Short abstract
Zika virus (ZIKV) is an arthropod-borne virus (arbovirus) member of the Flaviviridae family, which has been associated with the development of the congenital Zika syndrome (CZS). RNA viruses, such as flaviviruses, have been reported to exert a profound impact on host microRNAs (miRNAs). Cellular miRNAs modulated by ZIKV may help identify cellular pathways of relevance to pathogenesis. Here, we screened 754 human cellular miRNAs modulated by ZIKV infection (Brazilian PE strain) in a neuroblastoma cell line. Seven miRNAs (miR-99a*, miR-126*, miR-190b, miR-361-3p, miR-522-3p, miR-299-5p, and miR-1267) were downregulated during ZIKV infection, while miR-145 was upregulated. Furthermore, 11 miRNAs were exclusively expressed in ZIKV-infected (miR-148a, miR-342-5p, miR-598, and miR-708-3p) or mock cells (miR-208, miR-329, miR-432-5p, miR-488, miR-518b, miR-520g, and miR-767-5p). Furthermore, in silico analysis indicated that some central nervous system, cellular migration, and adhesion function-related biological processes were overrepresented in the list of target genes of the miRNAs regulated in ZIKV-infected cells, especially for miR-145 and miR-148a. The induction of miR-145 and miR-148a was confirmed in postmortem brain samples from stillborn with severe CZS. Finally, we determined the expression regulation of microcephaly related genes through RNA interference pathway caused by ZIKV directly on neuron cells.
Keywords: miRNAs, microcephaly, neuroblastoma, postmortem brain autopsies, Zika
Introduction
Zika virus (ZIKV) is an arthropod-borne virus member of the Flaviviridae family, Flavivirus genus. The family comprises viruses with positive-sense RNA genomes, and other family members include important human pathogens including Yellow Fever Virus (YFV), Dengue Virus (DENV), West Nile Virus (WNV), Japanese Encephalitis Virus (JEV), and Spondweni Virus (SPOV). Although ZIKV was repeatedly isolated from mosquitoes of the Aedes africanus species in Uganda’s Zika forest in 1947 (Dick, 1952), and the first evidence of human disease was reported in 1953 (MacNamara, 1954), the recent outbreak in the Americas and Pacific brought significant concern due to neurological complications associated with the infection, especially severe infections in newborns. ZIKV exposure during pregnancy can lead to the development of a neurological congenital syndrome in the fetus, including symptoms such as microcephaly, facial disproportion, hypertonia and spasticity, intracerebral calcifications, ventriculomegaly, arthrogryposis, and gyral malformations (Cavalheiro et al., 2016; Culjat et al., 2016; de Fatima Vasco Aragao et al., 2016; De Oliveira Melo et al., 2016; Driggers et al., 2016; Martines et al., 2016; van der Linden et al., 2016; Werner et al., 2016).
The ZIKV genome is approximately 11-kb in length and has a single open reading frame (ORF) which is flanked by two noncoding regions, named the 5′- and 3′-untranslated regions (5′- and 3′-UTRs) (Kuno and Chang, 2007). During virus replication, this ORF is translated into a single polyprotein that subsequently undergoes cleavage by viral and cellular proteins. The production of viral progeny represents a stress condition to the host cell, increasing IFN production, especially type I and III, controlling the virus replication (Rossi et al., 2016). The interaction between secreted IFNs and receptors on the cell surface promotes the transcription of the interferon-stimulated genes in an attempt to block viral infection and spread, by interfering in many steps of host and virus interactions such as protein synthesis, RNA replication and decay, virus assembly and release, culminating in apoptosis processes (Li et al., 2004). Animal models and transcriptome analysis have already shown the modulation of cell cycle, cytoskeleton pathways, and apoptosis process in ZIKV-infected cells (Cugola et al., 2016; Garcez et al., 2016, 2017; Souza et al., 2016). However, we still do not know how the virus modulates the expression of host proteins, and several steps of ZIKV replication are still unclear.
It is known that cellular stress, as the response to a virus infection, requires a complex level of gene regulation. MicroRNAs (miRNAs) can control the RNA accessibility to translation machinery bringing another level to control gene expression. These endogenous small noncoding RNAs have 21 to 24 nucleotides in length and are associated with the RNA-induced silencing complex (RISC). These miRNAs act like guide strands, targeting one or more specific sites at the mRNA 3′-UTR region that have partial, but sufficient, Watson-Crick base pairing complementarity with their sequences. Once the miRNA binds to its target, the gene silencing complex can lead the RNA to be cleaved or translationally repressed (Hutvágner & Zamore, 2002; Yekta et al., 2004).
MiRNAs play important regulatory roles in many physiological and disease processes, including viral infections. Eukaryotic miRNAs produced by the infected host cell can function as an antiviral mechanism. Differential expression of miRNAs has been described in various diseases, including flaviviruses infections. JEV infection upregulates miR-19b-3p leading to increased production of inflammatory cytokines and then modulates the virus-mediated inflammation by targeting the RING finger protein 11 (Ashraf et al., 2016). The cellular miR-126-5p is also modulated by DENV infection (Kakumani et al., 2016). MiRNAs could also have a pathologic effect, being regulated by the infectious agent for its own benefit. The endogenous miR-33a-5p is downregulated by JEV that targets the eukaryotic translation elongation factor 1A1, which stabilizes the components of the viral replicase complex promoting virus replication (Chen et al., 2016). Another example is the interaction between the liver-specific miRNA, miR-122, and hepatitis C virus (HCV). Usually, miRNAs downregulate their mRNA targets binding to the 3′-UTR region. MiR-122, in a nonclassical way, upregulates viral replication at the RNA level by targeting two adjacent sites in the 5′ end of the HCV genome (Jopling et al., 2005). This phenomenon is one of the first indications that, in certain cellular specific contexts, miRNAs could have RISC independent functions.
Identifying the profiles of cellular miRNAs modulated by the infection may reveal new cellular processes modulated by the viral infection. It is even more important when it comes to ZIKV, due to a lack of knowledge about its replicative cycle and the serious neuropathogenic effects caused by the infection. In this study, we performed a large screen of cellular miRNAs modulated by ZIKV replication in neuroblastoma cells infected with a Brazilian ZIKV strain and validated the expression of selected miRNAs directly in brain tissues from stillborn that died with congenital Zika syndrome (CZS).
Materials and Methods
Cells, Virus, and Infection
SH-SY5Y neuroblastoma cell line (ATCC CRL266) was cultured in six-well plates (Corning® Costar®) in glucose-enriched Dulbecco’s Modified Eagle Medium (DMEM)/F12 (Gibco) (final glucose concentration of 6 g/L) supplemented with 10% fetal bovine serum (FBS) (Gibco) at a density of 6 × 104 cells per well.
Just before the infection, the conditioned medium of each well was removed and centrifuged at 400 g for 8 min, in order to recover the cells in suspension. The supernatant was removed, and pelleted cells were resuspended with the ZIKV Brazilian strain PE (ZIKV PE/243 strain from Recife/Brazil, accession no: KX197192.1) at a MOI of 1. The PE strain was isolated from a patient with ZIKV infection symptoms from Pernambuco State, Brazil, during the 2015 outbreak (Donald et al., 2016). Virus suspension was used to infect the neuroblastoma cells in the absence of serum. Virus infection was performed for 1 h at 37°C in a humidified 5% CO2 atmosphere in a final volume of 500 µL. After infection, 1.5 mL of DMEM F12 supplemented FBS (10%) was added to each well. The infection was allowed to proceed for 72 h at 37°C and then cells were harvested using trypsin (Gibco), 1:4 diluted in PBS (Gibco).
The percentage of infected cells was monitored by flow cytometry (FACS). For this analysis, the cells were fixed using paraformaldehyde at 4%, permeabilized with Triton X-100 (0.1%), and stained with the 4G2 monoclonal antibody (that recognizes the flavivirus envelope protein) 1:5 diluted and the AlexaFluor® 488 Donkey Anti-Mouse antibody (LifeTechnologies—ThermoScientific), 1:1,000 diluted (LifeTechnologies—ThermoScientific). FACS analysis were performed with a BD Accuri™ C6 Flow Cytometer. About 10% of the harvested cells were used for the FACS analysis to investigate virus infectivity, and the remainder was stored at −80°C for further RNA isolation.
Stillborn Brain Samples
Five (n = 5) brain tissues of stillborn children with Zika Congenital Syndrome were obtained from two public health institutes in Brazil: Instituto Professor Joaquim Amorim Neto (IPESQ, Campina Grande/PB) and Instituto Fernandes Figueira (IFF, Rio de Janeiro/RJ).
The ZIKV-positive samples are from a cohort of pregnant women with symptoms of Zika exposition at distinct weeks of gestation from the northeast (Campina Grande, Paraiba state) and southeast regions of Brazil (Rio de Janeiro, Rio de Janeiro state). The respective pregnant women were referred to public healthcare with a diagnosis of rash or fetus central nervous system (CNS) abnormality confirmed by ultrasonography (USG) or magnetic resonance imaging (MRI), as well as postnatal physical examination in the newborn suggestive of microcephaly. These studies were already approved by the ethical internal review board from IPESQ and IFF institutes (52888616.4.0000.5693 and 52675616.0.000.5269, respectively). Informed consent was signed by mothers and fathers allowing autopsies procedures and postmortem samples collection.
Brains of ZIKV, CHIKV (Chikungunya virus), DENV, or STORCH (Syphilis, Cytomegalovirus, Herpes virus 1/2, Toxoplasma gondii, and Rubella virus) negative controls of the same gestational age (n = 3) were obtained from Instituto Estadual do Cérebro (Rio de Janeiro, Brazil) covered by the institutional review board–approved study (1705093). The causes of death of the ZIKV-negative control cases were acute perinatal anoxia or complications of prematurity.
Differential ZIKV Diagnostic on the Stillborn Brain Samples
ZIKV-positive diagnostic was confirmed during pregnancy by reverse transcription polymerase chain reaction (RT-PCR) targeting the env gene in different fluids and tissues from the embryo and their respective mothers (Lanciotti et al., 2008). ZIKV RNA was detected both in samples obtained during the gestational period (amniotic fluid, blood or blood cord, and urine) and in postnatal samples (autopsied placenta, brain, and other organs) (Table S5).
For these procedures, viral RNA was extracted from 140 μL of each fluid using QIAmp MiniElute Virus Spin (QIAGEN, Hilden, Germany), following manufacturer’s recommendations. For tissues, 50 mg were processed using the Tissuerupter ® (QIAGEN) using 325 μL of RTL buffer from RNEasy Plus Mini Kit (QIAGEN), following manufacturer’s protocol. RNA extraction was then performed using Rneasy Plus Mini Kit (QIAGEN), following recommendations of the manufacturer.
For fluids, ZIKV RNA detection was performed using One-Step TaqMan RT-PCR (Thermo Fisher Scientific, Waltham, MA, USA) on 7500 Real-time PCR System (Applied Biosystems, Foster City, CA, USA) using PCR conditions that were already described in the literature (Brasil et al., 2016). For tissues, real-time RT-PCR was performed using 1 μg of total tissue RNA as described earlier.
Dengue and Chikungunya virus were excluded either by RT-PCR using ZDC Trioplex kits (Bio-Manguinhos, Fiocruz, Rio de Janeiro, Brazil) or serological ELISA (Kit XGen, Biometrix, Brazil and Euroimmum kit, Lübeck, Germany). STORCH agents were also excluded using serological IgM ELISA (Dia. Pro Diagnostic Bioprobes, Italy), following manufacturer’s recommendations. For the ZIKV-infected stillborn, other microcephaly causes including congenital genetic diseases were investigated and ruled out.
RNA Isolation and Quantification
Total RNA was isolated from ZIKV-infected and uninfected mock cells using the mirVana RNA isolation kit (Ambion) according to manufacturer’s instructions. RNA concentration and integrity were determined using the RNA 6000 Nano kit, specifically optimized for the analysis of total RNA in the Agilent 2100 Bioanalyzer. RNA quality and integrity were assured through the determination of the RNA Integrity Number (RIN) and only samples with RIN values superior to 9.0 were considered for further analysis.
For the miRNA validation analysis in the brain samples, we obtained formalin-fixed paraffin-embedded (FFPE) tissue blocks from the already described stillborn. After their microtome sectioning of the tissue blocks, the slides were stored at −20°C until the RNA extraction procedures. To extract the miRNAs from the FFPE tissue sections, the samples were processed using the miRNeasy FFPE kit (QIAGEN). To preserve the RNA integrity as much as possible, the extraction was performed using the Deparaffinization Solution (QIAGEN) according to the manufacturer’s instructions. RNA was quantified using the Qubit fluorometer and Qubit RNA HS Assay Kit (Invitrogen).
Cellular miRNAs Screening
For analysis of the cellular miRNA profile induced by ZIKV infection, reverse transcription and preamplification steps were undertaken with Megaplex™ RT/PA primer pools using the manufacturer’s human TaqMan® OpenArray® MicroRNA Panel protocol (Life Technologies). Briefly, 100 ng of each RNA sample was reverse transcribed into complementary DNA (cDNA) using Megaplex™ RT Primers. To obtain a full miRNA profile, this reaction was run twice, using two different primer pools (Pools A and B) that targets in total 754 cellular miRNAs. Then, the cDNA products underwent the PA process in order to increase the quantity of the desired cDNA, using the Megaplex™ PreAmp Primers. Finally, the preamplification products were combined with TaqMan® OpenArray® Real-Time PCR Master Mix and loaded onto TaqMan® OpenArray® Human MicroRNA Panel (AccuFill™ system). The expression levels of miRNAs were evaluated by quantitative PCR (qPCR) reactions using the QuantStudio™ 12K Flex (Life Technologies) platform.
qPCR Array Data Analysis
For each qPCR reaction in the array, the exploratory data analysis was performed through the use of programming routines in R (R Development Core Team, 2016) for parsing the raw data derived from the OpenArray® Real-Time PCR System (Applied Biosystems).
The fluorescence background was corrected, and the amplification curves were recreated using the normalized reporter signal (Rn). In the relative quantification of the gene expression, the logistics function adjustment (sigmoid curve) of four parameters was used to represent each amplification curve using the qPCR package (Ritz and Spiess, 2008). The quantification cycle was determined as the cycle relative to the second derivative (maximum point) of the sigmoid adjusted curve (cpD2). The amplification efficiency was obtained by analyzing the exponential part of the curve, in the cpD2, and calculated as the ratio between the fluorescence at the cpD2 and the fluorescence at the immediately preceding cycle (cpD2-1). Each miRNA amplification efficiency was estimated by the mean of the calculated efficiencies among replicas. Endogenous small-nucleolar RNAs RNU 44, RNU 48, and U6 spliceosomal RNA were selected as normalization controls using geNorm method (Vandesompele et al., 2002).
Nonparametric permutation t test (B = 1,000 permutations) was used in order to compare the means of normalized expression values of each group. Then, the false discovery rate was estimated and used to adjust for multiple comparisons.
Gene Set Enrichment Analysis
Putative miRNA targets were identified based on the prediction of six online software and databases: Target Scan (http://www.targetscan.org/index.html), miRBase (http://www.mirbase.org/index.shtml), PicTar (http://pictar.bio.nyu.edu/), mirtarget2/miRDB (http://mirdb.org/miRDB/), miRanda (http://www.microrna.org/microrna/home), and TarBase (http://www.diana.pcbi.upenn.edu/tarbase). Any predicted gene was considered to be a putative target if it had been predicted in at least three of the six software and databases. However, for some interesting targets that could be involved in neurogenesis and cellular antiviral response, each online database or tool was consulted individually. All the functions mentioned in the results session are according to GeneCards® and NCBI Gene databases.
Furthermore, in order to identify the molecular mechanisms potentially associated with the miRNA modulation, a gene set enrichment analysis (GSEA) was performed (Alexa et al., 2006). The gene sets were defined as all the putative target genes that share the same ontology described in the Gene Ontology (GO) database (Harris et al., 2004). The overrepresentation was identified by the statistical significance based on the hypergeometric test (p ≤ .001).
miR-145 and miR-148a Expression Validation Through RT-qPCR
Reverse transcription was performed using miRNA first-strand cDNA synthesis Kit (Agilent Technologies) and qPCR reactions were made with high-specificity miRNA QPCR Core Kit (Agilent Technologies). Human U6 RNA forward primer (Agilent Technologies) was used as endogenous control and qPCR reaction was performed in 7500 Real-Time PCR System (Applied Biosystems). Cycling parameters were set according to the manufacturer’s instructions. The miRNA forward primers sequences are miR_145: 5′-CAGTTTTCCCAGGAATCCCT-3′ and miR_148a: 5′-TCAGTGCACTACAGAACTTTGT-3′. Quantification followed the same methodology described for the arrays, but normalization used solely the U6 spliceosomal RNA. Again, nonparametric permutation t test (B = 1,000 permutations) was used in order to compare the means of normalized expression values of each group.
Results
Cellular miRNAs Profile in ZIKV-Infected Neuroblastoma
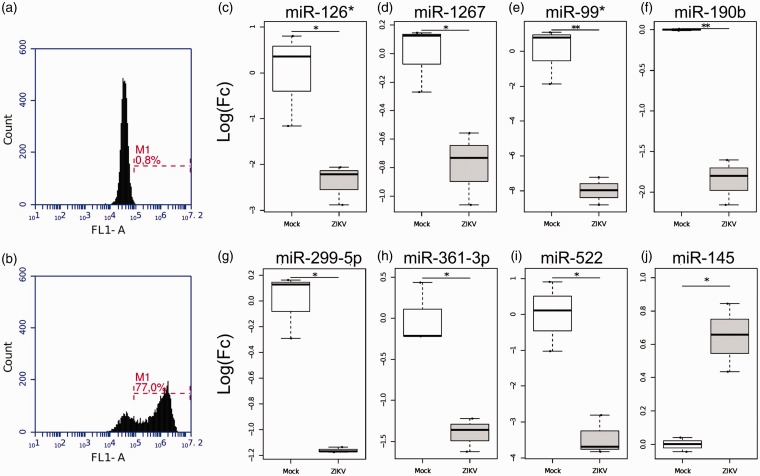
To define the miRNAs involved in ZIKV infection, we infected neuroblastoma cells (SH-SY5Y) with the Brazilian ZIKV strain PE, which belongs to the Asiatic lineage. Virus infectivity (up to 77% of cells) was confirmed 72 h postinfection (hpi) validating our model (Figure 1(a) and (b)).
Figure 1.
ZIKV infection modulates host cellular miRNAs. SH-SY5Y cells were infected with Brazilian ZIKV strain PE at MOI 1 and virus infectivity was evaluated by immunofluorescence using anti-env 4G2 antibody 72 hpi. (a) Mock noninfected cells. (b) ZIKV-infected cells. The dashed line (M1) represents the proportion of ZIKV positive cells (%). (c–j) Up- and downregulated miRNAs in ZIKV-infected neuroblastoma cells. Box plots representing relative miRNAs expression levels. The Quantstudio™ expression data were normalized using U6 snRNA, RNU44, and RNU48 as endogenous control genes. All the experiments were performed in triplicates and here we presented the median and its interquartile range for each group of noninfected (mock) and ZIKV-infected cells. *p ≤ .05; **p ≤ .01.
We next used the QuantStudio™ 12K Flex (Life Technologies) platform to characterize the miRNA expression profile of neuroblastoma cells infected with ZIKV. This platform screens for up to 754 human cellular miRNAs through RT-PCR using specific primers for each human inventoried miRNA. After the miRNA expression quantification, normalization and statistical analysis, we found that eight host miRNAs were significantly modulated during ZIKV infection (p ≤ .05), mainly downregulated (Figure 1(c)–(j)). The miR-99* was downregulated in ZIKV-infected cells reaching values of Log Fold Change (LogFC) = −7.99. The miR-145 was the only one upregulated in ZIKV-infected cells (LogFC = 0.64). Up to 137 miRNAs were not detected in neuroblastoma cells.
Eleven miRNAs were exclusively expressed in either ZIKV-infected or noninfected cells (mock). These miRNAs were miR-148a, miR-342-5p, miR-598, and miR-708-3p expressed only in ZIKV-infected cells and miR-208, miR-329, miR-432-5p, miR-488, miR-518b, miR-520g, and miR-767-5p expressed only in noninfected cells (Table 1). We proceed with all these 19 miRNAs for further bioinformatics analysis.
Table 1.
Exclusively Expressed miRNAs on ZIKV-Infected or Noninfected (Mock) SH-SY5Y Cells at 72 hpi.
| Only in ZIKV-infected cells | Only in mock cells |
|---|---|
| miR-148a | miR-208 |
| miR-342-5p | miR-329 |
| miR-598 | miR-432-5p |
| miR-708-3p | miR-488 |
| miR-518b | |
| miR-520g | |
| miR-767-5p |
Note. ZIKV = Zika virus.
Target Prediction and GSEA Analysis on the Putative Targets of the Differentially Expressed miRNAs
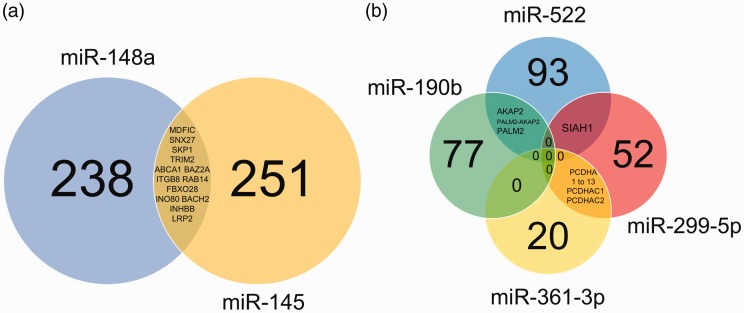
Cellular ontologies involved in ZIKV replication are poorly described. To evaluate them, we searched for target genes of all the exclusively/differentially expressed miRNAs modulated by ZIKV infection in six miRNA-target databases. Genes were selected as putative targets if they had been predicted in at least three of the six databases. Target prediction for the differentially expressed miRNAs resulted in 889 possible miRNA-target interactions that match our selection criteria. The results pointed out 502 potentially downregulated genes (577 transcripts) and 261 potentially upregulated genes (312 transcripts) assuming the classical mechanisms of miRNA posttranscriptional regulation (Figure 2, Supplemental Tables S1 and S2).
Figure 2.
Venn diagram of the putative gene targets predicted for up- and downregulated miRNAs in ZIKV infection. The circles represent the number of target genes for each miRNA that was predicted in at least three of the six software and databases investigated here. Genes that are predicted to be modulated by more than one miRNA are described in the intersections. (a) Venn diagram of upregulated/exclusively expressed miRNAs modulated by ZIKV (miR-145 and miR-148a). The complete list of the putative targets (including all the transcripts) is presented in Supplemental Table S1. (b) Venn diagram of downregulated miRNAs modulated by ZIKV (miR-190b, miR-299-5p, miR-361-3p, and miR-522). The complete list of the putative targets (including all the transcripts) is presented in Supplemental Table S2.
Selected Putative Targets From the ZIKV Upregulated miRNAs
Among the upregulated or exclusively expressed miRNAs (Supplemental Table S1 and Figure 2(a)), only miR-145 and miR-148a showed putative targets using the selected criteria before mentioned, presenting 577 putative targets, including all the transcripts. More specifically, these isoforms comprehend 251 genes exclusively targets of miR-145, 238 genes for miR-148 and 13 common targets for both miRNAs. Fundamental genes for the neuron system development could be found, such as ALCAM, LDB1, LOX, ERBB4, EFNA3, NRP1, ROBO2, XRN1, SIX4, and ZFHX4. Other two-neuron transcription factors include SOX9 and SOX11, members of the SRY-related HMG-box. The relevance of this gene family is due to its involvement in the regulation of embryonic development and in the determination of the cell commitment to neuron lineage (Xu et al., 2009).
Several of these possible downregulated genes can be targeted in two or more transcripts. These genes include important neuronal and neurogenic molecules such as ADCYAP1, CCKBR, CDH2, DYRK1A, INHBB, MPP5, MYO5A, NTN4, QKI, SRGAP1, SRGAP2, SRGAP2C, TRIM2, and ZNF423. Genes related to immune and antiviral responses were also found, such as BACH2, CD1D, FBXA11, and RNF216. Predicted cytoskeletal and motility-related genes include TMEM9B, ACTG1, ACTR3, CFL2, COL4A1, ELMO1, ITGA5, and STARD13.
Selected Putative Targets From the ZIKV Downregulated miRNAs
The possible upregulated targets (Supplemental Table S2 and Figure 2(b)) include 77 possible genes exclusively targeted by miR-190b, 52 by miR-299-5p, 20 by miR-361-3p, and 93 by miR-522. Several of these genes could be targeted in more than one transcript. A total of 15 common gene targets could be found between miR-361-3p and miR-299-5p, three between miR-190b and miR-522, and one between miR-299-5p and miR-522.
Again, we observed genes involved in CNS formation, including brain and eye development, such as CSPG5, DOCK7, FAM114A1, KAT6B, MELK, NEUROD1, NLGN1, PAX3, PAX6, ROBO1, STK38L, TCF4, and TSPAN2. Genes involved in neurophysiology also were observed, like ANO1, GLRA3, GPHN, KCNQ5, KCTD8, NAV3, SLC6A14, and SLC6A9. Remarkably, several genes related to calcium-dependent cell-adhesion protocadherins were found among the possible upregulated targets. A total of 46 transcripts of genes of the cadherin superfamily involved in neuronal connections in the brain seem to have their expression regulated by downregulated miRNAs, sometimes in a co-regulated way.
Not only the neurogenesis seems to be related with the predicted targets but also the whole organism development. Our analysis also showed genes involved with the Hippo/SWH signaling pathway (TEAD1) as well as genes required for the HSS/IHH signaling and regulation (KIF7). Targets involved in progression and control of the cell cycle are also modulated by the miRNAs including cyclin CCNJL, BACH2, PHLPP1, RASSF6, and TP53INP1.
Genes involved in antiviral and innate immune response could be also upregulated including DEFB4A, EMR3, FBXO4, FBXW2, HNRNPM, LAMP3, MAP3K8, and NFKBIZ. The CSDE1 gene found here, regulated by the miR-522, codes an RNA-binding protein that binds to DENV RNA in vitro and was also identified as an essential factor for viral replication, suggesting a canonical pathway modulated by flaviviruses (Ward et al., 2011).
Cellular Pathways Modulated by ZIKV Up- and Downregulated miRNAs
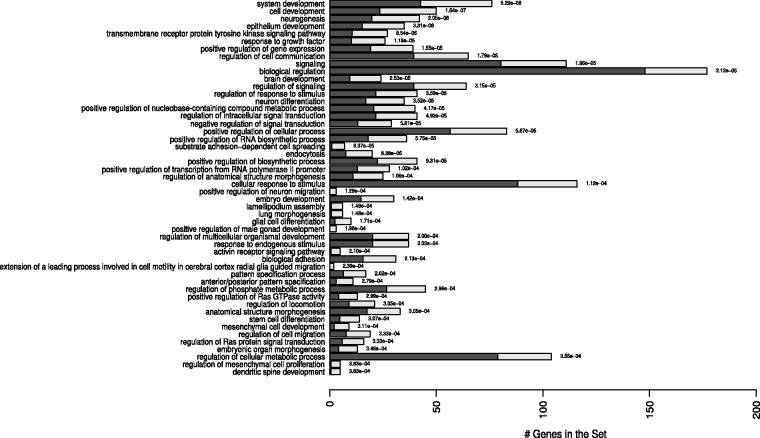
GSEA was performed on putative targets of both up- and downregulated miRNAs. Results revealed GOs enriched or overrepresented processes. Putative targets of upregulated miRNA in context of ZIKV infection, miR-145, were enriched in several biological processes (BPs) (Figure 3 and Supplemental Table S3). The same pattern was observed for the exclusively expressed in the ZIKV-infected cells, miR-148a (Supplemental Figure S1 and Supplemental Table S3).
Figure 3.
Graphical representation of the enriched biological processes (BPs) modulated by miR-145. Overrepresented pathways are depicted in the y-axis. The bars represent the expected (dark bars) versus the observed numbers of genes (gray light bars) for each BP enrichment with the statistical difference values alongside the bars. The overrepresentation was assessed with a statistical score based on hypergeometric tests with p ≤ .001. Complete list including the full names of the GO terms is presented in Supplemental Table S3.
Among them, it is possible to highlight very important BPs that could help to explain the malformations caused by ZIKV in the children, as system development (GO:0048731), neurogenesis (GO:0022008), brain development (GO:0007420), neuron differentiation (GO:0030182), regulation of anatomical structure morphogenesis (GO:0022603), positive regulation of neuron migration (GO:2001224), embryo development (GO:0009790), glial cell differentiation (GO:0010001), stem cell differentiation (GO:0048863), embryonic camera-type eye morphogenesis (GO:0048596), cell motility involved in cerebral cortex radial glia guided migration (GO:0021814), cell morphogenesis involved in neuron differentiation (GO:0048667), neuron projection morphogenesis (GO: 0048812), and cerebral cortex development (GO:0021987).
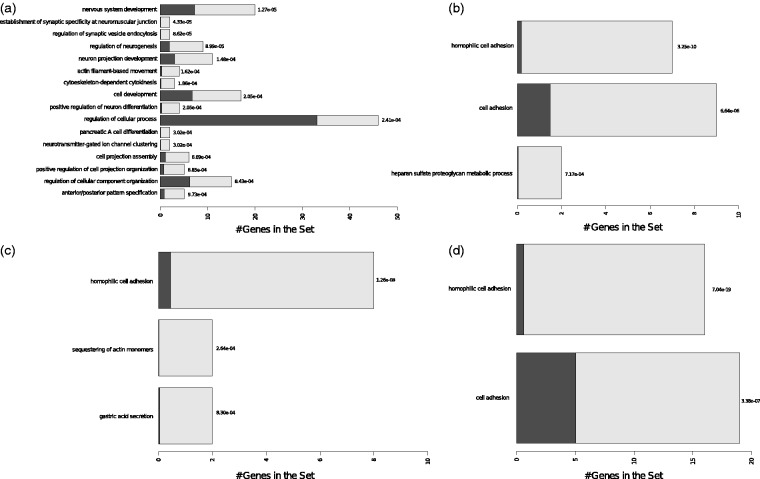
For the downregulated miRNAs, we observed BPs involved with neurogenesis, as neuron projection development (GO:0031175) and positive regulation of neuron differentiation (GO:0045666) for miR-190b. BPs related mainly to cell adhesion (GO:0007155) and homophilic cell adhesion (GO:0007156) were observed for the other three downregulated miRNAs (miR-299-5p, miR-361-3p, and miR-522) (Figure 4 and Supplemental Table S4).
Figure 4.
Graphical representation of the enriched biological processes (BPs) modulated by the downmodulated miRNAs. (a) BPs for miR-361-3p targets. (b) BPs for miR-190b targets. (c) BPs for miR-522 targets. (d) BPs for miR-299-5p targets. Overrepresented pathways are depicted in the y-axis. The bars represent the expected (dark bars) versus the observed numbers of genes (gray light bars) for each BP enrichment with the statistical difference values alongside the bars. The overrepresentation was assessed with a statistical score based on hypergeometric tests with p ≤ .001. Complete list including the full names of the GO terms is presented in Supplemental Table S4.
miR-145 and miR-148a Are Overexpressed in ZIKV-Infected Postmortem Brain Tissues
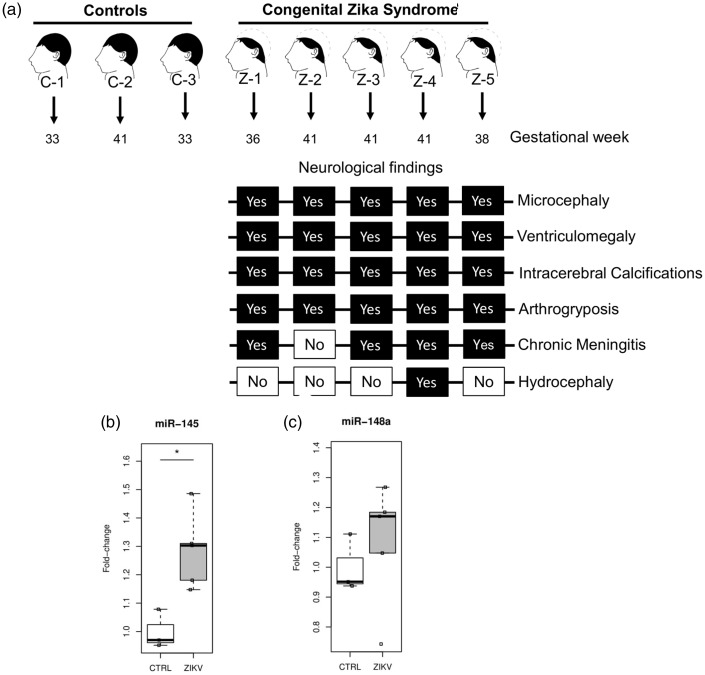
To validate the induction of miRNAs by ZIKV infection directly in brain tissues, we investigated the expression of miR-145 and miR-148a in postmortem brain autopsies of stillborn presenting CZS during pregnancy. We focused here on five neonates that had died in the first 48 h postpartum with severe microcephaly and intrauterine akinesia (arthrogryposis). ZIKV genome was detected in all cases during pregnancy by RT-PCR in clinical samples from mothers and the neonates such as urine, plasma, amniotic fluid, placenta, and umbilical cord. We also detected the virus genome through RT-PCR in fetal postmortem tissues (Figure 5(a) and Supplemental Table S5). Other microcephaly causes including congenital genetic diseases, infection with arboviruses that circulate in the same area (Dengue and Chikungunya) and teratogenic pathogens (STORCH) were all excluded (Supplemental Table S5). CZS was confirmed by USG or MRI, as well as postnatal physical examination. All the five cases presented classical CZS symptoms such as abnormalities in ventricular size (ventriculomegaly), gray and white matter volume loss, brainstem abnormalities, calcifications areas, and loss of cerebral gyrus (agyria or lissencephaly). We measured the expression of ZIKV induced miRNAs in brains of CZS compared with three brain controls samples from nonaffected children of the same gestational age (Figure 5(a) and Supplemental Table S5). We confirmed the upregulation of both miR-145 (FC = 1.28; p = .0065) and miR-148a (FC = 1.06; p = .57) in the postmortem brain tissues of children presenting CZS confirming the results obtained in the neuroblastoma cells (Figure 5(b) and (c)). Although we could not reach statistical significance for the upregulation of miR-148a, it is worth to note that four out the five ZIKV-infected children have expression levels of miR-148a above the highest level of this miRNA in noninfected children. This data corroborates the in vitro findings and suggests that these two miRNAs, especially miR-145 could potentially have a role during the development of congenital ZIKV syndrome.
Figure 5.
miR-145 and miR-148a are overexpressed in post mortem brain tissues from children with Congenital Zika syndrome. (a) Brief representative scheme of the stillborn enrolled in this study, including the gestational age and the neurological findings associated with CZS in each sample. (b) Upregulation of miR-145 in ZIKV-infected brain samples. (c) Upregulation of miR-148a in ZIKV-infected brain samples. Box plots representing relative mRNAs expression levels normalized by U6 snRNA and relative fold change over control samples. *p = .0065.
Discussion
In our work, a comprehensive screening was performed in order to understand the modulation of the miRNA profile induced by ZIKV infection. Among the modulated miRNAs during ZIKV infection, only one (miR-145) was found to be upregulated, while eight others were found to be downregulated in neuroblastoma-infected cells. A similar downregulation pattern was observed in the 11 miRNAs exclusively expressed in ZIKV-infected cells (4 upregulated against 7 downregulated). This global miRNA downregulation scenario promoted by flaviviruses infection has been previously described in mice brains infected with WNV (Kumar and Nerurkar, 2014). RNA virus replication generates subgenomic RNA (sfRNA) that suppress the RNAi machinery as a strategy to escape from cellular miRNAs that target the viral genome. This event was observed for other flaviviruses, such as DENV and Kunjin virus (Moon et al., 2015). Indeed, ZIKV was recently found to produce sfRNA (Akiyama et al., 2016; Donald et al., 2016). Moreover, previous reports showed the interaction of DICER1 transcripts and miR-148a and the downregulation of the RNAi pathway in human astrocytes and in liver, lung, and kidney cells infected with ZIKV (Ferreira et al., 2018; Kozak et al., 2017; Whisnant et al., 2013).
One of the downregulated miRNAs, miR-126* (or miR-126-5p), was also downregulated during DENV infection. Transfection of Huh-7 DENV-infected cells with a mimic of this miRNA increased the levels of miR-126-5p up to 3.41-fold and simultaneously reduced DENV RNA levels about 70% decreasing the virus infectivity (Kakumani et al., 2016). The same downregulation of its orthologs was found in the brain of mice infected with WNV (Kumar and Nerurkar, 2014). All these data suggest a role for miR-126-5p controlling flaviviruses replication in different models.
The relationship between ZIKV infection and development of neurological defects is now well accepted and established (Cugola et al., 2016; C. Li et al., 2016). Microcephaly is not the only neurological abnormality that can develop in ZIKV-exposed neonates during pregnancy. Several authors, including our group, reported a spectrum of changes, including other neurological and fetal development manifestations. Brain calcification is the most common feature of ZIKV-exposed babies. However, other observations include severe malformations such as reduction in cerebral volume, ventriculomegaly, cerebellar hypoplasia, the absence of the ridges or convolutions of the brain (lissencephaly) with hydrocephalus, and fetal akinesia deformation sequence (i.e., arthrogryposis) (Cavalheiro et al., 2016; Chimelli et al., 2017; Culjat et al., 2016; de Fatima Vasco Aragao et al., 2016; De Oliveira Melo et al., 2016; Driggers et al., 2016; Martines et al., 2016; Mlakar et al., 2016; Werner et al., 2016). All these abnormalities are associated with gene expression modulation involved in several cellular pathways including mitotic spindle orientation, premature chromosomal condensation, damaged DNA, deranged cytoskeleton and microtubule dynamics, transcriptional control, neuronal migration, or other mechanisms that regulate the number of neurons produced by neuronal precursor cells (Sansom et al., 2009). The balance between proliferative divisions, programmed cell death, and cellular migration are crucial during the earliest stages of forebrain development (Roberts and Frosch, 2016). Histopathological studies of ZIKV-infected brains showed common problems in neuronal migration and cell cycle division (De Oliveira Melo et al., 2016; Martines et al., 2016; Roberts and Frosch, 2016; Souza et al., 2016).
We hypothesize that ZIKV infection of the fetal brain modulates miRNAs involved in regulating microcephaly and forebrain development pathways leading to neurological disorders. We found genes related to microcephaly, lissencephaly, and other neuronal malformation listed as candidates of our miRNAs such as ACTG1, ACTB, CDK6, DAG1, LRP2, NEUROD1, PAX6, and TCF4 (Hussain et al., 2013; Romero et al., 2017; Sansom et al., 2009; Shalev et al., 2014). Most of them were predicted to be downregulated by miR-145, except by NEUROD1 and PAX6. We also observed the modulation of cellular miRNAs with targets in the cytoskeleton, adhesion, and extracellular matrix proteins, all of them being extremely important in the neuronal migration process. These proteins include the cadherin CDH2 (target of mir-145), protocadherins 1-13 (all targets of miR-299-5p and miR-361-3p), protocadherins gamma 1-12 (all targets of miR-522), actin beta and ARP3 actin-related protein (target of miR-145 and 190b), and integrin subunit beta 8 (target of miR-145). GO BPs related to cell migration and adhesion, especially neurons, were also found to be enriched in our analysis. Here, it is relevant to emphasize the possible modulation of the transcripts that give rise to the SLIT-ROBO Rho GTPase-Activating Proteins (SRGAP2 and SRGAP2C) that have fundamental roles during the development of the cerebral cortex, regulating neuron migration (Charrier et al., 2012). Altogether, these results reinforce our previous findings that ZIKV infection disturbs cell adhesion causing neuronal migration disturbances as evidenced by histopathological images of the same brains of stillborn reported here (Chimelli et al., 2017). We observed irregular clusters of immature neuron cells (for the gestational age) along the ventricular surface and toward the cortex, which could be explained by the downmodulation of cell-matrix adhesion proteins targeted by the miR-145 induced by the ZIKV infection.
Our findings confirm the induction of miR-145 by ZIKV in both models: neuroblastoma cells and brain tissues from stillborn with CZS. We also demonstrated that miR-148a was exclusively expressed in the ZIKV-infected neuroblastoma cells and, although its expression in the postmortem-infected samples did not reach statistical significance, it presented a tendency to be overexpressed. The role of both miR-145 and miR-148a as cellular migration inhibitors had already been explored in tumor cells. Indeed, miR-145 has been reported to affect cell migration, thus controlling invasion and metastasis, in a wide variety of tumors by targeting distinct cellular pathways (Gao et al., 2013; Sachdeva and Mo, 2010), and it is possible that miR-145 downregulation occurs early during tumorigenesis and it confers high metastatic potential to some types of aggressive tumors (Muti et al., 2014). Cellular migration assays using a trans-well approach demonstrated that ectopic expression of miR-145 reduces the migration ability of non-small cell lung carcinoma cells, while its depletion promotes an increase in this same feature (Donzelli et al., 2015). It was also found that restoration of miR-145 expression through demethylation of miR-145 locus also inhibits the migration of lung cancer cells (Donzelli et al., 2015). The same effect was observed for two distinct lineages of glioblastoma cells, U251 and U87, also measured using trans-well assay (Lee et al., 2013). Glioblastomas are well characterized by their ability to migrate and invade adjacent tissues, impairing tumor clearance and contributing to tumor recurrence (Giese et al., 2003; Lefranc et al., 2005). Transfection of miR-145 mimics significantly reduce cellular migration in 60% and 75%, respectively, for both lineages (U251 and U87) (Lee et al., 2013). As for miR-145, miR-148a has already been suggested as a tumor repressor and proved to impair tumor cell migration and invasion in gastric and breast cancer cells (Zhang et al., 2017; Zheng et al., 2011). Furthermore, miR-148a has already been proposed as a regulatory miRNA during neuronal development in a Zebrafish model. It was observed that knockdown of this miRNA compromised the development of the midbrain–hindbrain boundary and altered brain morphology (Hu et al., 2013). These findings allow us to trace a parallel between the effects of these two miRNAs in cellular migration potential in different cancer types and in ZIKV-infected neuron precursor cells, both in vitro and ex vivo models.
Previous work already applied some in silico strategy to study the role of the miRNAs during ZIKV infection. However, these studies applied different experimental strategies, models, and ZIKV strains (Azouz et al., 2019; Ferreira et al., 2018; Kozak et al., 2017; McLean et al., 2017; Zhang et al., 2018). In this work, the access to postmortem samples from CZS babies allowed to identify miRNAs modulated by ZIKV directly in human brain tissues. Perhaps, the most similar study was the evaluation of the miRNAs modulated by the expression of ZIKV E protein in primary cultures of human fetal neural stem cells (Bhagat et al., 2018). Here, we used the intact Brazilian ZIKV strain containing the whole genome. In common with our results, Bhagat et al. (2018) predicted the modulation of Wnt/β-catenin pathway as we found the prediction of WNT10B by miR-148a, VANGL1 by mir-299-5p, and TCF4 by miR-190b (Bhagat et al., 2018). Indeed, both groups found the BPs related to neurogenesis among the possible downregulated targets (Bhagat et al., 2018).
Overall, our results suggest that ZIKV may interfere in several biological pathways and processes by regulating cellular miRNA expression contributing to cause neuronal dysfunction and malformation.
Summary
We detected the microRNAs modulated in neuroblastoma cells infected with Zika virus. Their targets belong to pathways related to neurogenesis. We confirmed the upregulation of two miRNAs involved in cellular migration in brain samples from stillborn with congenital Zika syndrome.
Supplemental Material
Supplemental material, Supplemental Material1 for MicroRNAs 145 and 148a Are Upregulated During Congenital Zika Virus Infection by Fernanda L. Castro, Victor E. V. Geddes, Fábio L. L. Monteiro, Raphael M. D. T. Gonçalves, Loraine Campanati, Paula Pezzuto, Dominic Paquin-Proulx, Bruno L. Schamber-Reis, Girlene S. Azevedo, Alessandro L. Gonçalves, Daniela P. Cunha, Maria Elisabeth L. Moreira, Zilton F. M. Vasconcelos, Leila Chimeli, Adriana Melo Amilcar Tanuri, Douglas F. Nixon, Marcelo Ribeiro-Alves and Renato S. Aguiar in ASN Neuro
Supplemental Material
Supplemental material, Supplemental Material2 for MicroRNAs 145 and 148a Are Upregulated During Congenital Zika Virus Infection by Fernanda L. Castro, Victor E. V. Geddes, Fábio L. L. Monteiro, Raphael M. D. T. Gonçalves, Loraine Campanati, Paula Pezzuto, Dominic Paquin-Proulx, Bruno L. Schamber-Reis, Girlene S. Azevedo, Alessandro L. Gonçalves, Daniela P. Cunha, Maria Elisabeth L. Moreira, Zilton F. M. Vasconcelos, Leila Chimeli, Adriana Melo Amilcar Tanuri, Douglas F. Nixon, Marcelo Ribeiro-Alves and Renato S. Aguiar in ASN Neuro
Supplemental Material
Supplemental material, Supplemental Material3 for MicroRNAs 145 and 148a Are Upregulated During Congenital Zika Virus Infection by Fernanda L. Castro, Victor E. V. Geddes, Fábio L. L. Monteiro, Raphael M. D. T. Gonçalves, Loraine Campanati, Paula Pezzuto, Dominic Paquin-Proulx, Bruno L. Schamber-Reis, Girlene S. Azevedo, Alessandro L. Gonçalves, Daniela P. Cunha, Maria Elisabeth L. Moreira, Zilton F. M. Vasconcelos, Leila Chimeli, Adriana Melo Amilcar Tanuri, Douglas F. Nixon, Marcelo Ribeiro-Alves and Renato S. Aguiar in ASN Neuro
Supplemental Material
Supplemental material, Supplemental Material4 for MicroRNAs 145 and 148a Are Upregulated During Congenital Zika Virus Infection by Fernanda L. Castro, Victor E. V. Geddes, Fábio L. L. Monteiro, Raphael M. D. T. Gonçalves, Loraine Campanati, Paula Pezzuto, Dominic Paquin-Proulx, Bruno L. Schamber-Reis, Girlene S. Azevedo, Alessandro L. Gonçalves, Daniela P. Cunha, Maria Elisabeth L. Moreira, Zilton F. M. Vasconcelos, Leila Chimeli, Adriana Melo Amilcar Tanuri, Douglas F. Nixon, Marcelo Ribeiro-Alves and Renato S. Aguiar in ASN Neuro
Supplemental Material
Supplemental material, Supplemental Material5 for MicroRNAs 145 and 148a Are Upregulated During Congenital Zika Virus Infection by Fernanda L. Castro, Victor E. V. Geddes, Fábio L. L. Monteiro, Raphael M. D. T. Gonçalves, Loraine Campanati, Paula Pezzuto, Dominic Paquin-Proulx, Bruno L. Schamber-Reis, Girlene S. Azevedo, Alessandro L. Gonçalves, Daniela P. Cunha, Maria Elisabeth L. Moreira, Zilton F. M. Vasconcelos, Leila Chimeli, Adriana Melo Amilcar Tanuri, Douglas F. Nixon, Marcelo Ribeiro-Alves and Renato S. Aguiar in ASN Neuro
Supplemental Material
Supplemental material, Supplemental Material6 for MicroRNAs 145 and 148a Are Upregulated During Congenital Zika Virus Infection by Fernanda L. Castro, Victor E. V. Geddes, Fábio L. L. Monteiro, Raphael M. D. T. Gonçalves, Loraine Campanati, Paula Pezzuto, Dominic Paquin-Proulx, Bruno L. Schamber-Reis, Girlene S. Azevedo, Alessandro L. Gonçalves, Daniela P. Cunha, Maria Elisabeth L. Moreira, Zilton F. M. Vasconcelos, Leila Chimeli, Adriana Melo Amilcar Tanuri, Douglas F. Nixon, Marcelo Ribeiro-Alves and Renato S. Aguiar in ASN Neuro
Acknowledgments
The authors would like to thank to Biometrix (Dengue, Zika, and Chikungunya IgM and IgG ELISA) and Dia.Pro Diagnostic Bioprobes (STORCH IgM ELISA) for the donation of ELISA kits for this project.
Author Contributions
F. L. C., V. E. V. G., and R. S. A. contribute in the conceptualization of the study. The methodology was performed by F. L. C., V. E. V. G., F. L. L. M., R. M. D. T. G., P. P., B. L. S., G. S. A., and D. P. P. Formal data analysis were performed by F. L. C., M. R. R. S. A., A. T., and D. F. N. A. M., A. L. G., D. P. C., M. E. L. M., Z. F. M. V., and L. C. performed the clinical investigation and diagnosis. The data curation was made by F. L. C., M. R., A. T., R. S. A., and D. F. N. The writing—original draft preparation was made by F. L. C. and R. S. A. All authors reviewed and contributed to the final version of the manuscript. This study was supervised by R. S. A., who also administrated the project. The funding acquisition was obtained by R. S. A. and A. T. This manuscript is part of F. L. C.’s PhD thesis.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by National Council for Scientific and Technological Development (CNPq) grant 440900/2016-6, council for the Improvement of Higher Education (CAPES) grant 88881.130757/2016-01 and Fundação de Amparo a Pesquisa do Estado do Rio de Janeiro (FAPERJ) grant E-18/2015. F. L. C. and V. E. V. G. are funded by CAPES (grant 001).
Ethical Approval
Ethical approval was obtained from IPESQ and IFF covered by the institutional review board–approved study (52888616.4.0000.5693 and 52675616.0.000.5269, respectively).
References
- Akiyama B. M., Laurence H. M., Massey A. R., Constantino D. A., Xie X., Yang Y., Shi P. Y., Nix J. C., Beckham J. D., Kieft J. S. (2016). Zika virus produces noncoding RNAs using a multi-pseudoknot structure that confounds a cellular exonuclease. Science, 354, 1148–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexa A., Rahnenführer J., Lengauer T. (2006). Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics, 22, 1600–1607. [DOI] [PubMed] [Google Scholar]
- Ashraf U., Zhu B., Ye J., Wan S., Nie Y., Chen Z., Cui M., Wang C., Duan X., Zhang H., Chen H., Cao S. (2016). MicroRNA-19b-3p modulates Japanese encephalitis virus-mediated inflammation via targeting RNF11. J Virol, 90, 4780–4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azouz F., Arora K., Krause K., Nerurkar V. R., Kumar M. (2019). Integrated microRNA and mRNA profiling in Zika virus-infected neurons. Viruses, 11, E162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagat R., Prajapati B., Narwal S., Agnihotri N., Adlakha Y. K., Sen J., Mani S., Seth P. (2018). Zika virus E protein alters the properties of human fetal neural stem cells by modulating microRNA circuitry. Cell Death Differ, 25, 1837–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasil P., Pereira J. P., Raja Gabaglia C., Damasceno L., Wakimoto M., Ribeiro Nogueira R. M., Carvalho de Sequeira P., Machado Siqueira A., Abreu de Carvalho L. M., Cotrim da Cunha D., Calvet G. A., Neves E. S., Moreira M. E., Rodrigues Baião A. E., Nassar de Carvalho P. R., Janzen C., Valderramos S. G., Cherry J. D., Bispo de Filippis A. M., Nielsen-Saines K. (2016). Zika virus infection in pregnant women in Rio de Janeiro—Preliminary report. Obstet Gynecol Surv, 71, 331–333. [Google Scholar]
- Cavalheiro S., Lopez A., Serra S., Da Cunha A., da Costa M. D., Moron A., Lederman H. M. (2016). Microcephaly and Zika virus: Neonatal neuroradiological aspects. Childs Nerv Syst, 32, 1057–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier C., Joshi K., Coutinho-Budd J., Kim J. E., Lambert N., de Marchena J., Jin W. L., Vanderhaeghen P., Ghosh A., Sassa T., Polleux F. (2012). Inhibition of SRGAP2 function by its human-specific paralogs induces neoteny during spine maturation. Cell, 149, 923–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Ye J., Ashraf U., Li Y., Wei S., Wan S., Zohaib A., Song Y., Chen H., Cao S. (2016). miR-33a-5p modulates Japanese encephalitis virus replication by targeting eukaryotic translation elongation factor 1A1. J Virol, 90, 3722–3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimelli L., Melo A. S. O., Avvad-Portari E., Wiley C. A., Camacho A. H. S., Lopes V. S., Machado H. N., Andrade C. V., Dock D. C. A., Moreira M. E., Tovar-Moll F., Oliveira- Szejnfeld P. S., Carvalho A. C. G., Ugarte O. N., Batista A. G. M., Amorim M. M. R., Melo F. O., Ferreira T. A., Marinho J. R. L., Azevedo G. S., Leal J. I. B. F., da Costa R. F. M., Rehen S., Arruda M. B., Brindeiro R. M., Delvechio R., Aguiar R. S., Tanuri A. (2017). The spectrum of neuropathological changes associated with congenital Zika virus infection. Acta Neuropathol, 133, 983–999. [DOI] [PubMed] [Google Scholar]
- Cugola F. R., Fernandes I. R., Russo F. B., Freitas B. C., Dias J. L., Guimarães K. P., Benazzato C., Almeida N., Pignatari G. C., Romero S., Polonio C. M., Cunha I., Freitas C. L., Brandão W. N., Rossato C., Andrade D. G., Faria D. de P., Garcez A. T., Buchpigel C. A., Braconi C. T., Mendes E., Sall A. A., Zanotto P. M., Peron J. P., Muotri A. R., Beltrão-Braga P. C. (2016). The Brazilian Zika virus strain causes birth defects in experimental models. Nature, 534, 267–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culjat M., Darling S. E., Nerurkar V. R., Ching N., Kumar M., Min S. K., Wong R., Grant L., Melish M. E. (2016). Clinical and imaging findings in an infant with Zika embryopathy. Clin Infect Dis, 63, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Fatima Vasco Aragao M., van der Linden V., Brainer-Lima A. M., Coeli R. R., Rocha M. A., Sobral da Silva P., Durce Costa Gomes de Carvalho M., van der Linden A., Cesario de Holanda A., Valenca M. M. (2016). Clinical features and neuroimaging (CT and MRI) findings in presumed Zika virus related congenital infection and microcephaly: Retrospective case series study. BMJ, 353, i3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Oliveira Melo A. S., Aguiar R. S., Amorim M. M. R., Arruda M. B., Melo F. O., Ribeiro S. T., Batista A. G., Ferreira T., Dos Santos M. P., Sampaio V. V., Moura S. R., Rabello L. P., Gonzaga C. E., Malinger G., Ximenes R., de Oliveira-Szejnfeld P. S., Tovar-Moll F., Chimelli L., Silveira P. P., Delvechio R., Higa L., Campanati L., Nogueira R. M., Filippis A. M., Szejnfeld J., Voloch C. M., Ferreira O. C., Jr., Brindeiro R. M., Tanuri A. (2016). Congenital Zika virus infection: Beyond neonatal microcephaly. JAMA Neurol, 73, 1407–1416. [DOI] [PubMed] [Google Scholar]
- Dick G. W. A. (1952). Zika virus (I). Isolations and serological specificity. Trans R Soc Trop Med Hyg, 46, 509–520. [DOI] [PubMed] [Google Scholar]
- Donald C. L., Brennan B., Cumberworth S. L., Rezelj V. V., Clark J. J., Cordeiro M. T., Freitas de Oliveira França R., Pena L. J., Wilkie G. S., Da Silva Filipe A., Davis C., Hughes J., Varjak M., Selinger M., Zuvanov L., Owsianka A. M., Patel A. H., McLauchlan J., Lindenbach B. D., Fall G., Sall A. A., Biek R., Rehwinkel J., Schnettler E., Kohl A. (2016). Full genome sequence and sfRNA interferon antagonist activity of Zika virus from Recife, Brazil. PLoS Negl Trop Dis, 10, e0005048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donzelli S., Mori F., Bellissimo T., Sacconi A., Casini B., Frixa T., Roscilli G., Aurisicchio L., Facciolo F., Pompili A., Carosi M. A., Pescarmona E., Segatto O., Pond G., Muti P., Telera S., Strano S., Yarden Y., Blandino G. (2015). Epigenetic silencing of miR-145-5p contributes to brain metastasis. Oncotarget, 6, 35183–35201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driggers R. W., Ho C.-Y., Korhonen E. M., Kuivanen S., Jääskeläinen A. J., Smura T., Rosenberg A., Hill D. A., DeBiasi R. L., Vezina G., Timofeev J., Rodriguez F. J., Levanov L., Razak J., Iyengar P., Hennenfent A., Kennedy R., Lanciotti R., du Plessis A., Vapalahti O. (2016). Zika virus infection with prolonged maternal viremia and fetal brain abnormalities. N Engl J Med, 374, 2142–2151. [DOI] [PubMed] [Google Scholar]
- Ferreira R. N., Holanda G. M., Pinto Silva E. V., Casseb S. M. M., Melo K. F. L., Carvalho C. A. M., Lima J. A., Vasconcelos P. F. C., Cruz A. C. R. (2018). Zika virus alters the expression profile of microRNA-related genes in liver, lung, and kidney cell lineages. Viral Immunol, 31, 583–588. [DOI] [PubMed] [Google Scholar]
- Gao P., Xing A. Y., Zhou G. Y., Zhang T. G., Zhang J. P., Gao C., Li H., Shi D. B. (2013). The molecular mechanism of microRNA-145 to suppress invasion-metastasis cascade in gastric cancer. Oncogene, 32, 491–501. [DOI] [PubMed] [Google Scholar]
- Garcez P. P., Loiola E. C., Madeiro da Costa R., Higa L. M., Trindade P., Delvecchio R., Nascimento J. M., Brindeiro R., Tanuri A., Rehen S. K. (2016). Zika virus impairs growth in human neurospheres and brain organoids. Science, 352, 816–818. [DOI] [PubMed] [Google Scholar]
- Garcez P. P., Nascimento J. M., De Vasconcelos J. M., Madeiro da Costa R., Delvecchio R., Trindade P., Loiola E. C., Higa L. M., Cassoli J. S., Vitória G., Sequeira P. C., Sochacki J., Aguiar R. S., Fuzii H. T., de Filippis A. M., da Silva Gonçalves Vianez Júnior J. L., Tanuri A., Martins-de-Souza D., Rehen S. K. (2017). Zika virus disrupts molecular fingerprinting of human neurospheres. Sci Rep, 7, 40780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giese A., Bjerkvig R., Berens M. E., Westphal M. (2003). Cost of migration: Invasion of malignant gliomas and implications for treatment. J Clin Oncol, 21, 1624–1636. [DOI] [PubMed] [Google Scholar]
- Harris M. A., Clark J., Ireland A., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., Richter J., Rubin G. M., Blake J. A., Bult C., Dolan M., Drabkin H., Eppig J. T., Hill D. P., Ni L., Ringwald M., Balakrishnan R., Cherry J. M., Christie K. R., Costanzo M. C., Dwight S. S., Engel S., Fisk D. G., Hirschman J. E., Hong E. L., Nash R. S., Sethuraman A., Theesfeld C. L., Botstein D., Dolinski K., Feierbach B., Berardini T., Mundodi S., Rhee S. Y., Apweiler R., Barrell D., Camon E., Dimmer E., Lee V., Chisholm R., Gaudet P., Kibbe W., Kishore R., Schwarz E. M., Sternberg P., Gwinn M., Hannick L., Wortman J., Berriman M., Wood V., de la Cruz N., Tonellato P., Jaiswal P., Seigfried T., White R., & Gene Ontology Consortium. (2004). The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res, 32, D258–D261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C. W., Tseng C. W., Chien C. W., Huang H. C., Ku W. C., Lee S. J., Chen Y. J., Juan H. F. (2013). Quantitative proteomics reveals diverse roles of mir-148a from gastric cancer progression to neurological development. J Proteome Res, 12, 3993–4004. [DOI] [PubMed] [Google Scholar]
- Hussain M. S., Baig S. M., Neumann S., Peche V. S., Szczepanski S., Nürnberg G., Tariq M., Jameel M., Khan T. N., Fatima A., Malik N. A., Ahmad I., Altmüller J., Frommolt P., Thiele H., Höhne W., Yigit G., Wollnik B., Neubauer B. A., Nürnberg P., Noegel A. A. (2013). CDK6 associates with the centrosome during mitosis and is mutated in a large Pakistani family with primary microcephaly. Hum Mol Genet, 22, 5199–5214. [DOI] [PubMed] [Google Scholar]
- Hutvágner G., Zamore P. D. (2002). A microRNA in a multiple-turnover RNAi enzyme complex. Science, 297, 2056–2060. [DOI] [PubMed] [Google Scholar]
- Jopling C. L., Yi M., Lancaster A. M., Lemon S. M., Sarnow P. (2005). Molecular biology: Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science, 309, 1577–1581. [DOI] [PubMed] [Google Scholar]
- Kakumani P. K., Medigeshi G. R., Kaur I., Malhotra P., Mukherjee S. K., Bhatnagar R. K. (2016). Role of human GRP75 in miRNA mediated regulation of dengue virus replication. Gene, 586, 7–11. [DOI] [PubMed] [Google Scholar]
- Kozak R. A., Majer A., Biondi M. J., Medina S. J., Goneau L. W., Sajesh B. V., Slota J. A., Zubach V., Severini A., Safronetz D., Hiebert S. L., Beniac D. R., Booth T. F., Booth S. A., Kobinger G. P. (2017). MicroRNA and mRNA dysregulation in astrocytes infected with Zika virus. Viruses, 9, E297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Nerurkar V. R. (2014). Integrated analysis of microRNAs and their disease related targets in the brain of mice infected with West Nile virus. Virology, 452–453, 143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuno G., Chang G. J. J. (2007). Full-length sequencing and genomic characterization of Bagaza, Kedougou, and Zika viruses. Arch Virol, 152, 687–696. [DOI] [PubMed] [Google Scholar]
- Lanciotti R. S., Kosoy O. L., Laven J. J., Velez J. O., Lambert A. J., Johnson A. J., Stanfield S. M., Duffy M. R. (2008). Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis, 14, 1232–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. K., Bier A., Cazacu S., Finniss S., Xiang C., Twito H., Poisson L. M., Mikkelsen T., Slavin S., Jacoby E., Yalon M., Toren A., Rempel S. A., Brodie C. (2013). MicroRNA-145 is downregulated in glial tumors and regulates glioma cell migration by targeting connective tissue growth factor. PLoS One, 8, e54652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc F., Brotchi J., Kiss R. (2005). Possible future issues in the treatment of glioblastomas: Special emphasis on cell migration and the resistance of migrating glioblastoma cells to apoptosis. J Clin Oncol, 23, 2411–2422. [DOI] [PubMed] [Google Scholar]
- Li C., Xu D., Ye Q., Hong S., Jiang Y., Liu X., Zhang N., Shi L., Qin C. F., Xu Z. (2016). Zika virus disrupts neural progenitor development and leads to microcephaly in mice cell stem cell Zika virus disrupts neural progenitor development and leads to microcephaly in mice. Cell Stem Cell, 19, 120–126. [DOI] [PubMed] [Google Scholar]
- Li G., Xiang Y., Sabapathy K., Silverman R. H. (2004). An apoptotic signaling pathway in the interferon antiviral response mediated by RNase L and c-Jun NH2-terminal kinase. J Biol Chem, 279, 1123–1131. [DOI] [PubMed] [Google Scholar]
- MacNamara F. N. (1954). Zika virus: A report on three cases of human infection during an epidemic of jaundice in Nigeria. Trans R Soc Trop Med Hyg, 48, 139–145. [DOI] [PubMed] [Google Scholar]
- Martines R. B., Bhatnagar J., de Oliveira Ramos A. M., Davi H. P., Iglezias S. D., Kanamura C. T., Keating M. K., Hale G., Silva-Flannery L., Muehlenbachs A., Ritter J., Gary J., Rollin D., Goldsmith C. S., Reagan-Steiner S., Ermias Y., Suzuki T., Luz K. G., de Oliveira W. K., Lanciotti R., Lambert A., Shieh W. J., Zaki S. R. (2016). Pathology of congenital Zika syndrome in Brazil: A case series. Lancet, 388, P898–P904. [DOI] [PubMed] [Google Scholar]
- McLean E., Bhattarai R., Hughes B. W., Mahalingam K., Bagasra O. (2017). Computational identification of mutually homologous Zika virus miRNAs that target microcephaly genes. Libyan J Med, 12, 1304505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlakar J., Korva M., Tul N., Popović M, Poljšak-Prijatelj M., Mraz J., Kolenc M., Resman Rus K., Vesnaver Vipotnik T., Fabjan Vodušek V., Vizjak A., Pižem J., Petrovec M., Avšič Županc T. (2016). Zika virus associated with microcephaly. N Engl J Med, 374, 951–958. [DOI] [PubMed] [Google Scholar]
- Moon S. L., Dodd B. J. T., Brackney D. E., Wilusz C. J., Ebel G. D., Wilusz J. (2015). Flavivirus sfRNA suppresses antiviral RNA interference in cultured cells and mosquitoes and directly interacts with the RNAi machinery. Virology, 485, 322–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muti P., Sacconi A., Hossain A., Donzelli S., Ben, M.oshe N. B., Ganci F., Sieri S., Krogh V., Berrino F., Biagioni F., Strano S., Beyene J., Yarden Y., Blandino G. (2014). Downregulation of microRNAs 145-3p and 145-5p is a long-term predictor of postmenopausal breast cancer risk: The ORDET prospective study. Cancer Epidemiol Biomarkers Prev, 23, 2471–2481. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. (2016). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Ritz C., Spiess A. N. (2008). qpcR: An R package for sigmoidal model selection in quantitative real-time polymerase chain reaction analysis. Bioinformatics, 24, 1549–1551. [DOI] [PubMed] [Google Scholar]
- Roberts D. J., Frosch M. P. (2016). Zika and histopathology in first trimester infections. Lancet, 388, 847–849. [DOI] [PubMed] [Google Scholar]
- Romero D. M., Bahi-Buisson N., Francis F. (2017). Genetics and mechanisms leading to human cortical malformations. Semin Cell Dev Biol, 76, 33–75. [DOI] [PubMed] [Google Scholar]
- Rossi S. L., Tesh R. B., Azar S. R., Muruato A. E., Hanley K. A., Auguste A. J., Langsjoen R. M., Paessler S., Vasilakis N., Weaver S. C. (2016). Characterization of a novel murine model to study Zika virus. Am J Trop Med Hygiene, 94, 1362–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdeva M., Mo Y. Y. (2010). miR-145-mediated suppression of cell growth, invasion and metastasis. Am J Transl Res, 2, 170–180. [PMC free article] [PubMed] [Google Scholar]
- Sansom S. N., Griffiths D. S., Faedo A., Kleinjan D. J., Ruan Y., Smith J., van Heyningen V., Rubenstein J. L., Livesey F. J. (2009). The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genetics, 5, e1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalev S. A., Tenenbaum-Rakover Y., Horovitz Y., Paz V. P., Ye H., Carmody D., Highland H. M., Boerwinkle E., Hanis C. L., Muzny D. M., Gibbs R. A., Bell G. I., Philipson L. H., Greeley S. A. (2014). Microcephaly, epilepsy, and neonatal diabetes due to compound heterozygous mutations in IER3IP1: Insights into the natural history of a rare disorder. Pediatr Diabetes, 15, 252–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza B. S. F., Sampaio G. L. A., Pereira C. S., Campos G. S., Sardi S. I., Freitas L. A., Figueira C. P., Paredes B. D., Nonaka C. K., Azevedo C. M., Rocha V. P., Bandeira A. C., Mendez-Otero R., Dos Santos R. R., Soares M. B. (2016). Zika virus infection induces mitosis abnormalities and apoptotic cell death of human neural progenitor cells. Sci Rep, 6, 39775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Linden V., Filho E. L. R., Lins O. G., van der Linden A., Aragão M. F., Brainer-Lima A. M., Cruz D. D., Rocha M. A., Sobral da Silva P. F., Carvalho M. D., do Amaral F. J., Gomes J. A., Ribeiro de Medeiros I. C., Ventura C. V., Ramos R. C. (2016). Congenital Zika syndrome with arthrogryposis: Retrospective case series study. BMJ, 354, i3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol, 3, 34–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward A. M., Bidet K., Yinglin A., Ler S. G., Hogue K., Blackstock W., Gunaratne J., Garcia-Blanco M. A. (2011). Quantitative mass spectrometry of DENV-2 RNA-interacting proteins reveals that the DEAD-box RNA helicase DDX6 binds the DB1 and DB2 3’ UTR structures. RNA Biol, 8, 1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner H., Sodré D, Hygino C., Guedes B., Fazecas T., Nogueira R., Daltro P., Tonni G., Lopes J., Araujo Júnior E. (2016). First trimester intrauterine Zika virus infection and brain pathology: Prenatal and postnatal neuroimaging findings. Prenat Diagn, 36, 785–789. [DOI] [PubMed] [Google Scholar]
- Whisnant A. W., Bogerd H. P., Flores O., Ho P., Powers J. G., Sharova N., Stevenson M., Chen C. H., Cullen B. R. (2013). In-depth analysis of the interaction of HIV-1 with cellular microRNA biogenesis and effector mechanisms. MBio, 4, e000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N., Papagiannakopoulos T., Pan G., Thomson J. A., Kosik K. S. (2009). MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell, 137, 647–658. [DOI] [PubMed] [Google Scholar]
- Yekta S., Shih I. H., Bartel D. P. (2004). MicroRNA-directed cleavage of HOXB8 mRNA. Science, 304, 594–596. [DOI] [PubMed] [Google Scholar]
- Zhang H., Chang Y., Zhang L., Kim S. N., Otaegi G., Zhang Z., Nie Y., Mubarak T., Li C., Qin C. F., Xu Z., Sun T. (2018). Upregulation of microRNA miR-9 is associated with microcephaly and Zika virus infection in mice. Mol Neurobiol, 56, 4072–4085. [DOI] [PubMed] [Google Scholar]
- Zhang L., Xing M., Wang X., Cao W., Wang H. (2017). MiR-148a suppresses invasion and induces apoptosis of breast cancer cells by regulating USP4 and BIM expression. Int J Clin Exp Pathol, 10, 8361–8368. [PMC free article] [PubMed] [Google Scholar]
- Zheng B., Liang L., Wang C., Huang S., Cao X., Zha R., Liu L., Jia D., Tian Q., Wu J., Ye Y., Wang Q., Long Z., Zhou Y., Du C., He X., Shi Y. (2011). MicroRNA-148a suppresses tumor cell invasion and metastasis by downregulating ROCK1 in gastric cancer. Clin Cancer Res, 17, 7574–7583. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplemental Material1 for MicroRNAs 145 and 148a Are Upregulated During Congenital Zika Virus Infection by Fernanda L. Castro, Victor E. V. Geddes, Fábio L. L. Monteiro, Raphael M. D. T. Gonçalves, Loraine Campanati, Paula Pezzuto, Dominic Paquin-Proulx, Bruno L. Schamber-Reis, Girlene S. Azevedo, Alessandro L. Gonçalves, Daniela P. Cunha, Maria Elisabeth L. Moreira, Zilton F. M. Vasconcelos, Leila Chimeli, Adriana Melo Amilcar Tanuri, Douglas F. Nixon, Marcelo Ribeiro-Alves and Renato S. Aguiar in ASN Neuro
Supplemental material, Supplemental Material2 for MicroRNAs 145 and 148a Are Upregulated During Congenital Zika Virus Infection by Fernanda L. Castro, Victor E. V. Geddes, Fábio L. L. Monteiro, Raphael M. D. T. Gonçalves, Loraine Campanati, Paula Pezzuto, Dominic Paquin-Proulx, Bruno L. Schamber-Reis, Girlene S. Azevedo, Alessandro L. Gonçalves, Daniela P. Cunha, Maria Elisabeth L. Moreira, Zilton F. M. Vasconcelos, Leila Chimeli, Adriana Melo Amilcar Tanuri, Douglas F. Nixon, Marcelo Ribeiro-Alves and Renato S. Aguiar in ASN Neuro
Supplemental material, Supplemental Material3 for MicroRNAs 145 and 148a Are Upregulated During Congenital Zika Virus Infection by Fernanda L. Castro, Victor E. V. Geddes, Fábio L. L. Monteiro, Raphael M. D. T. Gonçalves, Loraine Campanati, Paula Pezzuto, Dominic Paquin-Proulx, Bruno L. Schamber-Reis, Girlene S. Azevedo, Alessandro L. Gonçalves, Daniela P. Cunha, Maria Elisabeth L. Moreira, Zilton F. M. Vasconcelos, Leila Chimeli, Adriana Melo Amilcar Tanuri, Douglas F. Nixon, Marcelo Ribeiro-Alves and Renato S. Aguiar in ASN Neuro
Supplemental material, Supplemental Material4 for MicroRNAs 145 and 148a Are Upregulated During Congenital Zika Virus Infection by Fernanda L. Castro, Victor E. V. Geddes, Fábio L. L. Monteiro, Raphael M. D. T. Gonçalves, Loraine Campanati, Paula Pezzuto, Dominic Paquin-Proulx, Bruno L. Schamber-Reis, Girlene S. Azevedo, Alessandro L. Gonçalves, Daniela P. Cunha, Maria Elisabeth L. Moreira, Zilton F. M. Vasconcelos, Leila Chimeli, Adriana Melo Amilcar Tanuri, Douglas F. Nixon, Marcelo Ribeiro-Alves and Renato S. Aguiar in ASN Neuro
Supplemental material, Supplemental Material5 for MicroRNAs 145 and 148a Are Upregulated During Congenital Zika Virus Infection by Fernanda L. Castro, Victor E. V. Geddes, Fábio L. L. Monteiro, Raphael M. D. T. Gonçalves, Loraine Campanati, Paula Pezzuto, Dominic Paquin-Proulx, Bruno L. Schamber-Reis, Girlene S. Azevedo, Alessandro L. Gonçalves, Daniela P. Cunha, Maria Elisabeth L. Moreira, Zilton F. M. Vasconcelos, Leila Chimeli, Adriana Melo Amilcar Tanuri, Douglas F. Nixon, Marcelo Ribeiro-Alves and Renato S. Aguiar in ASN Neuro
Supplemental material, Supplemental Material6 for MicroRNAs 145 and 148a Are Upregulated During Congenital Zika Virus Infection by Fernanda L. Castro, Victor E. V. Geddes, Fábio L. L. Monteiro, Raphael M. D. T. Gonçalves, Loraine Campanati, Paula Pezzuto, Dominic Paquin-Proulx, Bruno L. Schamber-Reis, Girlene S. Azevedo, Alessandro L. Gonçalves, Daniela P. Cunha, Maria Elisabeth L. Moreira, Zilton F. M. Vasconcelos, Leila Chimeli, Adriana Melo Amilcar Tanuri, Douglas F. Nixon, Marcelo Ribeiro-Alves and Renato S. Aguiar in ASN Neuro