Abstract
Owing to the availability of data of one biological phenomenon at different levels/scales, modelling of biological systems is moving from single level/scale to multiple levels/scales, which introduces a number of challenges. Coloured Petri nets (ColPNs) have been successfully applied to multilevel, multiscale and multidimensional modelling of some biological systems, addressing many of these challenges. In this article, we first review the basics of ColPNs and some popular extensions, and then their applications for multilevel, multiscale and multidimensional modelling of biological systems. This understanding of how to use ColPNs for modelling biological systems will assist readers in selecting appropriate ColPN classes for specific modelling circumstances.
Keywords: multilevel, multiscale and multidimensional modelling, coloured Petri nets, biological systems
Introduction
Systems biology [1, 2] studies the interactions between the components of a biological system and how the interactions produce the behaviour of that system. Mathematical and computational modelling plays a crucial role in achieving this goal. So far, a variety of modelling approaches, including Petri nets, Boolean networks and (ordinary or partial) differential equations, have been applied to a wide field of biological systems (see [3, 4] for reviews). Among them, Petri nets are particularly appropriate for describing and analysing the concurrent, asynchronous and dynamic behaviour of complex biological systems. Since Reddy et al. [5] introduced qualitative Petri nets to model metabolic pathways, different types of Petri nets [e.g. stochastic Petri nets (SPNs), timed Petri nets, continuous Petri nets (CPNs) and hybrid Petri nets] have been proposed for modelling biological systems [4, 6, 7]. However, as an unparameterized method, these standard Petri nets do not easily scale, and so they are usually applicable for representing smaller (biological) systems only.
In the past few years, because of the availability of data of one biological phenomenon at different levels/scales, modelling of biological systems has moved from single level/scale to multiple levels/scales [8]. Multilevel/multiscale modelling integrates information at different levels/scales into one model, which can more accurately describe a system and thus provide more insights into the system. Although ‘multi-level’ and ‘multiscale’ are often synonymously used, they are in fact distinct [9, 10]. In this article, we wish to distinguish them, but do not intend to provide a rigorous definition for them. Multilevel modelling considers dynamic processes at multiple levels (e.g. subcellular, cellular, tissue level) of biological systems, while multiscale modelling incorporates multiple different temporal and spatial scales in one model, regardless of whether the model has multiple levels. A multilevel model is not necessarily a multiscale model, and vice versa. However, multiple levels usually coincide with multiple spatial and temporal scales. Besides, apart from multilevel and multiscale aspects, a biological model could also be constructed as multidimensional [11]. For example, when studying reaction–diffusion processes, we can model this phenomenon in one-, two- or three-dimensional (shortly 1 D, 2 D or 3 D) space. The model involving more dimensions usually represents the system to be studied more accurately. Modelling beyond one level/scale introduces plenty of challenges, e.g. repetition of components (e.g. cells, tissues), (hierarchical) organization, communication or movement of components, differentiation, division or deletion of components or pattern formation of a biological system. To address these challenges, coloured Petri nets (ColPNs) have been used to construct multilevel, multiscale and multidimensional models, and gained increased popularity for a wide spectrum of applications [12, 13].
ColPNs [14, 15] are an extension of standard Petri nets, which were proposed to represent large complex systems. Using ColPNs, a group of similar components of a system can be represented as one component, each of which is encoded as a colour and thus distinguished by this colour. ColPNs offer parameterized and compact representations of complex systems, without losing the analysis capabilities of standard Petri nets thanks to automatic unfolding. Moreover, ColPNs provide the possibility to easily increase the size of a model consisting of many similar components just by adding new colours. ColPNs have been widely applied to modelling protocols and technical networks, software, workflows and business processes, hardware and manufacturing systems [16]. Recently, ColPNs have been used for modelling biological systems, e.g. in an early attempt, ColPNs were used for discriminating metabolites, which follow different T-invariants [17]. Later, a ColPN-based approach to multilevel/multiscale modelling of biological systems has been presented in [12], and some successful applications appeared, e.g. modelling multicellular systems [18] and spatial diffusion [11].
In summary, ColPNs have been proven to be appropriate to construct multilevel, multiscale and multidimensional models.
Multilevel modelling. The levels to be considered can be represented by the use of tuples within tuples. That is, each tuple encodes a level. For example, in the fly wing, we use a colour tuple (x, y) to represent the cell level, and another tuple (a, b) to represent each compartment of a cell. Thus, a nested tuple describes two levels of the fly wing model [19].
Multiscale modelling. Multiscale modelling is often accompanied with multilevelness. Thus, the encoding of multiscale models with colours is similar to that for multilevel modelling. The mapping functions between spatial scales can be implemented via media (auxiliary) nodes (places or transitions), which are then used by rate functions at different scales [20]. The mapping functions between temporal scales can be explicitly represented via hybrid Petri nets [21].
Multidimensional modelling. A multidimensional grid can be represented by the use of colour tuples, whose arity respects the number of dimensions: 1, 2 or 3. That is, a colour encodes a spatial locality of the grid in 1D, 2D or 3D space. For example, in a 2D grid, each grid cell can be defined as a colour tuple, e.g. (x, y), and the connectivity between cells can be defined as a neighbourhood function of colours [11]. Furthermore, tessellation of different shapes, e.g. hexagonal cells instead of rectangular cells, can also be easily defined [22].
In this article, we will review the basics and some extensions of ColPNs and also their applications for the modelling of biological systems in terms of the aforementioned three categories. We hope this review will open the door for a wide use of ColPNs in the systems biology area.
Coloured Petri nets
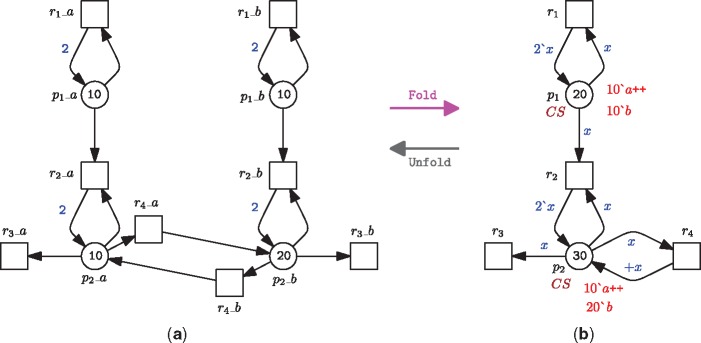
ColPNs offer a parameterized method for modelling a large system, where a group of similar components of the system is defined as and distinguished by a set of colours, thus presenting a compact representation of that system. For example, Figure 1B gives a ColPN by defining the left and right components (both components have the same structure) in Figure 1A as two colours.
Figure 1.
A ColPN example. (A) A prey–predator Petri net model with migration. (B) A ColPN model by folding the left and right components in (A). The declarations are as follows: CS=enumeration with a, b; variable x: CS. The successor operator ‘+’ in the arc expression returns the successor of x in an ordered finite colour set; if x is the last colour, then it returns the first colour. See [23] for the syntax of all declarations.
As standard Petri nets, ColPNs [12, 24] are directed bipartite multi-graphs and consist of places, transitions and arcs connecting places and transitions. In the biological scenario, places may represent any species or chemical compounds, such as genes, mRNAs, proteins, protein conformations or protein complexes, while transitions may represent chemical reactions (such as transcription and translation), molecular interactions or intramolecular changes. Additionally, a group of colour sets is defined for a ColPN. Each colour set is based on a data type, which is a set of values (colours) that obey some properties of a programming language [25]; common data types include integer, Boolean, string, enumeration and structure. Each place gets assigned a colour set and may contain distinguishable tokens, i.e. each token is associated with a specific colour. As there can be several tokens of the same colour on a given place, the tokens on the place are best described by a multiset over its colour set. A specific distribution of tokens on all places constitutes a marking of a ColPN. Each transition is associated with a guard, which is a Boolean expression over defined variables, constants and functions. The guard of a transition has to be evaluated to true for enabling the transition. The trivial guard ‘true’ is usually not explicitly given. Each arc gets assigned an expression; the result type of the expression is a multiset over the colour set of the connected place. In Table 1, we briefly compare properties of the elements in ColPNs and uncoloured Petri nets, taking the models in Figure 1 as an example.
Table 1.
A Comparison of properties of elements in ColPNs and uncoloured Petri nets
| Elements | ColPNs | Uncoloured Petri nets |
|---|---|---|
| Declaration | Colour sets, e.g. CS=enumeration with a, b | N/A |
| Variables, e.g. x: CS | N/A | |
| Place | A colour set, e.g. CS for p1 | N/A |
| Coloured tokens, e.g. 10`a++10`b on p1 | Black tokens, e.g. 10 on place | |
| Transition | A guard, e.g. ‘true’ for r1 | N/A |
| Arc | A multiset expression, e.g. 2`x on the arc (r1, p1) | A positive integer multiplicity, e.g. 2 on the arc |
| Marking | A vector of multiset expressions | A vector of non-negative integers |
Note: N/A: Not applicable.
Each colour of a place corresponds to a place instance when unfolded. Each transition is surrounded by a set of expressions, including its guard and the expressions on its adjacent arcs, which may involve a set of variables. Before the expressions are evaluated, the variables must be assigned values of suitable data types, which is called binding [24]. Each binding of a transition corresponds to a transition instance when unfolded. Enabling and firing of a transition instance are based on the evaluation of both its guard and related arc expressions. If the guard is evaluated to true and the preplaces have sufficient appropriately coloured tokens after the arc expressions were evaluated for a given binding, the transition instance that corresponds to the binding is enabled and may fire. When a transition instance fires, it removes appropriately coloured tokens from its preplaces and adds appropriately coloured tokens to its postplaces, i.e. it changes the current marking to a new reachable one. The colours of the tokens that are removed from preplaces and added to postplaces are decided by arc expressions. The set of markings reachable from the initial marking constitutes the state space of a given net. These reachable markings and transitions instances between them constitute the reachability graph of the net.
An uncoloured Petri net (Figure 1A) can be folded to a ColPN (Figure 1B), either manually or in a semi-automatic way [26]. Vice versa, a ColPN (Figure 1B) can be automatically unfolded to an uncoloured Petri nets (Figure 1A); afterwards all the simulation algorithms or analysis techniques for uncoloured Petri nets can be used for ColPNs [27].
Based on basic ColPNs, many extensions have been proposed for different purposes, e.g. arc extensions [coloured Petri nets with extended arcs (ColXPNs)], time extensions [coloured time and coloured stochastic Petri nets (ColSPNs)] and state space extensions [coloured continuous and coloured hybrid Petri nets (ColHPNs)] [28]. In the following, we briefly review the most important extensions, which have already been used or potentially could be used for the modelling of biological systems.
Coloured Petri nets with extended arcs
ColPNs have been extended to incorporate different special arc types such as read arcs (often also called test arcs), inhibitor arcs and reset arcs [12, 28]. These special arcs either make the model representation more compact while keeping the modelling power, or strictly extend the modelling power of the Petri net formalism. All these special arcs are only allowed to go from places to transitions. Read and inhibitor arcs add constraints on the firing of a transition, but the connected places are not affected on firing. A read arc allows to model that some resource (e.g. enzyme in a chemical reaction) is required, but not exclusively and it is not consumed on firing; hence, the same token could be used at the same time by more than one transition. An inhibitor arc reverses the logic of the enabling condition of a place, i.e. it imposes a constraint that a transition may only fire if the place contains less tokens than the weight that the arc indicates. A reset arc empties the place connected by this arc once the transition fires; the number of tokens on the place does not matter for enabling. Besides, ColPNs can be further enriched to include marking-dependent arcs, i.e. the arc multiplicities are allowed to be marking-dependent expressions of various types in terms of a transition’s preplaces [29], which facilitates the modelling of some special biological scenarios such as cell division [30, 31]. ColPNs and ColXPNs can be analysed using a variety of techniques, such as structural analysis (confined to models without special arcs extending the modelling power) [12] or state space analysis based on computational tree logic (CTL), which is a branching time temporal logic [32] matching the needs for analysing reachability graphs (model checking). See [12] for details on the use of these techniques for the analysis of ColPNs. ColPNs and ColXPNs have been widely used for modelling biological systems when kinetic data are not available.
Coloured timed Petri nets
There are many different types of coloured timed (or time) Petri nets (ColTPNs), but here we confine ourselves to the ColTPNs implemented in CPN tools [24], which have gained wider use in different fields. In a ColTPN, each token carries a second value called a time stamp (a non-negative integer) in addition to the token’s colour. The time stamp of a token tells us the time at which the token can be moved from its associated place. ColTPNs work in a similar way as event queues in many simulation engines of discrete event simulation. Using ColTPNs, performance measures of a system can be computed. In the biological area, early applications were usually done with ColPNs or ColTPNs supported by CPN tools or its predecessor Design/CPN [33]; see [34, 35].
Coloured stochastic Petri nets
ColSPNs are a coloured version of stochastic Petri nets (SPNs) [12]. A firing delay is introduced and associated with each transition, which is a random variable defined by an exponential probability distribution. Therefore, the semantics of a ColSPN is equivalent to a continuous time Markov chain (CTMC), which is constructed from the reachability graph of the underlying qualitative Petri net by labelling the arcs between states with the state transition rates. Thus, in addition to the analysis techniques given above, we can further use such quantitative analysis techniques as model checking continuous stochastic logic (CSL) [36], a probabilistic counterpart of CTL, or probabilistic linear-time temporal logic with numerical constraints (PLTLc) [37] for analysing ColSPNs. Besides, we can adopt a Gillespie stochastic simulation method [38] to simulate ColSPNs [28]. Nowadays, ColSPNs have been extended to incorporate all special arc types mentioned above, and also different kinds of transitions such as immediate, deterministic and scheduled transitions [12, 28]. These features make ColSPNs a powerful tool to model and analyse large-scale biological systems in a stochastic way.
Stochastic well-formed nets
Stochastic well-formed nets (SWNs) [39] were introduced by adding restrictions to general ColPNs to exploit symmetry properties to analyse large models. By restricting the syntax for the model definition, SWNs can be simulated or analysed more efficiently than general ColPNs with arbitrary and not well-formed colour functions. That is, SWNs obtain a gain in simulation and analysis speed at the cost of the loss in the modelling power and convenience, which, however, hinders the application of SWNs. Currently, SWNs are implemented in GreatSPN [40], and mainly used for modelling technical systems.
Coloured continuous Petri nets
Coloured continuous Petri nets (ColCPNs) are a coloured version of CPNs [12]. In ColCPNs, the discrete values on places are replaced with continuous real values, which describe the overall behaviour of species represented by places via concentrations. A deterministic rate is associated with each transition, which makes a CPN model representing a set of ordinary differential equations (ODEs). Contrary to discrete Petri nets, the state space of a ColCPN is continuous and linear, so we can analyse it using a linear temporal logic (LTL) [41], e.g. linear temporal logic with constraints (LTLc) in the manner of [42]. Of course, different ODE numerical algorithms can be used for simulating ColCPNs [28]. ColCPNs can be easily used to graphically model biological systems traditionally described by a set of ODEs, without having to write error-prone formulas, which should specifically be welcomed by biologists.
Coloured hybrid Petri nets
ColHPNs [43] represent a more recently introduced Petri net class, obtained by combining generalized hybrid Petri nets [30] with ColPNs. It supports both stochastic and deterministic processes in one model. The stochastic part can be explained as a CTMC, and the deterministic part as a set of ODEs. Efficient simulation algorithms for ColHPNs have been developed and implemented in Snoopy [28, 44]. ColHPNs are specifically appropriate to accommodate systems with multiple spatial and temporal scales. In [43], Herajy et al. illustrated the application of ColHPNs for modelling and analysing biological systems.
Moreover, a unifying ColPN framework has been developed and implemented in Snoopy, consisting of ColPNs, ColXPNs, ColSPNs, ColCPNs and ColHPNs [12, 28], which can be conveniently converted into each other. This allows us to investigate one and the same biological system with different modelling abstractions in various complementary ways. See [12, 28] for more details about the relationships among these net classes.
ColPN tools
There are a number of tools for ColPNs; Table 2 summarizes some popular ones and related modelling and analysis capabilities. Different tools vary a lot in the support of different net classes and their analysis techniques. For example, Snoopy supports many net classes, while CPN tools and GreatSPN only support one or two net classes. Moreover, both Snoopy and CPN tools support hierarchical modelling.
Table 2.
Selection of ColPN tools
| Name | Main functions (coloured-specific) | Website |
|---|---|---|
| Snoopy [28] | ColPN and ColXPN modelling, animation | http://www-dssz.informatik.tu-cottbus.de/DSSZ/Software/Snoopy |
| ColSPN modelling, animation, stochastic simulation | ||
| ColCPN modelling, deterministic simulation | ||
| ColHPN modelling, hybrid simulation | ||
| Hierarchical modelling | ||
| Export to external tools like Charlie and Marcie | ||
| CPN tools [24] | ColPN modelling, animation | http://cpntools.org/ |
| ColTPN modelling, simulation | ||
| Hierarchical modelling | ||
| State space analysis | ||
| GreatSPN [40] | SWN modelling | http://www.di.unito.it/greatspn/ |
| Coloured and symbolic reachability graph | ||
| Coloured and symbolic simulation |
As described above, ColPNs enjoy rich analysis techniques, which not only make use of all analysis capabilities of standard Petri nets by means of automatic unfolding but also have their own analysis techniques working directly on the coloured level. The animation is equipped by many tools, which enables us to experience the model behaviour by following the token flow, thus establishing initial confidence in the model. Further, simulation of each specific ColPN class (Table 2) strengthens this confidence by allowing us to investigate specific simulation traces. To gain deeper insights into the constructed models, formal analysis techniques and related tools, e.g. state space analysis in CPN tools, have to be adopted (see Table 3 for more details). For example, a ColPN model built in Snoopy can be automatically unfolded to an uncoloured Petri net and then fed into Charlie to obtain analysis results about its structural or behavioural properties. In a similar way, a ColSPN model can be subjected to CSL model checking by help of Marcie. For PLTLc model checking, we can export Snoopy’s simulation traces and feed them to MC2 to accomplish sophisticated analyses. A detailed description of the use of these analysis techniques for ColPNs can be found in [12, 45].
Table 3.
Some analysis tools of Petri nets, which can be used for coloured petri nets
| Name | Main functions | Website |
|---|---|---|
| Charlie [46] | Structural analysis of Petri nets | http://www-dssz.informatik.tu-cottbus.de/DSSZ/Software/Charlie |
| CTL/LTL model checking of Petri nets | ||
| Marcie [47] | CTL model checking of Petri nets | http://www-dssz.informatik.tu-cottbus.de/DSSZ/Software/Marcie |
| CSL model checking of Petri nets | ||
| PLTLc model checking of simulation traces | ||
| MC2 [37] | PLTLc model checking of simulation traces | http://www.brunel.ac.uk/__data/assets/file/0003/380820/MC2v2.0beta2.zip |
Early biological applications [34, 35, 48] of ColPNs and a few current ones [49] resort to Design/CPN or its successor CPN tools, which usually involve smaller models, and so the benefits of the application of ColPNs might not be always obvious. Moreover, neither tool was specifically designed with the requirements of systems biology in mind. Thus, they are not suitable in many aspects, e.g. they do not directly support stochastic or continuous modelling nor the simulative analysis of the models by stochastic or deterministic simulation. Keeping these drawbacks in mind, the ColPNs for systems biology were proposed and implemented in Snoopy, which are now widely used by biologists [12].
Applications
In this section, we will review applications of ColPNs for modelling biological systems in the aforementioned three categories.
Multilevel modelling
Multilevel modelling is still an open and challenging problem. A typical application of multilevel modelling is the multicellular pattern formation, which plays a central role in developmental biology [50]. Recently, ColPNs have been applied to address this issue and have shown promising results.
Liu et al. [51] used hierarchical ColPNs to build a multicellular model of Caenorhabditiselegans vulval development. Here, a colour set with six colours is defined, encoding six vulval precursor cells. They focused on illustrating the application of their ColPN framework for modelling and analysing complex biological systems. At first, the C. elegans vulval development issue is represented as a hierarchical ColXPN model, for which animation and structural analysis are done to assure the correctness of the model. Then, rate information is assigned to the ColXPN model, and thus the ColSPN and ColCPN models are obtained, depending on the interpretation of the rates (stochastic or deterministic). The ColSPN model is stochastically simulated and analysed by PLTLc model checking, while the ColCPN model is deterministically simulated and analysed by PLTLc model checking. Thus, a biological system can be explored from three different perspectives: qualitative, stochastic and continuous with this framework. Besides, they gave a detailed procedure for constructing a ColPN model for a biological system. This work is a good starting point for using ColPNs for the modelling and analysis of biological systems.
Gao et al. [18, 19, 52, 53] created a more complex multilevel model of the planar cell polarity in Drosophila wing with hierarchical ColSPNs and ColCPNs. The model considers an array of hexagonal epithelial cells in the wing tissue, and each cell is further divided into seven virtual compartments. This model uses a colour set of a two-layer hierarchy, which is implemented using two tuples. The first tuple represents the coordinates of a cell in the epithelial tissue, and the second tuple represents the position of a virtual compartment within that cell. The size of the model simulated in their paper [19] is a tissue of 400 cells, generating an underlying Petri net model of 164 000 places and 229 669 transitions. Such a huge model cannot be easily handled by traditional modelling approaches (if at all). Continuous and stochastic simulations together with numerical model checking are used for analysing the behaviour of the model. Note that this model illustrates many multilevel challenges such as repetition of cells, hierarchical organization of cells, communication between cells and pattern formation.
Carvalho et al. [54–56] created a multilevel ColPN model for reproducing the dynamics of the steps that are involved in the infection process and innate immune response. This model has a hierarchical structure with four different levels (molecular, intracellular, intercellular and the whole system). Three compound colour sets, Bacteria, Macrophage and Granuloma, are declared to represent the position and/or status of the individuals of Mycobacterium marinum bacteria, host macrophage immune cells and granuloma with a number of infected macrophages, respectively. Animation and stochastic simulation were used to analyse the model.
To facilitate the construction of ColPN models for complex biological systems, Liu et al. [57] presented two methods for automatically generating ColPN models for a given set of Petri net networks (solutions) that can be obtained be help of network reconstruction algorithms proposed in, e.g., [58]. In [26], Liu et al. describe a compositional ColPN approach to aid the automatic construction of multilevel models, where they defined several composition operations such as sequential, choice and synchronization operations. A ColPN model is obtained by iteratively applying these operations to the given components. Similarly, Gratie et al. [59, 60] proposed an algorithm for building ColPN models, given a reaction-based model with known compositional structure, which can be considered as a ColPN modelling method at the intracellular level. Besides, Pennisi et al. [61] gave a novel methodological approach mainly based on ColPNs, and illustrated their approach with the immune system response at the cellular level.
The applications summarized above illustrate that ColPNs are capable of multilevel modelling and thus have become a powerful tool for modelling and analysing multilevel systems. For example, [19] shows how a tissue with 400 cells can be easily modelled using ColPNs; however, [62] reports that it became extremely difficult to build a Petri net model with 60 cells.
Multiscale modelling
Models of multiple spatial scales can be constructed with any type of ColPNs. Moreover, ColHPNs offer a powerful tool for considering both multiple spatial and multiple temporal scales.
Liu et al. [20] used ColSPNs to model a large number of coupled Ca2+ channels at different spatial scales to explore the mechanisms of how coupled Ca2+ channels work in a stochastic way. They first constructed a ColSPN model for single clusters of coupled Ca2+ channels by arranging them in a regular or irregular lattice, each lattice site being a colour and representing a Ca2+ channel. They further created a ColSPN model for an array of clusters of coupled Ca2+ channels, where a tuple colour set is used to represent an array of clusters and another tuple colour set to represent the Ca2+ channels in each cluster. Here, a global place is used to count the number of activated channels (cluster scale), which is then used by the rate function of each channel (channel scale).
Heiner et al. [13] built a ColSPN model for the phase variation in bacterial colony growth. They represented a 3 D colony using a 2 D grid with a finite capacity on each grid position, and assumed an equal maximal height over the whole cell colony (i.e. all grid positions have the same capacity). The model highlights the following issues: multiple scales (from individual scale to colony scale), mobility of components and 2 D pattern formation. Further, Pârvu et al. [22, 63] explored the application of two different geometries, rectangular and circular, for modelling the phase variation patterning using ColSPNs, and developed techniques for analysing the properties of the patterns generated by phase variation. Besides, they developed a method to verify multiscale models with spatio-temporal model checking [64, 65].
Moreover, ColHPNs have been applied to the modelling of biological systems at multiple spatial and temporal scales. For example, Herajy et al. [43] demonstrated the application of ColHPNs using as example a synthetic circuit, the repressilator. Here, each gene is encoded as a colour. In the repressilator model, the protein part is considered as deterministic, but the gene part as stochastic. Although this model is simple, it illustrates an important issue, i.e. how to partition discrete and deterministic components of a biological system. To further illustrate the use of ColHPNs, Herajy et al. gave in [21] two more case studies, the bistable switch model based on multisite phosphorylation, and the dendritic spine model describing calcium dynamics. In the first model, the reactions guiding the perturbation of the system are modelled and simulated stochastically, and all others deterministically. In the second model, the diffusion reactions are treated deterministically, and the others stochastically. These works illustrate well the application of ColHPNs for modelling biological systems at multiple temporal scales. Besides, Herajy et al. [44] described all the hybrid simulation algorithms used for ColHPNs in Snoopy and illustrated the use of Snoopy to construct and analyse ColHPNs for biological systems. This article provides an overview of how the simulation of ColHPNs works.
Recently, whole-cell modelling has been presented, which is expected to have a great impact on systems biology [66]. Whole-cell modelling focuses on individual cells and thus needs to represent every known cellular and gene function in different compartments. However, so far, there are no well-established approaches for whole-cell modelling. Considering the features of whole-cell models, ColHPNs could be supportive for the systematic construction of whole-cell models at multiple spatial and temporal scales.
Multidimensional modelling
Some biological problems can be studied in different dimensional spaces. For example, reaction–diffusion is an important phenomenon in developmental processes, which can be considered in 1 D, 2 D or 3 D space. If the diffusion of species is fast, the involved biological processes can be seen as homogenous, and the spatial effects can be ignored; otherwise, the spatial distribution has to be taken into account [67]. Currently, widely used approaches include deterministic partial differential equations (PDEs) and spatial Gillespie’s stochastic simulation algorithm [68]. But ColPNs offer an alternative approach for the modelling of such systems with a user-friendly graphical interface.
Liu et al. [11] presented a ColPN approach for modelling and simulating reaction–diffusion systems. They divided the whole volume (the space that a biological system involves) into a number of subvolumes in 1 D, 2 D or 3 D space, and encoded each subvolume as a colour (coordinate). The diffusion among subvolumes is described as a corresponding neighbourhood function over colours, and finally a ColPN model at different dimensional space is obtained. Besides, a procedure for constructing a model for this type of systems is given with different net classes like ColSPNs, ColCPNs and ColHPNs. They also discussed how to map a set of PDEs to a ColCPN and illustrated their approach using the Brusselator example. This work shall serve as a tutorial for those who are interested in using ColPNs for modelling reaction–diffusion systems.
Besides, Blätke et al. [69, 70] briefly discussed the use of ColPNs to represent a 3 D lattice of space, which can be fitted with a topological 3 D model of an entire cell. In their approach, they simulate biochemical reactions in a reaction–diffusion system with the help of a localization component. Gilbert et al. [71] discussed in detail how to use ColPNs to encode 2 D space, which will be of much help to understand the coding mechanism of multidimensional modelling.
Discussion
An intuitive graphical representation with execution semantics is an invaluable asset in integrative research fields, typically involving professionals with a diverse background. However, while there are some approaches aiming at the support of modelling of biological systems at different levels or scales, to our knowledge none (besides Petri nets) come with a graphical representation. Of these, two approaches closely related to ColPNs are Chromar [72], which is based on multisets, and the rule-based approach described in [73].
ColPNs allow the modelling of large-scale systems in a parameterized and scalable way, and thus are capable of multilevel, multiscale and multidimensional modelling of biological systems. For each category, we first need to encode the biological components to be studied or the space, where a biological system evolves, as colours in a similar way. Thus, colours encode coordinates of components or spatial localities in 1 D, 2 D or 3 D grid or space; the connectivity between components is defined as a neighbourhood function over colours. After that, we obtain coloured models by applying the defined colours and colour functions. Considering their powerful representation capabilities, ColPNs have the power to become an ideal tool for multilevel, multiscale and multidimensional modelling of large-scale biological systems.
Moreover, a variety of ColPN extensions (from qualitative to stochastic, deterministic and hybrid) greatly expand the application ranges in the modelling and analysis of biological systems. Thus, the user can select the appropriate ColPN classes for specific modelling circumstances. Further, in Snoopy, different ColPN classes are integrated and can be conveniently converted into each other, which enables the investigation of a biological system using various complementary modelling abstractions. Besides, ColHPNs are likely to achieve more and more applications with the increasing interest in multilevel, multiscale and multidimensional modelling of biological systems.
Despite the potential and power of ColPNs for modelling large-scale systems, we have to acknowledge their limits and drawbacks, as they stand today. To improve the applicability of ColPNs, we think at least the following research directions need to be pursued.
Continue to improve the expressive power of ColPNs for addressing emerging issues in the systems biology area
To model biological systems characterized by highly diverse biological components, complex structures over levels, and interactions between components at the same level or across different levels, a more powerful and complex colour language is needed. However, this will make the analysis of the constructed models more difficult. To develop a more powerful colour language, but at the same time improve the efficiency of analytical techniques, we could adopt ideas from SWNs, and consider a constrained syntax that takes symmetry into account for colour expressions. It is unlikely that we could expect the whole model to be symmetric as they do in SWN models. Instead, a model could be divided into two parts, one obeying the rules of symmetric colours and the other not. We would expect that intra-level modelling belongs to the symmetric part because components at the same level usually share a similar structure, while inter-level relationships tend to exceed the expressive power of symmetric colour sets.
Moreover, there are some multiscale challenges such as compartment creation, division, merging or dissolving or cell differentiation, which cannot be well addressed by the current ColPNs with static colour sets. As discussed in [74], dynamic colour sets could be a good solution, but this issue needs further research.
Develop efficient analysis techniques
At present, to reuse existing analysis techniques of Petri nets, ColPNs have to be unfolded to uncoloured Petri nets. Thus, the unfolding efficiency heavily affects the efficiency of simulation or other analyses. Although we can improve the unfolding efficiency with a constraint satisfaction approach [27], parallel computing techniques [75] or interval decision diagrams [47], all three of which are supported by Snoopy, there is still room for improvement. We could exploit the solution at the coloured level, avoiding as much as possible the ‘brute force’ approach implied by the unfolding of the coloured model. This analysis can be done on the symmetric part of a model as described above. A group of biological components, each sharing exactly the same structure and distinguished by a colour, have similar or even the same structural or behavioural properties, such as P-invariants, T-invariants, reachability and boundedness [12]. In such a case, algorithms for structural analysis (as Kurt did in [76]) and model checking methods can be developed at the coloured level to perform an efficient analysis of large-scale models. SWNs represent a compromise between expressive power and computational complexity [39], and can be considered as a good basis for further research to improve the simulation efficiency of ColPNs without unfolding.
Strengthen statistical analysis of the simulation output
Although there are many analysis techniques for ColPNs, simulation is always the primary analysis technique used to evaluate multilevel and/or multiscale models. While simulation easily conveys the feeling that the model faithfully represents the real (biological) system, the statistical analysis of the simulation output needs to be improved and refined to deal (safely) with the huge state space characterizing the model; see for instance [77, 78] for statistical analysis techniques and according formal procedures for transit and steady-state analysis of simulation outputs.
Model checking based on PLTLc offers another way for analysing stochastic/deterministic/hybrid simulation traces [12, 37]. With PLTLc, we can analyse not only the properties of one simulation trace (time series of concentrations or reaction rates) but also the correlation or similarity of multiple traces. Thus, we can use PLTLc for analysing traces at one biological level or across different levels for a multilevel model. A general framework for model checking over multiple levels is described in [65].
Modelling over multiple scales
This is a challenging area because multiscale models by their very nature involve descriptions of systems, which are characterized by phenomena, which occur over different time and space scales and at different speeds. To integrate the components of such a model, some kind of interfacing mechanism must be used to map between the different time scales, often involving lumping parameters from lower scales to higher scales. This is exacerbated by the differing rates of observations that may be taken at the different scales, e.g. hourly at one scale as opposed to every millisecond at a lower scale, resulting in the need to interpolate data at higher scales. This results in difficulties in characterizing rate functions, and is especially problematic when modelling a transient system to predict transient behaviour. By their very nature, most biological systems are inherently multiscale, and hence, multiscale modelling of them is an important and challenging area for future research.
Acknowledgements
The authors would like to thank Mostafa Herajy, Christian Rohr and Martin Schwarick for their assistance in the related research on ColPNs. The authors also would like to thank the anonymous referees for their constructive comments.
Funding
Fei Liu has been supported by National Natural Science Foundation of China (grant number 61273226). Monika Heiner was partially funded by the Brunel University London, College of Engineering, Design and Physical Sciences via the SEED fund to promote multi-disciplinary research.
Biographies
Fei Liu is an Associate Professor at the School of Software Engineering, South China University of Technology. His research interests are modelling and simulation, Petri nets and systems biology.
Monika Heiner is a Professor at the Department of Computer Science, Brandenburg University of Technology Cottbus-Senftenberg. Her research interests include modelling and analysis of technical as well as biochemical networks using qualitative and quantitative Petri nets, model checking and simulation techniques.
David Gilbert is a Professor at the Department of Computer Science, Brunel University London. His research interests include Bioinformatics, Systems Biology, Synthetic Biology, multiscale modelling, model checking and computational methods for design of biological systems.
Key Points
Multilevel/multiscale modelling integrates information at different levels/scales into one model, which can more accurately describe a system and thus provide more insights into the system.
This article reviews applications of ColPNs for multilevel, multiscale and multidimensional modelling of biological systems.
This article discusses in-depth research directions that need to be pursued to improve the applicability of ColPNs.
This will assist readers in selecting appropriate ColPN classes for specific modelling circumstances.
References
- 1. Kitano H. Systems biology: a brief overview. Science 2002;295(5560):1662–4. 10.1126/science.1069492 [DOI] [PubMed] [Google Scholar]
- 2. Aderem A. Systems biology: its practice and challenges. Cell 2005;121(4):511–13. 10.1016/j.cell.2005.04.020 [DOI] [PubMed] [Google Scholar]
- 3. Heath AP, Kavraki LE.. Computational challenges in systems biology. Comput Sci Rev 2009;3(1):1–17. 10.1016/j.cosrev.2009.01.002 [DOI] [Google Scholar]
- 4. Baldan P, Cocco N, Marin A, et al. Petri nets for modelling metabolic pathways: a survey. Nat Comput 2010;9(4):955–89. 10.1007/s11047-010-9180-6 [DOI] [Google Scholar]
- 5. Reddy VN, Mavrovouniotis ML, Liebman MN.. Petri net representations in metabolic pathways In: Proceedings of the 1st International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Palo Alto, CA, 1993, 328–36. [PubMed] [Google Scholar]
- 6. Gilbert D, Heiner M. From Petri nets to differential equations – an integrative approach for biochemical network analysis. In: Proceedings of the 27th International Conference on Applications and Theory of Petri Nets and Other Models of Concurrency, Vol. 4024 of LNCS Springer, Berlin Heidelberg, 2006, 181–200.
- 7. Chaouiya C. Petri net modelling of biological networks. Brief Bioinform 2007;8(4):210–19. 10.1093/bib/bbm029 [DOI] [PubMed] [Google Scholar]
- 8. Dallon JC. Multiscale modeling of cellular systems in biology. Curr Opin Colloid Interface Sci 2010;15(1):24–31. 10.1016/j.cocis.2009.05.007 [DOI] [Google Scholar]
- 9. Uhrmacher AM, Degenring D, Zeigler B.. Discrete event multi-level models for systems biology In: Priami C. (ed). Transactions on Computational Systems Biology I, Vol. 3380 of LNCS. Springer, Berlin Heidelberg, 2005, 66–89. [Google Scholar]
- 10. Maus C. Toward accessible multilevel modeling in systems biology. PhD thesis, Department of Computer Science, University of Rostock, 2013.
- 11. Liu F, Blätke MA, Heiner M, et al. Modelling and simulating reaction-diffusion systems using coloured Petri nets. Comput Biol Med 2014;53:297–308. [DOI] [PubMed] [Google Scholar]
- 12. Liu F. Colored Petri nets for systems biology. PhD thesis, Department of Computer Science, Brandenburg University of Technology, 2012.
- 13. Heiner M, Gilbert D.. Biomodel engineering for multiscale systems biology. Prog Biophys Mol Biol 2013;111(2–3):119–28. [DOI] [PubMed] [Google Scholar]
- 14. Genrich HJ, Lautenbach K.. System modelling with high-level Petri nets. Theor Comput Sci 1981;13(1):109–35. 10.1016/0304-3975(81)90113-4 [DOI] [Google Scholar]
- 15. Jensen K. Coloured Petri nets and the invariant-method. Theor Comput Sci 1981;14(3):317–36. 10.1016/0304-3975(81)90049-9 [DOI] [Google Scholar]
- 16. Jensen K, Kristensen LM.. Coloured Petri Nets. Berlin Heidelberg: Springer-Verlag, 2009. [Google Scholar]
- 17. Heiner M, Koch I, Voss K. Analysis and simulation of steady states in metabolic pathways with Petri nets. In: Proceedings of the 3rd Workshop and Tutorial on Practical Use of Coloured Petri Nets and the CPN Tools. University of Aarhus, Aarhus, Denmark, 2001, 15–34.
- 18. Gao Q, Liu F, Gilbert D, et al. A multiscale approach to modelling planar cell polarity in Drosophila wing using hierarchically coloured Petri nets. In: Proceedings of the 9th International Conference on Computational Methods in Systems Biology, CMSB ’11. ACM, New York, NY, 2011, 209–18.
- 19. Gao Q, Gilbert D, Heiner M, et al. Multiscale modelling and analysis of planar cell polarity in the Drosophila Wing. IEEE/ACM Trans Comput Biol Bioinform 2013;10(2):337–51. 10.1109/TCBB.2012.101 [DOI] [PubMed] [Google Scholar]
- 20. Liu F, Heiner M.. Multiscale modelling of coupled Ca2+ channels using coloured stochastic Petri nets. IET Syst Biol 2013;7(4):106–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Herajy M, Liu F, Rohr C, et al. Coloured hybrid Petri nets: an adaptable modelling approach of multiscale biological networks. Comput Biol Chem 2017. Submitted. [DOI] [PubMed] [Google Scholar]
- 22. Pârvu O, Gilbert D, Heiner M, et al. Spatial-temporal modelling and analysis of bacterial colonies with phase variable genes. ACM Trans Model Comput Simul 2015;25(2):25. [Google Scholar]
- 23. Liu F, Heiner M, Rohr C. Manual for colored Petri nets in snoopy. Technical Report 02-12, Department of Computer Science, Brandenburg University of Technology Cottbus, 2012.
- 24. Jensen K, Kristensen LM, Wells L.. Coloured Petri nets and CPN tools for modelling and validation of concurrent systems. Int J Soft Tools Technol Transf 2007;9(3):213–54. [Google Scholar]
- 25. Cardelli L, Wegner P.. On understanding types, data abstraction, and polymorphism. ACM Comput Surv 1985;17(4):471–523. 10.1145/6041.6042 [DOI] [Google Scholar]
- 26. Liu F, Yang M.. Compositional colored Petri net approach to multiscale modeling for systems biology. Int J Model Simul Sci Comput 2014;05(4):1450017 10.1142/S1793962314500172 [DOI] [Google Scholar]
- 27. Liu F, Heiner M, Yang M.. An efficient method for unfolding colored Petri nets In: Proceedings of the 2012 Winter Simulation Conference. IEEE, Piscataway, NJ, 2012, 1–12. [Google Scholar]
- 28. Heiner M, Herajy M, Liu F, et al. Snoopy—a unifying Petri net tool. In: Proceedings of the 33rd International Conference on Application and Theory of Petri Nets and Concurrency, Vol. 7347 of LNCS Springer, Berlin Heidelberg, 2012, 398–407.
- 29. Ciardo G. Petri nets with marking-dependent arc cardinality: properties and analysis. In: Valette R (ed). Proceedings of the Application and Theory of Petri Nets 1994, Vol. 815 of LNCS Springer, Berlin Heidelberg, 1994, 179–98.
- 30. Herajy M, Heiner M.. Hybrid representation and simulation of stiff biochemical networks. Nonlinear Anal Hybrid Syst 2012;6(4):942–59. 10.1016/j.nahs.2012.05.004 [DOI] [Google Scholar]
- 31. Herajy M, Liu F, Heiner M.. Efficient modelling of yeast cell cycles based on multisite phosphorylation using coloured hybrid Petri nets with marking-dependent arc weights. Nonlinear Anal Hybrid Syst 2017;27:191–212. [Google Scholar]
- 32. Clarke EM, Grumberg O, Peled DA.. Model Checking. Cambridge: MIT Press, 2001. [Google Scholar]
- 33. Christensen S, Jørgensen JB, Kristensen LM. Design/CPN - a computer tool for coloured Petri nets. In: Proceedings of the 3rd International Workshop on Tools and Algorithms for Construction and Analysis of Systems, Vol. 1217 of LNCS Springer, Berlin Heidelberg, 1997, 209–23.
- 34. Täubner C, Mathiak B, Kupfer A, et al. Modelling and simulation of the TLR4 pathway with coloured Petri nets In: Proceedings of the 2006 International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE, Piscataway, NJ, 2006, 2009–12. [DOI] [PubMed] [Google Scholar]
- 35. Voss K, Heiner M, Koch I.. Steady state analysis of metabolic pathways using Petri nets. In Silico Biol 2003;3(3):367–87. [PubMed] [Google Scholar]
- 36. Kwiatkowska M. Model checking for probability and time: from theory to practice In: Proceedings of the 18th Annual IEEE Symposium of Logic in Computer Science. IEEE, Piscataway, NJ, 2003, 351–60. [Google Scholar]
- 37. Donaldson R, Gilbert D. A model checking approach to the parameter estimation of biochemical pathways. In: Heiner M, Uhrmacher AM (eds). Proceedings of the 6th International Conference on Computational Methods in Systems Biology, Vol. 5307 of LNCS. Springer, Berlin Heidelberg, 2008, 269–87.
- 38. Gillespie DT. Exact stochastic simulation of coupled chemical reactions. J Phys Chem 1977;81(25):2340–61. 10.1021/j100540a008 [DOI] [Google Scholar]
- 39. Chiola G, Dutheillet C, Franceschinis G, et al. Stochastic well-formed colored nets and symmetric modeling applications. IEEE Trans Comput 1993;42(11):1343–60. 10.1109/12.247838 [DOI] [Google Scholar]
- 40. Babar J, Beccuti M, Donatelli S, et al. GreatSPN enhanced with decision diagram data structures. In: Lilius J, Penczek W (eds). Proceedings of the 31st International Conference on Applications and Theory of Petri Nets, Vol. 6128 of LNCS Springer, Berlin Heidelberg, 2010, 308–17.
- 41. Pnueli A. The temporal logic of programs. In: Proceedings of the 18th Annual Symposium on Foundations of Computer Science, SFCS ’77. IEEE Computer Society, Washington, DC, 1977, 46–57.
- 42. Calzone L, Chabrier-Rivier N, Fages F, et al. Machine learning biochemical networks from temporal logic properties In: Priami C, Plotkin G (eds). Transactions on Computational Systems Biology VI, Vol. 4420 of LNCS. Springer, Berlin Heidelberg, 2006, 68–94. [Google Scholar]
- 43. Herajy M, Liu F, Rohr C. Coloured hybrid Petri nets for systems biology. In: Proceedings of the 5th International Workshop on Biological Processes & Petri Nets, satellite event of PETRI NETS 2014, Vol. 1159 of CEUR Workshop Proceedings. CEUR-WS.org, Aachen, 2014, 60–76.
- 44. Herajy M, Liu F, Rohr C, et al. Snoopy‘s hybrid simulator: a tool to construct and simulate hybrid biological models. BMC Syst Biol 2017;11(1):71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu F, Heiner M, Yang M.. Colored Petri nets for multiscale systems biology - current modeling and analysis capabilities in Snoopy In: Proceedings of the 7th International Conference on Systems Biology (ISB). IEEE, Piscataway, NJ, 2013, 24–30. [Google Scholar]
- 46. Heiner M, Schwarick M, Wegener JT.. Charlie-an extensible Petri net analysis tool In: Devillers R, Valmari A (eds). Proceedings of the International Conference on Applications and Theory of Petri Nets and Concurrency, Vol. 9115 of LNCS Springer, Berlin Heidelberg, 2015, 200–11. [Google Scholar]
- 47. Heiner M, Rohr C, Schwarick M.. MARCIE – model checking and reachability analysis done efficiently In: Colom JM, Desel J (eds). Proceedings of the International Conference on Applications and Theory of Petri Nets and Concurrency, Vol. 7927 of LNCS Springer, Berlin Heidelberg, 2013, 389–99. [Google Scholar]
- 48. LEE D, ZIMMER R, LEE S, et al. Colored Petri net modeling and simulation of signal transduction pathways. Metab Eng 2006;8(2):112–22. 10.1016/j.ymben.2005.10.001 [DOI] [PubMed] [Google Scholar]
- 49. Nickaeen N, Moein S, Heidary Z, et al. Colored Petri net modeling of small interfering RNA–mediated messenger RNA degradation. Adv Biomed Res 2016;5:100 10.4103/2277-9175.183660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bonzanni N, Krepska E, Feenstra KA, et al. Executing multicellular differentiation: quantitative predictive modelling of C. elegans vulval development. Bioinformatics 2009;25(16):2049–56. [DOI] [PubMed] [Google Scholar]
- 51. Liu F, Heiner M, Yang M.. Modeling and analyzing biological systems using colored hierarchical Petri nets, illustrated by C. elegans vulval development. WSPC J Biol Syst 2014;22(3):463–93. [Google Scholar]
- 52. Gao Q, Liu F, Gilbert D, et al. Multi-cell modelling using coloured Petri nets applied to planar cell polarity. In: Proceedings of the 2nd International Workshop on Biological Processes & Petri Nets, satellite event of PETRI NETS 2011, Vol. 724 of CEUR Workshop Proceedings. CEUR-WS.org, Aachen, 2011, 135–50.
- 53. Gao Q. A systems biology approach to multi-scale modelling and analysis of planar cell polarity in Drosophila Melanogaster Wing. PhD thesis, Department of Computer Science, Brunel University London, 2013.
- 54. Carvalho RV, Kleijn J, Meijer AH, et al. Modeling innate immune response to early mycobacterium infection. Comput Math Methods Med 2012; 2012:790482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Carvalho RV. Computational modeling of mycobacterium infection and innate immune reponse in zebrafish. PhD thesis, Leiden Institute of Advanced Computer Science (LIACS), Faculty of Science, Leiden University, 2015.
- 56. Carvalho RV, Heuvel J, Kleijn J, et al. Coupling of Petri net models of the mycobacterial infection process and innate immune response. Computation 2015;3(2):150–76. 10.3390/computation3020150 [DOI] [Google Scholar]
- 57. Liu F, Heiner M, Yang M.. Representing network reconstruction solutions with colored Petri nets. Neurocomputing 2016;174:483–93. 10.1016/j.neucom.2015.04.112 [DOI] [Google Scholar]
- 58. Wolfgang M, Wagler A, Weismantel R.. A mathematical approach to solve the network reconstruction problem. Math Methods Oper Res 2008;67(1):117–32. 10.1007/s00186-007-0178-5 [DOI] [Google Scholar]
- 59. Gratie DE, Gratie C.. Composition colored Petri nets for the refinement of reaction-based models. Electron Notes Theor Comput Sci 2016;326:51–72. 10.1016/j.entcs.2016.09.018 [DOI] [Google Scholar]
- 60. Diana-Elena G. Refinement of biomodels using Petri nets. PhD thesis, Faculty of Science and Engineering, Åbo Akademi University, 2016.
- 61. Pennisi M, Cavalieri S, Motta S, et al. A methodological approach for using high-level Petri nets to model the immune system response. BMC Bioinformatics 2016;17(S19):498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Matsuno H, Murakami R, Yamane R, et al. Boundary formation by notch signaling in Drosophila multicellular systems: experimental observations and gene network modeling by genomic object net In: Pacific Symposium on Biocomputing. Singapore: World Scientific Press, 2003, 152–63. [PubMed] [Google Scholar]
- 63. Pârvu O, Gilbert D, Heiner M, et al. Modelling and analysis of phase variation in bacterial colony growth. In: Gupta A, Henzinger TA (eds). Proceedings of the 11th International Conference on Computational Methods in Systems Biology, Vol. 8130 of LNCS. Springer, Berlin Heidelberg, 2013, 78–91.
- 64. Pârvu O, Gilbert D.. Automatic validation of computational models using pseudo-3D spatio-temporal model checking. BMC Syst Biol 2014;8(1):124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pârvu O, Gilbert D.. A novel method to verify multilevel computational models of biological systems using multiscale spatio-temporal meta model checking. PLoS One 2016;11(5):e0154847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Karr JR, Takahashi K, Funahashi A.. The principles of whole-cell modeling. Curr Opin Microbiol 2015;27:18–24. 10.1016/j.mib.2015.06.004 [DOI] [PubMed] [Google Scholar]
- 67. Elf J, Doncic A, Ehrenberg M.. Mesoscopic reaction-diffusion in intracellular signaling In: Proceedings of the 1st Symposium on Fluctuations and Noise in Biological, Biophysical, and Biomedical Systems, Vol. 5110 of SPIE, SPIE, Bellingham, WA, USA; 2003, 114–24. [Google Scholar]
- 68. Takahashi K, Arjunan SNV, Tomita M.. Space in systems biology of signaling pathways-towards intracellular molecular crowding in silico. FEBS Lett 2005;579(8):1783–8. 10.1016/j.febslet.2005.01.072 [DOI] [PubMed] [Google Scholar]
- 69. Blätke MA, Dittrich A, Rohr C, et al. JAK/STAT signalling - an executable model assembled from molecule-centred modules demonstrating a module-oriented database concept for systems and synthetic biology. Mol Biosyst 2013;9(6):1290–307. [DOI] [PubMed] [Google Scholar]
- 70. Blätke MA. BIOMODELKIT—a framework for modular biomodel engineering. PhD thesis, Faculty for Natural Sciences, Otto-von-Guericke University Magdeburg, 2017.
- 71. Gilbert D, Heiner M, Liu F, et al. Colouring space—a coloured framework for spatial modelling in systems biology. In: Colom JM, Desel J (eds). Proceedings of the 34th international conference on Application and Theory of Petri Nets and Concurrency, PETRI NETS 2013, Vol. 7927 of LNCS Springer, Berlin Heidelberg, 2013, 230–49.
- 72. Honorato-Zimmer R, Millar AJ, Plotkin GD, et al. Chromar, a language of parameterised objects. Theoretical Computer Science 2017. doi: 10.1016/j.tcs.2017.07.034. [Google Scholar]
- 73. Maus C, Rybacki S, Uhrmacher AM.. Rule-based multi-level modeling of cell biological systems. BMC Syst Biol 2011;5(1):166 10.1186/1752-0509-5-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Liu F, Heiner M.. Modeling membrane systems using colored stochastic Petri nets. Nat Comput 2013;12(4):617–29. 10.1007/s11047-013-9367-8 [DOI] [Google Scholar]
- 75. Grama A, Gupta A, Karypis G, et al. Introduction to Parallel Computing. Essex, UK: Addison Wesley, 2003. [Google Scholar]
- 76. Jensen K. How to find invariants for coloured Petri nets. In: Proceedings of the International Symposium on Mathematical Foundations of Computer Science, Vol. 118 of LNCS Springer, Berlin Heidelberg, 1981, 327–38.
- 77. Currie CSM, Cheng RCH.. A practical introduction to analysis of simulation output data In: Proceedings of the 2016 Winter Simulation Conference. IEEE, Piscataway, NJ, 2016, 118–32. [Google Scholar]
- 78. Law AM. Statistical analysis of simulation output data: the practical state of the art In: Proceedings of the 2015 Winter Simulation Conference. IEEE, Piscataway, NJ, 2015, 1810–19. [Google Scholar]