Significance
Plague is a devastating infectious disease that has caused three pandemics during the last millennia. Today, plague still causes sporadic cases every year and even some outbreaks. In this paper, we analyze how factors associated with climate change and globalization affect the spread of plague worldwide. Such information is important with respect to global disease prevention and control. For this purpose, we first constructed a global plague database of the Third Pandemic, and then analyzed the association of spatiotemporal environmental factors with spreading velocity. Our results provide insight into the global transmission and suggest strategies for preventing plague transmission under accelerated global change.
Keywords: Yersinia pestis, Third Pandemic, climate change, global transmission velocity, historical and genomic data
Abstract
Quantitative knowledge about which natural and anthropogenic factors influence the global spread of plague remains sparse. We estimated the worldwide spreading velocity of plague during the Third Pandemic, using more than 200 years of extensive human plague case records and genomic data, and analyzed the association of spatiotemporal environmental factors with spreading velocity. Here, we show that two lineages, 2.MED and 1.ORI3, spread significantly faster than others, possibly reflecting differences among strains in transmission mechanisms and virulence. Plague spread fastest in regions with low population density and high proportion of pasture- or forestland, findings that should be taken into account for effective plague monitoring and control. Temperature exhibited a nonlinear, U-shaped association with spread speed, with a minimum around 20 °C, while precipitation showed a positive association. Our results suggest that global warming may accelerate plague spread in warm, tropical regions and that the projected increased precipitation in the Northern Hemisphere may increase plague spread in relevant regions.
Plague is widely recognized as one of the greatest biological–environmental disasters in human history, having caused over 200 million deaths, greatly shaping the societies affected (1). There have been three pandemics of plague: the First Pandemic (sixth to seventh centuries, including the Justinian Plague of 541 to 542), the Second Pandemic (14th to 19th centuries, which started with the Black Death in 1348 to 1353), and the Third Pandemic (2). The Third Pandemic started in 1772 in Yunnan province, southwest China, and spread to other parts of the world from Hong Kong in 1894 (3). During the outbreak in Hong Kong, Alexandre Yersin discovered the plague bacillus Yersinia pestis (4). Currently, plague represents a significant threat, as it is considered a reemerging disease under accelerated climate change and globalization. The most recent outbreak of pneumonic plague occurred in Madagascar in 2017, with a total of 2348 suspected plague cases reported to the World Health Organization (WHO) and with a high risk of spread to the neighboring islands of the Indian Ocean and to other parts of the world (5). This recent outbreak underscores the urgency of understanding the natural and anthropogenic factors that influence the spread of plague—knowledge that can be gained by retrospective analysis of the transmission patterns of plague in the past.
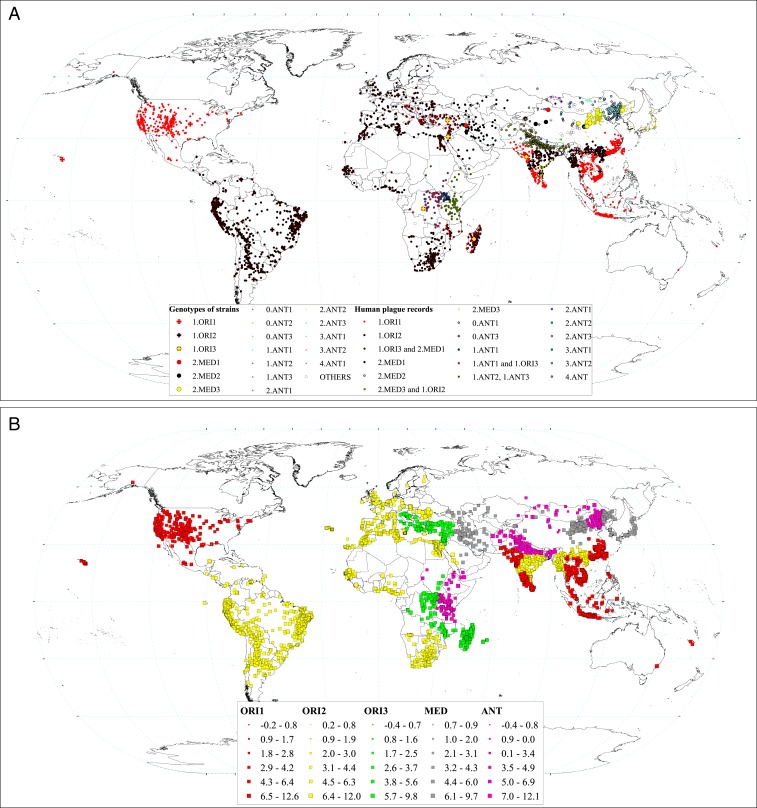
Plague is a zoonotic disease, with rodents and fleas playing key roles as major hosts and vectors in its transmission (6). Populations of Y. pestis, which are spatially separated depending on the spatial distribution and population dynamics of animal hosts, have led to the sustained prevalence of human plague from 1772 to 2014 (Fig. 1). Previous classifications of Y. pestis subdivided different populations into biovars (Antiqua, Medievalis, Orientalis, and Pestoides) based on their phenotypic characteristics (Pestoides is also designated as Microtus and has rarely infected humans) (7, 8). The prevalent forms assumed to cause the Justinian Plague, Black Death, and Third Pandemic were Antiqua, Medievalis, and Orientalis, respectively. However, this hypothesis has been challenged by recent molecular studies (2). Regardless, all known biovars were found in hosts (including humans) and vectors during the period of the Third Pandemic (7, 9). According to recent studies, which reconstructed the phylogeny of Y. pestis from the whole genomes of modern strains, the different populations can be classified into 19 subbranches (7). With reference to this phylogenetic subdivision, in this study’s analyses, we used only the most prevalent 15 subbranches (hereafter referred to as genotypes): 1.ORI1, 1.ORI2, 1.ORI3, 2.MED1, 2.MED2, 2.MED3, 0.ANT1, 0.ANT3, 1.ANT1, 1.ANT2, 1.ANT3, 3.ANT1, 3.ANT2, 3.ANT3, and 4.ANT (Fig. 1).
Fig. 1.
(A) Spatial distribution of plague spread during the Third Pandemic. Small points indicate locations with human plague appearance, with colors indicating suspected plague genotype. The suspected genotypes are listed in the legend. We identified the suspected genotype by matching with the nearest surveillance site with genotype data based on samples from patients, hosts, and/or vectors. Surveillance sites are shown by large points, circles, and triangles (see legend: Genotypes of strains). (B) Estimated spreading velocity of plague by using the g-NNA. Colors indicate 1.ORI.1, 1.ORI.2, 1.ORI.3, ANT, and 2.MED strains. The sizes of the symbols show the spreading velocity (kilometer per year in ln-transformed scale).
During the Third Pandemic, plague spread rapidly and caused outbreaks in many regions worldwide (10, 11) (SI Appendix, Fig. S1). Plague occurrences of the Third Pandemic were well recorded. However, spatial transmission and the influence of environmental and anthropogenic factors have been investigated in only a few countries (3, 12–14). Global plague transmission pattern has been previously proposed only on the basis of incomplete records (7, 9, 15, 16). Factors influencing global spatial transmission of plague have not been investigated. The increasing amount of genetic data available on Y. pestis populations (7–9, 17, 18) now makes it possible to evaluate the effects of environmental factors on the global transmission velocity of plague during the Third Pandemic, by integrating genetic data with historical records.
Estimation of Spread Velocity
The global spread of Y. pestis was reconstructed by assuming that the most likely origin of plague introduction into a new location was the geographically nearest location with a previous record of plague of the same genotype [genomic-constrained nearest neighbor approach (g-NNA), a reconstruction method that built upon our previous study (3); see Materials and Methods]. The transmission of 1.ORI.1 and 1.ORI.2 started in China. The transmission of 1.ORI3 started in India, and transmission of MED and ANT started in many parts of the world independently. The estimated spread velocity of all sites is shown in Fig. 1B. The median plague spread velocity was estimated to be 6.99 km⋅y−1 for all plague genotypes, 5.65 km⋅y−1 for 1.ORI1, 6.04 km⋅y−1 for 1.ORI2, 5.92 km⋅y−1 for 1.ORI3, 8.64 km⋅y−1 for ANT, and 41.04 km⋅y−1 for 2.MED. Note that these values do not necessarily reflect effect of genotype per se, as the genotypes occurred in different environments with potentially different conditions for plague transmission. We analyzed the spread velocity data using generalized additive models (GAMs; see Material and Methods) (19) that statistically accounted for spatiotemporal environmental influences, and found that 2.MED and 1.ORI3 spread significantly faster than the other genotypes, while 1.ORI2 spread significantly slower than others (Fig. 2G).
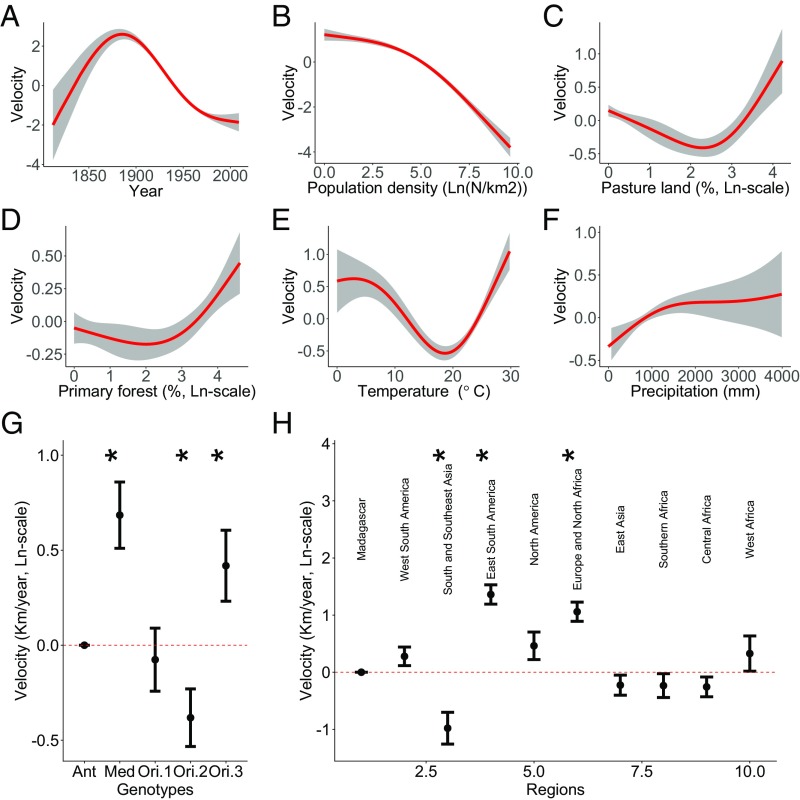
Fig. 2.
Effects of different environmental variables on velocity of plague spread based on results of the final model selected (SI Appendix, Table S1). (A–F) Partial effects of continuous predictor variables on plague spread velocity [ln(km⋅y−1)]. Gray bands are 95% nominal confidence bands. Red lines are statistically significant (P < 0.05). (G) Partial effects (±1 SE) of the categorical variable Genotypes, with ANT as reference level. (H) Partial effects of the categorical variable Regions, with Madagascar as reference level. Asterisks mark the significant genotypes and regions (P < 0.05) in G and H.
Identification of Influencing Factors on Spread Velocity
The GAM with the best combination of high out-of-sample predictive power and low model complexity (model 19, SI Appendix, Table S1) revealed that population density showed a significant negative association with spread velocity (Fig. 2B). Percentage of pastureland (Fig. 2C) and primary forest (Fig. 2D) showed nonlinear associations with spread velocity. Annual precipitation showed a significant positive association with spread velocity (Fig. 2F). Annual air temperature showed a nonlinear, U-shaped association with spread velocity, with a minimum around 20 °C (Fig. 2E). Finally, while accounting for these environmental influences, mean spread velocity differed over time, with a maximum around year 1900 (Fig. 2A) and between regions (Fig. 2H). The estimation of population and temperature effects appeared to be robust to whichever variables were included in the model (e.g., omitting year and region); effects of pastureland, primary forest, and precipitation were generally consistent among models that included time (year) and region effects (SI Appendix, Fig. S2).
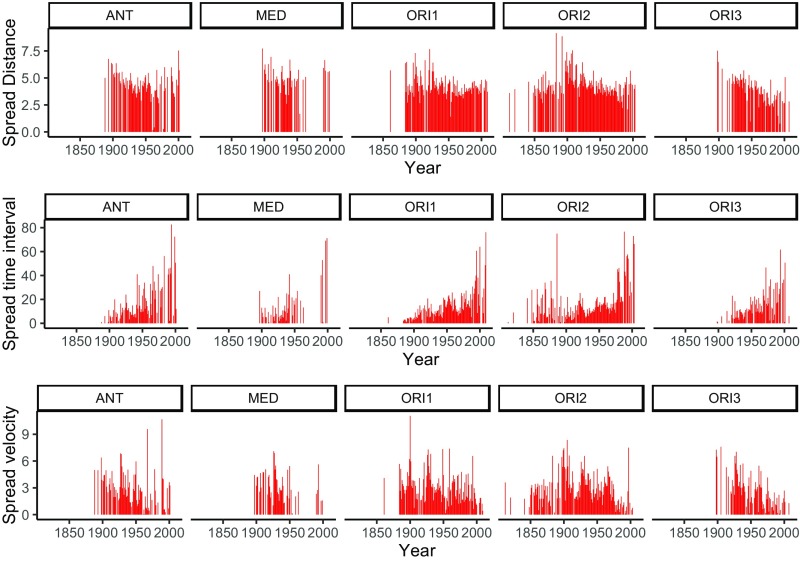
For the majority of plague genotypes, we found that the spread velocity of plague tended to decrease in time due to a decrease in spread distance and an increase in the interval between spreading events (Fig. 3). Overall, the median spread distance between two sites was 41.4 km, and the median time interval was 5.08 y. The spread velocity was strongly positively correlated with spread distance (SI Appendix, Fig. S3), meaning that long-distance spread was generally faster than short-distance links. The correlations between spread speed or distance and population density were negative (SI Appendix, Fig. S3), suggesting that plague spread slowly over shorter distances in densely populated places.
Fig. 3.
Yearly time series of spread distance (ln[km + 1], with 1 added before ln-transformation for presentational reasons), spread time interval (years), and spread velocity (ln[km·y-1 + 1]) between two connecting sites for each genotype.
Our results further suggested that in areas and/or periods with annual mean air temperatures around 20 °C, the speed of plague transmission is reduced compared with at either lower or higher temperatures (Fig. 2E). A more complex model with similarly high predictive power as that shown in Fig. 2 (model 24 in SI Appendix, Fig. S4 and Table S1) suggested that the nonlinear effect of temperature (as well as vegetation, see SI Appendix, Table S1) might be region specific; associations between temperature and transmission velocity were predominantly negative in regions colder than 20 °C and predominantly positive in regions warmer than 20 °C, but with regional differences in the strengths of these associations.
Discussion
Our study evaluated the impacts of environmental factors on spread velocity of plague during the Third Pandemic globally. We found that two strains (2.MED and 1.ORI.3) spread significantly faster than the other strains. Plague spread faster in less-populated regions than in densely populated ones. Temperature and vegetation showed region-dependent nonlinear effects on spread velocity, while precipitation showed an overall positive effect. Our results provide insight into the influencing factors in affecting global transmission velocity of plague, with implications for plague prevention and control.
The finding of the negative relationship between spread velocity and population density is unexpected. It is widely thought that disease would spread faster in densely populated areas because frequent contact between people and goods would facilitate disease transmission. One plausible explanation may be that, in densely populated regions (developed regions), human plague control interventions might be better than those in sparsely populated remote areas (undeveloped regions). Another explanation for this difference may be that plague in low-density areas is more frequently transmitted over long distances by trade or travel by infected humans, rather than short-distance house-to-house or neighborhood-to-neighborhood spread within villages or cities by rat–flea transmission. Our study suggests that although the number of human plague cases may be higher in densely populated rather than sparsely populated places, the spreading velocity is lower. The underlying mechanism needs to be further investigated. In addition, we found that the spread velocity of plague tended to decrease in time due to a decrease in spread distance and an increase in the interval between spreading events (Fig. 3), suggesting that increased human intervention, prevention, and medical treatment capacity contributed to a decrease in plague dissemination around the world.
The difference of spread velocity of different plague strains has not been evaluated in the previous literature. We found that 2.MED had the highest spread velocity; hypothetically, this is due to a higher proportion of the pneumonic form of plague (20, 21). The spread velocity of 1.ORI.3 was significantly higher than that of 1.ORI1 and 1.ORI2, suggesting that virulence of the 1.ORI genotype was enhanced during the evolutionary process.
Previous studies have indicated contrasting, and partly inconsistent, associations between spread velocity of plague and precipitation (3, 12). Differences may partly be caused by different regions and the temporal or spatial scales used in different studies (22). In this global-scale study, we found that high precipitation in infected locations showed a positive association with spread velocity of plague; it is likely that increased precipitation generally increased primary productivity, leading to increased rodent and, consequently, flea density and to increased plague transmission—a so-called trophic cascade (23, 24).
We speculate that the complex nonlinear association between temperature and plague spread is caused by the multiple effects that temperature has on the pathogen, vectors, hosts, and human behavior. For example, in the United States, percent flea transmission efficiencies for flea cohorts maintained at 10 and 15 °C also were higher than those for fleas held at 23 °C (25, 26), which is consistent with the findings that plague spread velocity decreased with temperatures in this temperature range (Fig. 2E) and within relatively cold regions, including North America (SI Appendix, Fig. S4J, although not statistically significant here), Europe and North Africa (SI Appendix, Fig. S4K), and central Africa (SI Appendix, Fig. S4N). In Madagascar, the developmental rate of two predominant rat flea species (Synopsyllus fonquerniei and Xenopsylla cheopis) were positively correlated to temperature, with large differences occurring around 21 °C (27), which is consistent with the findings that plague spread velocity increased with temperatures in this temperature range (Fig. 2E) and within relatively warm regions, including Madagascar (SI Appendix, Fig. S4F, although not statistically significant), south and Southeast Asia (SI Appendix, Fig. S4H), eastern Asia (SI Appendix, Fig. S4L), southern Africa (SI Appendix, Fig. S4M), and western Africa (SI Appendix, Fig. S4O). These observations suggest that temperature had a region-specific nonlinear effect on flea transmission efficiency.
Similarly to the effects of temperature, our results also suggested nonlinear associations of coverage of pastureland and forestland with spread velocity (Fig. 2 C and D, but see SI Appendix, Fig. S2 showing differences across model formulations in these effects). The selected model suggested an elevated risk of plague transmission when coverage of pasture or forest was high. This was possibly due to abundant populations of plague hosts and vectors in these habitats compared with cropland or urban areas (28) or to the areas having poor public health systems. Transportation has been found to facilitate plague transmission (3). The observed higher spread velocity of plague in Europe, North Africa, and eastern South America (Fig. 2H) was likely related to the more advanced road or water transportation systems in these regions, compared with the lower spread velocity in south and Southeast Asia (Fig. 2H).
Earth is facing accelerated climate warming and globalization, which may impose challenges in plague prevention and control. Our estimated median spread distance of plague between two sites was around 40 km, which could be a referenced distance for taking measures toward isolation and monitoring. Our climate is becoming warmer, thus it may accelerate plague spread (via flea transmission efficiency) in warm regions (e.g., Madagascar) but slow the spread in cold regions. Since climate warming would increase precipitation in the Northern Hemisphere (29), it may increase the risk of plague transmission (via benefiting hosts) in these regions. We should take into account the faster spread of plague in regions with low population density and a high proportion of pasture- or forestland compared with urban areas. Our estimated median time interval was around 5 y, which appears to be the typical period for plague to invade a new place. This suggests that plague would take some time to adapt to new environments. Therefore, extensive monitoring and control of rodents and fleas in newly invaded places is important for limiting plague occurrences. We call for more preventive monitoring and control of plague under accelerated global changes.
Materials and Methods
Data.
Human plague cases.
Data on human plague cases were mainly extracted from the WHO’s weekly reports from 1926 to 2002 (also including some earlier records) (https://www.who.int/wer/en/). Human plague cases of China from 1772 to 1964 were obtained from the Institute of Epidemiology and Microbiology, Chinese Academy of Medical Sciences (30) as described and analyzed by Xu et al. (3). Human plague data from the United States since 1900 were edited from the US Centers for Disease Control and Prevention (CDC) (12, 31). Earlier plague records from the United States were obtained from other literature [plague appeared in San Francisco in 1896 (10) and in Hawaii in 1899 (32, 33)]. Human plague data from Latin America from 1899 to 1936 were extracted from Pollitzer and WHO (10). Reported cases and deaths of plague in Europe from 1891 to 1950 were collected from the Morbidity and Mortality Weekly Reports of the CDC. Human plague cases from 1887 to 2008 in Africa were edited from Neerinckx et al. (34); these data, covering 26 countries in total, were compiled from published literature, WHO reports, and unpublished information obtained from African plague experts and national health authorities. Human plague data from Sydney from 1899 and 1900 were extracted from Turner (35) and Robinson (36). The spatial and temporal distributions of the final edited data for the Third Pandemic period are shown in Figs. 1 and 3.
Y. pestis genotypes.
Y. pestis genotypic data were edited from the literature (7–9, 37–39). Y. pestis strains can be classified into three main genovariants: 1.ORI, 2.MED, and ANT (9). The numbers indicate the branch on which the strains were placed on a phylogenetic tree. ANT populations are scattered on all of the five identified branches, from 0 to 4. In our study, we used data from three subtypes of 1.ORI (genotypes 1.ORI.1, 1.ORI.2, and 1.ORI.3), from three subtypes of 2.MED (2.MED1, 2.MED2, and 2.MED3), and from nine subtypes of ANT (0.ANT1, 0.ANT3, 1.ANT1, 1.ANT2, 1.ANT3, 3.ANT1, 3.ANT2, 3.ANT3, and 4.ANT); all genotypes were kept separated for the reconstruction of paths of spread. However, the genotypes of MED and ANT were grouped in the statistical analyses of spatial clusters and predictor effects on spread velocity (for parsimony). The classification of plague genotype was determined based on the multilocus variable-number tandem repeat analysis method, including genotype data from China (8), Mongolia (37, 38), India (39), and other parts of the world (7, 9). The genetic data of Y. pestis were mainly from samples of rodents and other nonhuman hosts, vectors, or accidental hosts of plague, while some genetic data were from patients. Previous studies reported that the genotype of Y. pestis from patients was highly related to other sources of the same region, suggesting that plague was a natural zoonosis that spilled over from animal reservoirs to humans (7, 8). Thus, we assigned to each reported human case the Y. pestis genotype of the geographically nearest strain found in patients, hosts, or vectors (Fig. 1). This assumption may cause biases if the genotype of the focal site was different from that of its nearest site; this can only be improved with more genomic data. In some sites, there were several genotypes of Y. pestis reported. We assigned these sites with multiple genotypes as reported, assuming that they might be infected by one of these suspected genotypes.
Historical human population density and pastureland coverage.
Historical data on human population density and pastureland coverage during our study period (1772 to 2014 AD) were obtained from the History Database of the Global Environment (https://themasites.pbl.nl/tridion/en/themasites/hyde/) (40). We assigned these data into our human plague case dataset (Dataset S1) by using both the geographical locations and the temporal period at a decadal resolution.
Historical forest coverage.
Historical forest coverage data from the Historical Land-Cover Changes and Land-Use Conversions Global Dataset were obtained from the University Corporation for Atmospheric Research Climate Data Guide (https://climatedataguide.ucar.edu/climate-data/) (41). The annual land-cover maps and land-use/cover conversion maps were divided into 0.5° × 0.5° grids, showing 28 land-cover types. We used three types of land cover as regrouped by Meiyappan and Jain (41): primary (old-growth) forest, secondary forest (i.e., regrowth forests after a disturbance such as timber harvest), and others.
Annual mean temperature and annual total precipitation.
Annual mean temperature and annual total precipitation during the study period (1772 AD to present) were extracted from two global datasets. For the period from 1901 to 2016 AD, the temperature and precipitation were from the observation-based Climatic Research Unit (University of East Anglia) TS4.01 dataset (42), which has a spatial resolution of 0.5° × 0.5°. For the period from 1772 to 1900 AD, the climate data were from a global climate model (Earth System Model developed by the Max Planck Institute for Meteorology) simulation for the last millennium (https://www.earthsystemcog.org/projects/cog/esgf_climate_model_metadata), which belongs to the Paleoclimate Modelling Intercomparison Project Phase III. Because the spatial resolution of these data were T63 (about 2° × 2°), we performed a bilinear interpolation of the data into 0.5° × 0.5° grids to match our data resolution for the later period of 1901 to 2016 AD.
Plague Spread Velocities.
Following our previous study (3), we used the NNA to reconstruct the transmission routes and spread velocity of plague during the Third Pandemic period. The routes were mapped based on the first appearance of human plague at different locations. The NNA assumed that the first plague case in a given invasion location i was transmitted from the spatially nearest location (source location j) where plague had been recorded earlier. Our previous study indicated that the NNA was robust to missing sites (3). As an improvement to the previous NNA method, in this study, we reconstructed the transmission routes constrained by genotype (g-NNA). The g-NNA method assumed that plague spread only within locations of the same Y. pestis genotype (i.e., the genotype of human plague was the same at locations i and j). Specifically, source location j of location i was found iteratively by finding the location j that gave
We calculated the ln-scale spread velocity Vi,j [ln(km⋅y−1)] by calculating the geographic distance between locations i and j [Geodis(locationi, locationj)] and dividing by the time interval (yearj − yeari) between the first record of plague at the two locations (3):
GAMs.
To estimate the effects of predictor variables on the speed of human plague spread, we analyzed spread velocity data (Vi,j) using GAMs (19) following Xu et al. (3, 20). The mgcv package (version 1.7-6) of the statistical programming environment R was used for these analyses (43). We used cross-validation to find the model formulation that provided the best out-of-sample predictive power (SI Appendix, Table S1). The initial candidate models of spread velocity included the following environmental predictor variables: population density [ln(number of people per square kilometer)], pastureland coverage (%), primary forest coverage (%), secondary forest coverage (%), annual mean temperature (°C), and annual total precipitation (mm). We also considered plague genotype (Genotypei) as a predictor variable and included year and geographic location (alternatively quantified as a tensor product smooth of longitude and latitude or as region identified by spatial clustering analysis; see SI Appendix, Fig. S1) as variables for accounting for temporal and spatial trends caused by factors not explicitly modeled by the environmental variables. We also tested whether effects of environmental variables were genotype or region specific. We got the following final model, based on model selection, which took into account both maximizing out-of-sample predictive power [lower cross-validation root-mean-squared prediction error (CV)] and minimizing model complexity [lower total degrees of freedom of predictor functions (df)] (SI Appendix, Table S1):
Where (or, equivalently, ) is the ln-scale plague spread velocity to location i (from location j), occurring at time t. Parameter a is the overall intercept. b(Yeart) is a smooth function of invasive time of plague into location i. is an effect of the categorical variable of genotype at location i. is an effect of the categorical variable of region as identified by spatial clustering without constraint by genotypes. , , , , and are, respectively, smooth functions of population density, pastureland coverage, primary forest coverage, and temperature and precipitation in location i and time t. All smooth functions are cubic regression spline functions with maximally 3 df. The model selection is summarized in SI Appendix, Table S1, and the fitted covariate effects under alternative model formulations are shown in SI Appendix, Fig. S2. Inspection of pairwise and multivariate associations between the predictor variables in the final model did not reveal serious identifiability problems, with the strongest correlations being between Pop and Pasture, between Prec and PriFor, and between Prec and Temp (SI Appendix, Fig. S5).
Supplementary Material
Acknowledgments
We thank Prof. Lars Walløe and Dr. Boris Schmid (Centre for Ecological and Evolutionary Synthesis, University of Oslo) for their valuable comments and suggestions; Sari Cunningham for linguistic copyediting of the manuscript; and Xu Xiaoqing, Guo Xue, Ma Tiantian, and others from Henan Normal University for their work digitizing the historical human plague record database. This study was supported by a Key Project for International Cooperation (Grant 31420103913); the China National Natural Science Foundation; the “Strategic Priority Research Program” (Grant XDB11050300); the External Cooperation Program of Bureau of International Cooperation, Chinese Academy of Sciences (Grant 152111KYSB20150023); the European Research Council under the FP7-IDEAS-ERC Program (Grant 324249); and the Centre for Ecological and Evolutionary Synthesis.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901366116/-/DCSupplemental.
References
- 1.Perry RD, Fetherston JD (1997) Yersinia pestis—Etiologic agent of plague. Clin Microbiol Rev 10:35–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stenseth NC, et al. (2008) Plague: Past, present, and future. PLoS Med 5:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu L, et al. (2014) Wet climate and transportation routes accelerate spread of human plague. Proc Biol Sci 281:20133159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kohn GC. (2008) Encyclopedia of Plague and Pestilence: From Ancient Times to the Present, Facts on File Library of World History (Facts on File, New York), 3rd Ed. [Google Scholar]
- 5.World Health Organization (2017) Plague Outbreak Madagascar. Available at https://www.who.int/csr/don/27-november-2017-plague-madagascar/en/. Accessed April 26, 2019.
- 6.Gage KL, Kosoy MY (2005) Natural history of plague: Perspectives from more than a century of research. Annu Rev Entomol 50:505–528. [DOI] [PubMed] [Google Scholar]
- 7.Morelli G, et al. (2010) Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nat Genet 42:1140–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui Y, et al. (2013) Historical variations in mutation rate in an epidemic pathogen, Yersinia pestis. Proc Natl Acad Sci USA 110:577–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Achtman M, et al. (1999) Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc Natl Acad Sci USA 96:14043–14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pollitzer R; World Health Organization (1954) Plague, World Health Organization Monograph Series No. 22 (WHO, Geneva). [Google Scholar]
- 11.Wu L, Chen YH, Pollitzer R, Wu CY (1936) Plague: Manual for Medical and Public Health Workers (Weishengshu National Quarantine Service, Shanghai, China: ). [Google Scholar]
- 12.Adjemian JZ, Foley P, Gage KL, Foley JE (2007) Initiation and spread of traveling waves of plague, Yersinia pestis, in the western United States. Am J Trop Med Hyg 76:365–375. [PubMed] [Google Scholar]
- 13.Christakos G, Olea RA, Yu HL (2007) Recent results on the spatiotemporal modelling and comparative analysis of Black Death and bubonic plague epidemics. Public Health 121:700–720. [DOI] [PubMed] [Google Scholar]
- 14.Snäll T, O’Hara RB, Ray C, Collinge SK (2008) Climate-driven spatial dynamics of plague among prairie dog colonies. Am Nat 171:238–248. [DOI] [PubMed] [Google Scholar]
- 15.Pollitzer R. (1951) Plague studies. 1. A summary of the history and survey of the present distribution of the disease. Bull World Health Organ 4:475–533. [PMC free article] [PubMed] [Google Scholar]
- 16.Prentice MB, Rahalison L (2007) Plague. Lancet 369:1196–1207. [DOI] [PubMed] [Google Scholar]
- 17.Achtman M. (2016) How old are bacterial pathogens? Proc Biol Sci 283:20160990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Achtman M, et al. (2004) Microevolution and history of the plague bacillus, Yersinia pestis. Proc Natl Acad Sci USA 101:17837–17842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hastie T, Tibshirani R (1990) Generalized Additive Models, Chapman & Hall/CRC Monographs on Statistics and Applied Probability Book 43 (Taylor & Francis, Abingdon, UK), 1st Ed. [DOI] [PubMed] [Google Scholar]
- 20.Xu L, et al. (2011) Nonlinear effect of climate on plague during the third pandemic in China. Proc Natl Acad Sci USA 108:10214–10219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dean KR, et al. (2018) Human ectoparasites and the spread of plague in Europe during the Second Pandemic. Proc Natl Acad Sci USA 115:1304–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ben-Ari T, et al. (2011) Plague and climate: Scales matter. PLoS Pathog 7:e1002160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Enscore RE, et al. (2002) Modeling relationships between climate and the frequency of human plague cases in the southwestern United States, 1960-1997. Am J Trop Med Hyg 66:186–196. [DOI] [PubMed] [Google Scholar]
- 24.Parmenter RR, Yadav EP, Parmenter CA, Ettestad P, Gage KL (1999) Incidence of plague associated with increased winter-spring precipitation in New Mexico. Am J Trop Med Hyg 61:814–821. [DOI] [PubMed] [Google Scholar]
- 25.Schotthoefer AM, et al. (2011) Effects of temperature on early-phase transmission of Yersina pestis by the flea, Xenopsylla cheopis. J Med Entomol 48:411–417. [DOI] [PubMed] [Google Scholar]
- 26.Williams SK, et al. (2013) Effects of low-temperature flea maintenance on the transmission of Yersinia pestis by Oropsylla montana. Vector Borne Zoonotic Dis 13:468–478. [DOI] [PubMed] [Google Scholar]
- 27.Kreppel KS, Telfer S, Rajerison M, Morse A, Baylis M (2016) Effect of temperature and relative humidity on the development times and survival of Synopsyllus fonquerniei and Xenopsylla cheopis, the flea vectors of plague in Madagascar. Parasit Vectors 9:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Tan J (2000) The Atlas of Plague and Its Environment in the People’s Republic of China (Science Press, Beijing; ). [Google Scholar]
- 29.IPCC (2013) Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change, eds Stocker TF, et al. (Cambridge Univ Press, Cambridge, UK: ). [Google Scholar]
- 30.Institute of Epidemiology and Microbiology, Chinese Academy of Medical Sciences (1980) Zhong Guo Shu Yi Liu Xing Shi [The History of Spread of Plague in China]. (People’s Medical Publishing House, Beijing). Chinese.
- 31.Ben Ari T, et al. (2008) Human plague in the USA: The importance of regional and local climate. Biol Lett 4:737–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carmichael DA. (1900) Hawaiian Islands. Plague at Honolulu. Public Health Reports (1896-1970) 15:44–46.
- 33.Barde R. (2003) Prelude to the plague: Public health and politics at America’s Pacific gateway, 1899. J Hist Med Allied Sci 58:153–186. [DOI] [PubMed] [Google Scholar]
- 34.Neerinckx S, Bertherat E, Leirs H (2010) Human plague occurrences in Africa: An overview from 1877 to 2008. Trans R Soc Trop Med Hyg 104:97–103. [DOI] [PubMed] [Google Scholar]
- 35.Turner AJ. (2011) Disease control during the colonial period in Australia. Aust Vet J 89:239–242. [DOI] [PubMed] [Google Scholar]
- 36.Robinson GA. (1992) A sop to the “oi polloi”: Capital, labor and the reform of New-South-Wales Maritime Administration 1867 to 1914. Aust J Public Adm 51:104–116. [Google Scholar]
- 37.Riehm JM, et al. (2012) Yersinia pestis lineages in Mongolia. Plos One 7:e30624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kukleva LM, et al. (2015) [Analysis of diversity and identification of the genovariants of plague agent strains from Mongolian foci]. Genetika 51:298–3 05. Russian. [PubMed] [Google Scholar]
- 39.Kingston JJ, Tuteja U, Kapil M, Murali HS, Batra HV (2009) Genotyping of Indian Yersinia pestis strains by MLVA and repetitive DNA sequence based PCRs. Antonie van Leeuwenhoek 96:303–312. [DOI] [PubMed] [Google Scholar]
- 40.Goldewijk KK, Beusen A, van Drecht G, de Vos M (2011) The HYDE 3.1 spatially explicit database of human-induced global land-use change over the past 12,000 years. Glob Ecol Biogeogr 20:73–86. [Google Scholar]
- 41.Meiyappan P, Jain AK (2012) Three distinct global estimates of historical land-cover change and land-use conversions for over 200 years. Front Earth Sci 6:122–139. [Google Scholar]
- 42.Harris I, Jones PD, Osborn TJ, Lister DH (2014) Updated high-resolution grids of monthly climatic observations—The CRU TS3.10 Dataset. Int J Climatol 34:623–642. [Google Scholar]
- 43.R Core Team (2018) R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna), Version 3.5.1. Available at https://www.R-project.org/. Accessed July 2, 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.