Abstract
The differentiation of fibroblasts into a transient population of highly activated extracellular matrix (ECM) producing myofibroblasts at sites of tissue injury is critical for normal tissue repair. Excessive myofibroblast accumulation and persistence, often as a result of a failure to undergo apoptosis when tissue repair is complete, leads to pathological fibrosis and is also a feature of the stromal response in cancer. Myofibroblast differentiation is accompanied by changes in cellular metabolism, including increased glycolysis, to meet the biosynthetic demands of enhanced ECM production. Here we show that transforming growth factor-β1 (TGF-β1), the key pro-fibrotic cytokine implicated in multiple fibrotic conditions, increased the production of activating transcription factor 4 (ATF4), the transcriptional master regulator of amino acid metabolism, in order to supply glucose-derived glycine to meet the amino acid requirements associated with enhanced collagen production in response to myofibroblast differentiation. We further delineate the signaling pathways involved and show that TGF-β1–induced ATF4 production was dependent on the cooperation between canonical TGF-β1 signaling through Smad3 and activation of the mechanistic target of rapamycin complex 1 (mTORC1) and eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1) axis. ATF4 in turn promoted a transcriptional enhancement of the de novo serine-glycine biosynthetic pathway, as well as the glucose transporter 1 (GLUT1). Our findings suggest that targeting the TGF-β1–mTORC1–ATF4 axis may represent a novel therapeutic strategy for interfering with myofibroblast function in fibrosis and potentially in other conditions, including cancer.
Introduction
Fibrosis is the concluding pathological outcome and major cause of morbidity and mortality in a number of common chronic inflammatory, immune-mediated and metabolic diseases (1). The perpetual and relentless deposition of a collagen-rich matrix is the cornerstone of the fibrotic response and eventually leads to organ failure and premature death. Despite the rising incidence of fibrotic disease and intense research efforts, there remains a paucity of effective treatment options. Idiopathic pulmonary fibrosis (IPF) represents the most rapidly progressive and lethal of all fibrotic diseases and is associated with a dismal median survival of 3 years from diagnosis (2, 3). Although the approval of pirfenidone and the small molecule tyrosine kinase inhibitor, nintedanib, for the treatment of IPF represented a watershed moment for the development of anti-fibrotic therapeutics, these agents slow but do not halt disease progression (4, 5). Therefore, there remains a pressing need to identify novel anti-fibrotic therapeutic strategies (6).
Myofibroblasts are the key effector cells responsible for the synthesis and deposition of a collagen-rich ECM during normal wound healing, as well as during the development of pathological tissue fibrosis (7). Myofibroblasts can be derived from multiple cell types, including tissue resident fibroblasts, and are characterized by the de novo production of α-smooth muscle actin (α-SMA) which assimilates into stress fibers giving rise to their contractile phenotype (reviewed in (8)). The excessive accumulation and persistence of these ECM producing myofibroblasts, as a result of a dysregulated wound healing response perpetuated by a network of proliferation and differentiation signals within a homeostatically dysregulated tissue microenvironment, represents a key common mechanism underlying the development of pathological fibrosis. Myofibroblasts are also integral to the epithelial-mesenchymal cross-talk as part of the stromal reaction in epithelial tumors (9) and current evidence suggests that the presence of stromal myofibroblasts is generally associated with poor prognosis in solid cancers (10).
Metabolic reprogramming is a hallmark of cancer but is now increasingly recognized as playing a key role in dictating cell fate and function in the context of inflammation and immunity (11). Evidence for altered metabolism in the context of fibrosis has also emerged. Indeed, 18F-fluorodeoxyglucose ([18F]-FDG) uptake by positron emission tomography (PET), a commonly used imaging marker of enhanced glycolysis in cancer, has been reported to be increased in IPF patients (12, 13) and is predictive of progression-free survival (14). Although multiple cell types might be responsible for the increased uptake of [18F]-FDG in IPF, glycolysis has been shown to be increased during TGF-β1-induced myofibroblast differentiation in vitro and targeting glycolysis attenuated experimental lung fibrosis (15, 16). However, the mechanisms that regulate metabolic reprogramming in fibrosis remain poorly understood.
The serine-threonine kinase, mechanistic target of rapamycin (mTOR), plays a key role in regulating cell metabolism, along with other key cellular processes, including cell cycle progression, proliferation, growth, autophagy and protein synthesis. mTOR is central to two distinct complexes, mTORC1 and mTORC2, which integrate critical environmental and intracellular cues provided by nutrients, energy, oxygen and growth factors (recently reviewed in (17)). mTOR signaling is commonly dysregulated in human disease and has been strongly implicated in coordinating metabolic reprogramming in cancer cells in order to optimize nutrient uptake and utilization, and meet the biosynthetic needs of proliferative cancer cells (18–20). This has led to major interest in targeting mTOR in the setting of cancer, as reflected by the growing number of ATP-competitive mTOR inhibitors that are either approved or in clinical development (17, 21, 22).
mTOR signaling has also been implicated in fibrosis (23–25); however, whether mTOR contributes to the development of fibrosis as a result of its well-recognized immuno-modulatory functions or by influencing fibroblast ECM deposition directly is still unclear. Studies from our laboratory provided strong scientific rationale for progressing the potent pan-PI3K-mTOR inhibitor, Omipalisib, to a first-in-human proof of mechanism trial in patients with IPF (clinicaltrials.gov/NCT01725139), based on the potential of this compound to interfere with fibroblast function, including collagen deposition induced in response to the potent fibrogenic mediator, TGF-β1 (26). We subsequently showed that the translational mTORC1-4E-BP1 axis is critical for TGF-β1–induced collagen synthesis in fibroblasts (27). However, the underlying mechanism(s) by which mTORC1 influences this response remains poorly understood.
In the current study, we used an unbiased bioinformatics approach to identify key transcriptional modules that are induced in response to TGF-β1 and modulated by mTOR inhibition. We report a critical mechanistic pathway linking mTORC1 activation to altered myofibroblast metabolism through the transcriptional master regulator of amino acid metabolism, activating transcription factor 4 (ATF4), in order to fuel ECM biosynthesis. These findings shed novel light on the signaling pathways by which TGF-β1 influences collagen deposition with important implications for the future development of novel therapeutic strategies for multiple conditions associated with the abnormal accumulation of myofibroblasts and excessive matrix deposition.
Results
mTOR enhances expression of glycine biosynthesis genes in TGF-β1–stimulated fibroblasts
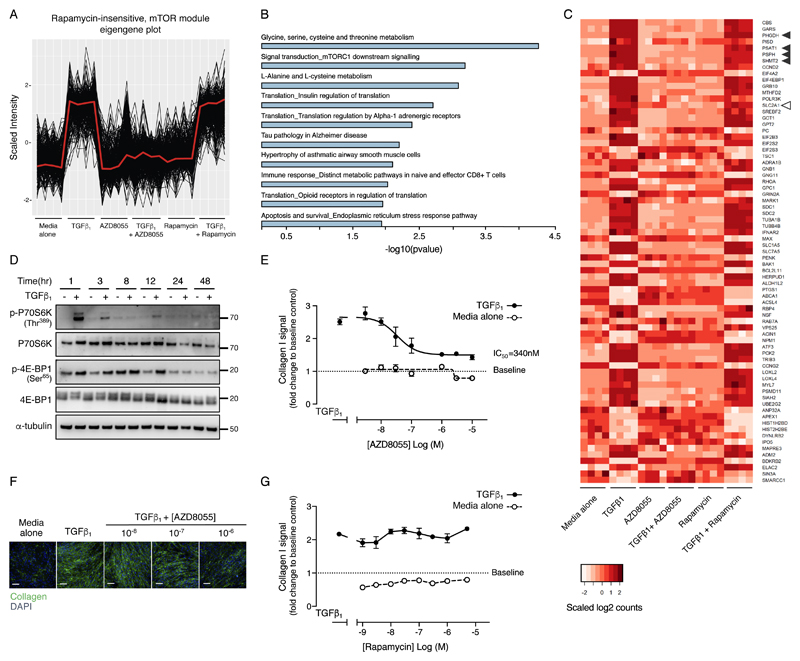
We have shown that rapamycin-insensitive mTORC1 signaling is critical for TGF-β1–induced collagen deposition (26, 27). To define the underlying mechanism, we applied weighted gene co-expression network analysis (WGCNA) using Metacore™ to interrogate our existing RNAseq (GSE102674) dataset, which compares the global transcriptomic effect of the highly-selective ATP-competitive mTOR inhibitor, AZD8055, to rapamycin in human lung fibroblasts exposed to TGF-β1.
WGCNA revealed 65 independent sets of highly correlated genes (also referred to as co-expression modules). For each of these modules, the measure of central tendency (the eigengene) correlated to a requisite profile showing complete reversal of the TGF-β1 response by AZD8055 and no reversal by rapamycin. This led to the identification of a single module with the highest correlation (r=0.99) (Fig. 1A). Pathway analysis revealed that this module was enriched for three amino-acid synthesis pathways (serine-glycine, alanine-cysteine and cysteine-glutamine) with the serine-glycine biosynthesis pathway representing the most enriched pathway (Fig. 1B and Table S1). Expression of the serine-glycine biosynthetic pathway genes, phosphoglycerate dehydrogenase (PHGDH), phosphoserine aminotransferase 1 (PSAT1), phosphoserine phosphatase (PSPH) and serine hydroxymethyltransferase (SHMT) 2 were all increased following TGF-β1 treatment. This increase was inhibited by AZD8055 treatment, whereas rapamycin had no effect (Fig. 1C), indicating that rapamycin-insensitive mTOR signaling may play a critical role in enhancing the expression of genes involved in serine and glycine biosynthesis in response to TGF-β1 stimulation.
Fig. 1. Global transcriptomic analysis of TGF-β1––stimulated fibroblasts by RNAseq analysis reveals a rapamycin-insensitive mTOR-dependent serine-glycine biosynthetic signature during TGF-β1––induced collagen deposition.
(A) Plot showing scaled gene expression intensities from the rapamycin-insensitive mTOR module eigengene as calculated by WGCNA. The module eigengene is defined as the first principal component of the genes contained within the module and is representative of the gene expression profiles in the module. All expression values have been z transformed and signals that are negatively correlated to the module eigengene have been inverted for plotting (N=4 biological replicates). (B) Bar plot showing the top 10 most significantly enriched pathways for the genes in the rapamycin-insensitive mTOR module. The serine-glycine biosynthesis pathway was most enriched (p-value = 5.45E-5). (C) Heatmap representing the genes from the top 20 most enriched pathways in the rapamycin-insensitive mTOR module, listed in order of the most to the least statistically significant. Genes that map to more than one pathway only appear for the pathway with the most significant p-value. Scaled counts were used to generate the heatmap, where darker red indicates higher number of counts. Black arrowheads indicate the genes belonging to the glycine metabolism pathway and the clear arrowhead indicates the SLC2A1 gene (N=4 biological replicates). (D) Confluent primary human lung fibroblasts (pHLFs) were stimulated with media alone or media plus TGF-β1 and immunoblotting was performed for the indicated proteins (N=3 experiments, representative data shown). (E) Confluent pHLFs were pre-incubated with media plus vehicle (DMSO) or AZD8055 and stimulated for 48 h with or without TGF-β1. Collagen I deposition was assessed by high content imaging. IC50 value was calculated using four-parameter non-linear regression. Each data point shown is mean +/- SEM of the fold change to baseline of 3 biological replicates per condition and data are representative of 3 independent experiments. (F) Immunofluorescence staining showing collagen I deposition in pHLFs treated as in (E), scale bar =100 µm. (G) Confluent pHLFs were pre-incubated with media plus vehicle (DMSO) or rapamycin and stimulated for 48 hours with or without TGF-β1. Collagen I deposition was assessed by high content imaging. Each data point shown is mean +/- SEM of the fold change relative to baseline of 3 biological replicates per condition and the data are representative of 3 independent experiments.
The original RNAseq dataset and our previous work demonstrating a key role for rapamycin insensitive mTORC1 signaling during TGF-β1–stimulated collagen deposition were generated in primary human lung fibroblasts (pHLFs) cultured in standard DMEM (25mM glucose; 2mM glutamine)(27). Because mTOR is sensitive to environmental signals, including energy and nutrient abundance and in view of the profound effects of mTOR inhibition on the transcriptome of multiple metabolic genes (reviewed in (28)), we performed all subsequent functional studies at physiological concentrations of glucose (5mM) and glutamine (0.7mM). We confirmed that under these conditions, TGF-β1 caused a marked increase in mTORC1 signaling in pHLFs as evidenced by p70S6K(Thr389) and 4E-BP1(Ser65) phosphorylation (Fig.1D and fig.S1, A and B). TGF-β1 also increased collagen deposition by 251 +/- 8.6%, as determined in a well-characterized quantitative assay based on high content imaging of collagen I immunostaining at 48 hours (26) (Fig. 1, E and F). ATP-competitive mTOR inhibition with AZD8055 attenuated TGF-β1–induced collagen deposition in a concentration-dependent manner with an IC50 of 340nM (Fig. 1E) whereas rapamycin had no effect (Fig. 1G). We also investigated whether mTOR regulated TGF-β1–induced myofibroblast differentiation, based on the assessment of α-SMA by high content imaging. As expected TGF-β1 increased fibroblast α-SMA production, but AZD8055 did not block this response (fig. S1, C and D).
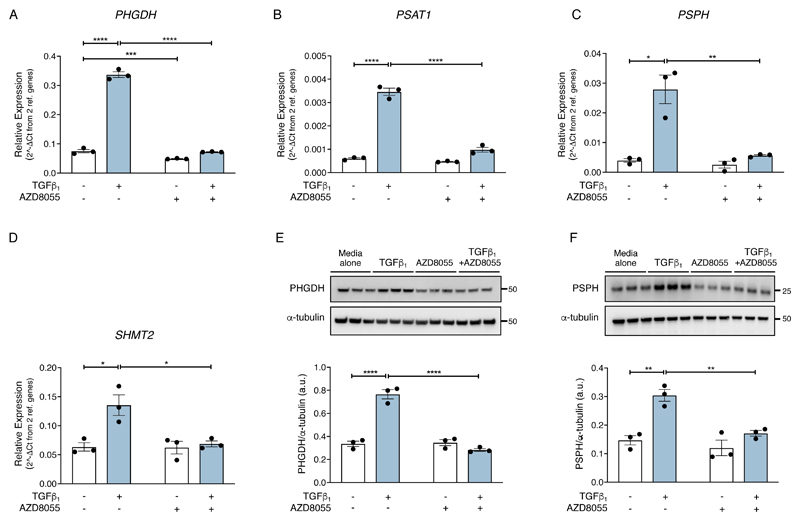
In agreement with our RNASeq dataset, under physiological glucose and glutamine concentrations, mRNA abundance of all the enzymes involved in serine-glycine biosynthesis, PHGDH, PSAT1, PSPH and SHMT2 were significantly increased in response to TGF-β1 stimulation at 24 hours (Fig. 2, A to D). In contrast, SHMT1, which preferentially converts glycine to serine (29), was not influenced by TGF-β1 treatment (fig. S1E). The TGF-β1–induced increase in mRNA abundance of the serine-glycine pathway enzymes was sensitive to inhibition by AZD8055 but insensitive to rapamycin treatment (fig. S1, F to I). Evaluation of the effect of TGF-β1 at the protein level confirmed that the rate-limiting enzymes, PHGDH and PSPH, were increased in response to TGF-β1–stimulation at 24 hours and that these increases were inhibited by AZD8055 (Fig. 2, E and F).
Fig. 2. TGF-β1 amplifies the serine-glycine pathway in an mTOR-dependent manner.
Confluent pHLFs were incubated in media alone or media plus TGF-β1 with AZD8055 or vehicle control (DMSO) for 24 hours. (A-D) PHGDH, PSAT1, PSPH and SHMT2 relative mRNA abundance was quantified by RT-qPCR. Data are presented as means ± SEM from 3 biological replicates. (E, F) Immunoblots of protein lysates and densitometric quantification of PHGDH (E) and PSPH (F) (N=3 biological replicates and data are representative of 3 independent experiments). Differences between groups were evaluated by two-way ANOVA test with Tukey post-hoc test. *p<0.05, **p<0.01, ***p< 0.001, ****p< 0.0001.
Overconnected node analysis reveals a key role for mTOR in promoting ATF4 protein production
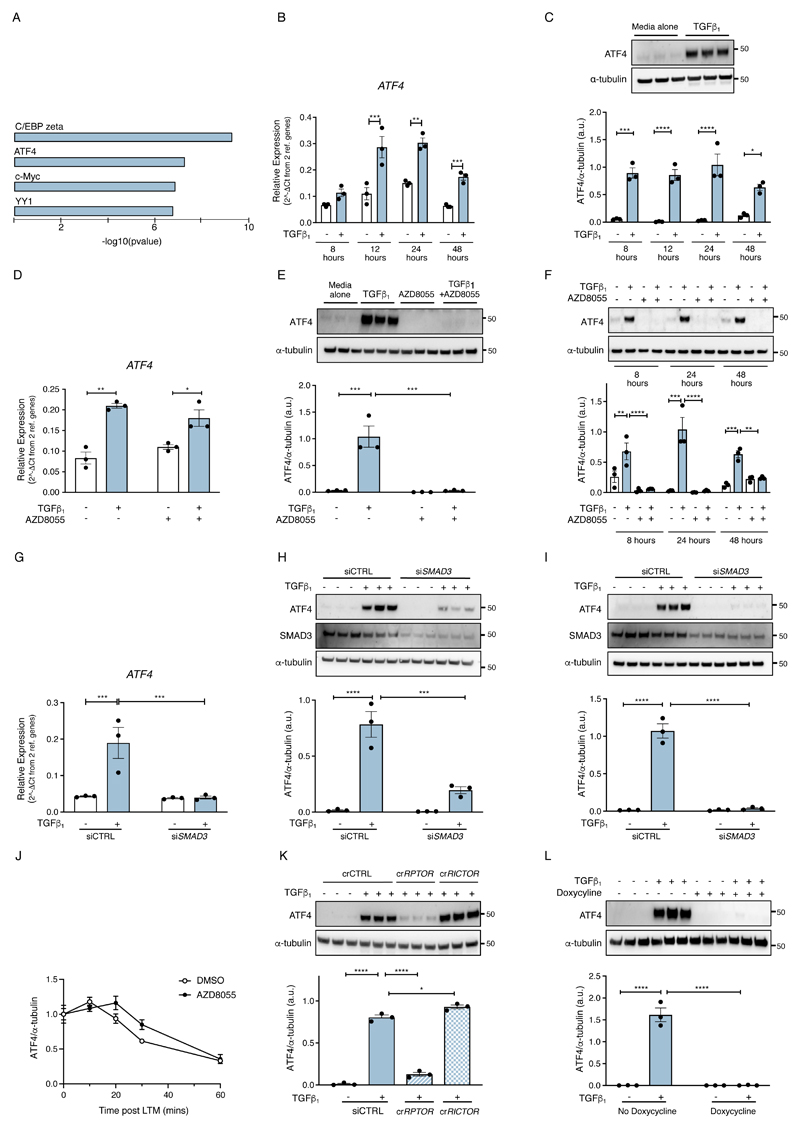
We next explored the potential mechanism by which rapamycin-insensitive mTOR promotes the de novo serine-glycine synthesis pathway in response to TGF-β1 stimulation. To this end, we first employed overconnected node analysis (OCN) based on the MetaCore™ database of curated literature gene and protein interactions in order to identify potential master regulators of gene expression. Using the principles of enrichment analysis (i.e. Fisher’s Exact Test), a p-value was calculated for each gene in the database by comparing the overlap of its known interactors with the gene list of interest. OCN of the RNAseq dataset revealed a cluster of transcription factors associated with the serine-glycine metabolism module of enriched mRNAs, including the transcription factor, ATF4 (Fig. 3A). ATF4 is a known mTOR-responsive gene and a key effector of the stress response that triggers increased gene transcription by binding to the C/EBP-ATF response element (CARE) in specific genes, including all enzymes of the de novo serine-glycine synthesis pathway (30, 31). We therefore next examined whether ATF4 was the downstream target of mTOR involved in mediating the TGF-β1––induced increase in de novo glycine biosynthesis and collagen deposition.
Fig. 3. Overconnected node analysis reveals a key role for mTOR in promoting ATF4 protein production.
(A) Overconnected node analysis of the RNAseq data revealed a cluster of transcription factors associated with the serine-glycine module of enriched mRNAs. (B) Confluent pHLFs were incubated with or without TGF-β1 and ATF4 relative mRNA abundance was measured by RT-qPCR over time (data are means ± SEM from 3 biological replicates). (C) Immunoblots and densitometric quantification of ATF4 protein abundance in pHLF lysates over time following TGF-β1 stimulation. Immunoblot shown represents ATF4 at 8 hours post TGF-β1 stimulation. Data are expressed as mean ± SEM from 3 biological replicates. (D) Relative ATF4 mRNA abundance measured at 24 hours post-TGF-β1 stimulation (N=3 biological replicates). (E) Immunoblot and densitometric quantification of ATF4 abundance at 24 hours post-TGF-β1 stimulation (N=3 biological replicates). (F) Immunoblot and densitometric quantification of ATF4 abundance at indicated times post-TGF-β1 stimulation (N=3 biological replicates). (G) Confluent pHLFs were transfected with scrambled control siRNA (siCTRL) or Smad3 siRNA (siSMAD3) and incubated with or without TGF-β1 and ATF4 relative mRNA abundance at 24 hours was measured by RT-qPCR (N=3 biological replicates). (H, I) ATF4 immunoblots and densitometric quantification represent samples analyzed at 8 hours (H) and 24 hours (I) (N=3 biological replicates). (J) pHLFs were pre-treated with TGF-β1 for 13 hours before treatment with lactimidomycin and vehicle (DMSO) or AZD8055. Lysates were harvested at indicated times post-treatment and ATF4 abundance measured by immunoblotting and densitometric quantification (N=3 biological replicates). (K) pHLFs were modified by CRISPR-Cas9 gene editing of RPTOR or RICTOR and stimulated with TGF-β1. Immunoblot for ATF4 and densitometric quantification were performed at 24 hours (N=3 biological replicates). (L) pHLFs expressing a 4E-BP1-4A dominant-negative phospho-mutant were induced with doxycycline or media alone for 24 hours prior to TGF-β1 stimulation. Immunoblot for ATF4 and densitometric quantification were performed at 18 hours post TGF-β1 stimulation (N=3 biological replicates). Differences between groups were evaluated by two-way ANOVA test with Tukey post-hoc test (B-I, L), repeated measures two-way ANOVA (J) or one-way ANOVA with Tukey post-hoc test (K). *p<0.05, **p<0.01, ***p< 0.001, ****p< 0.0001.
We first examined the relationship between ATF4 and TGF-β1, as ATF4 is not known to be a TGF-β1––responsive gene in fibroblasts. ATF4 mRNA abundance was significantly increased by 2.60 ± 0.37-fold and by 2.02 ± 0.12-fold in TGF-β1––stimulated cells relative to media alone-treated cells at 12 and 24 hours, respectively (Fig. 3B). At the protein level, ATF4 was barely detectable in media alone-treated cells by immunoblotting. In contrast, in TGF-β1–stimulated cells, ATF4 was abundant at all time points examined from 8 hours onwards (Fig. 3C).
We next examined whether TGF-β1––induced ATF4 was sensitive to ATP-competitive mTOR inhibition. AZD8055 had no effect on TGF-β1––induced ATF4 mRNA abundance (Fig. 3D) but completely blocked ATF4 protein production at all time points examined (Fig. 3, E and F). We also examined the contribution of canonical TGF-β1 signaling through the Smad pathway. Silencing Smad3 using siRNA completely inhibited the increase in ATF4 mRNA abundance at 24 hours as well as protein production assessed at 8 and 24 hours (Fig. 3, G to I, fig. S2A). Taken together these data led us to conclude that TGF-β1–induced ATF4 production is both Smad3 and mTOR dependent and further that mTOR is acting at the post-transcriptional level.
We next explored the possibility that mTOR might be influencing ATF4 protein stability. To this end, we assessed the impact of AZD8055 on the rate of decline of ATF4 abundance in the presence of the translation inhibitor, lactimidomycin (LTM). Both compounds were added at 13 hours post TGF-β1––stimulation to allow ATF4 protein to accumulate. ATF4 protein abundance was then monitored at different time points over an hour. These experiments revealed that AZD8055 did not influence the rate at which ATF4 protein levels declined in the presence of LTM and therefore allowed us to conclude that mTOR is not acting by promoting ATF4 protein stability (Fig. 3J and fig. S2B).
We also sought to identify the mTOR complex involved in mediating the TGF-β1 induced ATF4 response. To this end, we generated RPTOR and RICTOR-deficient fibroblasts by CRISPR-Cas9 gene editing to specifically disrupt either mTORC1 or mTORC2 signaling, respectively. Efficient gene-editing was confirmed by western blotting (fig. S2C). TGF-β1––induced ATF4 protein synthesis was significantly abrogated in RPTOR-edited fibroblasts but fully maintained in RICTOR-deficient fibroblasts, indicating that TGF-β1 exerts its stimulatory effects on ATF4 production exclusively through an mTORC1-dependent mechanism (Fig. 3K). We next interrogated the role of 4E-BP1 downstream of mTORC1, by generating pHLFs expressing a doxycycline-inducible 4E-BP1 dominant negative phospho-mutant (with mTORC1 phosphorylation sites replaced by alanine, abbreviated as 4E-BP1-4A) (27). We have previously confirmed that doxycycline treatment induces 4E-BP1-4A expression in transduced pHLFs and leads to inhibition of cap-dependent translation (27). We now show that TGF-β1–induced ATF4 protein production was completely abrogated in 4E-BP1-4A transduced pHLFs (Fig. 3L). These effects were not related to inhibition of ATF4 by doxycycline treatment since doxycycline treatment did not inhibit TGF-β1–induced ATF4 protein production in untransduced pHLFs (fig. S2D). Taken together, these data demonstrate that the translational mTORC1-4E-BP1 axis is critical for TGF-β1––induced ATF4 protein synthesis.
We also examined whether the ATF4 response to TGF-β1 was downstream of the classical stress response involving PERK activation and subsequent eIF2α phosphorylation. However, this possibility was ruled out on the evidence that TGF-β1 had no effect on eIF2α phosphorylation compared to unstimulated fibroblasts over 12 hours (fig. S2E). Furthermore, the PERK inhibitor, GSK2656157, did not inhibit TGF-β1––induced collagen deposition (fig. S2F).
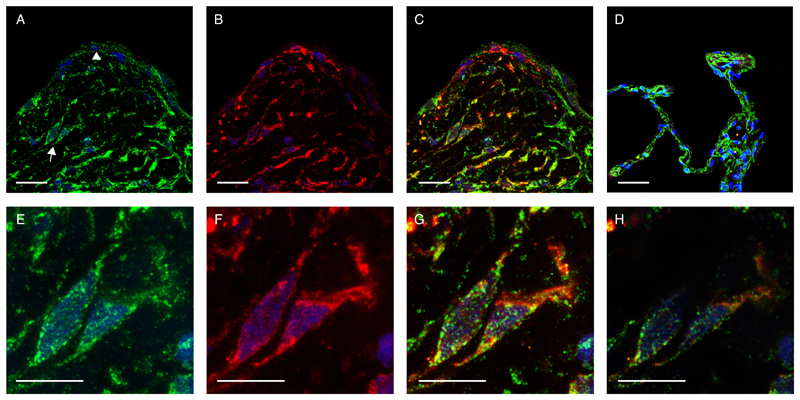
Having demonstrated that ATF4 is enhanced in TGF-β1–activated myofibroblasts in vitro, we questioned whether this observation holds potential translational significance in the clinical setting of fibrosis in humans. To this end we used double immunofluorescence to determine whether ATF4 and the myofibroblast differentiation marker, α-SMA were co-expressed by the same cells in IPF fibrotic foci, the cardinal lesions and leading edge of the fibrotic response. These studies revealed that ATF4 immunofluorescence was consistently and widely expressed in the IPF lung and was associated with α-SMA positive myofibroblasts within fibrotic foci, as well as with the hyperplastic alveolar epithelium overlying fibrotic foci (Fig. 4, A to C). In control lung parenchyma, ATF4 was largely found to be expressed by the alveolar epithelium (Figure 4D). Myofibroblasts are generally not found in normal lung but high magnification images of myofibroblasts within IPF fibrotic foci demonstrate that ATF4 in myofibroblasts was localized to both the cytoplasm and the nucleus (Figure 4, panels E to H).
Fig. 4. ATF4 co-localises with a-SMA-positive myofibroblasts within IPF fibrotic foci.
Immunofluoresence single staining for ATF4 (A, green), α-smooth muscle actin (B, α-SMA, red) and overlay of ATF4 and α-SMA (C, yellow) in a representative IPF fibrotic focus. (A) Arrow indicates myofibroblasts within the fibrotic focus and arrowhead points to the hyperplastic epithelium. Overlay of ATF4 and α-SMA in non-IPF lung tissue (D). (E-G) Corresponding high magnification images of myofibroblasts within the fibrotic focus (indicated by arrow in A) for ATF4(E), α-SMA(F) and overlay of ATF4 and α-SMA(G). (H) Mid-level non-composite confocal overlay image indicating nuclear localisation of ATF4 (green) in an α-SMA positive myofibroblast cell (<0.5 µm). All images counter-stained with DAPI (blue). Scale A-D = 50 µm, E-H = 25 µm, N=3 patients with IPF, N=2 control subjects and representative images shown.
ATF4-dependent modulation of the serine-glycine pathway is critical for TGF-β1–induced collagen deposition
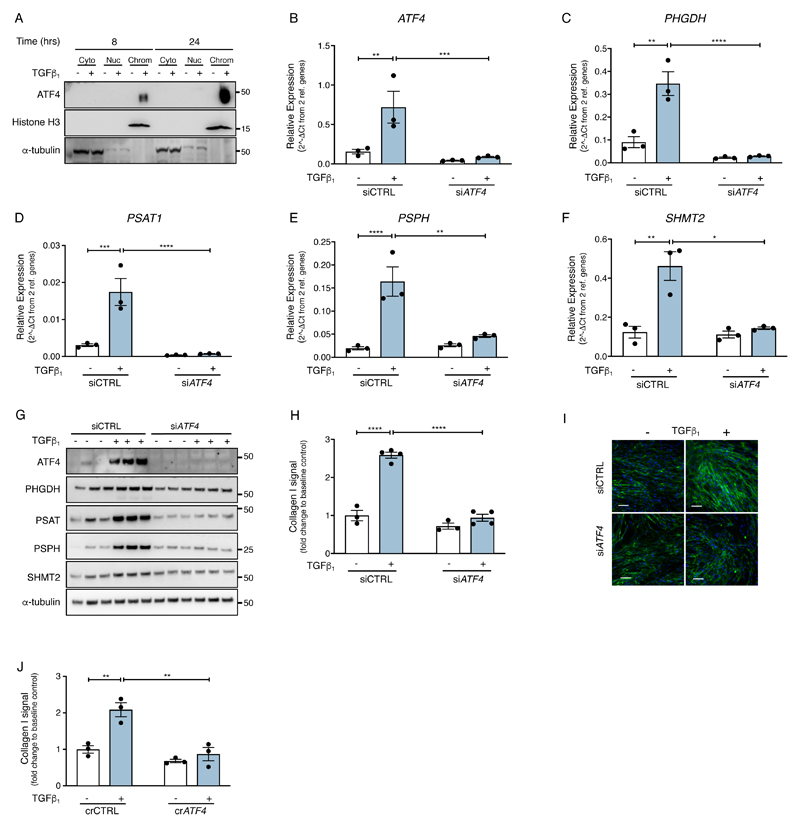
We next examined whether ATF4 plays a role in influencing glycine biosynthesis during TGF-β1––induced collagen deposition. Using chromatin-bound protein fractionation, we first confirmed that ATF4 was predominantly associated within the chromatin fraction in TGF-β1–– stimulated cells (Fig. 5A). Silencing ATF4 using siRNA (Fig. 5B) suppressed the TGF-β1––induced increase in PHGDH, PSAT1, PSPH and SHMT2 mRNA and protein abundance (Fig. 5, C to G and fig. S3, A to E) as well as TGF-β1–– induced collagen deposition (Fig. 5, H to I). In order to control for potential off-target effects with siRNA, we also used CRISPR-Cas9 gene editing to silence ATF4. This approach similarly demonstrated that ATF4-deficient cells were unable to increase collagen production in response to TGF-β1 stimulation (Fig. 5J and fig. S3F). Taken together these data suggest that ATF4 mediates the fibrogenic effects of TGF-β1 by enhancing the de novo serine-glycine pathway.
Fig. 5. ATF4-dependent modulation of the serine-glycine pathway is critical for TGF-β1–– induced collagen deposition.
(A) Confluent pHLFs were incubated with media plus TGF-β1 or media alone for 8 or 24 hours prior to cell lysis and separation into cytoplasmic (Cyto), nuclear (Nuc) and chromatin (Chrom) fractions. Immunoblots were then performed as indicated. (B-F) Confluent pHLFs were transfected with scrambled control siRNA (siCTRL) or ATF4 siRNA (siATF4) before exposure to media plus TGF-β1 or media alone and the relative mRNA abundance of ATF4, PHGDH, PSAT1, PSPH and SHMT2 at 24 hours was measured by RT-qPCR (N=3 biological replicates). (G) Representative immunoblots for protein lysates treated as indicated in (B-F) are shown (N=3 biological replicates and data are representative of 3 independent experiments). (H) Confluent pHLFs were transfected with scrambled control siRNA (siCTRL) or ATF4 siRNA (siATF4) before exposure to media plus TGF-β1 or media alone and collagen deposition assayed by high content imaging after 48 hours. (Each data point shown is mean +/- SEM of the fold change relative to baseline of 3-4 biological replicates per condition and data are representative of 3 independent experiments). (I) Representative immunofluorescence images presented in (H) are shown, scale bar =100 µm. (J) Confluent wildtype and ATF4-/- crispred pHLFs were exposed to media plus TGF-β1 or media alone and collagen deposition assayed by high content imaging after 48 hours (each data point shown is mean +/- SEM of the fold change relative to baseline of 3 biological replicates per condition). Differences between groups were evaluated by two-way (B-F, H, J) ANOVA test with Tukey post-hoc test. *p<0.05, **p<0.01, ***p< 0.001, ****p< 0.0001.
mTOR amplifies glucose metabolism during TGF-β1––induced fibroblast collagen synthesis through an ATF4-dependent mechanism
In addition to increased expression of genes encoding serine-glycine biosynthetic enzymes, increased substrate availability is also necessary for increased metabolic pathway activity. Glycine can be synthesised de novo from glucose (32) and in our RNAseq dataset, the gene SLC2A1, which encodes the facilitative glucose transporter, GLUT1, was present in the same WGCNA module as the serine-glycine pathway genes and followed the same pattern of induction with TGF-β1, inhibition of induction in the presence of AZD8055 and insensitivity to rapamycin (Fig.1C). We therefore next considered whether rapamycin-insensitive mTOR signaling downstream of TGF-β1 increases glucose uptake to support increased glycine biosynthesis during TGF-β1––induced collagen deposition.
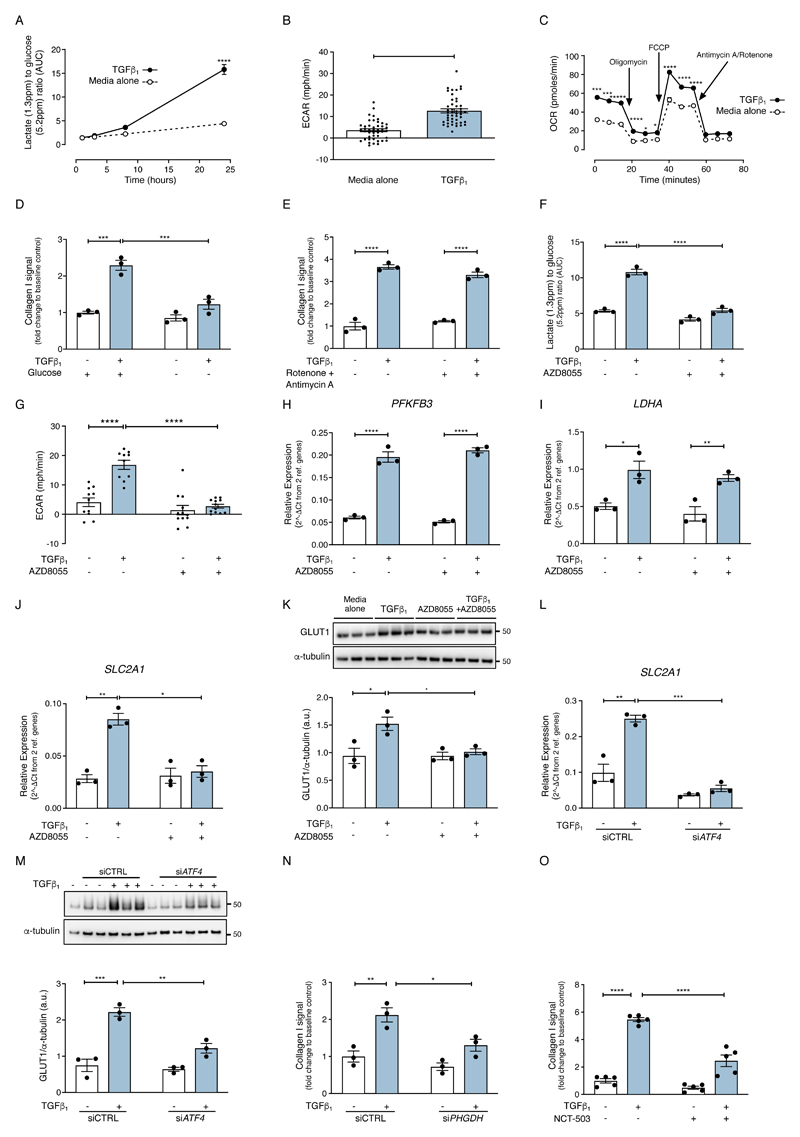
TGF-β1 stimulation of fibroblasts led to an increase in glycolytic flux, extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) (Fig. 6, A to C). This was associated with increased glucose uptake, SLC2A1 mRNA abundance, GLUT1 protein production and expression of genes encoding the glycolytic enzymes, PFKFB3 and LDHA (fig. S4, A to E). The TGF-β1––induced collagen response was decreased by 82 ± 11 % in media devoid of glucose (Fig. 6D) and by 90 ± 7% in the presence of the glycolytic inhibitor, 2-deoxyglucose (2DG) (fig. S4F), indicating that glycolysis is required for TGF-β1––induced collagen deposition. In contrast, the mitochondrial complex I and III inhibitors, rotenone and antimycin A, had no effect on TGF-β1–induced collagen deposition (Fig. 6E), despite inhibiting the TGF-β1––induced increase in OCR (Fig. 6C). Taken together, these data led us to conclude that glycolysis, but not mitochondrial respiration, is necessary for TGF-β1–induced collagen deposition.
Fig. 6. mTOR amplifies glucose metabolism during TGF-β1––induced fibroblast collagen synthesis through an ATF4-dependent mechanism.
Confluent pHLFs were exposed to media plus TGF-β1 or media only for 24 hours. (A) The AUC of lactate relative to AUC of glucose in cell supernatants was measured by NMR spectroscopy at the time points described (N=3 biological replicates and data are representative of 3 independent experiments). (B, C) Confluent pHLFs were exposed to media plus TGF-β1 or media only for 24 hours. Extracellular acidification rate and oxygen consumption rate were measured using the SeaHorse XF96e assay (N=46 biological replicates and data are representative of 3 independent experiments). (D-E) Confluent pHLFs were incubated in media deplete of glucose (D) or pre-incubated with rotenone and antimycin A (E) and stimulated for 48 hours with or without TGF-β1 and collagen deposition assessed by high content imaging. Each data point shown is mean +/- SEM of the fold change relative to baseline of 3 biological replicates and data are representative of 3 independent experiments. (F) Confluent pHLFs were pre-incubated with AZD8055 or vehicle control (DMSO) prior to exposure to media plus TGF-β1 or media alone. The AUC of the lactate peak relative to AUC of the glucose peak in cell supernatant was measured by NMR spectroscopy after 24 hours (N= 3 biological replicates and data are representative of 3 independent experiments). (G) The ECAR was assayed by SeaHorse XF96e after 24 hours with or without TGF-β1 stimulation (N=3 biological replicates and data are representative of 3 independent experiments). (H-K) Confluent pHLFs were pre-incubated with AZD8055 or vehicle control (DMSO) prior to TGF-β1 stimulation or media alone. Relative mRNA abundance of (H) PFKFB3, at 3 hours post TGF-β1 (I) LDHA and (J) SLC2A1, both at 24 hours post TGF-β1 was measured by RT-qPCR (N=3 biological replicates) and (K) immunoblot for GLUT1 and densitometric quantification were performed (N=3 biological replicates). (L) Confluent pHLFs were transfected with scrambled control siRNA (siCTRL) or ATF4 siRNA (siATF4) exposed to media plus TGF-β1 or media only and relative mRNA abundance of SLC2A1 at 24 hours was measured by RT-qPCR (N=3 biological replicates, data representative of 3 independent experiments). (M) Representative immunoblots of protein lysates derived from pHLFs treated in same conditions as (L) are shown (N=3 biological replicates). (N) Confluent pHLFs were transfected with scrambled control siRNA(siCTRL) or PHGDH siRNA (siPHGDH) and exposed to media plus TGF-β1 or media only for 48 hours and collagen deposition assessed by high content imaging (each data point shown is mean +/- SEM of the fold change relative to baseline of 3 biological replicates). (O) pHLFs were treated with PHGDH inhibitor, NCT-503 (or DMSO control) and stimulated with or without TGF-β1 for 48 hours. Collagen deposition was assessed by high content imaging, each data point represents mean +/- SEM of the fold change relative to baseline of 5 biological replicates per condition. Differences between groups were evaluated by unpaired t-test (B) or two-way ANOVA test with Tukey post-hoc test (A,C-O). *p<0.05, **p<0.01, ***p<0.001, ****p< 0.0001.
Next, we examined the effect of mTOR inhibition on TGF-β1––induced changes in glucose metabolism. ATP-competitive mTOR inhibition with AZD8055 significantly inhibited the TGF-β1––induced increase in glycolytic flux, represented by the lactate to glucose ratio measured in cell-conditioned media by 1H-NMR spectroscopy (Fig. 6F) and the TGF-β1––induced increase in ECAR (Fig. 6G). However, the TGF-β1––induced increase in gene expression of the key glycolytic genes LDHA and PFKFB3 (fig. S4, D and E) was insensitive to AZD8055 treatment (Fig. 6, H and I). In contrast, and in agreement with our RNAseq dataset (Fig. 1C), AZD8055 significantly inhibited the TGF-β1––induced increase in SLC2A1 mRNA abundance and GLUT1 protein abundance (Fig 6, J and K). The increase in SLC2A1 mRNA abundance was Smad3-dependent but rapamycin-insensitive (fig. S4, G and H), Finally, silencing ATF4 using siRNA inhibited the TGF-β1––induced increase in SLC2A1 mRNA abundance and GLUT1 protein abundance (Fig. 6, L and M).
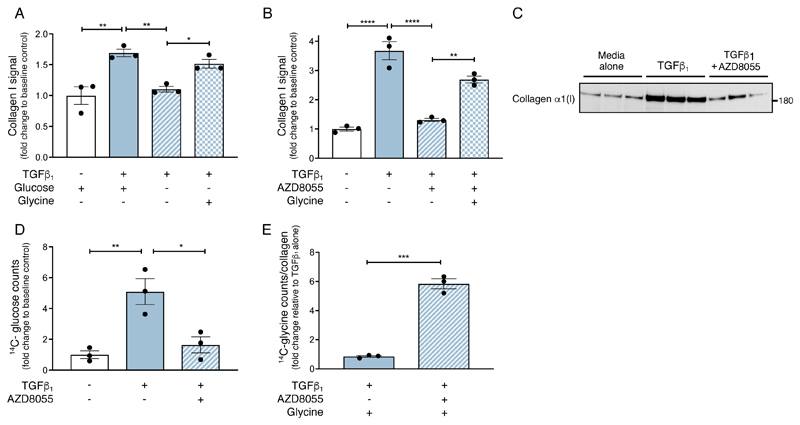
The observation that AZD8055 attenuates TGF-β1––induced collagen deposition, de novo serine-glycine pathway gene expression and glucose metabolism led us to speculate that mTOR signaling augments TGF-β1––induced collagen deposition by providing the glycolytic intermediates required for enhanced glycine and collagen synthesis. Phosphoglycerate dehydrogenase (PHGDH) is the first rate-limiting enzyme that catalyzes 3-phospho-glycerate (3-PG) from glycolysis into 3-phosphohydroxypyruvate (3-PHP). Disrupting PHGDH using siRNA or inhibition of PHGDH with a selective pharmacological inhibitor (NCT-503)(33) blocked TGF-β1––induced collagen deposition (Fig. 6, N and O and fig. S4I). Moreover, glycine supplementation rescued the inhibitory effect of glucose deprivation on TGF-β1–induced collagen deposition by 70 +/- 12 % (Fig. 7A). Furthermore, glycine supplementation rescued the inhibitory effect of AZD8055 on the TGF-β1–induced collagen response by 58 ± 5% (Fig. 7B). In contrast, supplementation with the glycine precursor, serine, did not rescue this inhibitory effect (fig. S5A).
Fig. 7. mTOR promotes glucose derived glycine biosynthesis to supply glycine during TGF-β1––induced collagen synthesis.
(A) Confluent pHLFs were deprived of glucose for 24 hours and incubated with or without glycine, followed by incubation with or without TGF-β1 for 48 hours before assessment of collagen deposition by high content imaging (each data point shown is mean +/- SEM of the fold change relative to baseline of N=3 biological replicates per condition and data are representative of 3 independent experiments). (B) Confluent pHLFs were incubated with AZD8055 and supplemented 30 minutes later with or without glycine, then incubated with or without TGF-β1 for 48 hours before assessment of collagen deposition by high content imaging. Each data point shown is mean +/- SEM of the fold change relative to baseline of 3 biological replicates and data are representative of 3 independent experiments. (C) Confluent pHLFs were incubated with AZD8055 or DMSO and exposed to media plus TGF-β1 or media alone in the presence of 12C-glucose for 48 hours. Collagen α1(I) was isolated by immunoprecipitation and immunoblotted. (D) Confluent pHLFs were incubated with AZD8055 or DMSO and exposed to media plus TGF-β1 or media alone in the presence of U-14C-glucose for 48 hours. U-14C-glucose incorporation into immunoprecipitated collagen α1(I) was assessed by scintillation counting (N=3 biological replicates). (E) Confluent pHLFs were incubated with AZD8055 or DMSO and stimulated with TGF-β1 in the presence of U-14C-glycine for 48 hours. U-14C-glycine incorporation into immunoprecipitated collagen α1(I) was assessed by scintillation counting (N=3 biological replicates) and expressed relative to immunoprecipitated collagen α1(I) protein abundance determined in a parallel immunoblot. Differences between groups were evaluated by unpaired t-test (E) or one way (A,B,D) ANOVA test with Tukey post-hoc test. *p<0.05, **p<0.01, ***p< 0.001, ****p< 0.0001.
We also performed experiments with exogenous radiolabeled glucose and glycine to determine whether they are incorporated into collagen α1(I) in response to TGF-β1 stimulation following isolation by immunoprecipitation (fig. S5B). Incorporation of either U-14C-glucose or U-14C-glycine was assessed by scintillation counting. The expected pattern of collagen α1(I) production in fibroblasts exposed to TGF-β1 and the impact of AZD8055 was observed following immunoprecipitation and visualization by western blot (Fig. 7C). Analysis of U-14C-glucose incorporation into collagen α1(I) mirrored this pattern, with a 5-fold increase in U-14C-glucose incorporation into isolated collagen α1(I) in TGF-β1–stimulated fibroblasts compared with media alone-treated fibroblasts (Fig. 7D). U-14C-glucose incorporation into collagen α1(I) was reduced to baseline values in TGF-β1–stimulated fibroblasts treated with AZD8055 (Fig. 7D). We also examined whether exogenous U-14C-glycine was incorporated into collagen α1(I) in conditions where the TGF-β1–induced serine-glycine biosynthesis pathway was inhibited by AZD8055. Exogenous U-14C-glycine incorporation into collagen α1(I) was low in TGF-β1–stimulated cells in the absence of AZD8055 but increased 6-fold in the presence of AZD8055, indicating that exogenous glycine is utilized by TGF-β1–stimulated fibroblasts to mount a collagen response when the de novo serine-glycine pathway was inhibited by AZD8055 (Fig. 7E).
Taken together, our data support the conclusion that the Smad3 signaling pathway and the mTORC1-4E-BP1 axis cooperate to enhance the production of ATF4 which in turn influences key ATF4 target genes in order to promote the biosynthesis of glycolysis-derived glycine to meet the collagen biosynthetic requirements of TGF-β1–activated myofibroblasts.
Discussion
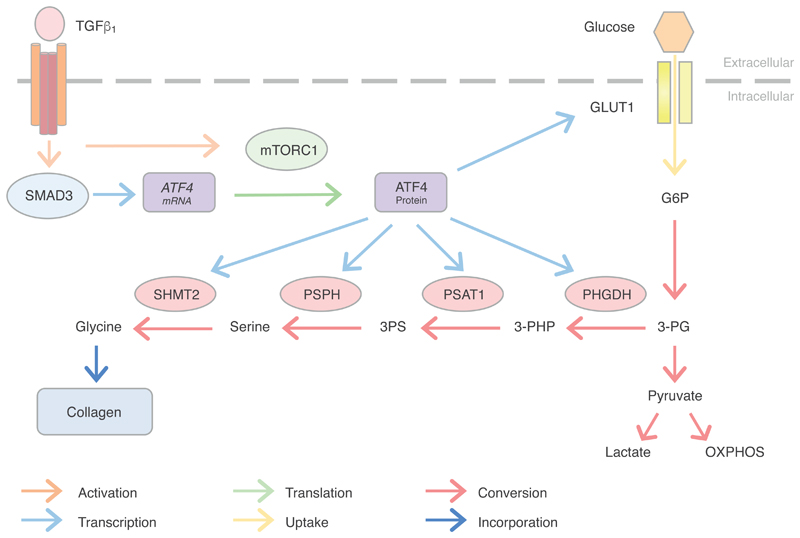
Excessive extracellular matrix deposition by myofibroblasts is a key pathogenic pathway leading to fibrosis and has also been implicated in promoting cancer progression. Previous data from our laboratory demonstrated a critical role for the mTORC1-4E-BP1 axis during TGF-β1–induced collagen deposition(27). In the present study, we now report a critical role for this axis, acting in cooperation with Smad3 to promote the production of ATF4 which in turn orchestrates the subsequent transcriptional amplification of the glucose-derived serine-glycine biosynthetic pathway (Fig. 8). As well as furthering our understanding of the signaling and transcriptional events by which TGF-β1 reprograms fibroblast metabolism to promote collagen biosynthesis, these observations have important implications for the development of future therapeutic strategies.
Fig. 8. Proposed mechanism by which ATF4 promotes the reconfiguration of myofibroblast metabolic and biosynthetic networks to support enhanced collagen biosynthesis in response to TGF-β1 stimulation.
TGF-β1 ligation of the TGF-β receptor complex leads to a Smad3 dependent increase in ATF4 mRNA abundance as well as mTOR activation. Activated mTORC1/4E-BP1 signaling in turn promotes ATF4 protein production through a translational mechanism. ATF4 subsequently promotes the transcriptional increase of SLC2A1/GLUT1 and key serine-glycine pathway genes. These then act together to promote glucose-derived glycine biosynthesis in order to support enhanced collagen synthesis rates in activated myofibroblasts. (TGF-β1: Transforming growth factor β1; mTORC1: mechanistic target of rapamycin complex 1; GLUT1: glucose transporter 1; G6P: glucose 6-phosphate; 3-PG: 3-Phosphoglycerate; PHGDH: phosphoglycerate dehydrogenase; 3-PHP: 3-phosphohydroxypyruvate; PSAT1: phosphoserine aminotransferase 1; 3PS: 3-phosphoserine; PSPH: phosphoserine phosphatase; SHMT2: serine hydroxymethyltransferase 2; OXPHOS: oxidative phosphorylation).
WGCNA analysis of our existing RNAseq dataset comparing the effect of ATP-competitive mTOR inhibition versus rapamycin on TGF-β1–induced human lung fibroblasts provided key insights into the potential mechanism by which mTOR influences collagen deposition. The initial motivation to exploit these pharmacological differences was predicated on previous studies that demonstrated that collagen deposition in activated fibroblasts and mesenchymal cells is sensitive to ATP-competitive mTOR inhibition but not to rapamycin treatment (27, 34, 35). These differences in pharmacological effect reflect the differential modes of mTOR inhibition by rapamycin and ATP-competitive mTOR inhibitors. Whereas the latter will block the phosphorylation sites on all downstream mTORC1 and mTORC2 substrates, rapamycin is a partial mTORC1 inhibitor which preferentially inhibits mTORC1 sites that are weakly phosphorylated by mTORC1 (p70S6K (Thr389) and 4E-BP1 (Ser65)). In contrast, mTORC1 substrates sites which are avidly phosphorylated by mTORC1 (4E-BP1 (Thr37/46) and 4E-BP1 (Thr70)) are largely insensitive to rapamycin treatment (27, 36).
The WGCNA module representing the profile of genes modulated by TGF-β1 treatment, sensitive to AZD8055 treatment but insensitive to rapamycin, comprised all four genes encoding the enzymes of the serine-glycine synthetic pathway (PHGDH, PSAT, PSPH and SHMT2) which were present in the top 20 most correlated genes overall. This finding merited further investigation given that glycine is present in every third amino acid position of the collagen molecule and is integral to the composition and stability of its triple helical structure (37). The observation that TGF-β1 stimulates the serine-glycine biosynthetic pathway to provide additional glycine needed for increased collagen synthesis is further in agreement with a recent report (38) and with the observation that pharmacologic inhibition of PHGDH attenuates fibrogenesis in vivo and also in the context of TGF-β1–induced collagen deposition reported herein (39).
In cancer cells, it is well-established that mTORC1 and its downstream substrate 4E-BP1, play critical roles in regulating metabolic pathways through the eIF4F complex-dependent translation of key transcription factors, including HIF1α and c-MYC (40). In our study, OCN identified several potential transcriptional regulators of interest, including notably ATF4. ATF4, a basic leucine zipper transcription factor, is a transcriptional master regulator of amino acid metabolism and stress responses and is classically enhanced as part of the integrated stress response (ISR) as a result of the activation of kinases, such as PERK and GCN2, which phosphorylate the translation factor eIF2α, leading to global reduction in mRNA translation except for select mRNAs, including ATF4 (41, 42). ATF4 in turn increases gene transcription by binding to the C/EBP–ATF response element (CARE) in the promoter of specific target genes (43–45). However, to the best of our knowledge, ATF4 had not previously been linked to TGF-β1-induced collagen deposition.
In our fibroblast studies, we now provide evidence that TGF-β1 increases ATF4 production through the cooperation of both canonical Smad3 and mTOR signaling pathways. Combining data obtained from kinetic studies of the signaling response with our data obtained in function blocking studies based on pharmacological inhibition (AZD8055), siRNA for Smad3 and ATF4, CRISPR-Cas9 gene editing of RICTOR and RPTOR and 4E-BP1 phospho-mutants, we propose a model in which the Smad3 pathway plays a critical role in regulating ATF4 mRNA abundance in an mTOR-independent manner; whereas the mTORC1-4E-BP1 axis is critical for ATF4 translation. Moreover, the early increase in ATF4 protein abundance is likely explained by the Smad3-dependent mTOR mediated translation of existing ATF4 mRNA and is consistent with our previous report that Smad3 is critical for TGF-β1-mediated mTORC1 activation (27). This model is further consistent with our subsequent in silico analysis of the ATF4 gene promoter for Smad binding sequences. Three different web-based platforms identified putative Smad2/3/4 binding elements in the ATF4 promoter with a predicted Smad3 binding site located at base pairs -283 to -275 from the transcription start site. Furthermore, the activation of ATF4 in the absence of the eIF2α kinase-mediated integrated stress response (ISR) has also been reported in mouse embryonic fibroblasts (46).
To the best of our knowledge, a key role for the mTORC1-4E-BP1 axis in mediating the stimulatory effects of TGF-β1 on ATF4 translation has not been previously reported but is in agreement with recent reports demonstrating a role for either mTORC1 (46), or both mTORC1 and 4E-BP1 in regulating the translation of ATF4 in other cell contexts (47). ATF4 gene disruption further demonstrated that ATF4 was necessary for TGF-β1–induced collagen deposition. Taken together with our recent report showing a role for mTORC1 and 4E-BP1 during TGF-β1–induced collagen deposition(27), the data herein now provides strong evidence for a key role for ATF4 downstream of the mTORC1-4E-BP1 axis in mediating the potent fibrogenic effects of TGF- β1.
In terms of potential relevance of our findings to the human disease setting, we further provide evidence that ATF4 is expressed by α-SMA positive myofibroblasts within IPF fibrotic foci, as well as by the epithelium. This latter observation is broadly in agreement with a previous report focusing on the role of ATF4 in the context of epithelial endoplasmic reticulum stress and apoptosis in IPF using a polyclonal antibody against ATF4 (48). The ATF4 staining pattern in control lung is predominantly epithelial and is in agreement with the pattern reported by Human Protein Atlas (https://www.proteinatlas.org/ENSG00000128272-ATF4/tissue/lung). Our data therefore now extends our current understanding of the role of ATF4 in the transcriptional regulation of the serine-glycine pathway beyond the cancer setting, where this pathway has been strongly implicated in supporting cell growth and proliferation, as well as clinical aggressiveness in non-small cell lung cancer (NSCLC) (31, 49).
We also herein provide evidence for the importance of both mTOR and ATF4 in regulating TGF-β1-induced increases in glycolysis. The first enzyme of the serine-glycine biosynthetic pathway (PHGDH) converts the glycolytic intermediate 3-phosphoglycerate (3PG) to 3-phosphohydroxypyruvate (3PHP). Previous studies have shown that glycolysis is elevated in myofibroblasts (16, 38) but whether mTOR signaling in response to TGF-β1 stimulation contributes to this glycolytic response in fibroblasts was not known. We confirmed that TGF-β1– stimulated myofibroblasts adopt a glycolytic phenotype and now provide further evidence that mTOR signaling plays a critical role in mediating these metabolic changes. However, although the mTOR-ATF4 axis was found to regulate the expression of SLC2A1 and GLUT1, we found no role for mTOR in regulating the expression of the TGF-β1–induced glycolytic genes, PFKFB3 and LDHA. In terms of linking enhanced glycolysis to TGF-β1–induced collagen deposition, extracellular glycine supplementation partially rescued the inhibitory effect of glucose deprivation, as well as mTOR inhibition, on TGF-β1-induced collagen deposition. An enhanced demand for glucose to meet the biosynthetic demands of collagen production following TGF-β1 stimulation is further supported by our observation that U-14C-glucose incorporation into collagen I is increased in TGF-β1-stimulated fibroblasts. U-14C-glycine tracing experiments further demonstrated that exogenous glycine enables fibroblasts to generate a TGF-β1–induced collagen response when the de novo serine-glycine pathway is inhibited by mTOR inhibition with AZD8055. Taken together, these data support the notion that mTOR promotes glycolysis to generate the necessary glycine to support enhanced collagen production. The observation that ATF4 mediates the stimulatory effects of TGF-β1 on SLC2A1 expression is further potentially in agreement with a previous study which identified two enhancer binding elements in the regulatory elements of the SLC2A1 gene which contains the same CRE consensus binding site shared by ATF4 (50).
It is also worth pointing out that the mechanism by which mTOR influences glycolysis in myofibroblasts contrasts with its role in cancer cells, where mTOR has been reported to promote the expression of multiple glycolytic genes by enhancing the translation of HIF1α and c-MYC (51–53). Although there is a degree of mechanistic overlap between cancer and fibrosis, the mutational landscapes are very different. mTOR is one of the most frequently mutated signaling hubs in cancer as a consequence of PI3K amplification/mutation, PTEN loss of function, AKT overexpression, and S6K1, 4E-BP1 and eIF4E overexpression(54). In contrast, mTOR pathway activation is likely to be a consequence of the altered tissue microenvironment in pathological fibrosis and in the stromal response in cancer, in that in both settings fibrogenesis is generally considered a reactive rather than a malignant process (55, 56). Of interest, although the mechanism is not known, IPF myofibroblasts have been reported to display evidence of PTEN deficiency (57).
It is also worth commenting that in contrast to an absolute requirement for extracellular glucose, mitochondrial respiration was found to be dispensable for TGF-β1-induced collagen deposition, suggesting that glycolytic flux in response to TGF-β1 stimulation is sufficient to meet both the biosynthetic and energetic requirements of the complex multistage process of collagen biosynthesis in primary adult lung fibroblasts over the course of our experiments (48 hours). Additionally, increased mitochondrial biogenesis has been reported to be important for the maintenance of the differentiated and contractile myofibroblast state in a fetal lung fibroblast line (IMR-90) (15). These studies report that the expression of the TGF-β1-induced matrix glycoprotein, fibronectin, was also not dependent on mitochondrial biogenesis. We further provide herein evidence that mTOR inhibition does not interfere with TGF-β1-induced myofibroblast differentiation which is also in agreement with a previous report(58). Taken together, a picture now seems to be emerging that TGF-β1–induced fibroblast differentiation and matrix synthesis, the two cardinal features that define the myofibroblast phenotype, may have distinct metabolic requirements that could be targeted separately or in combination to slowdown both the restrictive physiologic impairment and the gas exchange impairment that characterize the fibrotic lung in IPF.
In conclusion, we present a novel mechanistic model in which ATF4 reconfigures the fibroblast biosynthetic and metabolic network to meet the collagen synthetic demands of highly activated myofibroblasts following TGF-β1 stimulation. As well as providing novel insight into the signaling and transcriptional pathways by which the major pro-fibrotic mediator TGF-β1 exerts it pro-fibrotic effects, our findings suggest that targeting the mTORC1-ATF4 axis might represent a novel approach for the future development of anti-fibrotic strategies.
Materials and Methods
Primary human fibroblast culture
Primary human lung fibroblasts (pHLFs) were grown from explant cultures of healthy control lung tissue, as previously described (59). Human tissues were sourced ethically, and their research use was in accord with the terms of informed consents. Institutional research ethics committee approval for this work was obtained from the UCL Research Ethics Committee (12/EM/0058). To avoid exposing cells to supra-physiological glucose and glutamine concentrations in standard DMEM, pHLFs (passage 4-8) were cultured in glucose and glutamine-free DMEM (Thermo-Fisher) that was supplemented with 10% fetal calf serum (Simga-Aldrich), 5mM glucose, 0.7mM glutamine (Thermo-Fisher) and 1% Penicillin (100U/ml)/ Streptomycin (100 μg/ml)(Thermo-Fisher) (referred throughout the manuscript as “growth media”). All fibroblast cultures tested negative for mycoplasma.
A pHLF cell line expressing a dominant-negative mutant of 4E-BP1 was generated as described previously (27). Cells were selected with 2μg/ml puromycin (Sigma-Aldrich) for 4 days. Expression of the 4E-BP1 mutant was induced with 1 μg/ml doxycycline (Sigma-Aldrich) for 24 hour before treatment with or without TGF-β1. After 24 hours pHLF lysates were analyzed for ATF4 abundance by immunoblotting.
Real-time quantitative PCR (qPCR) analysis of gene expression
Total RNA was extracted from adherent cells using RNeasy Mini kit (Qiagen) according to the manufacturer’s instructions. Real-time PCR was performed using a Mastercycler Realplex ep gradient S (Eppendorf). Specific primers were designed to detect the expression of genes (Table S2). PCR amplification was carried out for 40 cycles at a melting temperature of 95°C for 15 seconds and an annealing temperature of 60°C for 60 sec. A dissociation curve was analyzed for each PCR experiment to assess primer-dimer formation or contamination. Relative expression was calculated using 2-ΔCt and ΔCt was calculated from the mean of two reference genes, ATP5B and β2 microglobulin (β2M) (PrimerDesign Ltd).
Immunoblotting
Adherent fibroblasts were washed with ice-cold PBS then lysed using Phosphosafe extraction reagent (Merck Millipore) supplemented with protease inhibitor (Merck Millipore). Equal protein quantities of lysate were separated by SDS-PAGE, transferred to nitrocellulose, and protein levels assessed by Western blotting with the following antibodies: P70S6K (CST #9202), Phospho-p70S6KThr389 (CST #9234), 4E-BP1 (CST #9644), Phospho-4E-BP1Ser65 (CST#9451), SMAD3 (CST #9523), Rictor (CST #2114), Raptor (CST#2280), GLUT1 (Abcam #EPR3915), PHGDH (CST #13428), PSAT (ThermoFisher #PA522124), PSPH (Insight Bio. #GTX109163-S), SHMT2 (CST #12762), ATF4 (CST #11815), α-tubulin (CST #9099), Histone H4 (Abcam #16483), Phospho-eIF2α(Ser51) (CST #3597) and eIF2α (#9722). The dilutions of primary and secondary antibodies were according to the manufacturer’s instructions. Tunicamycin (2µg/ml MP biomedicals) was used as a positive control for immunostaining of p-eIF2α. Protein band intensity was measured by densitometry in ImageQuant TL v8.1 (GE Healthcare) following electrochemiluminescence. All densitometries are presented relative to α-tubulin unless otherwise stated.
Determination of collagen type I deposition by high-content imaging
Extracellular collagen biosynthesis was measured in 96-well format by a high-content imaging based macro-molecular crowding assay as described previously (27). Briefly, confluent pHLFs were cultured in DMEM containing 0.4% FCS, 5 mM glucose, 0.7 mM L-glutamine and ascorbic acid (16.6µg/ml) in the presence of mixed Ficoll 70 and Ficoll 400 to facilitate molecular crowding. Following serum starvation for 24 hours, pHLFs were treated in the presence or absence of TGF-β1 (1 ng/ml) (R&D systems) with compounds AZD8055 (supplied by GSK, 1 µM used unless otherwise stated), Rapamycin (Millipore,100nm used unless otherwise stated), NCT-503 (20µM Sigma-Aldrich), GSK2656157(Cambridge Bioscience), 2-deoxy glucose (Sigma-Aldrich, 3mM used) or vehicle (0.1% DMSO). For rescue experiments, glycine (450 µM, Sigma Aldrich) and serine (Sigma-Aldrich) were used. After 48 hours, pHLFs were methanol-fixed and stained with antibodies to either human collagen I (C2456; Sigma) or α-smooth muscle actin (αSMA; (M085101-2, Thermo Fisher Scientific) followed by Alexa Fluor 488 secondary antibody (A11001, Life Technologies) and nuclear DAPI counterstain. Fluorescent signal was captured and quantified using ImageXpress Micro XLS high-content imaging system at 20x magnification (Molecular Devices), with 4 fields of view imaged per well and signal intensity normalized to cell count.
Histology and immunohistochemistry
All human samples were obtained with informed signed consent and with research ethics committee approval (10/H0504/9, 10/H0720/12 and 12/EM/0058). Patients with IPF were diagnosed in accordance with current international guidelines(60). Immunostaining for ATF4 and α-smooth-muscle actin was conducted on 10μm formalin-fixed paraffin-embedded serial sections of human lung biopsy material (N=3 patients with IPF and N=2 control subjects). Confocal dual-immunofluorescence for ATF4 (WH0000468M1, Sigma) was enhanced with streptavidin conjugated with Alexa Fluor® 488 (Thermo-Fisher) and co-localized with primary goat polyclonal α-smooth muscle actin (#PA5-18292, Thermo-Fisher) labeled with donkey anti-goat Alexa fluor® 647 (Abcam), and counterstained with DAPI. Digitized images were captured on a Leica DM6000 CS microscope fitted with a Leica TCS SP8 confocal head and the Leica LAS-X software suite (Leica Microsystems GmbH). All images are representative and final images were prepared with Image J.
NMR spectroscopy
pHLFs were seeded in 6-well plates at 2x105 cells/ml and incubated in growth media for 3 days. Media were then replaced with serum-free media and cells were further cultured for 24 hours, at which point they were treated with or without TGF-β1 at 1ng/mL. Cell supernatant was then collected and supplemented with 10% D2O for shift lock and 1mM TSP standard (Sigma). Using a 500MHz Bruker machine, 1H-spectra were obtained using a presaturation method after 128 scans. The resulting spectra were analyzed using ACD labs software (Advanced Chemistry Development UK Ltd) and integrals of the 5.2ppm peak and 1.3ppm peaks, corresponding to glucose and lactate respectively, were quantified relative to the internal TSP peak (0 ppm) standard.
3H-2-DG uptake
pHLFs were seeded in 6 well plates at 1 x105/ml and incubated in growth media for 3 days. Media were then replaced with serum free media for 24 hours. The next day, cells were treated with or without TGF-β1 at 1ng/mL. After 22 hours stimulation, media were changed to glucose-free media for 2 hours. Cells were then washed with 0.5ml of Krebs-Ringer-HEPES (KRH) buffer. Cells were incubated in 2ml/well of KRH buffer that contained 5mM 2-deoxyglucose and 0.625uCi/well 2-deoxy [3H] glucose (Perkin Elmer) for 5 mins. The cells were washed 4 times with KRH buffer and lysed with 0.05mM NaOH for 2 hours at 37°c. Incorporated radioactivity in the cell lysate was determined by liquid scintillation counting.
Assessment of extracellular acidification and oxygen consumption
The Seahorse Bioscience XF96e extracellular flux analyzer was used to measure oxygen consumption rate (OCR) and extracellular acidification rate (ECAR). Briefly, pHLFs were seeded in XF96e microplates at 1x105 per well and incubated for 3 days in growth media. Media were then replaced with DMEM containing 0.4% FBS for 24 hours. The next day, cells were treated with or without TGF-β1 at 1ng/mL, in the presence (or absence) of AZD8055. The following day, media were replaced with assay media pre-warmed to 37°C supplemented with 5mM glucose, 0.7 mM glutamine and 1mM pyruvate (Sigma S8636). Measurements of OCR and ECAR were performed after equilibration in assay medium (lacking supplemental CO2) for about 45 minutes. Oligomycin (Sigma O4876), FCCP (Sigma C2920), Antimycin A (Sigma A8674) and Rotenone (Sigma R8875) were added to perform a mitochondrial stress test as per the manufacturer’s instructions.
Transcriptomic analysis by RNA sequencing
Confluent fibroblast monolayers were untreated (“media alone”) or stimulated with TGF-β1 (and media) (1ng/ml, porcine origin, R&D Systems, USA) for 24 hours with and without AZD8055 (1μM, synthesized and provided by GSK) or rapamycin (100nM, Merck Chemicals) in standard DMEM (N=4 biological replicates per condition). Total RNA was extracted with miRCURY RNA Isolation Kit (Exiqon). Poly(A)-tailed RNA enrichment and library preparation were performed using the KAPA Stranded mRNA-Seq Kit with KAPA mRNA Capture Beads (KAPABiosystems, Wilmington, MA, USA). Paired-end RNA sequencing was performed with the NextSeq sequencing platform (Illumina, San Diego, CA, USA). Reads pre-processing and alignment to the genome (homo sapiens UCSC hg19) were performed using STAR aligner. Counts were normalised using the R package DeSeq2, then log2 transformed with an offset of 1 and filtered to remove genes with an average count <1 across all samples. This left 15959 genes that were then interrogated using the weighted gene co-expression network analysis (WGCNA) package in R. Briefly, gene co-expression similarity was estimated by generating an absolute Pearson product moment correlation matrix. Using a soft threshold of 20, an adjacency matrix was calculated and used to estimate the topological overlap and assign genes to modules. Highly correlated modules were merged using a cut-height of 0.06 on the topological matrix dendrogram. Sixty-five modules were returned and assigned colours as names. MetaCore™ (Thomson Reuter) was used to run pathway enrichment analyses and interaction network analyses on each of the 65 modules. Overconnected node analysis (OCN) was performed using the metabaseR R packages licensed from Clarivate Analytics and identifies one-step away direct regulators of the dataset that are statistically overconnected with the objects from the data set. The p-value of overconnectivity was calculated using hypergeometric distribution.
In-silico prediction of SMAD2/3/4 binding sites in the ATF4 promoter
The ATF4 promoter sequence was obtained from the eukaryotic promoter database (EPD) (Homo sapiens version 6, https://epd.vital-it.ch/human/human_database.php) (61). SMAD2/3/4 binding site prediction was performed using web-based applications: EPD Search Motif Tool (using a cut-off p-value of 0.001), PROMO (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3) (62) (selecting eukaryote for factor’s and site’s species), and ConTra V3 (http://bioit2.irc.ugent.be/contra/v3/#/step/1) (63) (selecting visualization as type of analysis, human as reference organism and ATF4 sequence under identification number NM_182810).
ATF4 protein stability
Lysates were prepared from pHLFs stimulated with TGF-β1 (1 ng/ml) for 13 hours before pre-treatment with DMSO or lactimidomycin (1 μM, Merck Millipore #506291) for 5 minutes, followed by exposure to DMSO or AZD8055 (1 μM) for 0/10/20/30/60 minutes before lysis. Immunoblotting was used to analyse the abundance of ATF4 normalised to α-tubulin.
Chromatin-bound protein fractionation
Cell lysis and cytosolic extraction were performed following the manufacturer’ instructions (NE-PER™, ThermoFisher). Nuclear extraction was performed by cell lysis in Buffer B (3mM EDTA, 0.2mM EGTA, 1mM DTT and protease inhibitor cocktail) for 30 minutes, followed by separation of soluble (nucleosoluble) and pellet (chromatin-bound) fractions by centrifugation at 1700 x g 4°C for 5 minutes. Chromatin-bound protein pellet was washed in Buffer B before resuspension in laemmli buffer (4% SDS, 20% Glycerol, 10% 2-ME, 0.004% bromophenol blue, 0.125M Tris HCl).
siRNA-mediated protein expression knock-down
For the determination of collagen deposition by molecular crowding assay, pHLFs were grown to confluence in our standard growth media in 96-well plates. During the serum-starvation period in growth media containing 0.4% FCS, cells were transfected with 10nM siRNAs (Dharmacon, SMARTpool) targeting ATF4, PHGDH and SMAD3 using RNAiMax lipofectamine (Invitrogen) according to the manufacturer’s instructions. The following day, the media were replaced by Ficoll-containing media and the macro-molecular crowding assay was performed as described above.
For western blotting and qPCR experiments, confluent pHLFs in 12-well plates were starved in serum-free media and transfected with 10nM siRNA (Dharmacon) using RNAiMax lipofectamine (Invitrogen) according to the manufacturer’s instructions. The following day, the media were replaced with fresh serum-free media with and without TGF-β1 (1 ng/ml) for the indicated time points.
CRISPR-Cas9 gene editing
pHLFs were electroporated with a CRISPR ribonucleoprotein (RNP) complex using the Lonza 4D Nucleofector™ system (Basel, Switzerland). The guide RNA (gRNA) sequence targeting ATF4 (AGGTCTCTTAGATGATTACCTGG), RPTOR exon 26 (CCGCGTCTACGACACAGAAGGATGG) or RICTOR exon 29 (AATATCGGCTCATCAAATTGGGG were designed using a combination of previously published work (47), the Deskgen design platform (https://www.deskgen.com/guidebook/advanced.html) and the Integrated DNA technology online tool (https://eu.idtdna.com/site/order/designtool/index/CRISPR_CUSTOM). A predesigned control gRNA sequence (Integrated DNA technology) was used to generate matched wildtype pHLFs.
Incorporation of U-14C-glucose and U-14C-glycine into collagen I
pHLFs were grown to confluence in 12-well plates and quiesced in serum-free media for 24 hours. Cells were then incubated in fresh serum-free media containing 5 mM 12C-glucose with or without 2 µCi U-14C-D-glucose (1 nM, Perkin Elmer). For glycine tracing studies, media containing 1 mM 12C-glycine, 1 mM 12C-glycine plus 2µCi U-14C-glycine (3.9 nM, Perkin Elmer) or media alone, was added to cells with either AZD8055 (1 µM) or vehicle (0.1% DMSO). For TGF-β1–treated cells, TGF-β1 was added at (1 ng/ml). After 48 hours cell layers were lysed (lysis buffer 300 mM NaCl, 10 mM Tris pH 7.4, 1% NP-40, protease-inhibitor) and pre-cleared with Protein G agarose beads (CST #37478) for 4 hours at 4°C. Cleared lysates were immunoprecipitated with 1 µg anti-collagen α1(I) primary antibody (CST #84336) followed by Protein G agarose bead incubation overnight at 4°C. Immunoprecipitated material was then eluted in LDS-containing sample buffer at 93°C for 5 minutes, and incorporated radioactivity in the eluate determined by liquid scintillation counting (scintillated in Beckman Coulter LS6500 using Ecoscint A (National diagnostics) scintillation fluid). pHLF lysates incubated with corresponding treatments in the presence of 12C-glucose or 12C-glycine were also subjected to SDS-PAGE, transferred to PVDF and immunoblotted to determine collagen α1(I) abundance. To demonstrate immunoprecipitation efficiency, representative input (10%) and unbound (10%) portions were immunoblotted for collagen α1(I) alongside immunoprecipitated protein, antibody alone (0.4µg) and beads alone (10% slurry).
Statistical analysis
All data are expressed as means ± SEM and figures constructed using GraphPad Prism version 7.00 and all experiments were repeated at least 3 times. Statistical differences between two groups were analyzed using a standard two-tailed t-test (assuming unpaired data sets and unequal variances). When more than two groups were compared, either one-way or 2-way ANOVA was used with post-hoc application of the Tukey method. Four-parameter non-linear regression analyses were used to generate IC50 values from concentration-response curves. The alpha level was set at 0.05 for all tests (GraphPad Prism).
Supplementary Material
Acknowledgments
The authors gratefully acknowledge our helpful discussions with Dr. Lindsay Edwards (GSK) and give thanks to Mr. Huw Jones (UCL Digital Media) for the production of the figures.
Funding: The authors gratefully acknowledge major funding support received for this work from the NIHR University College London Hospitals Biomedical Research Centre /NIMR (training fellowship to BS) and Fonds de la Recherche en Santé – Québec (training fellowship to IA). Support was also received from the Medical Research Council UK (training fellowship MR/K024078/1 to HVW) the Biotechnology and Biological Research Council (BBSRC) and GSK (iCASE studentships BB/M502844/1 and BB/P504440/1) awarded to RCC). RCC acknowledges funding from GlaxoSmithKline (GSK) under a collaborative framework agreement. DA’s lab is funded by the MRC (MC_UP_1202/1) and by the Francis Crick Institute which receives its core funding from Cancer Research UK, the UK Medical Research Council and the Wellcome Trust (FC001033).
Footnotes
Author contributions: Conceptualization, R.C.C., B.S, I.A., D.A.; Methodology; R.C.C., P.M., D.A., G.B., A.B, Investigation, B.S., I.A., M.P., D.G., H.V.W, G.C., E.J.F., P.F.D., M.R.; Writing – Original Draft, B.S. and I.A and R.C.C.; Writing – Review & Editing, R.C.C. and D.A.; Funding Acquisition, R.C.C., D.A., B.S. I.A., P.M., A.B., Data Curation, A.T. Resources R.C.C., G.B., Supervision, R.C.C. and D.A.
Competing interests: R.C.C. declares receiving research funding for some of this work from a collaborative framework agreement from GlaxoSmithKline. A.T., G.B. and A.D.B. are employees of GlaxoSmithKline with a commercial interest in the area of investigation.
Data and materials availability: The RNASeq dataset had been deposited and will be publically available at: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE102674 upon acceptance. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References and Notes
- 1.Wynn TA. Common and unique mechanisms regulate fibrosis in various fibroproliferative diseases. J Clin Invest. 2007;117:524–529. doi: 10.1172/JCI31487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coultas DB, Zumwalt RE, Black WC, Sobonya RE. The epidemiology of interstitial lung diseases. Am J Respir Crit Care Med. 1994;150:967–72. doi: 10.1164/ajrccm.150.4.7921471. [DOI] [PubMed] [Google Scholar]
- 3.King TE, et al. Idiopathic pulmonary fibrosis: relationship between histopathologic features and mortality. Am J Respir Crit Care Med. 2001;164:1025–32. doi: 10.1164/ajrccm.164.6.2001056. [DOI] [PubMed] [Google Scholar]
- 4.Richeldi L, et al. Efficacy and safety of nintedanib in idiopathic pulmonary fibrosis. N Engl J Med. 2014;370:2071–82. doi: 10.1056/NEJMoa1402584. [DOI] [PubMed] [Google Scholar]
- 5.King TE, et al. A phase 3 trial of pirfenidone in patients with idiopathic pulmonary fibrosis. N Engl J Med. 2014;370:2083–92. doi: 10.1056/NEJMoa1402582. [DOI] [PubMed] [Google Scholar]
- 6.Lee AS, Mira-Avendano I, Ryu JH, Daniels CE. The burden of idiopathic pulmonary fibrosis: An unmet public health need. Respir Med. 2014;108:955–967. doi: 10.1016/j.rmed.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 7.Klingberg F, Hinz B, White ES. The myofibroblast matrix: implications for tissue repair and fibrosis. J Pathol. 2013;229:298–309. doi: 10.1002/path.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Datta A, Scotton CJ, Chambers RC. Novel therapeutic approaches for pulmonary fibrosis. Br J Pharmacol. 2011;163:141–172. doi: 10.1111/j.1476-5381.2011.01247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Otranto M, et al. The role of the myofibroblast in tumor stroma remodeling. Cell Adh Migr. 2012;6:203–219. doi: 10.4161/cam.20377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu L, et al. Stromal Myofibroblasts Are Associated with Poor Prognosis in Solid Cancers: A Meta-Analysis of Published Studies. PLoS One. 2016;11:e0159947. doi: 10.1371/journal.pone.0159947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Neill LAJ, Kishton RJ, Rathmell JC. A guide to immunometabolism for immunologists. Nat Rev Immunol. 2016 doi: 10.1038/nrmicro.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Win T, et al. Areas of normal pulmonary parenchyma on HRCT exhibit increased FDG PET signal in IPF patients. Eur J Nucl Med Mol Imaging. 2014;41:337–342. doi: 10.1007/s00259-013-2514-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groves AM, et al. Idiopathic pulmonary fibrosis and diffuse parenchymal lung disease: implications from initial experience with 18F-FDG PET/CT. J Nucl Med. 2009;50:538–545. doi: 10.2967/jnumed.108.057901. [DOI] [PubMed] [Google Scholar]
- 14.Justet A, et al. [18F]FDG PET/CT predicts progression-free survival in patients with idiopathic pulmonary fibrosis. Respir Res. 2017;18:74. doi: 10.1186/s12931-017-0556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernard K, et al. J Biol Chem. doi: 10.1074/jbc.M115.646984. in press. [DOI] [Google Scholar]
- 16.Xie N, et al. Glycolytic reprogramming in myofibroblast differentiation and lung fibrosis. Am J Respir Crit Care Med. 2015;192:1462–1474. doi: 10.1164/rccm.201504-0780OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saxton RA, Sabatini DM. mTOR Signaling in Growth, Metabolism, and Disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soga T. Cancer metabolism: key players in metabolic reprogramming. Cancer Sci. 2013;104:275–81. doi: 10.1111/cas.12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beauchamp EM, Platanias LC. The evolution of the TOR pathway and its role in cancer. Oncogene. 2013;32:3923–3932. doi: 10.1038/onc.2012.567. [DOI] [PubMed] [Google Scholar]
- 20.Lien EC, Lyssiotis CA, Cantley LC. 2016. pp. 39–72. [DOI] [PubMed] [Google Scholar]
- 21.Benjamin D, Colombi M, Moroni C, Hall MN. Rapamycin passes the torch: a new generation of mTOR inhibitors. Nat Rev Drug Discov. 2011;10:868–880. doi: 10.1038/nrd3531. [DOI] [PubMed] [Google Scholar]
- 22.Sun S-Y. mTOR kinase inhibitors as potential cancer therapeutic drugs. Cancer Lett. 2013;340:1–8. doi: 10.1016/j.canlet.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shan L, et al. mTOR Overactivation in Mesenchymal cells Aggravates CCl4– Induced liver Fibrosis. Sci Rep. 2016;6:36037. doi: 10.1038/srep36037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshizaki AAAA, et al. Treatment with rapamycin prevents fibrosis in tight-skin and bleomycin-induced mouse models of systemic sclerosis. Arthritis Rheum. 2010;62:2476–87. doi: 10.1002/art.27498. [DOI] [PubMed] [Google Scholar]
- 25.Chen G, et al. Rapamycin Ameliorates Kidney Fibrosis by Inhibiting the Activation of mTOR Signaling in Interstitial Macrophages and Myofibroblasts. PLoS One. 2012;7:e33626. doi: 10.1371/journal.pone.0033626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mercer PF, et al. Exploration of a potent PI3 kinase/mTOR inhibitor as a novel anti-fibrotic agent in IPF. Thorax. 2016;71:701–711. doi: 10.1136/thoraxjnl-2015-207429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woodcock HV, et al. The mTORC1/4E-BP1 axis represents a critical signaling node during fibrogenesis. Nat Commun. 2019;10:6. doi: 10.1038/s41467-018-07858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laplante M, Sabatini DM. Regulation of mTORC1 and its impact on gene expression at a glance. J Cell Sci. 2013;126:1713–9. doi: 10.1242/jcs.125773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ducker GS, et al. Reversal of Cytosolic One-Carbon Flux Compensates for Loss of the Mitochondrial Folate Pathway. Cell Metab. 2016;23:1140–1153. doi: 10.1016/j.cmet.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adams CM. Role of the transcription factor ATF4 in the anabolic actions of insulin and the anti-anabolic actions of glucocorticoids. J Biol Chem. 2007;282:16744–53. doi: 10.1074/jbc.M610510200. [DOI] [PubMed] [Google Scholar]
- 31.DeNicola GM, et al. NRF2 regulates serine biosynthesis in non–small cell lung cancer. Nat Genet. 2015;47:1475–1481. doi: 10.1038/ng.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer. 2013;13:572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pacold ME, et al. A PHGDH inhibitor reveals coordination of serine synthesis and one-carbon unit fate. Nat Chem Biol. 2016;12:452–458. doi: 10.1038/nchembio.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker NM, et al. Mechanistic Target of Rapamycin Complex 1 (mTORC1) and mTORC2 as Key Signaling Intermediates in Mesenchymal Cell Activation. J Biol Chem. 2016;1 doi: 10.1074/jbc.M115.672170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang W, et al. A critical role for the mTORC2 pathway in lung fibrosis. PLoS One. 2014;9 doi: 10.1371/journal.pone.0106155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang SA, et al. mTORC1 Phosphorylation Sites Encode Their Sensitivity to Starvation and Rapamycin. Science (80-. ) 2013;341 doi: 10.1126/science.1236566. 1236566-1236566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mouw JK, Ou G, Weaver VM. Extracellular matrix assembly: a multiscale deconstruction. Nat Rev Mol Cell Biol. 2014;15:771–785. doi: 10.1038/nrm3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nigdelioglu R, et al. Transforming Growth Factor (TGF)-β Promotes de Novo Serine Synthesis for Collagen Production. J Biol Chem. 2016;291:27239–27251. doi: 10.1074/jbc.M116.756247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hamanaka RB, et al. Inhibition of Phosphoglycerate Dehydrogenase Attenuates Bleomycin-induced Pulmonary Fibrosis. Am J Respir Cell Mol Biol. 2018;58:585–593. doi: 10.1165/rcmb.2017-0186OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yecies JL, Manning BD. Transcriptional control of cellular metabolism by mtor signaling. Cancer Res. 2011;71:2815–2820. doi: 10.1158/0008-5472.CAN-10-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harding HP, et al. An Integrated Stress Response Regulates Amino Acid Metabolism and Resistance to Oxidative Stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 42.Ameri K, Harris AL. Activating transcription factor 4. Int J Biochem Cell Biol. 2008;40:14–21. doi: 10.1016/j.biocel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 43.Timosenko E, et al. Nutritional Stress Induced by Tryptophan-Degrading Enzymes Results in ATF4-Dependent Reprogramming of the Amino Acid Transporter Profile in Tumor Cells. Cancer Res. 2016;76:6193–6204. doi: 10.1158/0008-5472.CAN-15-3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao E, et al. KDM4C and ATF4 Cooperate in Transcriptional Control of Amino Acid Metabolism. Cell Rep. 2016;14:506–519. doi: 10.1016/j.celrep.2015.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kilberg MS, Shan J, Su N. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol Metab. 2009;20:436–443. doi: 10.1016/j.tem.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ben-Sahra I, Hoxhaj G, Ricoult SJH, Asara JM, Manning BD. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science. 2016;351:728–33. doi: 10.1126/science.aad0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park Y, Reyna-Neyra A, Philippe L, Thoreen CC. mTORC1 Balances Cellular Amino Acid Supply with Demand for Protein Synthesis through Post-transcriptional Control of ATF4. Cell Rep. 2017;19:1083–1090. doi: 10.1016/j.celrep.2017.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Korfei M, et al. Epithelial endoplasmic reticulum stress and apoptosis in sporadic idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2008;178:838–46. doi: 10.1164/rccm.200802-313OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ye J, et al. Pyruvate kinase M2 promotes de novo serine synthesis to sustain mTORC1 activity and cell proliferation. Proc Natl Acad Sci. 2012;109:6904–6909. doi: 10.1073/pnas.1204176109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murakami T, et al. Identification of two enhancer elements in the gene encoding the type 1 glucose transporter from the mouse which are responsive to serum, growth factor, and oncogenes. J Biol Chem. 1992;267:9300–6. [PubMed] [Google Scholar]
- 51.Aramburu J, Ortells MC, Tejedor S, Buxadé M, López-Rodríguez C. Transcriptional regulation of the stress response by mTOR. Sci Signal. 2014;7:re2. doi: 10.1126/scisignal.2005326. [DOI] [PubMed] [Google Scholar]
- 52.Goldberg AA, et al. J Biol Chem. doi: 10.1074/jbc.M116.771584. in press. [DOI] [Google Scholar]
- 53.Düvel K, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pópulo H, Lopes JM, Soares P. The mTOR signalling pathway in human cancer. Int J Mol Sci. 2012;13:1886–918. doi: 10.3390/ijms13021886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cool CD, et al. Fibroblast foci are not discrete sites of lung injury or repair: The fibroblast reticulum. Am J Respir Crit Care Med. 2006;174:654–658. doi: 10.1164/rccm.200602-205OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer. 2016;16:582–598. doi: 10.1038/nrc.2016.73. [DOI] [PubMed] [Google Scholar]
- 57.White ES, et al. Negative Regulation of Myofibroblast Differentiation by PTEN (Phosphatase and Tensin Homolog Deleted on Chromosome 10) Am J Respir Crit Care Med. 2006;173:112–121. doi: 10.1164/rccm.200507-1058OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ge J, et al. Glutaminolysis Promotes Collagen Translation and Stability via α-Ketoglutarate-mediated mTOR Activation and Proline Hydroxylation. Am J Respir Cell Mol Biol. 2018;58:378–390. doi: 10.1165/rcmb.2017-0238OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Keerthisingam CB, et al. Cyclooxygenase-2 Deficiency Results in a Loss of the Anti-Proliferative Response to Transforming Growth Factor-β in Human Fibrotic Lung Fibroblasts and Promotes Bleomycin-Induced Pulmonary Fibrosis in Mice. Am J Pathol. 2001;158:1411–1422. doi: 10.1016/s0002-9440(10)64092-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raghu G, et al. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med. 2011;183:788–824. doi: 10.1164/rccm.2009-040GL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dreos R, Ambrosini G, Cavin Périer R, Bucher P. EPD and EPDnew, high-quality promoter resources in the next-generation sequencing era. Nucleic Acids Res. 2013;41:D157–64. doi: 10.1093/nar/gks1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Messeguer X, et al. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18:333–4. doi: 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]
- 63.Kreft Ł, et al. ConTra v3: a tool to identify transcription factor binding sites across species, update 2017. Nucleic Acids Res. 2017;45:W490–W494. doi: 10.1093/nar/gkx376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.