Abstract
MicroRNA-519d (miR-519d) has been reported to play important roles in tumor development and progression in multiple cancers, either as tumor suppressor or tumor promotor. However, the expression level, biological function and molecular mechanisms of miR-519d in oral squamous cell carcinoma (OSCC) remain unclear. Therefore, the aims of this study were to investigate the functional role of miR-519d in OSCC and the possible underlying regulatory mechanism. In this study, we found that miR-519d was significantly downregulated in OSCC tissues and cell lines compared with normal oral mucosae and normal oral epithelial cells. Importantly, downregulation of miR-519d was closely correlated with the lymph node metastasis, advanced tumor stage and poor overall survival of OSCC patients. Furthermore, miR-519d significantly inhibited the migration and invasion of OSCC cells. Using bioinformatics and biological approaches, we showed that miR-519d directly targeted matrix metalloproteinase-3 (MMP3), which might account for the underlying mechanism involved in the miR-519d mediated suppression of OSCC progression. What is more, miR‐519d expression was inversely correlated with MMP3 expression in OSCC tissues, and high levels of MMP3 expression in OSCC tissues were also associated with the metastasis and poor prognosis of these patients. In addition, we further identified that miR-519d acted as a regulator of epithelial-mesenchymal transition (EMT) in OSCC cells. Overall, the present study highlighted miR-519d as a tumor suppressor in OSCC by targeting MMP3 and supported biological and clinical links between miR-519d-MMP3 and OSCC, thus indicating the potential therapeutic value of miR-519d for alleviating OSCC metastasis.
Keywords: miR-519d, oral squamous cell carcinoma, invasion, metastasis, MMP3, epithelial-mesenchymal transition (EMT)
Introduction
Head and neck squamous cell carcinoma (HNSCC) ranks sixth among all the cancers diagnosed worldwide, and is listed as the eighth most common cause of cancer-related death. As a common subtype of HNSCC, oral squamous cell carcinoma (OSCC) is a devastating disease with high morbidity and mortality and always seriously impacts the life quality of patients 1, 2. To be details, the incidence of OSCC in China is about 3-6/100,000 individuals and what is disappointing is the fact that the number extends an increasing trend every year. Furthermore, despite the constant improvement of multiple treatments, such as surgery, chemotherapy, radiotherapy, and anti-EGFR biotherapy, the 5-year survival rate of OSCC patients remained unchanged due to aggressive metastasis and recurrence 3.
Thus, in order to figure out an effective treatment of OSCC and to enhance the life quality of OSCC patients, a better understanding of the molecular mechanisms involved in OSCC progression is highly needed. In recent years, many novel biomarkers have been explored for efficient clinical applications. These include microRNAs (miRNAs) which can be utilized as an effective prognostic and treatment tool for different cancers 4.
MiRNAs are a class of small, noncoding RNA molecules, with approximately a length of 19-23 nucleotides 5. Numerous studies have highlighted the importance of miRNAs in the regulation of gene expression and cellular signaling transduction 6. In most cases, miRNAs regulate gene expression through direct binding to the matched sites of mRNAs, thus leading to rapid degradation of target mRNAs 7. It has been firmly established that miRNAs modulate many key cellular functions such as cell proliferation, differentiation, apoptosis, angiogenesis, and stress resistance 8, and are also involved in a variety of biological processes, such as organism growth, immune regulation and tumorigenesis 6, 9. Moreover, many studies revealed that miRNA expression patterns in tumor tissues enabled more reliable and accurate diagnosis and prognosis to be made in patients 10. Emerging evidences also illustrated aberrant expression of miRNAs, either overexpression or underexpression, can play critical roles in the occurrence and progression of various cancers. In addition, miRNAs may possess oncogenic or tumor-suppressive properties and the opposite effects of the same miRNA on different tumors may be dependent on the tumor type and the target genes 11.
Previous studies have reported that miR-519d is aberrantly expressed and modulates the cellular biological characteristics in sorts of human cancers. Meantime, many researchers have identified miR-519d as an effective mediator of cancer progression and take efforts to explore the underlying biological mechanisms of the regulatory process. For example, Yong Bai et al. found that miR-519d could inhibit both cell proliferation and invasion via the regulation of eIF4H in lung adenocarcinoma 12. Also, miR-519d was downregulated in hepatocellular carcinoma and suppressed the growth of tumor cells by targeting MKi67 13. In breast cancer, downregulation of miR-519d was found to be associated with cancer metastasis and advanced clinical stage 14. However, the expression levels and the biological functions of miR-519d in OSCC, as well as its underlying molecular mechanisms, have not been well elucidated till now.
Therefore, the present study aims to identify the expression of miR-519d in OSCC and tries to investigate the functional role of miR-519d in OSCC progression. Moreover, we also intend to identify the potential molecular mechanisms by which miR-519d modulate the cellular functions in OSCC cells.
Materials and Methods
Tissue samples
A total of 60 pairs of OSCC tissues and matched adjacent normal tissues were obtained from the patients who were diagnosed with primary OSCC and underwent initial surgery between October 2010 and September 2012 at the Department of Oral Maxillofacial‐Head and Neck Oncology, Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine (Shanghai, China). All of the tissue samples were frozen in liquid nitrogen for further experiments. Overall survival was defined as the time from start of treatment until death. Follow-up ended on December 2017 or at death. In this study, none of these OSCC patients received any preoperative cancer treatment and were histologically diagnosed as OSCC after operation. This study was approved by the Review Board of the Medical Ethics Committee of the Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine. Informed consent and approval were received from all patients. In this study, pathological differentiation and clinical stage were respectively determined according to World Health Organization Classification of Tumors and the TNM classification system of the International Union Against cancer (1988).
Cell culture
The human HN4 and HN30 cells were kindly provided by the University of Maryland Dental School, USA. 293T cells were purchased from the American Type Culture Collection (ATCC, USA). All the cells were cultured in DMEM (GIBCO-BRL, USA) supplemented with 10% fetal bovine serum (GIBCO-BRL, USA) and 1% penicillin and streptomycin at 37°C, 5% CO2 in a humidified atmosphere. In addition, normal oral epithelial cells were obtained from primary culture and were cultured in keratinocyte serum-free medium (KSF; GIBCO-BRL, USA) containing 0.2 ng/mL recombinant epidermal growth factor (rEGF; Invitrogen, USA).
RNA extraction and quantitative real-time PCR
Total RNA from the cells and tissues was extracted using the Trizol reagent (Takara, Japan) and complementary cDNA was synthesized by using a PrimeScript™ RT reagent kit (Takara, Japan). A miRcute Plus miRNA First-Strand cDNA Synthesis Kit (TIANGEN, China) was used to synthesize miRNA cDNA from total RNA. The real-time PCR for miR-519d was performed using a miRcute Plus miRNA qPCR Detection Kit (TIANGEN, China) with ABI StepOne Real-Time PCR System (Applied Biosystems, USA). The PCR condition was as follows: 95°C for 15 min, 40 cycles of 20 s at 94°C and 34 s at 60°C. The miR-519d expression levels were normalized to U6, a small nuclear RNA from the same sample. For mRNA expression, the real-time PCR was performed using a SYBR Premix Ex Taq Reagent Kit at the condition as followed: 95°C for 30 s, 40 cycles of 5 secs at 95°C, 30 s at 60°C. GAPDH was used as internal reference. All the primers were designed and synthetized by Sangon Biotech (Shanghai) and the detailed sequences for the primers were presented in the Table 1. The relative expression level was calculated by adopting the 2-ΔΔCt method and all experiments were repeated in triplicate.
Table 1.
The primers used for real-time PCR analysis.
| Genes | Primer sequences |
|---|---|
| Human miR-519d-3p | forward 5'-CAAAGTGCCTCCCTTTAGAGTG-3' |
| Human U6 snRNA | forward 5'-CTCGCTTCGGCAGCACA-3' |
| Human MMP3 | forward 5'-AGTCTTCCAATCCTACTGTTGCT-3' |
| reverse 5'-TCCCCGTCACCTCCAATCC-3' | |
| Human PTHLH | forward 5'-ATTTACGGCGACGATTCTTCC-3' |
| reverse 5'-GCTTGGAGTTAGGGGACACC-3' | |
| Human FNDC3B | forward 5'-TCTCGTTCAAGTTAATCCAGGTG-3' |
| reverse 5'-ACATGGCTGAGGGGTAGCTT-3' | |
| Human NETO2 | forward 5'-AGATGGGCCATTTGGTTTCTC-3' |
| reverse 5'-TGCTCGAAATCCCAGTCCTTC-3' | |
| Human GALNT6 | forward 5'-CTGTTCTCCATAAACCAGTCCTG-3' |
| reverse 5'-CGCTGGCAAAGGCATTGAAA-3' | |
| Human PANX1 | forward 5'-CCACGGAGTACGTGTTCTCG-3' |
| reverse 5'-CCGCCCAGCAATATGAATCC-3' | |
| Human APBB2 | forward 5'-ACTTGGGCATGTTACCTGTAGA-3' |
| reverse 5'-CACGACATTCCACTAAGACTTCC-3' | |
| Human KIF26B | forward 5'-GCTGGGAATAAAGAGAGGCTTG-3' |
| reverse 5'-ACTCCTCGTATGCTTTCCGGT-3' | |
| Human SOX4 | forward 5'-AGCGACAAGATCCCTTTCATTC-3' |
| reverse 5'-CGTTGCCGGACTTCACCTT-3' | |
| Human MMP2 | forward 5'-TACAGGATCATTGGCTACACACC-3' |
| reverse 5'-GGTCACATCGCTCCAGACT-3' | |
| Human STC1 | forward 5'-GTGGCGGCTCAAAACTCAG-3' |
| reverse 5'-GTGGAGCACCTCCGAATGG-3' | |
| Human AP2B1 | forward 5'-CTCTTTCCAGACGTAGTGAACTG-3' |
| reverse 5'-GGAGCGGCTCACAGAGATATT-3' | |
| Human SOD2 | forward 5'-GCTCCGGTTTTGGGGTATCTG-3' |
| reverse 5'-GCGTTGATGTGAGGTTCCAG-3' | |
| Human GAPDH | forward 5'-ACAACTTTGGTATCGTGGAAGG-3' |
| reverse 5'-GCCATCACGCCACAGTTTC-3' |
miRNA and siRNA transfection
A total of 2 × 105 cells cultured in a 6-well plate were transfected with synthetic miR-519d double-stranded mimics, miR-519d inhibitor or siRNA-MMP3 using Lipofectamine 2000 reagent (Invitrogen, USA) according to the manufacturer's instructions. Total RNA and protein were collected for experiments at 48 h and 72 h after transfection. The sequences of miR-519d mimics were as followed: sense: 5'-CAAAGUGCCUCCCUUUAGAGUG-3' and anti-sense: 5'-CACUCUAAAGGGAGGCACUUUG-3'. The sequences of miR-519d inhibitor were sense: 5'-CAAAGUGCCUCCCUUUAGAGUG-3' and anti-sense: 5'-CACTCTAAAGGGAGGCACTTTG-3'. The sequences of specific siRNAs for MMP3 were sense: 5'-AAAGGACAGUGGUCCUGUUGU tt--3' and anti-sense: 5'-ACAACAGGACCACUGUCCUUU tt -3'.
Methylation analysis, bisulphite treatment, methylation-specific PCR and demethylation
CpG islands locate upstream of the chromosome 19 miRNA cluster (C19MC) and in the proximity of the miR-519d precursor were predicted by the UCSC database (http://genome.UCSC.edu). Fortunately, a CpG island locating 17.6 kb upstream of C19MC was reported to be closely associated with the expression of miR-519d in previous study 15. Therefore, this CPG island was selected for further analysis. Genomic DNA of cells or tissue samples were isolated using TIANamp Genomic DNA Kit (TIANGEN, China) according to the manufacturer's protocol. About 1 µg DNA was subjected to bisulphite conversion by using a MethylCodeTM Bisulfite Conversion Kit (Invitrogen, USA) and Methylation-specific PCR (MSP) was carried out to detect the methylation status of the DNA region upstream of C19MC in cell and tissue samples. MSP was performed using specific M primers (forward primer: 5'-TGACGTCGTTACGTTTTGGA-3' and reverse primer: 5'-TAAACCGAAAAACGCGCG-3', 106 bp) and U primers (forward primer: 5'-TGATGTTGTTATGTTTTGGATTGAG-3' and reverse primer: 5'-AAACACACACCAACACCTCATAACA-3', 97 bp). The PCR products were then electrophoresed on a 3% agarose gel, stained with GeneGreen Nucleic Acid Dye (TIANGEN, China) and visualized under UV illumination. To further illustrate whether demethylation treatment induces miR-519d expression, HN4 and HN30 cells were treated with or without 10 μM 5-aza-2'-deoxycytidine (5-Aza-dc) for 72 h. Subsequently, these cells were collected, subjected to RNA isolation and used for quantitative analysis of miR-519d expression.
Cell proliferation assay
To evaluate the influence of miR-519d on the proliferation of OSCC cells, tumor cells transiently transfected with miR-519d mimics or miR-519d inhibitor were cultured in 96-well plates at the density of approximately 1000 cells/well in triplicate and analyzed for consecutive 6 days. For daily measurement, the cells were incubated with DMEM medium containing 50 µg/mL MTT (Sigma-Aldrich, USA) at 37 °C for 4 h. Then, the culture medium was removed and 100 µL dimethyl sulfoxide (DMSO) were added into each well. The 490 nm absorbance wavelength was read on a microplate reader to assess the cell viability. All the experiments were repeated for three times.
Wound healing assay
After being transfected, cells in the 6-well plates were cultured to achieve 90%-100% confluency. Then, cells were scraped with a P200 tip and incubated with serum-free medium. Five nonoverlapping pictures were captured at the time point of 0 h and 12 h under an inverted microscope.
Transwell migration and invasion assay
In order to investigate the role of miR-519d in the migration and invasion of OSCC cells, transwell chambers (pore 0.8 μm, Merck Millipore, USA) were coated with or without Matrigel (Corning, USA) on the upper chamber. A total of 40,000 tumor cells transfected with miR-519d mimics, miR-519d inhibitor, or NC controls in 350 μL serum-free DMEM were added into the upper chamber. The bottom chambers were filled with 700 μL DMEM containing 20% FBS as a chemoattractant. After being incubated for 48 h, cells that had migrated or invaded to the lower surface of the membrane were fixed with 4% paraformaldehyde, stained with 0.5% crystal violet and counted in five randomly selected nonoverlapping fields under a light microscope.
Plasmid construction and transfection
The cDNA of MMP3 was amplified by high-fidelity PCR (PrimeSTAR; Takara, Japan) with one set of primers (forward primer: 5'-TGAACCGTCAGATCGAATTCGCCACCATGAAGAGTCTTCCAATCC-3' and reverse primer: 5'-CCTTGTAGTCGAATTCACAATTAAGCCAGCTGTTAC-3') and subcloned into the EcoRI (4662) and EcoRI (6105) sites of H5188 vector (Obio Technology, China). To obtain the luciferase reporters, PCR-derived fragments from the 3' UTR of MMP3 were inserted into the MluI and HindIII sites of H306 pMIR-REPORT Luciferase vector (Obio Technology, China). The primers used for MMP3 3' UTR amplification were as follows: 5'-ATAGGCCGGCATAGACGCGTAAGAGATATGTAGAAGGCAC-3' (forward primer) and 5'-ATCCTTTATTAAGCTTAAGATAAAATAACTGACAAATC-3' (reverse primer). A PCR-based site-directed mutagenesis was performed to generate the mutant luciferase reporters.
The plasmid transfection was performed with LipofectamineTM 2000 reagent (Invitrogen, USA) following the manufacturer's protocol. A final concentration of 2 μg/mL of plasmid or control plasmid was used for each transfection. 24 h after transfection, the biological behavior of the cancer cells was detected.
Luciferase reporter assay
To further illustrate whether MMP3 is the direct target of miR-519d, 293T, HN4 and HN30 cells growing in 6-well plates were co-transfected with the 1 μg MMP3 3' UTR wild-type or mutant plasmid, 100 nM miR-519d mimics or miR-519d inhibitor and 100 ng Renilla luciferase vector (pRL-CMV; Genomeditech, China). About 24 h later, luciferase activity was determined using the Dual-Luciferase Reporter System (Promega, USA). Renilla luciferase activity was normalized against Firefly luciferase activity. All the experiments were performed in triplicate.
Western blot analysis
Proteins were extracted from the cells at indicated times using SDS lysis buffer (Beyotime, China). The protein samples (30 μg) were subjected to 4%-20% polyacrylamide gels (Genshare biological, China) to be electrophoresed and transferred to 0.22 μm PVDF membranes (Merck Millipore, USA) at 90 V for 1.5 h. After being blocked by the 5% skim milk for about 1 h, membranes were incubated with the primary antibodies against MMP3 (Proteintech, USA; 1:1,000), E-cadherin (Proteintech, USA; 1:1,000), N-cadherin (Proteintech, USA; 1:1,000), vimentin (Abcam, USA; 1:1,000) and GAPDH (Proteintech, USA; 1:10,000) at 4°C overnight. Thereafter, the membranes were probed by IR Dye-labeled secondary antibodies (LI-COR Bioscience, USA; 1:10,000) and the signals were observed using an Odyssey Infrared Imaging System (LI-COR Bioscience, USA).
Immunofluorescence
HN4 or HN30 were transfected as indicated, and the cells were inoculated into a 24-wel plate with prepared cell sheets. About 48 h later, the cells grown on cover slips were washed with PBS for three times, and then fixed with 4% paraformaldehyde for 30 min. Subsequently, they were permeabilized with 0.1% Triton X-100 for 10 min. After being washed for three times by PBS, cells were incubated with 3% H2O2 for 10 min and blocked with 10% goat serum plus 3% BSA for 30 min. The fixed cells were incubated overnight at 4 ˚C with primary antibodies against MMP3 (Proteintech, USA; 1:100), E-cadherin (Proteintech, USA; 1:100), N-cadherin (Proteintech, USA; 1:100), Vimentin (Abcam, USA; 1:100), and followed by incubation with secondary antibody: Alexa Fluor 488-conjugated goat anti-rabbit IgG, Alexa Fluor 594-conjugated goat anti-rabbit IgG (Invitrogen, USA; 1:200) for 1h at room temperature. The cover slips were stained with DAPI staining dilution (YEASEN, China; 1:500) for 3 min in the dark. The fluorescence images were finally observed and analyzed by a fluorescence microscope (ZEISS, Germany).
Statistical analysis
All numerical data were expressed as mean ± SD from triplicate experiments and statistical analysis was performed using SPSS 21.0 software. Comparisons between two groups were performed by Student's two-tailed t-test, and the significance among three or more groups was analyzed by one-way ANOVA. The associations between gene expression (miR-519d and MMP3) and clinicopathological characteristics of OSCC patients was analyzed with Mann-Whitney U-test and Kruskal-Wallis test. The correlation between miR-519d and MMP3 expression was examined by Pearson correlation analysis. The effect of clinical variables on patient survival was examined using cox proportional hazards regression analysis. Kaplan-Meier survival analysis was performed to compare OSCC patient survival based on dichotomized gene (miR-519d and MMP3) expression by log-rank test. P values less than 0.05 were considered statistically significant.
Results
Downregulation of miR-519d in OSCC indicates malignant transformation
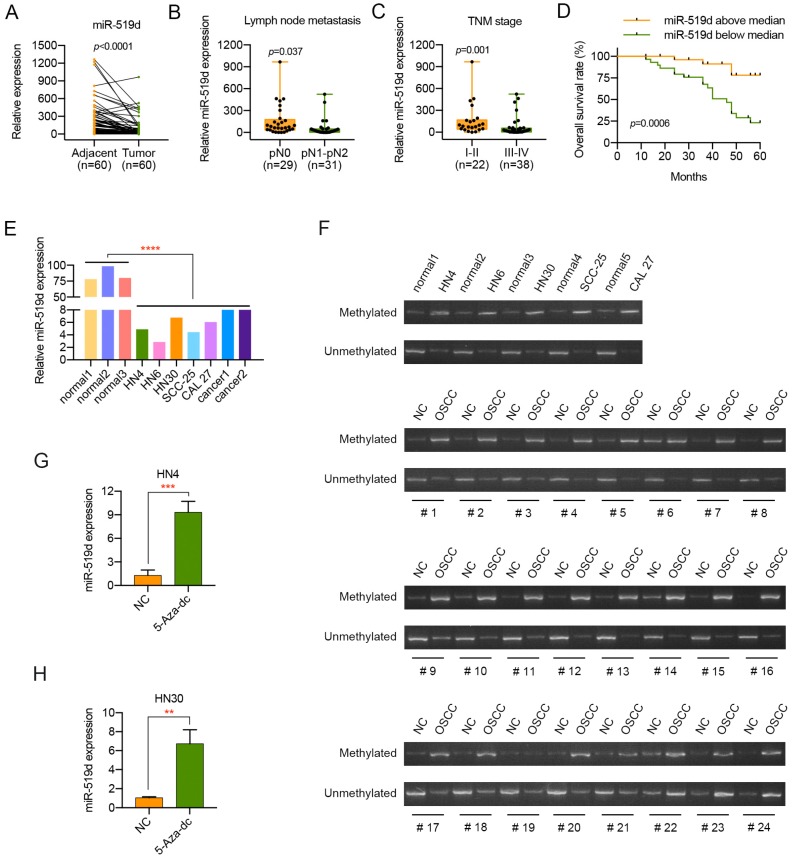
Although miR-519d has been detected to have an effect on some kinds of cancers, its impact on OSCC has not been well elucidated. Thus, the miR-519d expression level was examined in 60 pairs of OSCC samples and adjacent normal tissues. As suggested by Figure 1A, miR-519d expression in the OSCC tissues was significantly decreased compared with that in adjacent normal tissues. Moreover, lower miR-519d expression level was correlated with lymph node metastasis and advanced tumor stage (Figure 1B, 1C, Table 2). Meanwhile, Kaplan-Meier analyses manifested an overall better survival condition of patients with miR-519d levels above median (Figure 1D, Table 4). In addition, the expression level of miR-519d was also detected in normal oral epithelial cells, primary cultured OSCC cells and 5 OSCC cell lines. Consistently, compared with the normal oral epithelial cells, OSCC cells presented a significant lower expression of miR-519d (Figure 1E). The above data revealed that miR-519d might play an essential role in the progression of OSCC, especially tumor metastasis.
Figure 1.
Downregulation of miR-519d in OSCC indicates malignant transformation. (A) The expression level of miR-519d in 60 pairs of OSCC tissues and adjacent normal tissues (P<0.0001). (B, C) Low miR-519d expression was correlated with lymph node metastasis (P=0.037) and advanced TNM stage (P=0.001) in OSCC. (D) Kaplan-Meier analyses of overall survival. Patients with low miR-519d expression had a significantly lower overall survival rate than patients with high miR-519d expression (P=0.0006). (E) The miR-519d expression in normal oral epithelial cells, primary cultured cancer cells and 4 OSCC cell lines. (F) The methylation status of the DNA region located upstream of C19MC was detected in 5 OSCC cell lines and 24 pairs of OSCC samples and adjacent normal tissues by MSP. OSCC, oral squamous cell carcinoma; NC, matched adjacent normal tissue. (G, H) The expression of miR-519d in OSCC cells treated with or without the demethylating agent 5-Aza-dc (10 μM) for 72 h. **, P < 0.01. ***, P< 0.001. ****, P<0.0001.
Table 2.
Relationship between miR-519d level and clinicopathologic features (N = 60)
| Characteristics | No. of Patients | miR-519d Δ Cta | Non-parametric | P value | |
|---|---|---|---|---|---|
| No. | % | Mean ± SD | test value | ||
| Age (years) | |||||
| < 60 | 36 | 60.0 | 8.93 ± 2.94 | Z = -0.362 | 0.717 |
| ≥ 60 | 24 | 40.0 | 9.13 ± 3.01 | ||
| Gender | |||||
| Female | 19 | 31.7 | 8.91 ± 3.35 | Z = -0.358 | 0.721 |
| Male | 41 | 68.3 | 9.06 ± 2.78 | ||
| Smoking history | |||||
| Nonsmoker | 32 | 53.3 | 8.93 ± 3.35 | Z = -0.756 | 0.450 |
| Smoker | 28 | 46.7 | 9.10 ± 2.45 | ||
| Alcohol history | |||||
| Nondrinker | 38 | 63.3 | 8.97 ± 3.19 | Z = -0.337 | 0.736 |
| Drinker | 22 | 36.7 | 9.08 ± 2.54 | ||
| Tumor size (cm) | |||||
| ≤ 4 | 32 | 53.3 | 8.67 ± 3.24 | Z = -1.719 | 0.086 |
| > 4 | 28 | 46.7 | 9.40 ± 2.56 | ||
| Lymph node metastasis | |||||
| pN0 | 28 | 46.7 | 8.19 ± 2.47 | Z = -3.123 | 0.037 |
| pN1 to pN2 | 32 | 53.3 | 9.73 ± 3.17 | ||
| TNM stage | |||||
| I-II | 22 | 36.7 | 7.60 ± 2.24 | Z = -3.206 | 0.001 |
| III-IV | 38 | 63.3 | 9.83 ± 3.02 | ||
| Pathological differentiation | |||||
| Well | 25 | 41.7 | 8.64 ± 2.72 | Z = -0.787 | 0.431 |
| Moderately/poorly | 35 | 58.3 | 9.28 ± 3.10 | ||
| Disease Site | |||||
| Tongue | 25 | 41.7 | 8.44 ± 2.69 | H = 5.242 | 0.263 |
| Gingival | 8 | 13.3 | 11.09 ± 3.59 | ||
| Cheek | 9 | 15.0 | 9.82 ± 3.21 | ||
| Floor of Mouth | 12 | 20.0 | 8.50 ± 2.52 | ||
| Oropharynx | 6 | 10.0 | 8.41 ± 2.85 | ||
| Recurrence | |||||
| No | 50 | 22.5 | 9.02 ± 3.18 | Z = -0.635 | 0.526 |
| Yes | 10 | 77.5 | 8.96 ± 1.36 | ||
Abbreviations: SD, standard deviation; pN, pathological lymph node status; TNM stage, tumor-lymph node-metastasis stage.
aΔ Ct indicates the difference in the cycle number at which a sample's fluorescent signal passes a given threshold above baseline (Ct) derived from a specific gene compared with that of β-actin in tumor tissues.
Table 4.
Univariate and multivariate cox proportional hazards regression models for estimating overall survival (N = 60)
| Characteristics | HR | 95% CI | P |
|---|---|---|---|
| Univariate analysis | |||
| Overall survival | |||
| Age (< 60 y vs ≥ 60 y) | 0.488 | 0.192 to 1.238 | 0.131 |
| Gender (male vs female) | 1.043 | 0.429 to 2.538 | 0.924 |
| Smoking history (smoker vs nonsmoker) | 1.341 | 0.591 to 3.040 | 0.483 |
| Alcohol history (drinker vs nondrinker) | 1.038 | 0.439 to 2.451 | 0.932 |
| Tumor size (> 4 cm vs ≤ 4 cm) | 1.561 | 0.684 to 3.564 | 0.291 |
| Lymph node metastasis (pN1 to pN2 vs pN0) | 2.385 | 0.939 to 6.057 | 0.038 |
| TNM stage (III-IV vs I-II) | 2.631 | 0.971 to 7.130 | 0.017 |
| Pathological differentiation (Moderately to poorly vs Well) | 1.318 | 0.569 to 3.056 | 0.519 |
| Disease Site | 0.941 | 0.716 to 1.238 | 0.666 |
| Recurrence | 1.038 | 0.381 to 2.817 | 0.937 |
| miR-519d expression (high vs low) | 0.192 | 0.065 vs 0.564 | 0.003 |
| MMP3 expression (high vs low) | 2.615 | 1.104 to 6.193 | 0.029 |
| Multivariate analysis | |||
| Overall survival | |||
| Lymph node metastasis (pN1 to pN2 vs pN0) | 1.319 | 0.347 to 5.015 | 0.685 |
| TNM stage (III-IV vs I-II) | 1.172 | 0.330 to 4.167 | 0.806 |
| miR-519d expression (high vs low) | 0.215 | 0.058 to 0.805 | 0.023 |
| MMP3 expression (high vs low) | 1.403 | 0.430 to 4.581 | 0.575 |
Abbreviations: CI, confidence interval; HR, hazard ratio; T, tumor stage; pN, pathological lymph node status; TNM, tumor-lymph node-metastasis classification.
MiR-519d belongs to C19MC, which displays a very low expression in most human tissues 16. More importantly, a previous study has demonstrated that the aberrant methylation of the DNA region located upstream of C19MC contributes greatly to the dysregulation of miR-519d in hepatocellular carcinoma 15. Therefore, we tried to identify if the downregulation of miR-519d in OSCC resulted from the hypermethylation of the DNA region located upstream of C19MC. As shown in Figure 1F, five OSCC cell lines HN4, HN6, HN30, SCC-25 and CAL 27 presented a lower C19MC methylation status compared to normal oral epithelial cells. Moreover, in 19 of 24 (79.2%) cases the hypermethylation was associated with an downregulation of miR-519d in OSCC tissue. In addition, the expression of miR-519d was significantly increased in OSCC cells after 5-Aza-dc treatment (Figure 1G, 1H). All the findings above suggested that DNA methylation might function as a major factor in silencing miR-519d in OSCC.
miR-519d has no effect on the proliferation of OSCC cells
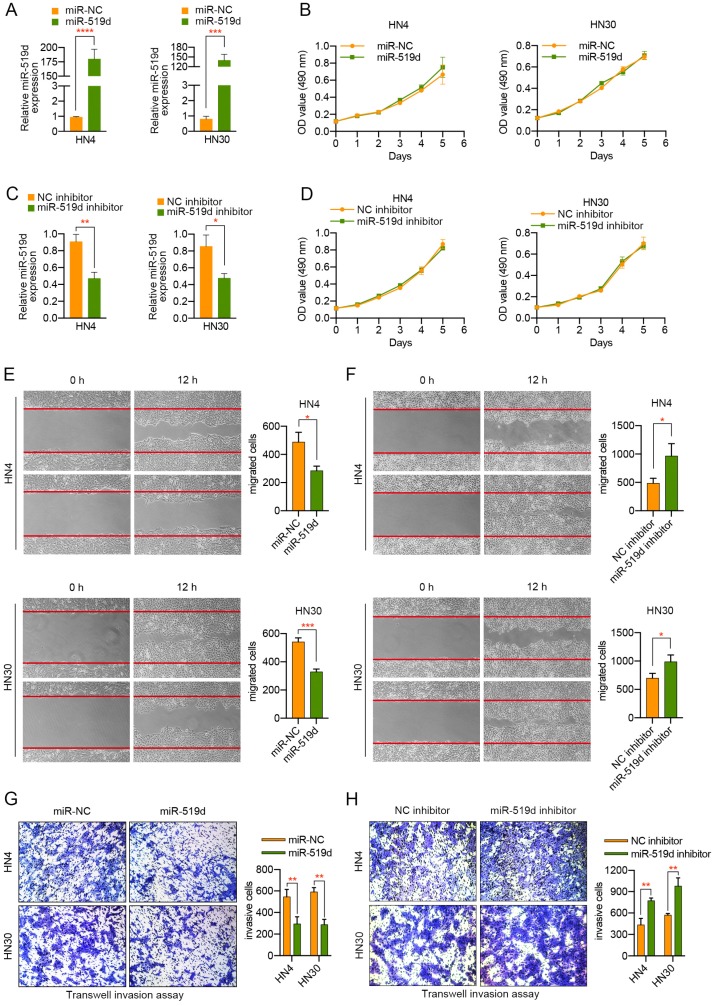
Due to the previous findings that miR-519d played a role in the cell growth of the hepatocellular carcinoma cell, ovarian cancer cell, and breast cancer cell, we tried to investigate whether it may affect the proliferation of OSCC cells. HN4 or HN30 cells were transfected with miR-519d or miR-NC, miR-519d inhibitor or NC inhibitor, and qPCR was then adopted to evaluate the transfection efficiency. As showed in Figure 2A, transfection of miR-519d mimics resulted in more than one hundred overexpression of miR-519d in HN4 and HN30 cells. On the contrary, miR-519d inhibitor led to more than a half decrease of miR-519d level in OSCC cells (Figure 2C). Thereafter, MTT experiment was conducted to evaluate its functions on cell proliferation. According to the proliferation curves by Figure 2B, 2D, the growth rates of HN4 and HN30 cells were not obviously changed after the overexpression or silencing of miR-519d. Hence, we came to the conclusion that miR-519d had no evident influences on the proliferation of OSCC cells.
Figure 2.
miR-519d significantly inhibits the migration and invasion of OSCC cells. (A) The expression of miR-519d in OSCC cells transfected with miR-519d mimics or miR-NC. (B) MTT experiment demonstrated that overexpression of miR-519d had no significant effect on the proliferation of OSCC cells. (C) The expression of miR-519d in OSCC cells transfected with miR-519d inhibitor or NC inhibitor. (D) MTT assay showed that silencing of miR-519d exhibited less effect on the proliferation of OSCC cells. (E, F) Dysregulation of miR-519d influenced the migration of OSCC cells, suggested by wound healing assay. (G, H) Transwell invasion assay revealed the suppressive role of miR-519d in the invasive abilities of OSCC cells. The fields of migrated and invasive cells on the membrane were captured (magnification × 100). Statistical analysis was performed using the t-tests. The data represent the mean values of three independent experiments. *, P < 0.05, **, P < 0.01. ***, P< 0.001. ****, P<0.0001.
miR-519d attenuates the migration and invasion of OSCC cells in vitro
As miR-519d expression in OSCC tissues was closely associated with lymph node metastasis of the patients, we tried to explore the potential role of miR-519d in the migration and invasion of OSCC cells. Transwell migration assay and wound healing assay were adopted to assess the migratory ability of OSCC cells. As presented in Figure 2E, overexpression of miR-519d remarkably inhibited the migratory capacity of both HN4 and HN30 cells. On the contrary, OSCC cells with miR-519d knockdown migrated more quickly compared to the control cells (Figure 2F). Furthermore, a transwell invasion assay was performed to explore the effect of miR-519d on the invasion activity of OSCC cells. As a result, compared to the control cells, HN4 or HN30 cells transfected with miR-519d displayed a less aggressive metastatic pattern, with weaker invasive abilities (Figure 2G). The opposite situation was observed when HN4 or HN30 cells transfected with miR-519d inhibitor (Figure 2H). In summary, the above results demonstrated that miR-519d play a suppressive role in the migration and invasion of OSCC cells.
miR-519d affects the migration and invasion of OSCC cells by directly targeting MMP3
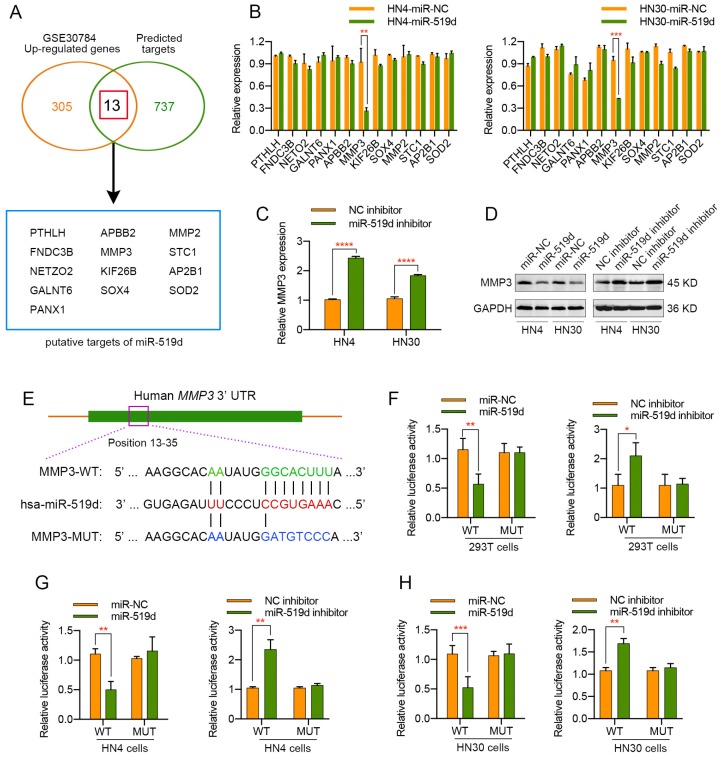
MiRNA-mediated gene expression silencing has previously been shown to be important in many physiological and pathological processes. To identify the potential target gene of miR-519d in OSCC, we applied 11 independent miRNA target prediction databases (Diana, MicroInspector, Miranda, MirTarget2, miTarget, NBmiRTar, PicTar, PITA, RNA22, RNAhybrid and TargetScan). Genes appeared in at least 4 out of the 11 databases were screened out. In order to narrow the candidate targets scope, we then made use of a OSCC gene chip data from GEO DataSets (GSE30784) and genes with upregulation of more than two fold were selected. An intersection of the predicted genes and the upregulated genes was assessed, and 13 candidate target genes were finally screened out (Figure 3A). Then, a real-time PCR analysis was performed to detect the expression changes of these potential target genes in HN4 and HN30 cells transfected with miR-519d mimics or miR-519d inhibitor. As presented in Figure 3B, MMP3 mRNA expression was significantly decreased in OSCC cells transfected with miR-519d mimics while other candidate targets displayed no evident changes. On the contrary, silencing of miR-519d in OSCC cells led to approximately two-fold increase of MMP3 mRNA levels (Figure 3C). Furthermore, western blot analysis revealed that overexpression or knockdown of miR-519d in HN4 and HN30 cells resulted in a significant lower or higher level of MMP3 correspondingly (Figure 3D). In addition, to further demonstrate that MMP3 acted as the direct target of miR-519d, a luciferase reporter assay was conducted. As illustrated by Figure 3F, overexpression of miR-519d in 293T cells expressing the wild type MMP3 3' UTR reporters resulted in approximately half reduction in luciferase activities, whereas mutations in the miR-519d-binding seed region of the MMP3 3' UTR abrogated the repressive effect of miR-519d. Conversely, reduction of miR-519d levels by miR-519d inhibitor in 293T cells expressing the wild type MMP3 3' UTR reporters presented much more luciferase activities. Simultaneously, similar results were obtained when the luciferase reporter assay was performed in HN4 and HN30 cells (Figure 3G, 3H). Therefore, we drew the conclusion that MMP3 was the direct target of miR-519d in OSCC cells and it might account for the biological effect of miR-519d on OSCC progression.
Figure 3.
MMP3 is a direct target of miR-519d in OSCC. (A) Venn diagrams of the predicted candidate target genes of miR-519d by bioinformatic method (miRecords) and mRNA microarray (GSE30784). (B) qPCR was adopted to validate the 13 potential targets in HN4 or HN30 cells after the transfection of miR-519d mimics or miR-NC. (C) The mRNA expression level of miR-519d in OSCC cells transfected with or without miR-519d inhibitor. (D) The protein level of MMP3 in HN4 or HN30 cells at 72 h after the transfection of miR-519d mimics or miR-519d inhibitor, as suggested by western blotting. (E) The predicted binding site of miR-519d and MMP3 and detailed sequences of WT-3′-UTR or MUT-3′-UTR of MMP3. (F, G and H) Luciferase reporter activity revealed the relative MMP3 reporter activities in 293T, HN4 or HN30 cells co-transfected with miR-519d mimics plus luciferase reporters or miR-519d inhibitor plus luciferase reporters. *, P < 0.05, **, P < 0.01. ***, P< 0.001. ****, P<0.0001
Dysregulation of MMP3 influenced the biological effects of miR-519d in OSCC
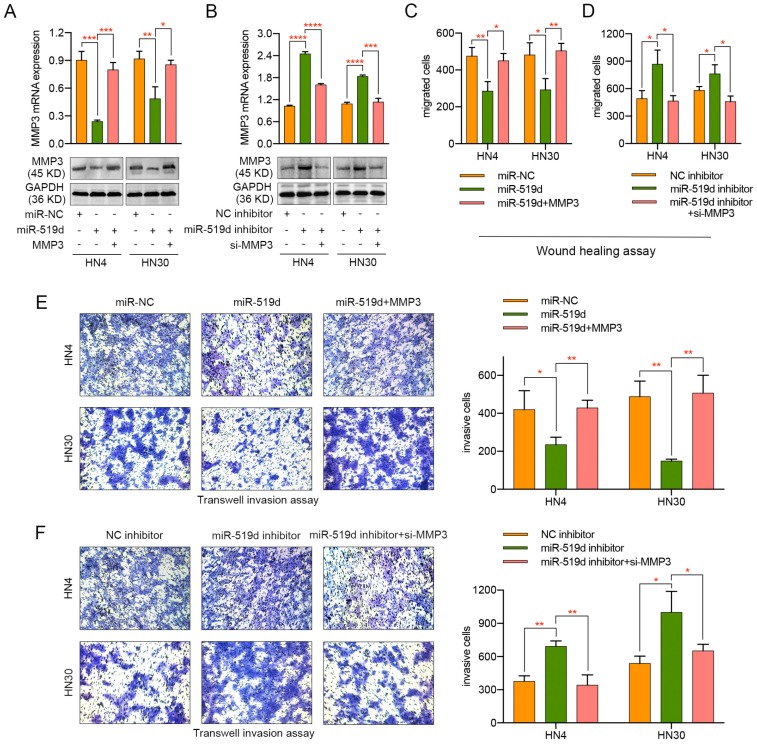
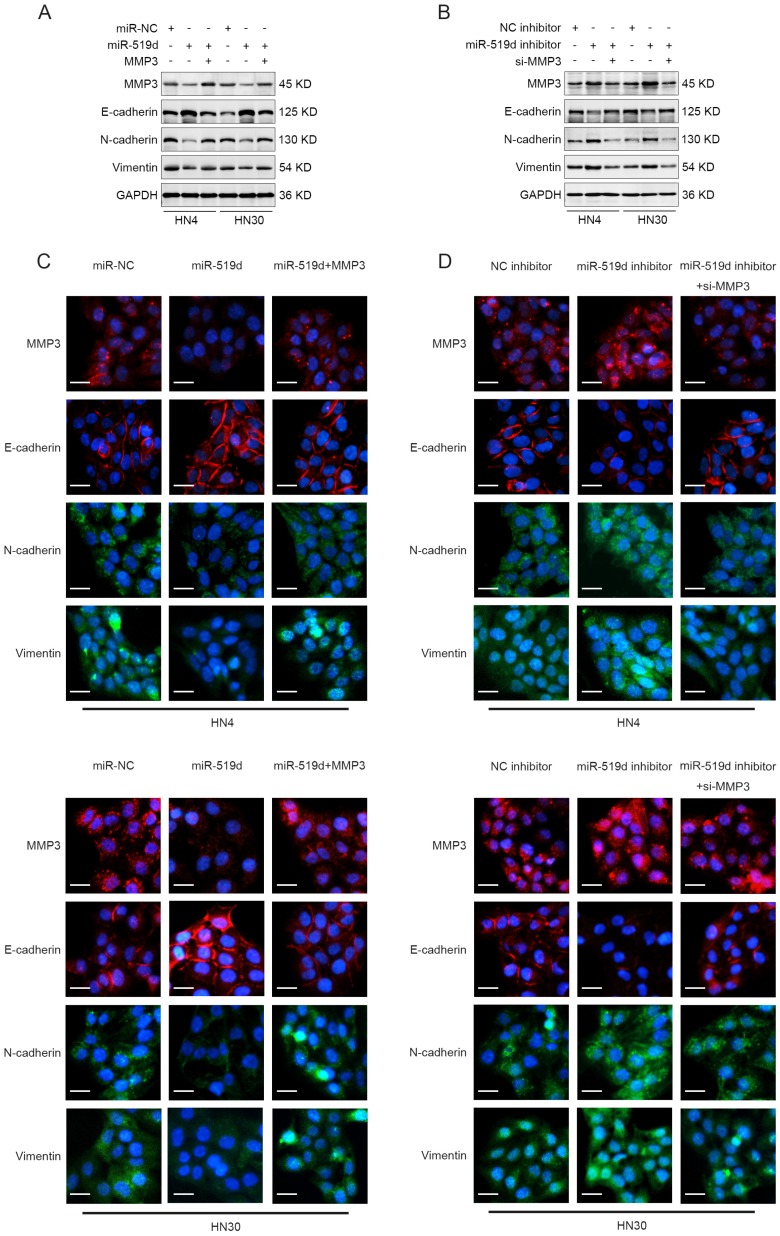
To further elucidate the function of miR-519d-induced inhibition of cell migration and invasion through MMP3 repression, HN4 and HN30 cells were transfected with miR-NC, miR-519d or miR-519d plus MMP3-expressing plasmid (without 3' UTR). As shown in Figure 4A, MMP3 was significantly downregulated upon miR-519d transfection but was rescued after transfection of MMP3-expressing plasmid both in mRNA and protein levels. In parallel, transfection of specific siRNA for MMP3 reversed the miR-519d inhibitor-mediated increase of MMP3 expression in OSCC cells (Figure 4B). Functionally, the miR-519d-induced inhibition of cell motility was abolished by MMP3 transfection, as revealed by both wound healing assay (Figure 4C) and transwell assay (Figure 4E, Figure S2C). Meanwhile, siRNA targeting MMP3 rescued the tumor promoting effect of miR-519d inhibitor on OSCC cells (Figure 4D, 4F, Figure S2D). These results demonstrated that MMP3 participated in the miR-519d-mediated suppression of migration and invasion in OSCC cells.
Figure 4.
Regulation of MMP3 influences the effect of miR-519d on the migration and invasion of OSCC cells. (A) The mRNA and protein expression of MMP3 in HN4 or HN30 cells after concurrent transfection with miR-519d or/and an exogenous MMP3 expression vector. (B) The mRNA and protein expression of MMP3 in OSCC cells were detected after the transfection of miR-519d inhibitor or/and MMP3 specific siRNA. (C) Wound healing assay revealed that miR-519d inhibited the migration of HN4 or HN30 cells while ectopic expression of MMP3 reversed this effect. (D) As presented by wound healing assay, inhibiting the expression of MMP3 by siRNA could rescue the miR-519d inhibitor-induced promotion of cell migration. (E) Transwell invasion assay performed in HN4 or HN30 cells showed that the miR-519d-induced inhibition of cell invasion was abolished by MMP3 transfection. (F) siRNA targeting MMP3 rescued the tumor promoting effect of miR-519d inhibitor on cell invasion in OSCC. *, P < 0.05, **, P < 0.01. ***, P< 0.001. ****, P<0.0001.
miR-519d is an important epithelial-mesenchymal transition (EMT) regulator in OSCC
As MMP3 was closely associated with cell migration and invasion 17 and was reported to be involved in EMT process 18, 19, we therefore detected if EMT associated markers (E-cadherin, N-cadherin and Vimentin) underwent changes after miR-519d dysregulation. The result of western blotting showed that OSCC cells with miR-519d overexpression expressed less N-cadherin and Vimentin, and more E-cadherin, while cotransfection of miR-519d and MMP3 reversed the expression patterns (Figure 5A). Moreover, OSCC cells transfected with miR-519d inhibitor resulted in the increased expression of N-cadherin and Vimentin, and decreased expression level of E-cadherin, whereas siRNA of MMP3 abolished this effect (Figure 5B). We further adopted the fluorescence assay to investigate the role of miR-519d in EMT process. As expected, similar results were obtained as described above (Figure 5C, 5D). These data suggested that miR-519d acted as an EMT regulator in OSCC progression and this process may be accomplished through targeting MMP3.
Figure 5.
miR-519d acts as a EMT regulator in OSCC. (A) HN4 or HN30 cells were transfected with miR-NC, miR-519d or miR-519d plus MMP3 plasmid. The protein levels of MMP3, E-cadherin, N-cadherin and Vimentin in these cells were measured by western blotting at 72 h after transfection. (B) HN4 or HN30 cells were transfected with NC inhibitor, miR-519d inhibitor or miR-519d inhibitor plus MMP3 siRNA. Western blotting showed the protein expression of MMP3, E-cadherin, N-cadherin and Vimentin in these cells at 72 h after transfection. (C) HN4 or HN30 cells were transfected with miR-NC, miR-519d or miR-519d plus MMP3 plasmid. Then immunofluorescence experiment was performed to detect the protein expression of MMP3, E-cadherin, N-cadherin and Vimentin in these cells (Scare bar: 20 μm). (D) OSCC cells (HN4 and HN30) were transfected with NC inhibitor, miR-519d inhibitor or miR-519d inhibitor plus MMP3 siRNA, and the expression of MMP3, E-cadherin, N-cadherin and Vimentin in tumor cells were measured using immunofluorescence (Scare bar: 20 μm).
Inverse correlation between the expression of miR-519d and MMP3 in OSCC
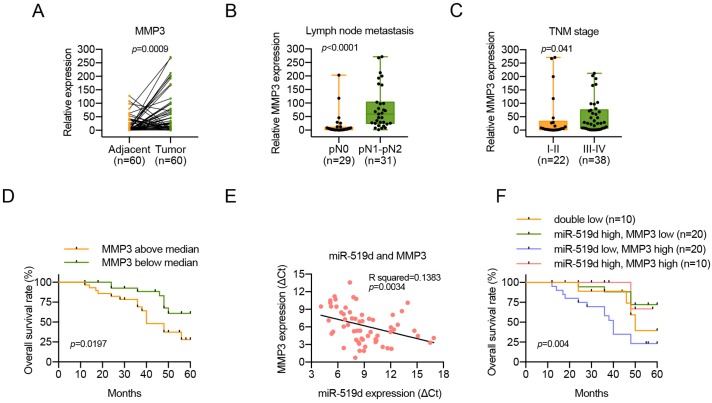
Since miR-519d was downregulated in OSCC, we determined to investigate the expression level of MMP3 in OSCC. The mRNA expression levels of MMP3 were measured in 60 pairs of OSCC tissues and adjacent normal tissues, and we found that MMP3 was upregulated in OSCC tissues (Figure 6A). Moreover, upregulation of MMP3 level in OSCC was positively associated with lymph node metastasis and advanced tumor stage (Figure 6B, 6C; Table 3). Importantly, the Kaplan-Meier survival curves show that patients with high expression levels of MMP3 had poorer overall survival than patients with low MMP3 expression (Figure 6D). Simultaneously, a significant inverse correlation was observed between miR-519d and MMP3 mRNA expression (R = -0.372, P = 0.0034; Figure 6E). In addition, OSCC patients with low miR-519d expression and high MMP3 expression always tended to have a more disappointing overall survival (Figure 6F), and the COX regression model revealed that miR-519d might serve as an independent factor for prognostic assessment (Table 4). These results indicated that the loss of miR-519d or overexpression of MMP3 might contribute to the metastatic potential and the poor prognosis of OSCC.
Figure 6.
Inverse correlation between the expression of miR-519d and MMP3, and the clinical implications of MMP3 in patients with OSCC. (A) MMP3 mRNA levels were determined by qPCR in 60 pairs of OSCC tumor samples and adjacent normal tissues. (P=0.0009). (B, C) The relative high expression level of MMP3 was correlated with lymph node metastasis (P<0.0001) and advanced TNM stage (P=0.041) in OSCC patients. (D) Survival curves revealed an overall better prognosis of patients with low MMP3 expression. (P=0.0197). (E) A negative correlation between miR-519d and MMP3 was found in OSCC clinical samples. (P=0.0034). (F) The Kaplan-Meier survival curves indicated that patients with a low miR-519d level and a high MMP3 level had the worst survival rate. (P=0.004).
Table 3.
Relationship between MMP3 level and clinicopathologic features (N = 60)
| Characteristics | No. of Patients | MMP3 Δ Cta | Non-parametric | P value | |
|---|---|---|---|---|---|
| No. | % | Mean ± SD | test value | ||
| Age (years) | |||||
| < 60 | 36 | 60.0 | 6.58 ± 2.93 | Z = -1.192 | 0.233 |
| ≥ 60 | 24 | 40.0 | 5.63 ± 2.78 | ||
| Gender | |||||
| Female | 19 | 31.7 | 6.26 ± 3.17 | Z = -0.087 | 0.930 |
| Male | 41 | 68.3 | 6.17 ± 2.79 | ||
| Smoking history | |||||
| Nonsmoker | 32 | 53.3 | 6.60 ± 3.10 | Z = -0.963 | 0.335 |
| Smoker | 28 | 46.7 | 5.75 ± 2.60 | ||
| Alcohol history | |||||
| Nondrinker | 38 | 63.3 | 6.59 ± 2.90 | Z = -1.350 | 0.177 |
| Drinker | 22 | 36.7 | 5.53 ± 2.79 | ||
| Tumor size (cm) | |||||
| ≤ 4 | 32 | 53.3 | 6.28 ± 3.06 | Z = -0.030 | 0.976 |
| > 4 | 28 | 46.7 | 6.11 ± 2.73 | ||
| Lymph node metastasis | |||||
| pN0 | 28 | 46.7 | 7.96 ± 2.75 | Z = -4.549 | 0.000 |
| pN1 to pN2 | 32 | 53.3 | 4.66 ± 2.00 | ||
| TNM stage | |||||
| I-II | 22 | 36.7 | 7.24 ± 3.23 | Z = -2.040 | 0.041 |
| III-IV | 38 | 63.3 | 5.60 ± 2.52 | ||
| Pathological differentiation | |||||
| Well | 25 | 41.7 | 7.09 ± 3.15 | Z = -1.897 | 0.058 |
| Moderately/poorly | 35 | 58.3 | 5.57 ± 2.54 | ||
| Disease Site | |||||
| Tongue | 25 | 41.7 | 6.19 ± 2.93 | H = 1.942 | 0.746 |
| Gingival | 8 | 13.3 | 5.70 ± 2.96 | ||
| Cheek | 9 | 15.0 | 5.44 ± 3.02 | ||
| Floor of Mouth | 12 | 20.0 | 6.62 ± 2.89 | ||
| Oropharynx | 6 | 10.0 | 7.21 ± 2.95 | ||
| Recurrence | |||||
| No | 50 | 22.5 | 6.42 ± 2.93 | Z = -1.309 | 0.190 |
| Yes | 10 | 77.5 | 5.10 ± 2.52 | ||
Abbreviations: SD, standard deviation; pN, pathological lymph node status; TNM stage, tumor-lymph node-metastasis stage.
aΔ Ct indicates the difference in the cycle number at which a sample's fluorescent signal passes a given threshold above baseline (Ct) derived from a specific gene compared with that of β-actin in tumor tissues.
Discussion
It is widely recognized that miRNAs play an important role in a variety of biological characteristics of cancers, including cell proliferation, apoptosis, migration and invasion. Accumulating studies have revealed that some kinds of miRNAs are closely associated with the initiation and progression of OSCC. For example, miRNA-21, which was upregulated in OSCC, could stimulate cell growth and indicate poor prognosis 20, 21. Also, by establishing transgenic mice, it was found that miR-211 enhanced the oncogenicity of carcinogen-induced oral carcinoma in vivo 22. On the contrary, certain kinds of miRNAs played a tumor suppressor role in OSCC and the underlying regulatory mechanisms involved differed. To be details, miR-9 could inhibit the proliferation of OSCC by suppressing CXCR4 via the Wnt/β-catenin signaling pathway 23. miR-145 inhibited tumor-initiating cells and IL-6-mediated paracrine effects in OSCC by targeting the SOX9/ADAM17 axis 24. What is more, some sorts of miRNAs even play crucial roles in chemoresistance and radioresistance in OSCC, thus providing a feasible and valid method for improving the therapeutic effect of patients 25-27. Apart from functioning as a tumor promotor or suppressor in OSCC, some kinds of miRNAs in saliva 28, 29, plasma 30 and tissues 31 can also be exploited as efficient biomarkers for OSCC diagnosis, certain miRNAs even act as guidance for the transformation of oral leukoplakia to OSCC 32, 33. Hence, it is worth investigating the potential regulatory mechanism of miRNAs in OSCC so as to discover promising strategies for treatment.
miR-519d has been identified as an important tumor related gene. Previous studies have reported the roles of miR-519d played in numerous cancers, including hepatocellular carcinoma 13, ovarian cancer 34, lung adenocarcinoma 12, gastric cancer 35 and so on. However, the regulatory roles of miR-519d in diverse tumor types presented different patterns, even to the contrary sometimes. To be details, miR-519d could serve as tumor suppressor in a great number of cancers, through inhibiting the proliferation 12-14, 34, 36-39, migration and invasion 12, 37, 38, and even EMT process 35. Nevertheless, regarding some kinds of cancers, miR-519d played a carcinogenic role, facilitating the progression and metastasis of cervical cancer 40, contributing to the tumorigenesis of colorectal cancer 41, and promoting melanoma progression 42. From our point of view, the opposite role of miR-519d in tumor progression might be due to the different genetic background and tumor origin and the biological function of miR-519d on tumor cells also may depend on the specific cellular context. It is also likely that miR-519d regulates distinct targets in different cancers, or that the function of miR-519d may be influenced by tumor microenvironment. Although the effect of miR-519d on tumor development is divergent, there is no denying that miR-519d plays an effective role in regulating the biological behaviors of tumor. In this study, we found that miR-519d was significantly downregulated in OSCC and was correlated with lymph node metastasis, advanced tumor stage and poor overall survival. Moreover, our gain-and loss-of-function experiments demonstrated that miR-519d inhibits the migration and invasion in OSCC.
In this study, we found that the miR-519d expression was significantly decreased in OSCC and one issue worth considering was how miR-519d was downregulated. Previous studies concluded that the possible causes of miRNA dysregulation include DNA copy number aberrations, mutations, epigenetic aberrations, dysregulation of transcription factors targeting miRNAs and alterations in the miRNA biogenesis pathway 43. Several studies also illustrated that miRNAs, including miR-519d, could be regulated by lncRNAs in a variety of cancers 44, 45. Furthermore, gain and loss of function of several canonical oncogenic and tumor suppressor pathways in cancer cells, such as myc, p53, NF-κB and Hippo signaling, contributes to miRNA dysregulation as well 46, 47. Some studies also illustrated that cytokines and growth factors, such as TGF-β1 and connective tissue growth factor, could modulate miRNA expression 48, 49. In addition, a great number of studies have identified that aberrant DNA methylation of miRNA genes could serve as an essential mechanism of miRNA dysregulation in cancers 50. Epigenetic gene silencing due to promoter hypermethylation is one of the most common mechanisms by which tumor suppressor genes are inactivated during tumorigenesis, including miRNAs as well 50. Considering the fact that abnormal methylation of the DNA region located upstream of C19MC is associated with the expression of miR-519, we have tried to identify the relationship between the methylation status of C19MC promoter and miR-519d expression in OSCC. Fortunately, we found that hypermethylation of the DNA region located upstream of C19MC was associated with the downregulation of miR-519d in OSCC. However, there still exist other factors which could modulate the expression of miR-519d in OSCC and more efforts should be taken to explain this problem in the future.
In addition, apart from illustrating the fundamental biological roles of miR-519d in OSCC, we also demonstrated MMP3 was the downstream target of miR-519d. MMP3, a member of the family of MMPs, has been reported to enhance the migratory and invasive abilities of tumor cells. The role of MMPs in cancers has been well elucidated; they can remarkably promote the malignancy of tumor cells by degrading the extracellular matrix (ECM), facilitating angiogenesis, tumor invasion, and metastasis 51. Meanwhile, previous studies described that dysregulation of MMP3 could be an important factor for mammary tumor cell progression to an invasive phenotype 52. Therefore, our explorations that MMP3 acted as a tumor promoter were in accordance with the previous findings. Furthermore, it was reported that MMP3 was closely associated with EMT characteristics, the acquisition of which facilitated tumor invasion and metastasis 19, 53. In addition, previous studies have illustrated that treatment of mouse mammary epithelial cells with MMP3 resulted in the induction of EMT, accompanied with a decrease of E-cadherin, increased motility and invasiveness 52, 53. Thus, we attempted to discover whether miR-519d could influence the EMT by degrading MMP3. Fortunately, we found that OSCC cells transfected with miR-519d underwent a less activity of EMT, while MMP3 plasmid transfection recovered this process, as suggested by the expression of EMT markers E-cadherin, N-cadherin and Vimentin. However, the specific mechanisms by which MMP3 or miR-519d induced EMT activity was not well elucidated. A published research has indicated that the induction of Snail by MMP3 could result in the downregulation of E-cadherin and the overexpression of Vimentin in various cancers, which suggested the functional role of MMP3 in the induction of EMT 54. Another study stated that MMP3-induced EMT conversion is accompanied by E-cadherin cleavage and a redistribution of B-catenin, where it can form a transcriptional complex and induces the transcription of some genes involved in EMT process 55, 56. However, whether the cleavage of E-cadherin by MMP3 was accomplished directly or indirectly remains unknown. Furthermore, a review concentrated on the regulation of EMT transitions by microRNAs concluded that microRNAs can efficiently mediate EMT by targeting multiple targets involved in transcription process or signaling pathways. Furthermore, the differences of targeted EMT genes by microRNAs may be attributed to diverse cell context and microRNAs may also function differently at various stages during physiological or pathological events 57. In this study, we found that miR-519d might modulate the EMT process in OSCC by targeting MMP3, but it did not exclude the existence of other mechanisms. Thus, further investigation of the precise underlying mechanisms is needed in the future study.
In conclusion, our study highlights the role of miR-519d in suppressing the migration and invasion of OSCC cells by targeting MMP3. Moreover, miR-519d may also act as a regulator of EMT in OSCC. Generally, OSCC patients with low miR-519d expression and high MMP3 expression tended to have the worst clinical outcome. Collectively, our results provide insights into the therapeutic value of miR-519d in reducing OSCC metastasis and suggest that miR-519d and MMP3 could serve as effective predictors for metastasis and prognosis in OSCC.
Supplementary Material
Supplementary figures.
Acknowledgments
This work was supported by National Key R&D Program of China (2017YFC 0840100, 2017YFC 0840110).
Abbreviations
- miR-519d
MicroRNA-519d
- OSCC
oral squamous cell carcinoma
- MMP3
matrix metalloproteinase-3
- EMT
epithelial-mesenchymal transition
- HNSCC
head and neck squamous cell carcinoma
- miRNAs
microRNAs
- DMSO
dimethyl sulfoxide
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Mao L. Oral squamous cell carcinoma - progresses from risk assessment to treatment. Chin J Dent Res. 2012;15:83–8. [PubMed] [Google Scholar]
- 3.Wang B, Zhang S, Yue K, Wang XD. The recurrence and survival of oral squamous cell carcinoma: a report of 275 cases. Chin J Cancer. 2013;32:614–8. doi: 10.5732/cjc.012.10219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yong-Deok K, Eun-Hyoung J, Yeon-Sun K, Kang-Mi P, Jin-Yong L, Sung-Hwan C. et al. Molecular genetic study of novel biomarkers for early diagnosis of oral squamous cell carcinoma. Med Oral Patol Oral Cir Bucal. 2015;20:e167–79. doi: 10.4317/medoral.20229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 6.Baltimore D, Boldin MP, O'Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008;9:839–45. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 7.Pereira DM, Rodrigues PM, Borralho PM, Rodrigues CM. Delivering the promise of miRNA cancer therapeutics. Drug Discov Today. 2013;18:282–9. doi: 10.1016/j.drudis.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 8.Zhan C, Li C, Zhang H, Tang H, Ji F, Qiao SC. et al. MicroRNA-150 upregulation reduces osteosarcoma cell invasion and metastasis by downregulating Ezrin. Oncol Lett. 2016;12:3457–62. doi: 10.3892/ol.2016.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plasterk RH. Micro RNAs in animal development. Cell. 2006;124:877–81. doi: 10.1016/j.cell.2006.02.030. [DOI] [PubMed] [Google Scholar]
- 10.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–22. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 11.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 12.Bai Y, Lu C, Zhang G, Hou Y, Guo Y, Zhou H. et al. Overexpression of miR-519d in lung adenocarcinoma inhibits cell proliferation and invasion via the association of eIF4H. Tumour Biol. 2017;39:1010428317694566. doi: 10.1177/1010428317694566. [DOI] [PubMed] [Google Scholar]
- 13.Hou YY, Cao WW, Li L, Li SP, Liu T, Wan HY. et al. MicroRNA-519d targets MKi67 and suppresses cell growth in the hepatocellular carcinoma cell line QGY-7703. Cancer Lett. 2011;307:182–90. doi: 10.1016/j.canlet.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 14.Li D, Song H, Wu T, Xie D, Hu J, Zhao J. et al. MiR-519d-3p suppresses breast cancer cell growth and motility via targeting LIM domain kinase 1. Mol Cell Biochem. 2018;444:169–78. doi: 10.1007/s11010-017-3241-4. [DOI] [PubMed] [Google Scholar]
- 15.Fornari F, Milazzo M, Chieco P, Negrini M, Marasco E, Capranico G. et al. In hepatocellular carcinoma miR-519d is up-regulated by p53 and DNA hypomethylation and targets CDKN1A/p21, PTEN, AKT3 and TIMP2. J Pathol. 2012;227:275–85. doi: 10.1002/path.3995. [DOI] [PubMed] [Google Scholar]
- 16.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O. et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–70. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 17.Cheng X, Wei L, Huang X, Zheng J, Shao M, Feng T. et al. Solute Carrier Family 39 Member 6 Gene Promotes Aggressiveness of Esophageal Carcinoma Cells by Increasing Intracellular Levels of Zinc, Activating Phosphatidylinositol 3-Kinase Signaling, and Up-regulating Genes That Regulate Metastasis. Gastroenterology. 2017;152:1985–97. doi: 10.1053/j.gastro.2017.02.006. e12. [DOI] [PubMed] [Google Scholar]
- 18.Sternlicht MD, Lochter A, Sympson CJ, Huey B, Rougier JP, Gray JW. et al. The stromal proteinase MMP3/stromelysin-1 promotes mammary carcinogenesis. Cell. 1999;98:137–46. doi: 10.1016/s0092-8674(00)81009-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radisky DC, Levy DD, Littlepage LE, Liu H, Nelson CM, Fata JE. et al. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature. 2005;436:123–7. doi: 10.1038/nature03688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mydlarz W, Uemura M, Ahn S, Hennessey P, Chang S, Demokan S. et al. Clusterin is a gene-specific target of microRNA-21 in head and neck squamous cell carcinoma. Clin Cancer Res. 2014;20:868–77. doi: 10.1158/1078-0432.CCR-13-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Huang H, Sun L, Yang M, Pan C, Chen W. et al. MiR-21 indicates poor prognosis in tongue squamous cell carcinomas as an apoptosis inhibitor. Clin Cancer Res. 2009;15:3998–4008. doi: 10.1158/1078-0432.CCR-08-3053. [DOI] [PubMed] [Google Scholar]
- 22.Chen YF, Yang CC, Kao SY, Liu CJ, Lin SC, Chang KW. MicroRNA-211 Enhances the Oncogenicity of Carcinogen-Induced Oral Carcinoma by Repressing TCF12 and Increasing Antioxidant Activity. Cancer Res. 2016;76:4872–86. doi: 10.1158/0008-5472.CAN-15-1664. [DOI] [PubMed] [Google Scholar]
- 23.Yu T, Liu K, Wu Y, Fan J, Chen J, Li C. et al. MicroRNA-9 inhibits the proliferation of oral squamous cell carcinoma cells by suppressing expression of CXCR4 via the Wnt/beta-catenin signaling pathway. Oncogene. 2014;33:5017–27. doi: 10.1038/onc.2013.448. [DOI] [PubMed] [Google Scholar]
- 24.Yu CC, Tsai LL, Wang ML, Yu CH, Lo WL, Chang YC. et al. miR145 targets the SOX9/ADAM17 axis to inhibit tumor-initiating cells and IL-6-mediated paracrine effects in head and neck cancer. Cancer Res. 2013;73:3425–40. doi: 10.1158/0008-5472.CAN-12-3840. [DOI] [PubMed] [Google Scholar]
- 25.Shiiba M, Shinozuka K, Saito K, Fushimi K, Kasamatsu A, Ogawara K. et al. MicroRNA-125b regulates proliferation and radioresistance of oral squamous cell carcinoma. Br J Cancer. 2013;108:1817–21. doi: 10.1038/bjc.2013.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu ZW, Zhong LP, Ji T, Zhang P, Chen WT, Zhang CP. MicroRNAs contribute to the chemoresistance of cisplatin in tongue squamous cell carcinoma lines. Oral Oncol. 2010;46:317–22. doi: 10.1016/j.oraloncology.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 27.Weng JH, Yu CC, Lee YC, Lin CW, Chang WW, Kuo YL. miR-494-3p Induces Cellular Senescence and Enhances Radiosensitivity in Human Oral Squamous Carcinoma Cells. Int J Mol Sci; 2016. p. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gai C, Camussi F, Broccoletti R, Gambino A, Cabras M, Molinaro L. et al. Salivary extracellular vesicle-associated miRNAs as potential biomarkers in oral squamous cell carcinoma. BMC Cancer. 2018;18:439. doi: 10.1186/s12885-018-4364-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Momen-Heravi F, Trachtenberg AJ, Kuo WP, Cheng YS. Genomewide Study of Salivary MicroRNAs for Detection of Oral Cancer. J Dent Res. 2014;93:86S–93S. doi: 10.1177/0022034514531018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pedersen NJ, Jensen DH, Lelkaitis G, Kiss K, Charabi BW, Ullum H. et al. MicroRNA-based classifiers for diagnosis of oral cavity squamous cell carcinoma in tissue and plasma. Oral Oncol. 2018;83:46–52. doi: 10.1016/j.oraloncology.2018.05.020. [DOI] [PubMed] [Google Scholar]
- 31.Troiano G, Mastrangelo F, Caponio VCA, Laino L, Cirillo N, Lo Muzio L. Predictive Prognostic Value of Tissue-Based MicroRNA Expression in Oral Squamous Cell Carcinoma: A Systematic Review and Meta-analysis. J Dent Res. 2018;97:759–66. doi: 10.1177/0022034518762090. [DOI] [PubMed] [Google Scholar]
- 32.Chen H, Liu X, Jin Z, Gou C, Liang M, Cui L. et al. A three miRNAs signature for predicting the transformation of oral leukoplakia to oral squamous cell carcinoma. Am J Cancer Res. 2018;8:1403–13. [PMC free article] [PubMed] [Google Scholar]
- 33.Philipone E, Yoon AJ, Wang S, Shen J, Ko YC, Sink JM. et al. MicroRNAs-208b-3p, 204-5p, 129-2-3p and 3065-5p as predictive markers of oral leukoplakia that progress to cancer. Am J Cancer Res. 2016;6:1537–46. [PMC free article] [PubMed] [Google Scholar]
- 34.Pang Y, Mao H, Shen L, Zhao Z, Liu R, Liu P. MiR-519d represses ovarian cancer cell proliferation and enhances cisplatin-mediated cytotoxicity in vitro by targeting XIAP. Onco Targets Ther. 2014;7:587–97. doi: 10.2147/OTT.S60289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yue H, Tang B, Zhao Y, Niu Y, Yin P, Yang W. et al. MIR-519d suppresses the gastric cancer epithelial-mesenchymal transition via Twist1 and inhibits Wnt/beta-catenin signaling pathway. Am J Transl Res. 2017;9:3654–64. [PMC free article] [PubMed] [Google Scholar]
- 36.Ye X, Lv H. MicroRNA-519d-3p inhibits cell proliferation and migration by targeting TROAP in colorectal cancer. Biomed Pharmacother. 2018;105:879–86. doi: 10.1016/j.biopha.2018.04.114. [DOI] [PubMed] [Google Scholar]
- 37.Yan CQ, Lu YH, Tang SM, Fan WX. MiR-519d inhibits prostate cancer cell proliferation, cycle and invasion via targeting NRBP1. Eur Rev Med Pharmacol Sci. 2018;22:2985–90. doi: 10.26355/eurrev_201805_15054. [DOI] [PubMed] [Google Scholar]
- 38.Li YY, Shao JP, Zhang SP, Xing GQ, Liu HJ. miR-519d-3p Inhibits Cell Proliferation and Invasion of Gastric Cancer by Downregulating B-Cell Lymphoma 6. Cytogenet Genome Res. 2018;154:12–9. doi: 10.1159/000487372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang L, Shi S, Shi Q, Zhang H, Xia Y, Zhong T. MicroRNA-519d-3p Inhibits Proliferation and Promotes Apoptosis by Targeting HIF-2alpha in Cervical Cancer Under Hypoxic Conditions. Oncol Res. 2018;26:1055–62. doi: 10.3727/096504018X15152056890500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou JY, Zheng SR, Liu J, Shi R, Yu HL, Wei M. MiR-519d facilitates the progression and metastasis of cervical cancer through direct targeting Smad7. Cancer Cell Int. 2016;16:21. doi: 10.1186/s12935-016-0298-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang X, Hu Y, Liu Y, Liu W, Zhao X, Liu M. et al. C14orf28 downregulated by miR-519d contributes to oncogenicity and regulates apoptosis and EMT in colorectal cancer. Mol Cell Biochem. 2017;434:197–208. doi: 10.1007/s11010-017-3049-2. [DOI] [PubMed] [Google Scholar]
- 42.Hua KT, Hong JB, Sheen YS, Huang HY, Huang YL, Chen JS. et al. miR-519d Promotes Melanoma Progression by Downregulating EphA4. Cancer Res. 2018;78:216–29. doi: 10.1158/0008-5472.CAN-17-1933. [DOI] [PubMed] [Google Scholar]
- 43.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–14. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang S, Lu W, Ge D, Meng N, Li Y, Su L. et al. A new microRNA signal pathway regulated by long noncoding RNA TGFB2-OT1 in autophagy and inflammation of vascular endothelial cells. Autophagy. 2015;11:2172–83. doi: 10.1080/15548627.2015.1106663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu H, Li C, Yang J, Sun Y, Zhang S, Yang J. et al. Long noncoding RNA CASC9/miR-519d/STAT3 positive feedback loop facilitate the glioma tumourigenesis. J Cell Mol Med. 2018;22:6338–44. doi: 10.1111/jcmm.13932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lotterman CD, Kent OA, Mendell JT. Functional integration of microRNAs into oncogenic and tumor suppressor pathways. Cell Cycle. 2008;7:2493–9. doi: 10.4161/cc.7.16.6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mori M, Triboulet R, Mohseni M, Schlegelmilch K, Shrestha K, Camargo FD. et al. Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer. Cell. 2014;156:893–906. doi: 10.1016/j.cell.2013.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang B, Komers R, Carew R, Winbanks CE, Xu B, Herman-Edelstein M. et al. Suppression of microRNA-29 expression by TGF-beta1 promotes collagen expression and renal fibrosis. J Am Soc Nephrol. 2012;23:252–65. doi: 10.1681/ASN.2011010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burns WC, Twigg SM, Forbes JM, Pete J, Tikellis C, Thallas-Bonke V. et al. Connective tissue growth factor plays an important role in advanced glycation end product-induced tubular epithelial-to-mesenchymal transition: implications for diabetic renal disease. J Am Soc Nephrol. 2006;17:2484–94. doi: 10.1681/ASN.2006050525. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki H, Maruyama R, Yamamoto E, Kai M. DNA methylation and microRNA dysregulation in cancer. Mol Oncol. 2012;6:567–78. doi: 10.1016/j.molonc.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lochter A, Srebrow A, Sympson CJ, Terracio N, Werb Z, Bissell MJ. Misregulation of stromelysin-1 expression in mouse mammary tumor cells accompanies acquisition of stromelysin-1-dependent invasive properties. J Biol Chem. 1997;272:5007–15. doi: 10.1074/jbc.272.8.5007. [DOI] [PubMed] [Google Scholar]
- 53.Lochter A, Galosy S, Muschler J, Freedman N, Werb Z, Bissell MJ. Matrix metalloproteinase stromelysin-1 triggers a cascade of molecular alterations that leads to stable epithelial-to-mesenchymal conversion and a premalignant phenotype in mammary epithelial cells. J Cell Biol. 1997;139:1861–72. doi: 10.1083/jcb.139.7.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Przybylo JA, Radisky DC. Matrix metalloproteinase-induced epithelial-mesenchymal transition: tumor progression at Snail's pace. Int J Biochem Cell Biol. 2007;39:1082–8. doi: 10.1016/j.biocel.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 55.Orsulic S, Huber O, Aberle H, Arnold S, Kemler R. E-cadherin binding prevents beta-catenin nuclear localization and beta-catenin/LEF-1-mediated transactivation. J Cell Sci. 1999;112( Pt 8):1237–45. doi: 10.1242/jcs.112.8.1237. [DOI] [PubMed] [Google Scholar]
- 56.Sternlicht MD, Bissell MJ, Werb Z. The matrix metalloproteinase stromelysin-1 acts as a natural mammary tumor promoter. Oncogene. 2000;19:1102–13. doi: 10.1038/sj.onc.1203347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lamouille S, Subramanyam D, Blelloch R, Derynck R. Regulation of epithelial-mesenchymal and mesenchymal-epithelial transitions by microRNAs. Curr Opin Cell Biol. 2013;25:200–7. doi: 10.1016/j.ceb.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures.