Abstract
Objective
To study the experimental influences to the measurement of cartilage thickness by magnetic resonance imaging (MRI).
Design
The complete thicknesses of healthy and trypsin-degraded cartilage were measured at high-resolution MRI under different conditions, using two intensity-based imaging sequences (ultra-short echo [UTE] and multislice-multiecho [MSME]) and 3 quantitative relaxation imaging sequences (T1, T2, and T1ρ). Other variables included different orientations in the magnet, 2 soaking solutions (saline and phosphate buffered saline [PBS]), and external loading.
Results
With cartilage soaked in saline, UTE and T1 methods yielded complete and consistent measurement of cartilage thickness, while the thickness measurement by T2, T1ρ, and MSME methods were orientation dependent. The effect of external loading on cartilage thickness is also sequence and orientation dependent. All variations in cartilage thickness in MRI could be eliminated with the use of a 100 mM PBS or imaged by UTE sequence.
Conclusions
The appearance of articular cartilage and the measurement accuracy of cartilage thickness in MRI can be influenced by a number of experimental factors in ex vivo MRI, from the use of various pulse sequences and soaking solutions to the health of the tissue. T2-based imaging sequence, both proton-intensity sequence and quantitative relaxation sequence, similarly produced the largest variations. With adequate resolution, the accurate measurement of whole cartilage tissue in clinical MRI could be utilized to detect differences between healthy and osteoarthritic cartilage after compression.
Keywords: MRI, cartilage, imaging sequence, loading, magic angle
Introduction
Articular cartilage is a thin layer of connective tissue that coats the end of long bones in a diarthrodial joint. The majority of cartilage is extracellular, which has mainly water, collagen fibers, and negatively charged glycosaminoglycans (GAG).1 The collagen fibers in cartilage commonly have different orientations in different depths of the tissue: parallel to the tissue at the articular surface (the superficial zone, SZ), randomly oriented in a layer deeper to the surface (the transitional zone, TZ), and perpendicular to the cartilage surface in the deep region (the radial zone, RZ).2-4 The GAG molecules are trapped inside the collagen matrix and interact with the water to generate a swelling pressure, which plays a key role in the viscoelastic properties of articular cartilage.5-9 High GAG content and an intact collagen architecture are essential for the mechanical functions of healthy cartilage and joints; while a loss in GAG would result in poor mechanical response and could be identified as an early sign in cartilage degradation.10,11
Magnetic resonance imaging (MRI) has been used extensively to evaluate cartilage degradation.12-20 In particular, T2 relaxation time can be quantified and made sensitive to the fibril orientations and water content.3,17 On T2 maps and, depending on echo time, T2-weighted images, cartilage could have a laminar appearance when the collagen fibers in RZ are parallel with the external magnetic field B0, but appears homogeneous, when oriented to the magic angle (~55° to B0), due to the minimization of the dipolar interaction.21 In addition to T2 relaxation, T1ρ relaxation has also been used for investigation of early cartilage degradation.17,22-25 In contrast, T1 relaxation is isotropic to the fibril orientation and has nearly uniform values over the tissue depth.3 All relaxation parameters in cartilage have been shown to be sensitive to the external loading of cartilage.26-29
Since the thickness of cartilage is an important factor in joint health in clinical MRI,30 this project investigated the influences of a number of experimental factors associated with the measurement of cartilage thickness. These factors included the external compression,31 2 proton-density imaging sequences (ultra-short echo [UTE], multislice-multiecho [MSME]), 3 quantitative relaxation imaging sequences (T1, T2, and T1ρ),24 2 soaking solutions (saline and phosphate buffered saline [PBS]),32 both healthy and trypsin-degraded cartilage,30 and different specimen orientations to the magnetic field.33,34 Each of these experimental factors, except for soaking in PBS, has an equivalent situation in clinical MRI of human joint. We hypothesized that appearance of articular cartilage and the measurement accuracy of cartilage thickness can be influenced significantly by a number of experimental factors in MRI.
Methods
Solutions of Saline and PBS
Solutions of saline and PBS were prepared in-house with 276 g of sodium phosphate monobasic (monohydrate) (Sigma, St. Louis, MO) dissolved in deionized water. After adjusting the pH value to 7.3 by NaOH (Sigma, St. Louis, MO), the volume of the phosphate buffer was finalized to 1 L. Then 9 g of NaCl was added to 50 mL of the phosphate buffer and diluted to 1 L using deionized water as a physiologic concentration. The final PBS solution contained a phosphate concentration of 100 mM with the pH corresponding to ~7.4.35 A saline solution was also prepared with 154 mM NaCl dissolved in per liter of deionized water to match the physiologic condition.
Specimen Preparation
Humeral heads were harvested shortly after the sacrifice of mature and healthy canines that were used for an unrelated research, which were approved by the institutional animal care and use committee (IACUC). A total of 12 cartilage-bone specimens were harvested from 4 humeral heads; each specimen was about 3.5 × 2.5 × 6 mm in size and contained the full-thickness cartilage still attached to the underlying bone. These 12 specimens were divided into 2 groups: native (n = 3) and degraded (n = 9). The native specimens were soaked in physiological saline with 1% protease inhibitor (Sigma, St. Louis, MO). The degraded specimens were soaked in 10 µg/mL trypsin solution (Sigma, St. Louis, MO) for more than 8 hours for the digestion of GAG. These degraded specimens were then soaked in saline with 1% protease inhibitor to remove any residual trypsin.24 This process can remove about 85% to 90% GAG in tissue, which has been documented in our previous study.26-29 None of the specimens were ever frozen.
Among the 12 specimens, 3 native specimens and 3 degraded specimens were used for the quantitative MRI relaxation experiments, where each specimen was imaged 3 times using the same sequence but under 3 different loading conditions (preloaded, loaded [~25% strain], after loading); and the other 6 degraded specimens were imaged by different MRI sequences (i.e., each specimen was imaged using 5 different pulse sequences).
Microscopic MRI (µMRI) Protocols
Both proton-intensity imaging sequences (UTE, proton density [MSME]) and quantitative relaxation imaging sequences (T1, T2, T1ρ) were used to image the cartilage specimens, which were without loading and loaded at about 25% strain. The tissue compression was performed with the use of a homemade unconfined loading device,26 where the strain values were determined in the intensity images by the difference in the total thickness of cartilage, which had an error about ±4%. Each specimen was allowed half an hour to reach equilibrium after loading.
All µMRI experiments were performed at room temperature on a Bruker AVANCE II 300 NMR spectrometer equipped with a 7-T/89-mm vertical-bore superconducting magnet and microimaging accessory (Bruker Instrument, Billerica, MA). A 5-mm solenoid coil was used for all µMRI experiments, which had a 90° hard pulse of 6.5 µs. The imaging experiments were carried out with an acquisition matrix of 256 × 128 (which was postreconstructed into a 256 × 256 matrix) and a slice thickness of 1 mm. The field of view (FOV) was 0.45 cm × 0.45 cm, resulting in the 2-dimensional (2D) in-plane pixel size of 17.6 µm. The repetition time (TR) was 2 seconds.
Quantitative relaxation imaging experiments were performed at different orientations with respect to the main magnetic field (0°, 55°, 90°) and followed the previously established protocols.24,25,36 Briefly, T1 mapping used an inversion-recovery magnetization-prepared sequence, with 5 inversion points (0, 0.4, 1.1, 2.2, 4.0 seconds); T2 mapping used a CPMG (Carr-Purcell-Meiboom-Gill) magnetization-prepared sequence, with 5 echo times (TEs) ranged from 2 to 140 ms (the echo times depending on the degradation, compression, and orientation of the tissue). The shortest range of 5 TEs (2, 4, 10, 20, and 50 ms) was for both native and degraded cartilage specimens with loading at a sample orientation of 0° to the B0 field; while the longest range of 5 TEs (2, 14, 36, 80, and 140 ms) was for the degraded cartilage specimens without loading at a sample orientation of 55° to the B0 field (the magic angle). The T1ρ mapping sequence used a preceding 90° hard pulse and a spin-lock pulse (the strength of the spin-lock was 1 kHz, which was calibrated by the strength of the 90º pulse). All relaxation maps were calculated by a single-component fit on a pixel-by-pixel basis using MATLAB (Mathworks, Natwick, MA). T1 experiments of both native tissues and degraded tissues before loading (preload), during loading (load), and after loading (postload recovery for 2 hours) were also acquired to study the compression recovery properties of the cartilage.
In addition to the quantitative MRI, the proton density images were produced by the commercial 2D UTE and MSME methods at various orientations with regard to the main magnetic field (0°, 30°, 55°, 90°, 125°, 150°, 180°, 210°, 235°, 270°, 305°, 330°, 360°). The echo time of UTE and MSME sequences was 200 µs and 3.0 ms, respectively. Other imaging parameters were identical to the relaxation mapping experiments.
Image and Statistical Analyses
The total cartilage thickness was measured from all images (both relaxation maps and proton-intensity maps) using a set of image intensity–based criteria in ImageJ, which is a public domain software developed at the National Institutes of Health. Specifically, the pixel location of the cartilage surface and cartilage/bone interface was visually determined in image analysis, by manually measuring the values of the image voxels. The determination of the cartilage surface was made if the cartilage intensity reached the level of the surrounding saline (in the case of uncompressed cartilage) or the background noise (in the case of compressed cartilage), as evident in Figure 1 . The determination of the cartilage-bone interface was made if the cartilage intensity reached the level of the background noise, since the calcified bone in the specimen had extremely short T2 hence appeared black in the images (cf. Fig. 1 ). The shortest difference between the 2 measurements was used as the total thickness of cartilage. Because of the high resolution in this imaging study, the pixel size can be considered as the uncertainty in the measurement of the total thickness. Consistent procedures were used for all measurements.
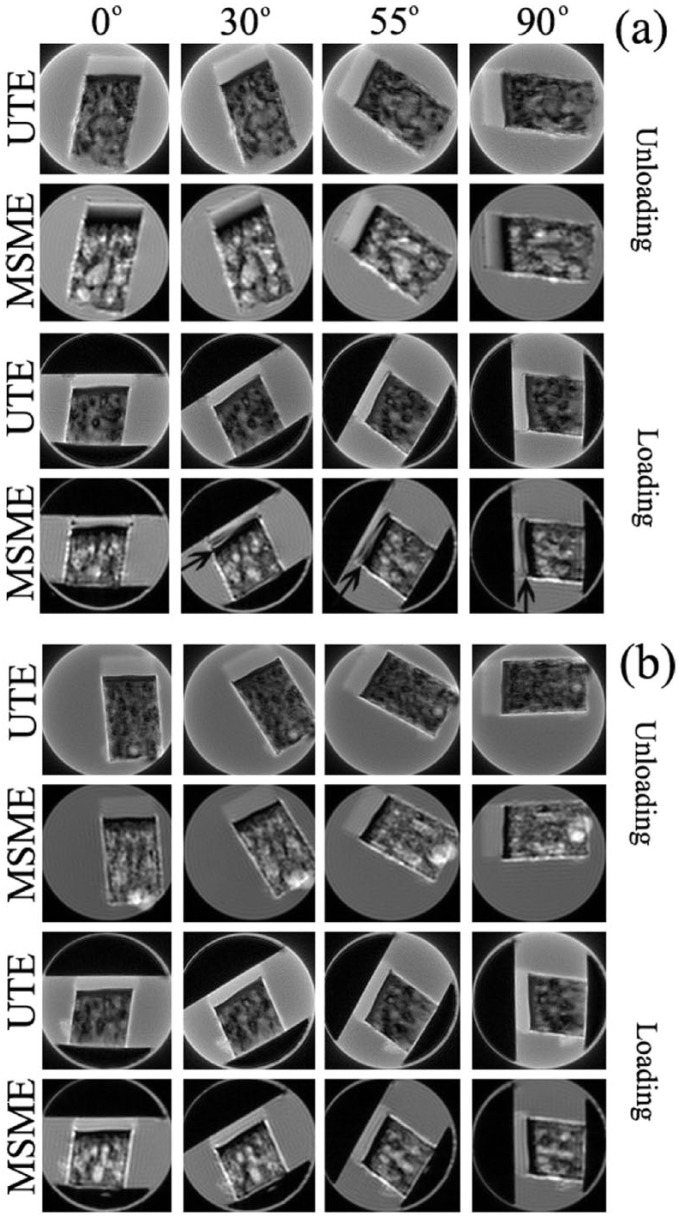
Figure 1.
The intensity images of degraded cartilage by the sequences of ultra-short echo (UTE) (echo time [TE] = 200 μs) and multislice-multiecho (MSME) (TE = 3 ms) in magnetic resonance imaging (MRI), both unloaded and loaded, in 4 of the 13 orientations with regard to B0, and soaked in saline (a) and phosphate buffered saline (PBS) (b). The arrows in the last row images in (a) point to the low-intensity layers in the deep cartilage, which appears when the degraded tissue is compressed while soaked in saline. All images were displayed with the same upper and lower limits.
The measured image data were evaluated for statistical differences using the commercial software KaleidaGraph (v. 4.0, Synergy Software, Reading, PA). One-way analysis of variance (ANOVA) test was performed to compare the T1, T2, or T1ρ values of degraded tissues after loading. The significance level between the data was set to P < 0.05.
Results
Intensity Images of Cartilage
The usual images of cartilage in MRI are the intensity images of protons, reflecting the contents of water molecule in the specimen. The appearance of one cartilage specimen in an image can be differ from one experiment to another, depending on the experimental parameters (in particular, the pulse sequence, the echo time, and the repetition time) as well as the specimen environment (in particular, the orientation of the specimen in the magnetic field, the amount of loading, and the disease).37-40 For normal (nondegraded) cartilage, a standard spin-echo-based imaging sequence (such as MSME) will produce either laminated or homogeneous tissue image, depending on the orientation of the specimen in the external magnetic field.38 For a trypsin-degraded cartilage, the tissue would appear visually similar to a saline-soaked or PBS-soaked normal cartilage, unless a careful comparison of the intensity profiles is made among the specimens. At the same image resolution and specimen orientation, minor differences could be observed in the intensity profiles in a depth-dependent manner.
The intensity images of a degraded cartilage by UTE (TE = 200 μs) and MSME (TE = 3 ms) sequences are shown in Figure 1, when the specimen was oriented at the first 4 of the 13 orientations with regard to B0, both uncompressed and compressed. (The other orientations not shown repeated the variations of the 4 selected-orientations.) In saline without compression, the MSME images of cartilage (the second row in Fig. 1a ) showed the usual laminar appearance at all orientations other than at the magic angles (55°); in contrast, the same cartilage in all UTE images (the first row in Fig 1a ) appeared homogenous and without any laminar layer regardless of the orientation. When the specimen was compressed (~25% strain), the MSME images of loaded tissue (the forth row in Fig 1a ) showed the low-intensity layer at the deep part of the tissue, which began to appear at 30° and became more evident at 55° and 90° ( Fig. 1a , arrows). These loading-induced orientation-dependent low-intensity layers were largely absent in the UTE images (the third row in Fig. 1a ). They were also absent in the images of native tissue, compressed or uncompressed (data not shown). When the same degraded specimens were immersed in the PBS solution ( Fig. 1b ), both UTE and MSME images showed an overall homogenous appearance, lacking any clear laminar appearance, with little orientation dependency for both unloaded and loaded specimens.
The intensity images in the T1, T2, and T1ρ relaxation weighted imaging experiments had a variable appearance of cartilage (data not shown), depending on the particular relaxation mechanism and the amount of weighting. When the relaxation weighting was minimal, cartilage in all intensity images looked very similar to the cartilage by MSME, since MSME is simply a spin-echo imaging sequence.
Measurements of Cartilage Thickness by Different Pulse Sequences
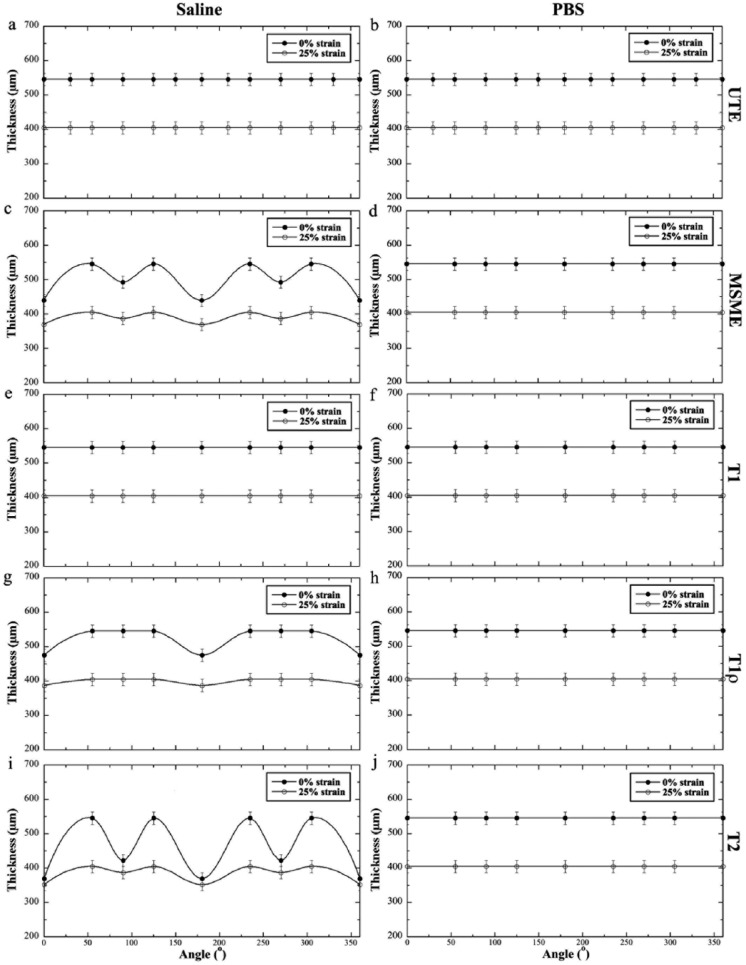
The total thickness of cartilage was measured for all images, both the proton-intensity images and the calculated relaxation maps, using a set of image intensity–based criteria, consistent for all images. For the T1, T2, and T1ρ relaxation maps, the concerned measurement is the total cartilage thickness, not the quantitative profiles (which were unrelated to the theme of this report and could be found elsewhere38). Figure 2 summarizes the quantitative measurements of the total thickness of cartilage, without and with loading (at ~25% strain) using all 5 pulse sequences (T1, T2, T1ρ, UTE, MSME) at all 13 sample orientations to the magnet field.
Figure 2.
Orientational dependent thickness of degraded cartilage, both unloaded and loaded, by different pulse sequences (T1, T2, T1ρ, ultra-short echo [UTE], and multislice-multiecho [MSME]) while soaked in saline or phosphate buffered saline (PBS). There were no variations in thickness measurement when the tissue was in PBS by any pulse sequence. The maximum variations in thickness (the difference between 55° and 0°/180°) were seen in saline by MSME and T2 sequences.
Several distinct characteristics could be identified when the specimens were soaked in PBS and saline. When the specimens were soaked in PBS, cartilage thickness measured by different pulse sequences remained constant and showed no variation at any orientation. In contrast, when the specimens were immersed in saline, only the thickness measurements from T1 and UTE sequences could give consistent results. The most striking orientational dependent variations in the thickness measurement were from the images by T2 or MSME, where the tissue thickness was always measured the thinnest at the 0° orientation. The thickness measurements from the T1ρ map showed less inconsistency, which should also be T1ρ-dispersion dependent.15,25,36 Some of the thickness variations are analyzed statistically in Table 1.
Table 1.
The Percent Thickness Variation (%) of Degraded Cartilage at 0% and 25% Strain.
| Strain |
||||||
|---|---|---|---|---|---|---|
| 0% | 25% | |||||
| Angle | 0° | 55° | 90° | 0° | 55° | 90° |
| Multilice-multiecho | 19.4 ± 3.1* | 0.0 ± 1.1 | 9.7 ± 2.4* | 8.7 ± 2.1* | 0.0 ± 1.9 | 4.3 ± 2.2* |
| T2 | 32.3 ± 3.5* | 0.0 ± 1.6 | 22.6 ± 3.7* | 13.0 ± 3.0* | 0.0 ± 2.2 | 0.0 ± 3.3 |
| T1ρ | 13.0 ± 2.7* | 0.0 ± 1.3 | 0.0 ± 2.1 | 4.3 ± 2.3* | 0.0 ± 2.0 | 0.0 ± 2.9 |
Indicates a significant difference between cartilage thickness change at certain orientation (0°, 90°) compared with the thickness change 55° (P < 0.05).
Compression Recovery of Cartilage Thickness
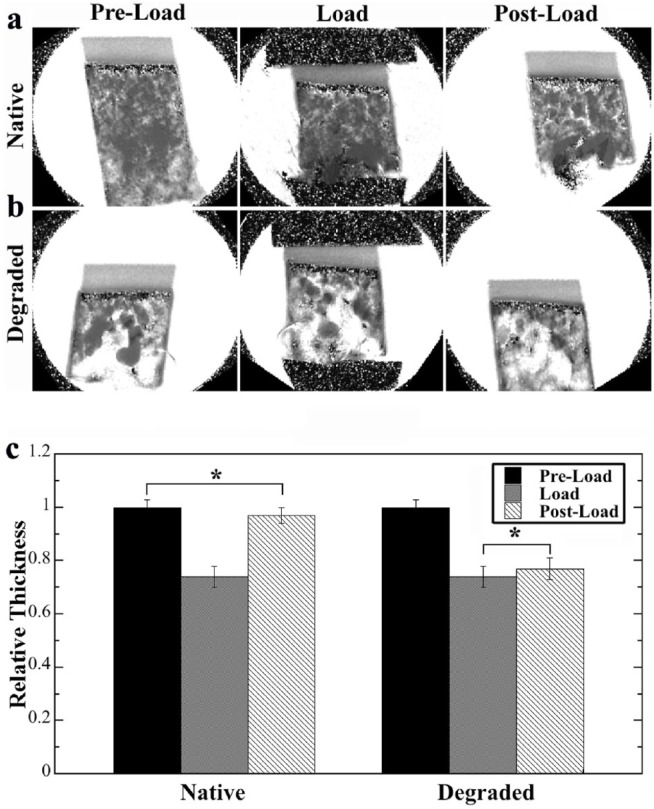
The recovery of the compressed cartilage was measured for both native and degraded cartilage specimens using the T1 mapping, where each specimen was measured 3 times: before loading (preload), while loaded (load), and after loading (resting for 2 hours in PBS solution). The results are summarized in Figure 3 . The images showed that both native and degraded tissues in PBS appeared to be homogeneous regardless of compression. The native specimens showed a complete recovery of the tissue thickness 2 hours postloading ( Fig. 3c left), where no significant thickness difference was found between preload and postload states. In contrast, the degraded specimens failed to recover from the compression—there was no significant difference between being loaded and post-load states after 2 hours of recovery ( Fig. 3c right).
Figure 3.
Magnetic resonance imaging T1 mapping of the compression recovery of native and degraded cartilage in 3 stages: preload, while loaded, 2-hour postloading: (a) native cartilage, (b) degraded cartilage, (c) the relative thickness among the 3 stages of compression recovery. For native tissue, the cartilage thickness showed full recovery after 2 hours postloading, with no significant difference in the thickness between the preload and postload. In contrast, the degraded tissue showed no signs of tissue recovery after 2-hour recovery (no significant difference between load and postload). *Indicates no significant thickness difference between 2 cartilage situations.
Discussion
Cartilage thickness is an important measure in diagnostic MRI of osteoarthritis. In the past, we have carried out repeated measures of cartilage thickness using various types of microscopic imaging techniques (MRI, optical imaging), which produced consistent results.2-4 This study concerned the measurement errors in the total cartilage thickness in MRI, in both proton-intensity images and relaxation maps. We showed the measurements could have complex variations, caused by the use of different sequences and different soaking solutions, and presence of external loading. These variations (i.e., measurement errors) are both orientation-dependent as well as loading dependent.
The Origins of Cartilage Thickness Variations by Different Pulse Sequences
The origin of the variation in cartilage thickness in MRI centers on the nature of the different pulse sequences and the zonal structure of the extracellular matrices in articular cartilage. Biologically, a layer of calcified cartilage provides the interface between the noncalcified cartilage and the subchondral bone plate. In clinical MRI, because of the large voxel size, this interface is not visible and likely grouped together with the bone plate. In high-resolution µMRI that uses the spin-echo based imaging sequences (non-UTE sequences in this study), the deepest noncalcified cartilage and the calcified cartilage appear low in signal, similar to background (because of their short T2), but occupy a few pixels in the high-resolution images. Hence the images in Fig. 1 have a black line or layer below the visible cartilage. The use of the UTE sequence enables a better visualization of these missing tissues and provides a more accurate measurement of the total cartilage thickness. As evident in this study, the current TE of 200 µs in the UTE experiments still cannot obtain signal from these tissues. Further development in the MRI technology for even shorter echo time could enable the study of the deepest noncalcified cartilage as well as the calcified cartilage, in particular, their roles in the disease development and during external loading.
From quantitative sequences of T2, T1, and T1ρ, it is well known that the cartilage T2 map has the most depth- and orientation-dependent variations due to the complex influence of dipolar interaction in articular cartilage.3,21,41 Several imaging parameters such as the echo time and signal-to-noise ratio also impact the measurement of the cartilage thickness. In contrast, the cartilage T1 map has the least variations due to the lack of sensitivity of T1 to the low-frequency molecular motion in cartilage.3 By comparison, cartilage appearance in the T1ρ sequence has the most complicated variations due to the T1ρ dispersion, which could be both depth and orientation dependent.24,36 Consequently, the total thickness of cartilage measured from the quantitative relaxation maps differ and the relaxation-weighted intensity images obtained by the sequences may show similar depth- and orientation-dependent appearances.
For the proton-intensity images, the striking difference between the MSME sequence and the UTE sequence illustrates the role of an experimental parameter, the echo time in MRI of cartilage. The MSME sequence is essentially a standard spin-echo imaging sequence, with a nominal echo time of several milliseconds. This echo time is sufficiently long to enable a depth-dependent T2 contrast in cartilage, which plays a similar role to the individual imaging by the T2 sequence. Hence the thickness variation in the MSME sequence was similar to the T2-weighted sequence, which had a maximum thickness variation of 32.3% between the thicknesses at 0° and 55°. In contrast, the most important feature of the UTE imaging sequence is the ultra-short echo time, which can be as short as a fraction of milliseconds (200 μs in our study). The dipolar influence in the UTE sequence is hence at minimal, which essentially made most tissues (including shorter T2 tissues) visible in imaging and resulted in a more consistent measurement of cartilage thickness. It is also clear that a 200 µm TE in our current UTE protocol is still long for the deepest noncalcified cartilage and the calcified cartilage, which therefore appear dark.
The Origin of Cartilage Thickness Variations by Saline and PBS
The variations of cartilage thickness in MRI also depended on the solution in which the tissue specimen was immersed—visible in saline and invisible in PBS. It was noticed in 1960s that different salt solutions could change the exchange rate between water molecules.42,43 For example, ammonium ions and phosphate ions were found to be the most effective salts for proton exchange, while some salts such as sodium chloride (NaCl) were not effective. The mechanism of proton exchange in tendon and cartilage had been studied when the tissue was soaked in different phosphate salts.25,32,44 The results in this project demonstrate the important role of the tissue soaking solution in the measurement of relaxation times (T2 and T1ρ). The use of PBS can eliminate the variations in cartilage thickness in all 5 pulse sequences tested in this study ( Fig. 2 ), which resulted a consistent measurement of cartilage thickness and overall visualization of all cartilage tissues. This result suggests the potential development of a decontrast agent in MRI, which increases the rate of proton exchange between the water molecules, thus reducing the apparent correlation time of proton pairs undergoing dipolar interaction. Such decontrast agent could enable the simultaneous visualization of all tissues in human joint with the use of common imaging sequences.
The Origins of Cartilage Thickness Variations due to External Loading
Since articular cartilage is a load-bearing tissue, it valuable to image cartilage while it is being loaded in MRI.31 In previous studies, both the intensity images of cartilage and the profiles of cartilage have been found to be very sensitive to the amount of external loading in MRI by different pulse sequences.9,28,29,35 In addition, the visualization and appearance of compressed cartilage can also be dependent on the status of cartilage health, where healthy and diseased cartilage appeared different.29 These loading dependencies of cartilage appearance and thickness are understandable, since MRI is sensitive to the molecular motions in the tissue, where the reduction of GAG in diseased cartilage changes the motional anisotropies of water molecules. In this project, the recovery of cartilage thickness was differentiated for the first time between healthy and diseased cartilage. The knowledge of this difference could be beneficial in clinical MRI of human joint, where a healthy cartilage can recover its thickness after the compression (due to walking or exercise) while a degraded cartilage cannot. Since the compressive modulus of cartilage is at least one order of magnitude smaller than the bone, the measured difference in tissue thicknesses under external loading should be attributed to the changes of cartilage thickness.
Limitations in Cartilage Thickness Measurement
Comparative studies need to be carried out to determine the actual thickness of articular cartilage, ideally done under in vivo situation with as little tissue processing as possible. The reasoning behind this is that any in vitro study, where biological tissue goes through a sequence of tissue processing procedures, could contain its own measurement errors, for example, the tissue shrinkage in a regular histology process. MRI as a noninvasive imaging tool should in principle result in more accurate measurement of tissue thickness, since it measures the native (i.e., unprocessed) tissue. At a 2D in-plane pixel size of 17.6 µm, this µMRI study likely has one of the most precise measurements of cartilage thickness. Despite of the high spatial resolution, the previous sections in this discussion illustrated various causes of measurement uncertainty in MRI. The cartilage thickness measured in this project is consistent with the results from several optical imaging reports that used nearly identical tissue.2-4 However, one must keep in mind that any experiment has measurement its own drawbacks, regardless of the imaging technique.
In conclusion, we have demonstrated that the measurement accuracy of the total cartilage thickness in MRI can be influenced by a number of experimental factors in ex vivo MRI, from the use of pulse sequence and soaking solutions to the health of the tissue. In particular, the T2-based sequences, both proton-intensity imaging sequence and quantitative imaging sequence, produced similar errors in the thickness measurement. In comparison, the sequences of UTE and T1 are capable of measuring precise tissue thickness without any orientation limitation, which should have technical implication in clinical MRI. Furthermore, the use of different soaking solutions, especially the PBS solution, in the ex vivo cartilage imaging could offer an additional avenue for better cartilage imaging. For example, it may be possible to develop a PBS-like solution as a contrast agent to increase the visualization of deep cartilage, which would be safer than the current MRI contrast agent based on heavy metal ions (e.g., gadolinium ions). At the present time, deep cartilage as well as the interface between cartilage and subchondral bone are mostly invisible in clinical MRI. With adequate resolution30,45 and better sequences, the visualization of the whole cartilage tissue could be utilized to detect localized lesions in deep cartilage, which may exist in trauma-induced osteoarthritis.
Footnotes
Acknowledgments and Funding: The authors thank Drs. Cliff Les and Hani Sabbah (Henry Ford Hospital, Detroit) for providing the canine specimens, Dr. Dieter Gross (Bruker Instrument, Rheinstetten, Germany) for helping the UTE sequence, and Ms. Carol Searight (Department of Physics, Oakland University) for editorial comments on the manuscript. The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Yang Xia is grateful to the National Institutes of Health for the R01 grants (AR052353 and AR069047).
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical Approval: Ethical approval for this study was obtained from the institutional animal care and use committee (IACUC), with an assurance number A3163-01.
Animal Welfare: National, and/or institutional guidelines for humane animal treatment and complied with relevant legislation.
References
- 1. Maroudas A. Biophysical chemistry of cartilaginous tissues with special reference to solute and fluid transport. Biorheology. 1975;12(3-4):233-48. [DOI] [PubMed] [Google Scholar]
- 2. Lehner KB, Rechl HP, Gmeinwieser JK, Heuck AF, Lukas HP, Kohl HP. Structure, function, and degeneration of bovine hyaline cartilage: assessment with MR imaging in vitro. Radiology. 1989;170(2):495-9. [DOI] [PubMed] [Google Scholar]
- 3. Xia Y. Relaxation anisotropy in cartilage by NMR microscopy (µMRI) at 14-µm resolution. Magn Reson Med. 1998;39(6):941-9. [DOI] [PubMed] [Google Scholar]
- 4. Xia Y, Moody JB, Burton-Wurster N, Lust G. Quantitative in situ correlation between microscopic MRI and polarized light microscopy studies of articular cartilage. Osteoarthritis Cartilage. 2001;9(5):393-406. [DOI] [PubMed] [Google Scholar]
- 5. Mow VC, Kuei SC, Lai WM, Armstrong CG. Biphasic creep and stress relaxation of articular cartilage in compression? Theory and experiments. J Biomech Eng. 1980;102(1):73-84. [DOI] [PubMed] [Google Scholar]
- 6. Xia Y, Zheng S, Bidthanapally A. Depth-dependent profiles of glycosaminoglycans in articular cartilage by microMRI and histochemistry. J Magn Reson Imaging. 2008;28(1):151-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moger CJ, Arkill KP, Barrett R, Bleuet P, Ellis RE, Green EM, et al. Cartilage collagen matrix reorientation and displacement in response to surface loading. J Biomech Eng. 2009;131(3):031008. [DOI] [PubMed] [Google Scholar]
- 8. Mayerhoefer ME, Welsch GH, Mamisch TC, Kainberger F, Weber M, Nemec S, et al. The in vivo effects of unloading and compression on T1-Gd (dGEMRIC) relaxation times in healthy articular knee cartilage at 3.0 tesla. Eur Radiol. 2010;20(2):443-9. [DOI] [PubMed] [Google Scholar]
- 9. Xia Y, Wang N, Lee J, Badar F. Strain-dependent T1 relaxation profiles in articular cartilage by MRI at microscopic resolutions. Magn Reson Med. 2011;65(6):1733-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Maroudas A, Venn M. Chemical composition and swelling of normal and osteoarthrotic femoral head cartilage. II. Swelling. Ann Rheum Dis. 1977;36(5):399-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rubenstein JD, Kim JK, Henkelman RM. Effects of compression and recovery on bovine articular cartilage: appearance on MR images. Radiology. 1996;201(3):843-50. [DOI] [PubMed] [Google Scholar]
- 12. Lesperance LM, Gray ML, Burstein D. Determination of fixed charge density in cartilage using nuclear magnetic resonance. J Orthop Res. 1992;10(1):1-13. [DOI] [PubMed] [Google Scholar]
- 13. Bashir A, Gray ML, Burstein D. Gd-DTPA2- as a measure of cartilage degradation. Magn Reson Med. 1996;36(5):665-73. [DOI] [PubMed] [Google Scholar]
- 14. Nieminen MT, Rieppo J, Toyras J, Hakumaki JM, Silvennoinen J, Hyttinen MM, et al. T2 relaxation reveals spatial collagen architecture in articular cartilage: a comparative quantitative MRI and polarized light microscopic study. Magn Reson Med. 2001;46(3):487-93. [DOI] [PubMed] [Google Scholar]
- 15. Akella SV, Regatte RR, Wheaton AJ, Borthakur A, Reddy R. Reduction of residual dipolar interaction in cartilage by spin-lock technique. Magn Reson Med. 2004;52(5):1103-9. [DOI] [PubMed] [Google Scholar]
- 16. Nag D, Liney GP, Gillespie P, Sherman KP. Quantification of T2 relaxation changes in articular cartilage with in situ mechanical loading of the knee. J Magn Reson Imaging. 2004;19(3):317-22. [DOI] [PubMed] [Google Scholar]
- 17. Li X, Benjamin Ma C, Link TM, Castillo DD, Blumenkrantz G, Lozano J, et al. In vivo T1ρ and T2 mapping of articular cartilage in osteoarthritis of the knee using 3 T MRI. Osteoarthritis Cartilage. 2007;15(7):789-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Regatte RR, Schweitzer ME. Ultra-high-field MRI of the musculoskeletal system at 7.0 T. J Magn Reson Imaging. 2007;25(2):262-9. [DOI] [PubMed] [Google Scholar]
- 19. Keenan KE, Besier TF, Pauly JM, Smith R L, Delp SL, Beaupre GS, et al. T1ρ dispersion in articular cartilage: relationship to material properties and macromolecular content. Cartilage. 2015;6(2):113-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Matzat SJ, McWalter EJ, Kogan F, Chen W, Gold GE. T2 relaxation time quantitation differs between pulse sequences in articular cartilage. J Magn Reson Imaging. 2015;42(1):105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xia Y, Farquhar T, Burton-Wurster N, Lust G. Origin of cartilage laminae in MRI. J Magn Reson Imaging. 1997;7(5):887-94. [DOI] [PubMed] [Google Scholar]
- 22. Du J, Carl M, Diaz E, Takahashi A, Han E, Szeverenyi NM, et al. Ultrashort TE T1ρ (UTE T1ρ) imaging of the Achilles tendon and meniscus. Magn Reson Med. 2010;64(3):834-42. [DOI] [PubMed] [Google Scholar]
- 23. Souza RB, Baum T, Wu S, Feeley BT, Kadel N, Li X, et al. Effects of unloading on knee articular cartilage T1ρ and T2 magnetic resonance imaging relaxation times: a case series. J Orthop Sports Phys Ther. 2012;42(6):511-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang N, Xia Y. Depth and orientational dependencies of MRI T2 and T1ρ sensitivities towards trypsin degradation and Gd-DTPA(2-) presence in articular cartilage at microscopic resolution. Magn Reson Imaging. 2012;30(3):361-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang N, Xia Y. Experimental issues in the measurement of multi-component relaxation times in articular cartilage by microscopic MRI. J Magn Reson. 2013;235:15-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Alhadlaq HA, Xia Y. The structural adaptations in compressed articular cartilage by microscopic MRI (µMRI) T2 anisotropy. Osteoarthritis Cartilage. 2004;12(11):887-94. [DOI] [PubMed] [Google Scholar]
- 27. Alhadlaq HA, Xia Y. Modifications of orientational dependence of microscopic magnetic resonance imaging T(2) anisotropy in compressed articular cartilage. J Magn Reson Imaging. 2005;22(5):665-73. [DOI] [PubMed] [Google Scholar]
- 28. Wang N, Badar F, Xia Y. MRI properties of a unique hypo-intense layer in degraded articular cartilage. Phys Med Biol. 2015;60(22):8709-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang N, Kahn D, Badar F, Xia Y. Molecular origin of a loading-induced black layer in the deep region of articular cartilage at the magic angle. J Magn Reson Imaging. 2015;41(5):1281-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chang G, Xia D, Sherman O, Strauss E, Jazrawi L, Recht MP, et al. High resolution morphologic imaging and T2 mapping of cartilage at 7 tesla: comparison of cartilage repair patients and healthy controls. MAGMA. 2013;26(6):539-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Greaves LL, Gilbart MK, Yung A, Kozlowski P, Wilson DR. Deformation and recovery of cartilage in the intact hip under physiological loads using 7 T MRI. J Biomech. 2009;42:349-54. [DOI] [PubMed] [Google Scholar]
- 32. Zheng S, Xia Y. Changes in proton dynamics in articular cartilage caused by phosphate salts and fixation solutions. Cartilage. 2010;1(1):55-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Garnova N, Gründerb W, Thörmera G, Trampelc R, Turnerc R, Kahna T, et al. In vivo MRI analysis of depth-dependent ultrastructure in human knee cartilage at 7 T. NMR Biomed. 2013;26(11):1412-9. [DOI] [PubMed] [Google Scholar]
- 34. Nemeth A, Di Marco L, Boutitie F, Sdika M, Grenier D, Rabilloud M, et al. Reproducibility of in vivo magnetic resonance imaging T1ρ and T2 relaxation time measurements of hip cartilage at 3.0 T in healthy volunteers. J Magn Reson Imaging. Epub 2017. June 26. doi: 10.1002/jmri.25799. [DOI] [PubMed] [Google Scholar]
- 35. Wang N, Chopin E, Xia Y. The effects of mechanical loading and gadolinium concentration on the change of T1 and quantification of glycosaminoglycans in articular cartilage by microscopic MRI. Phys Med Biol. 2013;58(13):4535-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang N, Xia Y. Orientational dependent sensitivities of T2 and T1ρ towards trypsin degradation and Gd-DTPA2- presence in bovine nasal cartilage. MAGMA. 2012;25(4):297-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xia Y. Contrast in NMR imaging and microscopy. Concepts Magn Reson. 1996;8(3):205-25. [Google Scholar]
- 38. Xia Y. MRI of articular cartilage at microscopic resolution. Bone Joint Res. 2013;2(1):9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zheng S, Xia Y. The influence of specimen and experimental conditions on NMR and MRI of cartilage. In: Xia Y, Momot KI, editors. Biophysics and biochemistry of cartilage by NMR and MRI. Cambridge, England: The Royal Society of Chemistry; 2017. p. 347-72. [Google Scholar]
- 40. Wang N, Xia Y. Loading-induced changes in cartilage studied by NMR and MRI. In: Xia, Momot KI, editors. Biophysics and biochemistry of cartilage by NMR and MRI. Cambridge, England: The Royal Society of Chemistry; 2017. p. 433-54. [Google Scholar]
- 41. Xia Y, Moody JB, Alhadlaq H. Orientational dependence of T2 relaxation in articular cartilage: a microscopic MRI (µMRI) study. Magn Reson Med. 2002;48(3):460-9. [DOI] [PubMed] [Google Scholar]
- 42. Luz Z, Meiboom S. Rate and mechanism of proton exchange in aqueous solutions of phosphate buffer. J Am Chem Soc. 1964;86(22):4764-6. [Google Scholar]
- 43. Berendsen HJ, Migchelsen C. Hydration structure of fibrous macromolecules. Ann N Y Acad Sci. 1965;125(A2):365-79. [Google Scholar]
- 44. Zheng S, Xia Y. Effect of phosphate electrolyte buffer on the dynamics of water in tendon and cartilage. NMR Biomed. 2009;22(2):158-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xia Y. Resolution ‘scaling law’ in MRI of articular cartilage. Osteoarthritis Cartilage. 2007;15(4):363-5. [DOI] [PMC free article] [PubMed] [Google Scholar]