Abstract
Background
In order to prepare the genome for gametogenesis, primordial germ cells (PGCs) undergo extensive epigenetic reprogramming during migration toward the gonads in mammalian embryos. This includes changes on a genome-wide scale and additionally in females the remodeling of the inactive X-chromosome to enable X-chromosome reactivation (XCR). However, if global remodeling and X-chromosomal remodeling are related, how they occur in PGCs in vivo in relation to their migration progress and which factors are important are unknown.
Results
Here we identify the germ cell determinant PR-domain containing protein 14 (PRDM14) as the first known factor that is instrumental for both global reprogramming and X-chromosomal reprogramming in migrating mouse PGCs. We find that global upregulation of the repressive histone H3 lysine 27 trimethylation (H3K27me3) mark is PRDM14 dosage dependent in PGCs of both sexes. When focusing on XCR, we observed that PRDM14 is required for removal of H3K27me3 from the inactive X-chromosome, which, in contrast to global upregulation, takes place progressively along the PGC migration path. Furthermore, we show that global and X-chromosomal reprogramming of H3K27me3 are functionally separable, despite their common regulation by PRDM14.
Conclusions
In summary, here we provide new insight and spatiotemporal resolution to the progression and regulation of epigenome remodeling along mouse PGC migration in vivo and link epigenetic reprogramming to its developmental context.
Electronic supplementary material
The online version of this article (10.1186/s13072-019-0284-7) contains supplementary material, which is available to authorized users.
Keywords: PRDM14, X-chromosome reactivation, PGCs, Epigenetic reprogramming, H3K27me3, Mouse
Background
The germ cell lineage has the unique function of transmitting genetic and epigenetic information from one generation to the next. In mice, germ cell development begins with the specification of PGCs, which then migrate through the hindgut in order to reach the genital ridges (future gonads), where they undergo meiosis and sex-specific differentiation into eggs and sperm. During migration and colonization of the gonads, the PGC epigenome is remodeled extensively at multiple levels, which includes global DNA-demethylation, erasure of genomic imprints, removal of histone H3 lysine 9 dimethylation (H3K9me2) and a global increase of the PRC2 (polycomb repressive complex 2)-associated H3K27me3 mark [1–7]. Around the same time, in female PGCs, the inactive X-chromosome is reactivated by XCR, which involves downregulation of the non-coding RNA Xist, the master regulator of X-inactivation, removal of repressive epigenetic marks like H3K27me3 and reactivation of X-linked genes [8–11]. Interestingly, H3K27me3 is therefore removed from the inactive X-chromosome, while it increases globally on autosomes. It is currently unknown, if these remodeling events are functionally linked or independent of each other.
XCR is a process linked to naïve pluripotency and germ cell fate [12, 13]. While XCR kinetics have been characterized during mouse and human germ cell development [8, 9, 11, 14, 15], the molecular mechanisms and factors, which are functionally important to reverse the inactive X-chromosome state, are largely unknown. Identifying such X-reactivation factors would be a key step toward revealing mechanistically how epigenetic memory can be erased in the germ line. We and others have shown that the transcriptional regulator PRDM14 plays a critical role in XCR in the mouse blastocyst epiblast in vivo and during induced pluripotent stem cell (iPSC) reprogramming [16] and epiblast stem cell (EpiSC) to embryonic stem cell (ESC) conversion [17] in vitro. In the absence of PRDM14, downregulation of Xist and removal of the H3K27me3 mark from the inactive X-chromosome are perturbed, likely due to repressive functions of PRDM14 on Xist and its activator Rnf12/Rlim [16, 18].
Besides being important for naïve pluripotency [19] and the associated XCR, PRDM14 is also critical for germ cell development. Prdm14-mutant mouse embryos show reduced PGC numbers [20], and overexpression of PRDM14 in epiblast-like cells (EpiLCs) is sufficient to induce germ cell fate in vitro [21]. Importantly, PRDM14 is a regulator of global epigenetic changes both in pluripotent stem cells and in PGCs [21, 22]. In particular, it has been shown that PRDM14 is responsible for the low global DNA-methylation levels characteristic of naïve pluripotent stem cells and PGCs, both by repressing DNA-methyltransferases and by recruiting DNA-demethylases of the TET family [19, 20, 23–26]. Besides controlling DNA-hypomethylation, PRDM14 is also important for global remodeling of histone marks. In migrating PGCs, Prdm14-mutant mouse embryos fail to downregulate the repressive H3K9me2 mark and its associated histone methyltransferase GLP/EHMT1 [20], most likely because Glp/Ehmt1 is a directly repressed target gene of PRDM14 [27]. The PGC-specific upregulation of H3K27me3 on a global level is also impaired in Prdm14−/− embryos [20] and genome-wide distribution of H3K27me3 is misregulated in Prdm14−/− ESCs [19]. However, it is not known, whether these epigenetic defects also occur in Prdm14+/− embryos.
Although the importance of PRDM14 for epigenetic remodeling in the germ cell lineage is known, how PRDM14 controls its different aspects in vivo is largely unresolved: For example, it remains unknown, if PRDM14 is important for X-chromosome reactivation in female PGCs, and if this is coordinated with genome-wide epigenetic changes occurring at that time. Furthermore, no information is available about the spatial distribution of germ cells along their migration path toward the gonads during epigenetic reprogramming. To address these questions, we here investigated epigenetic remodeling of the polycomb-associated H3K27me3 mark and its dependency on PRDM14 in migrating mouse PGCs. By using a whole mount embryo staining approach, we were able to collect spatiotemporal information about the epigenetic reprogramming process in mouse PGCs in relation to their migratory progress. We found that PRDM14 is critical both for X-chromosomal removal and for global upregulation of H3K27me3 in PGCs. We furthermore found differences in PRDM14-dosage dependence and kinetics of X-chromosomal and global H3K27me3 changes. While both events are controlled by PRDM14, they do not depend on each other and therefore must be regulated through different mechanisms. In summary, we describe here how PRDM14 regulates key aspects of epigenetic changes in germ cells and how this is linked with progression of germ cell migration and development.
Results
PRDM14 controls number and local distribution of PGCs during their migration
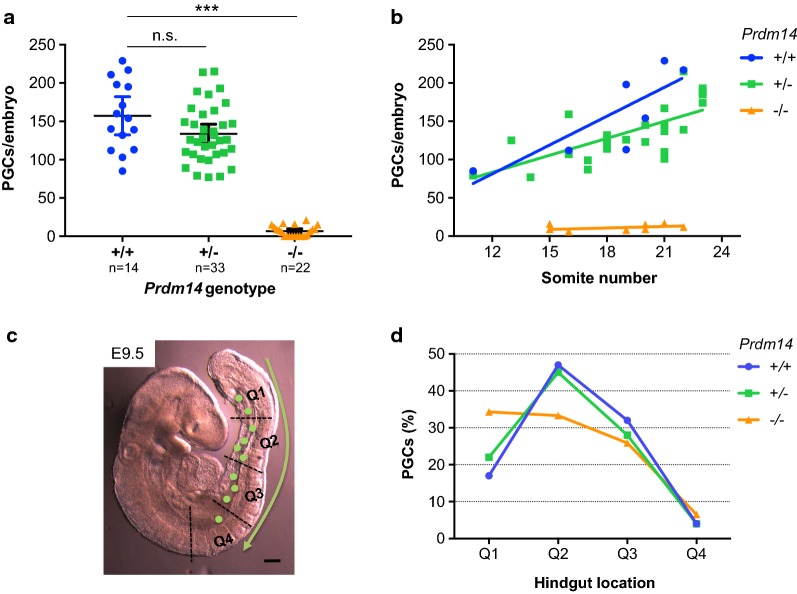
To address the function of PRDM14 for PGC number and migration, we immunostained whole mount embryonic day (E)9.5 mouse embryos from Prdm14+/− x Prdm14+/− heterozygous crosses for AP2γ, a specific marker and critical factor for PGC development [21, 28]. We found that migrating PGCs at this stage were spread throughout the hindgut and observed in Prdm14−/− embryos a strong depletion of PGCs or even their absence (9/22 embryos), when compared to wild type and heterozygous littermates (Fig. 1a, Additional file 1: Fig. S1, Additional file 2: Table S1), which is consistent with a previous study of this mouse line [20]. Generally, PGC numbers increased with developmental progression (somite number) in Prdm14+/+ and Prdm14+/−embryos, while they remained low in Prdm14−/− embryos (Fig. 1b). This is not due to a general developmental delay of Prdm14-mutant embryos at E9.5, as somite numbers were similar between different Prdm14 genotypes (Additional file 1: Fig. S2a, Additional file 2: Table S1).
Fig. 1.
Prdm14−/− E9.5 embryos have reduced PGC numbers with altered distribution along the hindgut. a Numbers of AP2γ-positive PGCs per embryo across Prdm14 genotypes. Means and 95% confidence intervals are shown as error bars. Kruskal–Wallis test was used for statistical comparison (p < 0.001). b Linear regression of AP2γ-positive PGC numbers versus developmental stage (somite number). R-square values for Prdm14+/+, +/− and −/− embryos were 0.659, 0.514 and 0.001, respectively. c Image of a E9.5 mouse embryo with PGCs (drawn green dots) migrating along the hindgut divided into four quadrants (Q1–Q4). Scale bar: 250 μm. d Distribution of PGCs along the hindgut across Prdm14 genotypes
In order to analyze the distribution of PGCs in more detail, we divided the migration path along the hindgut into 4 quadrants (Fig. 1c), starting with quadrant 1 (Q1) at the posterior end at the base of the allantois and finishing with quadrant 4 (Q4) near the area where PGCs would exit the hindgut and enter the genital ridges (future gonads). At E9.5, PGCs would be found mostly in Q2 and Q3 in Prdm14+/+ and Prdm14+/−embryos, while PGCs in Prdm14−/− embryos could be also frequently still found in Q1 (Fig. 1d, Additional file 1: Fig. S2b). This indicates that Prdm14-mutant PGCs might be slightly less efficient in initiating migration out of Q1, although they were also found in the other quadrants. In summary, Prdm14-mutant embryos show a strong reduction in total PGC numbers and some what reduced efficiency in PGC migration.
PRDM14 is required for global upregulation of H3K27me3 in migrating PGCs
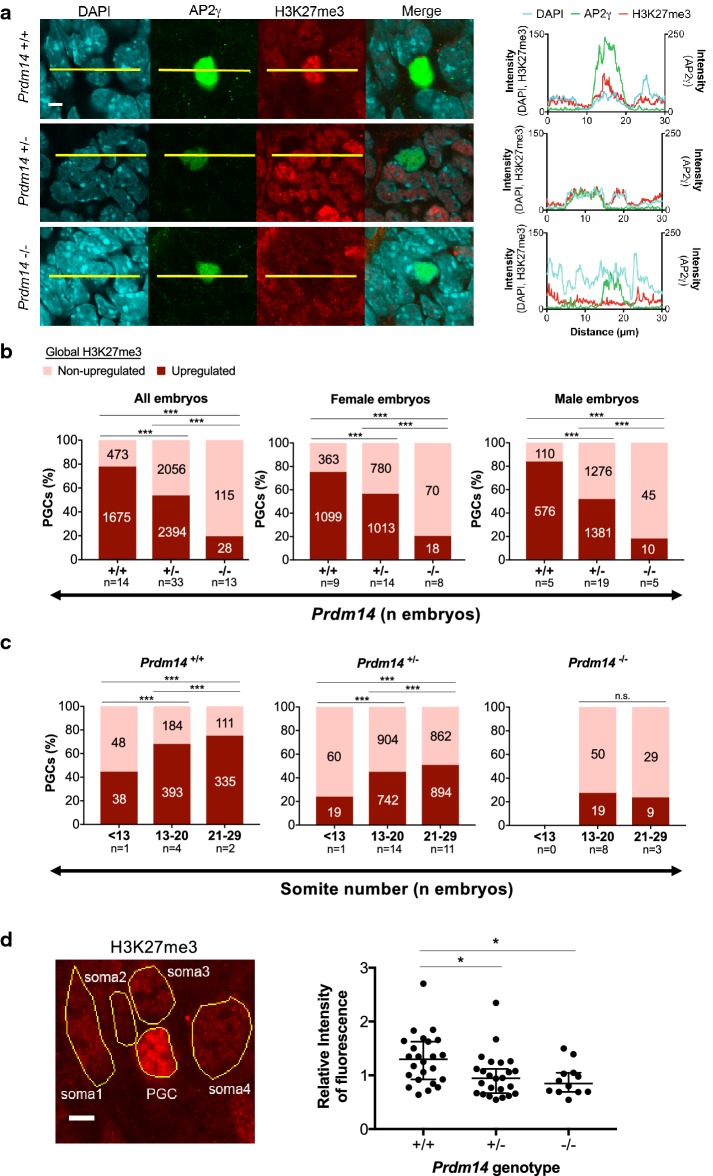
As PRDM14 has a role in the global epigenetic reprogramming occurring in PGCs [20], we wanted to investigate in more detail its function for upregulating the H3K27me3 mark, which is a hallmark of migrating PGCs [4–6]. We therefore costained E9.5 embryos of different Prdm14 genotypes with AP2γ and H3K27me3 antibodies (Fig. 2a, d). We scored H3K27me3 as being upregulated or non-upregulated in AP2γ-positive PGCs when compared to surrounding somatic cells. It thereby became apparent that H3K27me3 upregulation was in direct relation to PRDM14 dosage, as about 78% of wild-type PGCs had elevated H3K27me3 staining, while only 54% of heterozygote and only 20% of Prdm14-null PGCs did (Fig. 2b, d, Additional file 2: Table S1). These differences were observed to a similar degree both in male and in female embryos, indicating that PRDM14 controlled global H3K27me3 levels in a sex-independent manner.
Fig. 2.
PRDM14 dosage controls global H3K27me3 levels in migrating PGCs in both sexes. a Representative images of AP2γ-positive PGCs with upregulated and non-upregulated global levels of H3K27me3 across different Prdm14 genotypes. Scale bar: 5 µm. Intensity values for each channel along the yellow lines are plotted on the right. b Percentages of H3K27me3-upregulated and non-upregulated PGCs across Prdm14 genotypes in male and female embryos. c Percentages of H3K27me3 upregulated and non-upregulated PGCs at different developmental stages (somite number). Labels in each column indicate number of PGCs analyzed. Chi-square test was used for statistical comparisons (p < 0.001). d Representative image of a PGC and four neighboring somatic cells (same group of cells as shown in a Prdm14+/+ above). Their areas were selected to quantify the mean intensity of H3K27me3 signal. Scale bar: 5 µm. Relative intensity of H3K27me3 staining in PGCs in comparison with neighboring somatic cells across Prdm14 genotypes is plotted on the right. Medians and the 95% confidence intervals are shown as error bars. Kruskal–Wallis test was used for statistical comparison (p < 0.05)
When we assessed H3K27me3 upregulation in relation to developmental progression (somite number) at E9.5, we observed that H3K27me3 increased with somite number in Prdm14+/+ and Prdm14+/−, but not in Prdm14−/− embryos (Fig. 2c). Interestingly, H3K27me3 levels did not seem to vastly change in PGCs within different quadrants of their migration path, indicating that global H3K27me3 upregulation depends more on overall developmental progression of the embryo than on the position of the PGCs along the hindgut (Additional file 1: Fig. S3). Overall, we conclude that global H3K27me3 upregulation in PGCs is dependent on PRDM14 dosage and progresses with developmental stage at equal rate in both sexes.
X-chromosomal H3K27me3-erasure during XCR in female PGCs is dependent on PRDM14
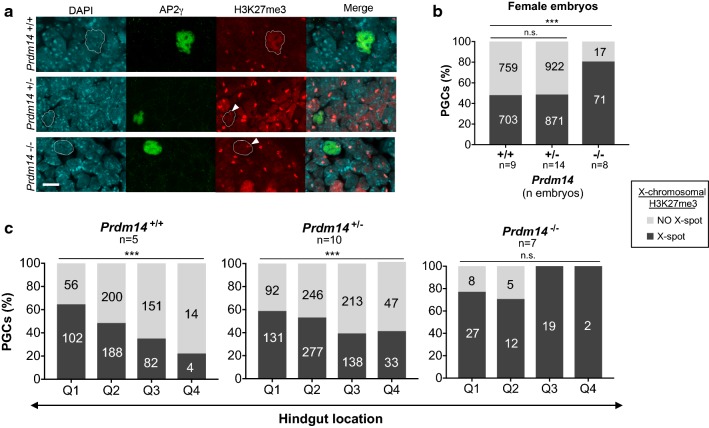
In contrast to the global increase of H3K27me3 stands the erasure of H3K27me3 from the inactive X-chromosome in female PGCs during the process of XCR [9, 11]. As these events occur around the same time and as XCR is controlled by PRDM14 in mouse blastocysts and during iPSC reprogramming [16], we wanted to know if PRDM14 is also required for XCR in PGCs. Thus, we scored the disappearance of the distinctive H3K27me3 accumulation from the inactive X-chromosome (“H3K27me3-spot”) in female PGCs in embryos of different Prdm14 genotypes (Fig. 3a). While about half of both Prdm14 wild-type (52%) and heterozygous (51%) PGCs have erased the H3K27me3-spot from the X, only around 19% of Prdm14-mutant PGCs have lost the spot at E9.5 (Fig. 3b, Additional file 2: Table S1). The erasure of the H3K27me3 spot occurred progressively in Prdm14+/+ and Prdm14+/− PGCs during their migration along the hindgut, but did not progress with migration in Prdm14−/− PGCs (Fig. 3c). In contrast to global H3K27me3 upregulation, removal of the X-chromosomal H3K27me3-spot therefore did not seem to be PRDM14-dose dependent and did not change substantially between E9.5 embryos of different somite number (Additional file 1: Fig. S4). Taken together, our results show that similar to its role in the pluripotent epiblast and during iPSC reprogramming [16], PRDM14 is also a key factor for XCR in PGCs by promoting the loss of the X-chromosomal H3K27me3 mark.
Fig. 3.
Erasure of H3K27me3-enrichment from the inactive X-chromosome in migrating female PGCs is dependent on PRDM14. a Representative images of AP2γ-positive PGCs with the presence or absence of H3K27me3 accumulation (H3K27me3 spot, white arrowheads) on the inactive X-chromosome. PGC nuclei are outlined in the H3K27me3 channel. Scale bar: 10 µm. b Percentage of H3K27me3 spot-positive and spot-negative PGCs across Prdm14 genotypes. Labels in each column indicate number of cells analyzed. c Erasure of the H3K27me3-spot in PGCs of different Prdm14 genotypes during their migration along the hindgut divided in four quadrants (Q1–Q4). Labels in each column indicate number of PGCs analyzed. Chi-square test was used for statistical comparisons (p < 0.001)
X-chromosomal and global H3K27me3 reprogramming occur independently in female PGCs
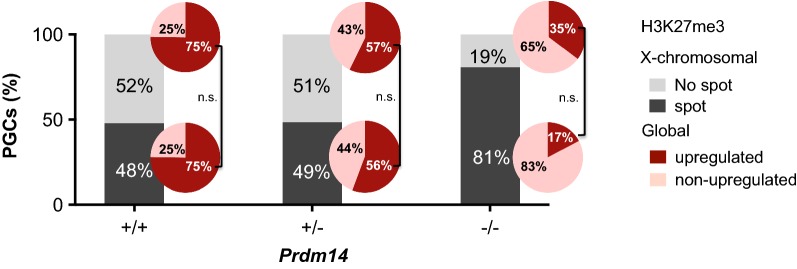
H3K27me3 and its associated enzymatic complex, PRC2, are strongly enriched on the inactive X-chromosome in female cells. Therefore, we hypothesized, if the X-chromosome could act as a “sink” for PRC2, whereby global upregulation of H3K27me3 in PGCs would require first stripping PRC2 off the X-chromosome through XCR to make it available for acting elsewhere in the genome. In order to test this hypothesis, we investigated the relationship between X-chromosomal downregulation and global upregulation of H3K27me3 in female PGCs (Fig. 4, Additional file 1: Fig. S5). If the inactive X were acting as a sink for PRC2, we would expect global H3K27me3 not to be upregulated in PGCs harboring an X-spot. However, when comparing within the different genotypes the fraction of PGCs which have lost or retained the X-chromosomal H3K27me3-spot, we did not detect any significant differences in global H3K27me3 upregulation (Fig. 4). This indicates that losing the X-spot is not a general prerequisite for global H3K27me3 upregulation in female PGCs. Also the fact that Prdm14+/+ and Prdm14+/− PGCs showed almost identical X-spot loss (52% in +/+ vs. 51% in +/−) but different global H3K27me3 upregulation levels (75% in +/+ vs. 56% in +/−) indicates that X-spot loss and global H3K27me3 upregulation are independent epigenetic events. In conclusion, while PRDM14 is both required for normal X-chromosomal and global H3K27me3 reprogramming in PGCs (Fig. 5), these events are mechanistically separable and do not depend on each other.
Fig. 4.
Changes in both X-chromosomal and global levels of H3K27me3 in female PGCs require PRDM14 but are independent of each other. Relationship between X-chromosomal (bar graphs, % of PGCs per genotype) and global (pie charts, % of PGCs per genotype within each H3K27me3 spot/no spot sub-category) levels of H3K27me3 in PGCs from female embryos of different Prdm14 genotypes. Chi-square test was used to compare dependency of global of H3K27me3 upregulation on X-chromosomal removal for each Prdm14 genotype (n.s., not significant; Prdm14+/+ p = 0.956, Prdm14+/− p = 0.499, Prdm14−/− p = 0.091)
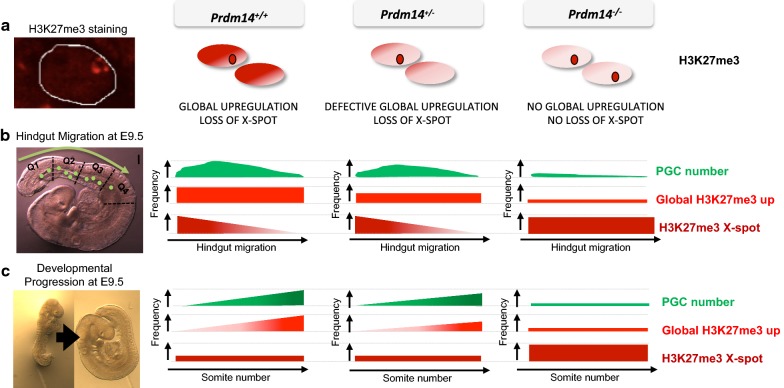
Fig. 5.
Summary of H3K27me3 dynamics in PGCs. a Schematic of H3K27me3 staining in representative female PGC nuclei of different Prdm14 genotypes. b PGC numbers and H3K27me3 changes during PGC migration along the hindgut at E9.5. PGCs are most frequently seen in quadrant 2 (Q2), and their number is strongly reduced in Prdm14−/− embryos. Global H3K27me3 levels are upregulated in most PGCs in Prdm14+/+ embryos throughout the hindgut, while it is less frequent in Prdm14+/− and rarely seen in Prdm14−/− embryos. The accumulation of H3K27me3 on the inactive X-chromosome (X-spot) is progressively lost during migration in both Prdm14+/+ and Prdm14+/− PGCs, but mostly maintained in Prdm14−/− PGCs. c PGC numbers and H3K27me3 changes in E9.5 embryos with different developmental progression (somite number). PGC numbers and global H3K27me3 levels increase with developmental progression in Prdm14+/+ and Prdm14+/− (reduced upregulation), but not in Prdm14−/− embryos. The X-chromosomal H3K27me3 spot is lost in both Prdm14+/+ and Prdm14+/− PGCs independently of developmental progression, while Prdm14−/− PGCs maintain the X-spot at all stages
Discussion
In this study, we have investigated the role of PRDM14 during epigenetic reprogramming in migrating mouse PGCs (summarized in Fig. 5). We thereby have uncovered multiple roles for PRDM14 during germ cell development. Most importantly, we have identified PRDM14 as the first factor with functional importance for XCR in the germ cell lineage. In particular, we found that erasure of H3K27me3 mark from the inactive X-chromosome, a key step during X-reactivation [9, 11, 29, 30], occurred progressively along the migration path of female PGCs and required PRDM14. This shows that PRDM14 has a universal role during X-reactivation whenever it is expressed; in the mouse blastocyst, during iPSC reprogramming [16] and in the germ cell lineage (this study).
Second, we uncovered that global H3K27me3 upregulation in PGCs is dependent on PRDM14 in a dosage-sensitive manner, as Prdm14+/− and Prdm14−/− PGCs showed a progressive defect. Interestingly, we observed that H3K27me3 upregulation seemed to mostly depend on the developmental stage of the embryo rather than the PGC position along the migratory path, which is in contrast to the kinetics of X-specific H3K27me3 removal.
Another key finding from our study is that the global upregulation and X-chromosomal depletion of the H3K27me3 mark seem to be controlled by distinct mechanisms despite their common dependency on PRDM14. This is based on several observations: First, global H3K27me3 upregulation is sensitive to PRDM14 dosage, while X-specific H3K27me3 removal is not. Second, global upregulation correlates with embryonic stage (somite number), while X-chromosomal H3K27me3 loss occurs progressively during PGC migration. Third, global H3K27me3 upregulation occurs with similar kinetics in male and female embryos and is affected to a similar extent by loss of PRDM14 in both sexes. Finally, X-chromosomal depletion of H3K27me3 does not seem to be required for global H3K27me3 upregulation on a single-cell level.
How then could PRDM14 control H3K27me3 remodeling on autosomes versus on the X-chromosome? On the global scale, previous studies suggested that PRDM14 could directly interact with the PRC2 complex in pluripotent stem cells and thereby facilitate its recruitment to PRDM14 target genes [19, 31] or that PRDM14 could activate expression of the PRC2 component Suz12 [24]. More recent studies, however, challenged this view and proposed that PRDM14 mainly acts though its binding partner and co-repressor CBFA2T2/MTGR1 [27, 32, 33], suggesting that PRC2 might be recruited secondarily to PRDM14 targets. Furthermore, PRDM14 is required for the low DNA-methylation levels in PGCs [24, 34]. Interestingly, PGC-like cells cultured in vitro lose DNA-methylation, while compensating by upregulating promoter H3K27me3 levels to ensure repression [34, 35], with BLIMP1 being a key factor in redistributing the H3K27me3 mark [36]. Future studies will need to address, if PRDM14 regulates global H3K27me3 upregulation in PGCs through direct or indirect mechanisms.
Regarding its X-chromosome-specific role, we have previously shown that PRDM14 binds to regulatory regions at Xist intron 1 and upstream of the Xist-activator Rnf12 [16] and thereby facilitates repression of Xist during XCR. As H3K27me3 accumulation on the X-chromosome is Xist-dependent [37–40], PRDM14 might thereby regulate X-chromosomal H3K27me3-depletion by downregulating Xist. The fact that H3K27me3 removal from the X-chromosome occurs progressively in PGCs along their migration path could speak in favor of a cell proliferation and replication dependent, passive dilution mechanism, similar as it has been proposed for global DNA-demethylation in PGCs [41, 42]. Nevertheless, also active mechanisms could play a role like removal of the mark by the H3K27me3-demethylase KDM6A/UTX, which has a partial effect on H3K27me3-demethylation during XCR in mouse blastocysts [30].
Finally, we have confirmed that Prdm14−/− embryos displayed severely compromised germ cell numbers [20] and observed for the first time their reduced efficiency in PGC migration. This is in line with a recent study [43], which found that genes implicated in cell migration are bound by PRDM14 in PGC-like cells derived in vitro, suggesting that PRDM14 might be potentially involved in controlling PGC migration.
Conclusions
In summary, here we have provided a first spatial roadmap of autosomal and X-chromosomal reprogramming of H3K27me3 during mouse PGC migration in vivo. We showed that X-chromosomal and global reprogramming occur independently of each other, but that they rely both on the key germ cell factor PRDM14. By adding to our understanding of the epigenetic reprogramming required for germ cell development in vivo, we provide a framework for assessing and improving the quality of in vitro-derived gametes.
Methods
Embryo isolation
Mouse care and procedures were conducted according to the protocols approved by the Ethics Committee on Animal Research of the Parc de Recerca Biomèdica de Barcelona (PRBB) and by the Departament de Territori i Sostenibilitat of the Generalitat de Catalunya.
Prdm14 mutant mice [20] were maintained in a predominant C57BL/6 strain background. Prdm14 heterozygous mice were mated, and resulting embryos were harvested from pregnant females at E9.5 and dissected from maternal tissues.
Whole mount immunostaining
The whole mount embryo immunostaining was performed as described in [20]. The primary antibodies used were rabbit polyclonal anti-AP2γ (TCFAP2C) (Santa Cruz Biotechnology sc-8977) and mouse monoclonal anti-H3K27me3 (Active Motif 61707, clone MABI0323). The secondary antibodies used were donkey anti-rabbit IgGAlexa Fluor 488 and goat anti-mouse IgGAlexa Fluor 555 (Molecular Probes A21206 and A21424).
After the immunostaining, the somite number of every embryo was counted under the stereomicroscope. Then the embryo was split in two parts: the head was used for genotyping, and the hindgut was dissected and mounted in Vectashield (Vector Laboratories) for observation with confocal microscopy.
Image capture and analysis
Bright-field images of the whole embryos were captured on a stereomicroscope using the Leica application suite software (Leica Microsystems, Wetzlar, Germany). Fluorescence imaging of the hindguts was performed on an inverted Leica TCS SP5 confocal microscope using the Leica Application Suite Advanced Fluorescence software. Z-stack images (1.5 μm intervals) of the full hindgut were acquired. Color setting and image processing were performed in Fiji [44] and Volocity (PerkinElmer) software.
The total length of the hindgut was divided in four quadrants (Q1–Q4 from the tail tip to the genital ridges, Fig. 1c) to ease the analysis of the location of AP2γ-positive PGCs. The global nuclear intensity of H3K27me3 staining and the presence of the X-spot were analyzed by visually inspecting all images from the confocal z-stack where a particular PGC was present in Volocity software. PGC showing a lower or similar nuclear H3K27me3 staining to the neighboring somatic cells in the same region/focal planes were categorized as “non-upregulated.” PGCs showing H3K27me3 nuclear staining levels more intense than the majority of neighboring somatic cells were categorized as “upregulated.” Image analysis was performed before Prdm14 genotyping to avoid bias. Alternatively, to quantitatively compare H3K27me3 staining levels of PGCs relative to somatic neighbors, a subset of PGCs from every Prdm14 genotype was analyzed by pixel intensity quantification with Fiji [44]. The median of the H3K27me3 average signal from four neighboring somatic cells was calculated and set as 1, and the H3K27me3 average signal from the PGC relative to this value was indicated (Fig. 2d).
Prdm14 and sex genotyping
Prdm14 genotype was determined by polymerase chain reaction after image analysis. Primer sequences are detailed in [20].
The sex of the embryo was determined by the presence of the H3K27me3 spot corresponding to the silent X-chromosome in somatic cells of female embryos [38].
Statistical analysis
Several E9.5 litters were collected, and the results from all embryos were pooled and analyzed with IBM SPSS and GraphPad Prism Statistics software. For qualitative variables Chi-square test was used, and for quantitative variables, the nonparametric Kruskal–Wallis test was used. Pairwise comparisons with Bonferroni’s correction were performed using the Mann–Whitney U test. For lineal regression modeling, R2 coefficient was calculated. In all cases, p < 0.05 was considered statistically significant.
Additional files
Additional file 1: Figures S1–S5. Figure S1 (related to Fig. 1). Migrating PGCs in E9.5 mouse embryos of different Prdm14 genotypes. Figure S2 (related to Fig. 1). Prdm14 genotype, developmental progression and PGC distribution during migration. Figure S3 (related to Fig. 2). Summary of global H3K27me3 levels in PGCs across somite number, migration progression and Prdm14 genotypes. Figure S4 (related to Fig. 3). Summary of the removal of X-chromosomal H3K27me3 accumulation (spot) in PGCs across somite number, migration progression and Prdm14 genotypes. Figure S5 (related to Fig. 4). Distribution of PGCs with different global and X-chromosomal H3K27me3 status dependent on Prdm14 genotype.
Additional file 2: Table S1. Summary table of embryos and PGCs analyzed in this study.
Acknowledgements
We thank members of the Payer laboratory for discussions and suggestions. We thank Mitinori Saitou for sharing the Prdm14-KO mouse strain and for advice on whole mount detection of PGCs by immunostaining. We also thank Guillaume Filion for critical reading and advice on statistical analysis. We thank the PRBB Animal Facility for mouse husbandry, the CRG Media kitchen for reagent preparation and the CRG Advanced Light Microscopy Unit for imaging support.
Abbreviations
- EpiLC
epiblast-like cell
- ESC
embryonic stem cell
- H3K27me3
histone H3 lysine 27 trimethylation
- iPSC
induced pluripotent stem cell
- PGC
primordial germ cell
- PRC2
polycomb repressive complex 2
- PRDM14
PR-domain containing protein 14
- XCR
X-chromosome reactivation
- Xist
X-inactive specific transcript
Authors’ contributions
AM, MG and BP contributed to conceptualization and project design; AM and MG helped with experimental execution; AM and MG collected the data; AM performed statistical analysis; AM and PB wrote the manuscript; BP performed supervision and funding acquisition. All authors read and approved the final manuscript.
Funding
This work has been funded by the Spanish Ministry of Science, Innovation and Universities (BFU2014-55275-P and BFU2017-88407-P), the AXA Research Fund and the Agencia de Gestio d’Ajuts Universitaris i de Recerca (AGAUR, 2017 SGR 346). We would like to thank the Spanish Ministry of Economy, Industry and Competitiveness (MEIC) to the EMBL partnership and to the “Centro de Excelencia Severo Ochoa.” We also acknowledge support of the CERCA Programme of the Generalitat de Catalunya.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information or are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Anna Mallol and Maria Guirola contributed equally to this work
References
- 1.Leitch HG, Tang WWC, Surani MA. Primordial germ-cell development and epigenetic reprogramming in mammals. Curr Top Dev Biol. 2013;104:149–187. doi: 10.1016/B978-0-12-416027-9.00005-X. [DOI] [PubMed] [Google Scholar]
- 2.Saitou M, Kagiwada S, Kurimoto K. Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development. 2012;139:15–31. doi: 10.1242/dev.050849. [DOI] [PubMed] [Google Scholar]
- 3.Reik W, Surani MA. Germline and Pluripotent Stem Cells. Cold Spring Harb Perspect Biol. 2015;7:1–24. doi: 10.1101/cshperspect.a019422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, Cesari F, et al. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452:877–881. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seki Y, Yamaji M, Yabuta Y, Sano M, Shigeta M, Matsui Y, et al. Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development. 2007;134:2627–2638. doi: 10.1242/dev.005611. [DOI] [PubMed] [Google Scholar]
- 6.Seki Y, Hayashi K, Itoh K, Mizugaki M, Saitou M, Matsui Y. Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev Biol. 2005;278:440–458. doi: 10.1016/j.ydbio.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 7.Prokopuk L, Stringer JM, Hogg K, Elgass KD, Western PS. PRC2 is required for extensive reorganization of H3K27me3 during epigenetic reprogramming in mouse fetal germ cells. Epigenetics Chromatin. 2017;10:7. doi: 10.1186/s13072-017-0113-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugimoto M, Abe K. X chromosome reactivation initiates in nascent primordial germ cells in mice. PLoS Genet. 2007;3:e116. doi: 10.1371/journal.pgen.0030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chuva de Sousa Lopes SM, Hayashi K, Shovlin TC, Mifsud W, Surani MA, McLaren A. X chromosome activity in mouse XX primordial germ cells. PLoS Genet. 2008;4:e30. doi: 10.1371/journal.pgen.0040030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Payer B. Developmental regulation of X-chromosome inactivation. Semin Cell Dev Biol. 2016;56:88–99. doi: 10.1016/j.semcdb.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 11.de Napoles M, Nesterova T, Brockdorff N. Early loss of Xist RNA expression and inactive X chromosome associated chromatin modification in developing primordial germ cells. PLoS ONE. 2007;2:e860. doi: 10.1371/journal.pone.0000860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pasque V, Plath K. X chromosome reactivation in reprogramming and in development. Curr Opin Cell Biol. 2015;37:75–83. doi: 10.1016/j.ceb.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Payer B, Lee JT. Coupling of X-Chromosome reactivation with the pluripotent stem cell state. RNA Biol. 2014;11:798–807. doi: 10.4161/rna.29779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang WWC, Dietmann S, Irie N, Leitch HG, Floros VI, Bradshaw CR, et al. A unique gene regulatory network resets the human germline epigenome for development. Cell. 2015;161:1453–1467. doi: 10.1016/j.cell.2015.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo F, Yan L, Guo H, Li L, Hu B, Zhao Y, et al. The transcriptome and DNA methylome landscapes of human primordial germ cells. Cell. 2015;161:1437–1452. doi: 10.1016/j.cell.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 16.Payer B, Rosenberg M, Yamaji M, Yabuta Y, Koyanagi-Aoi M, Hayashi K, et al. Tsix RNA and the germline factor, PRDM14, link X reactivation and stem cell reprogramming. Mol Cell. 2013;52:805–818. doi: 10.1016/j.molcel.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gillich A, Bao S, Grabole N, Hayashi K, Trotter MWB, Pasque V, et al. Epiblast stem cell-based system reveals reprogramming synergy of germline factors. Cell Stem Cell. 2012;10:425–439. doi: 10.1016/j.stem.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma Z, Swigut T, Valouev A, Rada-Iglesias A, Wysocka J. Sequence-specific regulator Prdm14 safeguards mouse ESCs from entering extraembryonic endoderm fates. Nat Struct Mol Biol. 2011;18:120–127. doi: 10.1038/nsmb.2000. [DOI] [PubMed] [Google Scholar]
- 19.Yamaji M, Ueda J, Hayashi K, Ohta H, Yabuta Y, Kurimoto K, et al. PRDM14 ensures naive pluripotency through dual regulation of signaling and epigenetic pathways in mouse embryonic stem cells. Cell Stem Cell. 2013;12:368–382. doi: 10.1016/j.stem.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 20.Yamaji M, Seki Y, Kurimoto K, Yabuta Y, Yuasa M, Shigeta M, et al. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat Genet. 2008;40:1016–1022. doi: 10.1038/ng.186. [DOI] [PubMed] [Google Scholar]
- 21.Nakaki F, Hayashi K, Ohta H, Kurimoto K, Yabuta Y, Saitou M. Induction of mouse germ-cell fate by transcription factors in vitro. Nature. 2013;501:222–226. doi: 10.1038/nature12417. [DOI] [PubMed] [Google Scholar]
- 22.Seki Y. PRDM14 is a unique epigenetic regulator stabilizing transcriptional networks for pluripotency. Front Cell Dev Biol. 2018;6:12. doi: 10.3389/fcell.2018.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okashita N, Kumaki Y, Ebi K, Nishi M, Okamoto Y, Nakayama M, et al. PRDM14 promotes active DNA demethylation through the ten-eleven translocation (TET)-mediated base excision repair pathway in embryonic stem cells. Development. 2014;141:269–280. doi: 10.1242/dev.099622. [DOI] [PubMed] [Google Scholar]
- 24.Grabole N, Tischler J, Hackett JA, Kim S, Tang F, Leitch HG, et al. Prdm14 promotes germline fate and naive pluripotency by repressing FGF signalling and DNA methylation. EMBO Rep. 2013;14:629–637. doi: 10.1038/embor.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leitch HG, Mcewen KR, Turp A, Encheva V, Carroll T, Grabole N, et al. Naive pluripotency is associated with global DNA hypomethylation. Nat Struct Mol Biol. 2013;20:311–316. doi: 10.1038/nsmb.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ficz G, Hore TA, Santos F, Lee HJ, Dean W, Arand J, et al. FGF signaling inhibition in ESCs drives rapid genome-wide demethylation to the epigenetic ground state of pluripotency. Cell Stem Cell. 2013;13:351–359. doi: 10.1016/j.stem.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tu S, Narendra V, Yamaji M, Vidal SE, Rojas LA, Wang X, et al. Co-repressor CBFA2T2 regulates pluripotency and germline development. Nature. 2016;534:387–390. doi: 10.1038/nature18004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weber S, Eckert D, Nettersheim D, Gillis AJM, Schäfer S, Kuckenberg P, et al. Critical function of AP-2 gamma/TCFAP2C in mouse embryonic germ cell maintenance. Biol Reprod. 2010;82:214–223. doi: 10.1095/biolreprod.109.078717. [DOI] [PubMed] [Google Scholar]
- 29.Mak W, Nesterova TB, de Napoles M, Appanah R, Yamanaka S, Otte AP, et al. Reactivation of the paternal X chromosome in early mouse embryos. Science. 2004;303:666–669. doi: 10.1126/science.1092674. [DOI] [PubMed] [Google Scholar]
- 30.Borensztein M, Okamoto I, Syx L, Guilbaud G, Picard C, Ancelin K, et al. Contribution of epigenetic landscapes and transcription factors to X-chromosome reactivation in the inner cell mass. Nat Commun. 2017;8:1297. doi: 10.1038/s41467-017-01415-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan Y-S, Göke J, Lu X, Venkatesan N, Feng B, Su I-H, et al. A PRC2-dependent repressive role of PRDM14 in human embryonic stem cells and induced pluripotent stem cell reprogramming. Stem Cells. 2013;31:682–692. doi: 10.1002/stem.1307. [DOI] [PubMed] [Google Scholar]
- 32.Nady N, Gupta A, Ma Z, Swigut T, Koide A, Koide S, et al. ETO family protein Mtgr1 mediates Prdm14 functions in stem cell maintenance and primordial germ cell formation. Elife. 2015;4:e10150. doi: 10.7554/eLife.10150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawaguchi M, Sugiyama K, Matsubara K, Lin C-Y, Kuraku S, Hashimoto S, et al. Co-option of the PRDM14-CBFA2T complex from motor neurons to pluripotent cells during vertebrate evolution. Development 2019;146. [DOI] [PubMed]
- 34.Shirane K, Kurimoto K, Yabuta Y, Yamaji M, Satoh J, Ito S, et al. Global landscape and regulatory principles of DNA methylation reprogramming for germ cell specification by mouse pluripotent stem cells. Dev Cell. 2016;39:87–103. doi: 10.1016/j.devcel.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 35.Ohta H, Kurimoto K, Okamoto I, Nakamura T, Yabuta Y, Miyauchi H, et al. In vitro expansion of mouse primordial germ cell-like cells recapitulates an epigenetic blank slate. EMBO J. 2017;36:1888–1907. doi: 10.15252/embj.201695862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurimoto K, Yabuta Y, Hayashi K, Ohta H, Kiyonari H, Mitani T, et al. Quantitative dynamics of chromatin remodeling during germ cell specification from mouse embryonic stem cells. Cell Stem Cell. 2015;16:517–532. doi: 10.1016/j.stem.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 37.Zhao J, Sun BK, Erwin JA, Song J-J, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 39.Silva JCR, Mak W, Zvetkova I, Appanah R, Nesterova TB, Webster Z, et al. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell. 2003;4:481–495. doi: 10.1016/S1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- 40.Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, et al. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 2006;25:3110–3122. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kagiwada S, Kurimoto K, Hirota T, Yamaji M, Saitou M. Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice. EMBO J. 2013;32:340–353. doi: 10.1038/emboj.2012.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, et al. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell. 2012;48:849–862. doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheetham Seth W., Gruhn Wolfram H., van den Ameele Jelle, Krautz Robert, Southall Tony D., Kobayashi Toshihiro, Surani M. Azim, Brand Andrea H. Targeted DamID reveals differential binding of mammalian pluripotency factors. Development. 2018;145(20):dev170209. doi: 10.1242/dev.170209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figures S1–S5. Figure S1 (related to Fig. 1). Migrating PGCs in E9.5 mouse embryos of different Prdm14 genotypes. Figure S2 (related to Fig. 1). Prdm14 genotype, developmental progression and PGC distribution during migration. Figure S3 (related to Fig. 2). Summary of global H3K27me3 levels in PGCs across somite number, migration progression and Prdm14 genotypes. Figure S4 (related to Fig. 3). Summary of the removal of X-chromosomal H3K27me3 accumulation (spot) in PGCs across somite number, migration progression and Prdm14 genotypes. Figure S5 (related to Fig. 4). Distribution of PGCs with different global and X-chromosomal H3K27me3 status dependent on Prdm14 genotype.
Additional file 2: Table S1. Summary table of embryos and PGCs analyzed in this study.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information or are available from the corresponding author on reasonable request.