Abstract
Enolase is an evolutionarily conserved enzyme involved in the processes of glycolysis and gluconeogenesis. Mycoplasma hyopneumoniae belongs to Mycoplasma, whose species are wall-less and among the smallest self-replicating bacteria, and is an important colonizing respiratory pathogen in the pig industry worldwide. Mycoplasma hyopneumoniae enolase (Mhp Eno) expression is significantly increased after infection and was previously found to be a virulence factor candidate. Our studies show that Mhp Eno is a cell surface-localized protein that can adhere to swine tracheal epithelial cells (STECs). Adhesion to STECs can be specifically inhibited by an Mhp Eno antibody. Mhp Eno can recognize and interact with plasminogen with high affinity. Here, the first crystal structure of the mycoplasmal enolase from Mycoplasma hyopneumoniae was determined. The structure showed unique features of Mhp Eno in the S3/H1, H6/S6, H7/H8, and H13 regions. All of these regions were longer than those of other enolases and were exposed on the Mhp Eno surface, making them accessible to host molecules. These results show that Mhp Eno has specific structural characteristics and acts as a multifunctional adhesin on the Mycoplasma hyopneumoniae cell surface.
Keywords: enolase, adhesin, Mycoplasma hyopneumoniae, plasminogen, structure
Introduction
Mycoplasma species are wall-less bacteria with minute genomes that are believed to have evolved by degenerative evolution from Firmicutes (Sladek, 1986; Razin et al., 1998). As the smallest self-replicating organisms, synthetic genome projects for Mycoplasma species have led to an unprecedented understanding of the minimal constituents of the genes enabling life (Glass et al., 2006; Gibson et al., 2010; Hutchison et al., 2016). Mycoplasmas are widespread in the natural world as important parasites of humans, mammals, reptiles, fish, arthropods, and plants (Razin et al., 1998). Among them, Mycoplasma hyopneumoniae (M. hyopneumoniae, Mhp) is the causative agent of enzootic pneumonia (EP), which is a chronic respiratory disease in pigs and causes significant economic loss in pig production globally (Maes et al., 2017). Intermittent and dry cough, labored breathing, anorexia, lethargy and emaciation are the characterized clinical signs of M. hyopneumoniae infection (Maes et al., 1996; Sibila et al., 2009). Lung lesions, composed of purple to gray consolidated areas in the apical, middle and cranial lung lobes in the infected animals, are typical pathological features of the disease (Garcia-Morante et al., 2016). In addition, affected pigs easily suffer from infections by other respiratory pathogens, including viruses, bacteria, and parasites, which can increase the severity of the illness (Li et al., 2015a; Kureljusic et al., 2016).
M. hyopneumoniae is mainly found on the mucosal surface of swine trachea, bronchi and bronchioles (Blanchard et al., 1992), inducing ciliostasis and loss of cilia (DeBey and Ross, 1994). The first step of infection caused by M. hyopneumoniae is due to successful adhesion to the cilia of the epithelium of the swine respiratory tract (DeBey and Ross, 1994). The P97/P102 paralogue families are recognized as the primary adhesins of M. hyopneumoniae (Adams et al., 2005). Repeat regions 1 and 2 are the most important areas of P97 for adhesion (Hsu and Minion, 1998; Minion et al., 2000). However, the host cell-binding region can also be expanded by posttranslational processing, which was recently confirmed by several proteomic studies of M. hyopneumoniae (Tacchi et al., 2016). P159 (Raymond et al., 2013), amino peptidases MHJ_0461 and MHJ_0125 (Robinson et al., 2013; Jarocki et al., 2015), EF-Tu (Yu et al., 2018a), and some other moonlighting proteins are newly determined adhesins. The genomics and comparative genomics of several strains of field and attenuated M. hyopneumoniae also provide clues to the pathogenicity of M. hyopneumoniae (Liu et al., 2013; Siqueira et al., 2013). Glycosaminoglycans, plasminogen, fibronectin, actin and some other molecules are the target molecules of M. hyopneumoniae adhesion in the swine respiratory tract epithelium (Jenkins et al., 2006; Deutscher et al., 2010; Jarocki et al., 2015; Raymond et al., 2018). However, few virulence factors of M. hyopneumoniae have been identified. Comparative proteomic analyses performed previously in our lab showed that six proteins, including enolase, had relatively higher expression in M. hyopneumoniae-infected swine tracheal epithelial cells (STECs) (Yu et al., 2018a).
Enolase (EC 4.2.1.11) is a conserved metalloenzyme present in almost all organisms of the phylogenetic tree (Ehinger et al., 2004; Kang et al., 2008; Sun et al., 2017) that plays a role as a phosphopyruvate hydratase in the reversible chemical reaction of 2-phosphoglycerate (2-PG) to phosphoenolpyruvate (PEP) in the glycolysis pathway (Wold and Ballou, 1957a,b; Reed et al., 1996). The localization of enolase to the cell surface, cytoplasm, and nuclei has been reported in prokaryotic and eukaryotic cells. Due to its short signaling sequence, the transport mechanism remains unknown (Seweryn et al., 2007). Surface enolase is recognized as a plasminogen receptor in several pathogens and is thought to be involved in extracellular matrix degradation and host cell invasion (Ehinger et al., 2004; Díaz-Ramos et al., 2012). Enolase is also reported to participate in the processing of the RNase E degradosome in Bacillus subtilis and Escherichia coli (Kühnel and Luisi, 2001; Newman et al., 2012; Bruce et al., 2018). The assembly of monomer enolase varies among species and is present either as a dimer or octamer but seldom both (Brown et al., 1998; Lu et al., 2012; Wu et al., 2015). To date, the crystal structures of enolase from various species, including human, flies, and bacteria, have been determined (Kang et al., 2008; Lu et al., 2012; Sun et al., 2017). However, the structure of the enolase of Mycoplasma, which is found in an independent evolutionary branch of bacteria, remains unknown.
Here, we reported the first three-dimensional (3D) structure of Mycoplasma enolase (Mhp Eno, short for Mycoplasma hyopneumoniae Enolase). The structure showed a unique Mycoplasma enolase with characteristic long loops in the S3/H1, H6/S6, H7/H8, and H13 regions. As a virulence factor candidate found in our previous studies, Mhp Eno showed a surface localization on M. hyopneumoniae cells as determined by flow cytometry and electron microscopy. Mhp Eno specifically adhered to STECs and recognized the host plasminogen. All of these results show the unique features of Mhp Eno.
Materials and Methods
Ethics Statement
All animal experiments were approved by the Committee on the Ethics of Animal Experiments and performed at Jiangsu Academy of Agricultural Sciences (License No. SYXK (Su) 2015-0019). The animal experiments were conducted following the guidelines of the Animal Regulations of Jiangsu Province (Government Decree No. 45) in accordance with international law, and the animals did not suffer excessively.
Bacterial Strains and Culture
All M. hyopneumoniae strains were maintained and cultured in our lab. M. hyopneumoniae strain 168 is a field strain initially isolated in Gansu Province, China (Liu et al., 2013). The pathogenic strains NJ and WX were isolated in Nanjing City and Wuxi City, respectively. KM2 cell-free liquid medium (a modified Friis medium) containing 20% (v/v) swine serum was used for M. hyopneumoniae cultures.
Protein Expression and Purification
Mhp Eno (MHP168_271) was synthesized by GenScript Biotech Corp. (Nanjing) and expressed using the pET21a vector in the BL21(DE3) E. coli strain. The bacterial cells were grown in LB medium at 37°C until the OD600 reached approximately 0.8. A final concentration of 0.25 mM IPTG was added to the culture for protein expression at 18°C. The cells were collected by centrifugation and resuspended in buffer A [30 mM Tris–HCl (pH 8.0), 300 mM NaCl, and 20 mM imidazole]. The cells were then lysed by sonication and centrifuged at 100,000 × g for 30 min. The soluble fraction was incubated with Ni Sepharose 6 FF resin (GE Healthcare) for 1 h at 4°C. Mhp Eno proteins were eluted in buffer A by adding 200 mM imidazole and concentrated by ultrafiltration using Centricons (Amicon). The proteins were dialyzed in buffer B [30 mM Tris–HCl (pH 8.0) and 300 mM NaCl]. Further purification was performed using a Superdex 200 Increase 10/300 GL column (GE Healthcare).
Crystallization and Structural Analyses
Size exclusion-purified Mhp Eno was concentrated and diluted to 10 mg/mL and 15 mg/mL for crystal screening. The crystallization experiment was performed using the sitting-drop vapor diffusion method at 291 K. Crystals of Mhp Eno grew in 0.056 M sodium phosphate monobasic monohydrate and 1.344 M potassium phosphate dibasic at pH 8.4 after 20 days. The Mhp Eno crystal refracted to 2.3 Å at beamline BL19U1 at the Shanghai Synchrotron Radiation Facility (SSRF). X-ray diffraction data were merged, integrated and scaled using HKL3000 software. The structure of Mhp Eno was solved by molecular replacement using the enolase molecule from Bacillus subtilis (Protein Data Bank [PDB] ID: 4A3R) as a reference model with Phaser in the CCP4 program suite (Hough and Wilson, 2018). REFMAC5 (Kovalevskiy et al., 2018) and COOT (Emsley and Cowtan, 2004) were used for initial restrained rigid-body refinement and manual model building, respectively. Further refinement was performed with Phenix (DiMaio et al., 2013). The stereochemical quality of the final model was further evaluated with the program PROCHECK. Structural analysis was performed using PyMOL software and the CCP4 program. Dali server was used for the structural similarity comparison.
Preparation of an Anti-Mhp Eno Polyclonal Antibody
A polyclonal antibody was raised against Mhp Eno by subcutaneously immunizing 1-month-old New Zealand white rabbits. Each rabbit was immunized three times with 1 mg of Mhp Eno emulsified in Freund's adjuvant (Sigma, USA) at 2-week intervals. Sera were collected 1 week after the third immunization.
Flow Cytometry
Flow cytometry was performed according to a previous study (Yu et al., 2018a). M. hyopneumoniae strains [1 × 108 color change units (CCUs)/mL for each) were incubated with anti-Mhp Eno serum at a 1:100 dilution (1:100 diluted preimmune serum was used as a negative control, and phosphate-buffered saline (PBS) was used as a blank control). M. hyopneumoniae cells were then stained with fluorescein isothiocyanate (FITC)-conjugated anti-IgG, and the fluorescence intensity was measured using a BD Accuri C6 flow cytometer. The assay was performed in triplicate, and data were analyzed with Student's t-tests using SPSS 20.0. For all tests, p ≤ 0.05 was considered statistically significant.
Immune Electron Microscopy
M. hyopneumoniae strains were grown to mid-log phase and harvested by centrifugation at 6,600 × g at 4°C. A bacterial suspension containing 1 × 108 CCUs was washed thrice with PBS and resuspended in 50 μL of PBS. Twenty microliters of each sample was added to 200-mesh formvar-coated nickel grids and allowed to stand for 5 min. The grids were subsequently fixed with 2% paraformaldehyde in PBS (pH 7.4) for 5 min at room temperature, blocked for 1 h using 1% normal rabbit serum and 1% BSA/PBS and incubated with 1:10 polyclonal rabbit antibody to Mhp Eno in the above-mentioned blocking buffer for 1 h (1:10 diluted preimmune serum was used as a negative control, and PBS was used as a blank control). The samples were washed five times for 5 min in blocking buffer and then incubated with secondary gold-conjugated antibodies at 1:20 (goat anti-rabbit IgG, 10-nm gold particles) for another 1 h. Before fixation for 5 min in 2% paraformaldehyde/PBS, the samples were washed five times with PBS for 5 min. The grids were then washed in distilled water eight times and stained for 15 s with 1% phosphotungstic acid (pH 6.5). The samples were observed with a Tecnai high-field transmission electron microscope after drying with an infrared lamp.
Indirect Immunofluorescence Assays
STECs were maintained and cultured in our lab (Yu et al., 2018a). After incubation with 100 μg of purified Mhp Eno at 37°C for 1 h, the cells were washed three times with PBS and incubated with the anti-Mhp Eno antibody at a 1:1,000 dilution and then with tetramethylrhodamine isothiocyanate (TRITC)-tagged anti-IgG (Proteintech, 1:500 dilution). Finally, the cell nuclei were stained with 6-diamidino-2-phenylindole (DAPI). The fluorescence was detected using a fluorescence microscope (Zeiss, Germany). BSA and its antibody were used instead of Mhp Eno and the anti-Mhp Eno antibody as negative controls.
Inhibition of Adherence Test
M. hyopneumoniae cells (1 × 107 CCUs/mL) were washed three times with PBS and preincubated with the polyclonal antibody against Mhp Eno or preimmune sera (1:20 dilution) at 37°C for 30 min. The bacteria suspended in RPMI-1640 medium were added to 24-well cell plates containing confluent STECs, and the plates were centrifuged at 800 × g for 10 min and incubated at 4°C for 2 hours. Following incubation, the wells were washed three times with PBS to remove unbound M. hyopneumoniae cells. The cells in the wells were treated with lysis buffer containing 0.1% trypsin and 0.025% (v/v) Triton X-100 and then subjected to bacterial genome extraction and real-time PCR for bacterial counting according to a previous method (Yu et al., 2018a). The assay was performed in triplicate, and the data were analyzed by Student's t-tests using SPSS 20.0.
Far-Western Blot (far-WB) Analysis
A 20-μg sample of Mhp Eno was separated by SDS-PAGE and transferred to a PVDF membrane (Li et al., 2015b). After blocking with 5% (w/v) skimmed milk, the membrane was incubated with 5 μg/mL fibronectin (Roche), complement factor H (Hycult Biotech) or plasminogen (Sigma) and then with anti-fibronectin, anti-factor H or anti-plasminogen antibody (Abcam; 1 μg/mL) as the primary antibody and horseradish peroxidase (HRP)-conjugated anti-IgG (Boster; 1:5,000 dilution) as the secondary antibody. Finally, the membrane was developed with electrochemiluminescence (ECL) substrate using a ChemiDoc XRS+ system (Bio-Rad). BSA was used instead of Mhp Eno as a negative control, and the polyclonal antibody against Mhp Eno was used as a positive control.
Surface Plasmon Resonance (SPR) Analysis
SPR was performed by a BIAcore X100 Plus instrument (GE Healthcare). Plasminogen, fibronectin and factor H were separately diluted to 50 μg/mL and covalently linked to the CM5 sensor chip as a ligand using an amine coupling kit (Biacore AB). The immobilization of soluble plasminogen, fibronectin, and factor H generated resonance units (RU) of ~2,000. The binding kinetics were measured with increasing concentrations (0–4,000 nmol/L) of the analytes (Mhp Eno) in running buffer (HBS-EP) consisting of 10 mM HEPES, 150 mM NaCl, 3 mM EDTA, and 0.05% (v/v) surfactant P20 (Biacore AB) at a flow rate of 30 μL/min for 180 s over immobilized Mhp Eno at 20°C. The dissociation phase was monitored for 1000 s by allowing buffer to flow over the chip. The association kinetics were analyzed manually using Biacore X100 Control software.
Enzymatic Activity Assays
Spectrophotometric assays were performed to detect the catalytic reaction at a temperature of 25°C. The reaction was followed by an increase in PEP, which exhibited UV absorbance at a wavelength of 240 nm. To determine the velocity of the enzyme-catalyzed reaction, measurements were performed at 0.5-min intervals for 5.5 min. The concentrations of 2-PGA (Sigma) and Mhp Eno used in the assay were 1 mmol/L and 1 μg/mL, respectively. The reaction buffer containing 50 mmol/L Tris–HCl (pH 8.0), 100 mmol/L NaCl, and 1.5 mmol/L MgSO4. 2-PGA was added immediately, and the mixture was mixed well before measurement. Enzymatic activity assays for yeast enolase (Sigma, the positive control) and BSA (the negative control) were performed using the same protocol. The specific activity was calculated using the following equation:
Protein Data Bank Accession Number
The coordinates and structural factors generated in this study were submitted to the Protein Data Bank (https://deposit-pdbj.wwpdb.org/deposition/) under the following accession number: 6J36.
Results
Surface Localization of Enolase on Mycoplasma hyopneumoniae Cells
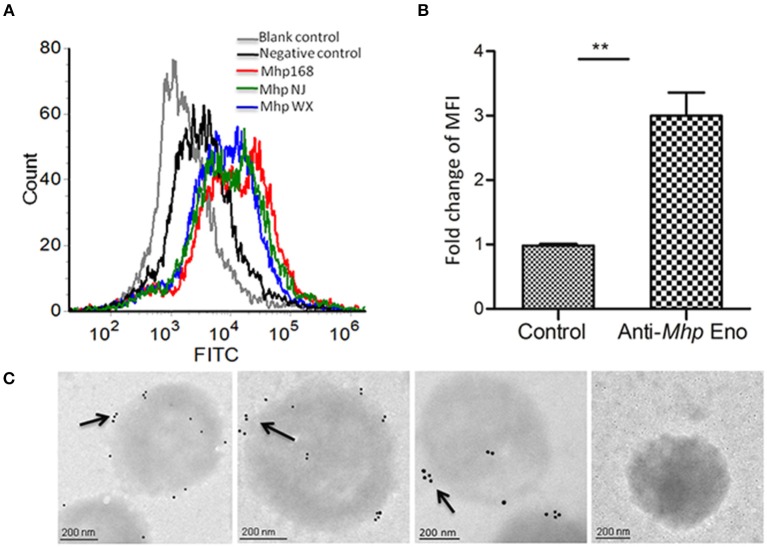
Our previous studies showed that Mhp Eno is a candidate virulence factor of M. hyopneumoniae (Yu et al., 2018a). Enolases of bacterial pathogens often have a surface localization, which contributes to mucosal surface colonization and host tissue invasion (Bergmann et al., 2001; Ehinger et al., 2004; Agarwal et al., 2008). To investigate the cell surface localization of enolase on M. hyopneumoniae, two approaches were used. A flow cytometry analysis showed that the mean fluorescence intensity (MFI) of M. hyopneumoniae strains 168, NJ and WX treated with anti-Mhp Eno serum was 3-fold higher than that of strains treated with preimmune serum (Figures 1A,B), which indicated that the surface of Mhp Eno was accessible to the Mhp Eno-specific antibody.
Figure 1.
Detection of Mhp Eno on the surface of M. hyopneumoniae by flow cytometry and immune electron microscopy. (A) Blank control, M. hyopneumoniae strain 168 treated with PBS; negative control, M. hyopneumoniae strain 168 treated with preimmune serum; M. hyopneumoniae strains 168, NJ, and WX treated with anti-Mhp Eno serum. (B) The MFI level of M. hyopneumoniae incubated with anti-Mhp Eno sera is expressed as the percentage of that found for the corresponding strain incubated with preimmune sera. The asterisks above the charts indicate statistically significant differences. (C) From left to right, the first three M. hyopneumoniae strains, 168, NJ and WX, were treated with anti-Mhp Eno serum and secondary gold-conjugated antibodies. The last column indicates the M. hyopneumoniae strain treated with preimmune serum and secondary gold-conjugated antibodies.
In addition, immune electron microscopy was further applied to study the surface localization of Mhp Eno. The results revealed that Mhp Eno was localized and restricted around the outer region of the bacteria cell for all three strains of M. hyopneumoniae used in this study, whereas the gold particles were not visible in the negative control group treated with preimmune sera (Figure 1C and Supplementary Figure 1). All of these results revealed the surface-accessible localization of Mhp Eno. We did not confirm the distribution of Mhp Eno in the cytoplasm of M. hyopneumoniae, but it is assumed that Mhp Eno also exists in the cytoplasm. First, enolase is a highly conserved enzyme in the glycolysis pathway. Second, the enzymatic activity of Mhp Eno is similar to that of natural yeast enolase (Supplementary Figure 2). Third, it is common for enolase to show multiple localizations at the cell surface, cytoplasm and nuclei (Kang et al., 2008).
Adherence of Mhp Eno to STECs
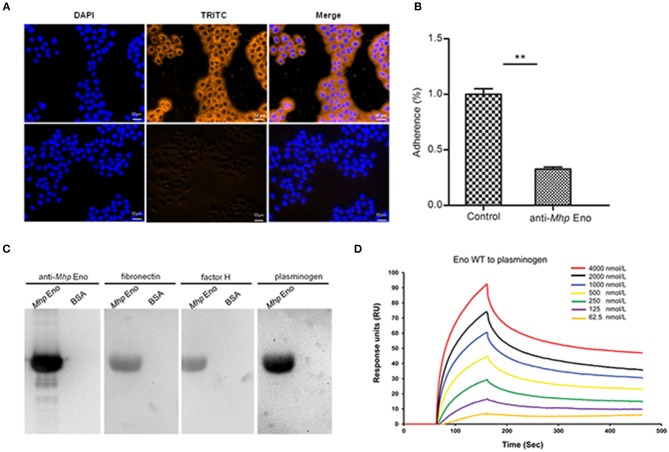
To explore the potential relationship between the surface localization of Mhp Eno and its function, the adhesive ability of Mhp Eno to STECs was studied by indirect immunofluorescence. Significant fluorescence was observed on the cell surface of STECs incubated with Mhp Eno. However, no specific fluorescence was detected around the DAPI-stained cell nuclei in the negative controls (Figure 2A). These results provided direct evidence showing that Mhp Eno binds specifically to the cell membranes of STECs.
Figure 2.
Role of Mhp Eno in the adhesion of M. hyopneumoniae to STECs. (A) Blue indicates STEC nuclei. The color orange in the first row indicates the adherence of Mhp Eno to STEC membranes, and the second row indicates the adherence of BSA (negative control) to STEC membranes. The white line indicates the scale. (B) Adhesion rate = (number of bacteria recovered from cells incubated with the anti-Mhp Eno sera / number of bacteria recovered in the group incubated with the preimmune sera) × 100. The data are expressed as the means ± SDs of at least three experiments with samples performed in triplicate. (C) Analysis of the interaction of Mhp Eno with fibronectin, factor H and plasminogen by far-WB. First lane: PVDF membrane with transferred Mhp Eno protein incubated with anti-Mhp Eno antibody as a positive control; second lane: PVDF membrane with transferred BSA (negative control) incubated with anti-Mhp Eno antibody; third lane: PVDF membrane with transferred Mhp Eno protein incubated with fibronectin and anti-fibronectin antibody; fourth lane: PVDF membrane with transferred BSA (negative control) incubated with fibronectin and anti-fibronectin antibody; fifth and sixth lanes: PVDF membrane with transferred Mhp Eno protein and BSA incubated with factor H and anti-factor H antibody; and seventh and eighth lanes: PVDF membrane with transferred Mhp Eno protein and BSA incubated with plasminogen and the anti-plasminogen antibody. The protein bands were visualized using the ECL substrate. (D) Gradient concentrations of Mhp Eno enolase ranging from 62.5 to 4,000 nmol/L flow through immobilized plasminogen in an SPR assay. The protein concentrations were consistent with the color lines. RU, resonance units.
Antibody inhibition assays were further performed to determine the contribution of surface-localized Mhp Eno to the adhesion function in M. hyopneumoniae strain 168. A polyclonal antibody against Mhp Eno was found to decrease the adherence of M. hyopneumoniae to STECs relative to that of the control group treated with preimmune sera (Figure 2B). The level of adherence inhibition is shown as percentages compared with the adherence of M. hyopneumoniae in the absence of antibody. Incubation with anti-Mhp Eno antibody resulted in a 68% (p < 0.05) reduction in the adherence efficiency of M. hyopneumoniae to STECs. This result confirmed that Mhp Eno plays an indispensable role in the adherence of M. hyopneumoniae to host cells.
Mhp Eno Exhibits High Affinity for Plasminogen-Binding Activities
To determine which components of STECs interact with Mhp Eno, the interactions between Mhp Eno and fibronectin, factor H and plasminogen were examined by far-WB analysis. Corresponding bands were observed in the reactions of Mhp Eno to the anti-Mhp Eno antibody (positive control) and to fibronectin, factor H and plasminogen, whereas no specific reaction was observed in the negative control. The analysis indicated that Mhp Eno specifically binds to fibronectin, factor H and plasminogen (Figure 2C). The real-time interactions between Mhp Eno and fibronectin, factor H and plasminogen were further investigated using SPR. However, no interactions were detected between Mhp Eno and fibronectin or factor H, indicating weak interactions. Mhp Eno was found to bind to plasminogen in a dose-dependent manner with an equilibrium dissociation constant (KD) value of 293.6 nmol. Only 62.5 nmol/L Mhp Eno yielded notable reflection signals (Figure 2D). All of these results indicated that Mhp Eno has multiple adhesive functions, and its highest affinity was to plasminogen.
Unique Structure of Mhp Eno
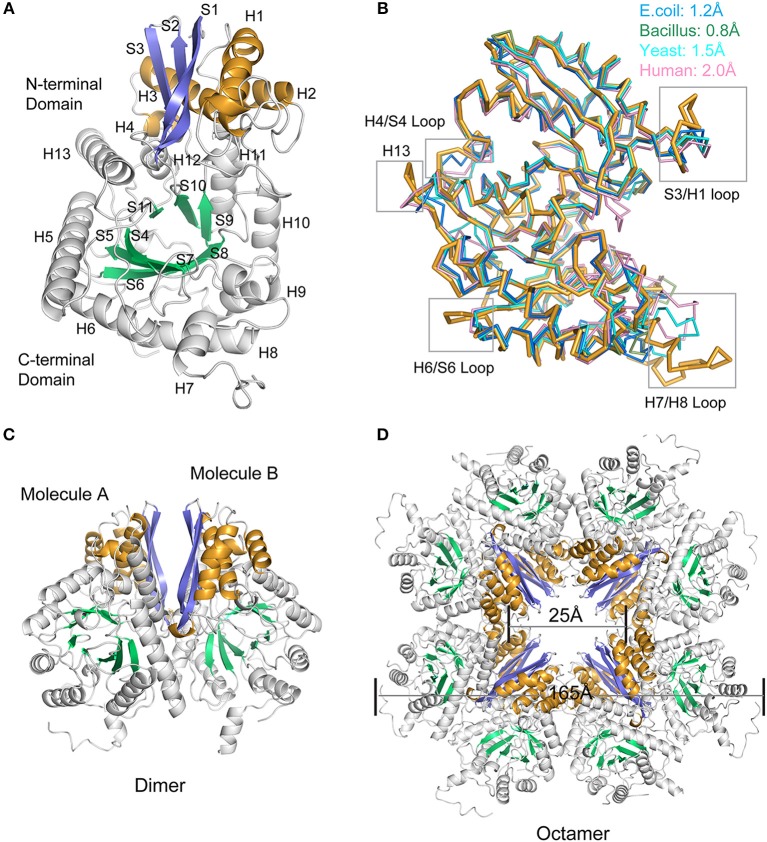
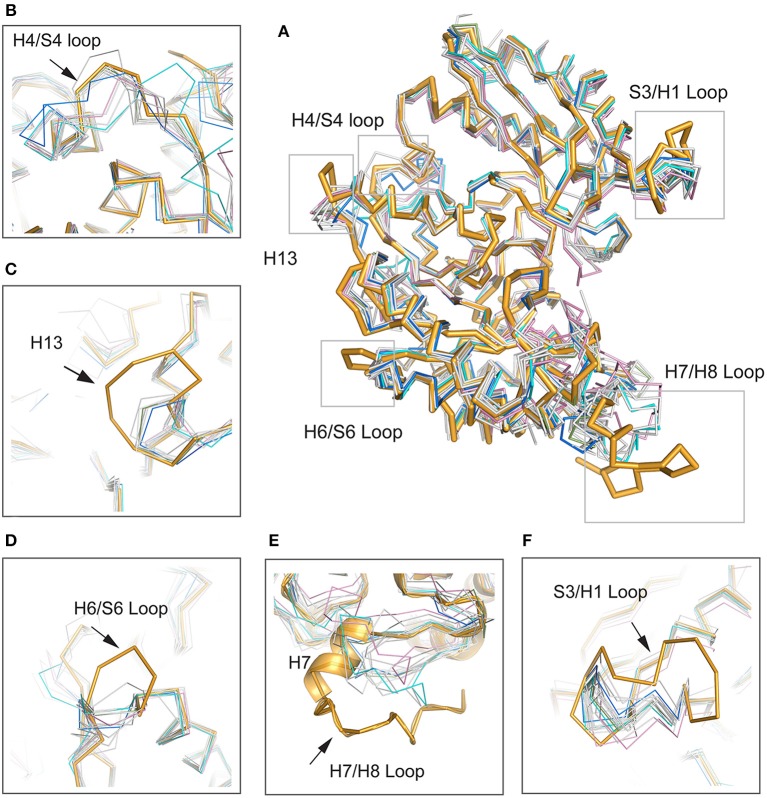
The oligomeric state of enolase, particularly enolases from prokaryotes, is always a topic of debate. Here, in Mycoplasma hyopneumoniae, an octamer form was determined for Mhp Eno (Figure 3A). The crystal structure was determined at a resolution of 2.3 Å (Figure 3 and Table 1). Two identical Mhp Eno monomers formed a “heart”-like or “butterfly”-like dimer (Figure 3C). Four dimers were then packed together to construct a ring-shaped octamer with a small tunnel in the center. Both octamer interfaces and dimer interfaces were observed in Mhp Eno. These structures were similar to those of other solved enolase structures (Supplementary Tables 1, 2). The diameters of the octamer Mhp Eno disc and the center tunnel were ~165 Å and ~25 Å, respectively (Figure 3D). Similar to enolases from other species, the overall structure of monomer Mhp Eno could be divided into an N-terminal domain and a C-terminal TIM-barrel domain. The N-terminal domain (residues 1–139) comprised an antiparallel three-stranded β-sheet (S1–S3) followed by four α-helices (H1– H4). The C-terminal domain (residues 140–452) was relatively larger and consisted of eight β-strands (S4–S11) and nine α-helices (H5–H13). Compared with other enolases, Mhp Eno had an additional α-helix, which was located on the site corresponding to the S6-H7 loop of other enolase structures, named H7 in Mhp Eno. The C-terminal domain topology consisted of a β2α2βα2(βα)5 pattern, which differed from either the traditional TIM-barrel domain or those of other enolases (Figure 3A).
Figure 3.
Overall structure of Mhp Eno. (A) Structure of the protomer Mhp Eno. The β-strands and α-helices are sequentially numbered; “S” stands for β-strand, and “H” stands for α-helix. The β strands of the N-terminal domain and C-terminal domain are indicated in slate (S1-S3) and green cyan (S4-S10) colors, respectively. The α-helices of the N-terminal domain and C-terminal domain are indicated in bright orange (H1-H4) and white (H5-H13), respectively. (B) Structural comparisons between Mhp Eno and enolases from human (pink, PDB ID: 3B97), yeast (cyan, PDB ID: 3ENL), E. coli (blue, PDB ID: 1E9I), and Bacillus subtilis (green, PDB ID: 4A3R). All structures are shown in a ribbon format. Mhp Eno is shown in bright orange. The structure deviations are indicated at the top. The regions of Mhp Eno that show noticeable differences are marked by gray boxes. (C) Overall structure of the Mhp Eno dimer unit. (D) Overall structure of the Mhp Eno octamer. The diameters of Mhp Eno and the tunnel are indicated.
Table 1.
Data collection and refinement statistics.
| Mhp Eno | |
|---|---|
| Data collection | |
| Beamline | SSRF BL19U1 |
| Space group | I4 |
| Cell Dimensions | |
| a, b, c (Å) | 193.154, 193.154, 63.943 |
| α, β, γ (°) | 90.0, 90.0, 90.0 |
| Wavelength (Å) | 0.97892 |
| Resolution (Å) | 48.29–2.30 (2.44–2.30) |
| Total no. of reflections | 241,925 |
| Rmerge (%) | 0.108 (0.597) |
| I/σI | 14.2 (3.07) |
| Completeness (%) | 99.8 (99.8) |
| Redundancy | 4.6 (4.7) |
| Refinement | |
| Resolution (Å) | 48.29–2.30 |
| No. reflections | 52,557 |
| Rwork/Rfree (%) | 0.1865/0.2257 |
| No. atoms | 7,101 |
| Protein | 7,000 |
| Water | 57 |
| Average B-factors (Å2) | |
| Protein | 31.3 |
| Water | 24.1 |
| Rmsd Values | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 0.886 |
| Ramachandran plot (%) | |
| Total favored | 96.44% |
| Total allowed | 3.11% |
| Outliers | 0.44% |
| Coordinate error (Å) | 0.29 |
The values in parentheses are for the highest-resolution shell.
Rmerge = ∑ | I−〈I〉| /∑I, where I is the integrated intensity of a given reflection.
R = ∑ || Fobs | – | Fcalc | |/∑hkl | Fobs |.
Rfree was calculated using 5% of the data omitted from the refinement.
I /σI = average (I /σI).
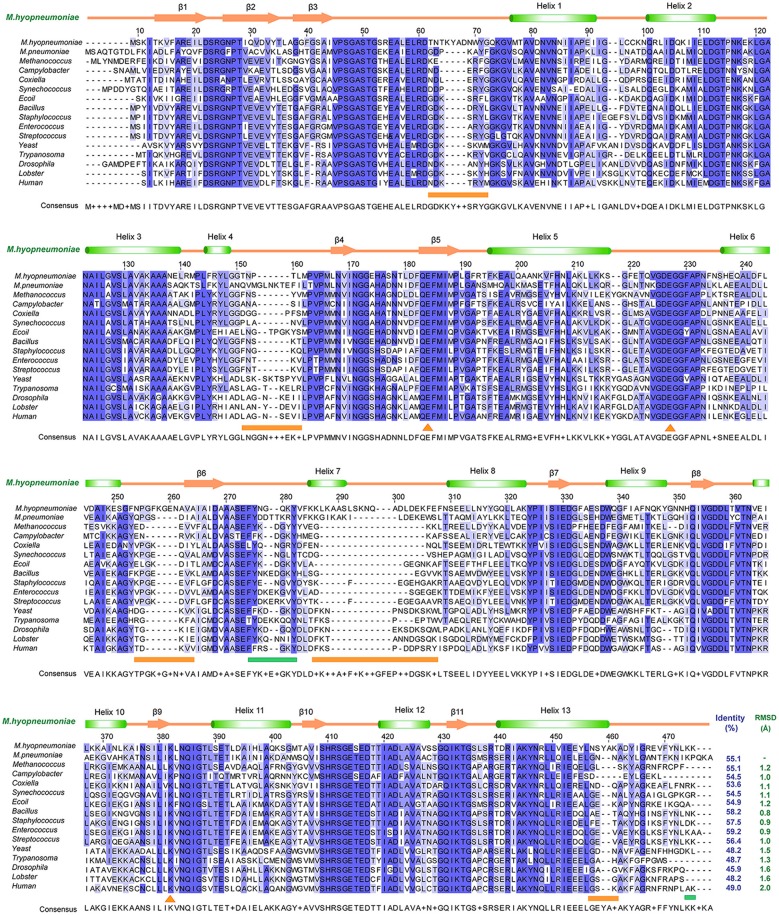
To determine the relationships between Mhp Eno and other enolases, we compared enolase at both the structural and primary sequence levels (Figures 4, 5). Overall, their similarities coincided with their evolutionary relationships. With the exception of Mycoplasma enolases, Mhp Eno showed the best identity with enolases of gram-positive bacteria. For example, Bacillus subtilis enolase had the most similar structure to that of Mhp Eno, with an RMSD of 0.8 Å. In addition, Enterococcus hirae enolase had the best sequence identity to Mhp Eno (59%). The similarities between Mhp Eno and gram-negative bacterial enolases were slightly worse than those of gram-positive bacterial enolases. Mhp Eno showed the worst structure and sequence identity with eukaryote enolases. For example, Mhp Eno showed only 46% sequence identity with Drosophila enolase and had a 2.0-Å RMSD with human enolase. The superimposition of enolases from different species was performed, and the results clearly showed five sites where Mhp Eno showed notable differences from other enolases, particularly the S3/H1 loop, H4/S4 loop, H6/S6 loop, H7/H8 loop, and H13 (Figures 3B, 5). These loops, especially the H7/H8 loop, of Mhp Eno were significantly longer than those of other enolases. Until now, with the exception of Mhp Eno, such large deviations in the enolase backbone structure have not been observed. All of these results indicated that M. hyopneumoniae or Mycoplasma has a very unique enolase structure.
Figure 4.
Structural and sequence alignments between Mhp Eno and other enolases. The sequence identities and structural deviations are shown at the end of each sequence. The second structure of Mhp Eno is above the alignment. The regions that show notable differences are indicated with orange lines at the bottom of the alignment. The plasminogen-binding regions are marked by green lines at the bottom of the sequence. The sequences or structures are from M. pneumoniae (Mycoplasma pneumoniae, GenBank ID: WP_010874963.1), Methanococcus (Methanococcus jannaschii, PDB ID: 2PA6), Campylobacter (Campylobacter jejuni, PDB ID: 3QN3), Coxiella (Coxiella burnetii, PDB ID: 3TQP), Synechococcus (Synechococcus elongatus, PDB ID: 4ROP), E. coli (PDB ID: 1E9L), Bacillus (Bacillus subtilis, PDB ID: 4A3R), Staphylococcus (Staphylococcus aureus, PDB ID: 5BOF), Enterococcus (Enterococcus hirae, PDB ID: 1LYX), Streptococcus (Streptococcus pneumonia, PDB ID: 1W6T), Yeast (PDB ID: 3ENL), Trypanosoma (Trypanosoma brucei, PDB ID: 1OEP), Drosophila (PDB ID: 3WRO), Lobster (PDB ID: 1PDZ), and Human (PDB ID: 3B97).
Figure 5.
Structural comparisons between Mhp Eno and other enolases. Mhp Eno is shown in bright orange. Enolases from human, yeast, E. coli and Bacillus subtilis are shown in pink, cyan, blue and green, respectively, and the other enolases are shown in white. The overall structure is shown in (A). (B–F) show enlarged pictures of the H4/S4 loop, H13, H6/S6 loop, H7/H8 loop, and S3/H1 loop regions. H7 is shown in both cartoon and ribbon forms in (E).
Characteristic Long Loops Are Exposed on the Surface of Mhp Eno
In addition to its enzymatic function in the glycolytic pathway, enolase has been implicated in numerous processes, such as DNA-binding and plasmin(ogen) receptor function. These additional functions are more significantly affected by 3D structures and surface properties rather than enzymatic activity (Ehinger et al., 2004; Bruce et al., 2018). To assess the specificity of long loops of Mhp Eno, we further compared these areas with those from other enolases. According to the primary sequence comparison, with the exception of M. hyopneumoniae, there were 4-5 residue deletions in the S3/H1 loop, 4-6 amino acid deletions in the H6/S6 loop, 8-18 residue deletions in the H7/H8 loop, and 2-5 amino acid deletion in the H13 relative to other enolases. Therefore, these regions were specific to M. hyopneumoniae enolase. For the H4/S4 loop, Mhp Eno, and most bacterial enolases had ~6 residue deletions, and eukaryote enolases shared a very similar pattern to those of bacterial enolases (Figures 3B, 5A). Therefore, the H4/S4 loop was not an M. hyopneumoniae-specific area (Figure 4).
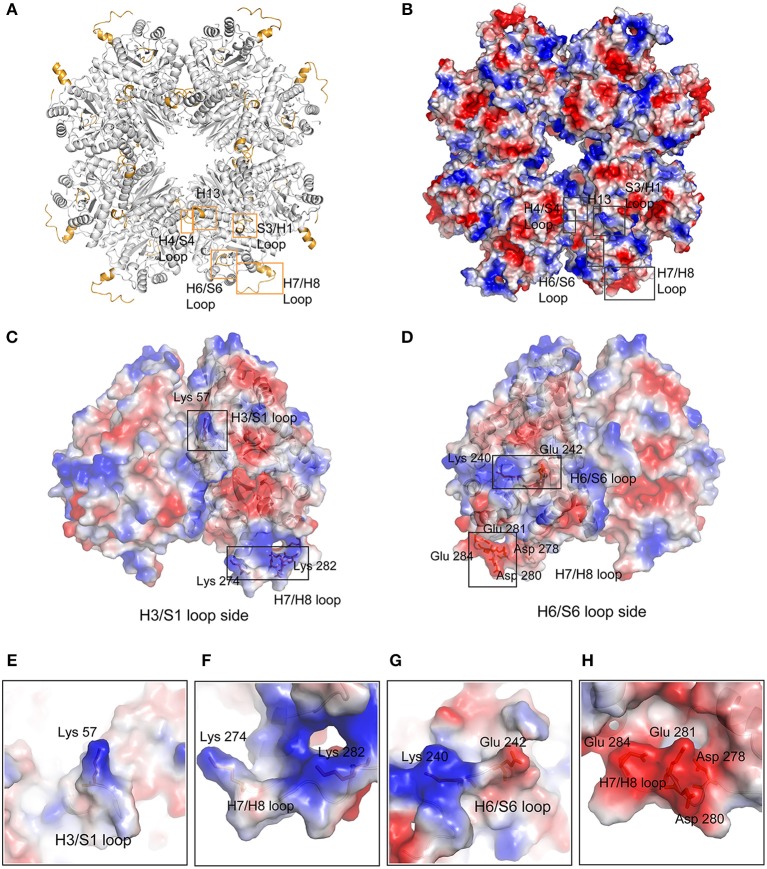
However, we do not know the relationship between Mhp Eno's characteristic long loops and its adhesion function. It is certain that only loops exposed on the surface can recognize host molecules. Clearly, with the exception of the H4/S4 loop, all regions were located on the surface of Mhp Eno (Figures 6A,B). The H4/S4 loop was buried in the dimer interface of Mhp Eno. Therefore, it was difficult for the H4/S4 loop to be in contact with other molecules. The H13 region and H6/S6 region were on one side of the disc-like octamer of Mhp Eno. The S3/H3 region was located at the opposite side of the octamer (Figures 6C,D). The S3/H3 region, which was on the top region of the disc, was the most accessible region (Figures 6C,E). The H7/H8 loop was at the edge of the octamer and had the largest exposed surface (Figures 6A–D). All of these parameters indicated that the two sides of the disc-like octamer Mhp Eno shared the same surface patterns. We further checked the electrostatic potentials of these regions. The H4/S4 and H13 regions showed no notable exposed charged areas. The H6/S6 surface had a positively charged K240 and a negatively charged E242 on (Figures 6D,G). A positively charged K57 was located in the S3/H3 region (Figures 6C,E). This result was interesting for the H7/H8 region because it consisted of a consecutive positively charged patch at 265–274 (KKLKAASLSK) and a negatively charged patch at 275–284 (NQADLDEKFE). However, these two areas were on opposite sides of the Mhp Eno disc (Figures 6C,D,F,H).
Figure 6.
Surface characteristics of featured regions of Mhp Eno. (A) The featured regions of Mhp Eno (bright orange) are shown in cartoon form. (B) Electrostatics of the Mhp Eno surface. The characteristic regions are boxed by rectangles. The S3/H1 loop region and the H13 region of one enolase molecule are located at the same face as the H6/S6 region of another enolase molecule. (C) The H3/S1 loop region side of the dimer unit of Mhp Eno. The charged surface residues in the featured regions (Lys57, Lys274, and Lys282) are shown in stick form. (D) The H6/S6 loop region side of the dimer unit of Mhp Eno. The charged surface residues in the featured regions (Lys240, Lys242, Asp278, Asp280, Glu281, Lys282, and Glu284) are shown in stick form. (E–H) The enlarged views of the residues mentioned in (C,D).
Discussion
Mycoplasma species are distinguished phenotypically from other bacteria by their minimal size and total lack of cell walls. In taxonomy, the lack of a cell wall is used to separate mycoplasmas from other bacteria in a class named Mollicutes (Razin et al., 1998). To date, various structures of enolases from different species, including human (Kang et al., 2008), Drosophila melanogaster (Sun et al., 2017), yeast (Stec and Lebioda, 1990), E. coli (Kühnel and Luisi, 2001), Staphylococcus aureus (Wu et al., 2015), and Streptococcus suis (Lu et al., 2012), have been determined. However, the structure of enolase from Mollicutes, which is one of the smallest self-replicating creatures, has never been solved. Here, the crystal structure of Mhp Eno was determined.
Enolase is a highly conserved enzyme throughout evolution both for its structure and its functions. Enolase catalyzes the conversion of 2-phosphoglycerate to phosphoenolpyruvate during both glycolysis and gluconeogenesis in all living beings (Seweryn et al., 2007). The enolase enzyme catalytic sites have two conformations, in which the ions or substrates are bound in a “closed” state or in an apo “open” state (Wu et al., 2015). Here, the Mhp Eno structure shows a “closed” state. Compared with the structure of PEP-EnoCa, the structure of Mhp Eno has all of the PEP-binding sites at E168, E209, R388, S389, and K359 (Supplementary Figure 3C). All residues are at almost the same sites and have the same conformations as those of PEP-EnoCa. Although we did not determine the PEP-Mhp Eno or PAG-Mhp Eno structures, structural superimposition indicates that Mhp Eno also has conserved enzyme catalytic functions. Furthermore, the enzymatic activity of recombinant Mhp Eno was confirmed by an enzymatic assay (Supplementary Figure 2), which indicated that recombinant Mhp Eno also has biological activity. Enolase is also involved in the formation of functional E. coli RNA degradosomes (Bruce et al., 2018). The complex structure of enolase and RNase E (EBS and AR2 regions) shows that the RNase E segment interacts with the intraprotomer cleft of the enolase dimer (Bruce et al., 2018). The superimposition of Mhp Eno on the E. coli enolase-RNase E complex showed good consistency with the interaction interface (Supplementary Figure 3D). Thus, Mhp Eno might also be included in the RNA metabolism of M. hyopneumoniae. These results show that Mhp Eno has the conserved functional regions of most enolases.
This study revealed that Mhp Eno is a multifunctional protein. This protein not only has conserved sites and structures for 2-phospho-D-glycerate hydrolase catalysis and RNaseE binding (Supplementary Figures 3C,D) but also recognizes fibronectin, factor H and plasminogen (Figure 2C). Mycoplasmas are genome-reduced bacteria that are deficient in many genes involved in metabolic pathways and biosynthesis processes. Amino acids, nucleotides, cholesterol, and other macromolecules need to be supplemented for Mycoplasma growth (Minion et al., 2004). To make the existing proteins more functionally efficient, Mycoplasmas have evolved to allow their proteins to have multiple functions. Adherence to mucosal cells is the first and most important step of M. hyopneumoniae infection. The use of M. hyopneumoniae proteins is maximized for adherence. The present study revealed professional adhesins, such as the P97 and P102 families, as well as large numbers of moonlighting proteins, such as MHJ_0125 (Robinson et al., 2013), MHJ_0461 (Jarocki et al., 2015), EF-Tu (Yu et al., 2018a), FBA (Yu et al., 2018b), Mhp Eno. Furthermore, the number of adhesins is enlarged by posttranslation processing (Deutscher et al., 2012). Unlike the limited numbers of receptors of other pathogens, there are various types of molecules for M. hyopneumoniae adherence. A promiscuous interaction mode has been found for the adhesion of M. hyopneumoniae to its host. In detail, one adhesin of M. hyopneumoniae can recognize more than one host target, and one target can also bind to more than one adhesin. The sites for catalytic function and RNA degradosome interaction of Mhp Eno are conserved and can be deduced by the complex structures of other enolases. However, the structural characteristics forming the basis of Mhp Eno adherence are unknown. Here, the crystal structure of Mhp Eno shows the unique characteristics of the Mycoplasma enolase structure. In our studies, Mhp Eno-specific features, such as the S3/H1 loop, H6/S6 loop, H7/H8 loop, and H13 regions (Figure 5), were found by both sequence alignment and structure comparisons (Figures 4, 5). These regions are located on the surface of the molecule and easily interact with host proteins. Thus, Mhp Eno-specific regions should be involved in newly determined Eno functions, such as interactions with fibronectin, factor H and plasminogen. The binding ability of Mhp Eno to glycosaminoglycans and actin from the host has not been tested. These regions might also be involved in adherence to glycosaminoglycans, actin or other unrecognized molecules that form the swine respiratory tract epithelium.
Most of the eukaryotic and prokaryotic α-enolases are supposed to bind plasmin(ogen) (Ehinger et al., 2004; Kang et al., 2008). The “FYDKERKVY” loop and two C-terminal lysine residues are believed to bind plasminogen in S. pneumoniae (Ehinger et al., 2004). In Mhp Eno, the C-terminal double lysine residues are conserved. The “FYDKERKVY” motifs are partially found in Mhp Eno at “FYNGQKY” sites. We mapped these regions on the octameric Mhp Eno. The “FYNGQKY” loop is located on the Mhp Eno surface and next to the H7/H8 loop (Supplementary Figures 3A,B). The C-terminal tail is found in the dimer interface, as observed in other enolases (Supplementary Figure 3B). However, according to the sequence alignment, both plasminogen-binding regions (the loop region and the C-terminal lysine residues) are not strictly conserved throughout evolution (Figure 4). Thus, enolases from different species might use different regions to recognize plasminogen. A salt bridge is one of the strongest noncovalent interactions between two proteins. For S. pneumoniae enolase, the charged amino acids D250, E252, K251, and K254 in the “FYDKERKVY” loop are critical for plasmin(ogen) binding(Ehinger et al., 2004). It is interesting to find that many charged sites exist in the featured loop regions, such as K240 and E242 in the H6/S6 loop, K57 in the S3/H3 region and a positively charged patch and a negatively charged patch in the H7/H8 loop. Furthermore, these sites are exposed on the surface of Mhp Eno, making them easily accessible to plasminogen. Thus, these sites are possibly involved in the interaction with plasminogen. However, an enolase-plasminogen complex structure is not currently available for determining detailed interaction features.
Because plasminogen is a common receptor of enolase (Bergmann et al., 2001; Ehinger et al., 2004; Seweryn et al., 2007; Agarwal et al., 2008), its activation triggers its conversion to plasmin, which is a serine protease that plays important roles in the maintenance of vascular patency and cell migration (Plow et al., 1995). In recent years, the mechanism through which human enolase induces tumor cell invasion and metastasis has been thought to be related to the activation of plasminogen (Godier and Hunt, 2013; Viedma-Rodríguez et al., 2018). For pathogens, host plasminogen is activated by enolase to function in the processes of tissue remodeling, extracellular matrix degradation and bacterial invasion (Bergmann et al., 2001, 2013; Agarwal et al., 2008; Feng et al., 2009). In M. hyopneumoniae infection, ciliostasis and the loss of cilia are thought to be the result of the activation of various proteases and amino peptidases, including plasminogen (Robinson et al., 2013). In some studies, M. hyopneumoniae was also detected in some nonrespiratory system organs, such as the liver and kidney (Razin et al., 1998). This result might be related to plasminogen activation by M. hyopneumoniae surface proteins, such as enolase. Additionally, in our previous studies, Mhp Eno was found to be an important virulence factor candidate whose expression level was significantly increased after infection. In this study, Mhp Eno showed strong binding to plasminogen, as verified by far-WB and SPR. This result indicated that Mhp Eno is involved in M. hyopneumoniae pathogenicity, which is induced by host plasminogen activation.
In conclusion, we have reported the first enolase structure from a Mycoplasma species. Structural and sequence analyses revealed that Mhp Eno has four characteristic regions that are different from those of other enolases. Mhp Eno is a cell-surface protein of M. hyopneumoniae and induces adhesion to STECs. In addition, Mhp Eno is a multifunction protein that not only has enzyme catalytic sites and RNase E-binding regions but also recognizes fibronectin, factor H and plasminogen. Our study will aid the investigation of enolase evolution and provide insights into the pathogenesis of M. hyopneumoniae.
Data Availability
The coordinates and structural factors generated in this study were submitted to the Protein Data Bank under the following accession number: https://www.rcsb.org/structure/6J36 (Supplementary Datasheet 2).
Author Contributions
RC, YY, RG, XX, ZZ, WW, and TR performed the experimental work. RC, YY, ZF, WW, QinX, WZ, QiyX, and GS performed the data analyses, and all research was conducted under the supervision of WW, WZ, and GS. The original draft of the manuscript was written by RC and YY, and revisions were made by TR, WZ, and GS.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Rong Xu and Fang Zhang of the Jiangsu Academy of Agricultural Sciences for their help with the SPR experiments. We also acknowledge the assistance of the staff at the SSRF synchrotron facility in Shanghai.
Footnotes
Funding. This work was supported by Programs of the National Natural Science Foundation of China (Grant Nos. 31800160, 31700158, 31770193, and 31800161) and the Natural Science Foundation of Jiangsu Province of China (BK20160583, BK20180297).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2019.00209/full#supplementary-material
References
- Adams C., Pitzer J., Minion F. C. (2005). In vivo expression analysis of the P97 and P102 paralog families of Mycoplasma hyopneumoniae. Infect. Immun. 73, 7784–7787. 10.1128/IAI.73.11.7784-7787.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal S., Kulshreshtha P., Bambah Mukku D., Bhatnagar R. (2008). alpha-Enolase binds to human plasminogen on the surface of Bacillus anthracis. Biochim. Biophys. Acta 1784, 986–994. 10.1016/j.bbapap.2008.03.017 [DOI] [PubMed] [Google Scholar]
- Bergmann S., Rohde M., Chhatwal G. S., Hammerschmidt S. (2001). alpha-Enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol. Microbiol. 40, 1273–1287. 10.1046/j.1365-2958.2001.02448.x [DOI] [PubMed] [Google Scholar]
- Bergmann S., Schoenen H., Hammerschmidt S. (2013). The interaction between bacterial enolase and plasminogen promotes adherence of Streptococcus pneumoniae to epithelial and endothelial cells. Int. J. Med. Microbiol. 303, 452–462. 10.1016/j.ijmm.2013.06.002 [DOI] [PubMed] [Google Scholar]
- Blanchard B., Vena M. M., Cavalier A., Le Lannic J., Gouranton J., Kobisch M. (1992). Electron microscopic observation of the respiratory tract of SPF piglets inoculated with Mycoplasma hyopneumoniae. Vet. Microbiol. 30, 329–341. 10.1016/0378-1135(92)90020-T [DOI] [PubMed] [Google Scholar]
- Brown C. K., Kuhlman P. L., Mattingly S., Slates K., Calie P. J., Farrar W. W. (1998). A model of the quaternary structure of enolases, based on structural and evolutionary analysis of the octameric enolase from Bacillus subtilis. J. Protein Chem. 17, 855–866. 10.1023/A:1020790604887 [DOI] [PubMed] [Google Scholar]
- Bruce H. A., Du D., Matak-Vinkovic D., Bandyra K. J., Broadhurst R. W., Martin E., et al. (2018). Analysis of the natively unstructured RNA/protein-recognition core in the Escherichia coli RNA degradosome and its interactions with regulatory RNA/Hfq complexes. Nucleic Acids Res. 46, 387–402. 10.1093/nar/gkx1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBey M. C., Ross R. F. (1994). Ciliostasis and loss of cilia induced by Mycoplasma hyopneumoniae in porcine tracheal organ cultures. Infect. Immun. 62, 5312–5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher A. T., Jenkins C., Minion F. C., Seymour L. M., Padula M. P., Dixon N. E., et al. (2010). Repeat regions R1 and R2 in the P97 paralogue Mhp271 of Mycoplasma hyopneumoniae bind heparin, fibronectin and porcine cilia. Mol. Microbiol. 78, 444–458. 10.1111/j.1365-2958.2010.07345.x [DOI] [PubMed] [Google Scholar]
- Deutscher A. T., Tacchi J. L., Minion F. C., Padula M. P., Crossett B., Bogema D. R., et al. (2012). Mycoplasma hyopneumoniae surface proteins Mhp385 and Mhp384 bind host cilia and glycosaminoglycans and are endoproteolytically processed by proteases that recognize different cleavage motifs. J. Proteome Res. 11, 1924–1936. 10.1021/pr201115v [DOI] [PubMed] [Google Scholar]
- Díaz-Ramos A., Roig-Borrellas A., García-Melero A., López-Alemany R. (2012). alpha-Enolase, a multifunctional protein: its role on pathophysiological situations. J. Biomed. Biotechnol. 2012:156795 10.1155/2012/156795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMaio F., Echols N., Headd J. J., Terwilliger T. C., Adams P. D., Baker D. (2013). Improved low-resolution crystallographic refinement with Phenix and Rosetta. Nat. Methods 10, 1102–1104. 10.1038/nmeth.2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehinger S., Schubert W. D., Bergmann S., Hammerschmidt S., Heinz D. W. (2004). Plasmin(ogen)-binding alpha-enolase from Streptococcus pneumoniae: crystal structure and evaluation of plasmin(ogen)-binding sites. J. Mol. Biol. 343, 997–1005. 10.1016/j.jmb.2004.08.088 [DOI] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. Section D Biol. Crystallogr. 60, 2126–2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Feng Y., Pan X., Sun W., Wang C., Zhang H., Li X., Tang J., et al. (2009). Streptococcus suis enolase functions as a protective antigen displayed on the bacterial cell surface. J. Infect. Dis. 200, 1583–1592. 10.1086/644602 [DOI] [PubMed] [Google Scholar]
- Garcia-Morante B., Segalés J., Fraile L., Pérez de Rozas A., Maiti H., Coll T. (2016). Assessment of Mycoplasma hyopneumoniae-induced pneumonia using different lung lesion scoring systems: a comparative review. J. Comp. Pathol. 154, 125–134. 10.1016/j.jcpa.2015.11.003 [DOI] [PubMed] [Google Scholar]
- Gibson D. G., Glass J. I., Lartigue C., Noskov V. N., Chuang R. Y., Algire M. A., et al. (2010). Creation of a bacterial cell controlled by a chemically synthesized genome. Science 329, 52–56. 10.1126/science.1190719 [DOI] [PubMed] [Google Scholar]
- Glass J. I., Assad-Garcia N., Alperovich N., Yooseph S., Lewis M. R., Maruf M., et al. (2006). Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. U.S.A. 103, 425–430. 10.1073/pnas.0510013103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godier A., Hunt B. J. (2013). Plasminogen receptors and their role in the pathogenesis of inflammatory, autoimmune and malignant disease. J. Thromb. Haemost. 11, 26–34. 10.1111/jth.12064 [DOI] [PubMed] [Google Scholar]
- Hough M. A., Wilson K. S. (2018). From crystal to structure with CCP4. Acta Crystallogr. Section D Struct. Biol. 74:67 10.1107/S2059798317017557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu T., Minion F. C. (1998). Identification of the cilium binding epitope of the Mycoplasma hyopneumoniae P97 adhesin. Infect. Immun. 66, 4762–4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison C. A., III., Chuang R. Y., Noskov V. N., Assad-Garcia N., Deerinck T. J., Ellisman M. H., et al. (2016). Design and synthesis of a minimal bacterial genome. Science 351:aad6253 10.1126/science.aad6253 [DOI] [PubMed] [Google Scholar]
- Jarocki V. M., Santos J., Tacchi J. L., Raymond B. B., Deutscher A. T., Jenkins C., et al. (2015). MHJ_0461 is a multifunctional leucine aminopeptidase on the surface of Mycoplasma hyopneumoniae. Open Biol. 5:140175 10.1098/rsob.140175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins C., Wilton J. L., Minion F. C., Falconer L., Walker M. J., Djordjevic S. P. (2006). Two domains within the Mycoplasma hyopneumoniae cilium adhesin bind heparin. Infect. Immun. 74, 481–487. 10.1128/IAI.74.1.481-487.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. J., Jung S. K., Kim S. J., Chung S. J. (2008). Structure of human alpha-enolase (hENO1), a multifunctional glycolytic enzyme. Acta Crystallogr. Section D Biol. Crystallogr. 64, 651–657. 10.1107/S0907444908008561 [DOI] [PubMed] [Google Scholar]
- Kovalevskiy O., Nicholls R. A., Long F., Carlon A., Murshudov G. N. (2018). Overview of refinement procedures within REFMAC5: utilizing data from different sources. Acta Crystallogr. Section D Struct. Biol. 74, 215–227. 10.1107/S2059798318000979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühnel K., Luisi B. F. (2001). Crystal structure of the Escherichia coli RNA degradosome component enolase. J. Mol. Biol. 313, 583–592. 10.1006/jmbi.2001.5065 [DOI] [PubMed] [Google Scholar]
- Kureljusic B., Weissenbacher-Lang C., Nedorost N., Stixenberger D., Weissenbock H. (2016). Association between Pneumocystis spp. and co-infections with Bordetella bronchiseptica, Mycoplasma hyopneumoniae and Pasteurella multocida in Austrian pigs with pneumonia. Vet. J. 207, 177–179. 10.1016/j.tvjl.2015.11.003 [DOI] [PubMed] [Google Scholar]
- Li B., Du L., Xu X., Sun B., Yu Z., Feng Z., et al. (2015a). Transcription analysis on response of porcine alveolar macrophages to co-infection of the highly pathogenic porcine reproductive and respiratory syndrome virus and Mycoplasma hyopneumoniae. Virus Res. 196, 60–69. 10.1016/j.virusres.2014.11.006 [DOI] [PubMed] [Google Scholar]
- Li Q., Liu H., Du D., Yu Y., Ma C., Jiao F., et al. (2015b). Identification of novel laminin- and fibronectin-binding proteins by far-western blot: capturing the adhesins of Streptococcus suis Type 2. Front. Cell. Infect. Microbiol. 5:82 10.3389/fcimb.2015.00082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Xiao S., Li M., Guo S., Li S., Luo R., et al. (2013). Comparative genomic analyses of Mycoplasma hyopneumoniae pathogenic 168 strain and its high-passaged attenuated strain. BMC Genomics 14:80 10.1186/1471-2164-14-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Lu H., Qi J., Lu G., Gao G. F. (2012). An octamer of enolase from Streptococcus suis. Protein Cell 3, 769–780. 10.1007/s13238-012-2040-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes D., Sibila M., Kuhnert P., Segalés J., Haesebrouck F., Pieters M. (2017). Update on Mycoplasma hyopneumoniae infections in pigs: knowledge gaps for improved disease control. Transbound Emerg. Dis. 65(Suppl. 1), 110–124. 10.1111/tbed.12677 [DOI] [PubMed] [Google Scholar]
- Maes D., Verdonck M., Deluyker H., de Kruif A. (1996). Enzootic pneumonia in pigs. Vet. Q. 18, 104–109. 10.1080/01652176.1996.9694628 [DOI] [PubMed] [Google Scholar]
- Minion F. C., Adams C., Hsu T. (2000). R1 region of P97 mediates adherence of Mycoplasma hyopneumoniae to swine cilia. Infect. Immun. 68, 3056–3060. 10.1128/IAI.68.5.3056-3060.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minion F. C., Lefkowitz E. J., Madsen M. L., Cleary B. J., Swartzell S. M., Mahairas G. G. (2004). The genome sequence of Mycoplasma hyopneumoniae strain 232, the agent of swine mycoplasmosis. J. Bacteriol. 186, 7123–7133. 10.1128/JB.186.21.7123-7133.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman J. A., Hewitt L., Rodrigues C., Solovyova A. S., Harwood C. R., Lewis R. J. (2012). Dissection of the network of interactions that links RNA processing with glycolysis in the Bacillus subtilis degradosome. J. Mol. Biol. 416, 121–136. 10.1016/j.jmb.2011.12.024 [DOI] [PubMed] [Google Scholar]
- Plow E. F., Herren T., Redlitz A., Miles L. A., Hoover-Plow J. L. (1995). The cell biology of the plasminogen system. FASEB J. 9, 939–945. 10.1096/fasebj.9.10.7615163 [DOI] [PubMed] [Google Scholar]
- Raymond B. B., Tacchi J. L., Jarocki V. M., Minion F. C., Padula M. P., Djordjevic S. P. (2013). P159 from Mycoplasma hyopneumoniae binds porcine cilia and heparin and is cleaved in a manner akin to ectodomain shedding. J. Proteome Res. 12, 5891–5903. 10.1021/pr400903s [DOI] [PubMed] [Google Scholar]
- Raymond B. B. A., Madhkoor R., Schleicher I., Uphoff C. C., Turnbull L., Whitchurch C. B., et al. (2018). Extracellular actin is a receptor for Mycoplasma hyopneumoniae. Front. Cell. Infect. Microbiol. 8:54 10.3389/fcimb.2018.00054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin S., Yogev D., Naot Y. (1998). Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62, 1094–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed G. H., Poyner R. R., Larsen T. M., Wedekind J. E., Rayment I. (1996). Structural and mechanistic studies of enolase. Curr. Opin. Struct. Biol. 6, 736–743. 10.1016/S0959-440X(96)80002-9 [DOI] [PubMed] [Google Scholar]
- Robinson M. W., Buchtmann K. A., Jenkins C., Tacchi J. L., Raymond B. B., To J., et al. (2013). MHJ_0125 is an M42 glutamyl aminopeptidase that moonlights as a multifunctional adhesin on the surface of Mycoplasma hyopneumoniae. Open Biol. 3:130017 10.1098/rsob.130017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seweryn E., Pietkiewicz J., Szamborska A., Gamian A. (2007). [Enolase on the surface of prockaryotic and eukaryotic cells is a receptor for human plasminogen]. Postepy Hig. Med. Dosw. 61, 672–682. [PubMed] [Google Scholar]
- Sibila M., Pieters M., Molitor T., Maes D., Haesebrouck F., Segalés J. (2009). Current perspectives on the diagnosis and epidemiology of Mycoplasma hyopneumoniae infection. Vet. J. 181, 221–231. 10.1016/j.tvjl.2008.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siqueira F. M., Thompson C. E., Virginio V. G., Gonchoroski T., Reolon L., Almeida L. G., et al. (2013). New insights on the biology of swine respiratory tract mycoplasmas from a comparative genome analysis. BMC Genomics 14:175 10.1186/1471-2164-14-175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sladek T. L. (1986). A hypothesis for the mechanism of mycoplasma evolution. J. Theor. Biol. 120, 457–465. 10.1016/S0022-5193(86)80039-X [DOI] [PubMed] [Google Scholar]
- Stec B., Lebioda L. (1990). Refined structure of yeast apo-enolase at 2.25 A resolution. J. Mol. Biol. 211, 235–248. 10.1016/0022-2836(90)90023-F [DOI] [PubMed] [Google Scholar]
- Sun C., Xu B., Liu X., Zhang Z., Su Z. (2017). Crystal structure of enolase from Drosophila melanogaster. Acta Crystallogr. Section F Struct. Biol. Commun. 73, 228–234. 10.1107/S2053230X17004022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacchi J. L., Raymond B. B., Haynes P. A., Berry I. J., Widjaja M., Bogema D. R., et al. (2016). Post-translational processing targets functionally diverse proteins in Mycoplasma hyopneumoniae. Open Biol. 6:150210 10.1098/rsob.150210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viedma-Rodríguez R., Martínez-Hernández M. G., Flores-López L. A., Baiza-Gutman L. A. (2018). Epsilon-aminocaproic acid prevents high glucose and insulin induced-invasiveness in MDA-MB-231 breast cancer cells, modulating the plasminogen activator system. Mol. Cell. Biochem. 437, 65–80. 10.1007/s11010-017-3096-8 [DOI] [PubMed] [Google Scholar]
- Wold F., Ballou C. E. (1957a). Studies on the enzyme enolase. II. Kinetic studies. J. Biol. Chem. 227, 313–328. [PubMed] [Google Scholar]
- Wold F., Ballou C. E. (1957b). Studies on the enzyme enolase. I. Equilibrium studies. J. Biol. Chem. 227, 301–312. [PubMed] [Google Scholar]
- Wu Y., Wang C., Lin S., Wu M., Han L., Tian C., et al. (2015). Octameric structure of Staphylococcus aureus enolase in complex with phosphoenolpyruvate. Acta Crystallogr. Section DBiol. Crystallogr. 71, 2457–2470. 10.1107/S1399004715018830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Liu M., Hua L., Qiu M., Zhang W., Wei Y., et al. (2018b). Fructose-1,6-bisphosphate aldolase encoded by a core gene of Mycoplasma hyopneumoniae contributes to host cell adhesion. Vet. Res. 49:114 10.1186/s13567-018-0610-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Wang H., Wang J., Feng Z., Wu M., Liu B., et al. (2018a). Elongation factor thermo unstable (EF-Tu) moonlights as an adhesin on the surface of Mycoplasma hyopneumoniae by binding to fibronectin. Front. Microbiol. 9:974 10.3389/fmicb.2018.00974 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates and structural factors generated in this study were submitted to the Protein Data Bank under the following accession number: https://www.rcsb.org/structure/6J36 (Supplementary Datasheet 2).