Abstract
Long reads obtained from third-generation sequencing platforms can help overcome the long-standing challenge of the de novo assembly of sequences for the genomic analysis of non-model eukaryotic organisms. Numerous long-read-aided de novo assemblies have been published recently, which exhibited superior quality of the assembled genomes in comparison with those achieved using earlier second-generation sequencing technologies. Evaluating assemblies is important in guiding the appropriate choice for specific research needs. In this study, we evaluated 10 long-read assemblers using a variety of metrics on Pacific Biosciences (PacBio) data sets from different taxonomic categories with considerable differences in genome size. The results allowed us to narrow down the list to a few assemblers that can be effectively applied to eukaryotic assembly projects. Moreover, we highlight how best to use limited genomic resources for effectively evaluating the genome assemblies of non-model organisms.
Keywords: de novo assembly, third-generation sequencing, single-molecule sequencing, PacBio SMRT, assembly evaluation
Introduction
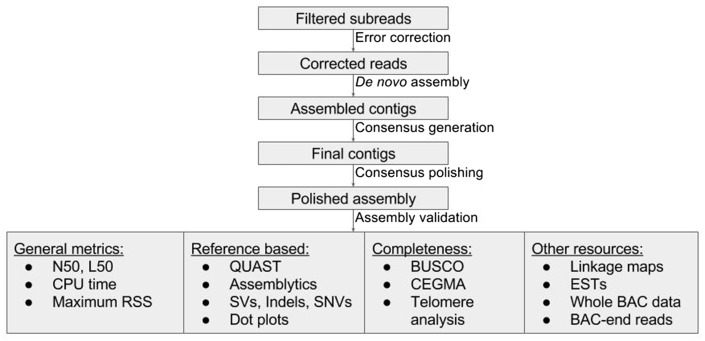
Pacific Biosciences (PacBio) single-molecule real-time (SMRT) and Oxford Nanopore sequencing technologies are the two widely used third-generation, single-molecule sequencing (SMS) technologies, which can generate average read lengths of several thousand base pairs. SMRT sequencing technology suffers from high error rates reaching up to 15% [1]; however, as these errors are random, high-quality error-corrected consensus sequences can be generated with sufficient coverage. Application of SMRT sequencing to eukaryotic genomes [2–18] has already demonstrated the obvious advantages provided by long reads in de novo assembly, such as higher contiguity, lesser gaps and fewer errors. The assembled contigs of recently assembled plant and animal genomes can be routinely seen to achieve an N50 of 1 Mb using SMS data. Hence, a significant rise in the number of genomes sequenced using SMS technologies is imminent, raising the need for evaluation of the available long-read assemblers. Large-scale evaluation studies such as GAGE [19], GAGE-B [20], Assemblathon1 [21] and Assemblathon2 [22] have been attempted with short-read assemblers, providing conclusions that serve as a useful guide for the de novo assembly of a given target organism. Although such evaluations have also been attempted for SMS data, these studies were either focused on bacterial and smaller eukaryotic genomes [23, 24] or were not sufficiently comprehensive to cover all of the available non-hybrid long-read assemblers [25–27], while others are already outdated because of continuous improvements in the technology [28, 29]. Also genome size was found to correlate with contiguity in long-read assemblies [17]; hence, diverse genome sizes can help differentiate the effect of the assemblers on each data set. In this study, we attempted to comprehensively evaluate three important features—contiguity, completeness and correctness [1]—of long-read assemblers (Table 1), using SMRT data of a bacterium (Escherichia coli, ∼5 Mb), protist (Plasmodium falciparum, ∼23 Mb), nematode (Caenorhabditis elegans, ∼105 Mb) and plant (Ipomoea nil, ∼750 Mb). We also designed a pipeline (Figure 1) for assembling the data and evaluating the results of different assemblers, which can be applied to both model organisms as well as to non-model organisms with limited genomic resources.
Table 1.
Summarized statistics of the assemblies
| Organism | #Contigs | Assembly size (Mb) | Longest contig (Mb) | N50 (Mb) | L50 | CPU time (hours) | Maximum RSS (GB) |
|---|---|---|---|---|---|---|---|
| Escherichia coli (4.6 Mb) | |||||||
| Maximum | 1 | 4.7 | 4.7 | 4.7 | 1 | 83.9 | 44.5 |
| Minimum | 1 | 4.6 | 4.6 | 4.6 | 1 | 2.2 | 3.6 |
| Mean | 1 | 4.7 | 4.7 | 4.7 | 1 | 19.4 | 15.7 |
| Plasmodium falciparum (23 Mb) | |||||||
| Maximum | 43 | 23.8 | 3.3 | 1.7 | 7 | 2012.6 | 43.9 |
| Minimum | 15 | 23.1 | 2.1 | 1.3 | 5 | 20.1 | 4.5 |
| Mean | 26.3 | 23.4 | 2.9 | 1.5 | 6.1 | 441.7 | 22.7 |
| Caenorhabditis elegans (105 Mb) | |||||||
| Maximum | 452 | 106.9 | 7.1 | 3.7 | 38 | 6733.8 | 251.7 |
| Minimum | 68 | 101.9 | 2.7 | 0.8 | 11 | 13.4 | 10.1 |
| Mean | 166.7 | 104.2 | 5.1 | 2.2 | 19.4 | 1221.4 | 56.9 |
| Ipomoea nil (750 Mb) | |||||||
| Maximum | 8751 | 752.7 | 11.5 | 1.8 | 1194 | 28 504.7 | 331.2 |
| Minimum | 1697 | 642 | 2.5 | 0.1 | 104 | 129.7 | 16.2 |
| Mean | 4288 | 702.7 | 6.2 | 0.7 | 439.4 | 10 065.8 | 78.2 |
Note: L50 and N50 represent the number of contigs and the length of the contig, respectively, crossing 50% mark of the assembly. Higher N50 and lower L50 values indicate highly contiguous assemblies. Max RSS represents the peak memory usage of the computational node.
Figure 1.
Evaluation pipeline.
Materials and methods
Long-read assembly pipelines
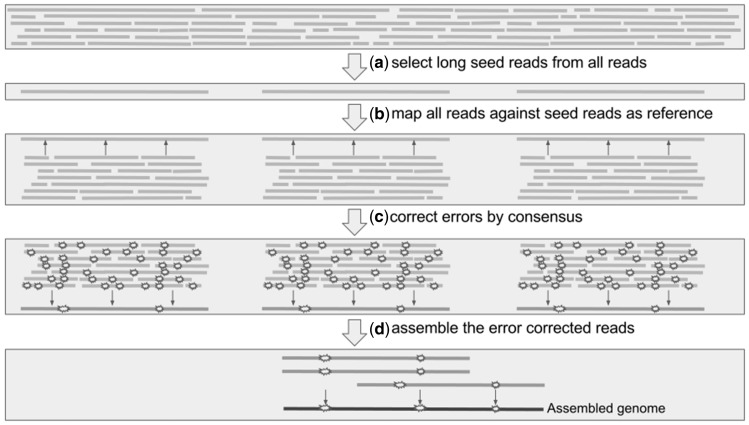
Overlap layout consensus (OLC) approach, de Bruijn graphs and string graphs are the commonly used algorithms for de novo assembly [30–33]. The advent of SMS data introduced a new challenge in de novo assembly because of the high error rates. Hence, application of de Bruijn graphs was rendered unfeasible [34], bringing back the OLC approach along with the string graphs to higher prominence. The longer the reads, the more efficient the assembly using the OLC approach, resulting in a linear increase in contiguity [35]. Although second-generation sequencing (SGS) reads were initially used for correcting long reads [36], most of the current long-read OLC pipelines follow a hierarchical approach (Figure 2), exclusively using SMS data as follows: (a) select a subset of longer reads as seed data; (b) use shorter reads to align against the longer seed data as reference, and correct sequencing errors by consensus of the aligned reads; (c) use the error-corrected reads for a draft assembly; and (d) obtain a polished consensus of the draft assembly [36, 37]. The procedure to identify overlaps has been the key difference in most long-read assemblers, and some of the overlap detection methods have been evaluated previously [38]. The long-read assemblers assessed in the present work are briefly summarized below.
Figure 2.
Hierarchical pipeline for OLC assembly approaches. Errors are displayed in Step C, which become reduced in number in the corrected reads. After assembly, a consensus polishing step, which is not shown in the figure, will also be performed as part of the hierarchical pipeline.
Hierarchical Genome Assembly Process
Hierarchical Genome Assembly Process (HGAP) [36] was one of the first hierarchical pipelines to exclusively use SMS reads for assembling a genome. Higher-quality preassembled reads with around 25–30× coverage are generated by aligning shorter reads against longer seed reads. The preassembled reads are then fed to the celera assembler [39] to obtain a draft assembly, followed by applying a consensus polishing procedure called quiver. BLASR [40] is used for aligning candidate overlaps, which are identified using an Ferragina–Manzini (FM)-index search and clustering of k-mer hits. The slower BLASR-based pipeline was replaced by FALCON in the latest version (v4). To distinguish between HGAP v3 and v4, the version used in the present evaluation is referred to as HGAP3.
PBcR
PBcR [41] also follows the hierarchical approach using MinHash Alignment Process (MHAP) for overlap detection. To identify k-mers shared between overlapping reads, without performing any alignments, k-mers of query reads are converted to integer fingerprints using multiple hash functions. The minimum values from the multiple hash functions are used to create a set called as MinHash sketch, for each read. MHAP then calculates the Jaccard similarity index by comparing the sketches of query reads to identify overlap candidates. Like HGAP3, the assembly of the corrected reads is performed using the celera assembler.
Canu
Canu [25] is a fork of the celera assembler and improves on the earlier PBcR pipeline into a single, comprehensive assembler. Highly repetitive k-mers, which are abundant in all the reads, can be non-informative. Hence, term frequency, inverse document frequency (tf-idf), a weighting statistic was added to MinHashing, giving weightage to non-repetitive k-mers as minimum values in the MinHash sketches, and sensitivity has been demonstrated to reach up to 89% without any parameter adjustment. By retrospectively inspecting the assembly graphs and also statistically filtering out repeat-induced overlaps, the chances of mis-assemblies are reduced.
FALCON
FALCON [42] is a hierarchical, haplotype-aware genome assembly tool. The sequence data are split into blocks for comparison using daligner [43]. Daligner first compiles a list of k-mers, along with their read identifiers and read coordinates, and then sorts them lexicographically. Identical k-mers from each block are merged into a new list containing both the query identifiers and their coordinates. A second sorting procedure, accounting for the query coordinates, places neighboring matches adjacent to each other, resulting in the identification of overlap candidates. A directed string graph is created from the alignment of the overlaps, with a collapsed diploid-aware layout, while maintaining the heterozygosity information.
HINGE
HINGE [34] is one of the few assemblers not requiring an error-correction step. Dalinger is used for overlap detection. The key innovation of this assembler is the placement of hinges to mark repeat regions that are not spanned by longer reads. Repeats are identified using the coverage gradients of the alignments, and an in-hinge and an out-hinge are marked on the reads, which are on the boundaries of unbridged repeats. Only two reads per repeat region, which have the longest overlap within the repeat, are chosen for placing the hinges. When a repeat is spanned by a completely bridged read, the other overlapping reads are marked as poisoned and not considered for hinge placing, thereby separating bridged repeats. Hinge-aided greedy graphs are used to resolve repeat junctions before obtaining a consensus.
Miniasm
Miniasm [37] was the first long-read assembler to not use error correction and hence is fast. Minimap is used for overlap detection, which indexes subsampled k-mers (minimizers [44]) from all the reads in a hash table, against which the query minimizers are then compared. The matches are sorted and clustered to find the longest collinear matching chains to identify overlap candidates. An assembly graph layout is subsequently constructed from the collinear matches and output as the assembled contigs, without building any consensus. Because error-correction and consensus procedures are not executed, the error rate of the final assembly is equivalent to that of the raw reads. To circumvent this, Racon [26], a consensus module, was shown to generate high-quality contigs within reasonable run times and is included in the present study as part of the miniasm pipeline.
SMARTdenovo
SMARTdenovo (https://github.com/ruanjue/smartdenovo) is another fast assembler, which can also work without error correction of the raw reads. Similar to minimap, SMARTdenovo searches subsampled query k-mers in indexed hash tables, which are then sorted and merged into collinear matches. Alignment using a dot-matrix alignment method is performed for adjacent matches, and the overlap candidates are subsequently input to a string graph layout. The consensus module can reach an accuracy of up to 99.7%, albeit taking up much of the entire computational time.
ABruijn
A de Bruijn graph [45] is a directed graph that is generally constructed from k−1 overlaps of adjacent k-mers. Rather, a set of solid strings (frequent k-mers), instead of all k-mers, is used to construct the ABruijn graphs because of the high error rates in SMS reads. A fast dynamic programming approach is used to find the longest common subpaths to obtain a rough estimate of the overlaps between two reads. Overlapping read vertices are added onto the graph, and the draft assembly is subsequently constructed. After aligning reads against the draft assembly, ABruijn graphs are constructed again to obtain a polished consensus assembly.
Wtdbg
Wtdbg (https://github.com/ruanjue/wtdbg) is another assembler that uses the framework of de Bruijn graphs. Unlike ABruijn graphs, overlapping k-mer hits are identified among the reads using a sorting approach similar to that adopted in minimap and SMARTdenovo, and the hits are used to add on and construct the fuzzy de Bruijn graphs. The resulting graphs, in comparison with ABruijn graphs, have reduced complexity and thereby consume lesser memory.
Mapping, Error Correction and de novo Assembly Tool
Mapping, Error Correction and de novo Assembly Tool (MECAT) [27] scans for identical k-mers, in blocks of sequences among query reads, to calculate distance difference factor (DDF) between neighboring k-mer hits. When the DDF is within a specified threshold, scores are assigned to the blocks of k-mers and extended to neighboring blocks. With the scoring mechanism, a large number of irrelevant read overlap candidates are filtered out, significantly reducing the computational time before alignment. After error correction, the corrected reads are pairwise-aligned and fed into a modified canu pipeline to construct contigs.
Data sets for evaluation
The evaluation data sets were broadly chosen in such a way that (i) data are available for public use, and (ii) genomes are of diverse sizes.
Initially, the standard bacterial model organism E.coli was chosen, and the sequence data (1 SMRT cell: ∼140× coverage) of P6-C4 chemistry (Supplementary Figure S1A) were downloaded from the PacBio DevNet website (https://github.com/PacificBiosciences/DevNet/wiki/Datasets).
Plasmodium falciparum (protist) is one of the few smaller eukaryotic genomes with long-read data available. Although the genome is only ∼23 Mb in length, it contains 14 chromosomes with a relatively high repeat content of 51.8% and a high AT% of 80.6% [46]. Plasmodiumfalciparum sequence data (9 SMRT cells: ∼180× coverage) of P6-C4 chemistry (Supplementary Figure S1B) were downloaded from the National Center for Biotechnology Information’s Sequence Read Archive (SRA360189) [47].
In contrast to P. falciparum, C.elegans (nematode) has a genome size of ∼105 Mb, but with only six, although much longer, chromosomes. The genome is also estimated to contain ∼20 000 genes making it more complex when compared with those of E. coli and P. falciparum, which have only ∼5000 genes each. There are also relatively fewer transposons (∼12%), although they are sufficiently long (1–3 kb) to confound the genome assembly [48]. Caenorhabditiselegans sequence data (11 SMRT cells: ∼45× coverage) of P6-C4 chemistry (Supplementary Figure S1C) were also downloaded from the PacBio DevNet website.
Next, we tackled the main challenge of focus for this evaluation using the genome of a non-model plant with a high repetitive content and longer repeats. For this purpose, I.nil (plant) data [2] of P5-C3 chemistry (Supplementary Figure S1A) were obtained based on our previous work (90 SMRT cells: ∼50× coverage; DRA002710). Ipomoeanil has a highly repetitive (64%) genome of an estimated size of 750 Mb, with limited available genomic resources, providing a good measure for similar repetitive plant genomes. To evaluate the correctness of the I. nil genome assemblies, restriction site-associated DNA (RAD)-seq (DRA002758), expressed sequence tags (ESTs; HY917605–HY949060) and bacterial artificial chromosome (BAC)-end data (GA933005–GA974698) were used.
PacBio RSII was the sequencer used in all cases. The P6-C4 chemistry, in comparison with P5-C3, has shown an increase in average read lengths and therefore the average read lengths of the I. nil data set are slightly shorter than those of the other data sets (Supplementary Figure S1). The reason for choosing only SMRT data for the present study is that one of the aims was to evaluate long-read assemblies without depending on SGS data, whereas the nonrandom errors of nanopore data may still have to rely on more accurate Illumina data [49, 50]. All four data sets were preprocessed using HGAP3 to obtain filtered subreads for assembly. Two rounds of consensus polishing were applied to all assemblies using quiver.
Criteria for evaluation
For assessing the assembly results, we considered various metrics (Figure 1). Apart from N50 and L50 measures, the average contigs-to-chromosomes (ctg/chr) ratio was calculated for assessing contiguity. For gene-level completeness, BUSCO [51] and CEGMA [52] were used. In eukaryotic contigs, the terminal regions were scanned using tandem repeats finder [53] for the presence of telomeres. Peak computational memory in the form of maximum resident set size (RSS) and CPU time were determined to compare computational requirements. When complete reference sequences were available, single-nucleotide variations (SNVs), indels and structural variations (SVs) were analyzed from QUAST [54] and Assemblytics [55] to evaluate correctness; unique SVs provided a relative measure of assembly errors. In addition, dot plots were visualized for rearrangements. The percentage of reference sequences covered by the assemblies was calculated using MUMmer [56] alignments.
For the non-model organism I. nil, linkage maps were constructed from RAD-seq [57] data using STACKS [58], to identify mis-assembled contigs. Because the marker density of the linkage maps was low, this also provided a good measure for contiguity, as larger contigs have a better chance of being incorporated in the linkage maps. ESTs and BAC-end reads were used for assessing completeness. Longer contigs had a better chance of concordantly mapping the 100 kb insert-sized BAC-end read pairs, whereas discordant mapping rates provided an indirect measure of mis-assemblies. Whole BAC sequences, of ∼100 kb in length, were used to assess contiguity and completeness, and also to identify SNVs and indels. Tpn transposons, a unique feature of I. nil flowers [2], were also considered to assess completeness.
Ranks were assigned for all the criteria, as listed in Supplementary Methods. The ranks for all criteria were summed up for each assembler. The summed score, in the decreasing order, was used for assigning an overall rank. Also, z-scores were calculated for all observed metrics, so that significant observations received rewards or penalties [22]. The average of the z-scores, from all metrics, for each assembler was plotted to observe z-score-based rankings, which displayed high and low scores for better and worse performances, respectively. For assemblies that failed during execution, either they were left out from the rankings or assigned arbitrary low rankings.
Versions of the tools and the commands used are detailed in the Supplementary Methods.
Results
Contiguity
All of the assemblers reported good contiguity (Table 1).
Escherichia coli
A single contig representing the complete bacterial genome was reconstructed by all the assemblers (Supplementary Table S1).
Plasmodium falciparum
Fewer number of contigs (15–43 contigs), high N50 values (1.2–1.7 Mb), low L50 values (5–7) and low ctg/chr ratios (1–2.27 ratios) were generally observed in all the assemblies, representing high level of contiguity, despite the repetitive nature of the genome. MECAT, in particular, reconstructed every chromosome in one piece, whereas miniasm, SMARTdenovo and wtdbg produced comparatively fragmented or redundant contigs (Supplementary Tables S2 and S3).
Caenorhabditis elegans
The N50 exceeded 1 Mb in all, but the PBcR assembly. Canu had the best N50 (3.6 Mb) and L50 (11) values, while PBcR had low N50 (847 kb) and high L50 (38) values. In general, six contigs, on an average, were found to be sufficient to represent a chromosome (Supplementary Tables S4 and S5).
Ipomoea nil
HGAP3 obtained the best contiguity (N50=1.53 Mb; L50=120) and was the only assembler to have contigs >10 Mb in length. Canu and FALCON shared the next best N50 (934 and 904 kb, respectively) and L50 values (191), while both wtdbg and miniasm had fragmented assemblies (Supplementary Table S6).
The shorter the genome, the lesser the differences observed in contiguity among the assemblers. However, with longer genomes, the contiguity profiles progressively started to differ among the assemblers (Supplementary Figures S2–S4).
Completeness
Escherichia coli
In all the cases, the assembly size was slightly larger than that of the reference genome, with 99.9% BUSCO completeness (Supplementary Table S1).
Plasmodium falciparum
On average, the contigs covered the 14 chromosomes in the range of 95.67–99.90% (Supplementary Table S7). Excluding ABruijn, the apicoplast genome was assembled by all the assemblers, while the mitochondrial genome was only present in the HGAP3 assembly. Canu was able to reconstruct 23 of the 28 telomeres, whereas the PBcR and wtdbg assemblies resolved <10 telomeres (Supplementary Table S8). Intriguingly, Miniasm was unable to resolve even a single telomere. BUSCO analysis showed 67.4–68.9% completeness for all the assemblies, while it should be noted that the original reference sequence also yielded only 68.8% completeness.
Caenorhabditis elegans
At least 99% of all the chromosomes were covered by the assembled contigs on average, excluding the wtdbg assembly (Supplementary Table S9). Canu and HGAP3 produced 10 of 12 telomeres, whereas wtdbg produced only a single telomere (Supplementary Table S10). All the assemblies also showed high BUSCO (97.2–99.2%) completeness ratios.
Ipomoea nil
Most of the assemblies fell short of the expected genome size of 750 Mb; however, BUSCO reported completeness ratios in the range of 92.9–94%. Most of the assemblies mapped around 99% of the ESTs and BAC-end reads (Supplementary Table S11). PBcR (314) and HGAP3 (311) resolved the largest number of Tpn transposons (Supplementary Table S11), followed by canu (307) and MECAT (307). MECAT (18), FALCON (16) and SMARTdenovo (16) were better at resolving telomeres (Supplementary Table S11).
Some smaller PBcR contigs were present redundantly and were covered within larger contigs with short overhangs. The high BUSCO and CEGMA ratios indicated that the gene regions were captured effectively, despite differences in the assembly sizes. The shorter, circular and high-copy nature of the mitochondrial genomes could have possibly confounded the assemblers and were largely unassembled.
Correctness
After two rounds of consensus polishing of the draft assemblies, the indel rates were drastically reduced.
Escherichia coli
Analysis using QUAST showed that all contigs had mis-assemblies. However, on closer inspection using Assemblytics, the source of the mis-assemblies reported by QUAST was revealed to be because of three SVs, which are likely strain-specific differences rather than mis-assemblies (Supplementary Table S1). For instance, in the ABruijn assembly, the contig length was equal to the reference length when the SVs were tallied. However, most other assemblies still had a large number of SVs (an average of 68.8 SVs compared with 9 SVs of ABruijn), even after two rounds of polishing.
Plasmodium falciparum
More than 5000 SVs were shared among all the assemblies. Wtdbg (6448) produced the largest number of unique SVs, whereas ABruijn (389), canu (384), MECAT (311) and PBcR (332) performed better by producing a relatively smaller share of the unique SVs (Supplementary Table S12). Dot plots were used for observing rearrangements, which displayed small rearrangements only in ABruijn and wtdbg assemblies. In other cases, an approximate straight diagonal line was observed with strong congruity.
Caenorhabditis elegans
A total of 17 893 SVs were shared among all the assemblies. Wtdbg (30 622) produced the largest number of unique SVs, whereas canu (2374), FALCON (3337), MECAT (2358) and PBcR (4179) produced a relatively smaller share of unique SVs (Supplementary Table S13). A single or a couple of mis-assembled contigs were visible in the dot plots of all assemblies, barring MECAT and SMARTdenovo.
Ipomoea nil
Miniasm (1.2 Mb) and wtdbg (5.8 Mb) assemblies had the shortest of the mis-assembled contigs, while HGAP3 (128 Mb) showed the largest share of mis-assembled data. HGAP3, FALCON and MECAT had >100 Mb of mis-assembled contigs, whereas canu offered the best balance in incorporating longer contigs (593.3 Mb) into the linkage maps, with shorter (20.9 Mb) mis-assemblies (Supplementary Table S14). Wtdbg (1.04%) and miniasm (2.53%) had the least discordantly mapping BAC-end read pairs. Surprisingly, FALCON (6.36%) had the highest discordant mapping rate (Supplementary Table S11). When BAC sequences were completely covered by contigs, the per-base accuracy was 99.9% in four of the five BAC sequences (Supplementary Table S15), while mismatched bases were almost nonexistent. Fragmented contigs were not considered for assessing per-base accuracy, as they had unresolved errors in overlapping terminal regions.
A lot of SVs were shared among all the assemblers, which may be actual variations rather than assembly errors. Unlike the SMRT data, the Illumina-based assembly was found to have large indels, and plenty of mismatches covering the five BAC sequences in I. nil [2]. The evaluated assemblers, which are based on the overlap information of the longer reads, had benefited not just in terms of contiguity but also in per-base accuracy for a repetitive genome like I. nil.
Circularity and overlapping fragmented contigs
With the application of Circlator [59], it was evident that the circularity of some of the E. coli assemblies was clearly not resolved, and hence the presence of additional base pairs, which were subsequently trimmed out. The increased indel rates were originally concentrated on the overlapping terminal ends of the circularly unresolved contigs. As a result, the indel rates became almost identical in all the circularly resolved assemblies (Supplementary Table S16). However, Circlator was unable to resolve the circularity for HGAP3, MECAT and wtdbg assemblies. Similarly, when the contigs were fragmented in repetitive regions, sometimes, the breakpoints happened in such a way that two nearby contigs shared considerable overlapping terminal ends. Consensus polishing did not have an impact in such overlapping regions leading to unresolved and high amount of indel errors.
Resource usage
Escherichia coli
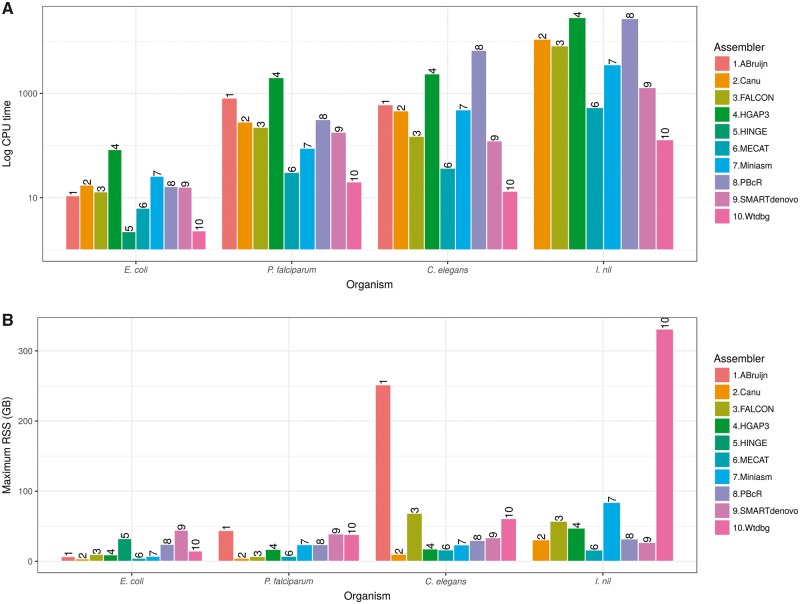
HINGE and wtdbg assemblies were quickly obtained, while HGAP3 was the slowest, as expected (Figure 3A). Miniasm was the fastest of all assemblers, and finished in about 16 min of CPU time; however, two rounds of RACON execution required 25.81 CPU h, making this pipeline the second slowest. SMARTdenovo consumed the maximum peak memory usage, while HGAP3 consumed the least amount of memory (Figure 3B).
Figure 3.
Computational resource requirements. Computational requirements are represented as (A) log CPU time and (B) maximum RSS, a measure of peak memory usage, for all assemblers. Refer to Supplementary Table S17 for actual CPU times. Failed assemblies (HINGE for the eukaryotic genomes, and ABruijn for the I. nil genome) are not plotted.
Plasmodium falciparum
Wtdbg was the quickest assembler, closely followed by MECAT. Other assemblers generally consumed hundreds of CPU hours, with HGAP3 being almost 100-fold slower compared with the speed of wtdbg (Figure 3A). ABruijn, SMARTdenovo and wtdbg were memory-intensive, whereas canu, FALCON and MECAT were memory-efficient (Figure 3B).
Caenorhabditis elegans
Wtdbg followed by MECAT were the fastest in producing assemblies, while PBcR was the slowest (Figure 3A). ABruijn consumed a huge amount of memory, while canu was the most memory-efficient, followed by MECAT and HGAP3 (Figure 3B).
Ipomoea nil
Wtdbg was again the fastest assembler (129.7 CPU h). It should be noted that HGAP3 took 83.9 CPU h even for a bacterial genome. MECAT was also fairly quick, while the celera-dependent pipelines were the slowest (Figure 3A). Wtdbg consumed 331.15 Gb of peak memory. MECAT was the best with respect to both CPU time and peak memory usage, while canu also showed a reasonable balance in resource usage (Figure 3B).
Ranking
Escherichia coli
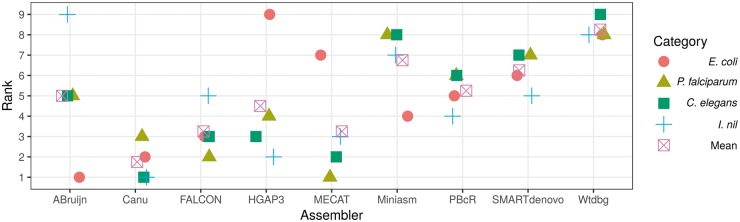
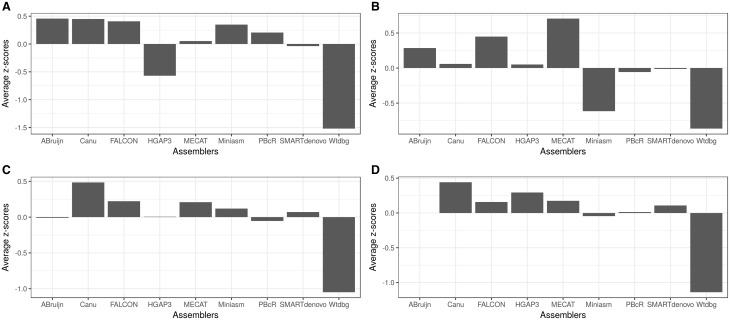
The assemblers ABruijn, canu and FALCON in the order were top-ranked in both the rankings (Figures 4 and 5A). The rankings were heavily influenced by whether the assemblies were circularly resolved, and hence, MECAT, HGAP3 and wtdbg were pushed to the bottom of the table.
Figure 4.
Rankings for all assemblies. The lower the rank, the better is the assembly.
Figure 5.
Z-score-based rankings. Average z-scores of all ranking metrics are plotted for (A) E. coli, (B) P. falciparum, (C) C. elegans and (D) I. nil. Higher the average z-value, the better is the assembly performance. The failed ABruijn assembly is left blank for I. nil data set.
Plasmodium falciparum
Although HGAP3 had the highest N50 value, it was not the top-ranked assembler (Figures 4 and 5B). Four assemblers in the order of MECAT, FALCON, ABruijn and canu were top-ranked according to their z-scores (Figure 5B), corroborating that N50 should not be the sole factor in choosing an assembly. HINGE assembly was excluded from the rankings, as it resulted in a segmentation fault and therefore was not tested for the other eukaryotic data sets too.
Caenorhabditis elegans
Canu ranked at the top, followed by FALCON and MECAT (Figure 5C). Although miniasm was eighth in the ranking (Figure 4), it surprisingly ranked fourth according to the z-scores, as a result of obtaining considerably high z-scores for contiguity metrics (Figure 5C). Without error correction, it would be difficult to distinguish duplications and repeats [37]; however, the repeat-sparse nature of the C. elegans genome likely contributed to the better contiguity achieved by miniasm.
Ipomoea nil
ABruijn assembly resulted in a segmentation fault and was not considered for evaluation. The highly repetitive nature and the shorter insert size of the I. nil data set prevented all of the assemblers from reaching a 1 Mb contig N50, excluding HGAP3. Nevertheless, canu ranked first, ahead of HGAP3, in either of the rankings (Figure 4 and 5D). If mis-assemblies were given additional penalties, the ranking of HGAP3 might come down further. For the first time, SMARTdenovo was ranked among the top five assemblers.
Mean ranking of the three eukaryotic assemblies
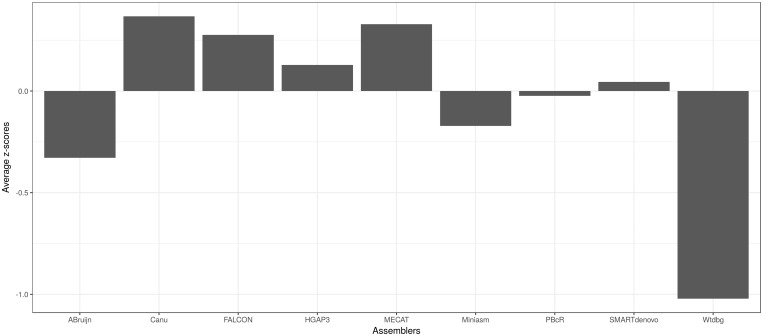
When the rankings of the eukaryotic assemblies were averaged (Figure 4), canu, MECAT, FALCON and HGAP3, in that order, were on the top of the rankings. Similarly, in the z-score-based mean rankings, canu, MECAT, FALCON and HGAP3, in that order, displayed better performances with positive mean z-scores (Figure 6).
Figure 6.
Mean z-score-based rankings. The mean scores of the individual average z-scores obtained from E. coli, P. falciparum, C. elegans and I. nil are plotted. Higher the average z-value, the better is the assembly performance.
Discussion
De novo genome assemblies using SMRT data, when compared with earlier versions, have been shown to increase contiguity by several hundred-folds [3, 6, 10], and resolve fragmented regions into contiguous, gapless sequences [6, 41]. The average and median contig N50 values of recently assembled plant and animal genomes using long reads are 6.24 and 3.60 Mb (Table 2), respectively. In the current study, the three important features—contiguity, completeness and correctness [1]—of long-read assemblers were evaluated.
Table 2.
A list of recently assembled genomes using PacBio’s SMRT data
| Organism | Technology | Assembly tool | Contig N50/NG50 (Mb) | Scaffold N50/NG50 (Mb) | Study |
|---|---|---|---|---|---|
| Taeniopygia guttata | PB | FALCON | 5.8 | NA | [3] |
| Calypte anna | PB | FALCON | 5.4 | NA | [3] |
| Drosophila serrata | PB | PBcR | 0.94 | NA | [4] |
| Utricularia gibba | PB | HGAP3 | 3.42 | NA | [5] |
| Arabidopsis thaliana | PB | PBcR | 11.16 | NA | [41] |
| Drosophila melanogaster | PB | Canu | 21.31 | NA | [25] |
| Homo sapiens CHM1 | PB | Canu | 21.95 | NA | [25] |
| Vitis vinifera | PB | FALCON | 2.39 | NA | [42] |
| Ipomoea nil | PB+Illumina+LM | HGAP3 | 1.87 | 2.88 | [2] |
| Vigna angularis | PB+Illumina+454 | Sprai, Celera | 0.8 | 2.95 | [7] |
| Oreochromis niloticus | PB+RH map+RAD map | Canu | 3.1 | NA | [8] |
| Gorilla gorilla | PB+BAC-end+Fosmid-end | FALCON | 9.56 | 23.14 | [6] |
| Lates calcalifer | PB+OM+LM | HGAP3 | 1.72 | 25.85 | [9] |
| Capra hircus | PB+OM+HiC | PBcR | 18.7 | 87.28 | [11] |
| Arabis alpina | PB+OM+HiC | PBcR, FALCON | 0.9 | 3.8 | [17] |
| Euclidium syriacum | PB+OM | PBcR, FALCON | 3.3 | 17.5 | [17] |
| Conringia planisiliqua | PB+OM | PBcR, FALCON | 3.6 | 8.9 | [17] |
| Corvus corone | PB+OM | FALCON | 8.91 | 18.36 | [10] |
| Zea mays | PB+OM | PBcR, FALCON | 1.19 | 9.56 | [13] |
| Homo sapiens NA12878 | PB+OM | PBcR, FALCON | 1.4 | 31.1 | [14] |
| Homo sapiens HX1 | PB+OM | FALCON | 8.3 | 22 | [12] |
| Oropetium thomaeum | PB+OM | HGAP3 | 2.4 | 7.1 | [16] |
| Oryza sativa indica | PB+Fosmids+OM+LM | PBcR | 4.43 | 1.22 | [15] |
| Homo sapiens NA19240 | PB+OM | FALCON | 7.25 | 78.6 | [18] |
PB, PacBio SMRT data; OM, Optical mapping data; LR, Linked reads; LM, Linkage maps.
Canu ranked the best in the average rankings of all the assemblies from all the data sets. Canu, because of its efficiency to handle repeats [25], had fewer assembly errors, sometimes trading contiguity for correctness. Indeed, it is essential to prioritize correctness rather than contiguity, which would otherwise defeat the purpose of building a reference genome for future studies.
Canu and MECAT showed the best balance in computational requirements. MECAT requires longer reads to effectively distinguish non-repetitive overlaps, and was found to underperform in the case of I. nil, whose transposons can be longer than the 7 kb average insert size of I. nil data.
FALCON, the only diploid-aware assembler, showed reasonable performance for genomes up to 100 Mb in length, similar to MECAT. The FALCON assembly was surprisingly filled with mis-assemblies for the I. nil data, probably because of the repeat filtering steps, leading to further loss of coverage in input data. An increase in insert sizes and coverage could yield better performance from both FALCON and MECAT.
HGAP3 was found to be the most contiguous assembler, but with the disadvantage of extremely slow computation times. Mis-assemblies were also most abundant in the HGAP3 assemblies, possibly because of the greedier nature of celera’s algorithm at the layout stage [36]. In addition, as previously observed for PBcR in the rice genome assembly [15], the celera-based assemblers, PBcR and HGAP3, were found to have redundant contigs.
PBcR is the second most widely used long-read assembler (Table 2); however, it is no longer maintained, as the focus has shifted to its successor canu, which seemed to outperform PBcR in almost every analysis.
SMARTdenovo, although not the best, produced moderately good results in all metrics and would be a suitable choice for obtaining larger genome assemblies quickly.
Leaving out the consensus module, miniasm was the fastest available assembler for all genomes evaluated, excluding I. nil. Miniasm requires as much as 13% divergence for repeat resolution, whereas canu and FALCON require only 3 and 5% divergence, respectively [25]. Hence, miniasm produced fragmented contigs for repeat-rich genomes, but obtained reasonable rankings otherwise.
HINGE may not be ideal for assembling large genomes, but would be a good choice for assembling highly repetitive bacterial genomes.
As observed in the assemblies of the slightly smaller yeast genome [24], ABruijn, despite its good contiguity, was chimeric. ABruijn failed to assemble the I. nil data set; however, when the error-corrected reads of canu were used, the assembly was possible but only after consuming almost 500 Gb of maximum RSS.
Similarly, wtdbg was also memory-intensive, and both the assemblers will need high-end servers for handling larger genomes. In the case of repetitive genomes, both assemblers could collapse repeats, leading to loss of information. In particular, the wtdbg assembly was found to be >100 Mb short of the expected genome size in I. nil. Wtdbg assemblies, which always ranked last, mostly because no consensus procedure was executed, would need additional rounds of consensus polishing to effectively compete with other assemblers. Wtdbg assemblies also had fragmented contigs.
Mitochondrial genomes were generally left unassembled. Hence, it might be necessary to either extract (i) reads that do not align to the assembled contigs, (ii) or reads that align to an available or a closely related mitochondrial genome. The extracted reads could be used to perform an additional round of assembly, for reconstructing extra-chromosomal genomes [47]. In addition, redundancy at the ends of contigs can be a major obstacle for polishing the genome, as it might become difficult for the reads to be aligned at such regions, leaving out errors stranded in the terminal portions of the contigs. Indeed, when whole BAC sequences of I. nil were covered by completely spanning contigs, the error rate was approximately homogenous across all the assemblers, whereas when contigs were in overlapping fragmented pieces, the terminal overlapping regions were found to have increased error rates. The same phenomenon was observed in redundant regions from circularly unresolved bacterial assemblies. Identifying such regions and trimming the redundant base pairs may lead to an improved overall per-base correctness.
Dot plots showed that many of the breakpoints in contig mis-assemblies originated from different locations for different assemblers. Contiguity profiles were also found to be different for FALCON and PBcR in plant genome assemblies, and a hybrid assembly using the different contiguity profiles was found to be highly successful [17]. Hence, an alternative solution to increasing the contiguity would be to combine different assemblies by using reconciliation tools such as quickmerge [60]. For example, miniasm had fewer contigs and breakpoints compared with MECAT for the C. elegans assemblies. Using miniasm assembly as a backbone for extending the MECAT assembly may result in longer and more accurate contigs in this case.
Similar to the evaluation of short read assemblers [19–22], the current study did not reveal a clear winner; a similar result was observed with evaluations of Nanopore sequencing data [24]. That is, an optimal assembler for one data set may not be optimal for a different data set. Hence, it would be ideal to try out a variety of assemblers, as performed in the Solanum pennelii genome project [49], and choose the best assembly based on various evaluation strategies. Any available resources such as BAC-end data, whole BAC sequences, previously annotated gene sets and similar resources could be effectively used for the purpose of evaluation, as demonstrated in this study.
Based on the results, we suggest that the best approach in handling larger genomes would be to generate assemblies from at least canu, FALCON, MECAT and SMARTdenovo, and basing the final decision on the assembler according to different evaluation metrics rather than on N50 alone. When time is not a limiting factor, HGAP3 could also be used, but care should be taken in recognizing mis-assembled and redundant contigs. Recently, scaffolding techniques, such as optical mapping, CHICAGO, Hi-C and linked reads, have been applied to correct mis-assemblies [10–18], which can also be used for achieving chromosome-scale assemblies.
Supplementary Material
Biographies
Vasanthan Jayakumar is a graduate student at the Department of Biosciences and Informatics, Keio University, Japan. His research interest is in Genomics and Bioinformatics.
Yasubumi Sakakibara is a Professor at the Department of Biosciences and Informatics, Keio University, Japan. His research interests are Bioinformatics including genome assembly, metagenome analysis and artificial intelligence.
Key Points
All non-hybrid long-read assemblers are good at producing excellent contiguity.
Considering correctness, computational time and memory requirements, canu, MECAT, FALCON and SMARTdenovo are recommended as minimum necessity for assembling third-generation, single-molecule sequencing (SMRT) data.
As observed in the previous evaluations for short-read assemblers [19–22], the assemblies should be carefully evaluated using different metrics before finalizing the assembly, without relying on just N50 metric.
Redundant base pairs in the overlapping terminal regions of fragmented contigs lead to unresolved errors, even after several rounds of consensus polishing.
High copy extrachromosomal genomes have a significant chance of being filtered out. To reconstruct mitochondrial genomes, it may be necessary to identify reads, which do not align to the assembled contigs, so that they can be separately assembled.
Funding
This work was supported by JSPS KAKENHI Grant Number 16H06279.
References
- 1. Lee H, Gurtowski J, Yoo S, et al. Third-generation sequencing and the future of genomics. bioRxiv 2016:048603. [Google Scholar]
- 2. Hoshino A, Jayakumar V, Nitasaka E, et al. Genome sequence and analysis of the Japanese morning glory Ipomoea nil. Nat Commun 2016;7:13295 10.1038/ncomms13295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Korlach J, Gedman G, Kingan S, et al. De novo PacBio long-read and phased avian genome assemblies correct and add to genes important in neuroscience research. Gigascience 2017;6:1–16. doi: 10.1093/gigascience/gix085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Allen SL, Delaney EK, Kopp A, Chenoweth SF.. Single-molecule sequencing of the Drosophila serrata genome. G3 2017;7(3):781–8. 10.1534/g3.116.037598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lan T, Renner T, Ibarra-Laclette E, et al. Long-read sequencing uncovers the adaptive topography of a carnivorous plant genome. Proc Natl Acad Sci USA 2017;114(22):E4435–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gordon D, Huddleston J, Chaisson MJ, et al. Long-read sequence assembly of the Gorilla genome. Science 2016;352(6281):aae0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sakai H, Naito K, Ogiso-Tanaka E, et al. The power of single molecule real-time sequencing technology in the de novo assembly of a eukaryotic genome. Sci Rep 2015;5(1):16780 10.1038/srep16780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Conte MA, Gammerdinger WJ, Bartie KL, et al. A high quality assembly of the Nile Tilapia (Oreochromis niloticus) genome reveals the structure of two sex determination regions. BMC Genomics 2017;18(1):341 10.1186/s12864-017-3723-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vij S, Kuhl H, Kuznetsova IS, et al. Chromosomal-level assembly of the Asian Seabass genome using long sequence reads and multi-layered scaffolding. PLoS Genet 2016;12:e1005954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Weissensteiner MH, Pang AW, Bunikis I, et al. Combination of short-read, long-read, and optical mapping assemblies reveals large-scale tandem repeat arrays with population genetic implications. Genome Res 2017;27(5):697–708. 10.1101/gr.215095.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bickhart DM, Rosen BD, Koren S, et al. Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat Genet 2017;49(4):643–50. 10.1038/ng.3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shi L, Guo Y, Dong C, et al. Long-read sequencing and de novo assembly of a Chinese genome. Nat Commun 2016;7:12065 10.1038/ncomms12065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jiao Y, Peluso P, Shi J, et al. Improved maize reference genome with single-molecule technologies. Nature 2017;546(7659):524–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pendleton M, Sebra R, Pang AW, et al. Assembly and diploid architecture of an individual human genome via single-molecule technologies. Nat Methods 2015;12(8):780–6. 10.1038/nmeth.3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Du H, Yu Y, Ma Y, et al. Sequencing and de novo assembly of a near complete indica rice genome. Nat Commun 2017;8:15324 10.1038/ncomms15324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. VanBuren R, Bryant D, Edger PP, et al. Single-molecule sequencing of the desiccation-tolerant grass Oropetium thomaeum. Nature 2015;527(7579):508–11. 10.1038/nature15714 [DOI] [PubMed] [Google Scholar]
- 17. Jiao WB, Accinelli GG, Hartwig B, et al. Improving and correcting the contiguity of long-read genome assemblies of three plant species using optical mapping and chromosome conformation capture data. Genome Res 2017;27(5):778–86. 10.1101/gr.213652.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Steinberg KM, Graves-Lindsay T, Schneider VA, et al. High-quality assembly of an individual of Yoruban descent. bioRxiv 2016:067447 doi: 10.1101/067447. [Google Scholar]
- 19. Salzberg SL, Phillippy AM, Zimin A, et al. GAGE: a critical evaluation of genome assemblies and assembly algorithms. Genome Res 2012;22(3):557–67. 10.1101/gr.131383.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Magoc T, Pabinger S, Canzar S, et al. GAGE-B: an evaluation of genome assemblers for bacterial organisms. Bioinformatics 2013;29(14):1718–25. 10.1093/bioinformatics/btt273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Earl D, Bradnam K, St John J, et al. Assemblathon 1: a competitive assessment of de novo short read assembly methods. Genome Res 2011;21(12):2224–41. 10.1101/gr.126599.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bradnam KR, Fass JN, Alexandrov A, et al. Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species. Gigascience 2013;2(1):10 10.1186/2047-217X-2-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sović I, Križanović K, Skala K, Šikić M.. Evaluation of hybrid and non-hybrid methods for de novo assembly of nanopore reads. Bioinformatics 2016;32(17):2582–9. [DOI] [PubMed] [Google Scholar]
- 24. Istace B, Friedrich A, d'Agata L, et al. De novo assembly and population genomic survey of natural yeast isolates with the Oxford Nanopore MinION sequencer. Gigascience 2017;6(2):1–13. 10.1093/gigascience/giw018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koren S, Walenz BP, Berlin K, et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 2017;27(5):722–36. 10.1101/gr.215087.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vaser R, Sović I, Nagarajan N, Šikić M.. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res 2017;27(5):737–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xiao CL, Chen Y, Xie SQ, et al. MECAT: an ultra-fast mapping, error correction and de novo assembly tool for single-molecule sequencing reads. Nat Methods 2017. doi: 10.1038/nmeth.4432. [DOI] [PubMed] [Google Scholar]
- 28. Cherukuri Y, Janga SC.. Benchmarking of de novo assembly algorithms for Nanopore data reveals optimal performance of OLC approaches. BMC Genomics 2016;17(S7):507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liao YC, Lin SH, Lin HH.. Completing bacterial genome assemblies: strategy and performance comparisons. Sci Rep 2015;5(1):8747 10.1038/srep08747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Myers EW. A history of DNA sequence assembly. Inf Technol 58:126–32. [Google Scholar]
- 31. Simpson JT, Pop M.. The theory and practice of genome sequence assembly. Annu Rev Genomics Hum Genet 2015;16:153–72. 10.1146/annurev-genom-090314-050032 [DOI] [PubMed] [Google Scholar]
- 32. Chen Q, Lan C, Zhao L, et al. Recent advances in sequence assembly: principles and applications. Brief Funct Genomics 2017, in press. doi: 10.1093/bfgp/elx006. [DOI] [PubMed] [Google Scholar]
- 33. Chaisson MJ, Wilson RK, Eichler EE.. Genetic variation and the de novo assembly of human genomes. Nat Rev Genet 2015;16(11):627–40. 10.1038/nrg3933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kamath GM, Shomorony I, Xia F, et al. HINGE: long-read assembly achieves optimal repeat resolution. Genome Res 2017;27(5):747–56. 10.1101/gr.216465.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Koren S, Schatz MC, Walenz BP, et al. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotechnol 2012;30:693–700. 10.1038/nbt.2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chin CS, Alexander DH, Marks P, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 2013;10(6):563–9. 10.1038/nmeth.2474 [DOI] [PubMed] [Google Scholar]
- 37. Li H. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics 2016;32(14):2103–10. 10.1093/bioinformatics/btw152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chu J, Mohamadi H, Warren RL, et al. Innovations and challenges in detecting long read overlaps: an evaluation of the state-of-the-art. Bioinformatics 2017;33:1261–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Myers EW, Sutton GG, Delcher AL, et al. A whole-genome assembly of Drosophila. Science 2000;287(5461):2196–204. 10.1126/science.287.5461.2196 [DOI] [PubMed] [Google Scholar]
- 40. Chaisson MJ, Tesler G.. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics 2012;13(1):238 10.1186/1471-2105-13-238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Berlin K, Koren S, Chin CS, et al. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat Biotechnol 2015;33(6):623–30. 10.1038/nbt.3238 [DOI] [PubMed] [Google Scholar]
- 42. Chin CS, Peluso P, Sedlazeck FJ, et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods 2016;13(12):1050–4. doi: 10.1038/nmeth.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Myers G. Efficient local alignment discovery amongst noisy long reads In: Brown D, Morgenstern B (eds). Algorithms in Bioinformatics. Lecture Notes in Bioinformatics, 8701 Berlin: Springer, 2014, 52–67. [Google Scholar]
- 44. Roberts M, Hayes W, Hunt BR, et al. Reducing storage requirements for biological sequence comparison. Bioinformatics 2004;20(18):3363–9. 10.1093/bioinformatics/bth408 [DOI] [PubMed] [Google Scholar]
- 45. Lin Y, Yuan J, Kolmogorov M, et al. Assembly of long error-prone reads using de Bruijn graphs. Proc Natl Acad Sci USA 113:E8396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Girgis HZ. Red: an intelligent, rapid, accurate tool for detecting repeats de-novo on the genomic scale. BMC Bioinformatics 2015;16(1):227 10.1186/s12859-015-0654-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vembar SS, Seetin M, Lambert C, et al. Complete telomere-to-telomere de novo assembly of the Plasmodium falciparum genome through long-read (>11 kb), single molecule, real-time sequencing. DNA Res 2016;23(4):339–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tyson JR, O’Neil NJ, Jain M, et al. Whole genome sequencing and assembly of a Caenorhabditis elegans genome with complex genomic rearrangements using the MinION sequencing device. bioRxiv 2017:099143. [Google Scholar]
- 49. Schmidt MH-W, Vogel A, Denton A, et al. Reconstructing the gigabase plant genome of Solanum pennellii using Nanopore sequencing. Plant Cell 2017. pii: tpc.00521.2017. doi: 10.1105/tpc.17.00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jain M, Koren S, Quick J, et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. bioRxiv 2017:128835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Simão FA, Waterhouse RM, Ioannidis P, et al. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015;31(19):3210–12. [DOI] [PubMed] [Google Scholar]
- 52. Parra G, Bradnam K, Korf I.. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 2007;23(9):1061–7. 10.1093/bioinformatics/btm071 [DOI] [PubMed] [Google Scholar]
- 53. Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 1999;27(2):573–80. 10.1093/nar/27.2.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gurevich A, Saveliev V, Vyahhi N, Tesler G.. QUAST: quality assessment tool for genome assemblies. Bioinformatics 2013;29(8):1072–5. 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nattestad M, Schatz MC.. Assemblytics: a web analytics tool for the detection of variants from an assembly. Bioinformatics 2016;32(19):3021–3. 10.1093/bioinformatics/btw369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kurtz S, Phillippy A, Delcher AL, et al. Versatile and open software for comparing large genomes. Genome Biol 2004;5(2):R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Baird NA, Etter PD, Atwood TS, et al. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One 2008;3(10):e3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Catchen JM, Amores A, Hohenlohe P, et al. Stacks: building and genotyping Loci de novo from short-read sequences. G3 2011;1:171–82. 10.1534/g3.111.000240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hunt M, Silva ND, Otto TD, et al. Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol 2015;16(1):294 10.1186/s13059-015-0849-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chakraborty M, Baldwin-Brown JG, Long AD, Emerson JJ.. Contiguous and accurate de novo assembly of metazoan genomes with modest long read coverage. Nucleic Acids Res 2016;44:e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.