Abstract
The spin-trap free radical scavenger N-tert-butyl-α-phenylnitron (PBN) ameliorated effects of several teratogens involving reactive oxygen species (ROS). We investigated for the first time whether PBN could ameliorate teratogenesis induced by a DNA hypomethylating hematological therapeutic 5-azacytidine (5azaC). At days 12 and 13 of gestation, Fisher rat dams were pretreated by an i.v. injection of PBN (40 mg/kg) and 1 h later by an i.p. injection of 5azaC (5mg/kg). Development was analyzed at gestation day 15 in embryos and day 20 in fetuses. PBN alone did not significantly affect development. PBN pretreatment restored survival of 5azaC-treated dams' embryos to the control level, restored weight of embryos and partially of fetuses, and partially restored crown-rump lengths. PBN pretreatment converted limb adactyly to less severe oligodactyly. PBN pretreatment restored global DNA methylation level in the limb buds to the control level. Cell proliferation in limb buds of all 5azaC-treated dams remained significantly lower than in controls. In the embryonic liver, PBN pretreatment normalized proliferation diminished significantly by 5azaC; whereas in embryonic vertebral cartilage, proliferation of all 5azaC-treated dams was significantly higher than in PBN-treated dams or controls. Apoptotic indices significantly enhanced by 5azaC in liver and cartilage were not influenced by PBN pretreatment. However, PBN significantly diminished ROS or reactive nitrogen species markers nitrotyrosine and 8-hydroxy-2′deoxyguanosine elevated by 5azaC in embryonic tissues, and, therefore, activity of this DNA hypomethylating agent was associated to the activation of free radicals. That pretreatment with PBN enhanced proliferation in the liver and not in immature tissue is interesting for the treatment of 5azaC-induced hepatotoxicity and liver regeneration.
Keywords: 5-azacytidine, N-tert–butyl-α-phenylnitron, embryo, teratogenesis, ROS/RNS
Introduction
We have previously shown that a DNA hypomethylating agent 5-azacytidine (5azaC), administered during the rat gestation in vivo, significantly impaired fetal growth, induced specific malformations, and disrupted placental development and its glycoprotein composition [1,2]. Although 5azaC negatively affects placenta, it may directly affect development of the embryo itself without any extraembryonic tissues in vitro [3,4]. We have also previously shown that the acetylsalicylic acid (ASA) ameliorated teratogenic impact on the offspring of 5azaC-treated dams in vivo [5,6]. ASA is a free radical scavenger and inhibitor of the peroxidase-cyclooxygenase component of the prostaglandin H synthetase. It has been effective in the inhibition of phenytoin-initiated oxidative damage to protein and lipids in embryonic tissues [7].
Another free radical scavenger the spin-trap, N-tert-butyl-α-phenylnitron (PBN), exerted similar activity as ASA [8], being protective against several teratogens that involve reactive oxygen species (ROS) such as phenytoin, benzo[a]pyrene, thalidomide, ethanol, and methanol [9,10]. PBN itself has been used for the detection and identification of many other short-lived free radicals that react covalently with PBN and form more stable adducts to be recognized by electron paramagnetic resonance spectroscopy [8]. The understanding of the full activity of PBNs is difficult because it also reacts with methyl, methanol sulfite, azidyl, cyanatyl, and cyanyl radicals and chlorine atoms [11], carotenoid radicals, retinil radicals etc. [12].
As the best known role of nitrones is to scavenge anion superoxide and hydroxyl radicals, they are sometimes classified as antioxidant therapeutics proposed for a wide variety of medicinal indications and have been used for therapy of various conditions such as neurodegeneration, cardiovascular disease, stroke, and cancer [13,14], in antipollution cosmetics, noise-induced neural degeneration, elimination of nociceptive behavior, and for the protection of retina from light-induced damage [14–17]. Differences in the activity of spin-traps and classical antioxidants (enzymatic or nonenzymatic) are that the latter may indiscriminately convert both normal oxygen and ROS molecules to water alone, leading to deep tissue hypoxia. Spin-traps react with the ROS by intercepting it before any damage is done and PBN seems to be the least toxic of nitrones [14]. PBN itself was able to act preventively and/or therapeutically in rat models of glioma, the choline deficiency liver cancer model, and the mouse APCMin/+ colon cancer model [13] and it can probably act similarly on other malignancies.
It must be stressed that the mechanism of therapeutic activity of nitrones is not yet clear. Nitrones act on a variety of pathways to change the redox state through scavenging ROS or reactive nitrogen species (ROS/RNS) but they are also involved in controlling signaling transduction and gene induction [12,18] through NO-releasing properties [19]. Nitric oxide reacts with superoxide (O2–) to form the strong oxidant peroxynitrite (ONOO–), and nitration on the three positions of tyrosine represents a major product of peroxynitrite attack on proteins that may disrupt phosphorylation that is crucial for signal transduction pathways [20].
Increase of 8-hydroxy-2′deoxyguanosine (8-OHdG), a marker of DNA oxidative damage, after experimental treatment of the kidney with carcinogen was partially prevented by PBN [21]. Importantly, 8-OHdG was correlated to methylation of 11 investigated tumor suppressor genes in the development of hepatic cancer [22]. However, DNA oxidative damage adducts may also decrease the binding capacity of DNA methyltransferases (DNMTs), leading to hypomethylation and genomic instability [23].
DNA methylation is an epigenetic mechanism that regulates gene expression [24] and, consequently, developmental processes during gestation or in the adult organism. If DNA methylation changes under the influence of the macro/microenvironment, deregulated developmental processes can lead to fetal malformations [25] or malignancy [26]. Oxidative stress represents such an important influence that has been associated with aberrant DNA methylation in reproduction [27] and carcinogenesis [28]. In carcinogenesis, oxidative stress was associated with both global hypomethylation [23,29] and hypermethylation of gene promotors, for example, involvement in myelodysplastic syndrome (MDS) [30]. Oxidative stress and DNA methylation have a common denominator: the one carbon cycle [27,31].
Necessary DNA methylation is acquired through DNMTs, and passive demethylation is acquired in the absence of DNMTs. Active demethylation proceeds through oxidative processes via the ten-eleven translocation methylcytosine dioxygenase (TET) enzymes [32,33] or by deamination of the amine group of 5mC by activity-induced deaminase/apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (AID/APOBEC) [34]. Aberrant hyper- or hypo-methylations are considered epimutations that can be heritable, at least in daughter cells. Unlike classical mutations, epimutations may be reversed by epigenetic therapeutics [35,36].
For example, DNA hypomethylating drug 5azaC and its deoxy-derivative, 5-aza-2′-deoxycytidine are the first-line treatment of MDSs characterized by immaturity of bone marrow cells that occurs especially in elderly patients and are also the treatment of acute myeloid leukemia [37]. 5azaC is a chemical analogue of the nucleoside cytidine that incorporates itself in both RNA and DNA, and 5-aza-2′-deoxycytidine incorporates itself in DNA, alone. By their DNA hypomethylating activity, achieved through inactivation of DNMT1, they are able to reactivate some genes, such as tumor suppressors. They also exert cytotoxic activity at higher doses [37]. DNA hypomethylating drugs are “genomic medicines” that act on the whole methylome [38] and, therefore, have numerous targets that can interact in many ways, some of which still have to be discovered and explained. Interestingly, the first global evaluation of the epigenetic effects of 5azaC on MDS bone marrow progenitor cells covering DNA methylation, repressive/permissive histone marks and gene expression, did not discover any major/consistent epigenetic changes, aside from the activation of endogenous retroviruses (ERVs) that may contribute to the clinical effects of 5azaC [39].
As mentioned earlier, proper genomic methylation is of utmost importance for normal development [40,41], whereas its aberrations caused by DNA hypomethylating drugs cause teratogenesis as a serious side-effect [1,2].
To assesses the possible beneficial effect of PBN pretreatment on embryonic development impaired by 5azaC, this research focused on the effects in offspring of pregnant dams treated on two consecutive days (days 12 and 13 of pregnancy), each time by a single dose of 5azaC. Apart from being characterized by extensive growth of embryos that may be influenced by extraneous factors, this period of development represents a sensitive time-window for induction of gross malformations, especially of the limbs because they start to develop, first the forelimb buds (day 12) and 1 day later (day 13) the hindlimb buds. Moreover, the liver starts to develop during that period and, therefore, should be susceptible to embryotoxic/teratogenic agents [42–44].
Our results suggest that the spin-trap PBN can ameliorate the most pronounced negative effects of 5azaC on the development of the offspring through scavenging of free radicals.
Materials and Methods
Ethical statement
All procedures on animals were conducted according to the Directive 2010/63/EU and Croatian Law on protection of experimental animals. The Ethical Committee of the School of Medicine, University of Zagreb, Croatia, approved them.
Animals
Three-month-old female Fisher inbred strain rat dams conventionally caged, with water and standard diet ad libitum were caged overnight with males and if sperm was found in the vaginal smear in the morning of the next day, it was assigned as gestation day 0. Pregnant female rats received 5azaC (Sigma A 2385) (5 mg/kg) dissolved in phosphate-buffered saline (PBS) on days 12 and 13 of gestation by the i.p. injection. PBN (Sigma B 7263) (40 mg/kg) was injected in the tail vein on days 12 and 13 of gestation as pretreatment 1 h before 5azaC. Control dams were injected with PBS on days 12 and 13 of gestation by i.p. or i.v. injections.
Sample isolation and processing
Pregnant females were anesthetized i.p. with 0.8 mL/kg of ketamine (Narketan®; Vétoquinol, Bern, Switzerland) and 0.6 mL/kg of xylazine (Xylapan®; Vétoquinol) on days 15 or 20 of gestation and their uteruses were isolated. Animals were sacrificed by this procedure. Embryos (day 15) and fetuses (day 20) were isolated according to the Witshi staging [42]. Embryonic and fetal crown-rump lengths (C-R length) and weights were measured. Samples were fixed in St. Marie solution (1% glacial acetic acid in 96% EtOH), dehydrated, and embedded in paraffin. Serial step sections (5 μm) were processed for routine histology or immunohistochemistry.
Resorptions and deaths in utero were recorded at day 15. The presence of malformations was determined under the dissecting microscope in fetuses at day 20. Differential staining of cartilage and bone was recorded after the alizarin red S and alcian blue staining for 4 days at 37°C, and it was further processed according to a modified Inouye method [45].
DNA isolation
At least six limb bud samples/group were deparaffinized by using xylene (2 × 5 min) followed by incubation in 100%–95%–70% ethanol (3 min each) in water. DNA was extracted in TE buffer pH9 with 0.1 μg/μL of Proteinase K and 0.25% of Nonidet P40 at 56°C for 24 h [46]. Next, samples were heated for 10 min at 95°C to inactivate Proteinase K, spun and the supernatant was frozen at −20°C. DNA concentrations and quality were measured with the NanoDrop ND-2000 spectrophotometer (NanoDrop Technologies, Wilmington, DE).
Bisulfite conversion and polymerase chain reaction
One thousand nanograms of unpurified isolated genomic DNA was used for bisulfite conversion by EpiTect Plus DNA Bisulfite Kit (#59124; Qiagen), which includes a clean-up step with no necessity for prior purification of DNA. PyroMark PCR Kit (#978703; Qiagen) was used for polymerase chain reaction (PCR) amplification in the following conditions: 95°C for 2 min, 43°C for 90 s, and 72°C for 60 s for 40 cycles with the following PCR primers: forward primer: 5′-GGGTTGGGGATTTAG-3′ and biotinylated reverse primer: 5′-AACCCAAAACCTTA-3′.
Global methylation analysis
Global methylation was measured by pyrosequencing. All the steps were performed as recommended by the manufacturer (Qiagen). Pyromark Q24 Advanced System with PyroMark Q24 CpG Advanced Reagents (#970922; Qiagen) was used for the pyrosequencing reaction; 5′-GGGGATTTAGTTTAGTGGT-3′ was the sequencing primer for the rat ID element [47]. DNA methylation data were obtained and analyzed by the PyroMark Q24 Advanced Software.
Immunohistochemistry
Primary mouse monoclonal antibody against proliferating cell nuclear antigen [monoclonal mouse anti-proliferating cell nuclear antigen (PCNA), clone PC 10, M0879 (Dako, Glostrup, Denmark)] was used (1:50) for the detection of proliferating cells. The antibody was incubated overnight at 4°C. The standard negative control reagent (V1617; Dako) was used to inhibit the appearance of endogenous signal. PCNA expression was visualized by using the LSAB2 System-HRP for use on Rat Specimens Kit (K0609; Dako) according to the manufacturer's instructions. Oxidative stress was detected by 8-OHdG (1:500, sc-66036) and nitrotyrosine (1:50, sc-32757), both from Santa Cruz Biotechnology, Inc. (Dallas, TX). The antibodies were incubated overnight at 4°C. Sections were treated with Dako REAL™ EnVision™/HRP, Rabbit/Mouse reagent (K5007; Dako; Agilent Technologies, Inc., Santa Clara, CA); 3,3′-diaminobenzidine-tetrahydrochloride (DAB) was used for signal staining, and hematoxylin was used for counterstaining.
Oxidative stress analysis
The expression of 8-OHdG and nitrotyrosine in single cells of embryonal tissues was used to calculate the histological score (H-score) [48,49]. In three specimens per group, the staining intensity (0, 1+, 2+, or 3+) was determined for each cell in a field of 500 cells. A score 0–300 was assigned to each specimen, based on multiplication of the percentage of cells with different staining intensity within the same field. The discriminatory threshold was set at 50, and samples were considered low (H-score <50) or high (≥50) [50].
Quantitative stereological analysis
Binocular light microscope “Nikon Alphaphot” × 400 for stereological analysis was used along with the Weibel's 42-points multipurpose test system (M42) [51]. The overall length of test lines (Lt) was 1.008 mm, and the area tested (At) was 0.0837 mm2 for each analyzed microscopic field. Sample size (no. of fields to be tested) was determined after analyzing samples on 10 fields with a 95% confidence interval by using the formula “n” = (200/y × s/x)2, where “n” marks the number of fields that need to be analyzed (at least 30 fields per sample), x is the arithmetic mean of the orientation sample, s is the standard deviation (SD) of the orientation sample, and y is the allowed variation from the arithmetic mean [52]. PCNA-positive cells were counted in samples and the numerical density (Nv) was determined by the formula Nv = N/At × D, where N is the number of PCNA-positive cells on the tested area. D is the mean tangential nucleus diameter calculated by light microscopy for 100 cells (for hepatocyte nuclei D = 0.0146 mm, for chondrocyte nuclei D = 0.0147, and for limb bud cell nuclei D = 0.076 mm), and At is 0.0837 mm2. The 3D Ellipse stereological program was used to measure PCNA-positive nucleus diameter.
Apoptotic index
Hematoxylin–eosin-stained specimens were used to measure the apoptotic index. Ten microscopic fields per sample were randomly selected. The histological analysis was performed at a high magnification of 1,000 × by counting apoptotic figures—cells with typical apoptotic morphology (condensed chromatin, cell shrinkage, fragmented nucleus, eosinophilic cytoplasm). The apoptotic index was determined as the number of apoptotic cells per 100 cells in a field [53]. Three specimens from different dams per group were analyzed.
Statistical methods/data analysis
The primary outcome measures were embryonic weights and C-R lengths differences between the control group and groups treated with 5azaC and/or PBN. Secondary outcome measures were the differences in the presence of PCNA-positive cells, differences in malformation rates between the different groups and the DNA methylation pattern between the treated groups and controls. Statistical evaluation of weights and lengths, PCNA numerical density, H-score, apoptotic indices, and global DNA methylation levels within groups were evaluated by using the Student's t-test, Mann–Whitney test, ANOVA with Holm–Sidak's multiple-comparison post hoc test, or Kruskal–Wallis test with Dunn's multiple-comparison post hoc test. Coefficient of variation was expressed as SD/mean × 100. Survival and malformation comparison were done by χ2 analysis. Before analyses, the descriptive statistics was done by the D'Agostino and Pearson test. GraphPad Prism software (version 6.0, GraphPad Software, Inc., San Diego, CA) was used for data analysis. Statistical significance was set at P ≤ 0.05.
Results
Survival
After treatment with PBN, 5azaC, or PBN and 1 h later with 5azaC on days 12 and 13 of pregnancy, all rat dams survived until day 20 of pregnancy with no visible side-effects on inspection.
On day 15 of gestation, the survival, intrauterine deaths, and resorption were assessed. Embryos assigned to the intrauterine death group were well formed although smaller and paler than controls, whereas those that were assigned to the resorption group were amorphic and much smaller. PBN alone had no impact on survival, which was comparable to the 100% survival rate in the control group. Survival in the group treated with 5azaC was poor (59%), resulting in intrauterine deaths and resorption. When PBN was administered 1 h before 5azaC, the survival rate was significantly better (85.5%) than affected by the treatment with 5azaC (Table 1).
Table 1.
Survival of Embryos Previously Treated with 5-Azacytidine and/or N-Tert-Butyl-α-Phenylnitron
| No. of embryos | Survival | Resorptions | Intrauterine deaths | |||||
|---|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | N | % | |
| Control | 43 | 100 | 43a,b | 100 | 0 | 0 | 0 | 0 |
| PBN (40 mg/kg) | 72 | 100 | 72 | 100 | 0 | 0 | 0 | 0 |
| 5azaC (5 mg/kg) | 64 | 100 | 38b,c | 59.4 | 15 | 39.47 | 11 | 28.9 |
| 5azaC (5 mg/kg) + PBN (40 mg/kg) | 83 | 100 | 71a,c | 85.5 | 10 | 14.08 | 2 | 2.8 |
χ2 = 6.8713; P = 0.008759216.
χ2 = 23.076; P = 0.0000001.
χ2 = 12.909; P = 0.0003.
5azaC, 5-azacytidine; PBN, N-tert-butyl-α-phenylnitron.
Overall growth
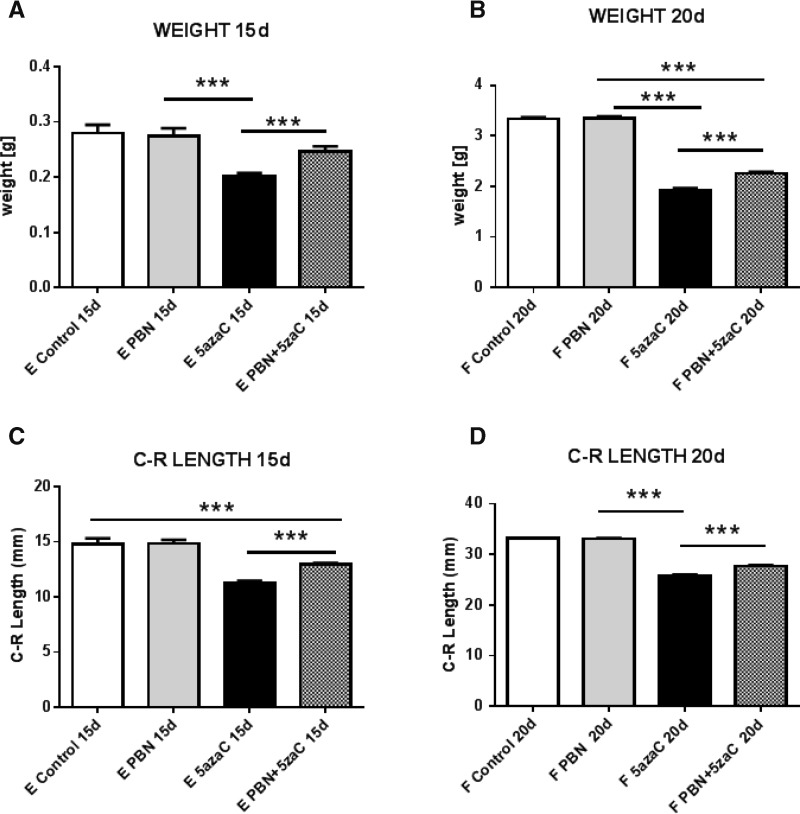
To assess whether overall growth was affected by treatments, both the weight and C-R length measurements were assessed at the embryonic and more advanced fetal stage of development. All surviving offspring were able to grow from day 15 until day 20 of the pregnancy, shown by substantially increased weights and C-R lengths (Fig. 1).
FIG. 1.
Overall growth of embryos (A, C) and fetuses (B, D) previously treated with 5azaC, PBN or pretreated with PBN and then with 5azaC was measured by body weight or by C-R length. Values represent mean ± SEM. ANOVA or Kruskall–Wallis and Mann–Whitney test. ***P < 0.001. 5azaC, 5-azacytidine; C-R length, crown-rump length; PBN, N-tert-butyl-α-phenylnitron; SEM, standard error of the mean.
Embryos derived from dams treated with PBN alone weighted as untreated controls and weighted significantly more than 5azaC-treated dams. Embryos from PBN-pretreated dams weighted significantly more than those from of 5azaC-treated dams, with no statistical difference in comparison to the control (Fig. 1A). Direct comparison of the group of fetuses from dams treated with 5azaC only and the group pretreated with PBN showed that fetuses in the pretreated group were significantly heavier than those treated only with 5azaC (Fig. 1B).
The C-R length of embryos and fetuses from dams treated with PBN showed no difference compared with the C-R length in the control groups. Embryos from dams treated only with 5azaC or pretreated with PBN were statistically smaller than controls. A direct comparison of two groups treated with 5azaC showed that pretreatment with PBN leads to a significant increase in size of the embryos (P < 0.001) (Fig. 1C, D).
Malformations
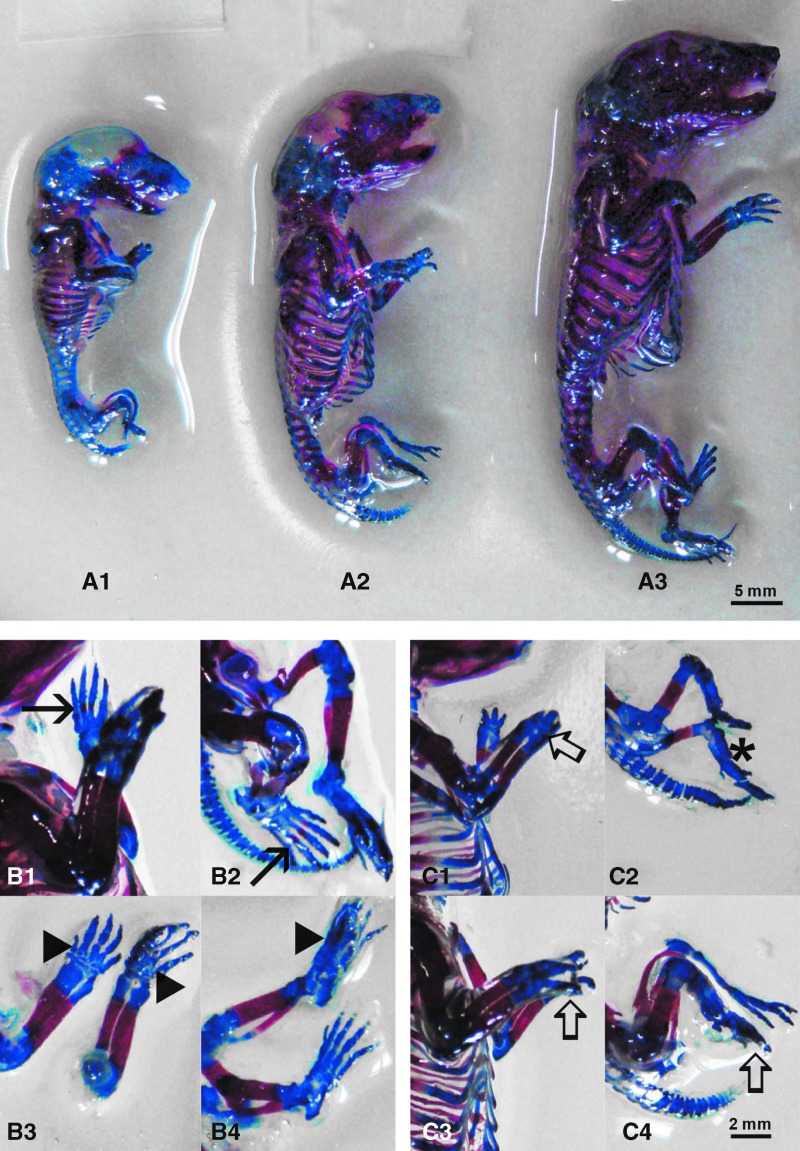
To determine whether fetuses exhibited any specific malformations, we inspected them under the dissecting microscope. The group of fetuses from dams treated with PBN alone showed no signs of malformations (Fig. 2A), the same as untreated controls (not shown). The only difference noted after PBN treatment of dams was the absence of palmar and plantar ossification nuclei (Fig. 2B). All fetuses from dams treated with 5azaC had malformations of forelimbs and hindlimbs (adactyly, oligodactyly) (Fig. 2C). Malformations were also present in 5azaC-treated fetuses from dams pretreated with PBN, and there was no statistical difference in the overall number of malformed fetuses as well as the number of limb malformations between the two groups. However, pretreatment of dams with PBN before 5azaC treatment significantly reduced the number of adactyly in the fetuses and significantly increased the number of fetal oligodactyly (Table 2).
FIG. 2.
Skeletal development and malformations in fetuses from dams previously treated with 5azaC and/or PBN: (A1) 5azaC-treated, (A2) PBN-pretreated and 5azaC treated, (A3) PBN-treated. Ossification: (B1) in control forelimb (arrow), (B2) in control hindlimb (arrow), (B3) PBN-treated forelimbs—missing ossification (arrowhead) and (B4) PBN-treated hindlimbs. (C1) forelimbs of fetuses from dams treated with 5azaC, oligodactyly (thick arrow), (C2) hindlimbs of fetuses from dams treated with 5azaC, adactyly (asterisk). (C3) forelimbs and (C4) hindlimbs of fetuses from dams treated with PBN and 5azaC, oligodactyly (thick arrow). Alizarin red S, Alcian blue, Inouye method. Color images are available online.
Table 2.
Malformations in Fore- and Hindlimb Buds of Fetuses Derived from Dams Previously Treated with 5-Azacytidine and/or N-Tert-Butyl-α-Phenylnitron
| 5azaC (5 mg/kg) | 5azaC (5 mg/kg) + PBN (40 mg/kg) | |||
|---|---|---|---|---|
| N | % | N | % | |
| Malformed/isolated fetuses | 38/38 | 100 | 69/71 | 97.18 |
| Forelimbs | ||||
| Malformed/isolated fetuses | 38/38 | 100 | 69/71 | 97.18 |
| Adactyly | 36a | 94.74 | 54a | 76.06 |
| Oligodactyly | 9b | 23.68 | 47b | 66.20 |
| Hindlimbs | ||||
| Malformed/isolated fetuses | 38/38 | 100 | 69/71 | 97.18 |
| Adactyly | 34c | 89.47 | 36c | 50.7 |
| Oligodactyly | 12d | 31.58 | 55d | 77.46 |
χ2 = 6.00135773; P < 0.02.
χ2 = 17.9080167; P < 0.001.
χ2 = 16.1914325; P < 0.001.
χ2 = 22.0040057; P < 0.001.
DNA methylation in limb buds
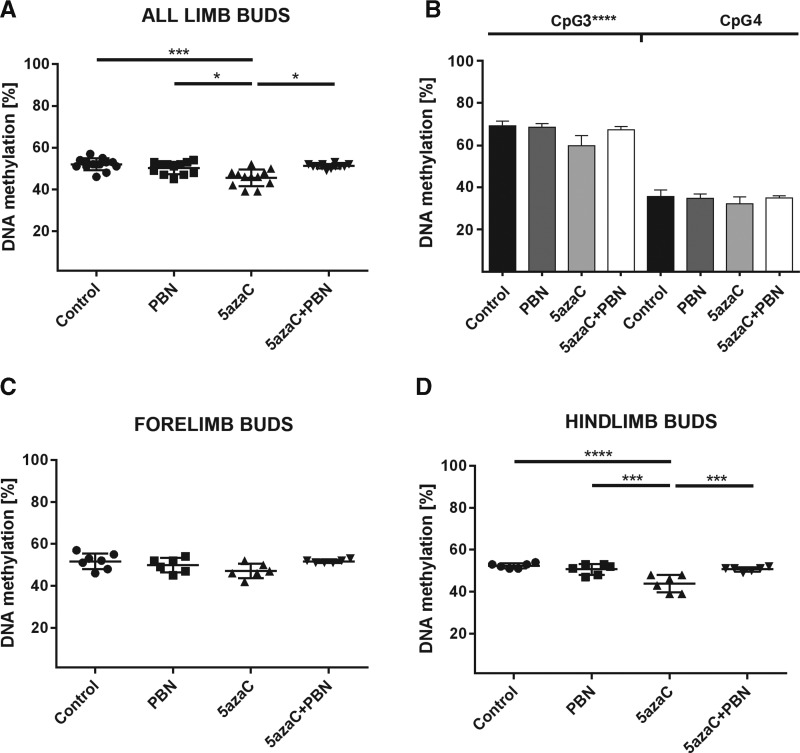
To determine whether the DNA demethylating agent 5azaC impaired DNA methylation in limb buds as the precursors of malformed limbs, we assessed global DNA methylation. That was done by using identifier (ID) element previously recommended for routine analysis of the global DNA methylation changes in rats for pharmaceutical treatment [47]. Our analysis was performed by bisulfite pyrosequencing assay that has been strongly recommended for the best all-round performance among other contemporary DNA methylation assays and is appropriate for sequencing short sequences [54] such as widely spread rat ID elements [47]. In embryonic limb buds from 5azaC-treated dams, statistically significant global DNA demethylating effect was found compared with the embryos from control group dams and PBN-treated dams (Fig. 3A). In limb buds of embryos from PBN-pretreated dams, DNA methylation levels were as in controls, suggesting that PBN prevented 5azaC-induced global demethylation in the developing limb buds. In addition, we discovered that DNA demethylation was significant in CpG3 of the ID element that was previously verified by whole genome amplification and 5azaC treatment in a cell line and not in CpG4 that was not previously verified as responsive to 5azaC treatment (Fig. 3B) [47].
FIG. 3.
Global DNA methylation in limb buds of embryos from dams previously treated with 5azaC and/or PBN. (A) 5azaC induced strong global DNA demethylation in limb buds that could be prevented by PBN. ANOVA. *P < 0.05, ***P < 0.001. (B) Statistically significant global demethylation was detected in CpG3 but not in CpG4 of the chosen ID element. ANOVA. ****P < 0.0001. (C) Methylation of forelimb buds and (D) hindlimb buds. Statistically significant decrease in global methylation appears only in the hindlimb buds. Kruskall–Wallis; Dunn's post hoc test. ***P < 0.001, ****P < 0.0001. ID, identifier.
However, when global DNA methylation was analyzed separately in fore- and hindlimb buds, statistically significant changes in global DNA methylation appeared to be induced in hindlimb buds only (Fig. 3C, D). Forelimb buds that are developmentally more advanced than hindlimb buds [55] were not as significantly affected by the treatment of their dams with 5azaC. Therefore, the intensity of 5azaC demethylating effect depended on the developmental stage of the limb buds.
Cell proliferation
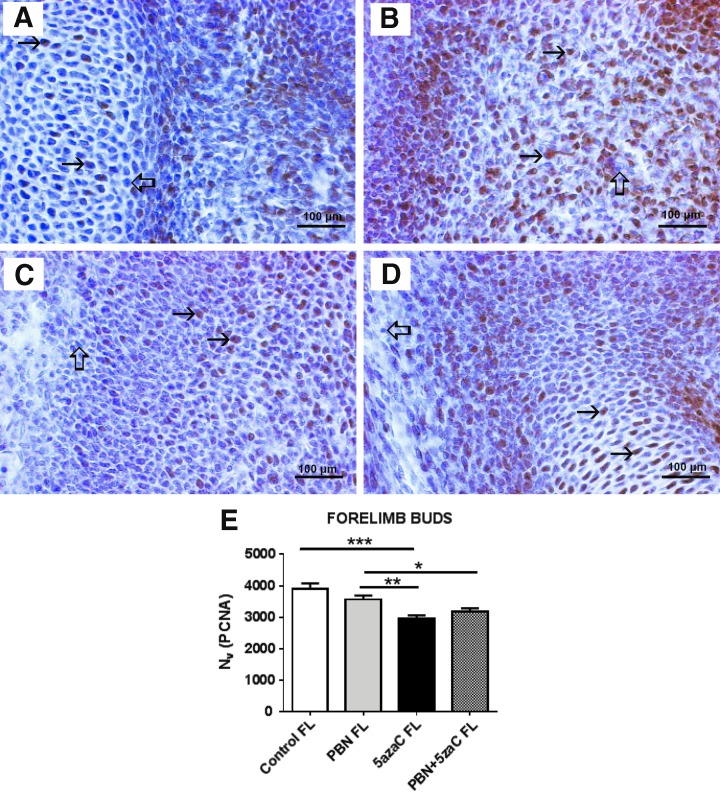
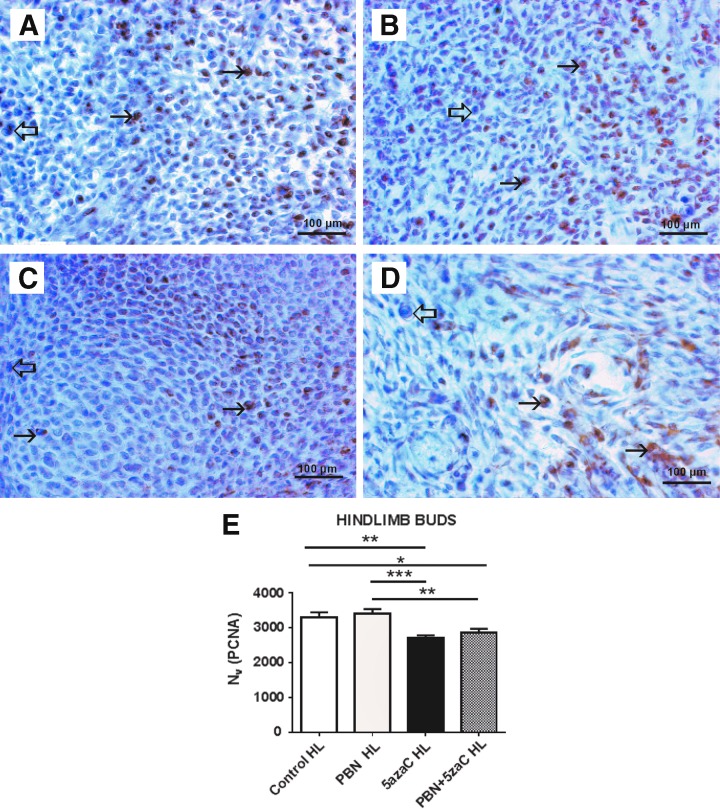
To determine whether the treatments of dams affected cell proliferation in their embryos, we again assessed PCNA expression at the single-cell level and positive signals were quantified by numerical density (Nv) (Figs. 4–7). First, we assessed cell proliferation within the embryonic fore- (Fig. 4) and hindlimb (Fig. 5) buds that subsequently developed into malformed limbs in fetuses. There was no significant difference between the numerical density (Nv) of PCNA when their dams were treated only with PBN or sham-treated controls. Significantly lower proliferation (Nv) in comparison to controls and PBN-treated embryos was found in limb buds treated only with 5azaC and those pretreated with PBN before 5azaC treatment (Figs. 4 and 5). Therefore, pretreatment of pregnant dams with PBN did not affect proliferation in limb buds of the 5azaC-treated dams' embryos.
FIG. 4.
PCNA expression (arrows) in forelimb buds of embryos derived from dams previously treated with 5azaC and/or PBN, detected by immunohistochemistry: (A) control; (B) PBN-treated; (C) 5azaC-treated; (D) PBN-pretreated and 5azaC-treated forelimb bud. DAB, hematoxylin counterstain. Thick arrows point to internal negative controls. Quantification of PCNA-positive signals (E) by the stereological measurement of numerical density (Nv). Nv values (columns) represent mean ± SEM. ANOVA, Holm–Sidak's multiple-comparison post hoc test. *P < 0.05, **P < 0.001, and ***P < 0.0001. DAB, diaminobenzidine-tetrahydrochloride; PCNA, proliferating cell nuclear antigen. Color images are available online.
FIG. 5.
PCNA expression (arrows) in hindlimb buds of embryos from dams previously treated with 5azaC and/or PBN, detected by immunohistochemistry: (A) control; (B) PBN-treated; (C) 5azaC-treated; (D) PBN-pretreated and 5azaC-treated forelimb bud. DAB, hematoxylin counterstain. Thick arrows point to internal negative controls. Quantification of PCNA-positive signals (E) by the stereological measurement of numerical density (Nv). Nv values (columns) represent mean ± SEM. ANOVA, Holm–Sidak's multiple-comparison post hoc test. *P < 0.05, **P < 0.001, and ***P < 0.0001. Color images are available online.
FIG. 6.
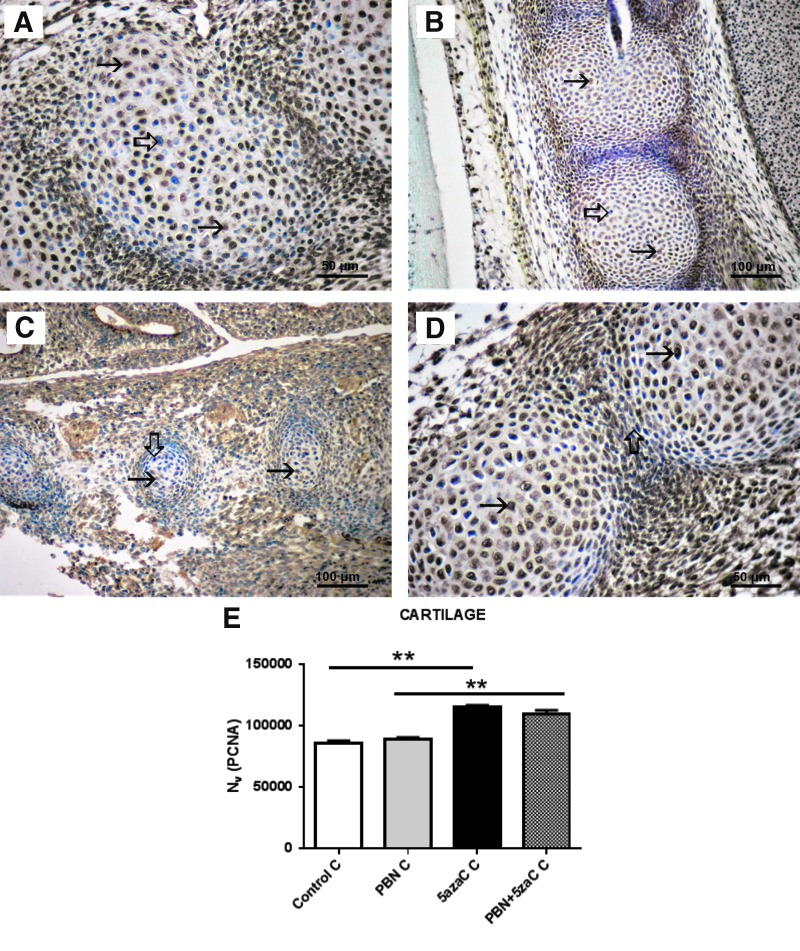
PCNA expression (arrows) in cartilage of embryos derived from dams previously treated with 5azaC and/or PBN, detected by immunohistochemistry: (A) control; (B) PBN-treated; (C) PBN-pretreated and 5azaC-treated; (D) 5azaC-treated cartilage. DAB, hematoxylin counterstain. Thick arrows point to internal negative controls. Quantification of PCNA-positive signals (E) by the stereological measurement of numerical density (Nv). Nv values (columns) represent mean ± SEM. Kruskall–Wallis; Dunn's post hoc test. **P < 0.001. Color images are available online.
FIG. 7.
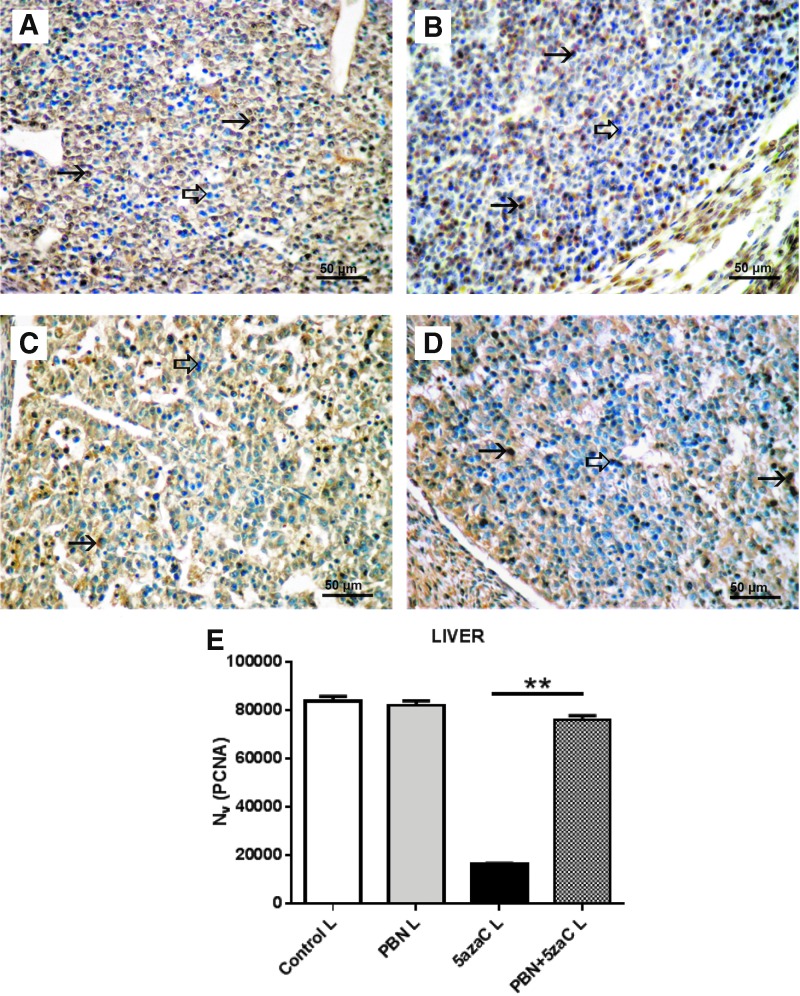
PCNA expression (arrows) in liver of embryos derived from dams previously treated with 5azaC and/or PBN, detected by immunohistochemistry: (A) control; (B) PBN-treated; (C) PBN-pretreated and 5azaC-treated; (D) 5azaC-treated cartilage. DAB, hematoxylin counterstain. Thick arrows point to internal negative controls. Quantification of PCNA-positive signals (E) by the stereological measurement of numerical density (Nv). Nv values (columns) represent mean ± SEM. Kruskall–Wallis; Dunn's post hoc test. **P < 0.001. Color images are available online.
To investigate whether other embryonic tissues responded in the same way to the treatment of dams as the developing limb buds, proliferation was assessed also in the cartilage of the vertebrae (Fig. 6) and in the liver (Fig. 7).
Contrary to the negative effect of the 5azaC detected in limb buds, PCNA expression in cartilage of the vertebrae was highest in both groups of embryos from dams treated with 5azaC, and it was significantly lower in all PBN-treated dams and controls (Fig. 6). Therefore, pretreatment of dams with PBN did not influence high cell proliferation in the embryonic cartilage.
Similarly as in limb buds, the numerical density (Nv) of PCNA expression in the liver of embryos from dams treated with 5azaC exhibited a statistically significant lower Nv of PCNA whereas in PBN pretreatment it did not differ from the PBN-treated dams or controls. Therefore, PBN pretreatment was able to abolish the negative impact of 5azaC on liver cells proliferation (Fig. 7).
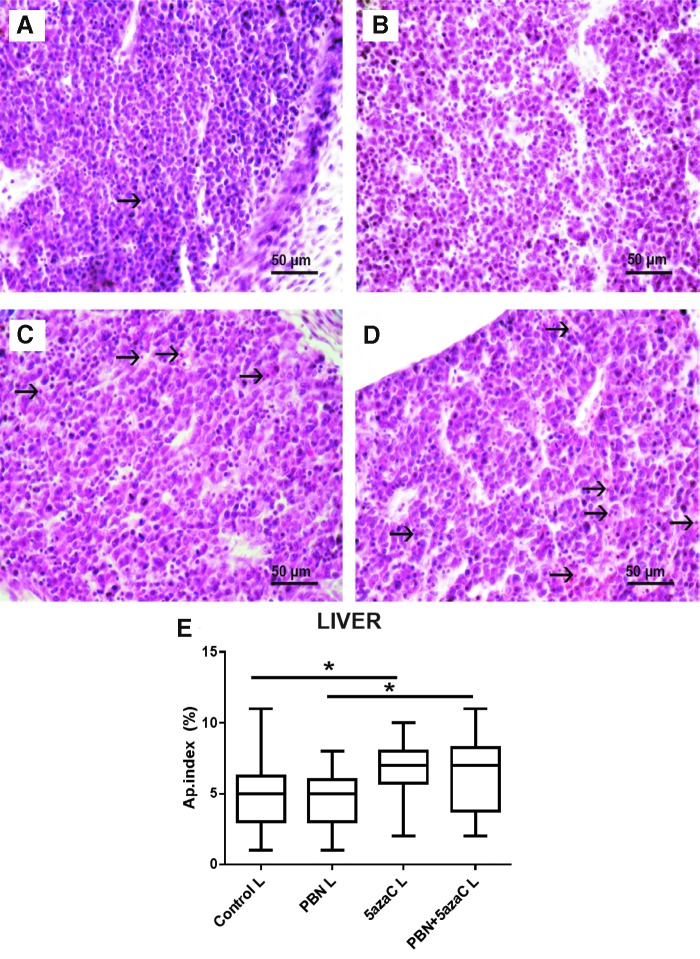
Apoptosis
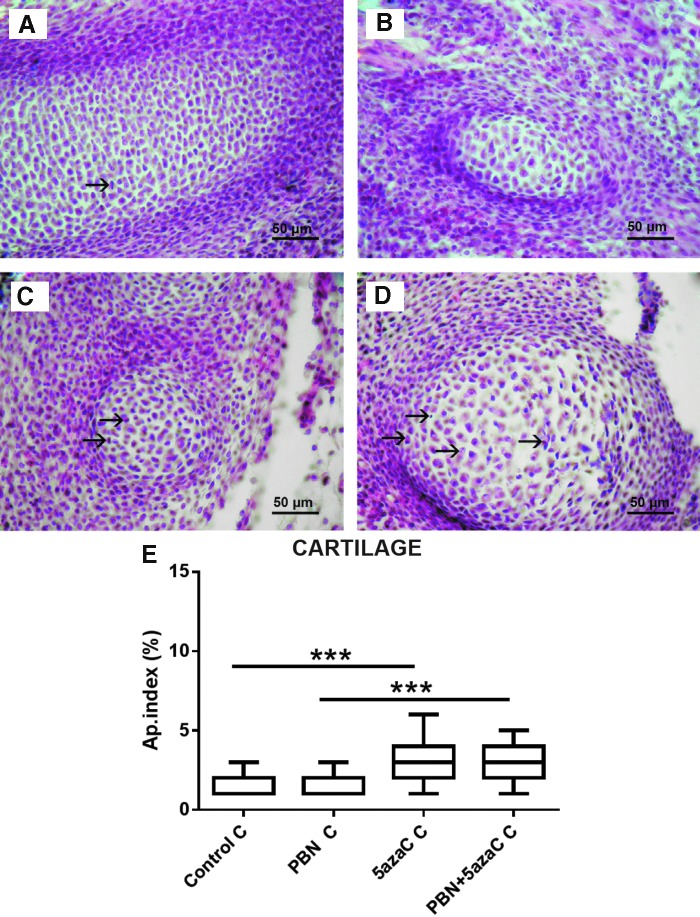
Apoptosis was detected in cartilage of the vertebrae (Fig. 8) and in embryonic liver (Fig. 9). A significantly increased apoptotic index in the embryonic liver and cartilage in both groups of embryos from dams treated with 5azaC in comparison to PBN treatment was determined (Figs. 8 and 9). Therefore, PBN pretreatment influenced the high level of apoptosis neither in the liver nor in the well-differentiated embryonic cartilage.
FIG. 8.
Apoptosis in the cartilage of embryos derived from dams, previously treated with 5azaC and/or PBN. Apoptotic figures (arrows) in (A) control; (B) PBN-treated; (C) PBN-pretreated and 5azaC-treated; (D) 5azaC-treated cartilage. Hematoxylin–eosin. Apoptotic index in embryonic cartilage (E). Boxplot shows apoptotic index distribution (box denotes median, and whiskers denote the smallest and the largest observation). Kruskall–Wallis ANOVA; post hoc Dunn's test. ***P < 0.0001. Color images are available online.
FIG. 9.
Apoptosis in liver tissue of embryos derived from dams, previously treated with 5azaC and/or PBN. Apoptotic figures (arrows) in (A) control; (B) PBN-treated; (C) PBN-pretreated and 5azaC-treated; (D) 5azaC-treated liver tissue. Hematoxylin–eosin. Apoptotic index in embryonic liver (E). Boxplot shows apoptotic index distribution (box denotes median, and whiskers denote the smallest and the largest observation). Kruskall–Wallis ANOVA; post hoc Dunn's test. *P < 0.05. Color images are available online.
Oxidative stress
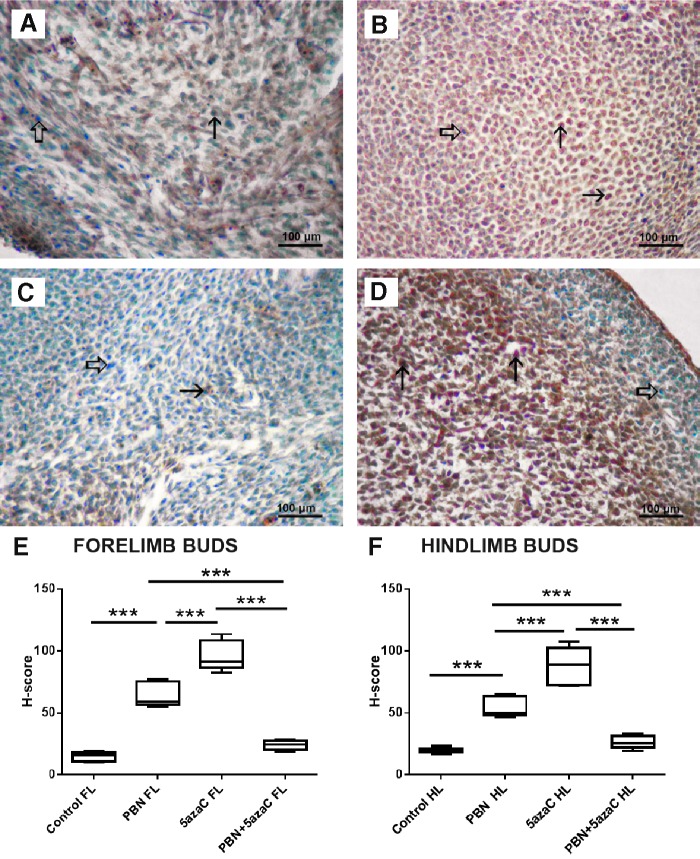
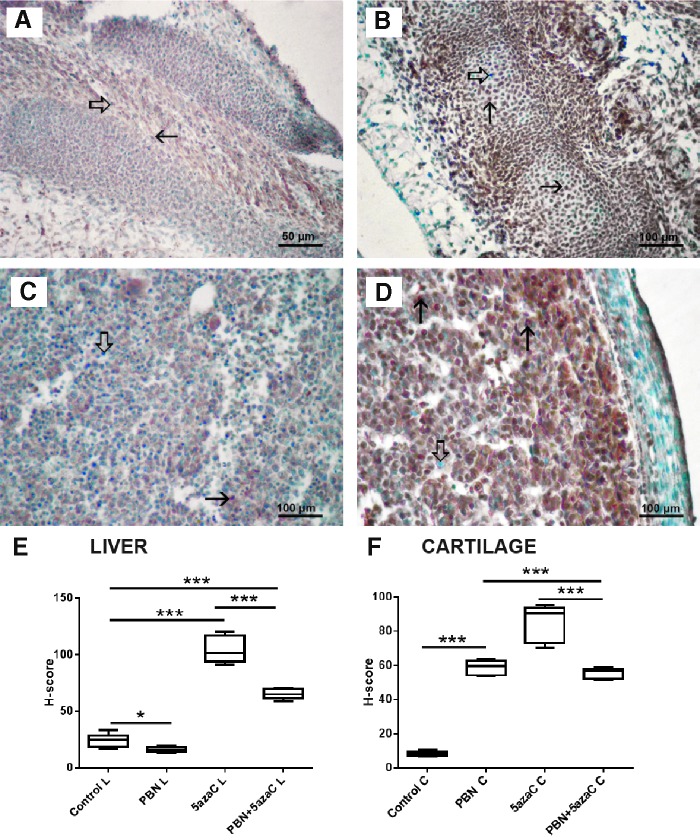
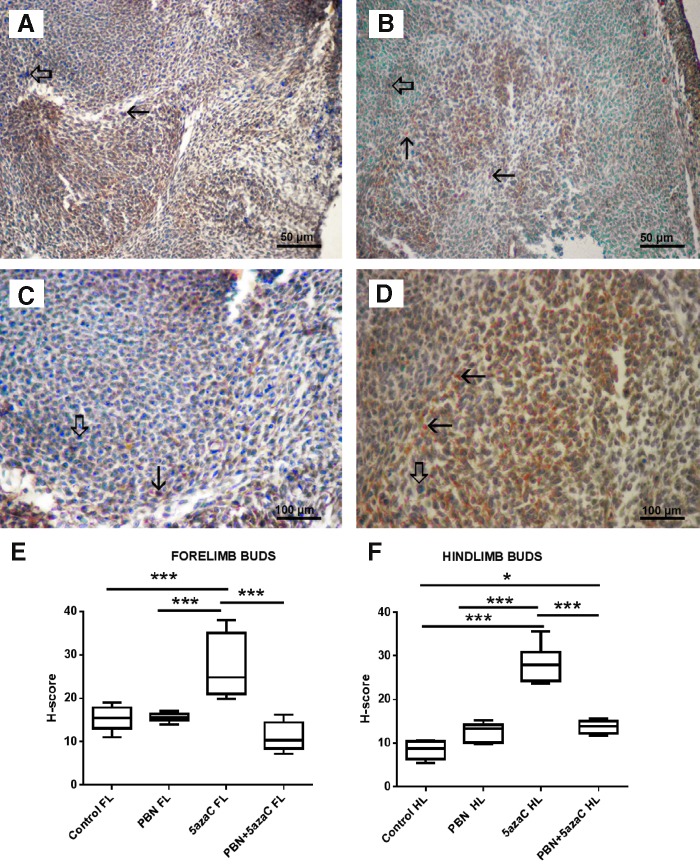
To determine whether oxidative stress took place in tissues of embryos after treatment of gestating dams with 5azaC with or without PBN pretreatment, we quantified expression of the nuclear oxidative stress-related marker 8-OHdG (Figs. 10 and 11) and the cytoplasmic oxidative stress marker nitrotyrosine (Figs. 12 and 13) in fore- and hindlimbs, cartilage, and liver.
FIG. 10.
8-OHdG expression (arrows) in fore- and hindlimb buds of embryos derived from dams previously treated with 5azaC and/or PBN, detected by immunohistochemistry: (A) forelimb bud, control; (B) forelimb bud, 5azaC; (C) hindlimb bud, control; (D) hindlimb bud, 5azaC. DAB, hematoxylin counterstain. Thick arrows point to internal negative control signal. Boxplot shows H-score distribution in fore- (E) and hindlimb buds (F) (box denotes median, and whiskers denote the smallest and the largest observation). ANOVA, Holm–Sidak's multiple-comparison post hoc test. ***P < 0.0001. Color images are available online.
FIG. 11.
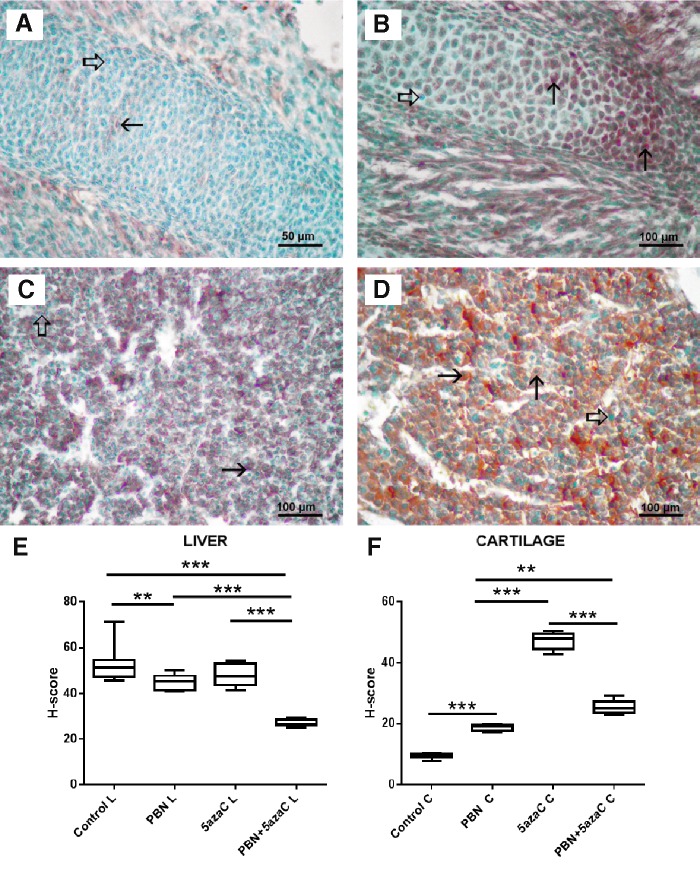
8-OHdG expression (arrows) in liver and cartilage buds of embryos derived from dams previously treated with 5azaC and/or PBN, detected by immunohistochemistry: (A) cartilage, control; (B) cartilage, 5azaC; (C) liver, control; (D) liver, 5azaC. DAB, hematoxylin counterstain. Thick arrows point to internal negative control signal. H-score in cartilage (E) and liver (F). Boxplot shows apoptotic index distribution (box denotes median, and whiskers denote the smallest and the largest observation). ANOVA, Holm–Sidak's multiple-comparison post hoc test. ***P < 0.0001. Color images are available online.
FIG. 12.
Nitrotyrosine expression (arrows) in fore- and hindlimb buds of embryos derived from dams previously treated with 5azaC and/or PBN, detected by immunohistochemistry: (A) forelimb bud, control; (B) forelimb bud, 5azaC; (C) hindlimb bud, control; (D) hindlimb bud, 5azaC. DAB, hematoxylin counterstain. Thick arrows point to internal negative control signal. H-score in fore- (E) and hindlimb buds (F). Boxplot shows apoptotic index distribution (box denotes median, and whiskers denote the smallest and the largest observation). ANOVA, Holm–Sidak's multiple-comparison post hoc test. *P < 0.05, and ***P < 0.0001. Color images are available online.
FIG. 13.
Nitrotyrosine expression (arrows) in cartilage and liver of embryos derived from dams previously treated with 5azaC and/or PBN, detected by immunohistochemistry: (A) cartilage, control; (B) cartilage, 5azaC; (C) liver, control; (D) liver, 5azaC. DAB, hematoxylin counterstain. Thick arrows point to internal negative control signal. H-score in fore- (E) and hindlimb buds (F). Boxplot shows apoptotic index distribution (box denotes median, and whiskers denote the smallest and the largest observation). ANOVA, Holm–Sidak's multiple-comparison post hoc test. **P < 0.001, ***P < 0.0001. Color images are available online.
5azaC treatment, in comparison to controls, always significantly elevated 8-OHdG expression level. Pretreatment with PBN always significantly diminished the 8-OHdG expression level in comparison to 5azaC (Figs. 10 and 11).
5azaC treatment, in comparison to controls, always (except in the liver) significantly elevated the expression of nitrotyrosine whereas PBN pretreatment always diminished nitrotyrosine expression level in comparison to 5azaC (Figs. 12 and 13).
Therefore, pretreatment of dams with PBN almost always diminished ROS activity elevated by 5azaC in tissues of their offspring.
Interestingly, PBN alone significantly diminished nitrotyrosine expression in the liver and elevated its expression in the cartilage in comparison to controls. Moreover, PBN alone was able to significantly increase 8-OHdG expression in limb buds and cartilage in comparison to the controls, although that was significantly less than with 5azaC and it diminished 8-OHdG expression in the liver. Therefore it seems that PBN alone was able to influence ROS/RNS.
Discussion
Our results have shown for the first time that pretreatment with a nitrone significantly ameliorated the major negative impact of the DNA hypomethylating drug 5azaC on the offspring of mothers treated only twice during gestation, because a statistically significant improvement in the survival and the gross phenotype, including overall growth and limb malformations, was detected with PBN pretreatment. We confirmed that 5azaC activated ROS, as was previously published, for example, for malignant cells [56] and that PBN diminished that activity in accordance with results obtained with other teratogens [27,28]. On the other hand, 5azaC sometimes enhanced antioxidant activity of the organism, such as in lipopolysaccharide-induced acute respiratory distress in mice [57] or during therapy of MDS by induction of the antioxidant pathway Nrf2 [58]. Therefore, the impact of 5azaC on oxidative stress probably depends on a complex biological/pathophysiological context [59,60]. That is also true for PBN that in our experiments alone sometimes even enhanced oxidative stress markers in the limb buds and cartilage, although that seemed to have only a minor impact on their ossification and neither significantly changed their overall growth nor induced major limb malformations. In comparison with our previous research where pretreatment with ASA abolished limb malformations in 46% of fetuses whereas in the rest only hindlimb oligodactyly was present [5], PBN pretreatment was less efficient despite the same dosage and timing of 5zaC application and we may suppose that the PBN concentration was suboptimal.
Global DNA methylation
We have been able to confirm the DNA hypomethylating effect of 5azaC in embryonic limb buds by pyrosequencing a widely spread global DNA methylation marker from the rat noncoding DNA [short interspersed nuclear element (SINE), rat ID element] [47]. To put that positive result in a proper context, it must be stressed that by platforms such as Illumina 450K, which were developed only for the human genome, the effects of 5azaC could not be associated with the DNA methylation levels of activated ERVs from the noncoding DNA, because the platform does not contain those annotations [39]. Treatment with 5azaC induced a statistically significant hypomethylation and pretreatment with PBN significantly normalized global DNA methylation only in developmentally less advanced embryonic hindlimb buds [42], although the improvement of malformations in fetuses was similar in both types of limbs. The reason for such improvement may be the known robustness to perturbations of the network structure that regulates early limb development [55]. Sometimes, 5azaC did not change DNA methylation level, although it changed developmental parameters such as proliferation [61]. Indeed, a recent review discusses many reasons why DNA methylation is still not a marker predictive of response to DNA hypomethylating therapy [62].
Cell proliferation
We now found that DNA hypomethylating agent 5azaC applied twice during pregnancy was able to diminish cell proliferation in limb buds of embryos of gestating dams treated in vivo, which confirmed our previous result on limb buds cultivated in vitro with 5azaC [63] or its activity discovered in cancer cells [64].
In contrast to the negative 5azaC activity on cell proliferation in the limb bud as an immature whole organ, we showed that proliferation of the vertebral cartilage was increased by 5azaC, similarly as in 17-day-old fetal epiglottis of the same strain of rats transplanted in vivo [65]. The 5azaC-induced hypomethylation also reversed the aged phenotype of the adult mesenchymal stem cells by enhancing cell proliferation [59].
These differences in the impact of 5azaC in various types of cells suggest that the effect of changes in DNA methylation on cell proliferation is organ and cell-type specific. In accordance to that is the recent genome-wide DNA methylation analysis by whole genome bisulfite sequencing that demonstrated no global changes or large-scale hypomethylated blocks in cell cycle phases of the human early passage primary dermal fibroblasts, although authors discuss that the situation might be different in more proliferative cells such as stem cells or cancer cell lines [66].
Importantly, although PBN pretreatment of 5azaC-treated dams neither improved proliferation in embryonic limb buds nor diminished apoptosis in the vertebral cartilage or extremely elevated apoptosis in the liver, it normalized proliferation in the liver that was significantly diminished by 5azaC. Although patients with MDS and hepatic impairment were excluded from the clinical trials on Vidaza [67,68], the FDA-approval summary on the MDS treatment with 5azaC (Vidaza) mentioned “liver function abnormalities in 16% of patients with hepatobiliary disorders and in two patients with previously diagnosed liver cirrhosis” [67,68]. Therefore, the possible beneficial activity of PBN pretreatment seems to be worth further investigation, and more so because it was found that 85% of MDS patients expressed high levels of ROS [69].
Conclusion
We discovered that the spin-trap free radical scavenger PBN significantly ameliorates the pronounced negative impact of a DNA hypomethylating drug 5azaC on the normal development of offspring during the mammalian gestation. Therefore, the mechanism of teratogenic activity of the 5azaC partially depends on activation of ROS/RNS. As PBN neither diminished apoptosis nor enhanced cell proliferation of immature tissue under the influence of 5azaC, its therapeutic impact on elevated free radical levels in MDS and related diseases may be of interest. That PBN restored proliferation in the liver may also be of interest for the treatment of hepatotoxicity and liver regeneration/transplantation strategies [44].
Acknowledgments
This work was supported by the Croatian Ministry of Science, Education and Sport under grant (no. 108-1080399-0335) “Experimental Embryonic Tumors and Development of Mammalian Embryo In Vitro and In Vivo”; University of Zagreb under supportive grants (no. 1.2.1.17, no. 1101310 BM1.22) and Scientific Center of Excellence for Reproductive and Regenerative Medicine, Republic of Croatia, and by the European Union through the European Regional Development Fund, under grant agreement no. KK.01.1.1.01.0008, project “Reproductive and Regenerative Medicine—Exploring New Platforms and Potentials.” The authors wish to thank Professor Barbara Knowles for critically reading the article, and Vedran Perić, Milan Kopač, and Mariana Dragojević for their technical assistance.
Author Disclosure Statement
No competing financial interests exist.
References
- 1. Serman L, Vlahović M, Sijan M, Bulić-Jakuš F, Serman A, Sinčić N, Matijević R, Jurić-Lekić G. and Katusic A. (2007). The impact of 5-azacytidine on placental weight, glycoprotein pattern and proliferating cell nuclear antigen expression in rat placenta. Placenta 28:803–811 [DOI] [PubMed] [Google Scholar]
- 2. Serman L, Sijan M, Kuzmić R, Sinčić N, Serman A, Bulić-Jakuš F, Vlahović M, Jurić-Lekić G. and Katušić A. (2008). Änderungen der expressionen von membranproteinen in der plazenta von ratten behandelt mit 5-azacytidine. Tierarztl Umsch 63:391–395 [Google Scholar]
- 3. Bulic-Jakus F, Vlahovic M, Juric-Lekic G, Crnek-Kunstelj V. and Serman D. (1999). Gastrulating rat embryo in a serum-free culture model: changes of development caused by teratogen 5-azacytidine. Altern Lab Anim 27:925–933 [DOI] [PubMed] [Google Scholar]
- 4. Bulic‐Jakus F, Katusic Bojanac A, Juric‐Lekic G, Vlahovic M. and Sincic N. (2016). Teratoma: from spontaneous tumors to the pluripotency/malignancy assay. WIREs Dev Biol 5:186–209 [DOI] [PubMed] [Google Scholar]
- 5. Sinčić N, Vlahović M, Bulić-Jakuš F, Serman L. and Serman D. (2002). Acetylsalycilic acid protects rat embryos from teratogenic effects of 5-azacytidine. Period Biol 104:441–444 [Google Scholar]
- 6. Vlahović M, Sinčić N, Bulić-Jakuš F, Serman L. and Serman D. (2001). Acetylsalicylic acid seems to diminish teratogenic effect of 5-azacytidine. Period Biol 103:343–346 [Google Scholar]
- 7. Liu L. and Wells PG. (1994). In vivo phenytoin-initiated oxidative damage to proteins and lipids in murine maternal hepatic and embryonic tissue organelles: potential molecular targets of chemical teratogenesis. Toxicol Appl Pharmacol 125:247–255 [DOI] [PubMed] [Google Scholar]
- 8. Williams HE, Claybourn M. and Green AR. (2007). Investigating the free radical trapping ability of nxy-059, s-pbn and pbn. Free Radic Res 41:1047–1052 [DOI] [PubMed] [Google Scholar]
- 9. Wells PG, Bhuller Y, Chen CS, Jeng W, Kasapinovic S, Kennedya JC, Kim PM, Laposa RR, McCallum GP, et al. (2005). Molecular and biochemical mechanisms in teratogenesis involving reactive oxygen species. Toxicol Appl Pharmacol 207:354–366 [DOI] [PubMed] [Google Scholar]
- 10. Miller-Pinsler L, Sharma A. and Wells PG. (2016). Enhanced NADPH oxidases and reactive oxygen species in the mechanism of methanol-initiated protein oxidation and embryopathies in vivo and in embryo culture. Arch Toxicol 90:717–730 [DOI] [PubMed] [Google Scholar]
- 11. Janzen EG, Stronks HJ, Nutter Jr DE, Davis ER, Blount HN, Poyer JL. and McCay PB. (1980). Spin trapping azidyl (N3•), cyanatyl (OCN•), cyanyl (•CN) radicals and chlorine atom (Cl•). Can J Chem 58:1596–1598 [Google Scholar]
- 12. Villamena FA, Das A. and Nash KM. (2012). Potential implication of the chemical properties and bioactivity of nitrone spin traps for therapeutics. Future Med Chem 4:1171–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Floyd RA, Chandru HK, He T. and Towner R. (2011). Anti-cancer activity of nitrones and observations on mechanism of action. Anticancer Agents Med Chem 11:373–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sawant PD. (2018). Potential use of spin traps to control ROS in antipollution cosmetics—a review. Cosmetics 5:8 [Google Scholar]
- 15. Choi SH. and Choi CH. (2015). Noise-induced neural degeneration and therapeutic effect of antioxidant drugs. J Audiol Otol 19:111–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Munoz FM, Gao R, Tian Y, Henstenburg BA, Barrett JE. and Hu H. (2017). Neuronal P2X7 receptor-induced reactive oxygen species production contributes to nociceptive behavior in mice. Sci Rep 7:3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stiles M, Moiseyev GP, Budda ML, Linens A, Brush RS, Qi H, White GL, Wolf RF, Ma JX, et al. (2015). PBN (phenyl-N-tert-butylnitrone)-derivatives are effective in slowing the visual cycle and rhodopsin regeneration and in protecting the retina from light-induced damage. PLoS One 22:10 :e0145305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Floyd RA. (2009). Serendipitous findings while researching oxygen free radicals. Free Radic Biol Med 46:1004–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Croitoru M, Petkes H, Fülöp I, Cotârlan R, Şerban O, Dogaru T, Gâz Florea ő, Tőkés B. and Majdik C. (2015). Nitrones: not only extraordinary spin traps, but also good nitric oxide sources in vivo. Acta Pharm 65, 4, 413–426 [DOI] [PubMed] [Google Scholar]
- 20. Viera L, Ye YZ. and Beckman JS. (2000). Anti-nitrotyrosine antibodies for immunohistochemistry. In: Septic Shock Methods and Protocols Vol 36. Evans TJ, ed. Methods in Molecular Medicine™, Humana Press, pp 159–170 [DOI] [PubMed] [Google Scholar]
- 21. Cadenas S. and Barja G. (1999). Resveratrol, melatonin, vitamin E, and PBN protect against renal oxidative DNA damage induced by the kidney carcinogen KBrO3. Free Radic Biol Med 26:1531–1537 [DOI] [PubMed] [Google Scholar]
- 22. Nishida N, Arizumi T, Takita M, Kitai S, Yada N, Hagiwara S, Inoue T, Minami Y, Ueshima K, Sakurai T. and Kudo M. (2013). Reactive oxygen species induce epigenetic instability through the formation of 8-hydroxydeoxyguanosine in human hepatocarcinogenesis. Dig Dis 31:459–466 [DOI] [PubMed] [Google Scholar]
- 23. Klaunig JE, Kamendulis LM. (2004). The role of oxidative stress in carcinogenesis. Annu Rev Pharmacol Toxicol 44:239–267 [DOI] [PubMed] [Google Scholar]
- 24. Serman A, Vlahović M, Serman L. and Bulić-Jakus F. (2006). DNA methylation as a regulatory mechanism for gene expression in mammals. Coll Antropol 30:665–671 [PubMed] [Google Scholar]
- 25. Vlahović M, Bulić-Jakus F, Jurić-Lekić G, Fučić A, Marić and D Šerman S. (1999). Changes in the placenta and in the rat embryo caused by the demethylating agent 5-azacytidine. Int J Dev Biol 43:843–846 [PubMed] [Google Scholar]
- 26. Buljubašić R, Buljubašić M, Katušić Bojanac A, Ulamec M, Vlahović M, Ježek D, Bulić-Jakuš F. and Sinčić N. (2018). Epigenetics and testicular germ cell tumors. Gene 661:22–33 [DOI] [PubMed] [Google Scholar]
- 27. Menezo YJ, Silvestris E, Dale B. and Elder K. (2016). Oxidative stress and alterations in DNA methylation: two sides of the same coin in reproduction. Reprod Biomed Online 33:668–683 [DOI] [PubMed] [Google Scholar]
- 28. Franco R, Schoneveld O, Georgakilas AG. and Panayiotidis MI. (2008). Oxidative stress, DNA methylation and carcinogenesis. Cancer Lett 266:6–11 [DOI] [PubMed] [Google Scholar]
- 29. Ziech D, Franco R, Pappa A. and Panayiotidis MI. (2011). Reactive oxygen species (ROS)—induced genetic and epigenetic alterations in human carcinogenesis. Mutation Res 711:167–173 [DOI] [PubMed] [Google Scholar]
- 30. Gonçalves AC, Cortesão E, Oliveiros B, Alves V, Espadana AI, Rito L, Magalhães E, Pereira S, Pereira A, et al. (2016). Oxidative stress levels are correlated with P15 and P16 gene promoter methylation in myelodysplastic syndrome patients. Clin Exp Med 16:333–343 [DOI] [PubMed] [Google Scholar]
- 31. Anderson OS, Sant KE. and Dolinoy DC. (2012). Nutrition and epigenetics: an interplay of dietary methyl donors, one-carbon metabolism and DNA methylation. J Nutr Biochem 23:853–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C. and Zhang Y. (2011). Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333:1300–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Taylor SE, Li YH, Smeriglio P, Rath M, Wong WH. and Bhutani N. (2016). Stable 5-hydroxymethylcytosine (5hmC) acquisition marks gene activation during chondrogenic differentiation. J Bone Miner Res 31:524–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moore LD, Le T. and Fan G. (2013). DNA methylation and its basic function. Neuropsychopharmacology 38:23–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hernandez-Vargas H, Sincic N, Ouzounova M. and Herceg Z. (2009). Epigenetic signatures in stem cells and cancer stem cells. Epigenomics 1:261–280 [DOI] [PubMed] [Google Scholar]
- 36. Sincic N. and Herceg Z. (2011). DNA methylation and cancer: searching for ghosts and angels above the genes. Curr Opin Oncol 23:69–76 [DOI] [PubMed] [Google Scholar]
- 37. Voso MT, Lo-Coco F. and Fianchi L. (2015). Epigenetic therapy of myelodysplastic syndromes and acute myeloid leukemia. Curr Opin Oncol 27:532–539 [DOI] [PubMed] [Google Scholar]
- 38. Jones PA, Issa JP. and Baylin S. (2016). Targeting the cancer epigenome for therapy. Nat Rev Genet 17:630–641 [DOI] [PubMed] [Google Scholar]
- 39. Tobiasson M, Abdulkadir H, Lennartsson A, Katayama S, Marabita F, De Paepe A, Karimi M, Krjutskov K, Einarsdottir E, et al. (2017). Comprehensive mapping of the effects of azacitidine on DNA methylation, repressive/permissive histone marks and gene expression in primary cells from patients with MDS and MDS-related disease. Oncotarget 8:28812–28825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Teperek-Tkacz M, Pasque V, Gentsch G. and Ferguson-Smith AC. (2011). Epigenetic reprogramming—is deamination key to active DNA demethylation? Reproduction 142: 621–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kohli RM, Zhang Y. (2013). TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502:472–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Witschi E. (1962). Development: Rat. In: Growth Including Reproduction and Morphological Development. Altman PL, Dittmer DS, eds. Federation of American Societies for Experimental Biology, Washington, DC, pp 304–314 [Google Scholar]
- 43. Kim M, Kim J, Park SR, Park DY, Kim YJ, Choi BH. and Min BH. (2016). Comparison of fetal cartilage-derived progenitor cells isolated at different developmental stages in a rat model. Dev Growth Differ 58:167–179 [DOI] [PubMed] [Google Scholar]
- 44. Gruppuso P, Sanders A. and Sanders JA. (2016). Regulation of liver development: implications for liver biology across the lifespan. Mol Endocrinol 56:115–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Inouye M. (1976). Differential staining of cartilage and bone in fetal mouse skeleton by alcian blue and alizarin red S. Cong Anom 16:171–173 [Google Scholar]
- 46. Vaissière T, Hung RJ, Zaridze D, Moukeria A, Cuenin C, Fasolo V, Ferro G, Paliwal A, Hainaut P, et al. (2009). Quantitative analysis of DNA methylation profiles in lung cancer identifies aberrant DNA methylation of specific genes and its association with gender and cancer risk factors. Cancer Res 69:243–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hwan-Hee K, Jung-Hoon P, Kyoung-Sin J. and Suman L. (2007). Determining the global DNA methylation status of rat according to the identifier repetitive elements. Electrophoresis 28:3854–3861 [DOI] [PubMed] [Google Scholar]
- 48. McCarty Jr KS, Miller LS, Cox EB, Konrath J. and Sr McCarty KS. (1985). Estrogen receptor analyses. Correlation of biochemical and immunohistochemical methods using monoclonal antireceptor antibodies. Arch Pathol Lab Med 109:716–721 [PubMed] [Google Scholar]
- 49. Parris TZ, Aziz L, Kovács A, Hajizadeh S, Nemes S, Semaan M, Chen CY, Karlsson P. and Helou K. (2014). Clinical relevance of breast cancer-related genes as potential biomarkers for oral squamous cell carcinoma. BMC Cancer 14:324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mazières J, Brugger W, Cappuzzo F, Middel P, Frosch A, Bara I, Klingelschmitt G. and Klughammer B. (2013). Evaluation of EGFR protein expression by immunohistochemistry using H-score and the magnification rule: re-analysis of the SATURN study. Lung Cancer 82:231–237 [DOI] [PubMed] [Google Scholar]
- 51. Weibel ER. (1979). Stereological methods. In: Vol. 1: Practical Methods for Biological Morphometry. Weibel ER, ed. Academic Press, London [Google Scholar]
- 52. Kalisnik M, ed. Basics of Stereology (In Croatian). (1985). Stereoloska sekcija Zveze drustev anatomov Jugoslavije, SFRJ, Ljubljana [Google Scholar]
- 53. Losa M, Barzaghi RL, Mortini P, Franzin A, Mangili F, Terreni MR. and Giovanelli M. (2000). Determination of the proliferation and apoptotic index in adrenocorticotropin-secreting pituitary tumors: comparison between micro- and macroadenomas. Am J Pathol 156:245–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. BLUEPR consortium INT. (2016). Quantitative comparison of DNA methylation assays for biomarker development and clinical applications. Nat Biotechnol 34:726–737 [DOI] [PubMed] [Google Scholar]
- 55. Petit F, Sears KE. and Nadav A. (2017). Limb development: a paradigm of gene regulation. Nat Rev Gen 18: 245–258 [DOI] [PubMed] [Google Scholar]
- 56. Tian E, Tang H, Xu R, Liu C, Deng H, Wang Q. (2013). Azacytidine induces necrosis of multiple myeloma cells through oxidative stress. Proteome Sci 11:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Huang X, Kong G, Li Y, Zhu W, Xu H, Zhang X, Li J, Wang L, Zhang Z, et al. (2016). Decitabine and 5-azacitidine both alleviate LPS induced ARDS through anti-inflammatory/antioxidant activity and protection of glycocalyx and inhibition of MAPK pathways in mice. Biomed Pharmacother 84:447–453 [DOI] [PubMed] [Google Scholar]
- 58. Nguyen LX, Troadec E, Kalvala A, Hoang DH, Viola D, Forman SJ, Marcucci G. and Pullarkat V. (2017). Inhibition of ROS-Induced Nrf2 antioxidant pathway activation may explain synergy between venetoclax and hypomethylating agents against acute myeloid leukemia. Blood 130:1367 [Google Scholar]
- 59. Kornicka K, Marycz K, Marezdziak M, Tomaszewski KA. and Nicpon J. (2017). The effects of the DNA methyltranfserases inhibitor 5-Azacitidine on ageing, oxidative stress and DNA methylation of adipose derived stem cells. J Cell Mol Med 21:387–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Miller L, Shapiro AM, Cheng J. and Wells PG. (2013). The free radical spin trapping agent phenylbutylnitrone reduces fetal brain DNA oxidation and postnatal cognitive deficits caused by in utero exposure to a non-structurally teratogenic dose of ethanol: a role for oxidative stress. Free Radic Biol Med 60:223–232 [DOI] [PubMed] [Google Scholar]
- 61. Fabijanovic D, ASerman, Jezic M, Katusic A, Sincic N, Curkovic-Perica M, Bulic-Jakus F, Vlahovic M, Juric-Lekic G. and Serman L. (2014). Impact of 5-azacytidine on rat decidual cell proliferation. Int J Exp Pathol 95:238–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Voso MT, Santini V, Fabiani E, Fianchi L, Criscuolo M, Falconi G, Guidi F, Hohaus S. and Leone G. (2014). Why methylation is not a marker predictive of response to hypomethylating agents. Haematologica 99:613–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mužić V, Katušić Bojanac A, Jurić-Lekić G, Himelreich M, Tupek K, Šerman L, Marn N, Sinčić N, Vlahović M. and Bulić-Jakuš F. (2013). Epigenetic drug 5-azacytidine impairs proliferation of rat limb buds in an organotypic model-system in vitro. Croat Med J 54:489–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Aykutluğ Ö, Develi S, Ozler M. and Korkmaz A. (2015). Effect of azacytidine and melatonin in prostate cancer cells. Acta Physiol (Oxf) 215:33 [Google Scholar]
- 65. Marinovic-Kulisic S, Juric-Lekic G, Vikic-Topic M, Lokosek V, Radujkovic V, Bulic-Jakus F, Katusic A, Vlahovic M, Serman L. and Sincic N. (2011). 5-azacytidine enhances proliferation in transplanted rat fetal epiglottis. Front Biosci (Elite Ed) 3:581–590 [DOI] [PubMed] [Google Scholar]
- 66. Vandiver AR, Idrizi A, Rizzardi L, Feinberg AP. and Hansen KD. (2015). DNA methylation is stable during replication and cell cycle arrest. Sci Rep 5:17911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. https://livertox.nih.gov/Azacitidine.htm (accessed September8, 2018).
- 68. Kaminskas E, Farrell AT, Wang YC, Sridhara R. and Pazdur R. (2005). FDA drug approval summary: azacitidine (5-azacytidine, Vidaza™) for injectable suspension. Oncologist 10:176–182 [DOI] [PubMed] [Google Scholar]
- 69. Saigo K, Takenokuchi M, Hiramatsu Y, Tada H, Hishita T, Takata M, Misawa M, Imoto S. and Imashuku S. (2011). Oxidative stress levels in myelodysplastic syndrome patients: their relationship to serum ferritin and haemoglobin values. J Int Med Res 39:1941–1945 [DOI] [PubMed] [Google Scholar]