Abstract
Fungus-growing termites engage in an obligate mutualistic relationship with Termitomyces fungi, which they maintain in monocultures on specialised fungus comb structures, without apparent problems with infectious diseases. While other fungi have been reported in the symbiosis, detailed comb fungal community analyses have been lacking. Here we use culture-dependent and -independent methods to characterise fungus comb mycobiotas from three fungus-growing termite species (two genera). Internal Transcribed Spacer (ITS) gene analyses using 454 pyrosequencing and Illumina MiSeq showed that non-Termitomyces fungi were essentially absent in fungus combs, and that Termitomyces fungal crops are maintained in monocultures as heterokaryons with two or three abundant ITS variants in a single fungal strain. To explore whether the essential absence of other fungi within fungus combs is potentially due to the production of antifungal metabolites by Termitomyces or comb bacteria, we performed in vitro assays and found that both Termitomyces and chemical extracts of fungus comb material can inhibit potential fungal antagonists. Chemical analyses of fungus comb material point to a highly complex metabolome, including compounds with the potential to play roles in mediating these contaminant-free farming conditions in the termite symbiosis.
Subject terms: Metabolomics, Evolutionary ecology, Metagenomics, Symbiosis
Introduction
Monoculture farming faces a number of challenges, including increased susceptibility to pathogens in genetically homogenous crop populations1,2, as exemplified by Ireland’s Great Famine caused by potato blight (Phytophthora infestans). While diverse crops may provide more robust defence against invasive disease3, two major farming symbioses in nature – the New World fungus-farming ants and the Old World fungus-farming termites – maintain basidiomycete fungal crops in monoculture4–7. These are expected to be disease-prone because plant substrates harvested to manure the fungal crop may contain potential antagonists that could spread throughout the genetically homogeneous fungal gardens. In the ants, specialised Escovopsis spp. (Ascomycota) mycoparasites infect ant cultivars with potentially devastating impact on colonies8, but with the exception of stowaway ascomycete fungi in the genus Pseudoxylaria (Ascomycota: Xylariales)9, farming termites do not appear to suffer from specialised diseases.
Farming in termites originated thirty million years ago in a monophyletic group of 11 genera (330 described species) of higher termites (subfamily Macrotermitinae)10–14. The lignocellulose-digesting fungus cultivar (Termitomyces; Basidiomycota) is a source of termite nutrition15,16, providing access to otherwise inaccessible plant resources. It is cultivated in fungal gardens (combs), which are built with foraged plant material and asexual Termitomyces spores (in fungal nodules) that are mixed during termite gut passage17,18. Combs provide an optimal medium for fungal growth16, with Termitomyces growing rapidly to produce nutrient- and spore-rich nodules that are ingested by termite workers19–21. Workers then deposit further plant-spore mixture as new fungus comb on the older comb in a continuation of the fungal growth cycle (Fig. 1).
Figure 1.
Right. A simplified schematic of the lifecycle of Macrotermes natalensis and Odontotermes sp. and O. cf. badius. In most termite species, Termitomyces spores are acquired from the environment (i) and brought back to incipient colonies, where spores fuse (ii) to form the dikaryotic mycelium characteristic of the growth within fungus combs. Left: Defence modes in farming termites. Termites forage for plant material (iii), which may contain potential antagonists such as Trichoderma or Beauveria (spore structures shown). Before being incorporated into the fungus comb, this plant material passes through the guts of young termite workers (iv). Any potential antagonists that pass through the gut unharmed must be suppressed through for example termite antimicrobial peptides, out-competition by Termitomyces, or antimicrobials of Termitomyces or bacterial origin.
The decaying plant substrate that the termites harvest to manure Termitomyces is expected to harbour fungi that could compete with or antagonise Termitomyces or the termites (Fig. 1); however, only few have been reported, and these appear only to emerge after the termites have died or abandoned the mound22–31. There is thus no evidence of persistent infections in active combs or indications of specialised diseases, implying highly effective defences. These may include contaminant avoidance32 or physical removal by the termite host, termite-derived antimicrobial peptides33,34, and/or gut and comb symbiont-derived antimicrobials28,29,35–37. Compounds from Termitomyces may also be implicated, as non-Termitomyces fungal growth within combs is delayed by days following termite removal38,39. To profile fungal communities within fungus combs, we employ a combination of culture-dependent and -independent characterisation of fungi within combs of healthy termite nests. Subsequently, we take a metabolomics approach to examine the fungus-comb chemical environment and test its potential for suppression of antagonists in vitro.
Results
Culture-dependent and -independent identification of fungi within fungus combs
Nodule inoculations yielded solely Termitomyces in 83.3% of cases, with the remaining inoculations resulting in 23 fungal isolates from nine different genera (Table S1). Six events of fungus comb infections of laboratory colonies produced four Trichoderma and one Fusarium isolate (Table S1). Amplicon analyses from combs of 19 colonies (eight M. natalensis, six Odontotermes sp. and five Odontotermes cf. badius; Table S1) produced 702,804 quality-filtered ITS1 gene sequences from 454 pyro-sequencing and 408,491 quality-filtered ITS2 gene sequences from Illumina MiSeq, with 12,570–53,562 reads per sample after 454 sequence splitting, and 5,652–55,281 reads from MiSeq (Table S1). Coverage was judged sufficient based on rarefaction analyses (Fig. S1). 99% similarity clustering analysis yielded 560 and 14,095 OTUs from 454 and MiSeq, respectively, with 19–167 OTUs per 454-sequenced and 181–1,527 OTUs per MiSeq-sequenced comb sample. Taxonomic assignments yielded two fungal genera in the 454 (99.9% Termitomyces) and 21 in the MiSeq (99.9% Termitomyces) analyses (Tables S2 and S3). Non-Termitomyces genera were thus present in extremely low abundance, with Preussia (<0.001% relative abundance) being the only other identified genus in the 454 dataset, and the remaining non-Termitomyces reads (0.11%) being unassigned. In the MiSeq dataset, 20 non-Termitomyces fungal genera accounted for 0.07% relative abundance. Pseudoxylaria was detected in one M. natalensis and three Odontotermes sp. combs but accounted for only 0.03% of reads. It should be noted that this metabarcoding approach is only semi-quantitative as polyploidy, multiple ITS copies, DNA extraction biases etc. may bias community analyses. Although our mock fungal community analyses indicated that the primers used would detect the expected range of fungi (Fig. S2), the bias towards Termitomyces might in reality be less extreme than what we observed.
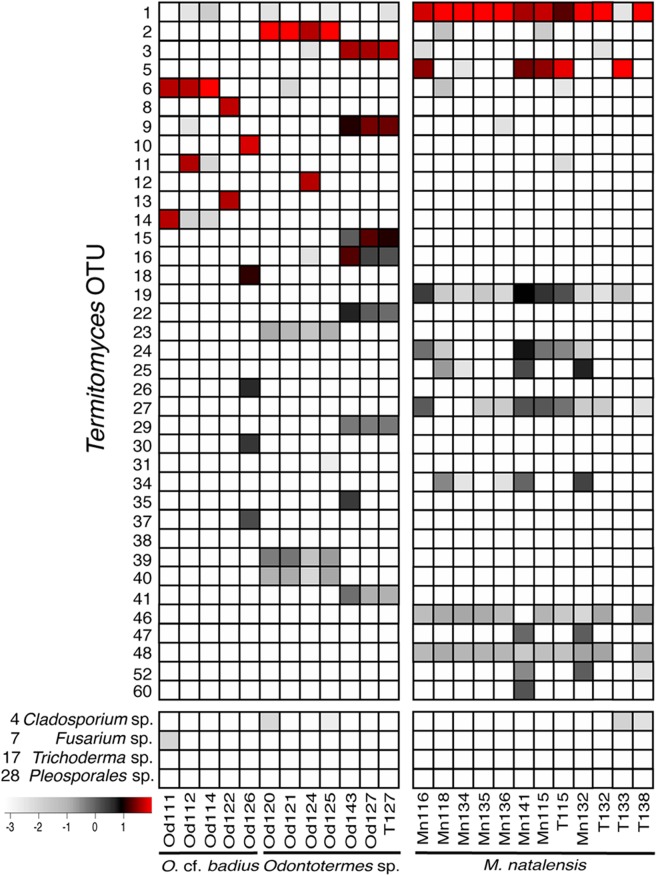
Among the 561 and 14,002 Termitomyces OTUs in the 454 and MiSeq analyses, respectively, the most abundant OTUs (>1% relative abundance across datasets) are presented in heatmaps in Figs 2, S2 and S3. As expected, the analyses identified different Termitomyces OTUs in combs from different termite species; however, differences in the most abundant OTU were also apparent between combs from the same termite species (Figs 2, S2–S4). For example, the dominant Termitomyces OTUs in Od122 and Od126 combs were absent from other O. cf. badius colonies. Interestingly, three abundant OTUs from Odontotermes sp. Od127 were equally abundant and similar to the pure Termitomyces culture from that nest (Figs 2, S2 and S3). Furthermore, it is noteworthy that variants deviating from the one or two main types were found, but usually at low frequencies.
Figure 2.
Heat maps showing the relative abundances of Termitomyces OTUs that accounted for >1% relative abundance per sample and the four other fungi identified in combs (fungus combs indicated with Od and Mn followed by colony code; pure cultures of Termitomyces indicated with T followed by colony code) in the MiSeq dataset for Odontotermes (left) and M. natalensis (right). The scale is a logarithmic calculation of the taxon read percentage out of the total number of quality-filtered and classified reads per sample.
Inter- and intra-colony variation of Termitomyces communities
Extraction and further analyses with 100% similarity clustering of Termitomyces MiSeq reads generated 47,492 Termitomyces variants across all fungus comb and pure culture samples (Table S3). At this threshold, the number of variants found ranged from 4,919 to 55,037 (average 22,427) per sample. While more Termitomyces variants were shared by fungus combs from colonies of different termite species, variants were apparent between conspecific colonies (Figs 2, S2 and S4; Table S3). For example, O. cf. badius colonies maintained different dominant Termitomyces variants. Differences between Termitomyces variants were present within single colonies. For example, the four most abundant of the 538 variants in the fungus comb from Mn115 had base pair differences at three positions, which were consistent with the findings for the pure culture (Table S3, Fig. S5). Similarly, Od127 showed differences between its eight dominant Termitomyces variants at 14 base pair positions, similar to the pure culture from that nest (Fig. S5), in which all but one of eight dominant Termitomyces variants were 100% identical to the dominant Termitomyces variants in the fungus comb.
The effect of crude and fractionated fungus comb extracts on fungal contaminant growth
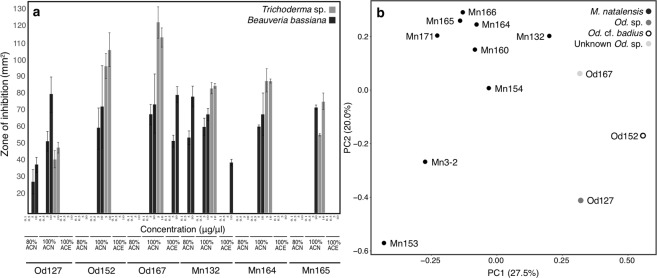
We evaluated the antifungal properties of extracts of comb material from six termite colonies (two of which were included in the fungal community analysis) against seven contaminants (four of which were detected at trace levels in the mycobiota analyses; Fig. 2). Five out of six crude acetonitrile (ACN) extracts inhibited the growth of Trichoderma sp. and three out of six Beauveria bassiana (Table S4, Fig. S7). Only the crude ACN extract from Od127 comb was inactive, while Od152 and Od167 extracts were active at 10 and 5 μg/μl (Table S4, Fig. S7). The crude Macrotermes ACN comb extracts showed varying degrees of activity at 10 and 5 μg/μl; in contrast, the six crude extracts prepared with a solution of acetonitrile/acetic acid (ACNAA) did not inhibit the growth of any of the tested fungi (Fig. S7; Table S4).
In order to narrow down the range of components potentially responsible for the observed activity, fractions from solid phase extraction (SPE) purification of the six tested extracts were also subjected to antifungal activity assays. Each of the active crude extracts also showed inhibition of Trichoderma sp. and B. bassiana in at least one of their corresponding fractions (Fig. 3; Table S4). The 100% ACN fraction of all comb extracts showed consistent inhibition of these two fungi at its highest concentration (10 μg/μl), followed by the next highest concentration (5 μg/μl) in all but one comb (Fig. 3; Table S4). While only the 100% ACN fractions inhibited Trichoderma sp., other fractions inhibited the entomopathogenic fungus B. bassiana (Fig. 3; Table S4). None of the crude comb extracts or fractions inhibited Termitomyces T112 or T115 (Table S4).
Figure 3.
Antimicrobial activity assays and LCMS-PCA of fractionated fungus comb extracts. (a) Mean (±SE; n = 3) zones of inhibition (mm2) of Trichoderma sp. (grey) and Beauveria bassiana (black) when exposed to different concentrations of fractionated fungus comb extracts. Fractions were obtained by SPE eluting with 80% and 100% acetonitrile (ACN) and 100% acetone (ACE). Colonies of origin of fungus combs are indicated at the bottom of each panel. (b) PCA plot (PC1 vs PC2) of LCMS data for fractionated fungus comb extracts from 12 colonies of M. natalensis, Odontotermes sp. and Odontotermes cf. badius. Colonies labelled with colony ID numbers.
LCMS/MS and principal component analysis (PCA) of fractionated fungus comb extracts
To analyse the overall metabolite composition and identify potential entities responsible for antimicrobial activity, tandem liquid chromatography-mass spectrometry (LCMS/MS) data were acquired for fractionated comb extracts prepared in the same manner, and for the same colonies, as those used for antifungal assays. These were supplemented with data from fractionated comb extracts for six additional M. natalensis colonies. Following data processing with MZmine 240, a total of 66 chemical features (unique combinations of m/z and retention time values) were detected under positive electrospray ionisation (Table S5). Of these, 35 were detected in combs from both termite genera, while 26 were unique to Macrotermes and five unique to Odontotermes. In negative ion mode, 64 chemical features were identified, of which 24 were found in combs from both termite genera, 27 were unique to Macrotermes and 13 unique to Odontotermes. Dereplication using AntiBase and an in-house database of commercial reference substances revealed a broad range of putative hits for about 60% of all identified features within the positive ion mode and about 80% for the negative ion mode leaving a significant amount of mostly termite genera-specific chemical features unidentified. The predicted identities belong to a broad range of compound classes assigned to primary and secondary metabolism of plants and microorganisms. These include important monosaccharides, metabolites of the shikimic pathway, small and long chain fatty acids, bacterial autoinducer and quorum sensing molecules, as well as terpenes, terpene-derived compounds, and sterols of microbial or plant origin. Furthermore, several phenolic and polyhydroxylated aromatic compounds were putatively assigned, which could be redox mediators of fungal origin (Beemelmanns, unpublished data). In addition, we identified several typical fungal metabolites, such as terpenes, sterols (e.g., ergosterol and derivatives, carboxysterols or andrastins) and PKS-derived cyclic lactones (e.g., phomolides). Principal component analysis (PCA) revealed clustering largely according to termite genus, with 47.5% of the variance captured within the first two PCA components (Fig. 3).
Inhibition of contaminant fungi by Termitomyces
To explore whether Termitomyces could be responsible for inhibition of other fungi, we conducted two in vitro interaction assays detailing inhibitory capabilities of a M. natalensis (T115) and an Odontotermes sp. (T112) Termitomyces (for details, see Supplementary Information). Most contaminant fungi grew less relative to controls when inoculated in direct contact with Termitomyces, with the exceptions of Trichoderma and Fusarium when tested against T115 (Fig. S8). Similar patterns were observed in the direct interactions with Termitomyces T112, but here Fusarium isolates never reached control growth levels, while a Cladosporium and a Pleosporales isolate grew more than twice as much in the presence of Termitomyces compared to controls (Fig. S8). In the indirect interaction assay, the Trichoderma isolates reached control rates in the presence of T115, a Fusarium isolate grew more than controls, while Umbelopsis and Cunninghamella had the same low growth rate as in the first assay (Fig. S8). In the T112 interactions, growth of one Fusarium isolate and both Cladosporium isolates was similar to in the direct assay; however, the other Fusarium isolate displayed increased growth and the Pleosporales sp. isolate exhibited reduced growth compared to the direct assay (Fig. S8).
Discussion
Termite fungus combs are essentially free of putative fungal pathogens
Non-Termitomyces fungi are extraordinarily rare within termite fungus combs, collectively accounting for <0.03% relative abundance based on the deep sequencing of mycobiotas from 19 nests. Only four of the contaminant isolates obtained from nodule and comb infections were detected in the amplicon sequencing, and even then, in extremely low abundances. Meta-barcoding data alone is only semi-quantitative as not all fungal species are picked up by this method to equal degrees and because polyploidy, heterokaryosis, differences in copy number of the ITS repeat and other factors may bias towards certain species. The mock community analyses to some extent help assess this potential bias, and we indeed amplified the four contaminant isolates that overlapped in our isolations and in the MiSeq of combs at expected high abundances in the mock communities. Several fungal genera, for example Cladosporium, Trichoderma and Umbelopsis, were obtained from isolations from multiple colonies of the three termite species, suggesting that they may enter combs relatively frequently, but likely only remain within combs as non-metabolising spores. These are frequently found in soil and decaying wood, and presumably enter the comb with the macerated plant substrate used to manure Termitomyces. Indeed, several have previously been reported from fungus-growing termites (e.g., Trichoderma harzianum28). Strikingly, Pseudoxylaria was encountered only at trace levels among amplicons. While Pseudoxylaria has not been reported from healthy functioning combs, it is often present in abandoned colonies22,41, and can usually readily be obtained if comb material is incubated in the absence of termites. Our findings thus support that co-occurring fungi such as Pseudoxylaria are stowaways that employ a sit-and-wait strategy42, emerging only when termites are absent.
Termitomyces diversity within individual termite colonies
Fungus combs of Macrotermitinae are optimised for Termitomyces growth, but with the exception of studies in M. natalensis4 and Odontotermes formosanus7, there has been little effort to determine the diversity of Termitomyces within colonies. Fungal symbiont acquisition is mainly horizontal in newly founded colonies43, apart from in Macrotermes bellicosus and the genus Microtermes, where Termitomyces transmission is vertical and uniparental via one of the two founding reproductive alates10,43. Despite horizontal transmission, there is broad co-cladogenesis between termite hosts and fungus crops, implying some degree of association specificity10. Whether acquired horizontally or vertically, Termitomyces is propagated asexually within termite colonies44, and competitive exclusion through frequency-dependent selection ensures monoculture within individual nests4. Our findings confirm that individual termite colonies maintain a single Termitomyces strain and are consistent with previous work showing that Termitomyces grows as a heterokaryotic mycelium with two genetically different nuclei within both fungus combs and pure cultures45,46.
Although most nests had combs with two dominant Termitomyces ITS2 variants, three dominant variants were observed in both comb and pure-culture samples of T127 from an Odontotermes sp. nest (Fig. S5). Given that this culture was obtained from a single nodule, and therefore is unlikely to consist of two fungal strains, either the strain has three nuclei with unique ITS2 variants or one of the nuclei has two distinct ITS2 variants. The most plausible explanation, which is also consistent with previous findings45, is that combs contain a dikaryotic mycelium with one nucleus having more than one ITS variant. This is also consistent with the finding of small percentages of ITS variants deviating at a few positions from the most dominant types in other strains (Tables S3 and S4; Figure S5). Those findings beg for more study on the organisation of the ITS variants in heterokaryotic strains, either between the repeat units within nuclei or between genetically different nuclei within a single heterokaryon. The generation of single-nucleus (homokaryotic) isolates using protoplasting would allow a determination of ITS variants within nuclei, and shed light on whether the consistent low-abundance presence of variants in fungus combs and pure cultures are within-nucleus variants that never dominate ITS compositions.
Termitomyces diversity between colonies
As expected, fungus combs from colonies of the same termite species harboured similar Termitomyces fungi, with our findings supporting the association of M. natalensis with a single Termitomyces species10,47. While most comb and isolate amplicon-sequencing data for M. natalensis indicated the presence of the same dominant ITS2 variant (OTU 1), one pure isolate was dominated by another variant (OTU 5). This variant was also abundant in several other M. natalensis nests and pure cultures, and almost always co-occurred with three other variants (OTUs 19, 24, and 27), which also supports that low-abundance variants are most likely present within single Termitomyces nuclei. As with M. natalensis, we found that the Termitomyces strains associated with Odontotermes sp. were dominated by the same ITS2 variants, suggesting that this termite species also likely associates with a single Termitomyces species, although this is based on only a few nests. If the unique ITS2 variant compositions are indeed indicative of different Termitomyces species, the five O. cf. badius species appear to associate with three different Termitomyces species, consistent with previous findings of low Termitomyces specificity in this termite species47.
Inhibitory effects of Termitomyces and comb-residing bacteria
To investigate the extent to which Termitomyces has antifungal activities, we explored direct and indirect interactions between Termitomyces and contaminant fungi. We found that Termitomyces can have a negative effect on the growth of certain but not all tested fungi, but there was quite some variation in the interactions observed between the two Termitomyces isolates (Fig. S8). The lack of a negative effect on Trichoderma implies that the observed activity against this fungus (and likely B. bassiana) of fungus comb extracts (see below) is unlikely to be attributable to Termitomyces alone, although culture-based assays may not mimic natural comb conditions. A more parsimonious explanation is thus that antagonistic fungi are suppressed by antifungals produced by bacteria residing within combs, with the most likely candidates being Actinobacteria, which are abundant within combs48 and well-known antimicrobial producers35,36,49.
The chemical environment of the fungus comb suppresses certain fungi
Compounds present in fungus combs could be responsible for suppression of potential antagonists. ACN extracts from five colonies showed antifungal activity against Trichoderma sp., which may compete with or antagonise Termitomyces, as well as the entomopathogenic fungus B. bassiana, suggesting that inhibitory properties may even target pathogens of the termite host. The comb chemical environment may thus suppress non-Termitomyces fungi, notably without negatively affecting Termitomyces (Table S4). To characterise this chemical environment, we performed LCMS/MS analyses complemented with PCA to assess the similarity of the chemical environments of fungus combs across different nests. Our analysis revealed some degree of clustering of colonies according to termite genera (Fig. 3), with the chemical profiles of samples collected from Odontotermes sp. and O. cf. badius nests resembling each other more closely than M. natalensis combs.
Careful examination of the LCMS/MS data revealed a complex assembly of detected chemical features in fungus combs. Subsequent comparison with the AntiBase natural compound database resulted in putative identifications of primary and secondary metabolites as shown in Table S5a–d. The identities of several metabolites (4-hydroxybenzaldehyde, azelaic acid, indole-3-carboxaldehyde, methylsuccinic acid, stearamide etc.) were verified using an in-house database of authentic commercial compounds, thereby comparing their retention times, high-resolution masses and fragmentation patterns. The most abundantly-detected features were universal primary metabolites (e.g., sugars, succinic acid derivatives), bacterial quorum sensing signals including several homoserine lactone derivatives, bacterial autoinducer signals, and plant and fungal derived fatty acids and sterols (e.g., cholesterols, ergosterols). While there is evidently an abundance of small primary metabolites present within fungal combs, several features attributed to secondary metabolism were also detected. A closer look at the assigned chemical features of comb extracts that inhibited fungal growth of contaminants revealed among others, the antioxidant ganosporeric acid and fungal-derived triterpenoid metabolites50, which have antibiotic properties. However, no general dominating antibiotic compounds present in all active comb samples could be identified.
In contrast, termite genera-specific chemical identities were often putatively assigned as secondary metabolites of diverse origin or were unassigned. Thus, it is tempting to speculate that genera-specific chemical features could be derived from differences in comb bacterial communities48 or Termitomyces associated with the termite genera. As our assay analysis revealed some degree of inhibition, we thus hypothesize that the observed antifungal activity is likely a result of the combination of bioactive compounds present within the comb material and cannot be attributed to single highly-abundant secondary metabolites. Overall the rate of true identification is illustrative of the challenges of metabolomics studies similar as the assignment of the observed activity to single compounds or compound groups. Pairing the metabolomic data with meta-transcriptomics data of healthy versus infected comb material or colonies directed at analysing secondary metabolite gene clusters and regulatory genes may help alleviate uncertainty surrounding compound identities.
The combined metabolome of the fungus comb may contribute to colony health not only by supporting Termitomyces growth but also suppressing antagonists within fungus combs, implying that the comb environment may act as a defence component in a series of consecutive defences that collectively allow the termites to sustainably maintain disease-free monoculture fungus farms. The termites themselves play key roles in keeping fungus combs healthy through hygienic practices, such as avoidance or physical removal of unwanted fungi and the secretion of antimicrobial peptides. Subsequent obligate gut passage of all incoming plant substrate for Termitomyces growth may provide an effective filter for removing substrate-dwelling competitors and antagonists, but unwanted microbes surviving gut passage should threaten comb health and select for the maintenance of comb defences. Collectively, the presence of sequential complementarity and multipartite defences in the symbiosis may be the key to the extremely robust broad-spectrum defence against a range of putative antagonists, but we have only just begun to unravel the relative importance of each of these defences and the antimicrobials involved.
Material and Methods
Culture-dependent isolation of cultivable fungi from fungus combs and PCR-based identification
Four Odontotermes cf. badius, three Odontotermes sp., one unknown Odontotermes sp., and four Macrotermes natalensis colonies were excavated from three sites in South Africa in 2011 (Table S1)48. Healthy fungus comb, workers, and soldiers were collected and brought back to the laboratory, where sub-colonies were set up in plastic containers at ambient humidity and 25 °C. For each nest, 12 nodules (asexual fruiting structures) of Termitomyces were aseptically placed on potato dextrose agar (PDA; 39 g/l) on the day of collection. Non-Termitomyces fungi growing from these inoculations were transferred to PDA, as were fungal infections of laboratory sub-colonies. These “contaminant” fungi have been deposited in the culture collection at the Department of Plant and Environmental Sciences, University of Copenhagen. We determined their identities by sequencing part of the Internal Transcribed Spacer (ITS) gene using ITS4 and ITS5 primers51.
Samples for high throughput sequencing and chemical extraction of fungus combs
To characterise the fungal community composition within the fungus combs, material from 20 colonies of two termite genera were collected (Table S1). Samples were stored in RNAlater (Ambion, Inc., USA) at −20 °C until further use. To investigate the antifungal properties and chemical environment of fungus combs, material from 12 colonies of the same two genera (Table S1) was collected and stored at −20 °C until extractions were performed.
ITS amplicon sequencing
DNA extraction and amplicon sequencing
The FastDNA SPIN Kit for Soil (MP Biomedicals, USA) was used for DNA extractions following the manufacturer’s instructions, with protocol modifications as in48. The ITS1 region was amplified for 454 pyrosequencing using ITS1-F (5′ CTTGGTCATTTAGAGGAAGTAA 3′) and ITS4-R (5′ TCCTCCGCTTATTGATATGC 3′) primers51,52 with additional sample specific multiplex identifier barcodes. PCR preparation, conditions and library preparations were performed as in48, with the exception that the annealing temperature was 58 °C. The ITS2 gene was chosen for Illumina MiSeq using ITS3-F (5′ GCATCGATGAAGAACGCAGC 3′) and ITS4-R (5′ TCCTCCGCTTATTGATATGC 3′) primers, and amplified using a dual-indexing sequencing strategy53. PCR preparation, conditions and library preparations were performed as for MiSeq pyrosequencing in48, with the exception that the annealing temperature was 56 °C. The use of both 454 pyrosequencing and Illumina MiSeq techniques reflects only the changing state of the art over the time period in which this work was performed. Five cultures of Termitomyces were included to compare the genetic makeup of pure cultures and natural fungus combs, as were three fungal community mock samples, each to test whether the ITS region adequately captured community diversity. Mock 1 had equal amounts of Cladosporium phaenocomae, Fusarium sp., and Trichoderma; Mock 2 had equal amounts of Cladosporium perangustum, a Pleosporales sp. and Alternaria; and Mock 3 had an equal mixture of Mock 1 and 2.
Bioinformatic analyses
The raw 454 flowgrams were fed into QIIME v. 1.8.054 while the MiSeq analysis was performed using MOTHUR v. 1.34.355. In QIIME, the multiplexed reads were assigned to samples based on unique barcodes and sequences were clustered into OTUs based on sequence similarity (99%). Representative sequences for each OTU were assigned to phylotypes using the UNITE database, and unassigned sequences were subjected to a BLASTn search against the non-redundant (NR) database in NCBI to determine their identity. Alpha diversity was estimated and represented as rarefaction curves. In MOTHUR, the standard operating procedure described at http://www.mothur.org/wiki/MiSeq_SOP was followed55. High quality sequences were aligned against the UNITE database. Alignments were assigned to taxa with a confidence threshold of 80% and operational taxonomic units (OTUs) were calculated at 3% species level classification. Finally, rarefaction curves based on 97% sequence similarity cut-off were generated using R v. 3.1.056. Community similarities based on Bray-Curtis distances were visualised in R v. 3.1.0. Further, Termitomyces MiSeq quality-filtered reads were extracted and fed into a similarity clustering analysis using ClustalW v. 2.157, with MOTHUR classification functions modified to a 100% base pair similarity threshold. For each comb or Termitomyces culture, all sequences that were 100% identical were pooled as one Termitomyces variant, after which variants accounting for >0.1% relative abundance per sample were aligned in Geneious v. 10.0.858.
Chemical extraction of fungus comb
First, for extraction of fungus combs in acetonitrile (ACN), comb from 12 colonies (Table S8) was pulverised and extracted with hexane (10 ml/g dry weight) overnight at room temperature (RT) to remove fatty acids and improve signal intensity. The hexane was filtered off and the comb extracted again with acetonitrile (ACN, 10 ml/g dry weight) overnight at RT. The ACN was concentrated under reduced pressure to yield crude comb extracts, six of which (Mn132, Mn164, Mn165, Od127, Od152 and Od167) were sampled (3 mg) for antifungal assays (see below). The remainders of these extracts, as well as the other six crude extracts, were dissolved in 80% aqueous (aq.) ACN (5 ml) using sonication and fractionated using a CHROMABOND C18 SPE column (500 mg sorbent capacity, Macherey Nagel GmbH & Co. KG). Metabolites were eluted with 80% aq. ACN (3 cv), 100% ACN (3 cv) and 100% acetone (3 cv) (Table S2). Second, for extraction of fungus combs in acetonitrile/acetic acid (ACNAA), comb material from six colonies (Table S2) was pulverised and extracted with 50% aq. ACN containing 1% acetic acid (10 ml/g dry weight) overnight at RT. The solvent was filtered off and concentrated under reduced pressure to yield crude extracts (ACNAA), which were sampled (3 mg) to perform antifungal bioassays.
In vitro antifungal bioassays of comb extracts
To assess the antifungal activity of crude and fractionated comb extracts, we performed growth inhibition assays against Cladosporium phaenocomae, Pleosporales sp., Trichoderma sp., Fusarium oxysporum, and Beauveria bassiana (Table S1) in addition to two Termitomyces strains (T112 from an O. cf. badius colony and T115 from a M. natalensis colony) to serve as controls. Target antagonist/competitor strains were chosen based on the detection of fungal OTUs belonging to these genera in low abundance in the mycobiota analyses. An exception was the entomopathogenic Beauveria bassiana, which was included to represent a potential antagonist of the termites themselves. Culturability was also a criterion in strain selection. Crude ACN and ACNAA extracts were dissolved in 50% aq. dimethyl sulfoxide (DMSO) to yield 10, 5, 1, 0.5, 0.3, 0.1 and 0.01 μg/μl solutions. SPE fractions of the ACN extracts were dissolved (50% DMSO) to yield 10, 5, 0.5 and 0.1 μg/μl solutions. In addition to a negative control of 50% DMSO against contaminant fungi, all comb extracts were also tested against Termitomyces sp. isolated from O. cf. badius colony Od112 (T112) and M. natalensis colony Mn115 (T115). For activity assays, 200 μl of a 7-day-old contaminant or Termitomyces broth was inoculated on PDA. After 20 min, 8 μl crude or fractionated extract were placed on the inoculated plates. All assays were performed in triplicate and were evaluated by measuring zones of inhibition using ImageJ59 two days after inoculation. This timepoint was considered to be ecologically relevant as, in a natural setting, termite behavioural defences (e.g., weeding to remove contaminant fungi32) would subsequently be expected to be in effect.
LCMS/MS and PCA of fractionated ACN fungus comb extracts
High-resolution LCMS/MS data were acquired for the SPE fractions of all 12 ACN fungus comb extracts and the extracts of Termitomyces cultures. Samples (2 mg/ml, 100% MeOH, 1 μL injection volume) were analysed using an Agilent UHPLCMS system, consisting of a 1290 Infinity UHPLC (Agilent Technologies, Torrance, CA, USA) equipped with a Poroshell 120 phenyl-hexyl column (250 mm × 2.1 mm, 2.7 μm particles). The column was eluted using a linear gradient consisting of A: HPLCMS grade H2O + 20 mM formic acid (FA) and B: HPLCMS grade ACN + 20 mM FA. The gradient ran from 10–100% B over 15 min, followed by 100% B for 2 min, returning to 10% B over 0.1 min and equilibrated for 1.9 + 2.0 (post run time) min prior to the next injection. A constant flow of 0.35 ml/min was used, and the column maintained at 60 °C. This was coupled to a 6545 QTOF-MS equipped with Agilent Dual Jet Stream electrospray ion source, in which samples were analysed in both negative and positive polarity. Mass spectra were recorded at 10, 20 and 40 eV as centroid data for m/z 85–1700 in MS mode and m/z 30–1700 in MS/MS mode, with an acquisition rate of 10 spectra/s. The ESI source parameters were: drying gas temperature, 250 °C; gas flow, 8 l/min; nebuliser gas pressure, 40 psig; sheath gas temperature, 300 °C; sheath gas flow, 12 l/min; capillary voltage, 4000 V; nozzle voltage, 500 V. Mass drift was corrected using a lock mass solution infused simultaneously with the sample using a secondary nebulizer, using m/z 922 (Agilent HP-0921) and m/z 186 (tributylamine) in positive polarity while m/z 966 (Agilent HP-0921 + formate) was used in negative polarity. Sample order was randomised.
Raw LCMS/MS data for fungus comb SPE fractions were converted to mzXML format using the ProteoWizard msconvert tool60 before pre-processing in MZmine 2 v. 2.3740 (parameters summarised in Table S7). Data were first baseline corrected, followed by peak detection, isotopic peak grouping, peak alignment, filtering and gap filling. Peaks were searched against the AntiBase database61 using the custom database search feature. To check which features were excluded by filtering for peaks with a corresponding MS2 scan, that otherwise would have been detected, both the positive and negative ESI datasets were also processed with this filter unchecked. This yielded no new features for the ESI + datasets, and only 20 additional features for the ESI- datasets. These are summarised in a separate spreadsheet within Table S5.
Peak lists (Table S5a–d) exported in csv format were edited to remove features that were also detected in the solvent or SPE blanks. Peak areas equal to zero (i.e., not detected) were replaced with a positive number being half of the value of the smallest non-zero peak area in the dataset, before a centred log ratio transformation was performed with the clr function from the chemometrics package in R v. 3.4.1. Principal component analysis (PCA) was then performed with the prcomp function (stats package) and results were visualised with the autoplot function (ggplot2 package).
In vitro effects of Termitomyces on contaminant growth
To explore the effect of Termitomyces on the growth of contaminant fungi, interactions were evaluated in all possible pairings between two Termitomyces isolates, one from M. natalensis Mn115 (T115) and one from Odontotermes sp. Od112 (T112), and 16 contaminant isolates, seven from M. natalensis nests and nine from Odontotermes nests, in two separate assays. A first assay tested direct interactions by growing contaminant fungi on Termitomyces, while a second assay tested indirect interactions by measuring growth of contaminant fungi on a flipped PDA plate of a lawn of Termitomyces. Termitomyces nodules were macerated in 500 μl 0.8% NaCl solution with a sterile pestle, and the suspension was homogenised. Pure contaminant cultures were inoculated in PDB (24 g/l) and incubated for three days. Termitomyces inoculations were done by spreading 20 μl of Termitomyces suspension on PDA plates, which were incubated for three days before day zero, where the contaminants were inoculated. In the first assay, 30 μl contaminant broth was placed in the middle and on top of the Termitomyces culture. In the second assay, on day zero, the three days old Termitomyces cultures were flipped to face the bottom of the plate and 30 μl of the contaminant broth was placed on the middle of the flipped PDA. For all combinations, we performed three replicates and growth was measured 5, 8, 11, 14, and 17 days after day zero using ImageJ59.
Supplementary information
Acknowledgements
We thank Z. Wilhelm de Beer, Michael J. Wingfield and the staff and students at the Forestry and Agricultural Biotechnology Institute, University of Pretoria, for hosting field work, previous and current members of the lab in Copenhagen for help with excavations, the Oerlemans family (Mookgophong) for permission to sample colonies at their farm, Rafael Rodrigues da Costa for preparation of figure 1, David R. Nash for valuable discussions about statistical analysis of LCMS/MS data, Christopher Phippen for assistance with LCMS/MS data acquisition, and Sylvia Mathiasen for laboratory assistance. We are grateful to Agilent Technologies for the Thought Leader Donation of the UHPLC-QTOF system. SO was supported by a PhD stipend from the Department of Biology, University of Copenhagen and the Danish National Research Foundation (DNRF57), DKA by The Netherlands Organisation for Scientific Research (VICI; NWO 86514007), CB by the Daimler Benz foundation and DFG (CRC 1127 ChemBioSys), and MP by a Villum Kann Rasmussen Young Investigator Programme (VKR10101).
Author Contributions
S.O. designed the study, performed DNA extractions, prepared libraries, performed bioinformatic analyses, interaction and activity assays, prepared first drafts of figures and the manuscript. V.L.C. performed chemical extractions, analysed the LCMS/MS data, and contributed to figure preparations. N.B.K. performed chemical extractions. SK acquired and interpreted LCMS/MS data. S.K.C. performed the antimicrobial activity assays and contributed to the interpretation of their results. L.L.M.L. was involved in training S.O. in fungal culturing and in the design of the in vitro interaction experiments. D.K.A. contributed to the interpretation of amplicon sequencing results. S.A.R. contributed to the acquisition and interpretation of the LCMS/MS data. C.B. designed the study, funded the experimental work, and contributed to the interpretation of results. M.P. designed the study, funded the experimental work, and contributed to the interpretation of results, and preparation of figures. All authors contributed to writing and revising the manuscript and gave their final approval for publication.
Data Availability
The datasets generated and analysed during the current study are available in the Supplementary Information files of this article, and in GenBank and Dryad repositories. ITS sequences have been deposited in GenBank (accession no. KJ817309-KJ817331), 454 and MiSeq data have been submitted to NCBI (SRR6856127-SRR6856172), and LCMS data in mzXML format are available through the Dryad data repository at datadryad.org (10.5061/dryad.t6t12).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Saria Otani and Victoria L. Challinor contributed equally.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-45364-z.
References
- 1.de Lapeyre de Bellaire L, Fouré E, Abadie C, Carlier J. Black leaf streak disease is challenging the banana industry. Fruits. 2010;65:327–342. doi: 10.1051/fruits/2010034. [DOI] [Google Scholar]
- 2.Piper JK. Density of Anomoea flavokansiensis on Desmanthus illinoensis in monoculture and polyculture. Entomol. Exp. Appl. 1996;81:105–111. doi: 10.1111/j.1570-7458.1996.tb02020.x. [DOI] [Google Scholar]
- 3.Lin BB. Resilience in agriculture through crop diversification: Adaptive management for environmental change. BioScience. 2011;61:183–193. doi: 10.1525/bio.2011.61.3.4. [DOI] [Google Scholar]
- 4.Aanen DK, et al. High symbiont relatedness stabilizes mutualistic cooperation in fungus-growing termites. Science. 2009;326:1103–1106. doi: 10.1126/science.1173462. [DOI] [PubMed] [Google Scholar]
- 5.Poulsen M, Boomsma JJ. Mutualistic fungi control crop diversity in fungus-growing ants. Science. 2005;307:741–744. doi: 10.1126/science.1106688. [DOI] [PubMed] [Google Scholar]
- 6.Mueller UG, Scott JJ, Ishak HD, Cooper M, Rodrigues A. Monoculture of leafcutter ant gardens. PLoS One. 2010;5(9):e12668. doi: 10.1371/journal.pone.0012668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katoh H, Miura T, Maekawa K, Shinzato N, Matsumoto T. Genetic variation of symbiotic fungi cultivated by the macrotermitine termite Odontotermes formosanus (Isoptera: Termitidae) in the Ryukyu Archipelago. Mol. Ecol. 2002;11:1565–72. doi: 10.1046/j.1365-294X.2002.01535.x. [DOI] [PubMed] [Google Scholar]
- 8.Currie CR, Mueller UG, Malloch D. The agricultural pathology of ant fungus gardens. Proc. Natl. Acad. Sci. USA. 1999;96:7998–8002. doi: 10.1073/pnas.96.14.7998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Visser AA, Kooij PW, Debets AJM, Kuyper TW, Aanen DK. Pseudoxylaria as stowaway of the fungus-growing termite nest: Interaction asymmetry between Pseudoxylaria, Termitomyces and free-living relatives. Fun. Ecol. 2011;4:322–332. doi: 10.1016/j.funeco.2011.05.003. [DOI] [Google Scholar]
- 10.Aanen DK, et al. The evolution of fungus-growing termites and their mutualistic fungal symbionts. Proc. Natl. Acad. Sci. USA. 2002;99:14887–14892. doi: 10.1073/pnas.222313099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aanen DK, Eggleton P. Fungus-growing termites originated in African rain forest. Curr. Biol. 2005;15:851–855. doi: 10.1016/j.cub.2005.03.043. [DOI] [PubMed] [Google Scholar]
- 12.Rouland-Lefevre C, Diouf MN, Brauman A, Neyra M. Phylogenetic relationships in Termitomyces (Family Agaricaceae) based on the nucleotide sequence of ITS: a first approach to elucidate the evolutionary history of the symbiosis between fungus-growing termites and their fungi. Mol. Phyl. Evol. 2002;22:423–429. doi: 10.1006/mpev.2001.1071. [DOI] [PubMed] [Google Scholar]
- 13.Roberts Eric M., Todd Christopher N., Aanen Duur K., Nobre Tânia, Hilbert-Wolf Hannah L., O’Connor Patrick M., Tapanila Leif, Mtelela Cassy, Stevens Nancy J. Oligocene Termite Nests with In Situ Fungus Gardens from the Rukwa Rift Basin, Tanzania, Support a Paleogene African Origin for Insect Agriculture. PLOS ONE. 2016;11(6):e0156847. doi: 10.1371/journal.pone.0156847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nobre, T., Fernandes, C., Boomsma, J. J., Korb, J. & Aanen, D. K. Farming termites determine the genetic population structure of Termitomyces fungal symbionts. Mol. Ecol.20, 2023–33 (2011). [DOI] [PubMed]
- 15.Eggleton, P. An introduction to termites: Biology, taxonomy and functional morphology in Biology of Termites: A Modern Synthesis (eds Bignell, D. E., Roisin, Y. & Lo, N.) 349–373 (Springer, 2011).
- 16.Nobre, T., Rouland-Lefevre, C. & Aanen, D. K. Comparative biology of fungus cultivation in termites and ants in Biology of Termites: A Modern Synthesis (eds Bignell, D. E., Roisin, Y. & Lo, N.) 193–210 (Springer, 2011).
- 17.Li H, et al. Lignocellulose pretreatment in a fungus-cultivating termite. Proc. Natl. Acad. Sci. USA. 2017;114:4709–4714. doi: 10.1073/pnas.1618360114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rouland-Lefevre, C. Symbiosis with fungi, in Termites: Evolution, Sociality, Symbioses, Ecology (eds Abe, T., Bignell, D. E. & Higashi, M) 289–306 (Kluwer Academic Press, 2000).
- 19.Badertscher S, Gerber C, Leuthold RH. Polyethism in food-supply and processing in termite colonies of Macrotermes subhyalinus (Isoptera) Behav. Ecol. Sociobiol. 1983;12:115–119. doi: 10.1007/BF00343201. [DOI] [Google Scholar]
- 20.Li H, et al. Investigation of age polyethism in food processing of the fungus-growing termite Odontotermes formosanus (Blattodea: Termitidae) using a laboratory artificial rearing system. J. Econ. Entomol. 2015;108:266–273. doi: 10.1093/jee/tou005. [DOI] [PubMed] [Google Scholar]
- 21.Rouland-Lefevre, C., Inoue, T. & Johjima, T. Termitomyces/termite interactions in Intestinal Microorganisms of Soil Invertebrates (eds König, H. & Varma, A.) 335–350 (Springer, 2006).
- 22.Batra, L. R. & Batra, S. W. T. Termite-fungus mutualism in Insect-Fungus Symbiosis:Nutrition, Mutualism, and Commensalism (ed. Batra, L. R.) 117–163 (Allaheld & Osmun, 1979).
- 23.Darlington, J. Nutrition and evolution in fungus-growing termites in Nourishment and Evolution in Insect Societies (eds Hunt, J. H. & Nalepa, C. A.) 105–130 (Westview Press, 1994).
- 24.Rogers JD. Thoughts and musings on tropical Xylariaceae. Myc. Res. 2000;104:1412–1420. doi: 10.1017/S0953756200003464. [DOI] [Google Scholar]
- 25.Rogers JD, Ju Y-M, Lehmann J. Some Xylaria species on termite nests. Mycologia. 2005;97:914–923. doi: 10.1080/15572536.2006.11832783. [DOI] [PubMed] [Google Scholar]
- 26.Sands, W.A. The association of termites and fungi in Biology of Termites (eds Krishna, A. & Weesner, F) 495–524 (Academic Press, 1969).
- 27.Visser AA, et al. Levels of specificity of Xylaria species associated with fungus-growing termites: a phylogenetic approach. Mol. Ecol. 2009;18:553–567. doi: 10.1111/j.1365-294X.2008.04036.x. [DOI] [PubMed] [Google Scholar]
- 28.Mathew GM, Ju YM, Lai CY, Mathew DC, Huang CC. Microbial community analysis in the termite gut and fungus comb of Odontotermes formosanus: the implication of Bacillus as mutualists. FEMS Microbiol. Ecol. 2012;79:504–517. doi: 10.1111/j.1574-6941.2011.01232.x. [DOI] [PubMed] [Google Scholar]
- 29.Um S, Fraimout A, Sapountzis P, Oh DC, Poulsen M. The fungus-growing termite Macrotermes natalensis harbors bacillaene-producing Bacillus sp. that inhibit potentially antagonistic fungi. Sci. Rep. 2013;3:1–7. doi: 10.1038/srep03250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shinzato N, Muramatsu M, Watanabe Y, Matsui T. Termite-regulated fungal monoculture in fungus combs of a macrotermitine termite Odontotermes formosanus. Zool. Sci. 2005;22:917–922. doi: 10.2108/zsj.22.917. [DOI] [PubMed] [Google Scholar]
- 31.Guedegbe HJ, et al. Occurrence of fungi in combs of fungus-growing termites (Isoptera: Termitidae, Macrotermitinae) Mycol. Res. 2009;113:1039–1045. doi: 10.1016/j.mycres.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 32.Katariya L, et al. Fungus-farming termites selectively bury weedy fungi that smell different from crop fungi. J. Chem. Ecol. 2017;43:986–995. doi: 10.1007/s10886-017-0902-4. [DOI] [PubMed] [Google Scholar]
- 33.Da Silva P, et al. Solution structure of termicin, an antimicrobial peptide from the termite Pseudacanthotermes spiniger. Prot. Sci. 2003;12:438–446. doi: 10.1110/ps.0228303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lamberty M, et al. Insect immunity. Constitutive expression of a cysteine-rich antifungal and a linear antibacterial peptide in a termite insect. J. Biol. Chem. 2001;276:4085–4092. doi: 10.1074/jbc.M002998200. [DOI] [PubMed] [Google Scholar]
- 35.Beemelmanns C, et al. Macrotermycins A-D, Glycosylated Macrolactams from a Termite-Associated Amycolatopsis sp. M39. Org. Lett. 2017;19:1000–1003. doi: 10.1021/acs.orglett.6b03831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo H, et al. Isolation, Biosynthesis and Chemical Modifications of Rubterolones A-F: Rare Tropolone Alkaloids from Actinomadura sp. 5-2. Chem. Eur. J. 2017;23:9338–9345. doi: 10.1002/chem.201701005. [DOI] [PubMed] [Google Scholar]
- 37.Beemelmanns C, Guo H, Rischer M, Poulsen M. Natural products from microbes associated with insects. Beilstein J. Org. Chem. 2016;12:314–327. doi: 10.3762/bjoc.12.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grassé PP, Noirot C. Le meule des termites champignonnistes et sa signification symbiotique. Ann. Sci. Nat. Zool. Biol. Anim. 1958;20:113–128. [Google Scholar]
- 39.Thomas RJ. Factors affecting the distribution and activity of fungi in the nests of Macrotermitinae (Isoptera) Soil Biol. Biochem. 1987;19:343–349. doi: 10.1016/0038-0717(87)90020-4. [DOI] [Google Scholar]
- 40.Pluskal T, Castillo S, Villar-Briones A, Oresic M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinf. 2010;11:395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wood, T. G. & Thomas, R. J. The mutualistic association between Macrotermitinae and Termitomyces in Insect–Fungus Interactions (eds Wilding, Collins, N. M., Hammond, P. M., Webber, J. F.) 69–92 (Academic Press, 1989).
- 42.Visser AA, Nobre T, Currie CR, Aanen DK, Poulsen M. Exploring the potential for Actinobacteria as defensive symbionts in fungus-growing termites. Micr. Ecol. 2012;63:975–985. doi: 10.1007/s00248-011-9987-4. [DOI] [PubMed] [Google Scholar]
- 43.Korb J, Aanen DK. The evolution of uniparental transmission of fungal symbionts in fungus-growing termites (Macrotermitinae) Behav. Ecol. Sociobiol. 2003;53:65–71. [Google Scholar]
- 44.Aanen DK. As you reap, so shall you sow: coupling of harvesting and inoculating stabilizes the mutualism between termites and fungi. Biol. Lett. 2006;2:209–212. doi: 10.1098/rsbl.2005.0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Fine Licht HH, Andersen A, Aanen DK. Termitomyces sp. associated with the termite Macrotermes natalensis has a heterothallic mating system and multinucleate cells. Myc. Res. 2005;109:314–318. doi: 10.1017/S0953756204001844. [DOI] [PubMed] [Google Scholar]
- 46.Nobre T, Koopmanschap B, Baars JJ, Sonnenberg AS, Aanen DK. The scope for nuclear selection within Termitomyces fungi associated with fungus-growing termites is limited. BMC Evol. Biol. 2014;14:121. doi: 10.1186/1471-2148-14-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aanen DK, et al. Patterns of interaction specificity of fungus-growing termites and Termitomyces symbionts in South Africa. BMC Evol. Biol. 2007;7:115. doi: 10.1186/1471-2148-7-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Otani S, Hansen LH, Sørensen SJ, Poulsen M. Bacterial communities in termite fungus combs are comprised of consistent gut deposits and contributions from the environment. Micr. Ecol. 2016;71:207–220. doi: 10.1007/s00248-015-0692-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benndorf R, et al. Natural products from Actinobacteria associated with fungus-growing termites. Antibiotics. 2018;7:83. doi: 10.3390/antibiotics7030083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gräfe U, et al. Structure of ergokonin C, a new carboxysterol antifungal antibiotic from a Tolypocladium inflatum mutant. Pharmazie. 1991;46:613–614. [PubMed] [Google Scholar]
- 51.White T.J., Bruns T., Lee S., Taylor J. PCR Protocols. 1990. AMPLIFICATION AND DIRECT SEQUENCING OF FUNGAL RIBOSOMAL RNA GENES FOR PHYLOGENETICS; pp. 315–322. [Google Scholar]
- 52.Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes - application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993;2:113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 53.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Env. Micr. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nature Meth. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schloss PD, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Env. Micr. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.R core Team, R: A language and environment for statistical computing. 2013, R Foundation for Statistical Computing: Vienna.
- 57.Thompson JD, Higgins DG, Gibson TJ. Clustal-W - Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucl. Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kearse M, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinf. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abràmoff MD, Magalhães PJ, Ram SJ. Image processing with ImageJ. Biophot. Intl. 2004;11:36–42. [Google Scholar]
- 60.Chambers MC, et al. A cross-platform toolkit for mass spectrometry and proteomics. Nature Biotech. 2012;30:918–20. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Laatsch, H. AntiBase: The Natural Compound Identifier (Wiley, 2017).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analysed during the current study are available in the Supplementary Information files of this article, and in GenBank and Dryad repositories. ITS sequences have been deposited in GenBank (accession no. KJ817309-KJ817331), 454 and MiSeq data have been submitted to NCBI (SRR6856127-SRR6856172), and LCMS data in mzXML format are available through the Dryad data repository at datadryad.org (10.5061/dryad.t6t12).