Abstract
Increased evidence reveals that miR-940 inhibits the migration and invasion of cancer cells. Considering transforming growth factor β (TGF-β) signaling is crucial to cellular epithelial-mesenchymal transition (EMT) process and metastasis of cancer, it is in urgent to explore whether and how miR-940 plays an essential role in regulating TGF-β-induced EMT in lung cancer progression. In the present study, we observed a reciprocal expression with down-regulated miR-940 and up-regulated Snail mRNA in non-small-cell lung cancer (NSCLC) tissues. we further found that the expression of miR-940 was decreased in NSCLC tissues with lymph node metastasis, advanced TNM stages and poor cell differentiation, in which, on the contrary, the expression of Snail was increased. Overexpression of miR-940 significantly inhibited Snail mRNA and protein expression in A549 and H226 cells. Mechanistically, Snail mRNA was identified as target of miR-940. In addition, miR-940 repressed TGF-β-induced EMT and further hampered the cell migration and invasion. Finally, siRNA-mediated knockdown of Snail copied the phenotype of miR-940 overexpression in A549 and H226 cells. Taken together, our study reveals that miR-940 can suppress TGF-β-induced EMT and cell invasion by targeting Snail 3'-UTR mRNA in NSCLC.
Keywords: NSCLC, EMT, transforming growth factor beta, microRNA, SNAI1
Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide 1. In China, lung cancer has been at the top of cancer morbidity and mortality for decades 2. Generally, 85% of all lung cancers are non-small-cell lung cancer (NSCLC) 3. Despite significant advances in early diagnosis and therapeutic approaches, poor survival for lung cancer patients remains with 5-year survival rate of approximately 10% 4. Over 90% cancer-related deaths from solid malignancies, including NSCLC, are reported to be caused by metastasis 5. Therefore, it is crucial and urgent to reveal the mechanisms underlying NSCLC metastasis.
Epithelial-mesenchymal transition (EMT), as an early event in tumor metastasis, is essential for the transformation of early tumors into aggressive malignancies 6. EMT is a process with particular phenotypic changes, through which epithelial cell lose their cell-cell adhesion and cell polarity, and gain migratory and invasive capabilities 7. During EMT, the epithelial marker E-cadherin is significantly downregulated while mesenchymal cell marker such as N-cadherin and Vimentin are upregulated 8, 9. Transforming growth factor β (TGF-β) is identified as a primary inducer of EMT in NSCLC cells, mainly by activating SMAD cascade signaling 10. Phosphorylated SMAD3 forms complex with SMAD4, and the complex get transported into the nuclear to regulate plenty of EMT-related genes, among which, Snail is established as a transcriptional factor that promote the repression of adhesion molecule E-cadherin to accelerate EMT 11, 12. Accumulating evidence indicates that numerous microRNAs (miRNAs) serve critical roles in cancer metastasis, and many miRNAs associated with tumor metastasis are supported to participate in TGF-β/SMAD signaling-mediated EMT process 13-16.
MiRNAs are a kind of noncoding RNAs containing 18 - 25 nucleotides, commonly presented in cells, that play a role in the precise control of various cell biological processes by their functions in RNA interfering and inhibition of post-transcriptional gene expression 17, 18. Recently, miR-940 has been identified as a prognostic biomarker in breast, thyroid and lung cancer, and a regulator for cellular migration in gastric cancer and hepatocellular carcinoma 19-22. In lung cancer, it has been reported that the patients with low expression of miR-940 show poor prognosis, and overexpression of miR-940 inhibits the proliferation of NSCLC cells 20. However, it remains unclear whether miR-940 is associated with NSCLC metastasis and influences TGF-β-induced EMT in NSCLC cells.
In the present study, for the first time, we found that miR-940 expression was negatively correlated with NSCLC metastasis and clinical stages. We elucidated a reciprocal relationship between the expression of miR-940 and Snail in NSCLCs, and confirmed a targeted inhibition for Snail expression from miR-940. Meanwhile, we found that overexpression of miR-940 obstructed TGF-β-induced EMT and cell migration and invasion in NSCLC cells.
Materials and Methods
Cell culture
Human NSCLC cells A549 (lung adenocarcinoma cell line) and H226 (lung squamous carcinoma cell line) from the Cell Bank of Chinese Academy of Sciences (Shanghai, China) were cultured in RPMI 1640 medium (HyClone, South Logan, UT, USA) containing 10% fetal bovine serum (FBS) (Gibco, Grand Island, NY, USA) at 37℃ in a 5% CO2 atmosphere.
NSCLC tissue samples collection
Ninety-one NSCLC tissues were obtained after informed consent from patients in the Second Affiliated Hospital of Soochow University. Pathological diagnostics for patients with NSCLC were assessed according to the Revised International System for Staging Lung Cancer. NSCLC patients had not received either chemotherapy or radiotherapy before surgical operation. All tissues were frozen in liquid nitrogen right after operation and stored at -80℃ in an ultra-deep freezer till use. This study was approved by the Ethics Review Board of Soochow University, and each patient had signed informed consent.
RNA extraction, cDNA synthesis, and real-time quantitative reverse transcriptase PCR (qRT-PCR)
Total RNA was extracted from cells and tissues using TRIzol (Invitrogen, USA) according to the manufacturer's protocol. The amount of RNA was measured on a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Synthesis of cDNA with reverse transcriptase (RT) was performed with a M-MLV First Strand Kit (Life Technologies, Gaithersburg, MD, USA). Primer sequences for miR-940 and U6 detection are listed in Table 1. The RT primer for mature miR-940 was designed according to the concept of a stem-loop RT primer 23. Primer sequences for Snail mRNA detection are as follows: 5'-CGAAAGGCCTTCAACTGCAAAT-3' (forward), 5'-ACTGGTACTTCTTGACATCTG-3' (reverse). Real-time PCR analysis was carried out using Platinum SYBR Green qPCR SuperMix-UDG kits (Life Technologies) according to the manufacturer's instructions. Real-time PCR was performed on an ABI Prism 7500 Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). The Ct values of Snail mRNA and miR-940 respectively take the β-actin mRNA and U6 as internal controls. ΔΔCt method was used to calculate the relative expression values.
Table 1.
Primers for reverse transcription or amplification of the mature miR-940 and U6
| Name | Sequence, 5'-3' |
|---|---|
| RT primers | |
| U6 | CGAGCACAGAATCGCTTCACGAATTTGCGTGTCAT |
| miR-940 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGGGGAGCG |
| qRT-PCR primers | |
| U6 | F: CGAGCACAGAATCGCTTCA; R: CTCGCTTCGGCAGCACATAT |
| miR-940 | F: CAGTGCAGGGTCCGAGGTA; R: GCATAAGGCAGGGCCCC |
F: forward; R: reverse
Construction of luciferase reporter plasmids, transient transfection and dual-luciferase assay
A psiCHECK-2 dual luciferase vector (Promega, Madison, WI, USA) was used to construct a plasmid, which containing the Snail 3'-untranslated region (3'-UTR) fused to the 3' end of the luciferase reporter. The wild type fragment containing predicted miR-940 target sites (positions 35-41) and mutant fragment were directly synthesized (GENEWIZ, Suzhou, China), and digested by XhoI and NotI (Fermentas, Glen Burnie, MD, USA), and then subcloned into the psiCHECK-2 vector to generate the psiCHECK-2-Snail-3'-UTR-wild type / psiCHECK-2-Snail-3'-UTR-mutant vectors.
Next, we inoculated A549 and H226 cells in a 24-well plate culture, and then co-transfected the cells with 50ng of psiCHECK-2-Snail-3'-UTR-wild type / psiCHECK-2-Snail-3'-UTR-mutant and 20nM of either miR-940 mimic (5'-AAGGCAGGGCCCCCGCUCCCC-3') or miRNA negative control (miR-NC). The sequence for miR-NC is 5'-UUCUCCGAACGUGUCACGUTT-3'. All transient transfections experiments were performed using Lipofectamine 3000 (Invitrogen, Carlsbad, California, USA). The cells were lysed after 48 hours, and luciferase activities were determined by the Dual-Luciferase Reporter Assay System (Promega) on a TD20/20 Luminometer (Turner Designs, Sunnyvale, CA, USA). Each experiment was repeated three times. Results were expressed as relative firefly luciferase activities, which were the ratio of Renilla luciferase activity to firefly luciferase activity.
Western blot assay
Cells and tissues were lysed in a RIPA buffer (Cell Signaling Technology, Danvers, MA, USA) with protease inhibitor and phosphatase inhibitor cocktail (Sigma-Aldrich, St. Louis, MO, USA) and centrifuged. The protein solution obtained by centrifugation were isolated by 10% SDS-PAGE electrophoresis and then transferred to nitrocellulose membranes (Millipore, Billerica, MA, USA). The membranes were sealed with 3% bovine serum albumin (BSA) dissolved in Tris-buffered saline buffer with 0.1% Tween-20 (TBST) for two hours, then incubated overnight at 4 ℃ with primary antibodies. After washing 3 times with TBST for 10 minutes each, the membranes were incubated at room temperature for 2 hours with the corresponding secondary antibody. Next, the membranes were washed with TBST for 3 times, 10 minutes each. The relative quantitative detection of protein was performed by using an electrochemiluminescence (ECL) kit (Pierce, Rockford, IL, USA). The antibodies used for the analysis were shown below: mouse anti-Snail (Cell Signaling Technology, Danvers, MA, USA), mouse anti-E-cadherin, anti-N-cadherin and anti-Vimentin (BD Biosciences, San Jose, CA, USA), mouse anti-β-actin and anti-mouse secondary antibodies (Santa Cruz Biotechnology, Santa Cruz, CA, USA).
RNA interference
Two pre-designed short interfering RNA (siRNA) sequences, which target different coding sequence regions of Snail, were directly synthesized (Ribo Bio Tech, Guangzhou, China). The sequences of the two siRNA were as follows: siRNA-Snail-1, 5'-ACTCAGATGTCAAGAAGTA-3'; siRNA-Snail-2, 5'-AGGCCTTCAACTGCAAATA-3'. Scrambled sequence was used as a negative control of siRNA (si-NC). A549 and H226 cells were transiently transfected with siRNA or si-NC using Lipofectamine 3000 (Invitrogen). After 48 hours of transfection, the cells were collected for further experiments.
Transwell migration and invasion assays
Transwell plates (BD Biosciences) were used for transwell experiments. According to the manufacturer's instructions, 5×104 cells and 1% fetal bovine serum were added to each upper compartment, and 20% fetal bovine serum was added to each lower compartment for transmembrane induction. Six hours later, TGF-β1 solution (5ng/ml) was added to the lower chambers. After 24 hours incubation at 37°C, taken out the upper compartment and the cells were fixed with methanol for 30 minutes, then the upper surface cells of the upper compartment were removed with cotton swabs. Cells invading to the lower side of the upper compartment were stained with 1% crystal violet. Cells in three different regions were photographed and counted under a microscope. All the experimental data were repeated three times.
Statistical analysis
In clinical samples of NSCLC, differences in miR-940 and Snail mRNA expression between NSCLC tissues (T) and adjacent noncancerous lung tissues (N) were analyzed using a paired t test (2-tailed). The expression differences of miR-940 and Snail between the two groups' cell lines were analyzed using an unpaired t test (2-tailed). The correlation between the levels of miR-940 (or Snail) and the clinicopathologic parameters of NSCLC patients were analyzed by non-parametric tests (Mann-Whitney U test for 2 groups, Kruskall-Wallis test for 3 or more groups). Spearman rank correlation test was used to analyze the correlation between miR-940 and Snail mRNA expression level ratios (T/N). The data from tissues are shown as box and whiskers with min to max, while the others are represented by mean ± SD. The statistical significance was accepted as P<0.05. All statistical analyses were performed using GraphPad Prism 7.01 (GraphPad, San Diego, CA, USA) and SPSS 17.0 software (SPSS, Chicago, IL, USA).
Results
miR-940 expression is down-regulated and inversely correlates with Snail expression in NSCLC tissues
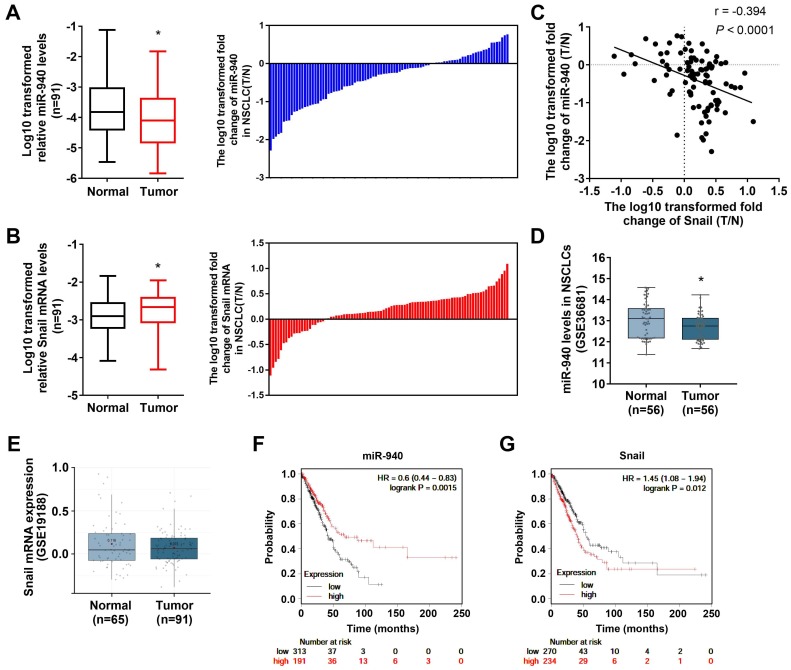
In order to determine the relationship between miR-940 and Snail mRNA, qRT-PCR was performed to detect their expression levels in 91 NSCLC tissues and paired noncancerous tissues. As shown in Fig. 1A and B, miR-940 levels were strongly downregulated (Fig. 1A) while Snail mRNA expressions were up-regulated (Fig. 1B) in NSCLC tissues compared with the paired noncancerous tissues. In addition, the differences in miR-940 and Snail mRNA levels were compared, respectively, when the NSCLC tissues were classified by various clinicopathologic characteristics (Table 2). The comparison results showed that the expression ratios (T/N) of miR-940 were markedly lower in NSCLC tissues with lymph node metastasis, advanced TNM stages and poor cell differentiation than in tissues with non-lymph node metastasis, earlier TNM stages, and high differentiation. On the contrary, the expression ratios of Snail were higher in NSCLC tissues with lymph node metastasis, advanced TNM stages and poor cell differentiation (Table 2). Interestingly, the ratio of miR-940 levels (T/N) were reversely correlated with that of Snail mRNA levels (T/N) in 91 paired tissues (Fig. 1C). In order to verify the results above, datasets from GEO were analyzed in NSCLC tissues. Data from GEO (GSE36681) show that the expression of miR-940 was downregulated in NSCLC tissues (Fig. 1D). However, there was no significant difference of the expression of Snail mRNA in NSCLC tissues compared with normal tissues from data of GSE19188 analyzed by Lung Cancer Explorer (http://lce.biohpc.swmed.edu/lungcancer/) as shown in Fig. 1E 24. But Fan et al. found that Snail mRNA was upregulated in NSCLC and high levels of Snail mRNA were associated with advanced TNM stages 25, which supports our present results. In addition, Survival analysis to TCGA dataset by KM plotter (http://kmplot.com/) 26 shows that low levels of miR-940 are associated with short survival in lung adenocarcinoma patients, and that high levels of Snail mRNA are related to poor prognosis in lung adenocarcinoma patients (Fig. 1F and G). These results support in a great extent our findings on the expression of miR-940 and Snail in NSCLC tissues. Taken together, the results provide a firm association between miR-940 and Snail expression, and imply a potential role for miR-940 in regulating Snail expression in NSCLCs.
Fig 1.
MiR-940 expression is down-regulated and inversely correlates with Snail expression in NSCLC tissues. (A) The left panel shows a comparison of the differential expression of miR-940 between 91 NSCLC tissues (T) and the paired noncancerous tissues (N). In the right panel, log10 transformed fold change (T/N) of miR-940 expression levels of each sample are illustrated. U6 was used as an internal control for miR-940. (B) The differential expression of Snail mRNA between T and N were comparatively analyzed (left panel). Log10 transformed fold change (T/N) of Snail mRNA levels of each sample are illustrated in the right panel. β-actin were used as an internal control for Snail. (C) Correlation between miR-940 levels and Snail mRNA expressions in 91 paired NSCLC tissues. X and Y axes represent the log10 transformed fold change of T/N expression ratios of Snail mRNA levels and miR-940, respectively. (D and E) The differential expression of miR-940 (D) and Snail (E) were analyzed utilizing the GEO data from GSE36681 and GSE19188, respectively. Data are shown as box and whiskers with min to max. (F and G) Survival analysis were performed using KM plotter database (http://kmplot.com/) to determine relationships of the expression levels of miR-940 (F) and Snail (G) to the overall survival of patients with LAD. *P < 0.05.
Table 2.
Differential expression ratios of miR-940 and Snail mRNA between various clinicopathologic parameters in NSCLC tissues.
| Parameter | n | miR-940 | Snail mRNA |
|---|---|---|---|
| Gender | |||
| Male | 60 | 0.866 ± 0.151 | 2.011 ± 0.284 |
| Female | 31 | 1.118 ± 0.230 | 1.252 ± 0.170 |
| P value | 0.348 | 0.071 | |
| Age | |||
| <65 | 49 | 1.009 ± 0.190 | 1.933 ± 0.345 |
| ≥65 | 42 | 0.886 ± 0.163 | 1.541 ± 0.155 |
| P value | 0.628 | 0.329 | |
| Histology | |||
| Adenocarcinoma | 57 | 0.984 ± 0.164 | 1.651 ± 0.252 |
| Squamous cell carcinoma | 29 | 0.855 ± 0.211 | 1.664 ± 0.319 |
| Others | 5 | 1.157 ± 0.632 | 3.415 ± 1.116 |
| P value | 0.880 | 0.057 | |
| Differentiation | |||
| Poor | 49 | 0.869 ± 0.195 | 2.233 ± 0.293 |
| Medium & High | 42 | 2.025 ± 0.575 | 1.357 ± 0.276 |
| P value | 0.047 | 0.034 | |
| Clinical stage | |||
| Ⅰ | 30 | 1.705 ± 0.304 | 0.925 ± 0.137 |
| Ⅱ | 24 | 0.807 ± 0.177 | 1.458 ± 0.227 |
| Ⅲ & Ⅳ | 37 | 0.436 ± 0.068 | 2.614 ± 0.415 |
| P value | 0.0006 | <0.0001 | |
| Lymph node metastasis | |||
| N=0 | 48 | 1.295 ± 0.211 | 1.251 ± 0.130 |
| N≥1 | 43 | 0.569 ± 0.102 | 2.312 ± 0.380 |
| P value | 0.004 | 0.007 |
The expression ratios (Tumor vs. Normal, T/N) of miR-940 and Snail mRNA were calculated and then analyzed according to various clinicopathologic parameters. Data are presented as mean ± SE.
Mann-Whitney U test for 2 groups. Kruskal-Wallis test for 3 or more groups.
MiR-940 inhibits Snail expression by targeting Snail 3'-UTR in NSCLC cells
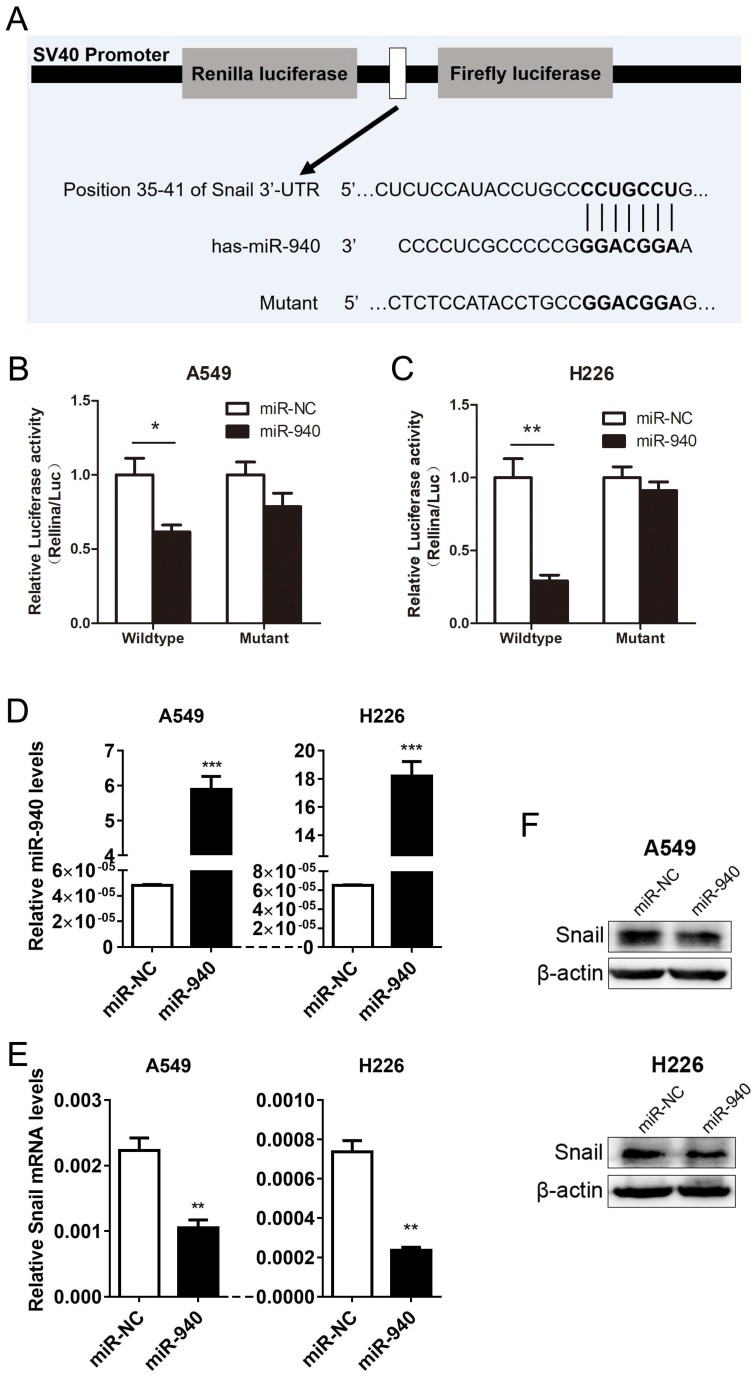
Based on the experiments above, we conjectured that miR-940 can repress Snail expression by directly binding to Snail 3'-UTR region. Using a target prediction program (TargetScan7.1), we found that miR-940 have got a potential to interact with Snail mRNA-3'UTR. To verify the prediction, we subcloned Snail 3'-UTR containing miR-940 binding site (wild type/mutant) into psiCHECK-2 vector (Fig. 2A) and transiently co-transfected the reporter construct with miR-940 mimics into A549 and H226 cells. Results showed that miR-940 significantly suppressed the luciferase activities in A549 cells transfected with the Snail 3'-UTR-wild type reporter, but not in A549 cells with mutant reporter (Fig. 2B). The same results were obtained in H226 cell lines (Fig. 2C). In addition, overexpression of miR-940 (Fig. 2D) markedly inhibited the expression levels of Snail mRNA (Fig. 2E) and protein (Fig. 2F) in both A549 and H226 cells. In conclusion, the results proved that miR-940 can negatively regulate Snail expression through the 3'-UTR-binding to Snail transcript.
Fig 2.
MiR-940 inhibits Snail expression by targeting Snail 3'-UTR in NSCLC cells. (A) A schematic drawing shows the cloning of the predicted miR-940 binding sites of Snail 3'-UTR in psiCHECK-2 luciferase construct. Predicted duplex formation between miR-940 and the wild-type/mutant of miR-940 binding sites are indicated. (B and C) Relative luciferase activities of the wild-type or mutant Snail 3'-UTR reporter gene in A549 and H226 cells transfected with negative control (miR-NC) or miR-940. Relative renilla luciferase activity was calculated after normalizing to the firefly luciferase activity. (D)The expression levels of miR-940 were detected by qRT-PCR in A549 and H226 cells after transfection with miR-NC or miR-940 mimics for 48 hours. (E and F) After transfection with miR-NC or miR-940 mimics in A549 and H226 cells for 48 hours, the expression of Snail mRNA (E) and Protein (F) were detected by qRT-PCR and western blot, respectively. * P < 0.05, ** P < 0.01, *** P < 0.001.
MiR-940 suppresses TGF-β-induced EMT and invasion in NSCLC cells
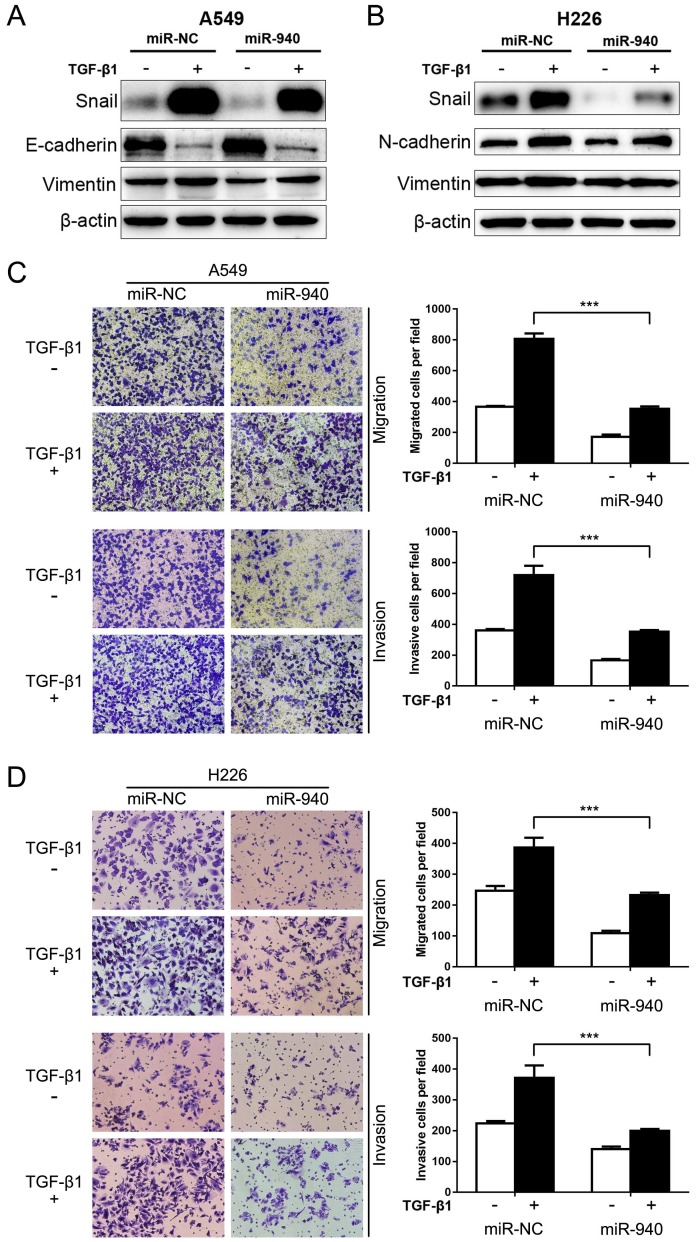
Since TGF-β signaling plays a crucial role in EMT and metastasis 27, we speculate that miR-940 can influence TGF-β-induced EMT and cell migration and invasion in NSCLC cells. To test this, miR-940 mimics were transfected into A549 and H226 cells, and then the protein levels of EMT markers including Snail, Vimentin, E-cadherin and N-cadherin were detected in the presence or absence of TGF-β1. As shown in Fig. 3A and B, TGF-β1 treatment markedly disturbed the expression of EMT markers in A549 and H226 cells, revealing down-regulated E-cadherin and up-regulated N-cadherin and Vimentin. Since E-cadherin nearly does not express in H226 cells, the expression of a mesenchymal marker, N-cadherin, was detected instead. More importantly, Overexpression of miR-940 significantly restrained TGF-β1-induced down-regulation of E-cadherin (determined in A549 cells), up-regulation of snail, N-cadherin (determined in H226 cells) and Vimentin in A549 and H226 cells (Fig. 3A and B). In addition, overexpression of miR-940 prevented the cells from migrating and invading in the presence of TGF-β1 (Fig. 3C and D). Taken together, our data suggest that overexpression of miR-940 can inhibit the expression of Snail and prevent TGF-β1-induced EMT and cell migration and invasion in NSCLC cells.
Fig 3.
MiR-940 inhibits TGF-β-induced EMT and invasion in NSCLC cells. (A) Western blot analysis for the expression of Snail, E-cadherin and Vimentin in A549 cells transfected with miR-NC or miR-940 mimics in the absence or presence of TGF-β1. (B) Expression of Snail, N-cadherin and Vimentin in H226 cells transfected with miR-NC or miR-940 mimics in the absence or presence of TGF-β1. β-actin was used as an internal control. (C) Transwell assays for A549 cells transfected with miR-NC or miR-940 mimics in the absence or presence of TGF-β1. Migrated and invaded cells were stained and counted in at least three microscopic fields (magnification ×100). (D) Transwell assays for H226 cells transfected with miR-NC or miR-940 mimics in the absence or presence of TGF-β1. *** P < 0.001.
Knockdown of Snail represses TGF-β-induced EMT and migration and invasion of NSCLC cells
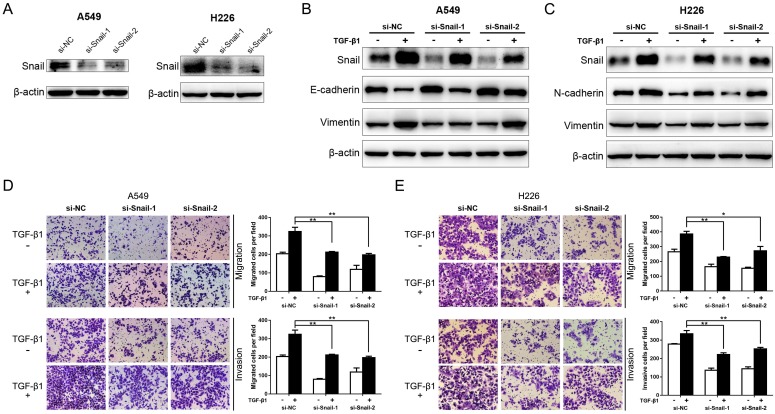
It's evident that Snail is an essential enhancer in TGF-β-induced EMT and cancer metastasis 11, 12, 28. To reconfirm this, the expression of Snail was suppressed by siRNA interference in A549 and H226 cells (Fig. 4A). Then, the protein levels of EMT markers aforementioned were detected in the presence or absence of TGF-β1. As shown in Fig. 4B and C, the expression of TGF-β1-induced Snail was significantly encumbered by Snail-siRNA transfection in A549 and H226 cells. In addition, knockdown of Snail significantly inhibits TGF-β1-induced down-regulation of E-cadherin (determined in A549 cells), and upregulation of N-cadherin (determined in H226 cells) and Vimentin in A549 and H226 cells. Moreover, the inhibitory effect on TGF-β1-induced EMT by knockdown of Snail expression was mirrored by suppression of TGF-β1-induced NSCLC cell migratory and invasive abilities (Fig. 4 D and E). These results copied the influence of miR-940 on TGF-β-induced EMT, suggesting that knockdown of Snail represses TGF-β-induced EMT and cell migration and invasion in NSCLC cells.
Fig 4.
Knockdown of Snail represses TGF-β-induced EMT and migration and invasion of NSCLC cells. (A) Western blot analysis for the expression of Snail in A549 and H226 cells transfected with si-NC, si-Snail-1 or si-Snail-2. β-actin was used as an internal control. (B) Expression of Snail, E-cadherin and Vimentin in A549 cells transfected with si-NC, si-Snail-1 or si-Snail-2 in the absence or presence of TGF-β1. (C) Expression of Snail, N-cadherin and Vimentin in H226 cells transfected with si-NC, si-Snail-1 or si-Snail-2 in the absence or presence of TGF-β1. (D) Transwell assays for A549 cells transfected with si-NC, si-Snail-1 or si-Snail-2 in the absence or presence of TGF-β1. Migrated and invaded cells were stained and counted in at least three microscopic fields (magnification ×100). (E) Transwell assays for H226 cells transfected with si-NC, si-Snail-1 or si-Snail-2 in the absence or presence of TGF-β1. * P < 0.05, ** P < 0.01.
Discussion
Metastasis is the leading cause of death in lung cancer patients 29. TGF-β-induced EMT is crucial to NSCLC cell invasion 30. Furthermore, Snail plays a pivotal role in TGF-β-induced EMT and tumor metastasis 28, 31. In this study, we investigate the miRNA-based mechanism underlying Snail regulation in TGF-β-induced EMT and invasion in NSCLC. Our findings reveal that miR-940 inhibits TGF-β-induced EMT and invasion by targeting Snail in NSCLC cells.
MiRNAs play important roles in the development and progression of tumors, and regulate cell proliferation, migration and other functions through the fine turning of various signal networks 32, 33. For instance, miR-122-5p inhibits cell invasion and growth by targeting ALDOA in bile duct carcinoma 34. In pancreatic cancer, miR-1179 suppresses proliferation, migration and invasion through repression of E2F transcription factor 535. Our previous research suggests that miR-150 may contribute to the malignant phenotype by regulating SIRT2/JMJD2A signaling pathway in NSCLC 36. EMT can be induced by TGF-β signaling in NSCLC cells 13, which is considered as an early stage in tumor metastasis 37. TGF-β is highly expressed in most human tumors and is a powerful upstream inducer of EMT 38. In tumor progression and metastasis, TGF-β signaling pathway plays a key role in EMT 27, 39. Numerous miRNAs are involved in TGF-β-mediated EMT process by interactions with key members of TGF-β/SMAD signaling 15, 16. In the present study, we found that miR-940 can inhibit TGF-β-induced EMT and cell migration and invasion in NSCLC cells.
Snail is a crucial EMT regulator that is involved in TGF-β signaling. In our present study, we found that Snail mRNA was upregulated in NSCLC tissues relative to the paired noncancerous tissues. Our present data supported a recent study in which Fan et al. reported that Snail mRNA was upregulated in NSCLC and high levels of Snail mRNA were associated with advanced TNM stages 25. Various signaling pathways, including RTKs, TGF-β, Notch, Wnt, TNF-α, and BMPs, activate Snail at the transcription level, leading to the induction of EMT 11. It has been observed that the expression of Snail is elevated in metastatic lesions in ovarian cancer 40. In addition to inducing of EMT, Snail also has the function of promoting tumor recurrence 41. We found that knockdown of Snail represses TGF-β-induced EMT and migration and invasion of NSCLC cells, which copied the functions of miR-940 in the present study.
In silico analysis reveals that Snail is a potential target of miR-940. Our luciferase reporter results confirmed this Gene-miRNA binding prediction, and we found that miR-940 significantly inhibited Snail expression in NSCLC cells. Importantly, in our study, siRNA-mediated knockdown of Snail duplicated the same experimental results of miR-940 overexpression in NSCLC cells, suggesting that miR-940 may attenuate TGF-β-induced EMT and NSCLC cell invasion via repression of Snail. Many studies revealed that several miRNAs could modulate Snail in cancer. Regalla et al. found that miR-30a targets Snail and inhibits migration, invasion and metastasis in NSCLC 42. A previous study showed that miR-137 and miR-34a act as suppressors of Snail in ovarian cancer 43. This is the first evidence of identifying that miR-940 suppresses TGF-β-induced EMT and invasion through repression of Snail in NSCLC cells, which consists with a reciprocal relationship between miR-940 and Snail expression in NSCLC patients, especially when samples were classified by characteristics of lymph node metastasis and TNM stages.
MiR-940 was found to play an anti-oncogenic role in many cancers, such as pancreatic ductal adenocarcinoma 44, prostate cancer 45, hepatocellular carcinoma 46, glioblastoma 47 and ovarian cancer 48, while some other researches reveal that miR-940 acts an oncogenic role in several cancers, including cervical cancer 49, bladder cancer 50, gastric cancer 19, 51 and osteosarcoma 52. This contradiction suggests that miR-940 plays different roles in various cancers. In the present study, we found that miR-940 can inhibit TGF-β-induced EMT and cell migration and invasion in NSCLC cells. These studies provide us with evidence that the effect of miR-940 can be mediated via alternative mechanisms in lung cancer.
In conclusion, in the present study, we found that miR-940 can directly target Snail and inhibit TGF-β-induced EMT and invasion in NSCLC cells. Our findings provide insights into the miRNA-based mechanism for controlling TGF-β-induced EMT and invasion of NSCLC cells and a strategy for targeted therapy of NSCLC.
Acknowledgments
We want to thank all the NSCLC patients for their participation and cooperation. This work was supported by the grants from National Natural Science Foundation of China (81502498 to Z. Lei, and 81802989 to M. Shen), Natural Science Foundation of Jiangsu Province (BK20161224 to Y. Chen), Jiangsu Provincial Commission of Health and Family Planning (H201521 to Y. Chen), the Youth Science and Technology Project of Suzhou Health and Family Planning Commission (KJXW2016016 to S. Duan), Science and Technology Research foundation of Suzhou Municipality (SYS2018063 to F. Zhang, and SYS2018064 to M. Shen).
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F. et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Kowal A, Wisniewski A, Kusnierczyk P, Jankowska R. Human leukocyte antigen (HLA)-G gene polymorphism in patients with non-small cell lung cancer. Thoracic cancer. 2015;6:613–9. doi: 10.1111/1759-7714.12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng J, Zhang X, Zhu H, Wang X, Ni S, Huang J. High expression of FoxP1 is associated with improved survival in patients with non-small cell lung cancer. Am J Clin Pathol. 2012;138:230–5. doi: 10.1309/AJCPDHQFNYJZ01YG. [DOI] [PubMed] [Google Scholar]
- 5.Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–95. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 6.Kang Y, Massague J. Epithelial-mesenchymal transitions: twist in development and metastasis. Cell. 2004;118:277–9. doi: 10.1016/j.cell.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 7.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 8.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C. et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–39. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 9.Yang H, Wang L, Zhao J, Chen Y, Lei Z, Liu X. et al. TGF-beta-activated SMAD3/4 complex transcriptionally upregulates N-cadherin expression in non-small cell lung cancer. Lung cancer. 2015;87:249–57. doi: 10.1016/j.lungcan.2014.12.015. [DOI] [PubMed] [Google Scholar]
- 10.Pirozzi G, Tirino V, Camerlingo R, Franco R, La Rocca A, Liguori E. et al. Epithelial to mesenchymal transition by TGFbeta-1 induction increases stemness characteristics in primary non small cell lung cancer cell line. PLoS One. 2011;6:e21548. doi: 10.1371/journal.pone.0021548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Shi J, Chai K, Ying X, Zhou BP. The Role of Snail in EMT and Tumorigenesis. Curr Cancer Drug Targets. 2013;13:963–72. doi: 10.2174/15680096113136660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong TS, Gao W, Chan JY. Transcription regulation of E-cadherin by zinc finger E-box binding homeobox proteins in solid tumors. Biomed Res Int. 2014;2014:921564. doi: 10.1155/2014/921564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li C, Wan L, Liu Z, Xu G, Wang S, Su Z. et al. Long non-coding RNA XIST promotes TGF-beta-induced epithelial-mesenchymal transition by regulating miR-367/141-ZEB2 axis in non-small-cell lung cancer. Cancer Lett. 2018;418:185–95. doi: 10.1016/j.canlet.2018.01.036. [DOI] [PubMed] [Google Scholar]
- 14.Zhang N, Liu Y, Wang Y, Zhao M, Tu L, Luo F. Decitabine reverses TGF-beta1-induced epithelial-mesenchymal transition in non-small-cell lung cancer by regulating miR-200/ZEB axis. Drug Des Devel Ther. 2017;11:969–83. doi: 10.2147/DDDT.S129305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng Y, Zhu J, Shen D, Qin H, Lei Z, Li W. et al. Repression of Smad4 by miR205 moderates TGF-beta-induced epithelial-mesenchymal transition in A549 cell lines. Int J Oncol. 2016;49:700–8. doi: 10.3892/ijo.2016.3547. [DOI] [PubMed] [Google Scholar]
- 16.Hu H, Xu Z, Li C, Xu C, Lei Z, Zhang HT. et al. MiR-145 and miR-203 represses TGF-beta-induced epithelial-mesenchymal transition and invasion by inhibiting SMAD3 in non-small cell lung cancer cells. Lung Cancer. 2016;97:87–94. doi: 10.1016/j.lungcan.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Q, Huang SX, Zhang F, Li SJ, Liu C, Xi YY, MicroRNAs: A novel potential biomarker for diagnosis and therapy in patients with non-small cell lung cancer. Cell Prolif; 2017. p. 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan JY, Zhang F, Sun CC, Li SJ, Li G, Gong FY. et al. miR-134: A Human Cancer Suppressor? Mol Ther Nucleic Acids. 2017;6:140–9. doi: 10.1016/j.omtn.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan Y, Che X, Hou K, Zhang M, Wen T, Qu X. et al. MiR-940 promotes the proliferation and migration of gastric cancer cells through up-regulation of programmed death ligand-1 expression. Exp Cell Res. 2018;373:180–7. doi: 10.1016/j.yexcr.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 20.Gu GM, Zhan YY, Abuduwaili K, Wang XL, Li XQ, Zhu HG. et al. MiR-940 inhibits the progression of NSCLC by targeting FAM83F. Eur Rev Med Pharmacol Sci. 2018;22:5964–71. doi: 10.26355/eurrev_201809_15927. [DOI] [PubMed] [Google Scholar]
- 21.Liu W, Xu Y, Guan H, Meng H. Clinical potential of miR-940 as a diagnostic and prognostic biomarker in breast cancer patients. Cancer Biomark. 2018;22:487–93. doi: 10.3233/CBM-171124. [DOI] [PubMed] [Google Scholar]
- 22.Hu J, Li C, Liu C, Zhao S, Wang Y, Fu Z. Expressions of miRNAs in papillary thyroid carcinoma and their associations with the clinical characteristics of PTC. Cancer Biomark. 2017;18:87–94. doi: 10.3233/CBM-161723. [DOI] [PubMed] [Google Scholar]
- 23.Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods. 2007;3:12. doi: 10.1186/1746-4811-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai L, Lin S, Girard L, Zhou Y, Yang L, Ci B. et al. LCE: an open web portal to explore gene expression and clinical associations in lung cancer. Oncogene. 2019;38:2551–64. doi: 10.1038/s41388-018-0588-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan MJ, Zhong YH, Shen W, Yuan KF, Zhao GH, Zhang Y. et al. MiR-30 suppresses lung cancer cell 95D epithelial mesenchymal transition and invasion through targeted regulating Snail. Eur Rev Med Pharmacol Sci. 2017;21:2642–9. [PubMed] [Google Scholar]
- 26.Nagy A, Lanczky A, Menyhart O, Gyorffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8:9227. doi: 10.1038/s41598-018-27521-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu RY, Zeng Y, Lei Z, Wang L, Yang H, Liu Z. et al. JAK/STAT3 signaling is required for TGF-beta-induced epithelial-mesenchymal transition in lung cancer cells. International journal of oncology. 2014;44:1643–51. doi: 10.3892/ijo.2014.2310. [DOI] [PubMed] [Google Scholar]
- 28.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–28. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 29.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 30.Wang L, Yang H, Lei Z, Zhao J, Chen Y, Chen P. et al. Repression of TIF1gamma by SOX2 promotes TGF-beta-induced epithelial-mesenchymal transition in non-small-cell lung cancer. Oncogene. 2016;35:867–77. doi: 10.1038/onc.2015.141. [DOI] [PubMed] [Google Scholar]
- 31.Vincent T, Neve EP, Johnson JR, Kukalev A, Rojo F, Albanell J. et al. A SNAIL1-SMAD3/4 transcriptional repressor complex promotes TGF-beta mediated epithelial-mesenchymal transition. Nature cell biology. 2009;11:943–50. doi: 10.1038/ncb1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun CC, Li SJ, Zhang F, Zhang YD, Zuo ZY, Xi YY. et al. The Novel miR-9600 Suppresses Tumor Progression and Promotes Paclitaxel Sensitivity in Non-small-cell Lung Cancer Through Altering STAT3 Expression. Mol Ther Nucleic Acids. 2016;5:e387. doi: 10.1038/mtna.2016.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun CC, Li SJ, Yuan ZP, Li DJ. MicroRNA-346 facilitates cell growth and metastasis, and suppresses cell apoptosis in human non-small cell lung cancer by regulation of XPC/ERK/Snail/E-cadherin pathway. Aging (Albany NY) 2016;8:2509–24. doi: 10.18632/aging.101080. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Xu Z, Liu G, Zhang M, Zhang Z, Jia Y, Peng L. et al. miR-122-5p Inhibits the Proliferation, Invasion and Growth of Bile Duct Carcinoma Cells by Targeting ALDOA. Cell Physiol Biochem. 2018;48:2596–606. doi: 10.1159/000492702. [DOI] [PubMed] [Google Scholar]
- 35.Lin C, Hu Z, Yuan G, Su H, Zeng Y, Guo Z. et al. MicroRNA-1179 inhibits the proliferation, migration and invasion of human pancreatic cancer cells by targeting E2F5. Chem Biol Interact. 2018;291:65–71. doi: 10.1016/j.cbi.2018.05.017. [DOI] [PubMed] [Google Scholar]
- 36.Jiang K, Shen M, Chen Y, Xu W. miR150 promotes the proliferation and migration of nonsmall cell lung cancer cells by regulating the SIRT2/JMJD2A signaling pathway. Oncol Rep. 2018;40:943–51. doi: 10.3892/or.2018.6487. [DOI] [PubMed] [Google Scholar]
- 37.Ding X, Park SI, McCauley LK, Wang CY. Signaling between transforming growth factor beta (TGF-beta) and transcription factor SNAI2 represses expression of microRNA miR-203 to promote epithelial-mesenchymal transition and tumor metastasis. The Journal of biological chemistry. 2013;288:10241–53. doi: 10.1074/jbc.M112.443655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Derynck R, Akhurst RJ. Differentiation plasticity regulated by TGF-beta family proteins in development and disease. Nat Cell Biol. 2007;9:1000–4. doi: 10.1038/ncb434. [DOI] [PubMed] [Google Scholar]
- 39.Bruna A, Darken RS, Rojo F, Ocana A, Penuelas S, Arias A. et al. High TGFbeta-Smad activity confers poor prognosis in glioma patients and promotes cell proliferation depending on the methylation of the PDGF-B gene. Cancer Cell. 2007;11:147–60. doi: 10.1016/j.ccr.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 40.Jin H, Yu Y, Zhang T, Zhou X, Zhou J, Jia L. et al. Snail is critical for tumor growth and metastasis of ovarian carcinoma. Int J Cancer. 2010;126:2102–11. doi: 10.1002/ijc.24901. [DOI] [PubMed] [Google Scholar]
- 41.Moody SE, Perez D, Pan TC, Sarkisian CJ, Portocarrero CP, Sterner CJ. et al. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell. 2005;8:197–209. doi: 10.1016/j.ccr.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 42.Kumarswamy R, Mudduluru G, Ceppi P, Muppala S, Kozlowski M, Niklinski J. et al. MicroRNA-30a inhibits epithelial-to-mesenchymal transition by targeting Snai1 and is downregulated in non-small cell lung cancer. Int J Cancer. 2012;130:2044–53. doi: 10.1002/ijc.26218. [DOI] [PubMed] [Google Scholar]
- 43.Dong P, Xiong Y, Watari H, Hanley SJ, Konno Y, Ihira K. et al. MiR-137 and miR-34a directly target Snail and inhibit EMT, invasion and sphere-forming ability of ovarian cancer cells. J Exp Clin Cancer Res. 2016;35:132. doi: 10.1186/s13046-016-0415-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song B, Zhang C, Li G, Jin G, Liu C. MiR-940 inhibited pancreatic ductal adenocarcinoma growth by targeting MyD88. Cell Physiol Biochem. 2015;35:1167–77. doi: 10.1159/000373941. [DOI] [PubMed] [Google Scholar]
- 45.Rajendiran S, Parwani AV, Hare RJ, Dasgupta S, Roby RK, Vishwanatha JK. MicroRNA-940 suppresses prostate cancer migration and invasion by regulating MIEN1. Mol Cancer. 2014;13:250. doi: 10.1186/1476-4598-13-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding D, Zhang Y, Yang R, Wang X, Ji G, Huo L. et al. miR-940 Suppresses Tumor Cell Invasion and Migration via Regulation of CXCR2 in Hepatocellular Carcinoma. Biomed Res Int. 2016;2016:7618342. doi: 10.1155/2016/7618342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi J, Wang YJ, Sun CR, Qin B, Zhang Y, Chen G. Long noncoding RNA lncHERG promotes cell proliferation, migration and invasion in glioblastoma. Oncotarget. 2017;8:108031–41. doi: 10.18632/oncotarget.22446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang F, Wang Z, Gu X, Cui J. miR-940 Upregulation Suppresses Cell Proliferation and Induces Apoptosis by Targeting PKC-delta in Ovarian Cancer OVCAR3 Cells. Oncol Res. 2017;25:107–14. doi: 10.3727/096504016X14732772150145. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49.Su K, Wang CF, Zhang Y, Cai YJ, Zhang YY, Zhao Q. miR-940 upregulation contributes to human cervical cancer progression through p27 and PTEN inhibition. Int J Oncol; 2017. [DOI] [PubMed] [Google Scholar]
- 50.Wang R, Wu Y, Huang W, Chen W. MicroRNA-940 Targets INPP4A or GSK3beta and Activates the Wnt/beta-Catenin Pathway to Regulate the Malignant Behavior of Bladder Cancer Cells. Oncol Res. 2018;26:145–55. doi: 10.3727/096504017X14902261600566. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Liu X, Ge X, Zhang Z, Zhang X, Chang J, Wu Z. et al. MicroRNA-940 promotes tumor cell invasion and metastasis by downregulating ZNF24 in gastric cancer. Oncotarget. 2015;6:25418–28. doi: 10.18632/oncotarget.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin ZW, Zhang W, Jiang SD, Wei WB, Li XF. Inhibition of microRNA-940 suppresses the migration and invasion of human osteosarcoma cells through the secreted frizzled-related protein 1-mediated Wnt/beta-catenin signaling pathway. J Cell Biochem; 2018. [DOI] [PubMed] [Google Scholar]