Abstract
In the early 1980s Jeff Hall and Michael Rosbash at Brandeis University and Mike Young at Rockefeller University set out to isolate the period (per) gene, which was recovered in a revolutionary genetic screen by Ron Konopka and Seymour Benzer for mutants that altered circadian behavioral rhythms. Over the next 15 years the Hall, Rosbash and Young labs made a series of groundbreaking discoveries that defined the molecular timekeeping mechanism and formed the basis for them being awarded the 2017 Nobel Prize in Physiology or Medicine. Here the authors recount their experiences as post-docs in the Hall, Rosbash and Young labs from the mid-1980s to the mid-1990s, and provide a perspective of how basic research conducted on a simple model system during that era profoundly influenced the direction of the clocks field and established novel approaches that are now standard operating procedure for studying complex behavior.
Keywords: Nobel Prize, Circadian rhythms, Clock genes, Feedback loop, Genetic screens, Behavioral neuroscience
Highlights
-
•
2017 Nobel Prize awarded to Hall, Rosbash and Young for circadian clock mechanisms.
-
•
Work on fruit flies in the 1980s and 1990s were key to deciphering clock mechanisms.
-
•
Authors recount their experiences as postdocs in the Hall, Rosbash and Young labs.
-
•
The broad impacts of basic research on fruit fly clock genes.
1. Introduction
On Oct. 2, 2017, the three of us awakened to the incredible news that the 2017 Nobel Prize in Physiology or Medicine was awarded to our postdoctoral mentors, Drs. Jeff Hall, Michael Rosbash and Mike Young, for “their discoveries of molecular mechanisms controlling the circadian rhythm” (Fig. 1). Our elation was amplified as each of us learned that the Nobel Assembly had included one of our papers on a list of key publications in the body of research that was being recognized. In this article, we reflect on our personal and scientific journeys as we were attracted to join these labs, and as we engaged with the challenges and joys of contributing to work that led to the Nobel Prize. We conclude with some perspectives on how this work in the Hall, Rosbash and Young labs laid the foundation for deciphering the molecular basis of clock function in all animals, pioneered approaches that are now common strategies for studying the neural mechanisms of complex behaviors and represents a powerful example of the value of basic research on model organisms.
Fig. 1.
The authors celebrating in Stockholm with their postdoctoral advisors, the 2017 Nobel laureates in Physiology or Medicine. Left, Paul Hardin and Michael Rosbash at the reception following the Nobel Lectures in Physiology or Medicine at the Karolinska Institute. Center, Jeff Hall and Kathy Siwicki at the reception honoring the 2017 Nobel Laureates at the Nordic Museum. Right, Mike Young and Jeff Price at the reception for Mike Young following the Nobel Prize ceremony at the Grand Hotel.
2. Kathy Siwicki
As I was finishing my Ph.D. in Neurobiology in the mid-1980s, studying lobster neuropeptides with Ed Kravitz at Harvard, I was attracted to Drosophila by the enormous potential of using flies to decipher the mechanisms of complex processes in animal biology. Major technical breakthroughs around that time allowed us, for the first time, to manipulate the fly genome, so biologists suddenly had unprecedented experimental power for studying how genes specify animal development and regulate physiology and behavior. As I looked for postdoc opportunities in labs where I could learn to use Drosophila to study how brain circuits are wired to produce complex behaviors, I discovered Konopka’s three remarkable period mutants (Konopka and Benzer, 1971). As is now well known, the perShort (perS) and perLong (perL) alleles changed the period of the fly’s daily rhythms in opposite directions, and the perZero (per0) mutant rendered flies completely arrhythmic. Even with no formal training in genetics, it was clear to me that the distinct phenotypes of these per mutant alleles were strong evidence that this gene was a key that could potentially unlock the mechanism of endogenous biological clocks. Since the gene had just been cloned and sequenced by groups at Brandeis and Rockefeller (Bargiello et al., 1984, Zehring et al., 1984), it seemed like a perfect time for a neurobiologist to begin investigating the cellular functions of the per gene product. I was particularly drawn to work with Jeff Hall because of his encyclopedic knowledge of neurogenetics, and the fact that his broad interests in fly brains and complex behavior were closely aligned with my own interests.
Thus, I joined Jeff Hall’s lab in 1985 with the modest aim of figuring out how the PER protein works to endow flies with endogenous daily rhythms. At that point, the basic question of how a specific mutant genotype produces a behavioral phenotype had been pioneered by a few labs studying Drosophila learning mutants or ion channel genes, areas of neuroscience where some of the relevant biology had been established through electrophysiological or biochemical approaches. By comparison, the circadian clock was a virtual black box. The sequence of the per gene offered no clues about the protein’s function – there were no similar sequence domains in the primitive databases of the time, and no other evidence to justify any specific hypothesis about its biochemical function. It was exciting to be joining a team that was forging into uncharted waters, supported by emerging molecular tools and our faith that the compelling phenotypes of Konopka’s mutants would lead us to novel discoveries.
I set out to develop an anti-PER antibody that I would use to find the fly’s “clock cells.” The intense competition between the Brandeis and Rockefeller teams at that time contributed to our sense of urgency. There were pressures to react and respond to rumors about progress in the Young lab by adopting new approaches. Nevertheless, after nearly a year of frustrated efforts, Jeff encouraged me to persist. I was raising and screening rabbit antibodies against a few synthetic peptides that we had selected simply by trying to divine insights about functional domains of the protein while staring at the primary sequence. Several months in, Jeff received an airmail package from Bambos Kyriacou (currently a Professor at the University of Leicester) containing new predictions about possible bends and loops in the protein (Thackeray, 1989). Jeff noticed that one predicted elbow included the just-identified site of the perS missense mutation (Yu et al., 1987), so he suggested that we add a 14-mer peptide version of this elbow to the pipeline. The first breakthrough occurred when an antibody to this “S-peptide” revealed nuclear staining of the photoreceptors in the compound eyes of wild-type flies and no staining in the control genotype (per deletion females). I also saw many small nuclei stained throughout the brain and optic lobes that appeared to be glial cells. My most memorable moment of discovery occurred shortly thereafter, upon seeing the famed “lateral neurons” for the first time and recognizing that these were very likely the cellular substrates of the fly’s circadian clock.
While it may seem obvious in retrospect that one should look for daily oscillations in the levels of a protein that was hypothesized to have a central function in circadian timekeeping, it was not so obvious at the time. Based on initial reports that per transcript levels were constant throughout the day/night cycle (Reddy et al., 1984, Young et al., 1985), we did not expect to find rhythms in PER protein staining, and we failed to fully recognize their significance when the data initially revealed them (Siwicki et al., 1988). Indeed, I recall being frustrated at first by the fact that some flies showed very strong staining for PER protein, while others seemed to have very little! When we recognized that those with the strongest staining had all been done first thing in the morning, I set up temporally controlled experiments and found clear evidence for daily and circadian cycling of PER in photoreceptors (Siwicki et al., 1988). As I was confirming and documenting these protein rhythms, I was also pregnant for the first time, and I was determined to finish experiments for this paper before the baby arrived. Although there were no official maternity leave policies for postdocs in 1987, and in spite of the competitive pressure to publish new findings promptly, Jeff emphatically encouraged me to take a three-month leave with full pay.
Soon thereafter I began working with Danielle Zerr, a talented Brandeis undergraduate (currently a Professor of Pediatrics at the University of Washington), to quantify the temporal features of rhythmic PER protein staining and the effects of per mutants on the phase and amplitude of the protein rhythms (Zerr et al., 1990). During this exciting and productive phase of the work, Jeff would often scribble new ideas for experiments on yellow post-it notes that I would find on my desk in the morning (a highly effective mode of communication in the days before email). Our findings that PER protein cycling in perS mutants was phase-advanced in LD and persisted with a short period in DD were especially important, as they indicated that the mutant PER protein defines the phase and period of its own molecular rhythms in parallel with its effects on behavioral rhythms (Zerr et al., 1990). As Danielle was writing up these results for her senior thesis in the spring of 1989, I had another baby and moved to Pennsylvania to join the faculty of Swarthmore College as a new Assistant Professor. Meanwhile, our evidence for PER protein rhythms in brain cells and photoreceptors led Paul Hardin to question earlier reports of “no cycling” of the per transcript, which had been based on RNA extracted from whole flies, and to re-assess the question specifically in head extracts. As he describes below, Paul found compelling evidence for robust daily and circadian cycling of head per mRNA (Hardin et al., 1990). Comparing the two sets of data, it was clear that the mRNA and the protein rhythms were nearly opposite in phase, leading to the breakthrough insight that PER protein negatively regulates the transcription of its own gene (Hardin et al., 1990).
In subsequent years, in parallel with advances in understanding the molecular gears of the clock, the details of neural circuits responsible for circadian rhythms emerged from neuroanatomical studies. Distinct clusters of per-expressing neurons were counted and described in increasing detail. John Ewer (currently a Professor at the University of Valparaiso) and Brigitte Frisch (who sadly passed away soon after this work) in the Hall lab described ventral and dorsal clusters of PER-expressing lateral neurons (LNds, LNvs), as well as distinct groups of dorsal neurons (DNs; (Ewer et al., 1992; Frisch et al., 1994)). At the same time, Charlotte Helfrich-Forster (currently Chair of Neurobiology and Genetics at the University of Wurzburg) discovered that the ventral group of PER-expressing LNs (the LNvs) stained with an antibody to the neuropeptide PDF (Helfrich-Forster and Homberg, 1993, Helfrich-Forster, 1995). This important discovery provided the first glimpse of the clock neural circuitry when anti-PDF revealed the processes of these cells. It revealed two subsets of LNvs, large and small, with distinctive morphologies: four small LNvs project to superior protocerebrum and arborize near the DNs, while 4–6 large LNvs arborize across the surface of the ipsilateral optic lobe medulla and also cross to the contralateral medulla. But most neurons in the clock circuit (defined as those that express both per and tim) do not express PDF, so the detailed morphology of the entire system was only revealed by using these clock gene promoters to drive the expression of reporters like GFP and tau. With this approach, Maki Kaneko (currently a genetic counselor at Children’s Hospital Los Angeles) produced detailed maps of the projections of the entire population of per/tim-expressing neurons and their development from larval, pupal, and adult brains (Kaneko and Hall, 2000). This comprehensive study provided the first hypothesized wiring diagram of the fly brain circuits responsible for circadian rhythms. Helfrich-Forster (2003) synthesized these findings, describing the putative clock neural circuitry with clear diagrams and stunning confocal images. Subsequent meticulous neuroanatomical studies identified a network of 150 per/tim-expressing neurons comprised of 7 spatially distinct sets (Helfrich-Forster, 2003, Shafer et al., 2006, Helfrich-Forster, 2014).
The hypothesis that the LNs were the most important subset of these neurons in controlling rhythms of eclosion and locomotor activity was initially based on evidence from conventional behavioral genetic approaches. For example, Charlotte Helfrich studied loss-of-function mutations that disrupted the visual system and found that photoreceptors were not necessary components of the circadian system (Helfrich and Engelmann, 1983, Helfrich, 1986). John Ewer’s heroic analysis of both neuroanatomy and behavior of per+/per0 mosaics confirmed this hypothesis, and focused attention on the central brain (Ewer et al., 1992). The disconnected mutation yielded intriguing evidence for the necessity of LNs. This variably expressed mutation disrupts development of optic lobes and adjacent neural structures, including the lateral regions of brain cortex where LN cell bodies are nestled. Most disco mutants have no detectable LNs and are behaviorally arrhythmic, in spite of apparently normal PER expression in DNs and glial cells (Dushay et al., 1989, Zerr et al., 1990, Helfrich-Forster, 1998). A per transgene lacking 5′ regulatory sequences provided the first compelling evidence for the sufficiency of LNs for behavioral rhythms. Brigitte Frisch in the Hall lab discovered that this transgene was expressed only in the LNs (both dorsal and ventral subsets) but not in DNs, photoreceptors or glial cells, and that this LN-only expression of per was sufficient to rescue circadian activity rhythms (Frisch et al., 1994). This study was a landmark demonstration of the power of genetic rescues to test the sufficiency of gene actions in spatially restricted brain areas. This strategy is a common standard of evidence in modern neuroscience, but the approach was unprecedented in 1994.
3. Paul Hardin
I entered the Rosbash lab in September of 1987. I received a Ph.D. in genetics from Indiana University, but my training was in molecular biology and gene regulation using sea urchins – a decidedly non-genetic model system. I wanted to use my formal training in genetics and practical training in molecular biology to study a novel problem using a tractable genetic system. My Ph.D. advisor Bill Klein knew Michael Rosbash (MR) from their days at CalTech. As I was starting to look for post-doc opportunities, Bill gave me some papers from the Rosbash lab about their work on circadian rhythms. I was fascinated by the concept that genes played such a prominent role in controlling behavior, and that the recent isolation of the per gene may provide an in road into the molecular mechanisms underlying daily rhythms in activity. I contacted MR, and we talked about my experience and interests, which were a good fit with his lab. Brandeis had an NIH post-doc training grant that MR was part of, and he offered me a slot on that training grant for the first year at ~ $16 K/year.
I arrived in the lab just after per coding sequences were defined and lesions that caused the original per mutants had been characterized (Jackson et al., 1986, Reddy et al., 1986, Baylies et al., 1987, Yu et al., 1987). Although PER contained a series of threonine-glycine (TG) repeats characteristic of proteoglycans (Jackson et al., 1986, Reddy et al., 1986), this analysis didn’t provide many clues as to how per contributed to circadian timekeeping. The only model for per function at the time stemmed from work in Mike Young and David Spray’s labs, which suggested that PER functions at the plasma membrane to regulate intercellular communication (Bargiello et al., 1987). However, experiments published in 1988 were not in line with this model for PER function. First, PER was a founding member of a group of proteins containing the PAS domain (Crews et al., 1988), of which all other members at the time were bHLH transcription factors. Even though PER lacked a bHLH motif, this domain linked PER to transcriptional regulation. Second, anti-PER antibodies raised by my co-commentator Kathy Siwicki and a PER-βGAL fusion protein generated by Xin Liu (currently an Associate Professor at UCLA) revealed that PER was expressed in a variety of tissues, was concentrated in head tissues such as brain and eyes, and localized subcellularly to nuclei in the visual system and some peripheral tissues (Liu et al., 1988, Siwicki et al., 1988). Third, Kathy Siwicki showed that anti-PER stained photoreceptors in the eyes strongly at night but weakly if at all during the day (Siwicki et al., 1988). Whether this rhythm in PER protein staining resulted from altered expression levels or some post-translational alteration was unknown, but the rhythm persisted in constant dark (DD), so it was clearly under clock control!
Since per copy number altered circadian period, my initial project in the Rosbash lab was to conduct an autosomal deficiency screen for other genes that altered behavioral rhythms. The screen was moving forward slowly, and I wanted a project where I could use my molecular biology and gene regulation experience. After discussions with MR I got the green light to test whether per mRNA cycling could account for the rhythms in PER staining. Previous tests of per mRNA levels over the day had been done on RNA extracted from whole animals; however no obvious changes were detected (Reddy et al., 1984, Young et al., 1985). In the fall of 1988 I tested whether per mRNA levels changed over the course of a day, but used fly heads rather than whole animals because the PER immunostaining and PER-βGAL reporter protein studies revealed that the gene was highly expressed in the head (Liu et al., 1988, Siwicki et al., 1988). It took some time to raise enough flies to obtain sufficient head RNA for northern blots, but the results clearly showed that per mRNA levels were low late at night and during the morning, and high during late day and early evening. This suggested that cycling in per mRNA levels may account for the changes in PER protein levels. I replicated this result and discussed the next set of experiments with MR, who was on sabbatical in France, as well as Jeff Hall and other post-docs and graduate students in the Rosbash and Hall labs. From these discussions, and a visit from MR over the holidays, it was apparent that to investigate per mRNA cycling in great detail (i.e. with multiple per mutants and more extensive timecourses), I would need to raise an enormous number of flies to get sufficient head mRNA for northern blots. This was not conducive to getting results quickly and efficiently; thus we abandoned northerns for RNAse protection assays (RPAs). RPAs were used to sensitively detect RNA species on the yeast mRNA splicing side of the Rosbash lab, and Lori Lorenz (currently an Assistant Professor at University of Massachusetts Medical School), who had used this technique in flies to measure a 0.9 kb transcript adjacent to per, taught me the method.
After developing an effective probe to measure per mRNA, I spent most of the spring and summer of 1989 collecting flies every one or two hours during a light:dark cycle or every four hours over multiple days in DD, rearing thousands of flies in a single incubator in a dark basement room called “The Pit”. This was a particularly anxious time for me as my wife Susan was pregnant with our first child and she was not too excited for me to be in lab for extended periods, particularly as the due date came near. Our son Ryan was almost two weeks late, but it was a great relief to have a healthy F1. As results came in showing that perL mRNA had a delayed phase compared to per+, perS mRNA had an advanced phase compared to per+, and per0 mRNA was arrhythmic, it was clear that these mutant PER protein alleles were influencing the levels of per mRNA. I thought, as did MR, Jeff and others in the two labs, that this feedback could be an important feature of the timekeeping mechanism, but there was more work to do to test this hypothesis.
In particular, I conducted experiments in DD showing the period of mRNA cycling in perS was ~ 20 h and that in per+ was 24 h. This was a challenge as income from two post-doc salaries couldn’t pay the high cost of daycare in Boston for 5 days a week; thus Susan and I took care of Ryan at home a couple of days a week, and took turns taking care of him at lab on Saturday and Sunday. Although some objected to having a baby in the lab during the weekend (you know who you are), the need to take weekdays off to care for Ryan subsided once our stipends increased and my support shifted to HHMI when MR became an HHMI investigator in 1989. During this time I tested whether feedback by PER was working in trans by determining if a per transgene that rescued behavior in per0 flies also rescued cycling of (arrhythmic) per0 mRNA. Indeed, per0 mRNA cycling was rescued by the transgene, demonstrating feedback by PER protein on per0 mRNA. Although it was clear PER fed back to control its own mRNA cycling, the level of control was unknown; thus the model for feedback was necessarily imprecise as PER could function directly, through intermediates or even through behavior to control mRNA cycling at the transcriptional or post-transcription levels. The paper was then written and submitted to Nature in November of 1989 and accepted just before Christmas (Hardin et al., 1990), thereby providing an incredible present. While I was working on per mRNA cycling, Kathy Siwicki and Danielle Zerr in Jeff Hall’s lab were analyzing PER cycling in fly heads. Their results clearly showed that PER staining cycled with a phase and period consistent with the per allele in LD and DD, and also showed that PER levels cycled with a peak almost antiphase to the mRNA (Zerr et al., 1990). This delay suggested that PER may function to reduce per mRNA levels.
During my time in his lab MR consistently pushed me to get results, which was fine as I was used to having a “hands-on” mentor in graduate school, yet I didn’t feel singled out as MR pushed everyone to get results! Although MR had a reputation as a tough mentor, and he did indeed provide harsh criticism at times, I don’t remember a single instance during my time in his lab where I was at the receiving end. I think a certain trust developed when MR was in Paris on sabbatical that I could make good progress while he was away. Whether he was away or at Brandeis, I appreciated MRs comments and suggestions because they were always on point and valuable. My experience in MRs lab greatly influenced how I organized lab operations and mentored trainees when I started my own lab and has served me well over the years.
Having demonstrated that PER feeds back to control per mRNA levels, I wanted to determine the level of regulation. At the same time, I decided (with a big push from Susan) that it was time to apply for faculty positions. Although I grew up in the Chicago area, my parents were both from Texas, where I spent many fun-filled summers visiting relatives, and I definitely wanted to find a position in a fair-weather climate. There were few positions available, but as luck would have it I applied for and was offered a tenure track Assistant Professor position at Texas A&M University. I happily accepted, but there was still plenty of work to finish up. RNAse protection was sensitive enough to detect per pre-mRNA, and the pre-mRNA cycled in phase with the mature per mRNA, suggesting that transcription was rhythmic. I generated a series of transgenes that fused different per promoter fragments to a reporter gene and found that a per upstream fragment that contained no coding sequences could drive mRNA cycling similar to endogenous per+ mRNA. This work showed that per participated in a transcriptional feedback loop and predicted that this feedback loop may control many rhythmic processes via rhythmic regulation of mRNAs. This work was submitted as I left with my son and pregnant wife in tow for a faculty position at Texas A&M University in 1991, and after some difficulty due to reviewers assuming regulation must be transcriptional, this work was published in PNAS in 1992 with Colin Pittendrigh as the communicating editor (Hardin et al., 1992). The transcriptional feedback loop (TFL) came to the forefront of models for circadian timekeeping. Importantly, in 1994 the Dunlap and Loros Labs at Dartmouth identified a similar TFL in Neurospora crassa that constituted the timekeeping mechanism (Aronson et al., 1994). The sole component of this feedback loop, called frequency (frq), was not orthologous to per, demonstrating that TFLs could be driven by different components, but represented a conserved mechanism for circadian timekeeping.
In the early 1990s per was the sole component of the TFL, but this was about to change. Continued genetic screening in the Young lab for autosomal clock mutants that altered the circadian period of behavior identified another clock gene, timeless (tim), in 1994. As a post-doc in the Young lab, Amita Sehgal (currently a Professor and HHMI investigator at the University of Pennsylvania School of Medicine) showed that the tim mutant was behaviorally arrhythmic, and also rendered per mRNA arrhythmic, indicative of a critical FBL component (Sehgal et al., 1994). The Young lab, in parallel with Chuck Weitz (currently a Professor at Harvard Medical School) and colleagues, isolated tim via different means (Gekakis et al., 1995, Myers et al., 1995), and Amita showed that tim mRNA also cycled in phase with per mRNA (Sehgal et al., 1995). How tim contributed to timekeeping was revealed in several papers showing that TIM stabilizes PER protein (Price et al., 1995), tim contributes to PER nuclear localization (Vosshall et al., 1994), and TIM is degraded in response to light (Hunter-Ensor et al., 1996, Myers et al., 1996, Zeng et al., 1996). This light-dependent TIM degradation was very exciting since it readily explained the phase advances and delays necessary for photic entrainment; TIM degradation early at night would lead to a delay in TIM accumulation because tim mRNA levels are high, whereas TIM degradation late at night would advance the cycle because more TIM could not be synthesized due to low tim mRNA levels. In 1994 Isaac Edery (currently a Professor at Rutgers University) had shown that PER was rhythmically phosphorylated, where phosphorylation was highest just before PER levels decreased (Edery et al., 1994). As part of the genetic screen for clock mutants in the Young lab, my co-commentator Jeff Price identified period-altering alleles of double-time (dbt) kinase (ortholog of Casein Kinase Iε) that alter the cycling of PER protein, and along with Brian Kloss (currently a Senior Scientist at the New York Structural Biology Center) and Justin Blau (currently a professor at New York University) found that DBT interacts with and phosphorylates PER (Kloss et al., 1998, Price et al., 1998). The discovery of tim and dbt revealed key determinants of period length through their impact on PER accumulation, stability and localization, thereby adding to the mechanistic detail of the TFL and its role in circadian timekeeping.
The work I did in the Rosbash lab provided a great foundation for building my own lab at Texas A&M. Initially I focused on determining how per was regulated and understanding the extent to which this regulation occurred in the fly. When dissecting the per promoter, we discovered that a CACGTG “E-box” sequence was necessary for rhythmically activating per transcription (Hao et al., 1997). While we were defining per regulatory sequences, the first mouse clock mutant, called Clock, was identified by Joe Takahashi (currently Chair of Neuroscience and an HHMI Investigator at University of Texas Southwestern Medical School) and colleagues (Vitaterna et al., 1994). The isolation and sequencing of mouse Clock revealed that it encoded a member of the bHLH-PAS family of transcription factors, which activate transcription by binding E-box sequences (King et al., 1997). They proposed a model in which a CLOCK ortholog activated per, and rising levels of PER repressed CLOCK activation perhaps via PAS domain interactions (King et al., 1997). Later in 1997 the first of three mouse per orthologs were isolated (Sun et al., 1997, Tei et al., 1997), leading to great excitement that key clock components were conserved, and may contribute to circadian timekeeping via a TFL similar to that in Drosophila. The next year Drosophila Clock (Clk) was identified in molecular screens for fly orthologs of Clock and genetic screens for arrhythmic fly mutants (Allada et al., 1998, Bae et al., 1998, Darlington et al., 1998), which also identified CYCLE (CYC) as the heterodimeric partner of CLK (Darlington et al., 1998, Rutila et al., 1998). Experiments from a collaborative effort between groups led by Steve Kay (currently a Professor at University of Southern California), Joe Takahashi, and Chuck Weitz showed that CLK-CYC heterodimers activated per-driven luciferase reporter gene expression via E-boxes (Darlington et al., 1998), thus closing the feedback loop in Drosophila. Likewise, collaborative work between groups headed by Chuck Weitz, Joe Takahashi and Fred Davis (currently a Professor at Northeastern University) provided strong evidence that the TFL was conserved in mice: CLOCK and its partner BMAL1 (a CYC ortholog) bound E-boxes to activate mouse Per transcription (Gekakis et al., 1998). In parallel, work by John Hogenesch (currently a Professor at University of Cincinnati College of Medicine), a student in the lab of Chris Bradfield (currently a Professor at the University of Wisconsin), showed that CLOCK-BMAL1 (referred to by them as CLOCK-MOP3) activated E-box transcription (Hogenesch et al., 1998), thus reinforcing the evidence for a TFL in mice.
Back in my lab I wanted to revisit an old problem: why is per mRNA cycling amplitude so low in whole flies? In my only single author research paper, I found that per mRNA cycled with high amplitude in the thorax and abdomen, which contained many per-expressing tissues (Liu et al., 1988, Siwicki et al., 1988), but constant high per mRNA levels in female ovaries masked per mRNA rhythms in these other tissues (Hardin, 1994). My demonstration of circadian clocks in Drosophila peripheral tissues was followed in spectacular fashion by work in the Kay and Hall labs showing the existence of light entrainable clocks in isolated fly peripheral tissues using a per-luciferase reporter gene (Plautz et al., 1997). The work in my lab remained focused (and still is!) on understanding how the timekeeping mechanism maintains a ~ 24 h rhythm, determining how the TFL controls rhythmic transcription, and deciphering how overt rhythms are regulated, all owing to the challenge and excitement stemming from my days as a post-doc in the Rosbash lab.
4. Jeff Price
My interest in circadian rhythms was generated while I was studying fish antifreeze protein gene expression during my Ph.D. studies in the Johns Hopkins University Biology Department. My graduate advisor Dr. Ru Chih Huang had undertaken an analysis of antifreeze protein genes in winter flounder, which express these proteins only during the winter to keep their blood from freezing. My work showed that low temperatures were needed to produce high levels of antifreeze protein mRNA in the liver, but additional work by others had shown that photoperiodic control, which typically operates through the circadian clock, was also a signal for induction of the mRNA. My interest in circadian rhythms was stimulated. I suspected that fundamental insights into the mechanism were likely to come from genetic analyses in Drosophila (Konopka and Benzer, 1971) or Neurospora (Feldman and Hoyle, 1973), since these analyses had identified single gene mutations that affected circadian period and rhythmicity.
At the time I began looking for postdoctoral positions, I was working with my Ph.D. advisor at the Institute of Molecular Biology in Taiwan, where she had taken a year’s sabbatical to direct it. I came back to the US briefly in the summer of 1988 with very limited time to interview for postdoctoral positions. Michael Rosbash was on sabbatical at the time and could not meet with me, but Michael Young agreed to meet with me. I was immediately taken with his approach to the problem. The per gene had been cloned, but the sequence of the protein had suggested nothing about its function in the circadian clock (it was a pioneer protein at the time). Genetic analysis is strongest when it examines the interactions of multiple genes, but the original genetic analysis by Konopka and Benzer had only focused on the X chromosome and had only revealed the per mutations. The 2nd and 3rd autosomal chromosomes had not been screened to isolate period-altering mutations, and the Young lab had initiated a screen for mutations produced by P element-mediated mutagenesis of these chromosomes. P elements are transposable elements that can be mobilized and then stabilized in novel locations with exquisite genetic precision. The Young lab was generating numerous P element insertions of its own and was also screening P element insertions generated by the enhancer trap approach (Bellen et al., 1989).
Therefore, when I became part of the Young lab in January of 1989, I became part of the genetic screen initiated by postdoctoral fellow Amita Sehgal and research assistant Bernice Man (currently a Professor of Clinical Medicine in the University of Illinois College of Medicine at Chicago). It is sometimes stated that most circadian clock mutants are isolated early in any screen (Konopka’s rule), and that was certainly true for me. The original timeless (tim) mutation was line fjy42 – my 42nd second chromosomal P element insertion line. The “fjy” was an abbreviation for “Fan Jin-Yuan,” my long-distance girlfriend whom I had met in Taiwan (She would later move to the US to undertake Ph.D. studies at Georgetown University, and we got married). This line scored strongly in the screen we were using at the time – the same one used by Ron Konopka to isolate the per mutants. While individual wild type flies eclose (emerge from pupa) only once in their lifetime, the eclosion is gated by the circadian clock, so that populations of flies of different ages produce daily bursts of eclosion activity around dawn. If one collects newly enclosed flies twice a day – once several hours before lights on and once several hours before lights off - most flies from wild type populations will eclose during the second collection (before lights off). Line fjy42 clearly produced equal numbers of flies in both collections. Amita and I performed detailed multiple time point analysis of the line’s eclosion activity showed that it was arrhythmic. Normally, wild type flies are active during subjective day (the time when lights were previously on) and relatively quiet during subjective night (the time when lights were previously off) in constant darkness, but the tim mutant showed no organized bouts of locomotor activity in DD. Clearly it was arrhythmic.
I undertook a recombination analysis of the tim line, and unfortunately this showed that the gene causing arrhythmic locomotor activity and eclosion was located elsewhere on the 2nd chromosome from the P element insertion site. So much for the immediate cloning opportunity that a P element insertion would afford! The recombination analysis indicated that the mutation causing the arrhythmic phenotypes segregated to the left arm of the 2nd chromosome.
In the mean time, the break-through papers from the Rosbash and Hall labs (produced by Paul Hardin and Kathy Siwicki, my co-commentators from the Rosbash and Hall labs, respectively) showed that the per protein (Siwicki et al., 1988, Zerr et al., 1990) and transcript (Hardin et al., 1990, Hardin et al., 1992) oscillated in the head with an ~ 24 h rhythm, and these rhythms were altered by the per mutations to periods that corresponded to the behavioral rhythms. Kathy’s immunostaining results showed that PER protein is concentrated in the nucleus. Could it be regulating its own transcription in a feedback loop? Paul Hardin subsequently showed that the per upstream region (the promoter) conferred per-dependent oscillations to a reporter gene. Clearly transcriptional control was involved in the oscillations, and this control might impact many other genes as well to generate the outputs of the clock (subsequent genome-scale approaches would confirm broad control of genes by the circadian clock). The transcriptional control was particularly unusual because nuclear PER protein only began to accumulate after the per mRNA had reached its peak, and then the mRNA began to decline, with decline of the protein coming much later. Clearly there were post-transcriptional delays in the feedback loop.
These were the first important insights on the circadian clock mechanism, and work from the Dunlap and Loros labs on the Neurospora frequency (frq) gene suggested that Neurospora used a similar mechanism (Aronson et al., 1994). While the Young lab was a bit chagrined that the insight had come from elsewhere, we joined forces as a lab to see if the tim gene might participate in this feedback loop. Amita Sehgal showed by RNAse protection assays (as Paul describes, a Herculean undertaking) that the tim mutant affected the per RNA oscillations like the original perO mutant; that is, per RNA expression was arrhythmic in the tim mutant. Graduate student Leslie Vosshall (currently Professor and HHMI Investigator at Rockefeller University) and I collaborated to address the role of the tim mutation on the post-transcriptional regulation of PER. Our studies were initially focused on a PER-βGAL fusion protein (ie, a fusion of the first half of PER to the βGAL protein of E. coli) expressed by a line given to us by the Rosbash lab. In the most exciting result from this analysis, Leslie found that the tim mutation produced cytosolic localization for PER-βGAL, rather than the nuclear localization found in wild type flies. The tim mutation did not affect nuclear localization of other reporters without this fragment of PER. Moreover, her assessment of endogenous PER with an antibody from our lab showed that nuclear PER was undetectable in the tim mutant. I attempted to assay the effects of the tim mutation on the daily oscillations of PER protein by assaying its effects on βGAL enzyme activity and by immunoblot with anti-βGAL antibodies. All of these assays showed that PER-βGAL did not oscillate even in wild type flies, and of course did not oscillate in the tim genotype. Evidently the βGAL fusion stabilized the fusion protein so that it was more stable than native PER and no longer responded to circadian oscillatory signals.
My problem with assessing the oscillations of PER alone was that our antibody did not detect PER on an immunoblot. The Rosbash lab had produced a PER antibody that did detect PER oscillations by immunoblot. Isaac Edery had used the antibody to show that PER protein did oscillate in level and mobility (Edery et al., 1994). The increase in total PER levels preceded the increase in nuclear PER, and the oscillations in mobility were due to changes in the phosphorylation status of PER. Michael Rosbash became interested in TIM and offered to let me assess the effects of the tim mutation on PER with their antibody. They had very limited amounts of this antibody, so I went to the Rosbash lab at Brandeis to perform the immunoblots. The results clearly showed that the tim mutation eliminated the rhythms in PER accumulation and phosphorylation. In fact, PER was constitutively low and resembled its state in constant light (a result which foreshadowed subsequent demonstrations that light eliminates TIM).
Taken together, these results suggested that TIM regulated both the nuclear localization and levels of PER. The stability of the PER-βGAL fusion protein, while it hampered my attempts to assess the daily oscillations of PER, produced a protein of equal abundance at all times and in both wild type and tim genotypes, allowing one to conclude that the difference in nuclear detection for PER-βGAL in the tim mutant and wild type was in fact a difference in localization rather than a difference in level. For endogenous PER the effects were mediated both by effects of TIM on PER levels and nuclear localization. The effects of TIM on PER nuclear localization and per mRNA oscillations were published in two papers (Sehgal et al., 1994, Vosshall et al., 1994), while the effect of TIM on PER levels was published by a joint publication from the Young and Rosbash labs (Price et al., 1995). The analysis of rhythms in flies now involved the interaction of two gene products. Subsequent work has supported a TIM-dependent gating of nuclear localization for PER, as well as the mechanism for TIM’s effects on PER stability (more on this below).
At this point, I decided to recuse myself from further analysis of TIM. The Drosophila genome had not yet been sequenced, and it seemed that it would take quite a bit of genome-walking to clone the tim gene. Amita Sehgal had accepted a job at the University of Pennsylvania and planned to focus on tim (as described by Paul Hardin, her subsequent work on entrainment of the clock by TIM degradation was seminal), and with the continued interest in the tim gene by the Young and Rosbash labs I felt the field had become rather crowded. Moreover, my salary was now covered by the NSF Center for Biological Timing operating through the University of Virginia, and I was tasked with screening the second chromosome for other circadian clock mutations. The Rosbash and Hall labs were also part of the center, and they were tasked with screening the third chromosome. Why not find additional circadian genes that would be as exciting as per and tim?
In the new genetic screens I was performing in the Young lab, we used a chemical mutagen (ethyl methanesulfonate; a.k.a. EMS) instead of P elements (They had not offered any benefit the first time for cloning, and my early luck in hitting a clock gene with them did not persist.), and we had expanded our screen to complete analysis of activity rhythms for all lines in constant darkness. The rationale was that locomotor activity analysis would allow detection of more subtle phenotypes than would the two-collection eclosion assay. The new approach did detect multiple new tim alleles, including period-altering alleles that were characterized by graduate student Adrian Rothenfluh (Currently Associate Professor, University of Utah School of Medicine). But the mutant that attracted my interest was one that showed variable phenotypes in different flies of the line, with periods ranging from 24 h, to 21 h, to 18 h. It turned out to be a single-gene short-period mutation on the third chromosome, which had not been isogenized in our crosses to generate homozygous second chromosome mutations and therefore existed in the wild type (24 h), heterozygous (21 h) and homozygous (18 h) state in the line. Adrian Rothenfluh subsequently identified a long–period mutant allele for the gene, and I mapped the short-period allele by standard recombination procedures and identified a P element insertion in the gene before leaving the lab to establish my own lab at West Virginia University.
During my initial time at West Virginia University, I was less able to devote extensive time to studies of the new gene (which we eventually called double-time, or dbt), with lab setup and new teaching duties in front of me. I was able to generate excisions of the P element and show that in some cases these reverted the mutant phenotype. In the Young lab, Brian Kloss determined that the P element was in fact inserted in an intron of a casein kinase I δ/ε ortholog (one of the last attempts to clone a gene prior to completion of the Drosophila genome sequence), while postdoctoral fellow Justin Blau determined that PER was constitutively high, nuclear and hypophosphorylated in the P-element-containing homozygous larval brains (the dbtP larvae did not survive to adulthood). In fact, the expression of PER resembled the expression of PER-βGAL fusion protein that Leslie and I had analyzed years before, and Justin’s analysis was critical for our models of DBT’s effects on PER oscillations. Clearly, DBT was needed to reduce PER levels in the absence of TIM, and as it was a protein kinase it most likely did so by phosphorylating PER. Lino Saez (who contributed for many years to the Young lab’s efforts but unfortunately passed away before the Nobel was awarded) demonstrated that DBT directly interacted with PER, and Adrian Rothenfluh showed that the long and short period alleles produced similar effects on the per and tim RNA oscillations as they did on behavior.
We proposed a model for the phase delays in transcription and nuclear accumulation/disappearance of PER. PER was phosphorylated during the day by DBT to keep it from accumulating as its mRNA accumulated. During the night, TIM accumulation stabilized PER against DBT mediated degradation (now known to be triggered by the SLIMB E3 ubiquitin ligase (Grima et al., 2002; Ko et al., 2002)). PER then moves into the nucleus and represses its own transcription (as well as that of tim) until light mediated degradation of TIM and a consequent slow phosphorylation program that triggers the phosphorylation of PER and its eventual degradation, leading to renewal of transcription. So in the end I think it was a wise career move for me to move on from tim. The work was reported in two papers (Kloss et al., 1998, Price et al., 1998).
Mike Young had an unusual but highly effective mentoring style. He almost never pushed us for results and left the day-to-day management of the projects in the hands of his students and postdoctoral fellows. Nevertheless, his students and postdoctoral fellows were competitive and worked long hours. I think he fostered this drive in his employees and felt that it was better for us to be self-motivated than pushed from above. He did become very involved in the interpretation of interesting results. Our results with the tim and dbt mutants were surprising but clear-cut, and I do not remember much disagreement about their interpretation. All of us (e.g., Amita Sehgal, Leslie Vossall, Brian Kloss, Justin Blau, Adrian Rothenfluh, Lino Saez, and Mike Young) were involved in their interpretation, and our opinions were mostly confluent. Mike Young often came up with the most compelling pitch for the journal article. In the end, I am happy that I was allowed to plot my own course in his lab, as it was good preparation for an independent faculty position.
For me, the excitement of this initial work is still present in the scope of my current work, which has followed up on our initial dbt discoveries. My lab (now at the University of Missouri – Kansas City), as well as Mike Young’s lab, showed that the dbtS and dbtL mutations both lower the kinase activity (Preuss et al., 2004, Kivimae et al., 2008), and my lab has therefore explored other ways in which these period-altering mutations may have opposite effects on circadian period. One way is by preferentially altering phosphorylation at different sites within PER – some of which lengthen period and some of which shorten period. Analysis of PER phosphorylation by other labs in both mammalian and Drosophila PER has produced evidence for such a mechanism. But another possibility is that the period-shortening dbt mutations, which alter residues on the surface of the protein, may affect interactions of DBT with other circadian regulators. These regulators may offer temporal control of DBT activity or access to PER, with some interactions lengthening period and others shortening period. Analyses in my own lab have discovered two such DBT interactors – one which we have called Bride of DBT (BDBT) (Fan et al., 2013) and the other already named Spaghetti (SPAG) (Means et al., 2015). Loss of both of these interactors lengthens rather than shortens period, but their discovery has taken us into new aspects of clock regulation - including links to apoptosis and Alzheimer’s Disease. My hope it that it will be possible to reconstitute a circadian post-translational oscillator in a test tube or at least a cell that is persistent – much like what has been done for the evolutionarily divergent cyanobacterial clock (Nakajima et al., 2005). We have yet to understand the animal clock until we can produce a persistent oscillation from scratch.
5. Conclusion and perspective
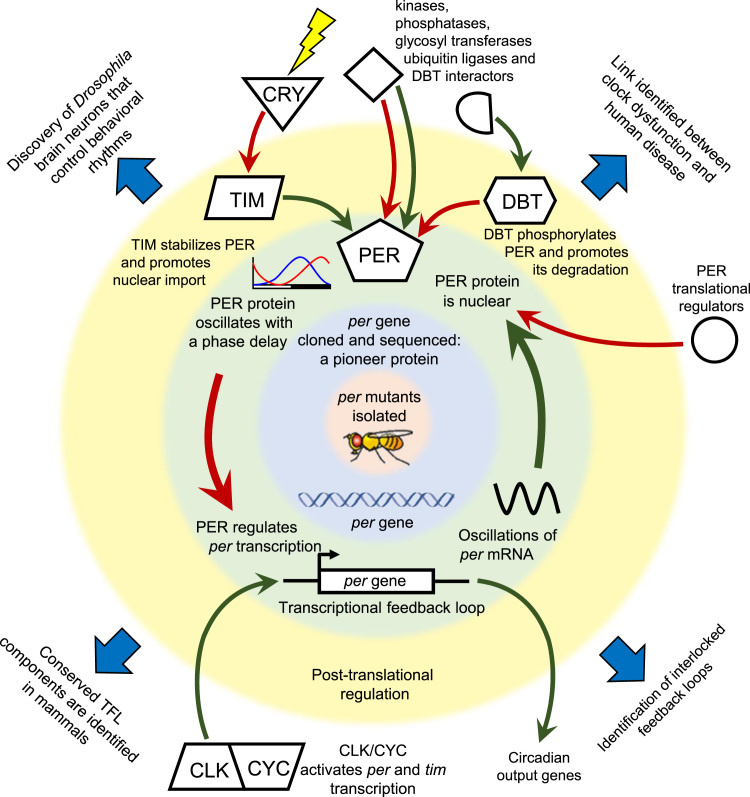
Work initiated in the Hall, Rosbash and Young labs during the 1980s and early 1990s laid the foundation for the “clockwork explosion” in the late 1990s (Reppert, 1998), which crystallized the TFL as the circadian timekeeping mechanism in flies and demonstrated that an analogous TFL largely consisting of the same components also kept circadian time in mammals. The Drosophila TFL provided a molecular basis for explaining core properties of animal clocks such as entrainment to environmental cues and regulation of overt rhythms. Importantly, Drosophila clock genes provided valuable reagents used to mark clock cells and to map neuronal circuits that control behavioral rhythms. Indeed, the impacts of research led by Hall, Rosbash and Young reach well beyond circadian biology, as the strategies used to decipher the mechanistic links between Drosophila clock genes and behavior have served as templates for modern neuroscientists who use genetic approaches to investigate the cellular and neural systems that mediate complex behavior. Here we highlight how discovery of the TFL enabled important advances in the field of biological rhythms, became a prototype for neurogenetic analysis of behavior, and provides a compelling illustration of the value of basic research on model organisms (Fig. 2).
Fig. 2.
Major conceptual advances in the analysis of theDrosophilaCircadian Clock. Sequential waves of progress are shown in concentric circles, with earlier advances in more central rings. Isolation of the original per mutants by Konopka and Benzer (1971) was followed by the cloning of the per gene in the Hall, Rosbash and Young labs. The Hall and Rosbash labs discovered the oscillations of per mRNA and protein and demonstrated that PER protein accumulates with a phase delay and regulates its own expression in a Transcriptional Feedback Loop (TFL). The Young lab discovered the post-translational PER regulators TIM and DBT, which positively (TIM) and negatively (DBT) regulate PER’s nuclear import and stability. Additional components of post-translational regulation are represented above the outer ring, while the activation of transcription by CLK-CYC is shown below. Beyond the circled regions, large arrows indicate related areas of significant progress, including mammalian clock mechanisms, interlocked feedback loops, fly clock neurons that control behavior, and links between clock dysfunction and human disease, as described in the text. Red arrows indicate inhibition and green arrows stimulation.
The TFL in Drosophila provided a fertile crescent of phenomena that were further exploited to reveal key features of clock function. For instance, knowing that TIM was degraded in response to light provided a molecular marker for clock entrainment. This observation, combined with screens for genes that disrupted molecular rhythms via a per-luciferase reporter and identification of plant cryptochrome photoreceptors, resulted in identification of CRYPTOCHROME (CRY) as a circadian photoreceptor (Emery et al., 1998, Stanewsky et al., 1998, Egan et al., 1999). Subsequent analysis revealed that light altered CRY structure so that it could bind TIM (Ceriani et al., 1999, Rosato et al., 2001, Busza et al., 2004, Dissel et al., 2004), thereby promoting degradation of TIM and CRY by JETLAG (JET) (Koh et al., 2006, Peschel et al., 2006, Peschel et al., 2009) to shift circadian phase (Emery et al., 2000). Although CRY is a key player in clock entrainment to light, traditional opsin based photoreceptors also contribute to light entrainment (Kistenpfennig et al., 2017, Ni et al., 2017), and CRY contributes to other aspects of physiology independent of light entrainment within and outside the circadian system (Agrawal et al., 2017, Fogle et al., 2015, Fogle et al., 2011, Krishnan et al., 2001, Levine et al., 2002).
Discoveries of post-translational modifications that regulate progression through the TFL were initiated with the discovery of dbt (Kloss et al., 1998, Price et al., 1998). Subsequent work in flies identified other kinases, phosphatases and even glycosylases that control the stability, subcellular localization and activity of core clock transcription factors (Tataroglu and Emery, 2015). Many of these regulatory enzymes and their clock protein targets are conserved in mammals, where the list of protein modifications has expanded to include acetylation, sumoylation and ubiquitylation (Mendoza-Viveros et al., 2017). Post-translational modifications of clock proteins profoundly influence feedback loop progression, and thus circadian period. Consequently, understanding the impact of progressive post-translational modifications on clock protein function remains an important goal of current research.
As described above, both PER and a PER-βGAL fusion protein were expressed in peripheral tissues (Liu et al., 1988, Siwicki et al., 1988), and rhythmic expression of per mRNA, PER protein, and per-luciferase reporter genes confirmed that these tissues harbored circadian clocks (Hardin, 1994, Emery et al., 1997, Plautz et al., 1997). These studies foreshadowed the discovery that many peripheral tissues and cell lines from mammals contain circadian clocks and regulate diverse physiological processes (Balsalobre et al., 1998, Balsalobre et al., 2000). The ability to readily identify, and ultimately to manipulate, the brain cells that express clock genes greatly advanced our understanding of the neural systems that control behavioral rhythms (Griffith, 2012). The GAL4/UAS system for targeting gene expression in flies (Brand and Perrimon, 1993) allowed for functional manipulation of subsets of clock neurons, revealing that distinct groups of cells in the clock neural system independently control the characteristic morning and evening activity bouts seen during light:dark cycles (Grima et al., 2004, Stoleru et al., 2004). These morning and evening cells function along with other defined subsets of per/tim-expressing neurons to endow this system with the ability to adapt rhythmic activity to different photoperiods and respond to different environmental cues (Yoshii et al., 2012). FBL components have similarly been used to mark and manipulate clock cells in the mammalian suprachiasmatic nucleus (SCN), revealing subregions responsible for receiving light input and driving downstream targets and defining signaling processes responsible for light entrainment, communication between clock neurons that regulate activity rhythms, and entrainment of peripheral tissues (Herzog et al., 2017).
The TFL model suggested that the clock regulates overt rhythms via rhythmic transcription of output genes. Indeed, the initial microarray studies identified hundreds of cycling “output gene” transcripts in fly heads over the course of a day (Claridge-Chang et al., 2001, McDonald and Rosbash, 2001, Lin et al., 2002, Ueda et al., 2002). Although the TFL directly controls many transcripts like per and tim that peak near dusk, other transcripts that peak near dawn are indirectly controlled by the TFL via an interlocked feedback loop driven by two transcription factors activated by CLK-CYC: vrille (vri) and Par Domain Protein 1 (Pdp1) (Blau and Young, 1999, Glossop et al., 1999, Cyran et al., 2003, Glossop et al., 2003). Interlocked feedback loops represent another conserved feature of the clock: two interlocked loops operate in mammals to control rhythmic transcription peaking at different phases of the circadian cycle (Mitsui et al., 2001, Preitner et al., 2002, Ueda et al., 2005). These feedback loops rhythmically transcribe a substantial proportion of mouse genes (5–20%) in a given tissue, with perhaps > 50% of genes transcribed under clock control (Takahashi, 2017). Thus, it is not surprising that the circadian clock plays such a pervasive role in regulating human physiology, metabolism and behavior. When the clock is chronically disrupted via mutations in clock genes, damage to light entrainment pathways or shift work, severe medical conditions can occur including increased risk for cancer, diabetes, sleep problems and mood disorders (Toh et al., 2001, Xu et al., 2005, Skene and Arendt, 2007, James et al., 2017). It is our hope that a broad understanding of the circadian system (i.e. input pathways, timekeeping mechanism, output pathways) will result in treatments for these medical conditions, which is an important practical application of work that was initiated simply to understand a fundamental biological process.
The 2017 Nobel Prize for Physiology or Medicine is a tribute to basic research and model organisms. Our efforts to understand how circadian rhythms are controlled were not driven by practical applications such as curing a disease, increasing agricultural production, or adapting to long-term space exploration, but rather by basic curiosity about how biological systems work. Our progress was facilitated by using Drosophila as a model system, not just because the per mutants had been identified, but also because existing genomic DNA libraries made it possible to isolate large chromosomal regions, techniques for generating transgenic flies had been established, and genetic reagents were available that facilitated gene isolation, analysis and screens for additional mutants. Another advantage was the quantifiable nature of the behavior itself: locomotor activity data are easy to collect and many features of rhythmic behavior can readily be measured and quantified. The efforts of multiple labs with complementary expertise (Hall and Young in Drosophila genetics and behavior, Rosbash slanted more towards biochemistry) led to integrative thinking and multidisciplinary approaches. Strong competition between labs greatly contributed to the rate of progress, because every new “advance” was subject to immediate critical analysis. Finally, because Hall, Rosbash and Young all strongly supported the careers of their former postdocs and graduate students, the field grew and spawned novel areas of research, like the neural circuitry underlying rhythms and the genetic analysis of sleep. We were incredibly fortunate to have joined their labs and benefited from their scientific expertise and intuitions at a time when biological timekeeping mechanisms were waiting to be deciphered.
Conflict of interest
The authors have no conflicts of interest or financial relationships to disclose.
References
- Agrawal P., Houl J.H., Gunawardhana K.L., Liu T., Zhou J. Drosophila CRY entrains clocks in body tissues to light and maintains passive membrane properties in a non-clock body tissue independent of light. Curr. Biol. 2017;27:2431–2441. doi: 10.1016/j.cub.2017.06.064. [DOI] [PubMed] [Google Scholar]
- Allada R., White N.E., So W.V., Hall J.C., Rosbash M. A mutant Drosophila homolog of mammalian clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- Aronson B.D., Johnson K.A., Loros J.J., Dunlap J.C. Negative feedback defining a circadian clock: autoregulation of the clock gene frequency. Science. 1994;263:1578–1584. doi: 10.1126/science.8128244. [DOI] [PubMed] [Google Scholar]
- Bae K., Lee C., Sidote D., Chuang K.Y., Edery I. Circadian regulation of a Drosophila homolog of the mammalian clock gene: PER and TIM function as positive regulators. Mol. Cell Biol. 1998;18:6142–6151. doi: 10.1128/mcb.18.10.6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsalobre A., Brown S.A., Marcacci L., Tronche F., Kellendonk C. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- Balsalobre A., Damiola F., Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- Bargiello T.A., Jackson F.R., Young M.W. Restoration of circadian behavioural rhythms by gene transfer in Drosophila. Nature. 1984;312:752–754. doi: 10.1038/312752a0. [DOI] [PubMed] [Google Scholar]
- Bargiello T.A., Saez L., Baylies M.K., Gasic G., Young M.W. The Drosophila clock gene per affects intercellular junctional communication. Nature. 1987;328:686–691. doi: 10.1038/328686a0. [DOI] [PubMed] [Google Scholar]
- Baylies M.K., Bargiello T.A., Jackson F.R., Young M.W. Changes in abundance or structure of the per gene product can alter periodicity of the Drosophila clock. Nature. 1987;326:390–392. doi: 10.1038/326390a0. [DOI] [PubMed] [Google Scholar]
- Bellen H.J., O’Kane C.J., Wilson C., Grossniklaus U., Pearson R.K. P-element-mediated enhancer detection: a versatile method to study development in Drosophila. Genes Dev. 1989;3:1288–1300. doi: 10.1101/gad.3.9.1288. [DOI] [PubMed] [Google Scholar]
- Blau J., Young M.W. Cycling vrille expression is required for a functional Drosophila clock. Cell. 1999;99:661–671. doi: 10.1016/s0092-8674(00)81554-8. [DOI] [PubMed] [Google Scholar]
- Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Busza A., Emery-Le M., Rosbash M., Emery P. Roles of the two Drosophila Cryptochrome structural domains in circadian photoreception. Science. 2004;304:1503–1506. doi: 10.1126/science.1096973. [DOI] [PubMed] [Google Scholar]
- Ceriani M.F., Darlington T.K., Staknis D., Mas P., Petti A.A. Light-dependent sequestration of Timeless by Cryptochrome. Science. 1999;285:553–556. doi: 10.1126/science.285.5427.553. [DOI] [PubMed] [Google Scholar]
- Claridge-Chang A., Wijnen H., Naef F., Boothroyd C., Rajewsky N. Circadian regulation of gene expression systems in the Drosophila head. Neuron. 2001;32:657–671. doi: 10.1016/s0896-6273(01)00515-3. [DOI] [PubMed] [Google Scholar]
- Crews S.T., Thomas J.B., Goodman C.S. The Drosophila single-minded gene encodes a nuclear protein with sequence similarity to the per gene product. Cell. 1988;52:143–151. doi: 10.1016/0092-8674(88)90538-7. [DOI] [PubMed] [Google Scholar]
- Cyran S.A., Buchsbaum A.M., Reddy K.L., Lin M.C., Glossop N.R. Vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell. 2003;112:329–341. doi: 10.1016/s0092-8674(03)00074-6. [DOI] [PubMed] [Google Scholar]
- Darlington T.K., Wager-Smith K., Ceriani M.F., Staknis D., Gekakis N. Closing the circadian loop: clock-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- Dissel S., Codd V., Fedic R., Garner K.J., Costa R. A constitutively active cryptochrome in Drosophila melanogaster. Nat. Neurosci. 2004;7:834–840. doi: 10.1038/nn1285. [DOI] [PubMed] [Google Scholar]
- Dushay M.S., Rosbash M., Hall J.C. The disconnected visual system mutations in Drosophila melanogaster drastically disrupt circadian rhythms. J. Biol. Rhythm. 1989;4:1–27. doi: 10.1177/074873048900400101. [DOI] [PubMed] [Google Scholar]
- Edery I., Zwiebel L.J., Dembinska M.E., Rosbash M. Temporal phosphorylation of the Drosophila period protein. Proc. Natl. Acad. Sci. USA. 1994;91:2260–2264. doi: 10.1073/pnas.91.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan E.S., Franklin T.M., Hilderbrand-Chae M.J., McNeil G.P., Roberts M.A. An extraretinally expressed insect Cryptochrome with similarity to the blue light photoreceptors of mammals and plants. J. Neurosci. 1999;19:3665–3673. doi: 10.1523/JNEUROSCI.19-10-03665.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery I.F., Noveral J.M., Jamison C.F., Siwicki K.K. Rhythms of Drosophila period gene expression in culture. Proc. Natl. Acad. Sci. USA. 1997;94:4092–4096. doi: 10.1073/pnas.94.8.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery P., So W.V., Kaneko M., Hall J.C., Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95:669–679. doi: 10.1016/s0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- Emery P., Stanewsky R., Hall J.C., Rosbash M. A unique circadian-rhythm photoreceptor. Nature. 2000;404:456–457. doi: 10.1038/35006558. [DOI] [PubMed] [Google Scholar]
- Ewer J., Frisch B., Hamblen-Coyle M.J., Rosbash M., Hall J.C. Expression of the period clock gene within different cell types in the brain of Drosophila adults and mosaic analysis of these cells' influence on circadian behavioral rhythms. J. Neurosci. 1992;12:3321–3349. doi: 10.1523/JNEUROSCI.12-09-03321.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J.Y., Agyekum B., Venkatesan A., Hall D.R., Keightley A. Noncanonical FK506-binding protein BDBT binds DBT to enhance its circadian function and forms foci at night. Neuron. 2013;80:984–996. doi: 10.1016/j.neuron.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman J.F., Hoyle M.N. Isolation of circadian clock mutants of Neurospora crassa. Genetics. 1973;75:605–613. doi: 10.1093/genetics/75.4.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogle K.J., Baik L.S., Houl J.H., Tran T.T., Roberts L. CRYPTOCHROME-mediated phototransduction by modulation of the potassium ion channel beta-subunit redox sensor. Proc. Natl. Acad. Sci. USA. 2015;112:2245–2250. doi: 10.1073/pnas.1416586112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogle K.J., Parson K.G., Dahm N.A., Holmes T.C. CRYPTOCHROME is a blue-light sensor that regulates neuronal firing rate. Science. 2011;331:1409–1413. doi: 10.1126/science.1199702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch B., Hardin P.E., Hamblen-Coyle M.J., Rosbash M., Hall J.C. A promoterless period gene mediates behavioral rhythmicity and cyclical per expression in a restricted subset of the Drosophila nervous system. Neuron. 1994;12:555–570. doi: 10.1016/0896-6273(94)90212-7. [DOI] [PubMed] [Google Scholar]
- Gekakis N., Saez L., Delahaye-Brown A.M., Myers M.P., Sehgal A. Isolation of timeless by PER protein interaction: defective interaction between timeless protein and long-period mutant PERL. Science. 1995;270:811–815. doi: 10.1126/science.270.5237.811. [DOI] [PubMed] [Google Scholar]
- Gekakis N., Staknis D., Nguyen H.B., Davis F.C., Wilsbacher L.D. Role of the Clock protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- Glossop N.R., Houl J.H., Zheng H., Ng F.S., Dudek S.M. Vrille feeds back to control circadian transcription of clock in the Drosophila circadian oscillator. Neuron. 2003;37:249–261. doi: 10.1016/s0896-6273(03)00002-3. [DOI] [PubMed] [Google Scholar]
- Glossop N.R., Lyons L.C., Hardin P.E. Interlocked feedback loops within the Drosophila circadian oscillator. Science. 1999;286:766–768. doi: 10.1126/science.286.5440.766. [DOI] [PubMed] [Google Scholar]
- Griffith L.C. Identifying behavioral circuits in Drosophila melanogaster: moving targets in a flying insect. Curr. Opin. Neurobiol. 2012;22:609–614. doi: 10.1016/j.conb.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima B., Chelot E., Xia R., Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature. 2004;431:869–873. doi: 10.1038/nature02935. [DOI] [PubMed] [Google Scholar]
- Grima B., Lamouroux A., Chelot E., Papin C., Limbourg-Bouchon B. The F-box protein slimb controls the levels of clock proteins period and timeless. Nature. 2002;420:178–182. doi: 10.1038/nature01122. [DOI] [PubMed] [Google Scholar]
- Hao H., Allen D.L., Hardin P.E. A circadian enhancer mediates PER-dependent mRNA cycling in Drosophila melanogaster. Mol. Cell Biol. 1997;17:3687–3693. doi: 10.1128/mcb.17.7.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin P.E. Analysis of period mRNA cycling in Drosophila head and body tissues indicates that body oscillators behave differently from head oscillators. Mol. Cell Biol. 1994;14:7211–7218. doi: 10.1128/mcb.14.11.7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin P.E., Hall J.C., Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- Hardin P.E., Hall J.C., Rosbash M. Circadian oscillations in period gene mRNA levels are transcriptionally regulated. Proc. Natl. Acad. Sci. USA. 1992;89:11711–11715. doi: 10.1073/pnas.89.24.11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich C. Role of the optic lobes in the regulation of the locomotor activity rhythm of Drosophila melanogaster: behavioral analysis of neural mutants. J. Neurogenet. 1986;3:321–343. doi: 10.3109/01677068609106857. [DOI] [PubMed] [Google Scholar]
- Helfrich C., Engelmann W. Circadian rhythm of the locomotor activity in Drosophila melanogaster and its mutants “sine oculis” and “small optic lobes”. Physiol. Entomol. 1983;8:257–272. [Google Scholar]
- Helfrich-Forster C. The period clock gene is expressed in central nervous system neurons which also produce a neuropeptide that reveals the projections of circadian pacemaker cells within the brain of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 1995;92:612–616. doi: 10.1073/pnas.92.2.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Forster C. Robust circadian rhythmicity of Drosophila melanogaster requires the presence of lateral neurons: a brain-behavioral study of disconnected mutants. J. Comp. Physiol. 1998;182:435–453. doi: 10.1007/s003590050192. [DOI] [PubMed] [Google Scholar]
- Helfrich-Forster C. The neuroarchitecture of the circadian clock in the brain of Drosophila melanogaster. Microsc. Res. Tech. 2003;62:94–102. doi: 10.1002/jemt.10357. [DOI] [PubMed] [Google Scholar]
- Helfrich-Forster C. From neurogenetic studies in the fly brain to a concept in circadian biology. J. Neurogenet. 2014;28:329–347. doi: 10.3109/01677063.2014.905556. [DOI] [PubMed] [Google Scholar]
- Helfrich-Forster C., Homberg U. Pigment-dispersing hormone-immunoreactive neurons in the nervous system of wild-type Drosophila melanogaster and of several mutants with altered circadian rhythmicity. J. Comp. Neurol. 1993;337:177–190. doi: 10.1002/cne.903370202. [DOI] [PubMed] [Google Scholar]
- Herzog E.D., Hermanstyne T., Smyllie N.J., Hastings M.H. Regulating the Suprachiasmatic Nucleus (SCN) circadian clockwork: interplay between cell-autonomous and circuit-level mechanisms. Cold Spring Harb. Perspect. Biol. 2017;9 doi: 10.1101/cshperspect.a027706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogenesch J.B., Gu Y.Z., Jain S., Bradfield C.A. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc. Natl. Acad. Sci. USA. 1998;95:5474–5479. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter-Ensor M., Ousley A., Sehgal A. Regulation of the Drosophila protein timeless suggests a mechanism for resetting the circadian clock by light. Cell. 1996;84:677–685. doi: 10.1016/s0092-8674(00)81046-6. [DOI] [PubMed] [Google Scholar]
- Jackson F.R., Bargiello T.A., Yun S.H., Young M.W. Product of per locus of Drosophila shares homology with proteoglycans. Nature. 1986;320:185–188. doi: 10.1038/320185a0. [DOI] [PubMed] [Google Scholar]
- James S.M., Honn K.A., Gaddameedhi S., Van Dongen H.P.A. Shift work: disrupted Circadian rhythms and sleep-implications for health and well-being. Curr. Sleep. Med. Rep. 2017;3:104–112. doi: 10.1007/s40675-017-0071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M., Hall J.C. Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J. Comp. Neurol. 2000;422:66–94. doi: 10.1002/(sici)1096-9861(20000619)422:1<66::aid-cne5>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- King D.P., Zhao Y., Sangoram A.M., Wilsbacher L.D., Tanaka M. Positional cloning of the mouse circadian clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistenpfennig C., Grebler R., Ogueta M., Hermann-Luibl C., Schlichting M. A new rhodopsin influences light-dependent daily activity patterns of fruit flies. J Biol. Rhythm. 2017;32:406–422. doi: 10.1177/0748730417721826. [DOI] [PubMed] [Google Scholar]
- Kivimae S., Saez L., Young M.W. Activating PER repressor through a DBT-directed phosphorylation switch. PLoS Biol. 2008;6:e183. doi: 10.1371/journal.pbio.0060183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss B., Price J.L., Saez L., Blau J., Rothenfluh A. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iepsilon. Cell. 1998;94:97–107. doi: 10.1016/s0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- Ko H.W., Jiang J., Edery I. Role for slimb in the degradation of Drosophila period protein phosphorylated by Doubletime. Nature. 2002;420:673–678. doi: 10.1038/nature01272. [DOI] [PubMed] [Google Scholar]
- Koh K., Zheng X., Sehgal A. Jetlag resets the Drosophila circadian clock by promoting light-induced degradation of Timeless. Science. 2006;312:1809–1812. doi: 10.1126/science.1124951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka R.J., Benzer S. Clock mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 1971;68:2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan B., Levine J.D., Lynch M.K., Dowse H.B., Funes P. A new role for cryptochrome in a Drosophila circadian oscillator. Nature. 2001;411:313–317. doi: 10.1038/35077094. [DOI] [PubMed] [Google Scholar]
- Levine J.D., Funes P., Dowse H.B., Hall J.C. Advanced analysis of a cryptochrome mutation’s effects on the robustness and phase of molecular cycles in isolated peripheral tissues of Drosophila. BMC Neurosci. 2002;3:5. doi: 10.1186/1471-2202-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Han M., Shimada B., Wang L., Gibler T.M. Influence of the period-dependent circadian clock on diurnal, circadian, and aperiodic gene expression in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 2002;99:9562–9567. doi: 10.1073/pnas.132269699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Lorenz L., Yu Q.N., Hall J.C., Rosbash M. Spatial and temporal expression of the period gene in Drosophila melanogaster. Genes Dev. 1988;2:228–238. doi: 10.1101/gad.2.2.228. [DOI] [PubMed] [Google Scholar]
- McDonald M.J., Rosbash M. Microarray analysis and organization of circadian gene expression in Drosophila. Cell. 2001;107:567–578. doi: 10.1016/s0092-8674(01)00545-1. [DOI] [PubMed] [Google Scholar]
- Means J.C., Venkatesan A., Gerdes B., Fan J.Y., Bjes E.S. Drosophila Spaghetti and Doubletime link the circadian clock and light to caspases, apoptosis and tauopathy. PLoS Genet. 2015;11:e1005171. doi: 10.1371/journal.pgen.1005171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Viveros L., Bouchard-Cannon P., Hegazi S., Cheng A.H., Pastore S. Molecular modulators of the circadian clock: lessons from flies and mice. Cell Mol. Life Sci. 2017;74:1035–1059. doi: 10.1007/s00018-016-2378-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui S., Yamaguchi S., Matsuo T., Ishida Y., Okamura H. Antagonistic role of E4BP4 and PAR proteins in the circadian oscillatory mechanism. Genes Dev. 2001;15:995–1006. doi: 10.1101/gad.873501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers M.P., Wager-Smith K., Rothenfluh-Hilfiker A., Young M.W. Light-induced degradation of TIMELESS and entrainment of the Drosophila circadian clock. Science. 1996;271:1736–1740. doi: 10.1126/science.271.5256.1736. [DOI] [PubMed] [Google Scholar]
- Myers M.P., Wager-Smith K., Wesley C.S., Young M.W., Sehgal A. Positional cloning and sequence analysis of the Drosophila clock gene, timeless. Science. 1995;270:805–808. doi: 10.1126/science.270.5237.805. [DOI] [PubMed] [Google Scholar]
- Nakajima M., Imai K., Ito H., Nishiwaki T., Murayama Y. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- Ni J.D., Baik L.S., Holmes T.C., Montell C. A rhodopsin in the brain functions in circadian photoentrainment in Drosophila. Nature. 2017;545:340–344. doi: 10.1038/nature22325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschel N., Chen K.F., Szabo G., Stanewsky R. Light-dependent interactions between the Drosophila circadian clock factors cryptochrome, jetlag, and timeless. Curr. Biol. 2009;19:241–247. doi: 10.1016/j.cub.2008.12.042. [DOI] [PubMed] [Google Scholar]
- Peschel N., Veleri S., Stanewsky R. Veela defines a molecular link between Cryptochrome and Timeless in the light-input pathway to Drosophila’s circadian clock. Proc. Natl. Acad. Sci. USA. 2006;103:17313–17318. doi: 10.1073/pnas.0606675103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plautz J.D., Kaneko M., Hall J.C., Kay S.A. Independent photoreceptive circadian clocks throughout Drosophila. Science. 1997;278:1632–1635. doi: 10.1126/science.278.5343.1632. [DOI] [PubMed] [Google Scholar]
- Preitner N., Damiola F., Lopez-Molina L., Zakany J., Duboule D. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Preuss F., Fan J.Y., Kalive M., Bao S., Schuenemann E. Drosophila doubletime mutations which either shorten or lengthen the period of circadian rhythms decrease the protein kinase activity of casein kinase I. Mol. Cell Biol. 2004;24:886–898. doi: 10.1128/MCB.24.2.886-898.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J.L., Blau J., Rothenfluh A., Abodeely M., Kloss B. double-time is a novel Drosophila clock gene that regulates period protein accumulation. Cell. 1998;94:83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- Price J.L., Dembinska M.E., Young M.W., Rosbash M. Suppression of PERIOD protein abundance and circadian cycling by the Drosophila clock mutation timeless. EMBO J. 1995;14:4044–4049. doi: 10.1002/j.1460-2075.1995.tb00075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy P., Jacquier A.C., Abovich N., Petersen G., Rosbash M. The period clock locus of D. melanogaster codes for a proteoglycan. Cell. 1986;46:53–61. doi: 10.1016/0092-8674(86)90859-7. [DOI] [PubMed] [Google Scholar]
- Reddy P., Zehring W.A., Wheeler D.A., Pirrotta V., Hadfield C. Molecular analysis of the period locus in Drosophila melanogaster and identification of a transcript involved in biological rhythms. Cell. 1984;38:701–710. doi: 10.1016/0092-8674(84)90265-4. [DOI] [PubMed] [Google Scholar]
- Reppert S.M. A clockwork explosion! Neuron. 1998;21:1–4. doi: 10.1016/s0896-6273(00)80234-2. [DOI] [PubMed] [Google Scholar]
- Rosato E., Codd V., Mazzotta G., Piccin A., Zordan M. Light-dependent interaction between Drosophila CRY and the clock protein PER mediated by the carboxy terminus of CRY. Curr. Biol. 2001;11:909–917. doi: 10.1016/s0960-9822(01)00259-7. [DOI] [PubMed] [Google Scholar]
- Rutila J.E., Suri V., Le M., So W.V., Rosbash M. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell. 1998;93:805–814. doi: 10.1016/s0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- Sehgal A., Price J.L., Man B., Young M.W. Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless. Science. 1994;263:1603–1606. doi: 10.1126/science.8128246. [DOI] [PubMed] [Google Scholar]
- Sehgal A., Rothenfluh-Hilfiker A., Hunter-Ensor M., Chen Y., Myers M.P. Rhythmic expression of timeless: a basis for promoting circadian cycles in period gene autoregulation. Science. 1995;270:808–810. doi: 10.1126/science.270.5237.808. [DOI] [PubMed] [Google Scholar]
- Shafer O.T., Helfrich-Forster C., Renn S.C., Taghert P.H. Reevaluation of Drosophila melanogaster’s neuronal circadian pacemakers reveals new neuronal classes. J Comp. Neurol. 2006;498:180–193. doi: 10.1002/cne.21021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siwicki K.K., Eastman C., Petersen G., Rosbash M., Hall J.C. Antibodies to the period gene product of Drosophila reveal diverse tissue distribution and rhythmic changes in the visual system. Neuron. 1988;1:141–150. doi: 10.1016/0896-6273(88)90198-5. [DOI] [PubMed] [Google Scholar]
- Skene D.J., Arendt J. Circadian rhythm sleep disorders in the blind and their treatment with melatonin. Sleep. Med. 2007;8:651–655. doi: 10.1016/j.sleep.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Stanewsky R., Kaneko M., Emery P., Beretta B., Wager-Smith K. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell. 1998;95:681–692. doi: 10.1016/s0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- Stoleru D., Peng Y., Agosto J., Rosbash M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature. 2004;431:862–868. doi: 10.1038/nature02926. [DOI] [PubMed] [Google Scholar]
- Sun Z.S., Albrecht U., Zhuchenko O., Bailey J., Eichele G. Rigui, a putative mammalian ortholog of the Drosophila period gene. Cell. 1997;90:1003–1011. doi: 10.1016/s0092-8674(00)80366-9. [DOI] [PubMed] [Google Scholar]
- Takahashi J.S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 2017;18:164–179. doi: 10.1038/nrg.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tataroglu O., Emery P. The molecular ticks of the Drosophila circadian clock. Curr. Opin. Insect Sci. 2015;7:51–57. doi: 10.1016/j.cois.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tei H., Okamura H., Shigeyoshi Y., Fukuhara C., Ozawa R. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature. 1997;389:512–516. doi: 10.1038/39086. [DOI] [PubMed] [Google Scholar]
- Thackeray, J., 1989. Molecular analysis of behavioural rhythms in Drosophila. In:Ph.D thesis, Leicester University.
- Toh K.L., Jones C.R., He Y., Eide E.J., Hinz W.A. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- Ueda H.R., Hayashi S., Chen W., Sano M., Machida M. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet. 2005;37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- Ueda H.R., Matsumoto A., Kawamura M., Iino M., Tanimura T. Genome-wide transcriptional orchestration of circadian rhythms in Drosophila. J Biol. Chem. 2002;277:14048–14052. doi: 10.1074/jbc.C100765200. [DOI] [PubMed] [Google Scholar]
- Vitaterna M.H., King D.P., Chang A.M., Kornhauser J.M., Lowrey P.L. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosshall L.B., Price J.L., Sehgal A., Saez L., Young M.W. Block in nuclear localization of period protein by a second clock mutation, timeless. Science. 1994;263:1606–1609. doi: 10.1126/science.8128247. [DOI] [PubMed] [Google Scholar]
- Xu Y., Padiath Q.S., Shapiro R.E., Jones C.R., Wu S.C. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature. 2005;434:640–644. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- Yoshii T., Rieger D., Helfrich-Forster C. Two clocks in the brain: an update of the morning and evening oscillator model in Drosophila. Prog. Brain Res. 2012;199:59–82. doi: 10.1016/B978-0-444-59427-3.00027-7. [DOI] [PubMed] [Google Scholar]
- Young M.W., Jackson F.R., Shin H.S., Bargiello T.A. A biological clock in Drosophila. Cold Spring Harb. Symp. Quant. Biol. 1985;50:865–875. doi: 10.1101/sqb.1985.050.01.104. [DOI] [PubMed] [Google Scholar]
- Yu Q., Jacquier A.C., Citri Y., Hamblen M., Hall J.C. Molecular mapping of point mutations in the period gene that stop or speed up biological clocks in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 1987;84:784–788. doi: 10.1073/pnas.84.3.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehring W.A., Wheeler D.A., Reddy P., Konopka R.J., Kyriacou C.P. P-element transformation with period locus DNA restores rhythmicity to mutant, arrhythmic Drosophila melanogaster. Cell. 1984;39:369–376. doi: 10.1016/0092-8674(84)90015-1. [DOI] [PubMed] [Google Scholar]
- Zeng H., Qian Z., Myers M.P., Rosbash M. A light-entrainment mechanism for the Drosophila circadian clock. Nature. 1996;380:129–135. doi: 10.1038/380129a0. [DOI] [PubMed] [Google Scholar]
- Zerr D.M., Hall J.C., Rosbash M., Siwicki K.K. Circadian fluctuations of Period protein immunoreactivity in the CNS and the visual system of Drosophila. J. Neurosci. 1990;10:2749–2762. doi: 10.1523/JNEUROSCI.10-08-02749.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]