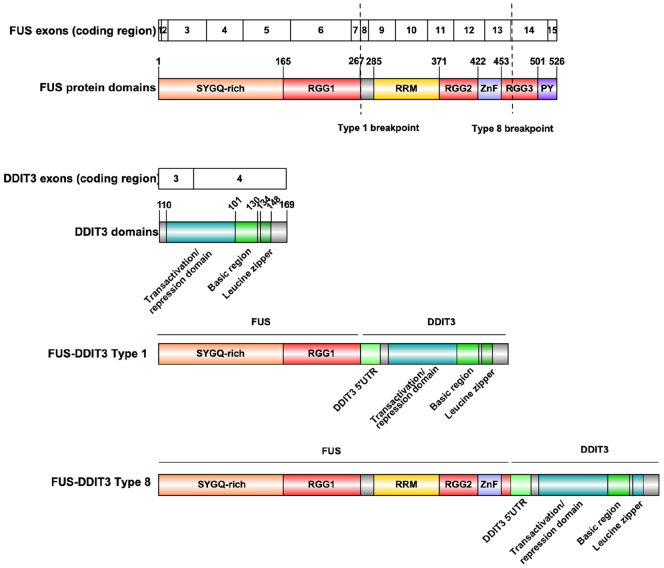
In myxoid liposarcoma, the FUS-DDIT3 fusion oncoprotein (Figure 1) contains at least part of the FUS N-terminus SYGQ-rich low complexity domain, fused to full length DDIT3, and acts as the central driver for myxoid liposarcoma. Early studies showed that FUS-DDIT3 is sufficient for in vitro transformation in ST-13 mouse pre-adipocytes and NIH3T3 mouse embryonic fibroblasts, a phenotype that requires the DNA binding domain of DDIT3 and the N-terminal domain of either FUS or EWSR1 [7], [8]. Other studies have suggested a role for FUS-DDIT3 as an aberrant transcription factor [9], [10].
Figure 1.
Structure and domains of FUS, DDIT3 and FUS-DDIT3. Wild type FUS contains the following protein domains: a low complexity serine/tyrosine/glycine/glutamine (SYGQ)-rich domain, three arginine- and glycine-rich RGG motif domains, a RNA recognition motif (RRM) domain, a zinc finger (ZnF) domain, and a non-classical proline-tyrosine (PY) nuclear localization signal. Wild type DDIT3 contains a transactivation/repression domain in the N-terminus followed by a basic leucine zipper in its C-terminus. The two FUS-DDIT3 fusion variants used in this study retain the SYGQ-rich and RGG1 domains of FUS, and also include the in frame amino acid sequence of a portion of the previously untranslated region (UTR) from DDIT3 exon 2. Schematic illustration of protein domain structure was generated with the tool Illustrator of Biological Sequences (IBS).
Although local control rates in myxoid liposarcoma are excellent with a combination of radiation therapy and surgery, chemotherapy is still the main treatment for unresectable and metastatic tumors, and disease-free survival is poor in the metastatic setting [11], [12]. Despite the presence of FUS-DDIT3 as a driver oncoprotein in myxoid liposarcoma, the exact mechanisms of action behind the capacity of FUS-DDIT3 for transformation are still unclear and represent part of the challenge in finding targeted therapies against this cancer of young adults.
Trabectedin, a chemotherapy drug that blocks the minor groove of DNA, has been recently approved as a sarcoma treatment. While trabectedin has been shown to reduce DNA binding of multiple transcription factors [13] including FUS-DDIT3 [10], [14], EWSR1-FLI1 [15], [16] and EWSR1-WT1 [17], no existing drugs specifically target FUS-DDIT3. An alternative strategy that might confer less toxicity would be to target functionally important protein interacting partners of FUS-DDIT3. To date, genomic profiling at the RNA expression or exome level has revealed a low frequency of secondary genetic alterations [18], [19], [20]. While there have been individual reports of FUS-DDIT3 interactors, including CCAAT/enhancer-binding protein β (C/EBPβ) [21], [22], cyclin-dependent kinase 2 (CDK2) [23], NFKB inhibitor zeta (NFKBIZ) [9], RNA polymerase II [24], and all three FET proteins [25], the lack of comprehensive data on the FUS-DDIT3 interactome represents one of the major gaps in knowledge behind the oncogenic functions of the fusion protein.
In this study, we used immunoprecipitation-mass spectrometry (IP-MS) for the unbiased identification of FUS-DDIT3 interactors, and observed the presence of several chromatin regulators in the FUS-DDIT3 interactome. Given the emerging evidence that other sarcoma fusion proteins function through epigenetic mechanisms of action, including EWSR1-FLI1 in Ewing sarcoma [26], [27], [28], SS18-SSX in synovial sarcoma [29], [30], [31], [32], and PAX3-FOXO1 in alveolar rhabdomyosarcoma [33], our findings support commonalities among fusion oncoprotein-associated sarcomas and provide a list of potentially targetable FUS-DDIT3 interactors in myxoid liposarcoma.
Materials and Methods
Mammalian Cell Culture
The following cell lines were kindly provided: myxoid liposarcoma cell lines 402-91 (Type 1 FUS-DDIT3, fusing FUS exon 7 to DDIT3 exon 2) and 1765-92 (Type 8 FUS-DDIT3, fusing FUS exon 13 to DDIT3 exon 2) by Dr. Pierre Aman (University of Gothenburg, Sweden) [1], and DL-221 (Type 1 FUS-DDIT3, fusing FUS exon 7 to DDIT3 exon 2) by Dr. Keila Torres (MD Anderson Cancer Center, Houston, TX, USA) [34]. Sarcoma cell lines were cultured in RPMI-1640 medium with 10% fetal bovine serum (Life Technologies). The non-sarcoma control cell line HeLa was maintained in DMEM medium with 10% fetal bovine serum. All cells were cultured at 37°C, 95% humidity, and 5% CO2.
Immunoprecipitation, Protein Clean Up and Digestion
Whole cell lysates were obtained by lysing cells in lysis buffer (1% Triton X-100, 15 mM Tris pH 8.0, 100 mM NaCl) supplemented with Roche cOmplete EDTA-free protease inhibitors and 1 U/mL benzonase (MilliporeSigma) for background reduction, with rotation for 1 hour in a cold room. Whole cell lysates were clarified by centrifugation at maximum speed for 10 min at 4°C, quantified using a BCA protein assay (Thermo Fisher Scientific), and stored on ice until ready for immunoprecipitation.
For preparation of nuclear lysates, cells were rinsed once with PBS on 15 cm dishes before trypsinization. Trypsinized cells were swelled under rotation for 15 min in the cold room with 5× packed cell volume of hypotonic lysis buffer (20 mM HEPES-KOH pH 7.5, 5 mM MgCl2, 5 mM CaCl2, 1 mM EDTA) supplemented with Roche cOmplete EDTA-free protease inhibitors, then lysed with 5 strokes of the dounce homogenizer on ice. Cell nuclei were washed once with PBS and collected by centrifugation at 2000 RPM at 4°C for 5 min, then lysed with Triton X-100 lysis buffer supplemented with Roche cOmplete EDTA-free protease inhibitors and 1 U/mL benzonase (MilliporeSigma) for 1 hour in the cold room with rotation. Nuclear lysates were clarified by centrifugation at 16,000 RPM at 4°C for 10 min, then quantified using a BCA protein assay (Thermo Fisher Scientific), and stored on ice until ready for immunoprecipitation.
Antibodies used for immunoprecipitations were: normal mouse or rabbit IgG (Santa Cruz), DDIT3 (L63F7) (Cell Signaling Technology, #2895), NONO (Abcam, ab70335), PSPC1 (Abcam, ab104238), and SFPQ (Novus Biologicals, NB-100-61045). Antibodies were first incubated with Dynabeads™ Protein G (Thermo Fisher Scientific) for 10 min at room temperature.
For non-mass spectrometry immunoprecipitations, the non-crosslinked antibody-Dynabead mixture was incubated with cell lysates overnight with rotation in the cold room. Beads were washed 3 times with cold lysis buffer, followed by 2 times with cold PBS, then eluted by boiling for 5 min in sample loading buffer.
For samples destined for mass spectrometry analysis, antibodies were crosslinked to Dynabeads with BS3 (bis(sulfosuccinimidyl)suberate) crosslinker (Thermo Fisher Scientific) according to the manufacturer protocol. The antibody-bead mixture was then pre-blocked with 10 mg/mL BSA in casein blocker (Thermo Fisher Scientific) with rotation at room temperature for 30 min, before being incubated with whole cell or nuclear lysates for 1 hour at room temperature, with rotation. Beads were washed 3 times with cold lysis buffer, followed by 2 times with cold PBS. Proteins were eluted by boiling for 5 min in the SP3 (below) elution buffer (4% SDS, 2% β-mercaptoethanol, 40 mM Tris pH 6.8).
Eluted immunoprecipitates were incubated at 45°C for 30 min, then alkylated with 400 mM iodoacetamide for 30 min at 24°C. Reactions were quenched with addition of 200 mM dithiothreitol. Eluted proteins were prepared for trypsin digestion using the single-pot solid-phase-enhanced sample preparation (SP3) paramagnetic bead cleanup protocol as previously described [35]. Acetonitrile was added to the SP3 bead-protein mixture to a final 50% vol/vol, and incubated for 8 min at room temperature. Using a magnetic rack, beads were washed two times with 200 μL 70% ethanol for 30 sec, and once with 180 μL 100% acetonitrile for 15 sec. For digestion, beads were reconstituted in 5 μL 50 mM HEPES pH 8.0 buffer containing trypsin/rLys-C enzyme mix (Promega) at a 1:25 enzyme to protein ratio, and incubated for 14 hour at 37°C. Digested peptides were recovered by removing the supernatant on a magnetic rack.
Tandem Mass Tag (TMT) Labeling
TMT 10-plex labeling kits were obtained from Pierce. Each TMT label (5 mg per vial) was reconstituted in 500 μL of acetonitrile and frozen at -80°C. TMT labels were removed from the freezer and allowed to equilibrate at room temperature. Labeling reactions were carried out through the addition of TMT label in two volumetrically equal steps to achieve a 2:1 (μg:μg) TMT label:peptide final concentration, 30 min apart. All incubations were carried out at room temperature. Reactions were quenched by addition of glycine. Labeled peptides were concentrated in a SpeedVac centrifuge to remove acetonitrile, combined, and desalted before MS analysis on the Orbitrap Fusion.
Mass Spectrometry Analysis
Mass spectrometry analysis of peptide samples was carried out on an Orbitrap Fusion Tribrid MS platform (Thermo Fisher Scientific). Samples were introduced using an Easy-nLC 1000 system (Thermo Fisher Scientific). Columns used for trapping and separations were packed in-house. Trapping columns were packed in 100 μm internal diameter capillaries to a length of 2.5 cm with C18 beads (Reprosil-Pur, Dr. Maisch, 3 μm particle size). Trapping was carried out for a total volume of 10 μL at a pressure of 400 bar. After trapping, gradient elution of peptides was performed on a C18 (Reprosil-Pur, Dr. Maisch, 1.9 μm particle size) column packed in-house to a length of 25 cm in 100 μm internal diameter capillaries with a laser-pulled electrospray tip and heated to 45°C using AgileSLEEVE column ovens (Analytical Sales & Service). Elution was performed with a gradient of mobile phase A (water and 0.1% formic acid) to 8% B (acetonitrile and 0.1% formic acid) over 5 min, to 25% B over 88 min, to 40% over 20 min, with final elution (80% B) and equilibration (5% B) using a further 7 min at a flow rate of 375 nL/min.
Data acquisition on the Orbitrap Fusion (control software version 2.1.1565.20) was carried out using a data-dependent method with multi-notch synchronous precursor selection MS3 scanning for TMT tags. Survey scans covering the mass range of 350 – 1500 were acquired at a resolution of 120,000 (at m/z 200), with quadrupole isolation enabled, an S-Lens RF Level of 60%, a maximum fill time of 50 ms, and an automatic gain control (AGC) target value of 4e5. For MS2 scan triggering, monoisotopic precursor selection was enabled, charge state filtering was limited to 2 – 4, an intensity threshold of 5e3 was employed, and dynamic exclusion of previously selected masses was enabled for 60 seconds with a tolerance of 20 ppm. MS2 scans were acquired in the ion trap in Rapid mode after CID fragmentation with a maximum fill time of 20 ms, quadrupole isolation, an isolation window of 1 m/z, collision energy of 30%, activation Q of 0.25, injection for all available parallelizable time turned OFF, and an AGC target value of 1e4. Fragment ions were selected for MS3 scans based on a precursor selection range of 400-1600 m/z, ion exclusion of 20 m/z low and 5 m/z high, and isobaric tag loss exclusion for TMT. The top 10 precursors were selected for MS3 scans that were acquired in the Orbitrap after HCD fragmentation (NCE 60%) with a maximum fill time of 90 ms, 50,000 resolution, 120-750 m/z scan range, ion injection for all parallelizable time turned OFF, and an AGC target value of 1e5. The total allowable cycle time was set to 4 seconds. MS1 and MS3 scans were acquired in profile mode, and MS2 in centroid format.
Protein Enrichment Score Calculation
The calculation of the protein enrichment score (PES) is adapted from the PSEA-Quant method which reflects the abundance and reproducibility of the abundance measurements across IP replicates [36]. The average peptide mass spectra value for each peptide is obtained from Proteome Discoverer to obtain the peptide-level signal value. These values are then normalized to the same median values within the control and DDIT3 IPs separately. For each peptide in each DDIT3 IP replicate, the ratio of the peptide-level signal value over the average value from all control IPs is obtained. The peptide-level ratios and coefficients of variation (CV) from each DDIT3 IP replicate are then aggregated to the protein-level. The protein level CV is transformed to the same scale as the average protein ratios (CV(transformed)) for easy visualization on a scatterplot. PES is calculated by:
where ratiomax is the largest average ratio in the dataset. Proteins with an average ratio below 1 (i.e. higher signal in control IPs than DDIT3 IPs) are then removed from the dataset. The bait protein should show the highest PES which reflects a high average protein ratio and low CV of the ratio across triplicate IPs. The PES values are then converted to z-scores to apply candidate selection cutoffs based on the number of standard deviations away from the mean.
Proximity Ligation Assay
Cells were grown in culture treated chamber slides for 24 hours, then fixed with 2% paraformaldehyde and permeabilized with 0.1% Triton X-100. Cells were blocked with Duolink blocking solution and incubated overnight at 4°C in the cold room with the following primary antibodies: DDIT3 (L63F7) (Cell Signaling Technology, #2895), MAP4K4 (Bethyl, A3011-502A). Proximity ligation was performed with the Duolink® In Situ Red Starter Kit Mouse/Rabbit (MilliporeSigma) according to the manufacturer’s instructions. Goat anti-rabbit Alexa Fluor 488 secondary antibody (Life Technologies) was added during signal amplification to confirm the presence of MAP4K4. Fluorescence was detected using a Zeiss Axiovert microscope at 63X.
Results
The FUS-DDIT3 Interactome From Whole Cell Lysates is Enriched in RNA Processing Proteins
Endogenous FUS-DDIT3 was immunoprecipitated from three myxoid liposarcoma cell lines (402-91, 1765-92 and DL-221) using an anti-DDIT3 antibody that recognizes the C-terminal portion of DDIT3 (retained in FUS-DDIT3) (Figure 1). In keeping with the reported function of DDIT3 as a stress sensor protein that is only expressed upon stress induction [6], the three myxoid liposarcoma cell lines do not express detectable wild type DDIT3 under standard cell culture conditions (Figure S1A). Even after treatment with tunicamycin to induce endoplasmic reticulum stress and wild type DDIT3 expression, the levels of FUS-DDIT3 expression far exceeded that of induced DDIT3 (Figure S1A). Therefore, immunoprecipitation (IP) with a DDIT3 antibody should specifically enrich for FUS-DDIT3 and its interactors, such as C/EBPβ [22] (Figure S1B). Specifically, no FUS-DDIT3 bands were observed at the expected molecular weight in negative control synovial sarcoma SYO-1 cells that do not express FUS-DDIT3 (Figure S1C).
Triplicate immunoprecipitations with the DDIT3 antibody were performed on whole cell lysates from each cell line, then analyzed by mass spectrometry. Western blot analysis of the three post-IP supernatants showed a reduction in FUS-DDIT3 levels in the IP:DDIT3 data compared to the no antibody or mouse IgG controls (Figure S2). To further confirm the immunoprecipitation of FUS-DDIT3, multiple DDIT3 peptides were detected in all triplicate IP:DDIT3 samples and in all cell lines (Table S1). FUS peptides were not used as an indication for successful enrichment of the fusion protein due to the presence of abundant wild type FUS in all cell lines, and the known association of wild type FUS with FUS-DDIT3 (Figure S1B) [25].
To curate the large number of MS-identified proteins for downstream data analysis, proteins were selected from each cell line that were (1) detected only in all three IP:DDIT3 experiments but not in the IP:IgG control, or (2) were present at a statistically significant (Student’s t-test, P < .05) positive average fold change in all IP:DDIT3 triplicates over IP:IgG. This analysis identified 12 proteins found in all three cell lines and 75 proteins that were common in at least two cell lines, and were selected for further analysis (Table S2).
The 12 proteins (including DDIT3) common in all three cell lines were ranked in Table S2 according to their total normalized abundance from all cell lines, followed by the remaining 63 proteins that were detected in any two of the three cell lines. C/EBPβ was detected, serving as a positive control for the presence of FUS-DDIT3 interactors and validated the methodology used (Table S2). However, C/EBPβ was only detected in the 402-91 and 1765-92 cell lines, likely due to the much lower expression of C/EBPβ in DL-221 (Figure S3) reducing the probability of its being detected in the mass spectrometry analysis.
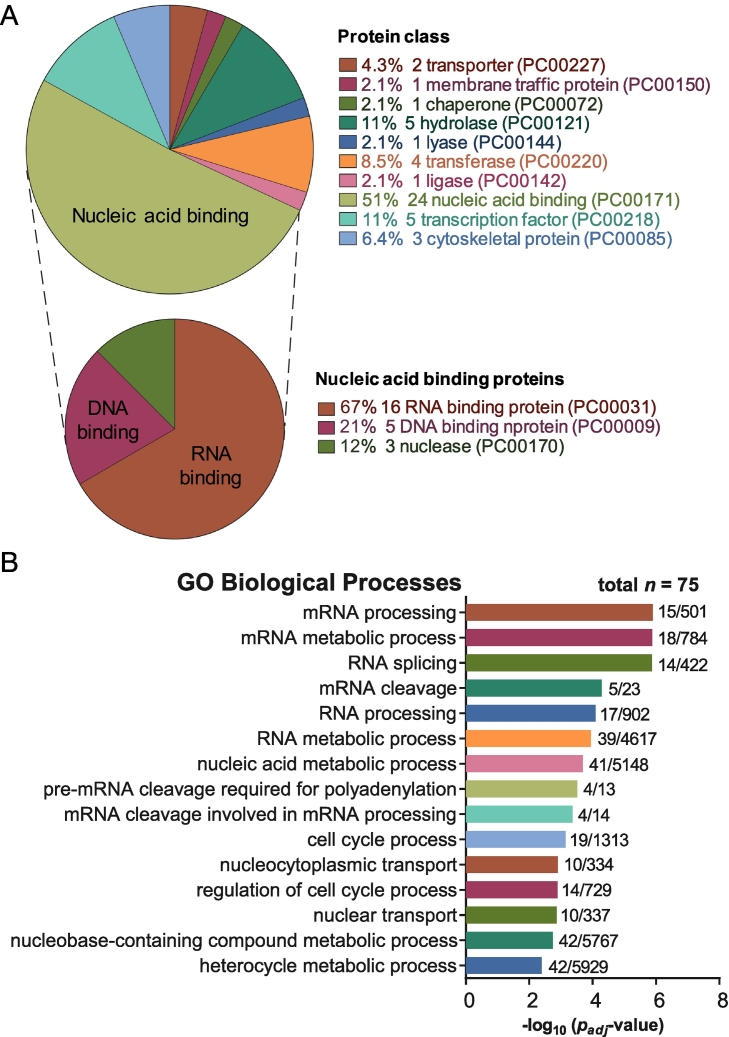
Analysis with the PANTHER database (Protein ANalysis THrough Evolutionary Relationships, http://pantherdb.org) showed that the largest protein class in the myxoid liposarcoma interactome are nucleic acid binding proteins (24 proteins), and within this category, two-thirds are RNA-binding proteins (16 proteins) (Figure 2A). Based on gene ontology (GO) classification of the FUS-DDIT3 interactome, eight of the top ten enriched biological processes are involved in RNA processing (Figure 2B). Together, these data suggest that the IP-MS screen successfully identified proteins within the FUS-DDIT3 interactome, and that the interactome so identified was enriched in proteins involved in RNA processing and splicing.
Figure 2.
Functional classification of the whole cell FUS-DDIT3 interactome. (A) The largest class of proteins in the FUS-DDIT3 interactome from whole cell lysates are the nucleic acid binding proteins. The PANTHER protein classification of the FUS-DDIT3 interactome shows the percentage of each protein class against the total number of proteins with a class hit (n = 47). The nucleic acid binding protein class (n = 24) is further broken down into three different sub-classes (DNA-binding, RNA-binding and nuclease). PC numbers refer to PANTHER class ID. (B) The FUS-DDIT3 interactome is enriched for RNA processing proteins, based on the Gene Ontology classification of biological processes enriched in the FUS-DDIT3 whole cell lysate interactome (n = 75).
Validation of IP-MS-Identified FUS-DDIT3 Interactors
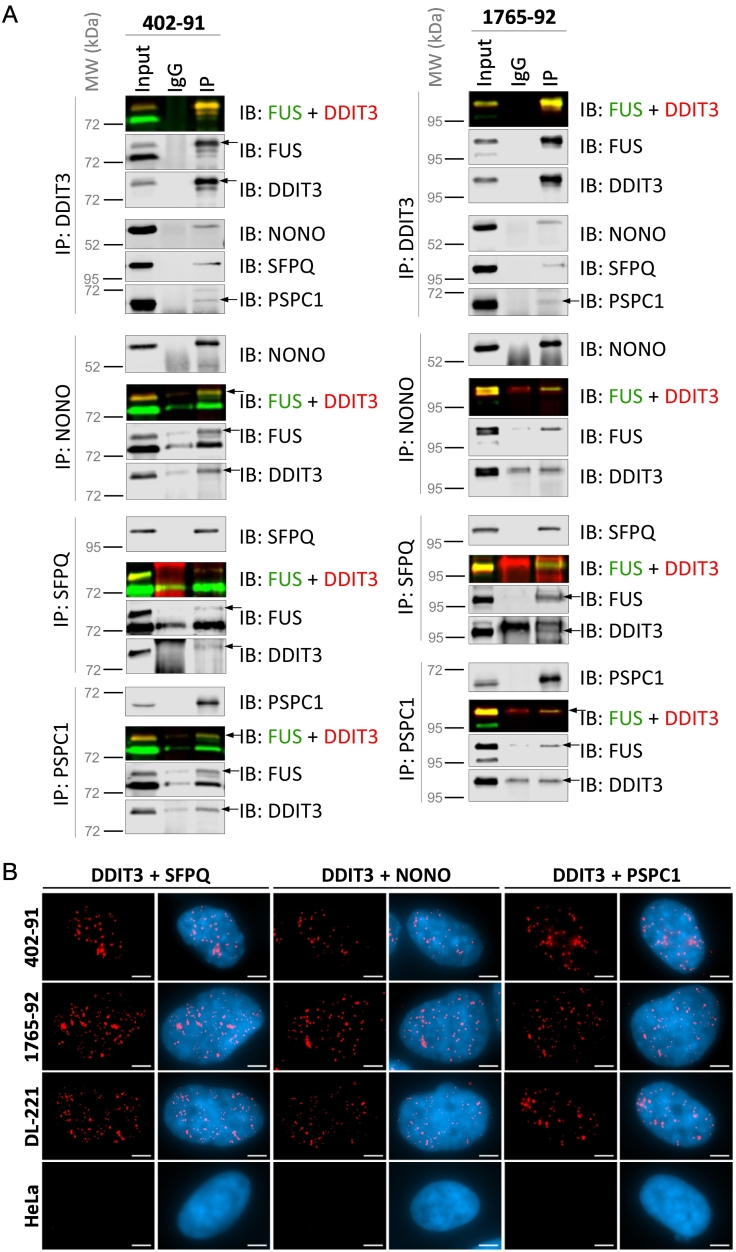
The top-ranked proteins in Table S2, non-POU domain containing octamer binding (NONO) and splicing factor proline and glutamine rich (SFPQ), were selected for validation of their associations with FUS-DDIT3. Additionally, although paraspeckle component 1 (PSPC1) was detected further down in the interactome ranking, it was chosen for validation as a member of the drosophila behavior/human splicing (DBHS) protein family that often functions in complex with NONO and SFPQ [37]. Results from reciprocal co-immunoprecipitation (Figures 3A and S4) and proximity ligation assays (Figure 3B) in myxoid liposarcoma cell lines verify the association of FUS-DDIT3 with NONO, PSPC1 and SFPQ.
Figure 3.
Reciprocal co-immunoprecipitation and proximity ligation assays validate FUS-DDIT3’s association with NONO, PSPC1 and SFPQ. (A) Whole cell lysates from two myxoid liposarcoma cell lines, 402-91 and 1765-92, each harboring a different variant of the FUS-DDIT3 fusion oncoprotein, were used to test for reciprocal co-immunoprecipitation of the Drosophila behavior/human splicing proteins NONO, PSPC1, and SFPQ with the FUS-DDIT3 oncoprotein. For color blots, FUS was detected in the green channel and DDIT3 in the red channel, and the FUS-DDIT3 band is indicated by an overlap of FUS and DDIT3 signal to form a merged yellow-colored band. Larger images of the same blots with visible IgG bands from each IP antibody are in Figure S4 to visualize the amount of antibody used. (B) Proximity ligation assay, a technique that amplifies a red signal when two proteins colocalize within 40 nm of each other in situ, was performed in myxoid liposarcoma cell lines 402-91, 1765-92, DL-221, and negative control cell line HeLa. Red signals indicate proximity between proteins of interest. Nuclei were counter-stained with DAPI (blue). Scale bar = 5 μm.
Reciprocal co-immunoprecipitation in myxoid liposarcoma cells 402-91 and 1765-92 also verified the association of FUS-DDIT3 with five other proteins identified by the IP-MS screen: nucleostemin / G protein nucleolar 3 (GNL3), mitogen-activated protein kinase kinase kinase kinase 4 (MAP4K4), astrin / sperm associated antigen 5 (SPAG5), zinc finger RNA binding protein (ZFR), and zinc finger protein 638 (ZNF638) (Figures S5 and S6). Four of the proteins, GNL3, SPAG5, ZFR and ZNF638, were among the top 12 interactors identified in the IP-MS screen. Although the fifth tested protein, MAP4K4, was ranked lower (66th out of 75 proteins), because of its role as a targetable kinase its association with FUS-DDIT3 was chosen for further verification by proximity ligation assay (Figure S7). The positive results for all tested validations suggest that the proteins detected by the IP-MS screen are likely real interactors of FUS-DDIT3.
The FUS-DDIT3 Nuclear Interactome Includes Multiple Chromatin Regulatory Complexes
The FUS-DDIT3 interactome identified from whole cell lysates of myxoid liposarcoma cell lines did not contain many transcription factors and regulators (Figure 2A and Table S2). The identification of transcriptional regulatory complexes associated with FUS-DDIT3 would be vital to understanding the role of FUS-DDIT3 as an aberrant transcription factor, and also to provide additional potential therapeutic targets that are important to the oncogenic function of FUS-DDIT3. However, transcription factors are notoriously difficult to detect via mass spectrometry due to their low abundance within highly complex eukaryotic proteomes [38]. To bypass these limitations, two strategies were adopted to increase identification of transcription factors and regulators in the FUS-DDIT3 interactome: target enrichment through the immunoprecipitation of FUS-DDIT3 from nuclear instead of whole cell lysates [38], and label-based relative quantification using tandem mass tags (TMT) to reduce variances and improve quantification accuracy for lower abundance proteins [39].
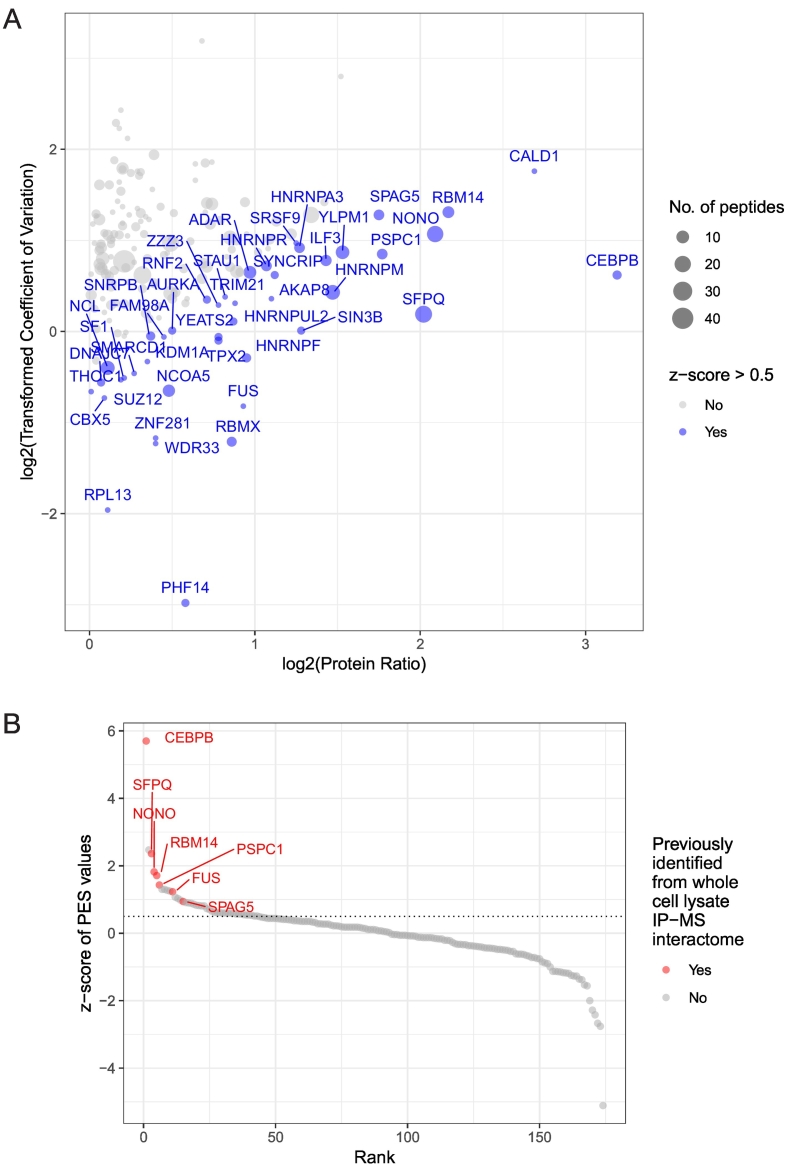
Immunoprecipitation (IP) of endogenous FUS-DDIT3 was performed on nuclear lysates from the 402-91 myxoid liposarcoma cell line. The IPs were carried out in four sets of replicates, with the first three sets of replicates used for TMT labeling and mass spectrometry analysis. The fourth set of replicates was used for Western blot analysis to confirm immunoprecipitation of FUS-DDIT3 in the experiment (Figure S8A). Following TMT labeling and mass spectrometry, 174 proteins with an average ratio of enrichment over control IPs greater than 1 were selected for further analysis (Table S3). The selected proteins were visualized on a scatterplot showing their relative enrichment ratio in the DDIT3 IPs and the coefficient of variation (CV) of this ratio across triplicates (Figure 4A).
Figure 4.
The FUS-DDIT3 nuclear interactome. (A) Scatterplot of the FUS-DDIT3 nuclear interactome showing proteins with an average enrichment ratio of >1 in DDIT3 vs control IPs (n = 174). Each circle represents a putative FUS-DDIT3 nuclear interactor, with circle size corresponding to the number of peptides detected in the mass spectrometry analysis. Each protein is plotted by the log2 values of its average enrichment ratio (log2(Protein Ratio)) and the transformed coefficient of variation for the average ratio across triplicate IPs. The protein enrichment score (PES, see Methods) is calculated to take into account the abundance measurement for each protein (enrichment ratio) and the reproducibility of this measurement through the CV values (Table S3). A cutoff PES z-score ≥ 0.5 was applied to identify 43 top ranking-proteins in the FUS-DDIT3 nuclear interactome (highlighted in blue). (B) The same 174 proteins from the FUS-DDIT3 nuclear interactome are plotted by their z-scores and ranking of their PES values (higher PES value = higher rank). The seven proteins previously validated from the whole cell lysate IP-MS screen are highlighted in red. The PES z-score = 0.5 is indicated by a dotted line.
A protein with a higher protein ratio and lower CV would be considered a higher confidence FUS-DDIT3 interactor. This measure of confidence was quantified by calculating the protein enrichment score (PES), and used to rank the candidate interactors (Table S3), with a higher ranking protein showing a higher protein ratio and a lower CV on the scatterplot (Figure 4A). Seven proteins (C/EBPβ, FUS, NONO, PSPC1, RBM14, SFPQ, SPAG5) that were identified in both the nuclear and whole cell lysate interactomes (Figure S8B) also ranked high in the nuclear interactome with a PES z-score > 0.5 (Figure 4B). Apart from RBM14, these proteins had all been validated using reciprocal co-IP and/or proximity ligation (Figures 2 and S1B, S5-7). These observations support the successful identification of putative FUS-DDIT3 interactors from nuclear lysates of 402-91 myxoid liposarcoma cells.
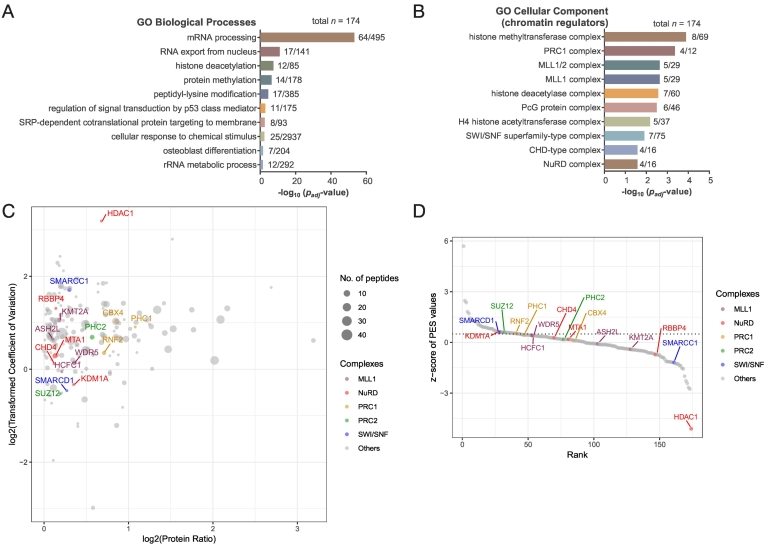
To identify proteins in previously annotated complexes that are enriched in the FUS-DDIT3 nuclear interactome, the CORUM database (comprehensive resource of mammalian protein complexes: http://mips.gsf.de/genre/proj/corum/index.html) was used to analyze the 43 proteins with PES z-score > 0.5 (Table S3). The enriched complexes containing these 43 proteins were similar to the ones found in the whole cell lysate interactome, and included the SFPQ/NONO complex, and mRNA and miRNA processing complexes (Figure S9). Analysis of the full list of 174 nuclear proteins with an FUS-DDIT3 IP enrichment ratio of >1 without applying the PES z-score cut-off revealed a longer list of CORUM protein complexes, which included several chromatin regulatory complexes such as the nucleosome remodeling and deacetylation (NuRD) complex, mixed-lineage leukemia 1 (MLL1) COMPASS-like complex, and polycomb repressive complex 1 (PRC1) (Figure S10).
While the top gene ontology biological process in the full list of 174 nuclear interactors was mRNA processing (Figure 5A), which was also observed in the whole cell lysate interactome (Figure 2B), several chromatin modification processes including histone deacetylation, protein methylation, and peptidyl-lysine modification were also enriched in the nuclear interactome (Figure 5A). Similarly, gene ontology cellular component analysis identified a number of chromatin regulatory complexes (Figure 5B) among a longer list of enriched cellular components (Table S4). A closer look at the full list of 174 nuclear interactors (Table S3) identifies 16 proteins belonging to various chromatin regulatory complexes, including NuRD, SWItch/Sucrose non-fermentable (SWI/SNF), PRC1 and PRC2, and MLL1 (Figure 5C and D).
Figure 5.
The FUS-DDIT3 nuclear interactome contains mRNA processing proteins and chromatin regulators. Enriched gene ontology (A) biological processes or (B) cellular components in the FUS-DDIT3 nuclear interactome (n = 174). Numbers after each bar indicate the number of group members found in interactome / total number of group members in database. For enriched cellular components, only chromatin regulatory complexes are presented here; the full list can be found in Table S4. (C) Proteins with an average enrichment ratio of >1 in DDIT3 vs control IPs are presented (n = 174). Components of various chromatin regulatory complexes are indicated. Each circle represents a putative FUS-DDIT3 nuclear interactor, with circle size corresponding to the number of peptides detected in the mass spectrometry analysis. Each protein is plotted by the log2 values of its average enrichment ratio (log2(Protein Ratio)) and the transformed coefficient of variation for the average ratio across triplicate IPs. (D) The same proteins are plotted by the z-scores and rank of their PES values (higher PES value = higher rank). The PES z-score = 0.5 is indicated by a dotted line.
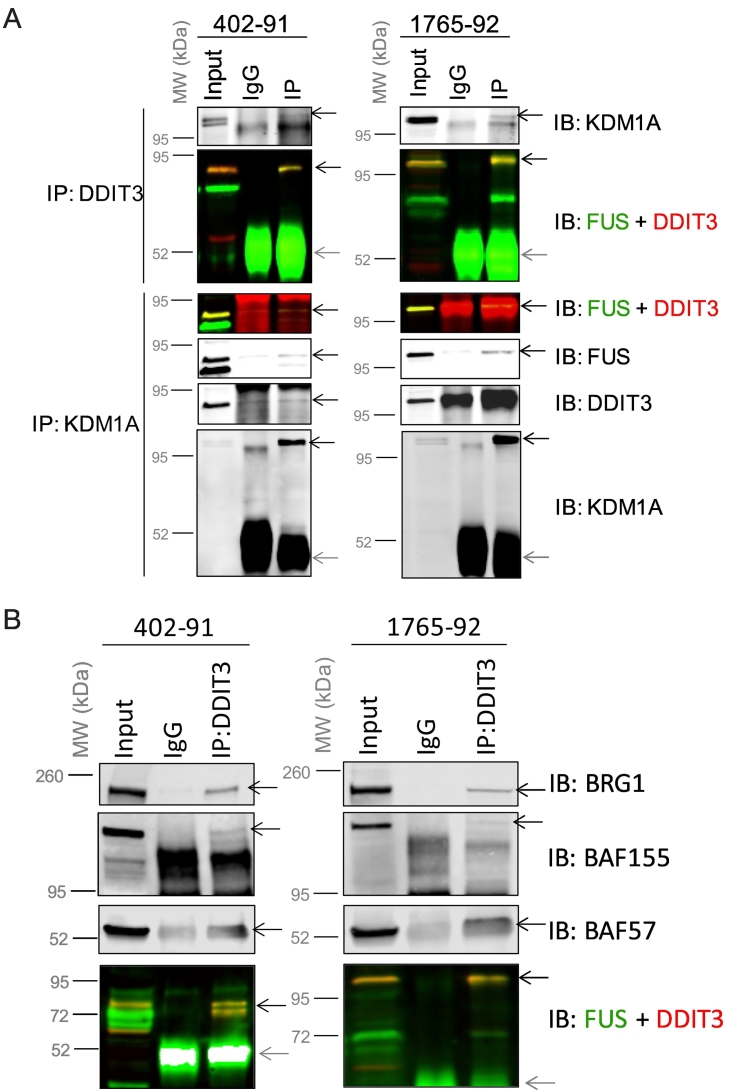
To validate the association of FUS-DDIT3 with chromatin regulators, co-IP experiments were carried out. Lysine demethylase 1 (KDM1A) was the top ranked chromatin regulator in the interactome (Figure 5D), and reciprocal co-IP verified the association of KDM1A with FUS-DDIT3 in myxoid liposarcoma cells (Figure 6A). Similarly, FUS-DDIT3 was also shown by co-IP to associate with SWI/SNF core components BRG1 (SMARCA4), BAF155 (SMARCC1), and accessory protein BAF57 (SMARCE1), although the co-IP was stronger for BRG1 than BAF155 and BAF57 (Figure 6B). These results from bioinformatic and co-IP analyses suggest that the FUS-DDIT3 interactome includes multiple chromatin regulatory complexes.
Figure 6.
Co-immunoprecipitation (co-IP) validates FUS-DDIT3’s interactions with KDM1A, BRG1, BAF155 and BAF57 in 402-91 and 1765-92 myxoid liposarcoma cells.
FUS was detected in the green channel, DDIT3 in the red channel. A merged yellow band represents FUS-DDIT3. Black arrows on blots point to proteins detected by the indicated Western blot antibodies. Grey arrows point to IgG chains from IP antibody. (A) Reciprocal co-IPs of FUS-DDIT3 and KDM1A were carried out in whole cell lysates. (B) Co-immunoprecipitation with FUS-DDIT3 was observed for SWI/SNF components BRG1 (SMARCA4), BAF155 (SMARCC1) and BAF57 (SMARCE1) in nuclear lysates. White bands indicate scan signal saturation for IgG heavy chain of IP antibody.
Discussion
Myxoid liposarcoma is driven by the fusion oncoprotein FUS-DDIT3, but the exact mechanism of action behind its capacity for transformation and tumor maintenance is unclear. Identification of the FUS-DDIT3 interactome is crucial to gaining a better understanding of its mechanism of action, and would also provide functionally relevant secondary targets against which rational therapeutic strategies could be designed, given the lack of any available agents confirmed to specifically target FUS-DDIT3.
While a previous study has identified the interactome of the N-terminal portions of the FET proteins that are often retained in sarcoma fusion oncoproteins [25], this is the first report on the FUS-DDIT3 interactome in myxoid liposarcoma based on comprehensive proteomics, and presents the following novel findings: (1) A large portion of the interactome consists of proteins involved in RNA processing and splicing, and that (2) FUS-DDIT3 associates with chromatin regulators such as KDM1A and members of the SWI/SNF chromatin remodeling complex (BRG1/SMARCA4, BAF155/SMARCC1, BAF57/SMARCE1).
The FUS-DDIT3 interactome identified from whole cell lysates of myxoid liposarcoma cell lines provides a valuable starting point for systematic evaluation of whether and how the interactors contribute to the oncogenic function of FUS-DDIT3. For example, SFPQ, one of the top hits in the interactome, is known to function as a transcriptional repressor by recruiting Sin3A [40]. Incidentally, HDAC inhibition has shown promise as a targeted therapy in myxoid liposarcoma - a HDAC inhibitor (pracinostat) trial that included five myxoid liposarcoma patients showed stable disease in three of the four assessable patients [41].
The large number of RNA processing proteins identified in the FUS-DDIT3 interactome raises the question as to whether FUS-DDIT3 plays a role in deregulating normal RNA processing, as wild type FUS is also involved in regulating RNA splicing [42], [43]. Even though many FUS-DDIT3 fusion variants have lost the RNA-binding domain of FUS, the fusion protein retains the ability to bind wild type FUS [25], and by extension, the ability to associate with FUS interactors. Since our interactome data only identifies proteins that are present in the same complex as FUS-DDIT3, future validation may be useful in determining whether FUS-DDIT3 interacts directly with these RNA processing proteins. Nonetheless, FUS-DDIT3 has been reported to localize to splicing factor compartments [44]. The functional implication of RNA processing proteins in the FUS-DDIT3 interactome is unknown, but our findings in the FUS-DDIT3 interactome are reminiscent of the large number of RNA splicing and processing proteins found in the Ewing sarcoma EWSR1-FLI1 interactome, in which the spliceosomal network of EWSR1-FLI1 differed from EWSR1, and EWSR1-FLI1 was found to interfere with the alternative splicing profile [45].
Since the RNA-binding domains of FUS are not retained in most FUS-DDIT3 variants, the fusion protein is not expected to retain the same RNA processing functions of wild type FUS. It is possible that the loss of one normal FUS allele due to the translocation event could impact normal FUS-mediated RNA functions in the cell. However, based on the demonstration of increased mRNA and protein expression of the remaining FUS allele from an unknown compensatory mechanism in myxoid liposarcoma primary cell lines [46], loss of function due to reduced wild type FUS expression appears unlikely.
Another possible impact of the oncoprotein on RNA processing could come if FUS-DDIT3 exerted a dominant negative effect on the RNA processing activity of wild type FUS. How FUS-DDIT3 would effect a dominant negative function is unclear, but could occur through the loss of C-terminal FUS interactors, one of which is YB-1. YB-1 was reported to mediate alternative splicing of adenovirus E1A pre-mRNA by FUS, but this activity was blocked by the presence of FUS-DDIT3, which does not interact with YB-1 [24]. Alternatively, the interaction of FUS-DDIT3 with multimers of wild type FUS [25] might result in improper sequestration and functional inactivation of FUS.
There is also increasing evidence that altered epigenetic control of developmentally regulated genes is the major mechanism of action of the fusion proteins in other translocation-associated sarcomas [26], [27], [28], [29], [30], [31], [32]. The identification of multiple chromatin regulatory complexes in the FUS-DDIT3 interactome, even those that possess opposing activities such as SWI/SNF and PRC, was therefore not surprising. Some sarcoma fusion proteins have been reported to interact with and to function through more than one chromatin regulatory complex — EWSR1-FLI1 with NuRD/KDM1A and SWI/SNF in Ewing sarcoma [26], [28], and SS18-SSX with PRC1.1/PRC2 and SWI/SNF in synovial sarcoma [30], [31].
Although there has been no published studies (i.e. ChIP-seq) on direct gene targets of FUS-DDIT3, it would be interesting to investigate FUS-DDIT3 regulation of histone modifications at the promoters or cis regulatory elements of important target genes that might be revealed in future epigenomic studies. More importantly, further investigation into the occupancy of chromatin regulators will determine whether any changes to histone modifications are the result of FUS-DDIT3 modulating the recruitment and/or activity of chromatin regulatory complexes at its targets.
The association of FUS-DDIT3 with the SWI/SNF complex is particularly interesting. Recent evidence points to strong SWI/SNF regulation of lineage-specific distal enhancers through direct interactions with p300 and acetylation of H3K27 [47], [48], [49]. This has implications for tumorigenesis, as deregulation of SWI/SNF function would likely disrupt the tightly-regulated expression of genes involved in differentiation and development. It is possible that FUS-DDIT3 could perturb normal SWI/SNF function by redirecting distribution of the chromatin remodeling complex to FUS-DDIT3 targets. Such a mechanism of action has been reported in Ewing sarcoma, where EWSR1-FLI1 recruits and redistributes SWI/SNF to tumor-specific enhancers that otherwise have no regulatory functions [28], and also in synovial sarcoma where SS18-SSX redirects SWI/SNF to KDM2B [31] and polycomb targets [32].
Determining whether FUS-DDIT3 also redistributes SWI/SNF would require detailed ChIP-seq analyses to identify the DNA regulatory elements and/or genes targeted by FUS-DDIT3, along with changes to the distribution of histone marks, SWI/SNF and other chromatin regulators, in the presence or absence of the fusion oncoprotein. Such data would aid in further clarifying how FUS-DDIT3 affects chromatin regulators to drive tumorigenesis, and perhaps identify key components whose activity can be reversed by emerging epigenetic drugs.
Conclusions
Our study reports the interactome of endogenous FUS-DDIT3 in myxoid liposarcoma cell lines, which includes RNA processing and splicing proteins, as well as members of multiple chromatin regulatory complexes.
Funding
This research was funded by the Canadian Cancer Society Research Institute (grant number 705615), the Terry Fox Research Institute (grant number 1082), and the Liddy Shriver Sarcoma Initiative (grant name “A ‘Bedside to Bench’ Investigational Platform for the Study of Myxoid Liposarcoma").
Acknowledgements
The authors would like to thank Angela Goytain and Jenny Wang for technical assistance, and Drs. Pierre Åman, Alexander Lazar, Neeta Somaiah, Hannah Beird, Chia Chin Wu, and Judith Bovée for helpful discussion.
Footnotes
Declaration of interest: The authors declare that they have no competing interests.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2019.05.004.
Contributor Information
Jamie S.E. Yu, Email: jamiesyu@mail.ubc.ca.
Shane Colborne, Email: seccolborne@gmail.com.
Christopher S. Hughes, Email: chughes@bcgsc.ca.
Gregg B. Morin, Email: gmorin@bcgsc.ca.
Torsten O. Nielsen, Email: torsten@mail.ubc.ca.
Appendix A. Supplementary data
Supplementary material
References
- 1.Aman P, Ron D, Mandahl N, Fioretos T, Heim S, Arheden K, Willén H, Rydholm A, Mitelman F. Rearrangement of the transcription factor gene CHOP in myxoid liposarcomas with t(12;16)(q13;p11) Genes Chromosom Cancer. 1992;5(4):278–285. doi: 10.1002/gcc.2870050403. [DOI] [PubMed] [Google Scholar]
- 2.Panagopoulos I, Höglund M, Mertens F, Mandahl N, Mitelman F, Aman P. Fusion of the EWS and CHOP genes in myxoid liposarcoma. Oncogene. 1996;12(3):489–494. [PubMed] [Google Scholar]
- 3.Powers MP, Wang W-L, Hernandez VS, Patel KS, Lev DC, Lazar AJ, López-Terrada DH. Detection of myxoid liposarcoma-associated FUS-DDIT3 rearrangement variants including a newly identified breakpoint using an optimized RT-PCR assay. Mod Pathol. 2010;23(10):1307–1315. doi: 10.1038/modpathol.2010.118. [DOI] [PubMed] [Google Scholar]
- 4.Schwartz JC, Cech TR, Parker RR. Biochemical properties and biological functions of FET proteins. Annu Rev Biochem. 2014;84(1) doi: 10.1146/annurev-biochem-060614-034325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dormann D, Haass C. Fused in sarcoma (FUS): an oncogene goes awry in neurodegeneration. Mol Cell Neurosci. 2013;56:475–486. doi: 10.1016/j.mcn.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 6.Yang Y, Liu L, Naik I, Braunstein Z, Zhong J, Ren B. Transcription factor C/EBP homologous protein in health and diseases. Front Immunol. 2017;8:1612. doi: 10.3389/fimmu.2017.01612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zinszner H, Albalat R, Ron D. A novel effector domain from the RNA-binding protein TLS or EWS is required for oncogenic transformation by CHOP. Genes Dev. 1994;8(21):2513–2526. doi: 10.1101/gad.8.21.2513. [DOI] [PubMed] [Google Scholar]
- 8.Kuroda M, Ishida T, Takanashi M, Satoh M, Machinami R, Watanabe T. Oncogenic transformation and inhibition of adipocytic conversion of preadipocytes by TLS/FUS-CHOP type II chimeric protein. Am J Pathol. 1997;151(3):735–744. [PMC free article] [PubMed] [Google Scholar]
- 9.Göransson M, Andersson MK, Forni C, Ståhlberg A, Andersson C, Olofsson A, Mantovani R, Aman P. The myxoid liposarcoma FUS-DDIT3 fusion oncoprotein deregulates NF-kappaB target genes by interaction with NFKBIZ. Oncogene. 2009;28(2):270–278. doi: 10.1038/onc.2008.378. [DOI] [PubMed] [Google Scholar]
- 10.Uboldi S, Bernasconi S, Romano M, Marchini S, Fuso Nerini I, Damia G, Ganzinelli M, Marangon E, Sala F, Clivio L. Characterization of a new trabectedin resistant myxoid liposarcoma cell line that shows collateral sensitivity to methylating agents. Int J Cancer. July 2011 doi: 10.1002/ijc.26340. [DOI] [PubMed] [Google Scholar]
- 11.Katz D, Boonsirikamchai P, Choi H, Lazar AJ, Wang W-L, Xiao L, Park MS, Ravi V, Benjamin RS, Araujo DM. Efficacy of first-line doxorubicin and ifosfamide in myxoid liposarcoma. Clin Sarcoma Res. 2012;2(1):2. doi: 10.1186/2045-3329-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffman A, Ghadimi MPH, Demicco EG, Creighton CJ, Torres K, Colombo C, Peng T, Lusby K, Ingram D, Hornick JL. Localized and metastatic myxoid/round cell liposarcoma: clinical and molecular observations. Cancer. 2013;119(10):1868–1877. doi: 10.1002/cncr.27847. [DOI] [PubMed] [Google Scholar]
- 13.Bonfanti M, La Valle E. Fernandez Sousa Faro JM, Faircloth G, Caretti G, Mantovani R, D'Incalci M. Effect of ecteinascidin-743 on the interaction between DNA binding proteins and DNA. Anticancer Drug Des. 1999;14(3):179–186. [PubMed] [Google Scholar]
- 14.Forni C, Minuzzo M, Virdis E, Tamborini E, Simone M, Tavecchio M, Erba E, Grosso F, Gronchi A, Aman P. Trabectedin (ET-743) promotes differentiation in myxoid liposarcoma tumors. Mol Cancer Ther. 2009;8(2):449–457. doi: 10.1158/1535-7163.MCT-08-0848. [DOI] [PubMed] [Google Scholar]
- 15.Grohar PJ, Griffin LB, Yeung C, Chen Q-R, Pommier Y, Khanna C, Khan J, Helman LJ. Ecteinascidin 743 interferes with the activity of EWS-FLI1 in Ewing sarcoma cells. Neoplasia. 2011;13(2):145–153. doi: 10.1593/neo.101202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harlow ML, Maloney N, Roland J, Guillén-Navarro MJ, Easton MK, Kitchen-Goosen SM, Boguslawski EA, Madaj ZB, Johnson BK, Bowman MJ. Lurbinectedin inactivates the Ewing sarcoma oncoprotein EWS-FLI1 by redistributing it within the nucleus. Cancer Res. 2016;76(22):6657–6668. doi: 10.1158/0008-5472.CAN-16-0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uboldi S, Craparotta I, Colella G, Ronchetti E, Beltrame L, Vicario S, Marchini S, Panini N, Dagrada G, Bozzi F. Mechanism of action of trabectedin in desmoplastic small round cell tumor cells. BMC Cancer. 2017;17(1):107. doi: 10.1186/s12885-017-3091-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barretina J, Taylor BS, Banerji S, Ramos AH, Lagos-Quintana M, Decarolis PL, Shah K, Socci ND, Weir BA, Ho A. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat Genet. 2010;42(8):715–721. doi: 10.1038/ng.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joseph CG, Hwang H, Jiao Y, Wood LD, Kinde I, Wu J, Mandahl N, Luo J, Hruban RH, Diaz LA. Exomic analysis of myxoid liposarcomas, synovial sarcomas, and osteosarcomas. Genes Chromosom Cancer. 2014;53(1):15–24. doi: 10.1002/gcc.22114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofvander J, Viklund B, Isaksson A, Brosjö O, Vult von Steyern F, Rissler P, Mandahl N, Mertens F. Different patterns of clonal evolution among different sarcoma subtypes followed for up to 25 years. Nat Commun. 2018;9(1):3662. doi: 10.1038/s41467-018-06098-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ron D, Habener JF. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes Dev. 1992;6(3):439–453. doi: 10.1101/gad.6.3.439. [DOI] [PubMed] [Google Scholar]
- 22.Crozat A, Aman P, Mandahl N, Ron D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature. 1993;363(6430):640–644. doi: 10.1038/363640a0. [DOI] [PubMed] [Google Scholar]
- 23.Bento C, Andersson MK, Aman P. DDIT3/CHOP and the sarcoma fusion oncoprotein FUS-DDIT3/TLS-CHOP bind cyclin-dependent kinase 2. BMC Cell Biol. 2009;10:89. doi: 10.1186/1471-2121-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rapp TB, Yang L, Conrad EU, Mandahl N, Chansky HA. RNA splicing mediated by YB-1 is inhibited by TLS/CHOP in human myxoid liposarcoma cells. J Orthop Res. 2002;20(4):723–729. doi: 10.1016/S0736-0266(02)00006-2. [DOI] [PubMed] [Google Scholar]
- 25.Thomsen C, Grundevik P, Elias P, Ståhlberg A, Aman P. A conserved N-terminal motif is required for complex formation between FUS, EWSR1, TAF15 and their oncogenic fusion proteins. FASEB J. August 2013 doi: 10.1096/fj.13-234435. [DOI] [PubMed] [Google Scholar]
- 26.Sankar S, Bell R, Stephens B, Zhuo R, Sharma S, Bearss DJ, Lessnick SL. Mechanism and relevance of EWS/FLI-mediated transcriptional repression in Ewing sarcoma. Oncogene. 2013;32(42):5089–5100. doi: 10.1038/onc.2012.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riggi N, Knoechel B, Gillespie SM, Rheinbay E, Boulay G, Suvà ML, Rossetti NE, Boonseng WE, Oksuz O, Cook EB. EWS-FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma. Cancer Cell. 2014;26(5):668–681. doi: 10.1016/j.ccell.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boulay G, Sandoval GJ, Riggi N, Iyer S, Buisson R, Naigles B, Awad ME, Rengarajan S, Volorio A, McBride MJ. Cancer-specific retargeting of BAF complexes by a prion-like domain. Cell. August 2017 doi: 10.1016/j.cell.2017.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su L, Sampaio AV, Jones KB, Pacheco M, Goytain A, Lin S, Poulin N, Yi L, Rossi FM, Kast J. Deconstruction of the SS18-SSX fusion oncoprotein complex: insights into disease etiology and therapeutics. Cancer Cell. 2012;21(3):333–347. doi: 10.1016/j.ccr.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kadoch C, Crabtree GR. Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell. 2013;153(1):71–85. doi: 10.1016/j.cell.2013.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banito A, Li X, Laporte AN, Roe J-S, Sanchez-Vega F, Huang C-H, Dancsok AR, Hatzi K, Chen C-C, Tschaharganeh DF. The SS18-SSX oncoprotein hijacks KDM2B-PRC1.1 to drive synovial sarcoma. Cancer Cell. 2018;33(3):527–541. doi: 10.1016/j.ccell.2018.01.018. e528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McBride MJ, Pulice JL, Beird HC, Ingram DR, D'Avino AR, Shern JF, Charville GW, Hornick JL, Nakayama RT, Garcia-Rivera EM. The SS18-SSX fusion oncoprotein hijacks BAF complex targeting and function to drive synovial sarcoma. Cancer Cell. 2018;33(6):1128–1141.e7. doi: 10.1016/j.ccell.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Böhm M, Wachtel M, Marques JG, Streiff N, Laubscher D, Nanni P, Mamchaoui K, Santoro R, Schäfer BW. Helicase CHD4 is an epigenetic coregulator of PAX3-FOXO1 in alveolar rhabdomyosarcoma. J Clin Invest. 2016;126(11):4237–4249. doi: 10.1172/JCI85057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Graaff MA, Yu JSE, Beird HC, Ingram DR, Nguyen T, Juehui Liu J, Bolshakov S, Szuhai K, Aman P, Torres KE. Establishment and characterization of a new human myxoid liposarcoma cell line (DL-221) with the FUS-DDIT3 translocation. Lab Investig. June 2016 doi: 10.1038/labinvest.2016.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hughes CS, Foehr S, Garfield DA, Furlong EE, Steinmetz LM, Krijgsveld J. Ultrasensitive proteome analysis using paramagnetic bead technology. Mol Syst Biol. 2014;10(10):757. doi: 10.15252/msb.20145625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lavallée-Adam M, Rauniyar N, McClatchy DB, Yates JR. PSEA-Quant: a protein set enrichment analysis on label-free and label-based protein quantification data. J Proteome Res. 2014;13(12):5496–5509. doi: 10.1021/pr500473n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knott GJ, Bond CS, Fox AH. The DBHS proteins SFPQ, NONO and PSPC1: a multipurpose molecular scaffold. Nucleic Acids Res. 2016;44(9):3989–4004. doi: 10.1093/nar/gkw271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simicevic J, Deplancke B. Transcription factor proteomics-Tools, applications, and challenges. Proteomics. 2017;17(3-4) doi: 10.1002/pmic.201600317. [DOI] [PubMed] [Google Scholar]
- 39.Lindemann C, Thomanek N, Hundt F, Lerari T, Meyer HE, Wolters D, Marcus K. Strategies in relative and absolute quantitative mass spectrometry based proteomics. Biol Chem. 2017;398(5-6):687–699. doi: 10.1515/hsz-2017-0104. [DOI] [PubMed] [Google Scholar]
- 40.Mathur M, Tucker PW, Samuels HH. PSF is a novel corepressor that mediates its effect through Sin3A and the DNA binding domain of nuclear hormone receptors. Mol Cell Biol. 2001;21(7):2298–2311. doi: 10.1128/MCB.21.7.2298-2311.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chu QS-C, Nielsen TO, Alcindor T, Gupta A, Endo M, Goytain A, Xu H, Verma S, Tozer R, Knowling M. A Phase II study of SB939, a novel pan-histone deacetylase inhibitor, in patients with translocation-associated recurrent/metastatic sarcomas -NCIC-CTG IND 200. Ann Oncol. January 2015 doi: 10.1093/annonc/mdv033. [DOI] [PubMed] [Google Scholar]
- 42.Lagier-Tourenne C, Polymenidou M, Hutt KR, Vu AQ, Baughn M, Huelga SC, Clutario KM, Ling S-C, Liang TY, Mazur C. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat Neurosci. 2012;15(11):1488–1497. doi: 10.1038/nn.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rogelj B, Easton LE, Bogu GK, Stanton LW, Rot G, Curk T, Zupan B, Sugimoto Y, Modic M, Haberman N. Widespread binding of FUS along nascent RNA regulates alternative splicing in the brain. Sci Rep. 2012;2(1):603. doi: 10.1038/srep00603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Göransson M, Wedin M, Aman P. Temperature-dependent localization of TLS-CHOP to splicing factor compartments. Exp Cell Res. 2002;278(2):125–132. doi: 10.1006/excr.2002.5566. [DOI] [PubMed] [Google Scholar]
- 45.Selvanathan SP, Graham GT, Erkizan HV, Dirksen U, Natarajan TG, Dakic A, Yu S, Liu X, Paulsen MT, Ljungman ME. Oncogenic fusion protein EWS-FLI1 is a network hub that regulates alternative splicing. Proc Natl Acad Sci U S A. 2015;112(11):201500536–E201501316. doi: 10.1073/pnas.1500536112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spitzer JI, Ugras S, Runge S, Decarolis P, Antonescu C, Tuschl T, Singer S. mRNA and protein levels of FUS, EWSR1, and TAF15 are upregulated in liposarcoma. Genes Chromosom Cancer. 2011;50(5):338–347. doi: 10.1002/gcc.20858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alver BH, Kim KH, Lu P, Wang X, Manchester HE, Wang W, Haswell JR, Park PJ, Roberts CWM. The SWI/SNF chromatin remodelling complex is required for maintenance of lineage specific enhancers. Nat Commun. 2017;8 doi: 10.1038/ncomms14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mathur R, Alver BH, San Roman AK, Wilson BG, Wang X, Agoston AT, Park PJ, Shivdasani RA, Roberts CWM. ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. Nat Genet. 2017;49(2):296–302. doi: 10.1038/ng.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X, Lee RS, Alver BH, Haswell JR, Wang S, Mieczkowski J, Drier Y, Gillespie SM, Archer TC, Wu JN. SMARCB1-mediated SWI/SNF complex function is essential for enhancer regulation. Nat Genet. 2017;49(2):289–295. doi: 10.1038/ng.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material