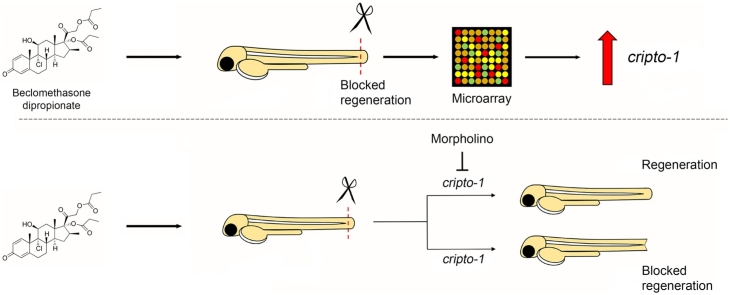
Graphical abstract
Abbreviations: AEC, apical epithelial cap; BDP, beclomethasone dipropionate; DMSO, dimethyl sulfoxide; dpa, days post-amputation; dpf, days post-fertilization; EB, embryoid body; ECM, extracellular matrix; EMT, epithelial-to-mesenchymal transition; ERK, extracellular signal-regulated kinase; eSC, embryonic stem cell; FGF, fibroblast growth factor; GC, glucocorticoid; GR, glucocorticoid receptor; hpa, hours post-amputation; hpf, hours post-fertilization; ISH, in situ hybridization; MIAME, Minimum Information About a Microarray Experiment; mLIF, murine leukemia inhibitory factor; MO, morpholino oligonucleotide; RA, retinoic acid; SEM, standard error of the mean; TGF-β, transforming growth factor beta; qRT-PCR, quantitative reverse transcription polymerase chain reaction; zf, zebrafish
Keywords: Zebrafish, Epimorphic regeneration, Beclomethasone dipropionate, Glucocorticoids, Cripto-1, One-eyed pinhead
Highlights
-
•
cripto-1 increased expression in regenerating zebrafish fins during BDP exposure.
-
•
Morpholino knockdown of Cripto-1 allowed regeneration during BDP exposure.
-
•
Induction of cripto-1 following BDP exposure is conserved in mammalian stem cells.
Abstract
We previously used a chemical genetics approach with the larval zebrafish to identify small molecule inhibitors of tissue regeneration. This led to the discovery that glucocorticoids (GC) block early stages of tissue regeneration by the inappropriate activation of the glucocorticoid receptor (GR). We performed a microarray analysis to identify the changes in gene expression associated with beclomethasone dipropionate (BDP) exposure during epimorphic fin regeneration. Oncofetal cripto-1 showed > eight-fold increased expression in BDP-treated regenerates. We hypothesized that the mis-expression of cripto-1 was essential for BDP to block regeneration. Expression of cripto-1 was not elevated in GR morphants in the presence of BDP indicating that cripto-1 induction was GR-dependent. Partial translational suppression of Cripto-1 in the presence of BDP restored tissue regeneration. Retinoic acid exposure prevented increased cripto-1 expression and permitted regeneration in the presence of BDP. We demonstrated that BDP exposure increased cripto-1 expression in mouse embryonic stem cells and that regulation of cripto-1 by GCs is conserved in mammals.
1. Introduction
Some vertebrates, such as zebrafish (Danio rerio), have the capacity to fully restore complex tissues such as limbs, appendages, and organs in a process called epimorphic regeneration. Zebrafish caudal fin regeneration is a useful model of this process for several reasons including the fast restoration of resected fin tissue (2–3 weeks), tractability of the zebrafish genome, high survival rate, and ease of husbandry, observation, and data acquisition [1,2]. The deterministic mechanism(s) leading to epimorphic regeneration versus scarring is unknown, but it is understood that regeneration is divided into four major phases: wound healing, blastema formation, regenerative outgrowth, and termination. Each phase requires a tightly coordinated sequence of molecular events involving several signaling pathways. Fibroblast growth factor, Wnt, Activin, and others are among the first activated pathways following injury [1,3]. During wound healing, epithelial cells migrate to form a wound epidermis from which the apical epithelial cap (AEC) is derived. Epithelial-mesenchymal interactions involving the AEC cause the underlying mesenchyme to dedifferentiate into a multipotent mass of cells called a blastema. Once established, the blastema undergoes rapid proliferation and differentiation to restore all damaged or lost structures. This process terminates once the damaged tissue is fully regenerated to its original structure [1].
Like adults, larval zebrafish also undergo epimorphic regeneration following fin amputation, and the molecular and structural processes are fundamentally similar [2,4,5]. Larval fin regeneration offers several advantages over adults, the most notable being rapid rate of regeneration (3 days), amenability to transient knockdown of gene expression using antisense repression, and small size of the organism. These characteristics make the larval regeneration model amenable to high-throughput screening of chemicals that can modulate epimorphic regeneration [6]. This chemical genetics approach is guided by the hypothesis that compounds inhibiting regeneration do so by perturbing specific signaling events required for the regenerative process. This makes chemical genetics a useful tool for providing mechanistic insight into regeneration. Likewise, such a screen could also identify novel effects of chemicals within the regenerative framework [6]. For example, we previously used this larval regeneration approach to identify a novel link between Wnt and Aryl hydrocarbon receptor signaling via R-spondin 1 [7].
We previously performed a blinded screen of a 2000-member library of FDA-approved chemicals to identify compounds that modulate larval zebrafish fin regeneration [6]. Among the chemical classes inhibiting regeneration was glucocorticoids (GCs). GCs modulate several biological processes including energy metabolism, immunity, development, and wound healing [[8], [9], [10], [11], [12]]. Endogenous GCs such as cortisol and exogenous GCs such as dexamethasone act primarily through the glucocorticoid receptor (GR), a nuclear receptor that can potentially transactivate or transrepress thousands of genes [8].
Beclomethasone dipropionate (BDP) had the greatest potency to inhibit regeneration and this inhibition was GR-dependent. This BDP regenerative inhibition, however, was independent of anti-inflammatory effects on neutrophil and macrophage recruitment to the wound site. Finally, the inhibitory effects of BDP occurred within a narrow 4 h critical window following amputation indicating that the upstream GR target was present during the early stages of regeneration [6]. We performed microarray analysis (unpublished until now) on regenerating caudal fins exposed to BDP and discovered that BDP increased the expression of the oncofetal gene cripto-1, also known as one-eyed pinhead (oep) in zebrafish and teratoma-derived growth factor 1 (tdgf1) in humans. Using qRT-PCR, we found that GR activity (based on anxa1b repression) was similar between GCs that inhibited or permitted regeneration [13]. Those that inhibited regeneration had increased cripto-1 expression similar to BDP, whereas those that permitted regeneration did not increase cripto-1 expression.
In vertebrates, cripto-1 is a required co-factor in Nodal signaling as well as an antagonist of Activin signaling [[14], [15], [16]]. The importance of functional cripto-1 in zebrafish development was demonstrated in oep loss-of-function mutants, which developed cyclopia and died as larvae due to impaired Nodal signaling [17]. Antisense knockdown of cripto-1 expression using translation-blocking MOs produced identical effects [18]. As an oncogene controlling cellular stemness, cripto-1 is expressed in various cancer types [19] and modulation of its expression could be a desirable therapeutic strategy. In human and murine teratocarcinoma cells, cripto-1 expression is downregulated in response to compounds that induce cellular differentiation such as retinoic acid [20], indicating that its expression can be influenced by chemical exposure.
Increased abundance of Cripto-1 in regenerating tissue could potentially interfere with critical regenerative pathways such as Activin [15]. We therefore hypothesized that BDP inhibited regeneration by modulating the expression of a gene (cripto-1) that other GCs do not. In the current study, we present the original microarray data demonstrating that BDP increased the expression of cripto-1. We then conclusively demonstrate that a GR-dependent increase in cripto-1 expression following BDP exposure was responsible for inhibited zebrafish fin regeneration, and that the effect of BDP on cripto-1 expression is conserved in murine stem cells.
2. Materials and methods
2.1. Ethics statement
All experiments were performed according to the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The Oregon State University Institutional Animal Care and Use Committee reviewed and approved the animal care and use protocols (internal approval number 3903). Tricaine mesylate (MS-222) was used to anesthetize animals during the amputation procedures, and every effort was implemented to curtail pain and suffering.
2.2. Chemical exposures
All chemical exposures started at 2 days post-fertilization (dpf) immediately following fin amputation. Static exposure to beclomethasone dipropionate (BDP) (Sigma; ≥99% pure) was performed at a final concentration of 1 μM using 0.1% DMSO (vehicle). Static exposure to SB431542 (Sigma; ≥98% pure) was performed at a final concentration of 100 μM. Others have reported the use of <100 μM SB431542 [3] in adult zebrafish and our most recent data (not shown) indicate that 50 μM is sufficient to inhibit fin regeneration in the larval model. Exposure to all-trans retinoic acid (RA) (Sigma; ≥ 98%) was performed at a concentration of 0.1 μM for 8 h followed by static co-exposure to a lower RA concentration of 0.01 μM as well as BDP at a final concentration of 1 μM. Dimethyl sulfoxide (DMSO) was used to prepare all chemical stocks.
2.3. Zebrafish embryos and larvae
Standard husbandry procedures were used for all embryos [21]. Microarray experiments were performed using AB strain embryos, while Tropical 5D strain embryos were used to validate the response to beclomethasone dipropionate (BDP) in subsequent experiments. Excluding microarray and qRT-PCR procedures (described below), a sample size of n = 12 was used for all experiments. Caudal fin amputations were performed according to previously described procedures [6,22,23].
2.4. Fin RNA isolation
Larvae were subject to caudal fin amputation at 2 dpf and were immediately exposed to either 1 μM BDP or vehicle control (0.1% DMSO) as described above. The regenerating fin tissues were amputated a second time at 24 h post-amputation (hpa) and collected for total RNA extraction using the RNAqueous Micro kit (Ambion). Each biological replicate consisted of pooled fin tissue from 150 larvae, and each treatment consisted of three replicates. The quality and quantity of isolated total RNA was determined using UV absorbance analysis. Electropherogram patterns were analyzed for degradation and ribosomal RNA abundance using the 2100 Bioanalyzer and RNA 6000 Nano chips (Agilent Technologies).
2.5. Affymetrix microarray processing
Microarray preparation and processing were performed for the Affymetrix platform at the Center for Genome Research and Biocomputing (CGRB), Oregon State University. Single-stranded cDNA was synthesized using 100 ng total RNA from larval regenerating fin tissue (AB strain) with Superscript II reverse transcriptase and T7-(dT)24 primer (Invitrogen). A second round of cDNA synthesis was performed to generate double-stranded cDNA. This cDNA template was used for generating biotinylated cRNA using biotin-conjugated pseudouridine and T7 polymerase (Affymetrix). Following quantification, 10 μg of the purified and fragmented biotinylated cRNA was hybridized to zebrafish genome arrays (Zebrafish430_2) as specified in the Affymetrix GeneChip Expression Analysis Technical Manual (7010201 Rev.5). Affymetrix Scanner 3000 was used to scan arrays. Each array was visualized to screen for non-specific signals from debris, scratches, or other artefacts. Microarray experiments were certified under Minimum Information About a Microarray Experiment (MIAME) standards.
Genespring software (Agilent Technologies) was used to analyze Affymetrix CEL files generated from the microarray. Background signal was removed using gene chip-robust multiarray processing. Each transcript was normalized based on median signal thus allowing comparisons to be made between arrays on a relative scale for each gene. To identify significant (p < 0.05) differential expression of genes following BDP exposure, one-way analysis of variance (ANOVA) assuming equal variance was performed comparing BDP- and DMSO-treated experimental groups. Further analysis focused primarily on genes that had differential expression of at least 2-fold. These genes were annotated by comparing sequence similarity of the respective Affymetrix probe set with known mammalian proteins based on the Sanger database (http://www.sanger.ac.uk/Projects/D_rerio/). Annotation was confirmed using the Genbank and Ensembl (http://uswest.ensembl.org/Danio_rerio/Info/Index) databases. Raw data were uploaded to the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) under the accession number GSE10766. A heatmap of bi-hierarchical clusters were generated using MultiExperiment Viewer (MEV) from which a gene list was created. Human orthologs of these genes were identified using ZFIN (https://zfin.org/) and Ensembl databases.
2.6. Quantitative real time reverse transcriptase polymerase chain reaction (qRT PCR)
As previously described, total RNA was isolated from larval regenerating fin tissue exposed to either 1 μM BDP or 0.1% DMSO. Biological replicates consisted of pooled fin samples of n = 60 with three replicates per experimental group. cDNA was generated from 1 μg total RNA using Superscript II (Life Technologies) and oligo(dT) primers. qRT-PCR was performed using gene-specific primers (Table S6) in the Opticon 2 real time PCR detection system (MJ Research) with the SYBR green qPCR kit (Finnzymes). Samples were normalized to endogenous β-actin quantity. Formation of the anticipated PCR products was verified using agarose gel electrophoresis and melt curve analysis. Sigmastat software (Systat Software) was used to identify statistically significant differences in mRNA abundance by one-way ANOVA with Tukey’s post-hoc test (p < 0.05) on log10-transformed data.
2.7. Oligonucleotides
Primers were designed according to Affymetrix probe target sequences and are listed in Table S6. Forward and antisense reverse primers are prefixed with F and R, respectively.
2.8. In situ hybridization
Localization of transcripts within larvae at 24 hpa was performed using in situ hybridization according to published methods. Probes for dlx5a, mvp, smarca4, and ilf2 were generously gifted by Atsushi Kawakami (Tokyo Institute of Technology, Yokahama, Japan).
2.9. Morpholinos
Knockdown of Cripto-1 and the glucocorticoid receptor (GR) was performed using morpholino oligos (MOs) (Gene Tools) that blocked the translation start site of cripto-1 and the splice junction of exons 7 and 8 of the GR. Both the cripto-1 MO (5′ GCCAATAAACTCCAAAACAACTCGA 3′) [18] and the GR MO (5′ - CGGAACCCTAAAATACATGAAGCAG - 3′) [6] were fluorescein tagged and targeted zebrafish-specific transcript sequences. MO controls consisted of a standard MO control sequence (5′ CTCTTACCTCAGTTACAATTTATA 3′). MOs were reconstituted to a stock concentration of 3 mM in 1x Danieau’s solution (58 mM NaCl, 0.7 mM KCl, 0.4 mM MgSO4, 0.6 mM Ca(NO3)2, 5 mM HEPES, pH 7.6). 1–4 cell stage embryos were injected with either 1.2 mM cripto-1 MO or 3 mM GR MO, and controls were injected with a matching concentration of standard control MO. Uniform distribution of morpholino was evaluated at 1 dpf by screening for fluorescence using a GFP filter. Morphant larvae underwent caudal fin amputation as previously described and then were statically exposed to 1 μM BDP or 0.1% DMSO. Larvae were grown out to 3 days post-amputation (dpa) as previously described in standard regeneration protocols [6].
2.10. Embryonic stem cell (eSC) treatment
Murine D3 embryonic stem cells (eSCs) (ATCC, Rockville, MD) were sustained in Dulbecco’s modified Eagle’s medium (Invitrogen, Breda, The Netherlands) at 37 °C and 5% CO2. Cells were routinely subcultured every 2–3 days. Medium was supplemented with 20% heat inactivated fetal calf serum (Hyclone, Thermo-Fisher Scientific, Etten-Leur, The Netherlands), 2 mM glutamine (Invitrogen), 50 U/ml penicillin (Invitrogen), 50 μg/ml streptomycin (Invitrogen), 1% non-essential amino acids (Invitrogen), and 0.1 mM β-mercaptoethanol (Sigma-Aldrich, Zwijndrecht, The Netherlands). Pluripotency was maintained by addition of murine leukemia inhibitory factor (mLIF) (Chemicon, Amsterdam, The Netherlands) to cell cultures at a final concentration of 1000 U/mL [24]. D3 eSCs were induced to differentiate into cardiac cells using routine culture medium (without mLIF) as previously described. On culture day 0, 20 μL of eSC suspension (3.75 × 104 cells/mL) was translocated to the inner side of the lid of a 10 cm Petri dish (Greiner) containing 5 mL of PBS. eSCs were incubated in a humidified atmosphere at 37 °C and 5% CO2. The resultant embryoid bodies (EBs) were transferred to bacteriological Petri dishes (Greiner). The differentiation cultures were exposed from culture day 3 onward to BDP and retinoic acid (Fluka, Buchs, Switzerland). Two parallel cultures were performed for each of the compounds. After 24 h of exposure (culture day 4), EBs from one culture were collected and directly stored in RNA Protect at −20 °C to stabilize RNA (Qiagen, Venlo, The Netherlands).
2.11. Promoter analysis
The promoters of zebrafish, mouse, and human cripto-1 were analyzed for transcription factor binding sites using MatInspector release professional 8.4.1 (Genomatix Software Suite v3.10) [25]. The genomic region 3000 bp upstream of the zebrafish (GRCz11), mouse (GRCm38.p6), and human (GRCh38.p12) cripto-1 transcriptional start sites were retrieved using Ensembl (release 95). These sequences were input into MatInspector and analyzed for general core promoter element and vertebrate element matrices from Matrix Library 11.0. The core/matrix similarity thresholds were set at 0.75/optimized.
3. Results
3.1. Gene expression analysis identified cripto-1 as a potential GR target in regenerating tissue
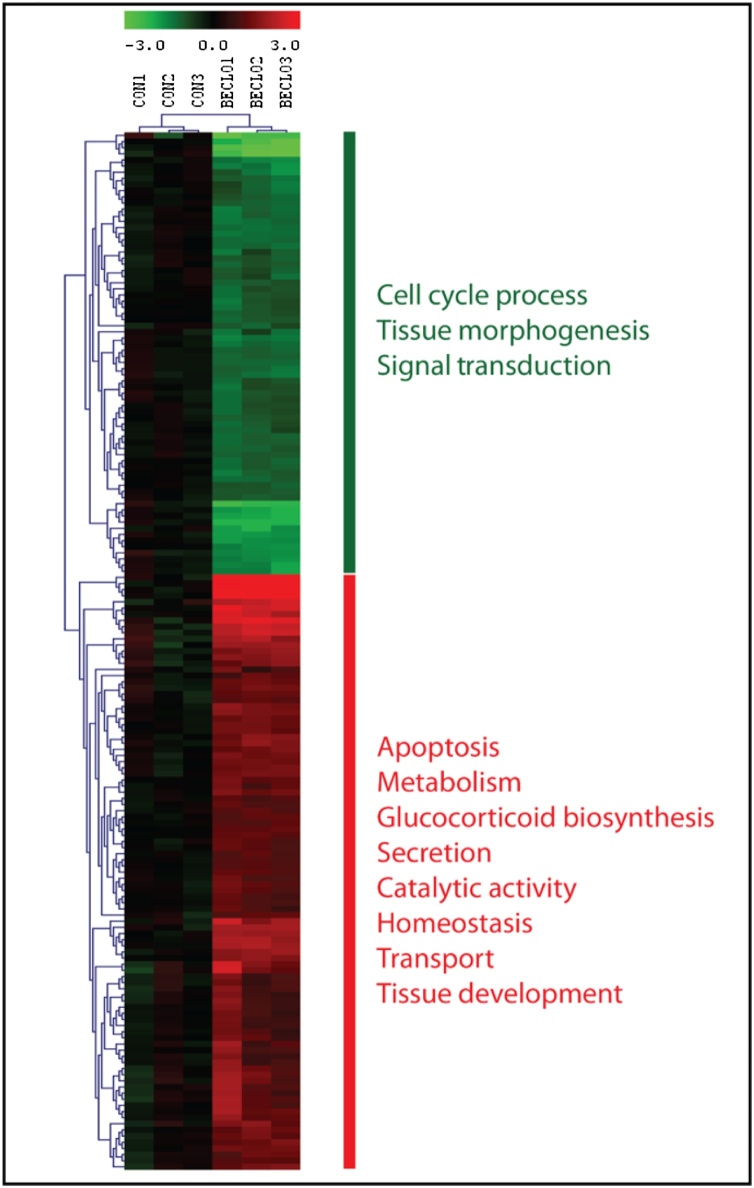
To identify the early gene expression changes associated with the inhibition, we performed global gene expression analysis in the regenerating fin tissue exposed to vehicle (0.1% DMSO) or 1 μM BDP. Only genes that were at least 2-fold differentially expressed in comparison to the control were considered (Fig. 1).
Fig. 1.
Gene expression changes in larval regenerating fin tissue after exposure to BDP. Heat map demonstrates bi-hierarchical clustering of 169 statistically significant (p < 0.05) transcripts that are at least two-fold differentially expressed.
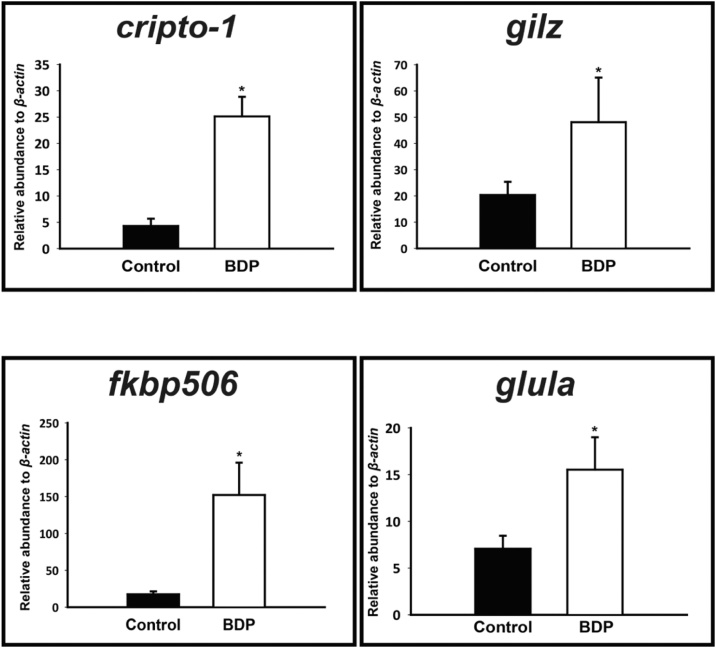
The heat map illustrated two major clusters of differentially expressed mRNAs. Statistical significance determined by one-way ANOVA revealed 169 transcripts with greater than a 2-fold change (p < 0.05) (Table S1). These transcripts were analyzed based on sequence homology and further grouped by function (Table S1). Most of the transcripts were involved in wound healing, extracellular matrix (ECM) remodeling and metabolism. Other known GR target genes such as gilz [26] and fkbp506 [27] were also significantly affected (Fig. 2). Notably, cripto-1 was expressed 8.5-fold higher in the regenerating fin tissue upon BDP exposure (Table S1). qRT-PCR analysis confirmed this induction in the regenerating fin tissue as well as in the whole embryo (Fig. 2).
Fig. 2.
qRT-PCR analysis of BDP-enhanced transcripts in DMSO or BDP treated larval fin tissue at 1 dpa. The expression of cripto-1, gilz, fkbp506, and glula following beclomethasone dipropionate (BDP) exposure at one day post-amputation (dpa) are illustrated as relative abundance to β-actin mRNA levels. Gene specific primers were used to quantify mRNAs using real time qRT-PCR. Data presented as mean ± SEM (n = 3). One-way ANOVA was conducted to determine differences in expression. Asterisks indicate significant difference between vehicle and treatment (p < 0.05).
3.2. Activin signaling is important for larval caudal fin regeneration
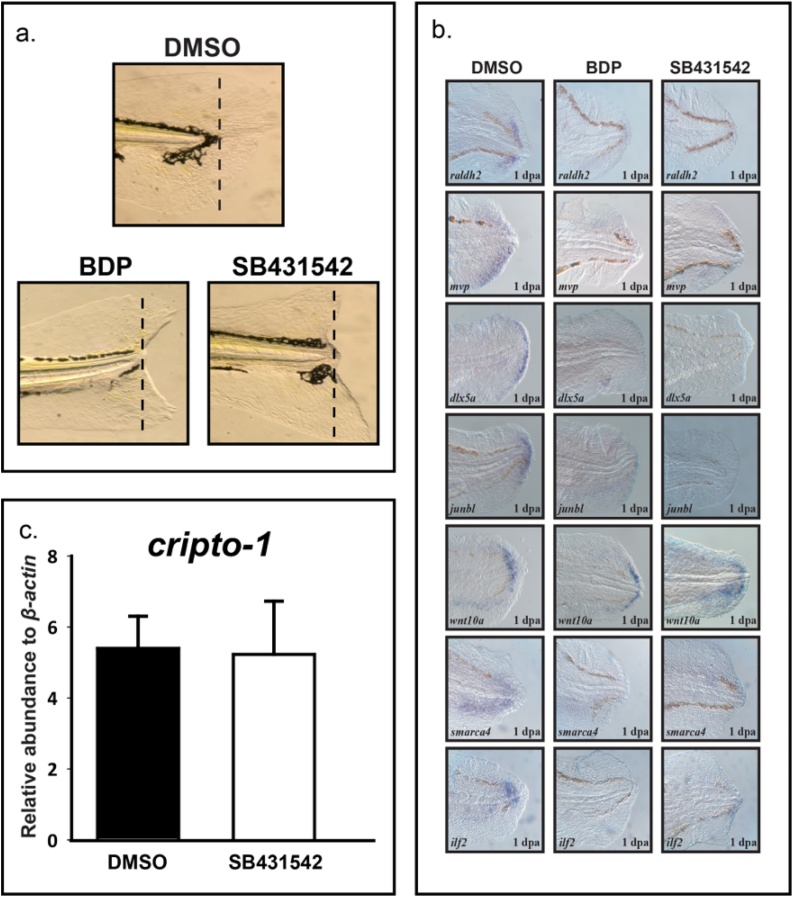
As a first step to understand the downstream effectors of activated GR, we explored the role of Activin signaling specifically for larval tissue regeneration. SB431542 is a specific inhibitor of endogenous Activin and Transforming growth factor beta (TGF-β) signaling [[28], [29], [30]]. Exposure of amputated larvae to 100 μM SB431542 from 0 to 72 h post-amputation (hpa) completely impaired regeneration producing the characteristic “V” shape observed in BDP-exposed larvae (Fig. 3a). Regeneration was characterized by the expression of specific transcripts in the wound epithelium and blastema. Comparative in situ analysis between BDP- and SB431542-exposed fish revealed loss of dlx5a expression in the wound epithelium, signifying absence of apical epithelial cap [4,31]. Moreover, the expression of raldh2, mvp, junbl, smarca4, wnt10a, and ilf2 in the blastemal cells were similarly misexpressed in BDP- and SB431542-exposed larvae compared to control animals. Exposure to SB431542 did not change cripto-1 expression (Fig. 3c).
Fig. 3.
BDP and SB431542 impact larval regeneration. a) Caudal fins of 2 dpf (days post-fertilization) larvae were amputated and exposed to vehicle (0.1% DMSO), 1 μM beclomethasone dipropionate (BDP), or 100 μM SB431542. Regenerative progression was evaluated and pictures were taken at 3 dpa (days post-amputation). b) In situ localization of dlx5a, junbl, wnt10a, ilf2, smarca4, raldh2 and mvp in larvae exposed to BDP and SB431542 demonstrated a similar expression pattern. c) Exposure to SB431542 did not significantly alter expression of cripto-1.
3.3. GR-mediated misregulation of cripto-1 is required for inhibition of regeneration
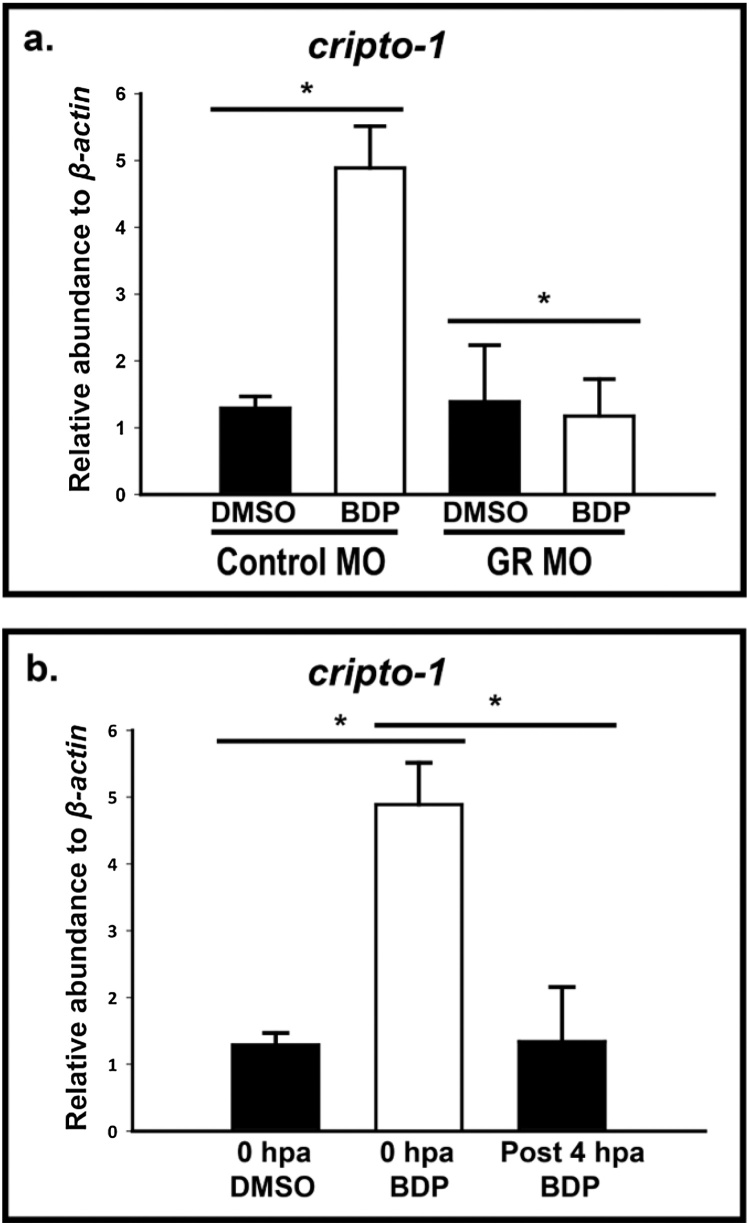
An initial goal was to determine if cripto-1 induction is GR dependent. Quantitative analysis of cripto-1 transcript in GR-MO injected larvae (morphants) exposed to BDP showed significantly reduced cripto-1 expression compared to control morphants exposed to BDP (Fig. 4a). When BDP exposure was initiated immediately after amputation (0 hpa), cripto-1 expression was significantly induced by 1 dpa. When BDP exposure was initiated after the 4 -h window (4 hpa), cripto-1 expression was not induced by 1 dpa (Fig. 4b).
Fig. 4.
Inappropriate GR activation during early stages of regeneration leads to induced cripto-1 expression. a) Glucocorticoid receptor (GR) splice variant morpholino oligo (MO) transiently knocked down GR compared to standard control MO-injected embryos. The amputated control and GR morphants were exposed to DMSO or beclomethasone dipropionate (BDP). The abundance of cripto-1 transcript estimated by qRT-PCR at 1 day post-amputation (dpa) in the whole embryo indicate significantly reduced expression in the BDP exposed morphants. The respective values represent the mean ± SEM and the asterisks indicate statistical significance (One-way ANOVA, n = 3, p < 0.05). b) 2 days post-fertilization (dpf) embryos were amputated and exposed to three different groups; DMSO at 0 h post-amputation (hpa), BDP at 0 hpa and BDP post 4 hpa. The expression of cripto-1 was evaluated in whole embryos at 1 dpa. cripto-1 expression was significantly up-regulated when exposed at 0 hpa, but BDP exposure didn’t induce cripto-1 when exposed after 4 hpa. The respective values represent the mean ± SEM and the asterisks indicate statistical significance (One-way ANOVA, n = 3, p < 0.05).
3.4. Cripto-1 is required for the BDP-inhibition of fin regeneration
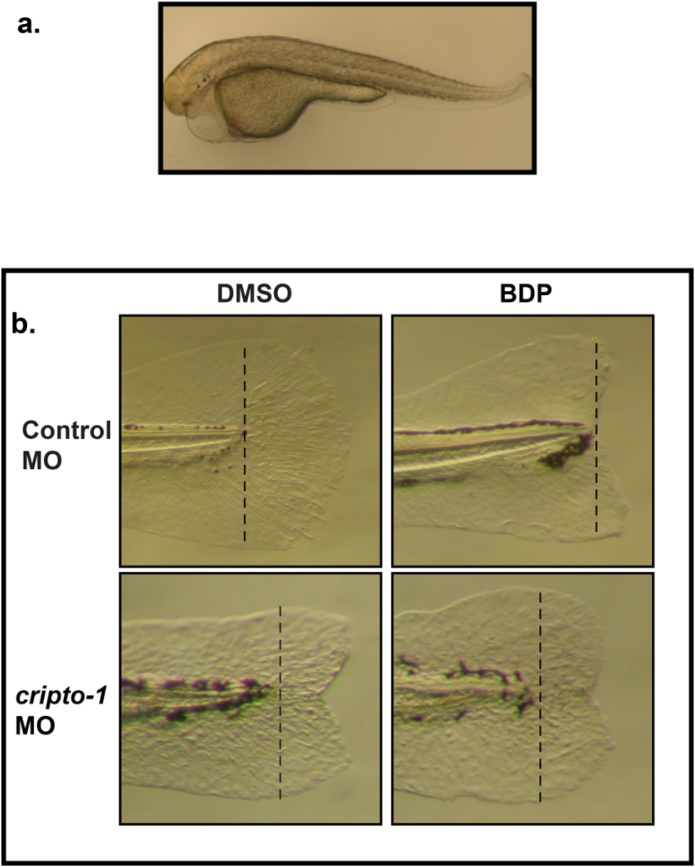
To determine the role of cripto-1 in regeneration, we utilized a cripto-1 translation blocking morpholino. When Cripto-1 expression was completely repressed, larvae exhibited the one eyed pinhead phenotype (Fig. 5a), and lethality by 5 dpf (days post fertilization) [18]. To avoid lethality, we injected 2–4 cell-staged embryos with reduced volumes of fluorescein tagged morpholino. The morphants were screened for uniform fluorescence at 24 hpf, and both control and cripto-1 morphants were exposed to vehicle and BDP following amputation at 2 dpf. The transient antisense repression allowed us to avoid lethality and show that the regenerative response was completely abrogated in control morphants exposed to BDP. cripto-1 morphants exposed only to vehicle were able to regenerate except for the presence of a characteristic notch on the fin (Fig. 5b). In nearly 80% of the cripto-1 morphants exposed to BDP, regeneration progressed similar to the untreated control larvae (Fig. 5b). BDP exposure did not block regeneration when Cripto-1 levels were repressed.
Fig. 5.
Partial antisense repression of Cripto-1 rescues inhibition of regeneration by BDP. a) Translation-blocking cripto-1 morpholino oligo (MO) transiently knocked down Cripto-1. The morphants developed the characteristic one-eyed pinhead phenotype by 2 days post-fertilization (dpf). b) The control and cripto-1 morphants were exposed to DMSO or beclomethasone dipropionate (BDP) at 2 dpf following amputation. The dotted lines mark the plane of amputation. Regenerative progression was evaluated and pictures were taken at 3 days post-amputation (dpa). In each replicated experiment approximately 80% of the cripto-1 morphants were resistant to inhibition of regeneration by BDP exposure.
3.5. Suppression of cripto-1 by retinoic acid rescues BDP-inhibited regeneration
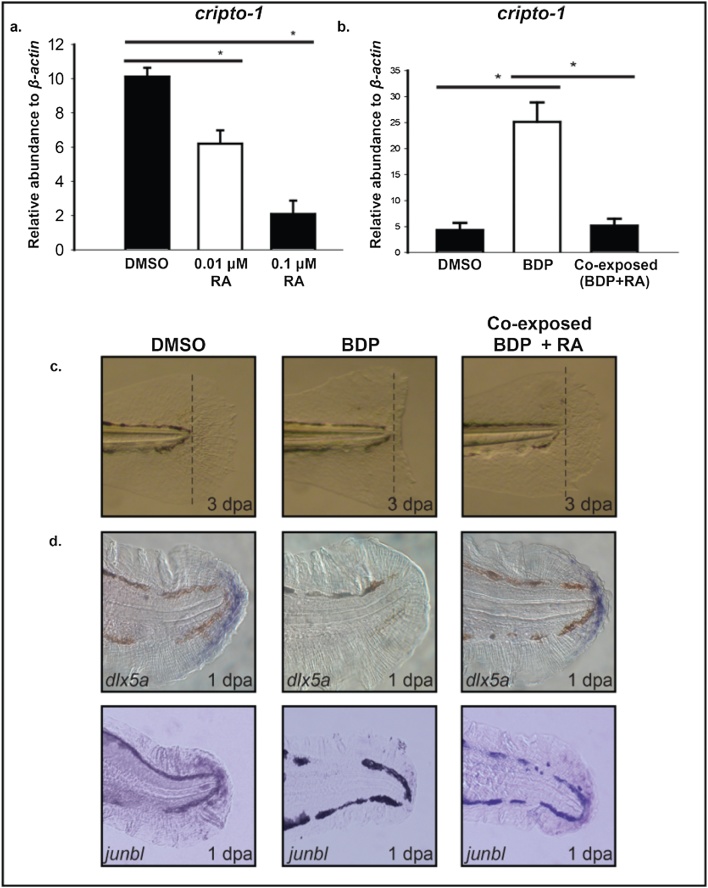
We used RA treatment to transiently suppress cripto-1 expression. Analysis of cripto-1 expression in whole embryos exposed to 0.1 and 0.01 μM RA at 1 dpa, revealed concentration-dependent suppression of cripto-1 expression (Fig. 6a). Co-exposure to BDP and 0.01 μM RA did not rescue regeneration. An extended exposure to RA > 0.01 μM inhibited regeneration (data not shown). Pre-exposure of 2 dpf larvae to 0.1 μM RA for 8 h prior to amputation, then co-exposure to BDP and 0.01 μM RA post-amputation was tested. RA pre-exposure alone did not affect regeneration, but RA pre-exposure followed by co-exposure to 0.01 μM RA during the BDP treatment rescued regeneration in approximately 75% of amputated larvae. RA exposure reduced BDP-induced cripto-1 expression down to the baseline level of vehicle control larvae (Fig. 6b). The presence of dlx5a and junbl in co-exposed larvae suggested normal regenerative signaling (Fig. 6c), and the animals completed fin regeneration similar to the controls (Fig. 6d).
Fig. 6.
RA exposure suppress cripto-1 expression in the fin tissue and rescues BDP impaired regeneration. a) Caudal fin of 2 days post-fertilization (dpf) embryos were amputated and exposed to DMSO or either 0.01 μM or 0.1 μM retinoic acid (RA). The abundance of cripto-1 was estimated in the regenerating fin tissues. The expression of cripto-1 was significantly reduced in a concentration-dependent manner compared to the control. The respective values represent the mean ± SEM and the asterisks indicate statistical significance (One-way ANOVA, n = 3, p < 0.05). b.) 2 dpf larvae were exposed to 0.1 μM RA for 8 h followed by amputation and co-exposure with 0.01 μM RA and DMSO or beclomethasone dipropionate (BDP). The abundance of cripto-1 was evaluated at 1 day post-amputation (dpa) in the regenerating fin tissue by qRT-PCR. Co-exposure with RA significantly suppressed cripto-1 expression compared to BDP exposure. The respective values represent the mean ± SEM and the asterisks indicate statistical significance (One-way ANOVA, n = 3). c). Amputated 2 dpf larvae pre-exposed with 0.1 μM RA were exposed to DMSO, BDP or co exposed with BDP and 0.01 μM RA. Regenerative progression was monitored and pictures were taken at 3 dpa. In each replicated experiment regeneration was restored in approximately 75% embryos co exposed to BDP and RA compared to BDP alone. d) In situ localization of dlx5a and junbl in the regenerating fin tissue at 1 dpa demonstrated restoration of regeneration markers in the caudal fin of larvae co-exposed with BDP and RA.
3.6. BDP induces cripto-1 expression in mouse embryonic stem cells
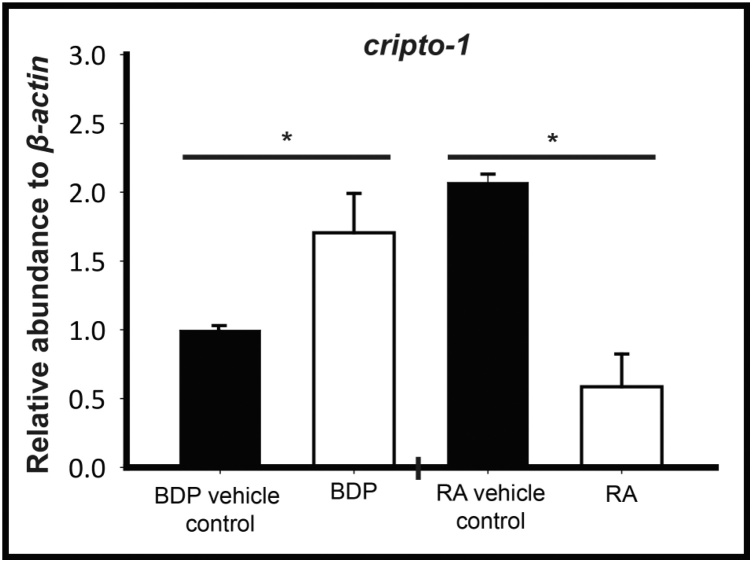
To understand whether the regulatory role of GR activation is conserved in other biological systems, we analyzed the expression of cripto-1 in mouse embryonic stem cells (eSCs). BDP exposure at 1 μM for 24 h induced cripto-1 expression in differentiating mouse eSCs, indicating that activated GR can modulate cripto-1 expression across species (Fig. 7). We also observed RA suppression of cripto-1 expression in mouse eSCs.
Fig. 7.
BDP induces Cripto-1 expression in the mouse ES cells. Embryoid bodies cultured from mouse D3 embryonic stem cells were exposed to beclomethasone dipropionate (BDP). The relative abundance of cripto-1 was quantified by qRT-PCR. Exposure to 1 μM BDP significantly induced while retinoic acid (RA) exposure suppressed cripto-1 expression. The respective values represent the mean ± SEM and the asterisks indicate statistical significance compared to the respective vehicle controls (One-way ANOVA on ranks, n = 3, p < 0.05).
To identify possible conserved transcriptional mechanisms of cripto-1 induction, we performed promoter analysis of the zebrafish, mouse, and human cripto-1 promoters 3000 bp upstream of the transcriptional start site. Using a list (Table S2) of differentially expressed transcription factors (TFs) from the microarray data plus the GR, we searched a database of predicted response elements in the zebrafish, mouse, and human promoter regions. Only the TFs having predicted response elements in all three species were considered for further analysis (Tables S3-S5). We found predicted repressive glucocorticoid response elements (GREs) in the cripto-1 promoter. We also identified potential binding sites for ISL1, HMX3, POU3F3, MYT1, MAFK, EN1, and MYBL2.
4. Discussion
Understanding tissue regeneration requires having the critical signaling targets in hand to experimentally manipulate and assemble a mechanism. In a microarray analysis, we found that exposure to beclomethasone dipropionate (BDP) severely inhibited zebrafish’s natural capacity for regeneration of amputated caudal fin tissue, and that the inhibition was closely accompanied by strong induction of the cripto-1 transcript. This induction following BDP exposure was also observed in a separate study [32]. Cripto-1 is an evolutionarily conserved regulator of cell function and proliferation [33]. Preventing cripto-1 induction with either retinoic acid (RA) exposure or antisense knockdown rescued fin regeneration in the presence of BDP. This demonstrated that induction of cripto-1 was required for BDP to block fin regeneration. The regenerative block and increased expression of cripto-1 were glucocorticoid receptor (GR)-dependent, indicating that modulation of cripto-1 expression occurred downstream of the GR. cripto-1 was induced when BDP exposure occurred within 4 h of fin amputation, correlating with the 4 h critical window previously established for BDP-mediated disruption of regeneration [6]. These results, along with previous work, demonstrate a novel link between the GR and cripto-1 induction, and that inappropriate activation of the GR blocks fin regeneration in a cripto-1-dependent manner. Importantly, we further demonstrated that glucocorticoid (GC)-mediated expression of cripto-1 is conserved in mammals.
During vertebrate development, Cripto-1 functions primarily as a required co-factor for Transforming growth factor beta (TGF-β) signaling, specifically for Nodal signaling [14]. During gastrulation, Cripto-1-dependent Nodal signaling at the primitive streak facilitates epithelial-to-mesenchymal transition (EMT) and regulates cell migration, allowing the embryo to form multiple tissue layers [34,35]. As an oncofetal gene, cripto-1 is expressed during development but largely absent in adult tissues except during tumorigenesis or carcinogenesis [33]. It is overexpressed in triple negative breast cancer, colon carcinoma, squamous cell carcinoma, lung adenocarcinoma among several others [[36], [37], [38], [39]]. Cripto-1 promotes tumor cell survival, proliferation, and migration by interacting with proteins that affect signaling pathways beyond TGF-β [40]. There is no evidence to our knowledge that cripto-1 expression is endogenously modulated by the GR during development or carcinogenesis.
We showed that cripto-1 expression increased in larval fin tissue if zebrafish were exposed to BDP within four hours of amputation. After 4 hpf, the fin tissue may undergo a change that no longer supports GR-dependent induction of cripto-1. Notably, the lack of increased dlx5a expression demonstrated that BDP exposure prevented formation of the apical epithelial cap (AEC), despite formation of the wound epithelium [6]. Several signaling pathways are active prior to and after AEC formation including Fibroblast growth factor (FGF), Wnt, RA, and Activin/Transforming growth factor beta (TGF-β) [3,41,42]. Previous work in adult zebrafish has shown that epimorphic fin regeneration requires TGF-β/Activin signaling within the first day of regeneration [3]. By inhibiting type I receptors using SB431541, we demonstrated that TGF-β/Activin signaling is also required during the early stages of larval fin regeneration.
Given our results and Cripto-1’s known role of antagonizing Activin, we conjectured that increased expression of cripto-1 following BDP exposure prevents fin regeneration by a similar mechanism. The identical expression patterns of AEC and blastema markers in the fin tissue exposed to BDP and SB431542 suggested disrupted Activin signaling in BDP-exposed larvae. Since Cripto-1 suppressed Activin signaling in multiple cell types by binding Activin ligands at the protein level [15,16,[43], [44], [45]], we expect that inhibition of Activin signaling has no effect on cripto-1 transcription. This was demonstrated to be the case in our model, indicating that cripto-1 is upstream of Activin signaling.
Cripto-1 is a pleiotropic protein that could also interfere with other signaling pathways including Wnt (canonical and planar cell polarity pathways) [19,46], c-Src [47], and Notch [48]. Graphical summaries of these interactions can be found in reviews such as Strizzi et al. (2005) [19], Nagaoka et al. (2012) [49], and Klauzinska et al. (2014) [40]. Additionally, a recent proteomic analysis of the Cripto-1 interactome identified 51 Cripto-1 binding proteins from human epithelial cells including regulators of extracellular exosomes, myosin II complexes, and the cytoskeleton. The myosin II activity was shown to regulate subcellular localization of Cripto-1 in epithelial and mesenchymal stem cell populations and to function cooperatively with zebrafish Cripto-1 to promote caudal fin regeneration [50]. Although we were unable to localize cripto-1 expression in the fin, these new data suggest that Cripto-1 may be expressed in both epithelial and mesenchymal cells following BDP exposure. Furthermore, their study identified an intrinsic role for increased cripto-1 expression during the outgrowth phase of regeneration, which may explain why oep morphants had notched fins following regeneration. The impacts of increased cripto-1 expression during the initiation of fin regeneration appear quite complex.
Since RA co-exposure rescues fin regeneration, and raldh2 is not detectable by ISH at 1 dpa following BDP exposure, Cripto-1 likely acts upstream of raldh2 induction and normal RA signaling [5]. We did not observe any chemical precipitation during BDP and RA co-exposures. Since RA concentrations greater than 0.01 μM during the pre-exposure did not result in a higher incidence of regeneration, the uptake of BDP by the animals was likely not affected by the presence of RA in the media. In human teratocarcinoma cells, the addition of exogenous RA to cell culture induces germ cell nuclear factor which directly represses cripto-1 expression [51]. It is possible that this mechanism occurs in the regenerating zebrafish fin during BDP and RA co-exposure. However, RA co-exposure also rescues regeneration when fish are exposed to FGF and ERK1/2 signaling inhibitors [5]. Its rescuing effect on regeneration might therefore not be specific for its effect on cripto-1 induction. Another possibility is that RA co-exposure could be “overriding” Cripto-1’s effect on upstream pathways by stimulating downstream signaling events mediated by RA.
We show conclusively for the first time that activation of the GR by BDP (and other specific GCs) is sufficient to increase cripto-1 expression to levels that exert physiological consequence in vivo. cripto-1 induction would seem an important conserved signaling step between taxa. We note that differences have been observed in xenobiotic responses between murine and non-human primate eSCs [52], indicating that further work is needed to demonstrate a link between cripto-1 induction and the GR in humans. If this response is conserved in humans, it would have implications for therapeutic GC treatment whether for immunosuppression or cancer treatment. Modulation of Cripto-1 expression via the GR could be useful in the laboratory setting. For example, cripto-1 induction promotes cardiomyogenesis whereas suppression promotes neural differentiation [53,54]. Certain GCs may be useful for directing cell fate commitment in vitro. Considering the importance of Cripto-1 in ES cells, development, cancer, and regenerative medicine, GCs have the potential to offer novel modes of manipulating Cripto-1 for therapeutic benefit.
Funding
This work was supported by the National Institute of Environmental Health SciencesT32 ES07060 and National Science Foundation grant #0641409.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgements
We thank Michael Simonich for his manuscript edits, Anne-Marie Girard (Center for Gene Research and Biocomputing Oregon State University) for her valuable assistance in performing the microarray study, and the Staff of the Sinnhuber Aquatic Research Laboratory for their technical assistance.
References
- 1.Tal T.L., Franzosa J.A., Tanguay R.L. Molecular signaling networks that choreograph epimorphic fin regeneration in zebrafish – a mini-review. Gerontology. 2010;56(2):231–240. doi: 10.1159/000259327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iovine M.K. Conserved mechanisms regulate outgrowth in zebrafish fins. Nat. Chem. Biol. 2007;3(10):613–618. doi: 10.1038/nchembio.2007.36. [DOI] [PubMed] [Google Scholar]
- 3.Jazwinska A., Badakov R., Keating M.T. Activin-betaA signaling is required for zebrafish fin regeneration. Curr. Biol. 2007;17(16):1390–1395. doi: 10.1016/j.cub.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 4.Kawakami A., Fukazawa T., Takeda H. Early fin primordia of zebrafish larvae regenerate by a similar growth control mechanism with adult regeneration. Dev. Dyn. 2004;231(4):693–699. doi: 10.1002/dvdy.20181. [DOI] [PubMed] [Google Scholar]
- 5.Mathew L.K. Comparative expression profiling reveals an essential role for raldh2 in epimorphic regeneration. J. Biol. Chem. 2009;284(48):33642–33653. doi: 10.1074/jbc.M109.011668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mathew L.K. Unraveling tissue regeneration pathways using chemical genetics. J. Biol. Chem. 2007;282(48):35202–35210. doi: 10.1074/jbc.M706640200. [DOI] [PubMed] [Google Scholar]
- 7.Mathew L.K. Crosstalk between AHR and Wnt signaling through R-Spondin1 impairs tissue regeneration in zebrafish. FASEB J. 2008;22(8):3087–3096. doi: 10.1096/fj.08-109009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weikum E.R. Glucocorticoid receptor control of transcription: precision and plasticity via allostery. Nat. Rev. Mol. Cell Biol. 2017;18(3):159–174. doi: 10.1038/nrm.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dallman M.F. Feast and famine: critical role of glucocorticoids with insulin in daily energy flow. Front. Neuroendocrinol. 1993;14(4):303–347. doi: 10.1006/frne.1993.1010. [DOI] [PubMed] [Google Scholar]
- 10.Cain D.W., Cidlowski J.A. Immune regulation by glucocorticoids. Nat. Rev. Immunol. 2017;17(4):233–247. doi: 10.1038/nri.2017.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bird A.D. Glucocorticoid regulation of lung development: lessons learned from conditional GR knockout mice. Mol. Endocrinol. 2015;29(2):158–171. doi: 10.1210/me.2014-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jozic I. Stress signals, mediated by membranous glucocorticoid receptor, activate PLC/PKC/GSK-3beta/beta-catenin pathway to inhibit wound closure. J. Invest. Dermatol. 2017;137(5):1144–1154. doi: 10.1016/j.jid.2016.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sengupta S. Alternate glucocorticoid receptor ligand binding structures influence outcomes in an in vivo tissue regeneration model. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2012;156(2):121–129. doi: 10.1016/j.cbpc.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schier A.F. Nodal morphogens. Cold Spring Harb. Perspect. Biol. 2009;1(5):a003459. doi: 10.1101/cshperspect.a003459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gray P.C., Harrison C.A., Vale W. Cripto forms a complex with activin and type II activin receptors and can block activin signaling. Proc. Natl. Acad. Sci. U. S. A. 2003;100(9):5193–5198. doi: 10.1073/pnas.0531290100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray P.C. Cripto binds transforming growth factor beta (TGF-beta) and inhibits TGF-beta signaling. Mol. Cell. Biol. 2006;26(24):9268–9278. doi: 10.1128/MCB.01168-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gritsman K. The EGF-CFC protein one-eyed pinhead is essential for nodal signaling. Cell. 1999;97(1):121–132. doi: 10.1016/s0092-8674(00)80720-5. [DOI] [PubMed] [Google Scholar]
- 18.Feldman B., Stemple D.L. Morpholino phenocopies of sqt, oep, and ntl mutations. Genesis. 2001;30(3):175–177. doi: 10.1002/gene.1058. [DOI] [PubMed] [Google Scholar]
- 19.Strizzi L. Cripto-1: a multifunctional modulator during embryogenesis and oncogenesis. Oncogene. 2005;24(37):5731–5741. doi: 10.1038/sj.onc.1208918. [DOI] [PubMed] [Google Scholar]
- 20.Ciccodicola A. Molecular characterization of a gene of the’ EGF family’ expressed in undifferentiated human NTERA2 teratocarcinoma cells. EMBO J. 1989;8(7):1987–1991. doi: 10.1002/j.1460-2075.1989.tb03605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Westerfield M. 4th ed. University of Oregon; Eugene, OR: 2000. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio) [Google Scholar]
- 22.Mathew L.K., Andreasen E.A., Tanguay R.L. Aryl hydrocarbon receptor activation inhibits regenerative growth. Mol. Pharmacol. 2006;69(1):257–265. doi: 10.1124/mol.105.018044. [DOI] [PubMed] [Google Scholar]
- 23.Poss K.D. Mps1 defines a proximal blastemal proliferative compartment essential for zebrafish fin regeneration. Development. 2002;129(22):5141–5149. doi: 10.1242/dev.129.22.5141. [DOI] [PubMed] [Google Scholar]
- 24.Genschow E. Validation of the embryonic stem cell test in the international ECVAM validation study on three in vitro embryotoxicity tests. Altern. Lab. Anim. 2004;32(3):209–244. doi: 10.1177/026119290403200305. [DOI] [PubMed] [Google Scholar]
- 25.Cartharius K. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21(13):2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 26.Wang J.C. Chromatin immunoprecipitation (ChIP) scanning identifies primary glucocorticoid receptor target genes. Proc. Natl. Acad. Sci. U. S. A. 2004;101(44):15603–15608. doi: 10.1073/pnas.0407008101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phuc Le P. Glucocorticoid receptor-dependent gene regulatory networks. PLoS Genet. 2005;1(2):e16. doi: 10.1371/journal.pgen.0010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Callahan J.F. Identification of novel inhibitors of the transforming growth factor beta 1 (TGF-beta 1) type 1 receptor (ALK5) J. Med. Chem. 2002;45(5):999–1001. doi: 10.1021/jm010493y. [DOI] [PubMed] [Google Scholar]
- 29.Laping N.J. Inhibition of transforming growth factor (TGF)-beta 1-induced extracellular matrix with a novel inhibitor of the TGF-beta type I receptor kinase activity: SB-431542. Mol. Pharmacol. 2002;62(1):58–64. doi: 10.1124/mol.62.1.58. [DOI] [PubMed] [Google Scholar]
- 30.Inman G.J. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 2002;62(1):65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- 31.Schebesta M. Transcriptional profiling of caudal fin regeneration in zebrafish. Transfus. Apher. Sci. 2006;6:38–54. doi: 10.1100/tsw.2006.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chatzopoulou A. Glucocorticoid-induced attenuation of the inflammatory response in zebrafish. Endocrinology. 2016;157(7):2772–2784. doi: 10.1210/en.2015-2050. [DOI] [PubMed] [Google Scholar]
- 33.Bianco C. Role of Cripto-1 in stem cell maintenance and malignant progression. Am. J. Pathol. 2010;177(2):532–540. doi: 10.2353/ajpath.2010.100102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rangel M.C. Role of Cripto-1 during epithelial-to-mesenchymal transition in development and cancer. Am. J. Pathol. 2012;180(6):2188–2200. doi: 10.1016/j.ajpath.2012.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou X. Nodal is a novel TGF-β-like gene expressed in the mouse node during gastrulation. Nature. 1993;361(6412):543. doi: 10.1038/361543a0. [DOI] [PubMed] [Google Scholar]
- 36.Rangel M.C. Developmental signaling pathways regulating mammary stem cells and contributing to the etiology of triple-negative breast cancer. Breast Cancer Res. Treat. 2016;156(2):211–226. doi: 10.1007/s10549-016-3746-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bianco C. Identification of Cripto-1 as a novel serologic marker for breast and colon cancer. Clin. Cancer Res. 2006;12(17):5158–5164. doi: 10.1158/1078-0432.CCR-06-0274. [DOI] [PubMed] [Google Scholar]
- 38.Yoon H.J. The role of Cripto-1 in the tumorigenesis and progression of oral squamous cell carcinoma. Oral Oncol. 2011;47(11):1023–1031. doi: 10.1016/j.oraloncology.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 39.Zhang H. Clinical significance of cripto-1 expression in lung adenocarcinoma. Oncotarget. 2017;8(45):79087–79098. doi: 10.18632/oncotarget.15761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klauzinska M. The multifaceted role of the embryonic gene Cripto-1 in cancer, stem cells and epithelial-mesenchymal transition. Semin. Cancer Biol. 2014;29:51–58. doi: 10.1016/j.semcancer.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poss K.D. Roles for Fgf signaling during zebrafish fin regeneration. Dev. Biol. (Basel) 2000;222(2):347–358. doi: 10.1006/dbio.2000.9722. [DOI] [PubMed] [Google Scholar]
- 42.Kawakami Y. Wnt/beta-catenin signaling regulates vertebrate limb regeneration. Genes Dev. 2006;20(23):3232–3237. doi: 10.1101/gad.1475106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shani G. GRP78 and cripto form a complex at the cell surface and collaborate to inhibit transforming growth factor beta signaling and enhance cell growth. Mol. Cell. Biol. 2008;28(2):666–677. doi: 10.1128/MCB.01716-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeo C., Whitman M. Nodal signals to Smads through Cripto-dependent and Cripto-independent mechanisms. Mol. Cell. 2001;7(5):949–957. doi: 10.1016/s1097-2765(01)00249-0. [DOI] [PubMed] [Google Scholar]
- 45.Kelber J.A. Cripto is a noncompetitive activin antagonist that forms analogous signaling complexes with activin and nodal. J. Biol. Chem. 2008;283(8):4490–4500. doi: 10.1074/jbc.M704960200. [DOI] [PubMed] [Google Scholar]
- 46.Nagaoka T. Cripto-1 enhances the canonical Wnt/β-catenin signaling pathway by binding to LRP5 and LRP6 co-receptors. Cell. Signal. 2013;25(1):178–189. doi: 10.1016/j.cellsig.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bianco C. A Nodal- and ALK4-independent signaling pathway activated by Cripto-1 through Glypican-1 and c-Src. Cancer Res. 2003;63(6):1192–1197. [PubMed] [Google Scholar]
- 48.Watanabe K. Enhancement of Notch receptor maturation and signaling sensitivity by Cripto-1. J. Cell Biol. 2009;187(3):343–353. doi: 10.1083/jcb.200905105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nagaoka T. An evolving web of signaling networks regulated by Cripto-1. Growth Factors. 2012;30(1):13–21. doi: 10.3109/08977194.2011.641962. [DOI] [PubMed] [Google Scholar]
- 50.Hoover M. Identification of myosin II as a cripto binding protein and regulator of cripto function in stem cells and tissue regeneration. Biochem. Biophys. Res. Commun. 2019;509(1):69–75. doi: 10.1016/j.bbrc.2018.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hentschke M. Germ cell nuclear factor is a repressor of CRIPTO-1 and CRIPTO-3. J. Biol. Chem. 2006;281(44):33497–33504. doi: 10.1074/jbc.M606975200. [DOI] [PubMed] [Google Scholar]
- 52.Walker L. Non-human primate and rodent embryonic stem cells are differentially sensitive to embryotoxic compounds. Toxicol. Rep. 2015;2:165–174. doi: 10.1016/j.toxrep.2014.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu C. Specific arrest of cardiogenesis in cultured embryonic stem cells lacking Cripto-1. Dev. Biol. 1998;196(2):237–247. doi: 10.1006/dbio.1998.8862. [DOI] [PubMed] [Google Scholar]
- 54.Parisi S. Nodal-dependent Cripto signaling promotes cardiomyogenesis and redirects the neural fate of embryonic stem cells. J. Cell Biol. 2003;163(2):303–314. doi: 10.1083/jcb.200303010. [DOI] [PMC free article] [PubMed] [Google Scholar]