Abstract
Excessive alcohol use is extremely prevalent in the United States, particularly among trauma-exposed individuals. While several studies have examined genetic influences on alcohol use and related problems, this has not been studied in the context of trauma-exposed populations. We report results from a genome-wide association study of alcohol consumption and associated problems as measured by the alcohol use disorders identification test (AUDIT) in a traumaexposed cohort. Results indicate a genome-wide significant association between total AUDIT score and rs1433375 [N = 1036, P = 2.61 × 10–8 (dominant model), P = 7.76 × 10–8 (additive model)], an intergenic single-nucleotide polymorphism located 323 kb upstream of the sodium channel and clathrin linker 1 (SCLT1) at 4q28. rs1433375 was also significant in a meta-analysis of two similar, but independent, cohorts (N = 1394, P = 0.0004), the Marine Resiliency Study and Systems Biology PTSD Biomarkers Consortium. Functional analysis indicated that rs1433375 was associated with SCLT1 gene expression and cortical-cerebellar functional connectivity measured via resting state functional magnetic resonance imaging. Together, findings suggest a role for sodium channel regulation and cerebellar functioning in alcohol use behavior. Identifying mechanisms underlying risk for problematic alcohol use in traumaexposed populations is critical for future treatment and prevention efforts.
Keywords: alcohol consumption, alcohol use disorder, AUDIT, expression QTL, fMRI, genome-wide association study
INTRODUCTION
Excessive alcohol use is the third leading cause of preventable deaths in the United States (Jonas et al. 2012; Centers for Disease Control and Prevention 2012). A large literature has demonstrated that excessive alcohol use and related disorders are particularly prevalent among traumaexposed individuals, such as those living in neighborhoods with high rates of assaultive and interpersonal violence (Stockdale et al. 2007) and among military personnel (Allen, Crawford, & Kudler 2016). While several studies have examined genetic influences on alcohol involvement, there is a limited understanding of genetic risk for drinking in populations with high levels of trauma exposure.
While several large-scale genetic studies of alcohol dependence (AD) have been conducted over the past two decades, few replicable genetic markers have been identified. Notable exceptions include findings from the Study of Addiction: Genetics and Environment and the Collaborative Study on the Genetics of Alcoholism, which report risk variants for AD in novel loci including KIAA0040 (Zuo et al. 2012), the PHF3-PTP4A1 gene complex (Zuo et al. 2011, 2014), rs1437396 at chromosomal location 2p16.1 and other variants in the alcohol dehydrogenase (ADH) gene cluster (Gelernter et al. 2014). However, with the exception of the ADH gene cluster, genetic variants (and corresponding biological pathways) identified in previous genetic studies of AD have not yet pointed to tractable mechanisms underlying genotype-phenotype associations that intersect with current neurobiological understanding of the effects of alcohol on behavior.
Previous studies have indicated that genetic analyses of quantitative phenotypes that precede AD, such as frequency and quantity of alcohol consumption, may improve power to detect variants with small effects (Agrawal et al. 2009). For example, genome-wide association studies (GWASs) of alcohol consumption have implicated several genome-wide significant variants (e.g. Schumann et al. 2011; Clarke et al. 2017; Webb et al. 2017). Reviews of the current state of GWAS findings for alcohol use behaviors indicate that larger sample sizes and quantitative measures of alcohol use increase the likelihood of identifying genome-wide significant findings via an increase in statistical power. For this reason, conducting GWAS on quantitative measures of alcohol use has gained popularity (Agrawal et al. 2009; Baik et al. 2011; Heath et al. 2011; Schumann et al. 2011; Takeuchi et al. 2011; Agrawal et al. 2013; Kapoor et al. 2013; Wang et al. 2013; Xu et al. 2015; Clarke et al. 2017; Webb et al. 2017).
In this study, we aimed to increase understanding of genetic risk for alcohol use behavior by conducting a GWAS in the Grady Trauma Project (GTP), a large, primarily African-American community cohort study initially designed to assess post-traumatic stress disorder (PTSD; Binder et al. 2008; Gillespie et al. 2009; Ressler et al. 2011; Almli et al. 2013). The GTP sample was recruited from the general medical clinics of the Grady Memorial Hospital, a publicly funded hospital that serves economically disadvantaged individuals in Atlanta, Georgia. To validate GWAS findings from the GTP, we also analyzed the findings in a similar sample consisting of 1394 men and women from two independent cohorts with high rates of trauma exposure. Finally, we characterized GWAS findings in the discovery (GTP) sample using both gene expression and brain imaging data available on a subset of the population. Given that the executive control network (ECN) has been repeatedly associated with alcohol use behavior, and in particular, fronto-cerebellar connectivity (Chanraud et al. 2010; Herting, Fair, & Nagel 2011; Sutherland et al. 2012; Fryer et al. 2013; Krmpotich et al. 2013; Moulton et al. 2014), we examined the influence of genetic risk variants implicated in alcohol use behavior on this neural network.
METHODS AND MATERIALS
Discovery sample and assessment
The GTP cohort was collected to investigate the genetic and environmental predictors of psychiatric disorders in a population of urban men (28.2 percent) and women (71.8 percent). The mean (SD) age in the sample was 39.5 (13.4) years, ranging from 18 to 65 years (Table S1). Screening interviews were completed on participants approached while in the waiting rooms of primary care or obstetrical-gynecological clinics of the Grady Memorial Hospital in Atlanta, Georgia. Participant’s demographic information (e.g. self-identified race, sex and age) and substance use behaviors were assessed by verbal interview ($15 for participation in this phase of the study). Further details regarding the GTP dataset can be found in Gillespie et al. (2009). Written and verbal informed consent was obtained for all participants, and all procedures in this study were approved by the institutional review boards of Emory University School of Medicine and Grady Memorial Hospital, Atlanta, Georgia.
Alcohol consumption and associated behaviors were measured using alcohol use disorders identification test (AUDIT), which measures both frequency of use and consequences accompanying excessive use (Saunders et al. 1993; Babor et al. 2001); the total score was obtained by summing the responses from the 10 questions on the AUDIT survey. Each item of the AUDIT is rated on a scale of 0–4, with higher scores reflecting more problematic alcohol drinking. Thus, our composite score is a quantitative measure of alcohol consumption and related hazardous behaviors (Babor et al. 2001; Iacono et al. 1999; Reinert & Allen 2002, 2007). Of the individuals included in the GTP genetic sample (N = 3340, Table S1), 55 percent had AUDIT data (N = 1829). Of those with AUDIT data, 57 percent reported drinking in the last year (N = 1036, 373 men and 663 women). To avoid confounding our measure of problematic drinking behavior with the decision to initiate alcohol use (Sartor et al. 2013), only individuals who had at least one drink during the last year (57 percent) were included in the GWAS (N = 1036). While the AUDIT is typically used as a screening tool in clinical settings, two recent studies have validated the use of AUDIT scores as a useful tool to examine genetic risk for alcohol use behavior in samples with lower levels of overall alcohol consumption and alcohol use problems as is observed in the GTP (Mbarek et al. 2015; Xu et al. 2015).
Genetic data: quality control and analyses
Genotyping
Grady Trauma Project participants provided a saliva sample and/or blood sample. With Oragene saliva kits (DNA Genotek Inc., Ottawa, Ontario, Canada), DNA was extracted from saliva in Oragene collection vials (DNA Genotek Inc.) using the DNAdvance kit (Beckman Coulter Genomics, Danvers, MA), while DNA from blood was extracted using either the E.Z.N.A. Mag-Bind Blood DNA Kit (Omega Bio-Tek, Inc., Norcross, GA) or ArchivePure DNA Blood Kit (5 Prime, Inc., Gaithersburg, MD). Genome-wide single-nucleotide polymorphism (SNP) genotyping was conducted on 4622 subjects using Illumina’s (San Diego, CA) HumanOmniExpress (N = 280) and Omnil-Quad BeadChip (N = 4342). The HumanOmniExpress interrogates 730 525 individual SNPs per sample, while the Omnil-Quad BeadChip interrogates 1 011 219 individual SNPs. Genotypes were called using Illumina’s GENOMESTUDIO software. PLINK (Purcell et al. 2007) was utilized to perform quality control (QC) analyses on the genetic data from each chip separately and then after the data from each chip was combined (Supporting Information Fig. S1). For each chip, we first removed individuals with greater than 5 percent missing data. We further identified and removed related individuals by using PLINK to estimate the proportion of identity by descent (IBD) for each pair of individuals. Among pairs of individuals with an IBD proportion >0.12 (indicating cousins or a closer relation), we removed the individual in each pair with the higher rate of missing genotype data. We restricted SNPs to autosomes, those with greater than a 95 percent call rate, a frequency of less than 0.01 and Hardy-Weinberg equilibrium (HWE) P-value < 1 × 10–5. We next performed principal component analysis (PCA) to infer axes of genetic ancestry. Prior to PCA, we used PLINK to prune the data in windows of 50 base pairs, removing one SNP from each pair of SNPs with r2 > 0.05 to obtain a set of roughly independent SNPs. Based on PCA, we retained those individuals who fell within three standard deviations of the medians of the first and second principal components of self-reported African Americans in order to minimize genetic differences due to genetic ancestry. Supporting Information Fig. S2 shows that these individuals cluster with Africans in a plot of the first and second principal components anchored by three HapMap populations [CEU, YRI and Asian (CHB and JPT combined)]. After completion of QC and PCA, we merged the cleaned data from the two chips and performed more stringent QC filters; SNPs with call rates less than 98 percent, a frequency of less than 0.1 and a HWE < 1 × 10–5 were removed. After conducting a PCA on the combined chip data, we removed another 99 subjects who fell outside of our criteria (i.e. three standard deviations from the medians of the first and second principal components of self-reported African Americans); thus, the genetic sample consisted of 3340 African-American individuals genotyped for 501 278 SNPs. When comparing the genotyped individuals to the non-genotyped individuals, the proportion of men to women was different with a greater proportion of men in the analytic sample (Table S1). We note that to address this potential confound, we used sex as a covariate throughout the discovery and follow-up analyses.
Gene expression
A subset of the participants following the screening interview returned for more detailed interviews and a blood draw. The methodology for assessment of gene expression has been previously described by Mehta et al. (2011, 2013). Briefly, whole blood was collected between 8 and 9 A.M. in Tempus RNA tubes. Whole-genome expression profiles were generated as follows: RNA was isolated using the Versagene kit (Gentra Systems, Big Lake, MN), quantified using the NanoPhotometer and checked for quality on the Agilent (Santa Clara, CA) Bioanalyzer. Total RNA (250 ng) were reverse transcribed to cDNA, converted to biotin-labeled cRNA using the Ambion kit (AMIL1791, Applied Biosystems, Foster City, CA). cRNAs (750 ng) were hybridized to Illumina HT-12 v3.0 or v4.0 arrays (Illumina) and incubated overnight for 16 hours at 55°C. Arrays were washed, stained with Cy3-labeled streptavidin, dried and scanned on the Illumina BeadScan confocal laser scanner. A total of 21,394 transcripts were on both the v3.0 and v4.0 arrays and were significantly expressed above background levels (detection P < 0.05) in at least 10 percent of subjects and were thus eligible for further analysis. Briefly, raw microarray scan files from Illumina HT-12 v3.0 arrays were exported using the Illumina BEADSTUDIO program and loaded into R for downstream analysis (www.R-pro-ject.org). Evaluation of the different microarray steps was performed using the Illumina internal controls. The data were transformed and normalized using variance-stabilizing normalization (Huber et al. 2002). A total of 15, 877 probes passing the filter criteria of Illumina probe detection P-value of <0.01 in at Least 5 percent of the individuals were used for subsequent analysis. To correct for confounding due to batch effects, data were normalized using an empirical Bayes method for batch correction (Johnson, Li, & Rabinovic 2007). Reproducibility of gene expression data was assessed using six pairs of technical replicates, yielding average Pearson correIations of 0.996. Only the nearest gene to the most significant GWAS variant was investigated.
Statistical analyses for genetic data in discovery sample
Using the statistical package PLINK (Purcell et al. 2007), we regressed the quantitative outcome, total AUDIT score (among drinkers only, N = 1036), on allele count assuming an additive model (zero, one or two copies of risk allele), including sex, chip type (Illumina OmniExpress or Omnil-Quad) and 10 principal components (Price et al. 2006; Lin & Zhoa 2009) as covariates. Each marker was run separately; thus, to account for the multiple testing, a threshold of 9.97 × 10–8 (Bonferroni correction = 0.05/501 278 markers analyzed; note that this differs from the standard GWAS threshold of 5 × 10–8) was required to meet genome-wide significance. Quantile-quantile (QQ) plots and Manhattan plots were generated using R (R development team, 2011). Regional association plots were generated using Locus Zoom (Pruim et al. 2010). To ensure that our test results did not depend on linearity or distributional assumptions (Supporting Information Fig. S3), we verified key results with PLINK’s max(T) permutation (1 × 10–8). To test the model of best fit for the top SNP (e.g. additive or dominant), we used Clarke’s method, which tests the hypothesis that both models are equally distant from the true model (Clarke 2007). To determine if any individual AUDIT item was driving the overall association effect, the significant variant was subsequently tested for association with the individual AUDIT items. To examine the specificity of effects on alcohol use problems, the significant variant was also tested for association with co-morbid conditions of drug abuse, PTSD, depression and anxiety. Gene expression analyses were conducted in R by regressing SNP genotype on transformed and normalized gene signal intensities (Mehta et al. 2013) using both additive and dominant models. Gene-based tests of association were conducted using Multi-marker Analysis of GenoMic Annotation (de Leeuw et al. 2015), using sex, chip type and 10 principal components as covariates. To account for the number of genes tested, a P-value threshold of 3.31 × 10–5 was considered genome-wide significant (0.05/1508 genes tested).
Confirmation analyses in similar cohorts
Parallel SNP-based association analyses were conducted in the Marine Resiliency Study (MRS) and Systems Biology PTSD Biomarkers Consortium (SBPBC) military samples for all variants meeting genome-wide significance criteria. These military cohorts have higher than average trauma levels (Supporting Information, Table 2). The all-male MRS (Baker et al. 2012; Mustapic et al. 2014; Nievergelt et al. 2014) is a prospective study of Marines recruited from four infantry battalions of the First Marine Division stationed at the Marine Corps Air-Ground Combat Center, 29 Palms, or Camp Pendleton, both in California (N = 1275). The SBPBC military cohort was recruited as part of a larger study that was designed as a longitudinal examination of male and female combat veterans (Yan et al. 2013; Yehuda et al. 2014), and participants were recruited at two sites in New York City (N = 119). Analyses were run parallel to those described in the GTP. Briefly, using the statistical package PLINK, we regressed total AUDIT score on allele count (zero, one or two risk alleles) as above, with sex (for SBPBC), study information (for MRS: MRSI/MRSII) and principal components (five for MRS and three for SBPBC) as covariates, thus controlling for ancestry across the analyses. Using inverse-variance meta-analysis, in R (cite), we assessed the significance of the association between SNP and total AUDIT score when combined across two independent cohorts (the SBPBC cohort plus MRS, which includes four separate self-identified racial/ethnic groups), using the R package Metafor (Viechtbauer 2010).
Post hoc, we examined whether there were associations between total AUDIT score and published SNPs previously associated with AD or alcohol consumption (Bierut et al. 2010; Gelernter et al. 2014; Xu et al. 2015); linear regressions were conducted as described above (Supporting Information).
Imaging data
Data acquisition and preprocessing
Imaging data were obtained on a subset of the GTP sample including 67 African-American women between 27 and 45 years of age (mean = 40.6, SD = 12.8; 52 women had GG genotype ‘resilient’ and 15 had AA/AG ‘risk’ genotype of rs14333 75, Supporting Information). Exclusion criteria for the GTP imaging sample included current psychotropic medication use; medical or physical conditions that preclude magnetic resonance imaging scanning; a history of bipolar disorder, schizophrenia or other primary psychotic disorder; current alcohol or substance dependence; medical conditions that contribute significantly to psychiatric symptoms; history of head injury or loss of consciousness for >5 minutes; or history of neurological disease. Scanning took place on a research-dedicated Siemens 3-T TIM-Trio scanner (Siemens Medical Solutions, Malvern, PA) using a 12-channel head coil at Emory University Hospital. Structural images were acquired using a three-dimensional T1-weighted magnetization-prepared rapid gradient echo imaging sequence [176 slices, 1 × 1 × 1 mm3, repetition time (TR) = 2600 milliseconds, echo time (TE) = 3.02 milliseconds, flip angle = 8°]. Resting state functional images were acquired using the Z-SAGA pulse sequence (Heberlein & Hu 2004) to recover signal loss due to susceptibility artifacts. Participants were instructed to focus on the cross in the middle of the screen while staying very still and awake during the resting state scan. Volumes were acquired axially, parallel to the anteriorposterior commissure line (scan time = 7 minutes 23 seconds, 150 volumes, 30 slices, 3.44 × 3.44 × 4 mm3, TR = 2950 milliseconds, TE1 = 30 milliseconds, TE2 = 67 milliseconds, flip angle = 90°).
Data preprocessing was accomplished using a combination of tools from AFNI (Cox 1996) and FSL (Smith et al. 2004) packages. Structural images were skull stripped, segmented and then registered to the standard Montreal Neurological Institute (MNI) space (AFNI’s 3dQwarp). Functional image preprocessing begins with despiking, slice timing correction, motion correction and spatial smoothing (full width at half maximum = 6 mm). The time series were additionally processed to minimize artifacts from head motion, respiration, cardiac pulsation and hardware using the ANATICOR method (Jo et al. 2013) by regressing out motion parameters and averaged signal from eroded local white matter mask as well as band-pass filtering between 0.01 and 0.1 Hz. Finally, functional images were registered to the MNI space via co-registration to the structure image using boundary-based registration (Greve & Fischl 2009) within the FSL package.
Functional connectivity analyses
The ECN includes the ventrolateral prefrontal cortex (VLPFC), the superior parietal cortex, the angular gyrus and cerebellum crus regions (Seeley et al. 2007; Krienen & Buckner 2009). To specifically derive fronto-cerebellar connectivity, we conducted functional connectivity analyses to examine the contralateral fronto-cerebellar circuit using previously defined right and left cerebellum crus regions (Krienen & Buckner 2009) (MNI = −36, −70, −46 and 36, −68, −44 with 6-mm-diameter spheres). First, the preprocessed time series were averaged across these seed regions. Pearson’s correlation coefficient was calculated between the seed time series and other time series within the cerebellum ROI. Then, these correlation coefficients were converted to z-values using Fisher’s transformation. Two-sample t-tests were used to examine quantitative group difference in functional connectivity. To correct for multiple comparisons, Monte Carlo simulations were performed using AFNI’s 3DCLUSTSIM program with whole-brain mask.
RESULTS
The demographic characteristics of the GTP subjects are shown in Table S1, including a comparison between the full genetic sample and the analytic sample. Note that there was a greater proportion of men in the analytic sample compared to the full genetic sample. Sixty-seven percent of those who consumed alcohol reported drinking less than two alcoholic drinks per day, with male drinkers consuming more alcohol per day than female drinkers. Among alcohol consumers, the total AUDIT score had an overall mean of 7.0 (SD = 7.3) and was higher among men (male: mean = 9.6, SD = 8.0; female: mean = 5.6, SD = 6.5).
Genome-wide association study in discovery sample
Analyses included 501 278 SNPs genotyped on Illumina’s Omni1-Quad and OmniExpress arrays. There was no evidence to suggest inflation of the association test statistics in the discovery (GTP) sample (genomic inflation factor λ = 0.98; Supporting Information Fig. S4). Following a Bonferroni multiple-test correction, rs14333 75, an intergenic SNP residing upstream of SCLT1 at chromosomal position 4q28, was significantly associated with total AUDIT score [minor allele frequency of 0.17; assuming an additive model: N = 1036, β (SE) = 2.21 (0.41); P = 7.76 × 10–8; Fig. 1a, Table 1; permuted P = 1.2 × 10–7]. The means of total AUDIT score by genotype are as follows: AA 8.6, AG 8.6 and GG 6.3. Further, five SNPs in linkage disequilibrium with rs14333 75 also associated with total AUDIT score (Fig. 1b). Of note are two sub-threshold SNPs in UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 6 (B4GALT6) (Table 1).
Figure 1.
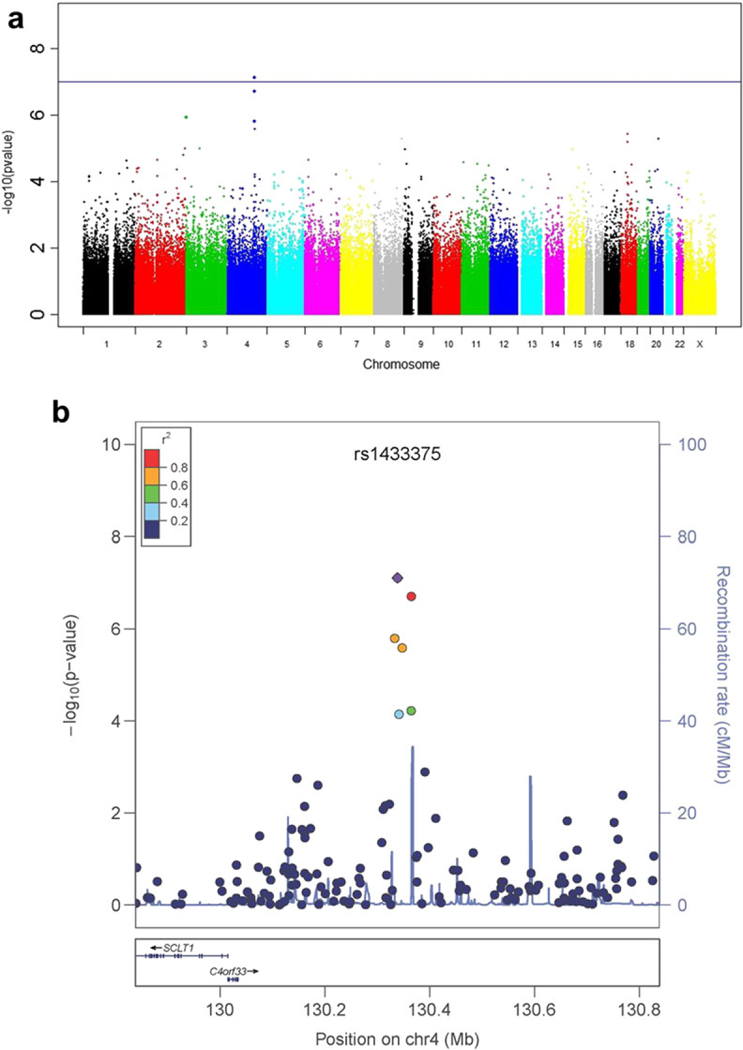
Genome-wide association study (GWAS) for problematic alcohol use and behavior in the GTP discovery sample. (a) Manhattan plot of the GWAS for total alcohol use disorders identification test score (N = 1036, rsl433375 P = 7.76 × 10–8, black line is Bonferroni-adjusted threshold for genome-wide significance). (b) Regional plot of single-nucleotide polymorphism (SNP) associations within 500 kb of rsl433375 (purple triangle) on chromosome 4. Several SNPs are ir linkage disequilibrium with the top SNP
Table 1.
Top 10 variants from SNP-based analysis of alcohol consumption and behavior (alcohol use disorders identification test score)
| Chr | Gene | Approximate distance to downstream gene | SNP | BP location | MAF | β (SE) | P-value |
|---|---|---|---|---|---|---|---|
| 4 | Intergenic | 323 kb, SCLT1 | rs1433375 | 130338059 | A (0.17) | 2.21 (0.41) | 7.76 × 10−8a |
| 4 | Intergenic | 350 kb, SCLT1 | rs995637 | 130364383 | T (0.16) | 2.22 (0.42) | 1.96 ×10−7 |
| 3 | Intergenic | 149 kb, CHL1 | rs991639 | 89511 | A (0.13) | 2.24 (0.46) | 1.22 × 10−6 |
| 4 | Intergenic | 318 kb, SCLT1 | rs6837783 | 130332644 | A (0.13) | 2.32 (0.48) | 1.60 × 10−6 |
| 4 | Intergenic | 332 kb, SCLT1 | rs1816429 | 130347005 | G (0.21) | 1.80 (0.38) | 2.58 × 10−6 |
| 18 | B4GALT6 | Intragenic | rs1667284 | 29226208 | A (0.14) | 2.20 (0.47) | 3.63 × 10−6 |
| 8 | Intergenic | 151 kb, ST3GAL1 | rs7843718 | 134735220 | G (0.18) | 1.86 (0.41) | 5.05 × 10−6 |
| 20 | PTPRT | Intragenic | rs11086862 | 41754601 | C (0.15) | 2.00 (0.44) | 5.14 × 10−6 |
| 18 | Intergenic | 15 kb, TTR | rs1375445 | 29156999 | C (0.26) | 1.64 (0.36) | 6.27 × 10−6 |
| 18 | B4GALT6 | Intragenic | rs1791161 | 29204210 | G (0.15) | 2.04 (0.45) | 6.42 × 10−6 |
SNPs on chromosome 4 are in linkage disequilibrium (shown in Fig. 1). Abbreviations: Chr, chromosome; BP, base-pair position; SE, standard error; MAF, minor allele frequency; SNP, single-nucleotide polymorphism.
Bonferroni-adjusted significance in the additive model, genome-wide significant in the dominant model [β(SE) = 2.69 (0.48); P = 2.61 × 10−8]
We conducted several post hoc analyses. Clarke’s method indicated that the dominant model was the preferred model (P = 2.1 × 10–14). Under a dominant model, rs1433375 was genome-wide significant [β (SE) = 2.69 (0.48); P = 2.61 × 10–8, Table 1]. In addition to the total AUDIT score, rs1433375 was associated with individual AUDIT items and related measures of substance and alcohol use behavior (e.g. ‘Have you had a problem with alcohol or drugs within the last year’; Supporting Information Table S3) but was not associated with co-morbid conditions of drug abuse, PTSD, depression and anxiety (Supporting Information). Total AUDIT score was not associated with published SNPs previously associated with AD or alcohol consumption (Supporting Information Table S4). No gene-based test of association withstood a genome-wide significance threshold; however, genes meeting a nominal level of significance are shown in Supporting Information Table S5.
Additional genetic analyses in similar samples
Next, rs1433375 was tested for association in two similar cohorts assessed with the total AUDIT score. Using an inverse-variance meta-analysis of two independent cohorts (N = 1394), the MRS and SBPBC military cohorts, additional evidence for the association of the A allele of rs1433375 and total AUDIT score was observed[β(SE) = 0.60 (0.17), P = 0.0004, Fig. 2, Table 2]. Thus, in both the discovery sample and the extension sample, the A allele of rs1433375 is associated with increased total AUDIT score. Note that results were driven by the SBPBC military cohort and by the MRS participants who identified as ‘Hispanic and Native American’.
Figure 2.
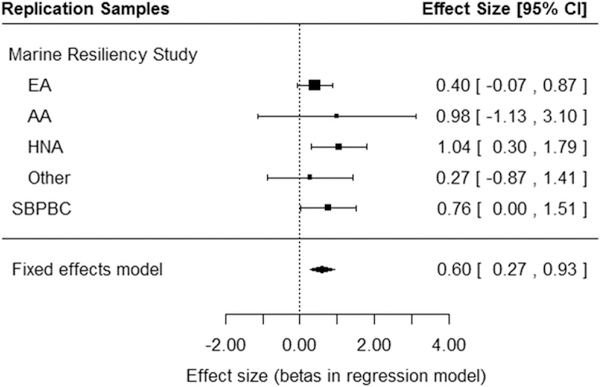
Meta-analysis of two independent cohorts extends the findings of association between single-nucleotide polymorphism (SNP) rsl433375 and problematic alcohol use and behavior. Forest plot showing the meta-analysis of SNP rs1433375 with total AUDIT score ir the additional samples (N = 1394, P = 0.0004). Abbreviations: EA, European Americans; AA, African Americans; HNA, Hispanic/Native Americans; SBPBC, Systems Biology PTSD Biomarkers Consortium
Table 2.
Sample characteristics for discovery and extension samples for association between rs1433375 and total alcohol use disorders identification test (AUDIT) score
| Discovery cohort | Gender |
Total sample (N) |
MAF | β | SE | P | |
|---|---|---|---|---|---|---|---|
| Male | Female | ||||||
| Grady Trauma Project | 373 | 663 | 1036 | A (0.17) | 2.21a | 0.41 | 7.76 × 10–8 |
| Additional Cohorts | |||||||
| Marine Resiliency Study | |||||||
| European American | 790 | — | 790 | G (0.42) | −0.40b | 0.24 | 0.10 |
| African American | 82 | — | 82 | G (0.70) | −0.98b | 1.08 | 0.36 |
| Hispanic, Native American | 236 | — | 236 | G (0.39) | − 1.04b | 0.38 | 0.006 |
| Other | 167 | — | 167 | G (0.46) | −0.27b | 0.58 | 0.64 |
| Systems Biology PTSD Biomarkers Consortium | 119 | 0.76a | 0.39 | 0.05 | |||
| Hispanic | 39 | 7 | 46 | A (0.5) | — | — | — |
| Non-Hispanic Asian | 6 | 0 | 6 | A (0.42) | — | — | — |
| Non-Hispanic Black | 23 | 3 | 26 | A (0.19) | — | — | — |
| Non-Hispanic Other | 3 | 0 | 3 | A (0.13) | — | — | — |
| Non-Hispanic White | 34 | 4 | 38 | G (0.46) | — | — | — |
Note that in all cohorts the A allele is positively associated with the AUDIT score and the ‘G’ allele is negatively associated with the AUDIT score.
Reference allele A.
Reference allele ‘G’.
Functional analyses: SCLT1 gene expression
To determine the functional basis of the GWAS risk variant, we examined whether rs1433375 influences expression levels of SCLT1, which was the closest gene passing QC. Furthermore, rs1433375 is reportedly a suggestive expression quantitative trait locus for SCLT1 in the frontal cortex (BRAINEAC, Trabzuni et al. 2011). In a subset of the GTP discovery sample, with peripheral blood RNA available, the normalized signal intensities of SCLT1 (Illumina probe ILMN_1797107; accession NM_144643.2) were used for expression analysis (N = 330). Findings indicated that rs1433375 genotype significantly predicted messenger RNA (mRNA) expression [N = 330, β (SE) = 0.047 (0.02), P = 0.047]. Under the dominant model [N = 330, β (SE) = 0.059 (0.03), P = 0.026], we show that A-allele carriers of rs1433375 exhibited higher mRNA expression (i.e. higher signal intensities; Fig. 3a).
Figure 3.
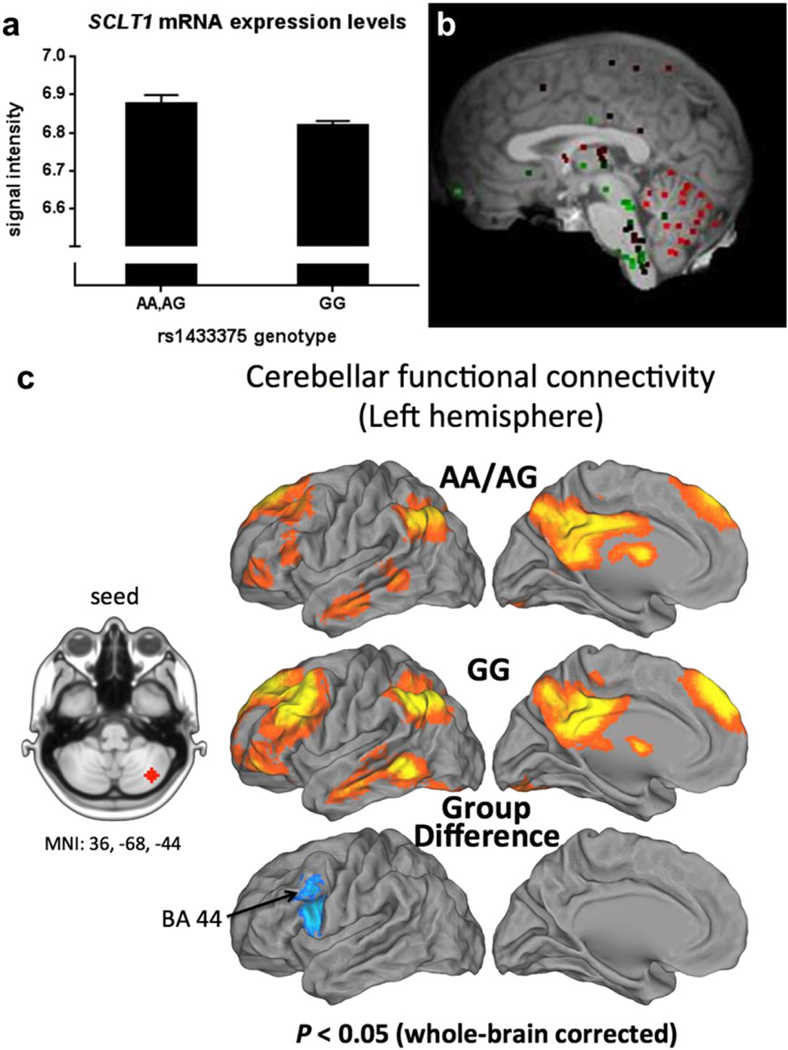
Functional analyses of rsl433375 in Grady Trauma Project discovery sample. (a) rsl433375 associates with gene SCLTI as shown by significant differences in SCLTI messenger RNA expression levels ir blood (probe signal intensities, mean + standard error of the mean) between genotypes (N = 330, P = 0.026). (b) Allen Brain Atlas image shows high SCLT1 expression (red dots) in the cerebellum. (c) Effect of genotype on resting state functional connectivity in executive control network using the right seed: the risk group (A-allele carriers, N =15) showed decreased fronto-cerebellar connectivity relative to the resilient group (GG genotype, N = 52), between the left cerebellum crus region and a cluster in the left ventrolateral prefrontal cortex (Brodmann area 44, BA 44)
Functional analyses: neuroimaging
We hypothesized that rs1433375 would influence fronto-cerebellar connectivity, an intermediate phenotype previously associated with alcohol use disorders. Functional connectivity analyses were performed using seed regions for the left and right cerebellum crus regions, the areas of highest SCLT1 gene expression (Fig 3b). We found that the right-seeded contralateral fronto-cerebellar connectivity was significantly decreased in the risk group (P < 0.05, whole-brain correction), relative to the resilient group, in the left VLPFC (Brodmann area 44; Fig. 3c). Although there was no cortical-cerebellar difference when using the left seed, we found that the risk group showed decreased cerebellar connectivity relative to the resilient group between the left cerebellum crus regions (Supporting Information Fig. S5).
DISCUSSION
This study reports a novel association between intergenic variant rs1433375 (upstream of SCLT1) and alcohol use behavior in a community cohort of African Americans with high levels of trauma exposure. Two independent populations of mixed ancestry provide additional support for this association. In addition, rs14333 75 associates with SCLT1, which is expressed most highly in the cerebellum, a brain area that has been repeatedly associated with alcohol involvement (Baker et al. 1999; Andersen 2004; Manzardo et al. 2005; Hill et al. 2011; Fitzpatrick & Crowe 2013). Finally, we examined resting state brain activity (functional magnetic resonance imaging) in a subset of the GTP to determine whether human cerebellar function was differentially associated with the rs14333 75 polymorphism and found altered connectivity in the ECN in carriers of the ‘A’ risk allele. Converging evidence from genotypic, expression-based and functional neuroimaging data presented in this study provides support for the role of SCLT1 in alcohol use and related problems, particularly among highly trauma-exposed populations.
Findings from this study point to novel genetic pathways and mechanisms, in addition to providing support for previously identified pathways. Results from both the discovery (GTP) and additional (SBPBC and MRS) samples indicate that rs1433375, an SNP residing upstream of SCLT1 at chromosomal position 4q28, and five correlated (in linkage disequilibrium) SNPs significantly associated with total AUDIT score. Further, we found that rs1433375 was associated with related measures of substance use and behavior in the full GTP cohort, suggesting that this association is robust to different measures of alcohol use behavior. In addition, the finding of rs1433375 in two independent cohorts of mixed ancestry suggests that this variant may not be unique to populations of African ancestry. We also note that two SNPs located within B4GALT6 (Table 1) associate with total AUDIT score but do not meet genome-wide significance. However, B4GALT6 has previously been associated with alcohol-related phenotypes, including alcohol consumption during pregnancy (Guasch, Renau-Piqueras, & Guerri 1992; Segui et al. 1996). This converging evidence suggests that variants in both SCLT1 and B4GALT6 should be investigated in future genetic association studies of alcohol use disorders.
While no gene-based test of association withstood a genome-wide significance threshold, several important genes met nominal levels of significance below genome-wide threshold (detailed in Supporting Information Table S5), including genes that have previously been associated with alcohol-related phenotypes. For example, HTR1A and HTR1B have been implicated in several genetic association studies of alcohol and drug dependence, including studies of co-morbid AD and depression (Huang et al. 2003; Sinha, Cloninger, & Parsian 2003; Wrzosek et al. 2011; Contini et al. 2012; Cao, LaRocque, & Li 2013; Zuo et al. 2013). In addition, PER2 has been shown to influence alcohol consumption and related problems in human and animal studies (Spanagel et al. 2005; Yuferov, Bart, & Kreek 2005; Partonen 2015).
The AUDIT was designed for the World Health Organization to screen for hazardous as well as harmful drinking in various cultural settings. There have been a number of studies reflecting potential benefits of using different cutoff points for identification of at-risk drinking (Reinert & Allen 2007). Prior studies have evaluated the test-retest reliability of the AUDIT when it is scored dichotomously to classify patients as at risk. Three prior studies (Selin 2003; Dybek et al. 2006; Rubin et al. 2006) were conducted with general population samples and reported ks of 0.70, 0.86, and 0.89, respectively, at the standard AUDIT cut-point of 8. Together, such data suggest that while a variety of cut-points may be used, an AUDIT score of 8 is generally considered a good marker of risk (Reinert & Allen 2007). Given that we found means of total AUDIT score by genotype of AA/AG = 8.6, whereas GG = 6.3, we believe that these are likely clinically significant differences representing differential at-risk alcohol drinking behavior.
Heritability estimates based on twin data suggest moderately high heritability at 60 percent for AUDIT (Mbarek et al. 2015). AUDIT score represents the composite effects of alcohol consumption and related behaviors; this variable is quite distinct from maximum number of drinks (as reported in Gelernter et al. 2014; Xu et al. 2015) and the case-control AD (as reported in Bierut et al. 2010). Previously published variants conferring risk for heavy alcohol consumption and/or AD were not associated with AUDIT scores in the GTP cohort. We hypothesize that this lack of replication is due to several aspects of the GTP population and study design. Given the known heterogeneity of genetic influences on typical alcohol use, risk drinking and AD (Kendler et al. 2011), it is likely that different risk variants may influence maximum drinks consumed in a 24-hour period, AD diagnoses and AUDIT scores for example. In addition, the characteristics of the GTP study population differ in several ways from previous studies of alcohol use behavior and problems including genetic ancestry, patterns of substance use, demographic characteristics and relatively high levels of trauma exposure, PTSD and depression co-morbidity. For example, the majority of previous studies have been conducted in samples of European ancestry, in which general patterns of alcohol use are greater, levels of SES are higher and exposure to assaultive trauma is lower.
rs1433375 is located within a distal upstream enhancer region for SCLT1 transcriptional regulation. Interestingly, SCLT1 produces a linker-like protein between the voltage-gated sodium channel Na(v)1.8 (SCN10A) and clathrin (CLTC), a ubiquitous protein involved in receptor endocytosis and recycling (Liu et al. 2005). Sodium channel activity, particularly Na(v)1.8, has been associated with the neuronal effects of ethanol exposure (Mullin & Hunt 1987; Harris & Allan 1989; Horishita & Harris 2008) and linked to alcohol use behavior (Botta et al. 2010). Moreover, the interaction with sodium channels has been hypothesized as a potentially robust target of novel drugs for the treatment of alcohol use disorders (Rezvani et al. 2012). We note that risk allele carriers of rs14333 75 exhibited higher SCLT1 mRNA expression levels in this study. The mechanism for which ‘overexpression’ of SCLT1 can lead to pathological conditions is beyond the scope of this study; however, examples of this relationship can be found in Prelich (2012).
Because SCLT1 is highly expressed in the cerebellum, we used cerebellar seed regions for connectivity analyses. We hypothesized that variation in rs1433375 would influence connectivity between the cerebellum and prefrontal cortex, components of the ECN. Significant differences in connectivity emerged between participants with and without the risk genotype for rs1433375 (‘A’ carriers); risk allele carriers demonstrated lower fronto-cerebellar connectivity compared to those without the risk allele (Figure 3c).
These patterns of neural connectivity suggest decreased functional interactions between prefrontal brain regions controlling complex decision making and the cerebellum, which is thought to be associated with motoric and habitual behavior. Both the prefrontal cortex and cerebellum are thought to play a role in executive control functions, particularly in the presence of emotionally salient stimuli (Konarski et al. 2005). One particularly salient control function is the ability to refrain from maladaptive, but inherently rewarding behaviors, such as excessive alcohol consumption. Earlier studies have shown that individuals with alcohol use problems have demonstrated deficits in executive control (Chanraud et al. 2010; Maurage et al. 2014) as well as diminished activation in prefrontal brain regions (Forbes et al. 2014) and lower frontal-cerebellar connectivity (Rogers et al. 2012) during performance of reward-related tasks.
Given that the genotype groups were well matched in terms of alcohol use, the present findings suggest that those individuals who carry the rs1433375 risk allele may be more susceptible to problematic alcohol use. This may be secondary to alterations in connectivity in the ECN, including cerebellar regulation of responses mediated by the frontal cortex. Thus, altered fronto-cerebellar connectivity may represent an intermediate phenotype for risky alcohol use. Because SCLT1 is expressed highly in the cerebellum (Fig. 3b), we hypothesized that variation in rs1433375 would influence fronto-cerebellar connectivity. Functional connectivity analyses for the cerebellum (i.e. an area of high SCLT1 gene expression) indicated that there were significant differences in connectivity between the cerebellar crus and VLPFC for those with and without the risk genotype (rs1433375 ‘A’ allele carriers). Given that the VLPFC has an established role in impulse control (Sebastian et al. 2014), this may suggest a potential neural substrate for problematic alcohol use patterns for carriers of this risk genotype.
Results from this study should be interpreted in light of several key limitations. First, sample sizes in both the discovery (GTP) and additional (MRS and SBPBC) samples were limited; however, the use of a quantitative measure of alcohol use problems, the cross-methodology convergence, as well as the extension of the findings in an independent sample lessens this concern. Second, we note the lack of replication in an independent sample of African ancestry, owing to limitations in availability of samples with the same phenotype. Such replications represent an important next step in understanding these findings. Third, the present analysis uses data available at a single timepoint, using retrospective reports of alcohol use behavior. Previous studies have found that individuals typically underreport their drinking consumption (e.g. Ekholm, Strandberg-Larsen, & Gronbaek 2011), which would likely diminish the association effects observed in this study. However, recent studies directly comparing retrospective reports to observational data conclude that this bias is significantly less pronounced in regular drinkers (Ekholm et al. 2011). Further, the assessment of alcohol use behavior used in this study, total AUDIT score, has excellent psychometric properties and has been used widely in prior health research (Saunders et al. 1993; Babor et al. 2001; Reinert & Allen 2002, 2007; Carey, Carey, & Chandra 2003; Cassidy, Schmitz, & Malla 2008).
This study has provided evidence of association between alcohol involvement and a novel genetic variant located upstream of SCLT1, a gene that has been associated with the regulation of sodium channel functioning, a putative target of ethanol action. Importantly, rs14333 75 was also associated with differential cortical–cerebellar functioning, a neural circuit that has been hypothesized to be mechanistically associated with alcohol use disorders. Converging data for the role of a sodium channel regulator from the genotypic, expression-based and functional neuroimaging findings presented in this study provide support for these mechanistic pathways in alcohol use and related behaviors.
Supplementary Material
Acknowledgements
This work was primarily supported by the National Institutes of Mental Health (MH071537 and MH096764 to K.J.R., T32 MH87977–5 to L.M.A. and MH093500 to C.M.N.), the National Institutes of Drug Abuse (K01 DA037914 to J.L.M) and the Department of Defense (DoD: W81XWH-09–2-0044 to C.R.M. and W911NF-09–1-0298 to R.Y.). Support was also received from the Emory and Grady Memorial Hospital General Clinical Research Center, NIH National Centers for Research Resources (M01RR00039), the Howard Hughes Medical Institute (K.J.R.) and the Steven and Alexandra Cohen Foundation (C.R.M.). We are grateful for the staff and participants from the GTP for their time and effort in supporting this research. We thank Murray Stein, Chia-Yen Chen and Army STARRs researchers for their helpful comments on this research study.
Footnotes
FINANCIAL DISCLOSURE
The authors declare no conflict of interest, financial or otherwise.
SUPPORTING INFORMATION
Additional Supporting Information may be found online in the supporting information tab for this article.
References
- Agrawal A, Grant JD, Littlefield A, Waldron M, Pergadia ML, Lynskey MT, Madden PA, Todorov A, Trull T, Bucholz KK, Todd RD, Sher K, Heath AC (2009) Developing a quantitative measure of alcohol consumption for genomic studies on prospective cohorts. J Stud Alcohol Drugs 70:157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Wetherill L, Bucholz KK, Kramer J, Kuperman S, Lynskey MT, Nürnberger JI Jr, Schuckit M, Tischfield JA, Edenberg HJ, Foroud T, Bierut LJ (2013) Genetic influences on craving for alcohol. Addict Behav 38:1501–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JP, Crawford EF, Kudler H (2016) Nature and treatment of comorbid alcohol problems and post traumatic stress disorder among American military personnel and veterans. Alcohol Res 38:133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almli LM, Mercer KB, Kerley K, Feng H, Bradley B, Conneely KN, Ressler KJ (2013) ADCYAP1R1 genotype associates with posttraumatic stress symptoms in highly traumatized African-American females. Am J Med Genet B Neuropsychiatr Genet 162B:262–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen BB (2004) Reduction of Purkinje cell volume in cerebellum of alcoholics. Brain Res 1007:10–18. [DOI] [PubMed] [Google Scholar]
- Babor TF, Higgins-Biddle JC, Saunders JB, Monteiro MG (2001) AUDIT-The Alcohol Use Disorders Identification Test: Guidelines for use inprimary care. Geneva: World Health Organization. [Google Scholar]
- Baik I, Cho NH, Kim SH, Han BG, Shin C (2011) Genome-wide association studies identify genetic loci related to alcohol consumption in Korean men. Am J Clin Nutr 93:809–816. [DOI] [PubMed] [Google Scholar]
- Baker DG, Nash WP, Litz BT, Geyer MA, Risbrough VB, Nievergelt CM, O’Connor DT, Larson GE, Schork NJ, Vasterling JJ, Hammer PS, Webb-Murphy JA, Team MRS (2012) Predictors of risk and resilience for posttraumatic stress disorder among ground combat Marines: methods of the Marine Resiliency Study. Prev Chronic Dis 9:E97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker KG, Harding AJ, Halliday GM, Kril JJ, Harper CG (1999) Neuronal loss in functional zones of the cerebellum of chronic alcoholics with and without Wernicke’s encephalopathy. Neuroscience 91:429–438. [DOI] [PubMed] [Google Scholar]
- Beck AT, Ward CH, Mendelson M, Mock J, Erbaugh J (1961) An inventory for measuring depression. Arch Gen Psychiatry 4:561–571. [DOI] [PubMed] [Google Scholar]
- Bernstein DP, Stein JA, Newcomb MD, Walker E, Pogge D, Ahluvalia T, Stokes J, Handelsman L, Medrano M, Desmond D, Zule W (2003) Development and validation of a brief screening version of the Childhood Trauma Questionnaire. Child Abuse Negl 27:169–190. [DOI] [PubMed] [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, Fisher S, Fox L, Howells W, Bertelsen S, Hinrichs AL, Almasy L, Breslau N, Culverhouse RC, Dick DM, Edenberg HJ, Foroud T, Grucza RA, Hatsukami D, Hesselbrock V, Johnson EO, Kramer J, Krueger RF, Kuperman S, Lynskey M, Mann K, Neuman RJ, Nothen MM, Nurnberger JI Jr, Porjesz B, Ridinger M, Saccone NL, Saccone SF, Schuckit MA, Tischfield JA, Wang JC, Rietschel M, Goate AM, Rice JP, Gene EASC (2010) A genome-wide association study of alcohol dependence. Proc Natl Acad Sci U S A 107:5082–5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder EB, Bradley RG, Liu W, Epstein MP, Deveau TC, Mercer KB, Tang Y, Gillespie CF, Heim CM, Nemeroff CB, Schwartz AC, Cubells JF, Ressler KJ (2008) Association of FKBP5 polymorphisms and childhood abuse with risk of posttraumatic stress disorder symptoms in adults. JAMA 299:1291–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botta P, de Souza FM, Sangrey T, De Schutter E, Valenzuela CF (2010) Alcohol excites cerebellar Golgi cells by inhibiting the Na+/K+ ATPase. Neuropsychopharmacology 35:1984–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremner JD, Bolus R, Mayer EA (2007) Psychometric properties of the Early Trauma Inventory-Self Report. J Nerv Ment Dis. 195:211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremner JD, Vermetten E, Mazure CM (2000) Development and preliminary psychometric properties of an instrument for the measurement of childhood trauma: the Early Trauma Inventory Depression and anxiety. 12:1–12. [DOI] [PubMed] [Google Scholar]
- Cao J, LaRocque E, Li D (2013) Associations of the 5-hydroxytryptamine (serotonin) receptor 1B gene (HTR1B) with alcohol, cocaine, and heroin abuse. Am J Med Genet B Neuropsychiatr Genet 162B:169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey KB, Carey MP, Chandra PS (2003) Psychometric evaluation of the alcohol use disorders identification test and short drug abuse screening test with psychiatric patients in India. J Clin Psychiatry 64:767–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy CM, Schmitz N, Malla A (2008) Validation of the alcohol use disorders identification test and the drug abuse screening test in first episode psychosis. Canadian Journal of Psychiatry/Revue canadienne de psychiatrie 53:26–33. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (2012) FastStats: Alcohol Use. Centers for Disease Control and Prevention: Atlanta, GA. [Google Scholar]
- Chanraud S, Pitel AL, Rohlfing T, Pfefferbaum A, Sullivan EV (2010)Dual tasking and working memory in alcoholism: relation to frontocerebellar circuitry. Neuropsychopharmacology 35:1868–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke K (2007) A simple distribution-free test for nonnested hypotheses. Political Analysis 15:347–363. [Google Scholar]
- Clarke T-K, Adams MJ, Davies G, Howard DM, Hall LS, Padmanabhan S, Murray AD, Smith BH, Campbell A, Hayward C, Porteous DJ, Deary IJ, McIntosh AM (2017) Genome-wide association study of alcohol consumption and genetic overlap with other health-related traits in UK Biobank (N = 112,117) 00:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey SF, Dansky BS, Falsetti SA, Saladin ME, Brady KT (1998) Screening for PTSD in a substance abuse sample: psychometric properties of a modified version of the PTSD Symptom Scale Self-Report. Posttraumatic stress disorder. J Trauma Stress 11:393–399. [DOI] [PubMed] [Google Scholar]
- Contini V, Bertuzzi GP, Polina ER, Hunemeier T, Hendler EM, Hutz MH, Bau CH (2012) A haplotype analysis is consistent with the role of functional HTR1B variants in alcohol dependence. Drug Alcohol Depend 122:100–104. [DOI] [PubMed] [Google Scholar]
- Cox RW (1996) AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res 29:162–173. [DOI] [PubMed] [Google Scholar]
- de Leeuw CA, Mooij JM, Heskes T, Posthuma D (2015) MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol 11: e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dybek I, Bischof G, Grothues J, Reinhardt S, Meyer C, Hapke U, John U, Broocks A, Hohagen F, Rumpf HJ (2006) The reliability and validity of the alcohol use disorders identification test (AUDIT) in a German general practice population sample. J Stud Alcohol 67:473–481. [DOI] [PubMed] [Google Scholar]
- Ekholm O, Strandberg-Larsen K, Gronbaek M (2011) Influence of the recall period on a beverage-specific weekly drinking measure for alcohol intake. Eur J Clin Nutr 65:520–525. [DOI] [PubMed] [Google Scholar]
- Falsetti SA, Resnick HS, Resick PA, Kilpatrick D (1993) The Modified PTSD Symptom Scale: a brief self-report measure of posttraumatic stress disorder. The Behavior Therapist 16:161–162. [Google Scholar]
- Fitzpatrick LE, Crowe SF (2013) Cognitive and emotional deficits in chronic alcoholics: a role for the cerebellum? Cerebellum 12:520–533. [DOI] [PubMed] [Google Scholar]
- Foa EB, Tolin DF (2000) Comparison of the PTSD Symptom Scale-interview version and the Clinician-Administered PTSD scale. J Trauma Stress 13:181–191. [DOI] [PubMed] [Google Scholar]
- Forbes EE, Rodriguez EE, Musselman S, Narendran R (2014) Prefrontal response and frontostriatal functional connectivity to monetary reward in abstinent alcohol-dependent young adults. PLoS One 9: e94640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer SL, Jorgensen KW, Yetter EJ, Daurignac EC, Watson TD, Shanbhag H, Krystal JH, Mathalon DH (2013) Differential brain response to alcohol cue distractors across stages of alcohol dependence. Biol Psychol 92:282–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH, Anton R, Preuss UW, Ridinger M, Rujescu D, Wodarz N, Zill P, Zhao H, Farrer LA (2014) Genome-wide association study of alcohol dependence: significant findings in African-and European-Americans including novel risk loci. Mol Psychiatry 19:41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie CF, Bradley B, Mercer K, Smith AK, Conneely K, Gapen M, Weiss T, Schwartz AC, Cubells JF, Ressler KJ (2009) Trauma exposure and stress-related disorders in inner city primary care patients. Gen Hosp Psychiatry 31:505–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greve DN, Fischl B (2009) Accurate and robust brain image alignment using boundary-based registration. Neuroimage 48:63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guasch R, Renau-Piqueras J, Guerri C (1992) Chronic ethanol consumption induces accumulation of proteins in the liver Golgi apparatus and decreases galactosyltransferase activity. Alcohol Clin Exp Res 16:942–948. [DOI] [PubMed] [Google Scholar]
- Harris RA, Allan AM (1989) Alcohol intoxication: ion channels and genetics. FASEB Journal 3:1689–1695. [DOI] [PubMed] [Google Scholar]
- Heath AC, Whitfield JB, Martin NG, Pergadia ML, Goate AM, Lind PA, McEvoy BP, Schrage AJ, Grant JD, Chou YL, Zhu R, Henders AK, Medland SE, Gordon SD, Nelson EC, Agrawal A, Nyholt DR, Bucholz KK, Madden PA, Montgomery GW (2011) A quantitative-trait genome-wide association study of alcoholism risk in the community: findings and implications. Biol Psychiatry 70:513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberlein KA, Hu X (2004) Simultaneous acquisition of gradient-echo and asymmetric spin-echo for single-shot z-shim: Z-SAGA. Magn Reson Med 51:212–216. [DOI] [PubMed] [Google Scholar]
- Herting MM, Fair D, Nagel BJ (2011) Altered fronto-cerebellar connectivity in alcohol-naive youth with a family history of alcoholism. Neuroimage 54:2582–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SY, Wang S, Carter H, Tessner K, Holmes B, McDermott M, Zezza N, Stiffler S (2011) Cerebellum volume in high-risk offspring from multiplex alcohol dependence families: association with allelic variation in GABRA2 and BDNF. Psychiatry Res 194:304–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horishita T, Harris RA (2008) n-Alcohols inhibit voltage-gated Na+ channels expressed in Xenopus oocytes. J Pharmacol Exp Ther 326:270–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 5: e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YY, Oquendo MA, Friedman JM, Greenhill LL, Brodsky B, Malone KM, Khait V, Mann JJ (2003) Substance abuse disorder and major depression are associated with the human 5-HT1B receptor gene (HTR1B) G861C polymorphism. Neuropsychopharmacology 28:163–169. [DOI] [PubMed] [Google Scholar]
- Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M (2002) Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18:S96–104. [DOI] [PubMed] [Google Scholar]
- Iacono WG, Carlson SR, Taylor J, Elkins IJ, McGue M (1999) Behavioral disinhibition and the development of substance-use disorders: findings from the Minnesota Twin Family Study. Dev Psychopathol 11:869–900. [DOI] [PubMed] [Google Scholar]
- Jo HJ, Gotts SJ, Reynolds RC, Bandettini PA, Martin A, Cox RW, Saad ZS (2013) Effective preprocessing procedures virtually eliminate distance-dependent motion artifacts in resting state FMRI. J Appl Math 2013:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A (2007) Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8:118–127. [DOI] [PubMed] [Google Scholar]
- Jonas DE, Garbutt JC, Amick HR, Brown JM, Brownley KA, Council CL, Viera AJ, Wilkins TM, Schwartz CJ, Richmond EM, Yeatts J, Evans TS, Wood SD, Harris RP (2012) Behavioral counseling after screening for alcohol misuse in primary care: a systematic review and meta-analysis for the U.S. Preventive Services Task Force. Ann Intern Med 157:645–654. [DOI] [PubMed] [Google Scholar]
- Kapoor M, Wang JC, Wetherill L, Le N, Bertelsen S, Hinrichs AL, Budde J, Agrawal A, Bucholz K, Dick D, Harari O, Hesselbrock V, Kramer J, Nurnberger JI Jr, Rice J, Saccone N, Schuckit M, Tischfield J, Porjesz B, Edenberg HJ, Bierut L, Foroud T, Goate A (2013) A meta-analysis of two genome-wide association studies to identify novel loci for maximum number of alcoholic drinks. Hum Genet 132:1141–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Aggen SH, Knudsen GP, Roysamb E, Neale MC, Reichborn-Kjennerud T (2011) The structure of genetic and environmental risk factors for syndromal and subsyndromal common DSM-IV axis I and all axis II disorders. Am J Psychiatry 168:29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarski JZ, McIntyre RS, Grupp LA, Kennedy SH (2005) Is the cerebellum relevant in the circuitry of neuropsychiatric disorders? Journal of psychiatry & neuroscience :JPN 30:178–186. [PMC free article] [PubMed] [Google Scholar]
- Krienen FM, Buckner RL (2009) Segregated fronto-cerebellar circuits revealed by intrinsic functional connectivity. Cereb Cortex 19:2485–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krmpotich TD, Tregellas JR, Thompson LL, Banich MT, Klenk AM, Tanabe JL (2013) Resting-state activity in the left executive control network is associated with behavioral approach and is increased in substance dependence. Drug Alcohol Depend 129:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Zhoa H (2009) Handbook on Analyzing Human Genetic Data: Computational Approaches and Software. Springer: New York. [Google Scholar]
- Liu C, Cummins TR, Tyrrell L, Black JA, Waxman SG, Dib-Hajj SD (2005) CAP-1A is a novel linker that binds clathrin and the voltage-gated sodium channel Na(v)1.8. Mol Cell Neurosci 28:636–649. [DOI] [PubMed] [Google Scholar]
- Majer M, Nater UM, Lin JM, Capuron L, Reeves WC (2010) Association of childhood trauma with cognitive function in healthy adults: a pilot study. BMC Neurol 10:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzardo AM, Penick EC, Knop J, Nickel EJ, Hall S, Jensen P, Gabrielli WF Jr (2005) Developmental differences in childhood motor coordination predict adult alcohol dependence: proposed role for the cerebellum in alcoholism. Alcohol Clin Exp Res 29:353–357. [DOI] [PubMed] [Google Scholar]
- Maurage P, de Timary P, Billieux J, Collignon M, Heeren A (2014) Attentional alterations in alcohol dependence are underpinned by specific executive control deficits. Alcohol Clin Exp Res 38:2105–2112. [DOI] [PubMed] [Google Scholar]
- Mbarek H, Milaneschi Y, Fedko IO, Hottenga JJ, de Moor MH, Jansen R, Gelernter J, Sherva R, Willemsen G, Boomsma DI, Penninx BW, Vink JM (2015) The genetics of alcohol dependence: Twin and SNP-based heritability, and genome-wide association study based on AUDIT scores. Am J Med Genet B Neuropsychiatr Genet 168:739–748. [DOI] [PubMed] [Google Scholar]
- Mehta D, Gonik M, Klengel T, Rex-Haffner M, Menke A, Rubel J, Mercer KB, Pütz B, Bradley B, Holsboer F, Ressler KJ, Müller-Myhsok B, Binder EB (2011) Using polymorphisms in FKBP5 to define biologically distinct subtypes of posttraumatic stress disorder: evidence from endocrine and gene expression studies. Arch Gen Psychiatry 68:901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta D, Klengel T, Conneely KN, Smith AK, Altmann A, Pace TW, Rex-Haffner M, Loeschner A, Gonik M, Mercer KB, Bradley B, Muller-Myhsok B, Ressler KJ, Binder EB (2013) Childhood maltreatment is associated with distinct genomic and epigenetic profiles in posttraumatic stress disorder. Proc Natl Acad Sci U S A 110:8302–8307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minassian A, Maihofer AX, Baker DG, Nievergelt CM, Geyer MA, Risbrough VB, Marine Resiliency Study T (2015) Association of Predeployment Heart Rate Variability With Risk of Postdeployment Posttraumatic Stress Disorder in Active-Duty Marines. JAMA Psychiatry 72:979–986. [DOI] [PubMed] [Google Scholar]
- Moulton EA, Elman I, Becerra LR, Goldstein RZ, Borsook D (2014) The cerebellum and addiction: insights gained from neuroimaging research. Addict Biol 19:317–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullin MJ, Hunt WA (1987) Effects of ethanol on the functional properties of sodium channels in brain synaptosomes. Recent Dev Alcohol 5:303–311. [DOI] [PubMed] [Google Scholar]
- Mustapic M, Maihofer AX, Mahata M, Chen Y, Baker DG, O’Connor DT, Nievergelt CM (2014) The catecholamine biosynthetic enzyme dopamine beta-hydroxylase (DBH): first genome-wide search positions trait-determining variants acting additively in the proximal promoter. Hum Mol Genet 23:6375–6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nievergelt CM, Wineinger NE, Libiger O, Pham P, Zhang G, Baker DG, Marine Resiliency Study I, Schork NJ (2014) Chipbased direct genotyping of coding variants in genome wide association studies: utility, issues and prospects. Gene 540:104–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partonen T (2015) Clock genes in human alcohol abuse and comorbid conditions. Alcohol 49:359–365. [DOI] [PubMed] [Google Scholar]
- Prelich G (2012) Gene overexpression: uses, mechanisms, and interpretation. Genetics 190:841–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38:904–909. [DOI] [PubMed] [Google Scholar]
- Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ (2010) LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26:2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2011) R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Reinert DF, Allen JP (2002) The alcohol use disorders identification test (AUDIT): a review of recent research. Alcohol Clin Exp Res 26:272–279. [PubMed] [Google Scholar]
- Reinert DF, Allen JP (2007) The alcohol use disorders identification test: an update of research findings. Alcohol Clin Exp Res 31:185–199. [DOI] [PubMed] [Google Scholar]
- Reiss S, Peterson RA, Gursky DM, McNally RJ (1986) Anxiety sensitivity, anxiety frequency and the prediction of fearfulness. Behav Res Ther 24:1–8. [DOI] [PubMed] [Google Scholar]
- Ressler KJ, Mercer KB, Bradley B, Jovanovic T, Mahan A, Kerley K, Norrholm SD, Kilaru V, Smith AK, Myers AJ, Ramirez M, Engel A, Hammack SE, Toufexis D, Braas KM, Binder EB, May V (2011) Post-traumatic stress disorder is associated with PACAP and the PAC1 receptor. Nature 470:492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezvani AH, Lawrence AJ, Arolfo MP, Levin ED, Overstreet DH (2012) Novel medication targets for the treatment of alcoholism: preclinical studies. Recent Pat CNS Drug Discov 7:151–162. [DOI] [PubMed] [Google Scholar]
- Rogers BP, Parks MH, Nickel MK, Katwal SB, Martin PR (2012) Reduced fronto-cerebellar functional connectivity in chronic alcoholic patients. Alcohol Clin Exp Res 36:294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin A, Migneault JP, Marks L, Goldstein E, Ludena K, Friedman RH (2006) Automated telephone screening for problem drinking. J Stud Alcohol 67:454–457. [DOI] [PubMed] [Google Scholar]
- Sartor CE, Nelson EC, Lynskey MT, Madden PA, Heath AC, Bucholz KK (2013) Are there differences between young African-American and European-American women in the relative influences of genetics versus environment on age at first drink and problem alcohol use? Alcohol Clin Exp Res 37:1939–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders JB, Aasland OG, Babor TF, de la Fuente JR, Grant M (1993) Development of the alcohol use disorders identification test (AUDIT): WHO collaborative project on early detection of persons with harmful alcohol consumption. II. Addiction 88:791–804. [DOI] [PubMed] [Google Scholar]
- Schumann G, Coin LJ, Lourdusamy A, Charoen P, Berger KH, Stacey D, Desrivieres S, Aliev FA, Khan AA, Amin N, Aulchenko YS, Bakalkin G, Bakker SJ, Balkau B, Beulens JW, Bilbao A, de Boer RA, Beury D, Bots ML, Breetvelt EJ, Cauchi S, Cavalcanti-Proenca C, Chambers JC, Clarke TK, Dahmen N, de Geus EJ, Dick D, Ducci F, Easton A, Edenberg HJ, Esko T, Fernandez-Medarde A, Foroud T, Freimer NB, Girault JA, Grobbee DE, Guarrera S, Gudbjartsson DF, Hartikainen AL, Heath AC, Hesselbrock V, Hofman A, Hottenga JJ, Isohanni MK, Kaprio J, Khaw KT, Kuehnel B, Laitinen J, Lobbens S, Luan J, Mangino M, Maroteaux M, Matullo G, McCarthy MI, Mueller C, Navis G, Numans ME, Nunez A, Nyholt DR, Onland-Moret CN, Oostra BA, O’Reilly PF, Palkovits M, Penninx BW, Polidoro S, Pouta A, Prokopenko I, Ricceri F, Santos E, Smit JH, Soranzo N, Song K, Sovio U, Stumvoll M, Surakk I, Thorgeirsson TE, Thorsteinsdottir U, Troakes C, Tyrfingsson T, Tonjes A, Uiterwaal CS, Uitterlinden AG, van der Harst P, van der Schouw YT, Staehlin O, Vogelzangs N, Vollenweider P, Waeber G, Wareham NJ, Waterworth DM, Whitfield JB, Wichmann EH, Willemsen G, Witteman JC, Yuan X, Zhai G, Zhao JH, Zhang W, Martin NG, Metspalu A, Doering A, Scott J, Spector TD, Loos RJ, Boomsma DI, Mooser V Peltonen L, Stefansson K, van Duijn CM, Vineis P, Sommer WH, Kooner JS, Spanagel R, Heberlein UA, Jarvelin MR, Elliott P (2011) Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc Natl Acad Sci U S A 108:7119–7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz AC, Bradley RL, Sexton M, Sherry A, Ressler KJ (2005) Posttraumatic stress disorder among African Americans in an inner city mental health clinic. Psychiatr Serv 56:212–215. [DOI] [PubMed] [Google Scholar]
- Sebastian A, Jung P, Krause-Utz A, Lieb K, Schmahl C, Tuscher O (2014) Frontal dysfunctions of impulse control—a systematic review in borderline personality disorder and attention-deficit/ hyperactivity disorder. Front Hum Neurosci 8:698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley WW, Menon V, Schatzberg AF, Keller J, Glover GH, Kenna H, Reiss AL, Greicius MD (2007) Dissociable intrinsic connectivity networks for salience processing and executive control. J Neurosci 27:2349–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segui JM, Guasch R, Azorin I, Guerri C, Renau-Piqueras J (1996) Alcohol consumption during pregnancy alters glycoconjugates, galactosyltransferase activity and beta-COP proteins in the Golgi apparatus of hepatocytes in developing liver. Int J Dev Biol 285S–286S. [PubMed] [Google Scholar]
- Selin KH (2003) Test-retest reliability of the alcohol use disorder identification test in a general population sample. Alcohol Clin Exp Res 27:1428–1435. [DOI] [PubMed] [Google Scholar]
- Sinha R, Cloninger CR, Parsian A (2003) Linkage disequilibrium and haplotype analysis between serotonin receptor 1B gene variations and subtypes of alcoholism. Am J Med Genet B Neuropsychiatr Genet 121B:83–88. [DOI] [PubMed] [Google Scholar]
- Skinner HA (1982) The drug abuse screening test. Addict Behav 7:363–371. [DOI] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM, Matthews PM (2004) Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage 23:S208–S219. [DOI] [PubMed] [Google Scholar]
- Spanagel R, Pendyala G, Abarca C, Zghoul T, Sanchis-Segura C, Magnone MC, Lascorz J, Depner M, Holzberg D, Soyka M, Schreiber S, Matsuda F, Lathrop M, Schumann G, Albrecht U (2005) The clock gene Per2 influences the glutamatergic system and modulates alcohol consumption. Nat Med 11:35–42. [DOI] [PubMed] [Google Scholar]
- Stockdale SE, Wells KB, Tang L, Belin TR, Zhang L, Sherbourne CD (2007) The importance of social context: neighborhood stressors, stress-buffering mechanisms, and alcohol, drug, and mental health disorders. Soc Sci Med 65:1867–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland MT, McHugh MJ, Pariyadath V, Stein EA (2012) Resting state functional connectivity in addiction: lessons learned and a road ahead. Neuroimage 62:2281–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi F, Isono M, Nabika T, Katsuya T, Sugiyama T, Yamaguchi S, Kobayashi S, Ogihara T, Yamori Y, Fujioka A, Kato N (2011) Confirmation of ALDH2 as a major locus of drinking behavior and of its variants regulating multiple metabolic phenotypes in a Japanese population. Circ J 75:911–918. [DOI] [PubMed] [Google Scholar]
- Trabzuni D, Ryten M, Walker R, Smith C, Imran S, Ramasamy A, Weale ME, Hardy J (2011) Quality control parameters on a large dataset of regionally dissected human control brains for whole genome expression studies. J Neurochem 119:275–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viechtbauer W (2010) Conducting meta-analyses in R with the metafor package. Journal of Statistical Software 36:1–48. [Google Scholar]
- Vogt D, Smith BN, King DW, King LA (2012) Manual for the Deployment Risk and Resilience Inventory-2 (DRRI-2): A Collection of Measures for Studying Deployment-Related Experiences of Military Veterans. Boston, MA: National Center for PTSD. [Google Scholar]
- Wang JC, Foroud T, Hinrichs AL, Le NX, Bertelsen S, Budde JP, Harari O, Koller DL, Wetherill L, Agrawal A, Almasy L, Brooks AI, Bucholz K, Dick D, Hesselbrock V, Johnson EO, Kang S, Kapoor M, Kramer J, Kuperman S, Madden PA, Manz N, Martin NG, McClintick JN, Montgomery GW, Nurnberger JI Jr, Rangaswamy M, Rice J, Schuckit M, Tischfield JA, Whitfield JB, Xuei X, Porjesz B, Heath AC, Edenberg HJ, Bierut LJ, Goate AM (2013) A genome-wide association study of alcohol-dependence symptom counts in extended pedigrees identifies C15orf53. Mol Psychiatry 18:1218–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb BT, Edwards AC, Wolen AR, Salvatore JE, Aliev F, Riley BP Sun C, Williamson VS, Kitchens JN, Pedersen K, Adkins A, Cooke ME, Savage JE, Neale Z, Cho SB, Dick DM, Kendler KS (2017) Molecular genetic influences on normative and problematic alcohol use in a population-based sample of college students. Front Genet 8:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzosek M, Lukaszkiewicz J, Wrzosek M, Serafin P, Jakubczyk A, Klimkiewicz A, Matsumoto H, Brower KJ, Wojnar M (2011) Association of polymorphisms in HTR2A, HTR1A and TPH2 genes with suicide attempts in alcohol dependence: a preliminary report. Psychiatry Res 190:149–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Kranzler HR, Sherva R, Sartor CE, Almasy L, Koesterer R, Zhao H, Farrer LA, Gelernter J (2015) Genomewide association study for maximum number of alcoholic drinks in European Americans and African Americans. Alcohol Clin Exp Res 39:1137–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Brown AD, Lazar M, Cressman VL, Henn-Haase C, Neylan TC, Shalev A, Wolkowitz OM, Hamilton SP, Yehuda R, Sodickson DK, Weiner MW, Marmar CR (2013) Spontaneous brain activity in combat related PTSD. Neurosci Lett 547:1–5. [DOI] [PubMed] [Google Scholar]
- Yehuda R, Flory JD, Bierer LM, Henn-Haase C, Lehrner A, Desarnaud F, Makotkine I, Daskalakis NP, Marmar CR, Meaney MJ (2014) Lower methylation of glucocorticoid receptor gene promoter 1 in peripheral blood of veterans with posttraumatic stress disorder. Biol Psychiatry 77:356–364. [DOI] [PubMed] [Google Scholar]
- Yudko E, Lozhkina O, Fouts A (2007) A comprehensive review of the psychometric properties of the Drug Abuse Screening Test. J Subst Abuse Treat 32:189–198. [DOI] [PubMed] [Google Scholar]
- Yuferov V, Bart G, Kreek MJ (2005) Clock reset for alcoholism. Nat Med 11:23–24. [DOI] [PubMed] [Google Scholar]
- Zuo L, Gelernter J, Zhang CK, Zhao H, Lu L, Kranzler HR, Malison RT, Li CS, Wang F, Zhang XY, Deng HW, Krystal JH, Zhang F, Luo X (2012) Genome-wide association study of alcohol dependence implicates KIAA0040 on chromosome 1q. Neuropsychopharmacology 37:557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Wang K, Wang G, Pan X, Zhang X, Zhang H, Luo X (2014) Common PTP4A1-PHF3-EYS variants are specific for alcohol dependence. Am J Addict 23:411–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Zhang CK, Wang F, Li CS, Zhao H, Lu L, Zhang XY, Lu L, Zhang H, Zhang F, Krystal JH, Luo X (2011) A novel, functional and replicable risk gene region for alcohol dependence identified by genome-wide association study. PLoS One 6: e26726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Zhang XY, Wang F, Li CS, Lu L, Ye L, Zhang H, Krystal JH, Deng HW, Luo X (2013) Genome-wide significant association signals in IPO11-HTR1A region specific for alcohol and nicotine codependence. Alcohol Clin Exp Res 37:730–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


