Abstract
Background/Purpose
To search for a transmissible agent involved in lupus pathogenesis, we investigated the faecal microbiota of patients with systemic lupus erythematosus (SLE) for candidate pathobiont(s) and evaluated them for special relationships with host immunity.
Methods
In a cross-sectional discovery cohort, matched blood and faecal samples from 61 female patients with SLE were obtained. Faecal 16 S rRNA analyses were performed, and sera profiled for antibacterial and autoantibody responses, with findings validated in two independent lupus cohorts.
Results
Compared with controls, the microbiome in patients with SLE showed decreased species richness diversity, with reductions in taxonomic complexity most pronounced in those with high SLE disease activity index (SLEDAI). Notably, patients with SLE had an overall 5-fold greater representation of Ruminococcus gnavus (RG) of the Lachnospiraceae family, and individual communities also displayed reciprocal contractions of a species with putative protective properties. Gut RG abundance correlated with serum antibodies to only 1/8 RG strains tested. Anti-RG antibodies correlated directly with SLEDAI score and antinative DNA levels, but inversely with C3 and C4. These antibodies were primarily against antigen(s) in an RG strain-restricted pool of cell wall lipoglycans. Novel structural features of these purified lipoglycans were characterised by mass spectrometry and NMR. Highest levels of serum anti-RG strain-restricted antibodies were detected in those with active nephritis (including Class III and IV) in the discovery cohort, with findings validated in two independent cohorts.
Conclusion
These findings suggest a novel paradigm in which specific strains of a gut commensal may contribute to the immune pathogenesis of lupus nephritis.
Keywords: commensal, microbiome, autoantibodies, pathobiont, gut barrier
Key messages.
What is already known about this subject?
Patients with lupus have been reported to have restricted gut microbiome diversity and general patterns of dysbiosis but few patients with active disease have been previously characterised.
What does this study add?
Patients with lupus were found to have characteristic patterns of gut microbiome dysbiosis that directly paralleled disease activity.
Patients with systemic lupus erythematosus commonly displayed signs of impaired gut barriers that may result in immune exposure to gut commensal bacteria.
Intestinal expansions of Ruminococcus gnavus (RG) were directly proportional to overall disease activity and most pronounced in those with lupus nephritis. Lupus faecal samples displayed increases in sIgA-coated RG bacteria. In three independent cohorts, patients with lupus nephritis displayed elevated serum IgG predominantly to RG strain-restricted cell wall lipoglycan antigens.
How might this impact on clinical practice or future developments?
Identification of R. gnavus as a candidate pathobiont opens new areas of investigation of the mechanistic basis by which these outgrowths may affect the overall pathogenesis of lupus and the immune complex-mediated pathogenesis of lupus nephritis.
These findings may lead to the development of bioassay(s) with prognostic value for the risk of lupus nephritis.
Introduction
Systemic lupus erythematosus (SLE) is an inflammatory autoimmune disease with hallmarks of B-cell abnormalities, circulating autoantibodies to nuclear antigens and immune-complex formation.1 The heterogeneity of disease presentation and organ involvement in different individuals, and the variability of disease activity from remission to exacerbations and progression, all contribute to clinical challenges for diagnosis and effective management. Indeed, such heterogeneity suggests that SLE may not represent a single disease but rather several.
Serum autoantibodies to native DNA are a specific diagnostic criterion for SLE,2 and a prognostic factor for the development of lupus nephritis (LN) that affects 30%–60% of patients.3 However, the earliest reports of antibody responses to nucleic acids/nucleoproteins were documented in association with clinically apparent bacterial infections.4–6 Yet two decades later autoantibodies to nuclear antigens were recognised to be a common feature of SLE.7–9 Indeed, some DNA-reactive autoantibodies are directly nephritogenic in animal models.10 Conversely, only ~20% of the IgG eluted from lupus kidneys is DNA-reactive,11 suggesting that other antibody reactivities may also contribute to the pathogenesis of LN.12
While a transmissible agent has long been suspected in lupus pathogenesis, only recently has suitable technology become available that enable in-depth consideration of the potential roles of the immense dynamic communities of commensal microorganisms that coevolved with our species. The largest microbiome community resides within our gut, where these microbes play essential roles, including for the early priming of our immune systems13 and subsequent immune regulation.14 Mounting evidence has implicated imbalances within these gut microbial communities, also termed dysbioses, in the autoimmune pathogenesis of several diseases: inflammatory bowel disease (IBD), type 1 diabetes, multiple sclerosis and rheumatoid arthritis.15 Yet, there have only been a few reports on the human lupus microbiome, in small cohorts that have included only a few active patients.16–18
In the present study, we investigated the gut microbial communities in a cross-sectional cohort of 61 female patients with lupus heterogeneous for ethnicity/race, disease activity and organ involvement and immune profiles. Key findings were then evaluated in two independent lupus cohorts.
Methods
Ethics statement
The study was conducted according to the Declaration of Helsinki. Before study inclusion, written informed consent, approved by the NYU IRB, was obtained from all subjects for research use and publication of their data.
Study design
Patients were consecutively recruited from the NYU Langone Medical Center and Bellevue Hospital. All patients fulfilled the American College of Rheumatology Criteria for the diagnosis of SLE.2 Further details on trial-specific inclusion and exclusion criteria, clinical evaluations, sampling and 16S rRNA amplicon analyses are described in the supplementary data. Sections on immunoassays, as well as culture of bacterial strains and purification of R. gnavus lipoglycans (LGs) are also provided (see supplementary materials).
annrheumdis-2018-214856supp001.docx (53.4KB, docx)
Results
Patients with lupus have distinctive patterns of dysbiosis that parallel disease activity
In the discovery phase of our studies, we analysed the faecal microbiota of 61 female patients with lupus in a cross-sectional urban cohort and 17 female healthy controls (online supplementary table 1). Our patients displayed great heterogeneity in their organ involvement, and in disease activity from clinical remission to highly active, that was scored using the composite SLE disease activity index (SLEDAI).19
annrheumdis-2018-214856supp002.xlsx (19.7KB, xlsx)
To define the richness of alpha diversity in gut communities, Chao1 estimates of the total number of operational taxonomic units20 demonstrated that the overall biodiversity of the intestinal microbiota in patients with lupus was significantly restricted compared with unaffected female adult controls (Mann-Whitney, p=0.038) (figure 1A). Among patients with lupus, there was a numerical trend towards an inverse correlation of Chao1 estimates of alpha diversity with SLEDAI score (Mann-Whitney, p=0.08) (data not shown). Moreover, after empirically considering all possible cut-off values to dichotomise patients based on SLEDAI score, those with high disease activity (defined as SLEDAI ≥8) displayed significantly restricted microbiota diversity compared with controls (Mann-Whitney, p=0.003) (figure 1A).
Figure 1.
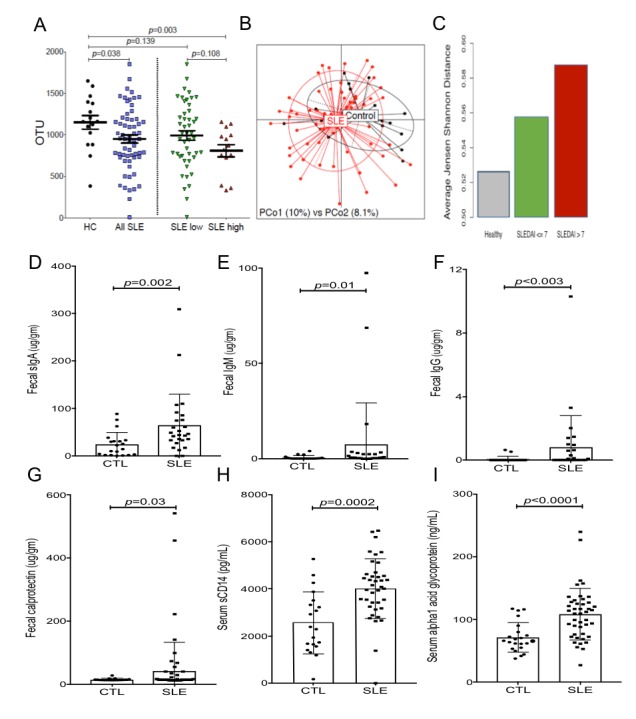
Patients with SLE have altered faecal communities of commensal taxa, elevated faecal immunoglobulin levels and biomarker evidence of altered gut barrier function. (A) Chao1 estimates of alpha diversity represent the total expected number of OTUs that represent quasi-species identified from 16S rRNA amplicon sequencing surveys, with larger values representing higher diversity. Patients with SLE (n=61) have less diverse faecal microbiota than healthy controls (n=17), indicating that SLE commonly have intestinal dysbioses. At far right, the distribution of Chao1 values in patients with SLE with high activity (ie, SLEDAI≥8) was significantly contracted compared with healthy adult controls (HC), with a trend towards more limited diversity in SLE with high disease activity when compared with SLE with low disease activity. We used a cut-point that was associated with greatest statistically significant differences. The SLE low disease activity group had a range of 0–7 SLEDAI scores (n=47), and SLE high disease activity group had scores of 8–18 (n=14). p values were based on the Mann-Whitney test. (B) PcoA showed that the beta diversity within bacterial communities in the faecal microbiomes in healthy controls was less different than the communities from patients with SLE low disease activity (PERMANOVA, p=0.02). Furthermore, healthy subjects were more like one another than were patients with SLE. (C) Results indicate average within group beta diversity differences in the three different groups, with comparisons for Control-SLEDAIhi-SLEDAIlow, p=0.002. The binary comparison, control vs SLE had p=0.02. Both comparisons were done using PERMANOVA test. Examinations of faecal extract demonstrated patients with SLE commonly display: (D) elevated faecal sIgA levels, (E) elevated faecal IgM levels, (F) elevated faecal IgG levels, and (G) elevated faecal calprotectin. Patients with SLE also displayed: (H) elevated serum sCD14 levels and (I) elevated serum α1-acid glycoprotein levels, as measured by commercial assay. Results are for the NYU cohort of adult female patients with SLE, were compared to adult female controls without inflammatory or autoimmune disease (CTL). (D–F) and (H–I) using unpaired two-tailed t test with Welch’s correction, (G) used Mann-Whitney test. Significance for p<0.05. OTUs, operational taxonomic units; PCoA, principal coordinates analysis; SLE, systemic lupus erythematosus; SLEDAI, SLE disease activity index.
The human gut microbiome is dominated by four bacterial phyla—Firmicutes, Bacteroidetes, Actinobacteria and Proteobacteria—with thousands of different species and countless strains in complex dynamic relationships within individual gut communities.21 22 To visualise community variation, we performed principal coordinates analyses that confirmed that taxonomic distributions within patients with SLE were significantly different than in controls (PERMANOVA, p=0.02) (figure 1B). When the patients with SLE were stratified by high or low disease activity, the beta diversity differences were even more pronounced (PERMANOVA, p=0.002, data not shown). Furthermore, the variability in beta diversity of healthy subjects was lower than in patients with lupus (figure 1B, C). The within group variability between communities in patients with lupus with low disease activity was also lower than that in patients with high disease activity (figure 1C). However, the severity of the detected disease-associated dysbiosis did not correlate with disease duration (online supplementary figure 1A).
annrheumdis-2018-214856supp003.pdf (1.7MB, pdf)
With 16S rRNA amplicon sequence analysis, the taxa in each community with the most highly abundant (>1%) representation at all phylogenetic levels were identified by univariate Kruskal Wallis ANOVA analysis (Table 1 and online supplementary table 2). At the species level, patients with SLE displayed a mean 5-fold overabundance of an anaerobic Gram-positive taxon in the Firmicutes phylum and Lachnospiraceae family, identified as Ruminococcus gnavus 23 (RG) (range 0.00%–10.79%, mean±SD 1.35%±2.01%) compared with controls (0.00%–1.27%, 0.25%±0.39%, Mann-Whitney, p=0.01). Strikingly, RG relative abundance correlated with lupus disease activity, as even patients with SLE with low disease activity had a mean 4-fold RG overrepresentation, while those with high disease activity had >8 fold greater RG abundance (Mann-Whitney, p=0.01) (table 1, complete data in online supplementary table 2). Compared with those without a history of renal disease, patients with a history of renal involvement displayed an even greater abundance of a RG specific amplicon sequence variant (ASV) (Mann-Whitney, p=0.04), as well as several other ASVs that included two assigned to the Veillonella genus (online supplementary table 3). In an individual community these Veillonella species were highly correlated with each other (CLR-transformed, r=0.89, p<10–16) although less strongly with RG (39% V. parvula, 43% V. dispar, p<0.005). Hence, the microbiota in patients with lupus commonly displayed concurrent expansions of both RG and Veillonella species.
Table 1.
Shifts in taxonomic abundance in SLE by disease activity
| Taxonomic level | Taxonomy | Healthy (%) | SLEDAIlow (%) | SLEDAIhigh(%) | P value |
| Class | Erysipelotrichi | 1.37 | 1.44 | 5.92 | 0.086 |
| Order | Erysipelotrichales | 1.37 | 1.44 | 5.92 | 0.086 |
| Family | Erysipelotrichaceae | 1.37 | 1.44 | 5.92 | 0.086 |
| Family | Veillonellaceae | 1.58 | 3.41 | 12.27 | 0.008 |
| Family | Ruminococcaceae | 25.66 | 11.68 | 15.11 | 0.045 |
| Genus | Blautia | 2.23 | 4.10 | 3.59 | 0.077 |
| Genus | Ruminococcus | 0.53 | 1.76 | 3.15 | 0.007 |
| Species | Ruminococcus gnavus | 0.25 | 1.19 | 2.11 | 0.013 |
| Species | Bacteroides uniformis | 2.11 | 0.87 | 0.34 | 0.014 |
| Species | Bacteroides fragilis | 1.17 | 2.41 | 2.20 | 0.081 |
Results from univariate Kruskal-Wallis ANOVA analysis of 16S rRNA abundance for all taxa across all subjects for p<0.10 and not FDR corrected. The same bacterial set is identified as Erysipelotrichi, Erysipelotrichales and Erysipelotrichaceae. SLEDAIlow patients have SLEDAI scores of less than 8. For the complete analysis, see online supplementary table 2.
ANOVA, analysis of variance; SLE, systemic lupus erythematosus; SLEDAI, SLE disease activity index.
annrheumdis-2018-214856supp004.docx (24.3KB, docx)
annrheumdis-2018-214856supp005.xlsx (13KB, xlsx)
As an alternative approach, DADA224 was used to assign ASVs (ie, quasi-species): patients with a history of renal disease displayed increased representation of ASV31 (Mann-Whitney, p<0.05) (online supplementary tables 2-4). In fact, this ASV identifies RG and has the predictive effect size of AUC=0.65 for renal involvement (see Ref. 25 for description of predictive effect size analysis protocol), although this study was not adequately powered for multiple comparison correction. Nonetheless, these findings collectively indicate that patients with LN commonly have intestinal expansions of the RG species.
The lupus microbiome is associated with altered dynamic relationships between species
Within microbiome communities, we also looked for evidence of coordinated shifts in the representation of species within individual lupus communities, and outgrowths of RG were found to be commonly associated with reciprocal reductions of the Bacteroides uniformis species (r=−0.43, Pearson correlation for CLR-transformed abundances, p<0.001). Indeed, lupus patients with the highest disease activity scores also had the lowest abundance of B. uniformis (Mann-Whitney, p=0.01) (table 1), suggesting that these species may be in dynamic reciprocal relationships. Notably, as B. uniformis conveys anti-inflammatory properties that can ameliorate metabolic abnormalities in diabetes-prone mice,26 a paucity in active lupus patients could also be relevant to understanding the drivers of autoimmune pathogenesis.
Evidence of in vivo gut recognition of RG and altered gut barrier function
To assess in vivo local recognition of specific gut taxa, we evaluated the representation of secretory IgA (sIgA)-coated faecal bacteria recovered from 15 unaffected controls and 23 patients with SLE (online supplementary figure 1B). Overall, RG bacteria were prominent among sIgA-coated subcommunities by 16S rRNA analysis, and lupus samples displayed a 2-fold greater abundance of RG among sIgA-coated faecal bacteria than healthy controls, However, in these limited surveys, the numerical differences did not attain significance (Wilcoxon rank sum, p=0.14, not significant) (online supplementary figure 1C).
In healthy individuals, an intact intestinal barrier prevents leakage of luminal contents out of, and also systemic IgG from entering into, the gut lumen. In these faecal samples, patients with lupus had 2.6-fold overall sIgA elevations vs controls (Welch test, p=0.002) (figure 1D). Furthermore, the elevations of faecal IgM in patients with lupus, as well as the increases in faecal IgG, provide additional circumstantial evidence of an impairment of the epithelial barrier in these patients (figure 1E, F).
To further search for evidence of impaired gut barriers, we measured faecal levels of calprotectin, an accepted biomarker for intestinal barrier defect.27 In these pilot studies, we foud that 12/61 patients with lupus exceeded the level in any of the tested control subjects (Mann-Whitney, p=0.03) (figure 1G). Furthermore, patients with lupus also displayed raised serum soluble CD14 (Welch test, p=0.0002) and raised α1-acid glycoprotein levels (Welch test, p<0.0001) (figure 1H, I), which in other conditions have been attributed to gut bacterial translocation.28 29 Taken together, these findings suggest that patients with lupus may at times experience impaired gut barrier function, which potentially may enable commensals, or their components, to escape the intestinal lumen.
Lupus serum IgG antibodies react with a RG2 strain-associated non-protein antigen
To investigate for a special immune relationship in patients with SLE with their candidate pathobionts, we electrophoretically separated the components in extracts of eight monocultured RG strains (online supplementary table 5) after treatment with lysozyme and endonuclease. Immunoblots revealed that many patients with lupus and healthy adult controls had IgG reactivity with antigenic bands, common to a range of RG strains (figure 2A–D), and these were protease-sensitive (figure 2E). In contrast, lupus serum antibodies were not generally reactive with commensal strains P. copri or B. thetaiotaomicron (figure 2A–C). Notably, serum IgG antibodies of lupus patients with SLEDAI activity scores of ≥8 were often highly reactive with specific conserved protease-resistant bands in the extract of the R. gnavus strain CC55_001C, which is here termed RG2 (online supplementary table 5). These conserved bands in the RG2 strain migrated with ~20–30 kDa protein markers, but such bands were not detected in the seven other RG strains tested (figure 2E).
Figure 2.
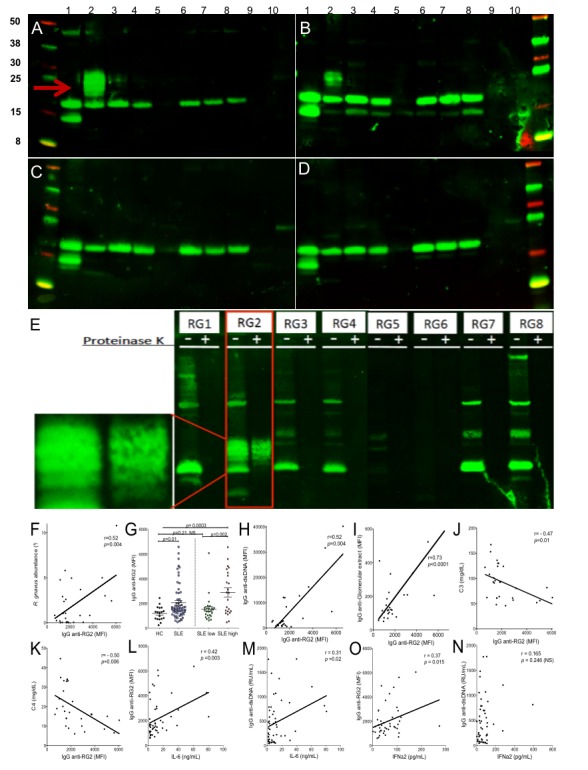
Patients with LN have serum IgG that recognise RG2 strain-restricted non-protein antigen(s) and that levels correlate with scores forlupus disease activity. Immunoblot results for IgG from (A) patient with LN (S-134), (B) patients with LN (S-047), (C) patient with non-active lupus (S-096) and (D) healthy control (CTL23). Bacterial extracts were prepared with nuclease and lysozyme treatment; lanes 1–8 represent R. gnavus RG1-RG8 strains (see online supplementary table 4), and lanes 9 and 10 contain extracts from gut commensal species, Bacteroides thetaiotaomicron and Prevotella copri. After electrophoretic transfer, replicate membranes were incubated with different sera at 1:100 dilution, with detection for human IgG deposition. Only patients with highly active lupus had serum IgG that recognised the oligomeric antigenic band in RG2 (red arrow) that was not detected in other RG strains. (E) Immunoblot of human gut isolated strains of R. gnavus, RG1-RG8. For each strain, molecular species were electrophoretically separated side-by-side, after nuclease and lysozyme treatment (left lane) and after additional thorough proteinase K treatment (right lane). Reactivity for serum IgG from patient S-134 with active LN is shown, with the serum tested at 1:100. Inset shows magnified view of non-protein oligomeric bands in RG2 extract, which are also seen in panels A and B. (F) In patients with SLE, RG faecal abundance by 16S rRNA analysis correlates with the levels of serum IgG anti-RG2 antibody. (G) Levels of serum IgG anti-RG2 in individual patients with lupus in the NYU cohort were compared with unaffected adults (see Methods section). Patients with SLE with high disease activity (SLEDAI ≥8) had higher levels of anti-RG2 IgG antibodies than healthy controls and than SLE with low disease activity. (H) Levels of serum IgG antinative DNA directly correlate with IgG anti-RG2 antibodies. Results from multiplex assay using extracts treated with lysozyme and a broad endonuclease, which was separately coupled to a set of paramagnetic beads (Luminex) in parallel with other antigens. (I) Levels of IgG anti-RG2 directly correlate with IgG to an extract of human glomeruli. Results from multiplex assay. (J) Levels of serum C3 inversely correlated with levels of serum lupus IgG anti-RG2 antibodies in patients with SLE. (K) Levels of serum C4 inversely correlated with levels of serum lupus IgG anti-RG2 antibodies in patients with SLE. (L) IgG anti-RG2 levels strongly correlate with serum IL-6 levels. (M) Levels of IgG anti-dsDNA, determined by commercial ELISA (INOVA), have only a modest correlation with serum IL-6 levels. (N) IgG anti-RG2 levels correlate with IFNα2 levels, determined by commercial bead-based assay (Luminex). (O) Levels of IgG anti-dsDNA antibodies do not correlate with serum IFNα2 levels. IgG anti-DNA and RG2 levels were measured by in-house custom bead-based assays. C3 and C4 were measured with commercial ELISA, while cytokines were measured by commercial bead-based assays. Significance was based on Mann-Whitney test or Spearman correlations. LN, lupus nephritis; SLE, systemic lupus erythematosus; SLEDAI, SLE disease activity index.
annrheumdis-2018-214856supp007.pdf (19.4KB, pdf)
annrheumdis-2018-214856supp006.pdf (1.2MB, pdf)
High antibody responses to the candidate RG pathobiont correlate with high disease activity
Given the common overabundance of RG in patients with active SLE, we investigated for special host-pathobiont relationships within the systemic immune systems of affected individuals. Serum IgA-anti-RG2 antibody responses were significantly higher in patients with SLE than controls, even after normalisation for total IgA levels (Welch test, p=0.01) (online supplementary figure 1D, E). Strikingly, serum IgG anti-RG2 responses were also increased in patients with SLE, and in matched lupus samples we found a significant direct correlation between the relative faecal abundance of the candidate RG pathobiont with the levels of serum IgG anti-RG2 antibodies (Spearman, p=0.002) (figure 2F) but not with the VPI C7-9 strain, here termed RG1 (online supplementary table 5 and data not shown), however such a relationship was also not found in studies of control subjects (not shown).
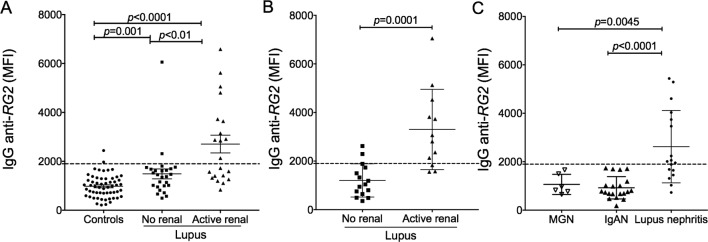
Based on a cut-off defined by values from unaffected control subjects (see Methods section), significant elevations of serum IgG anti-RG2 antibodies were detected in 22.9% of patients with SLE in the NYU cross-sectional cohort (figure 2G). Furthermore, serum levels of IgG anti-RG2 strain antibodies directly correlated with SLEDAI disease score (Spearman, p=0.04) but not with the serum IgG antibodies to the RG1 strain (online supplementary figure 1F). When patients were dichotomised based on disease activity the statistical differences between these patient subsets were most pronounced, as patients with SLEDAI scores≥8 had significantly higher IgG antibody levels to RG2 than patients who were quiescent or had only low disease activity (Mann-Whitney, p<0.001) (figure 2G), suggesting that the IgG anti-RG2 antibody level could represent a surrogate disease activity marker.
Serum levels of IgG anti-RG2 strain antibodies correlated with anti-native DNA autoantibodies (Spearman, p<0.0001) (figure 2H), which are commonly found in patients with LN.2 30 IgG antibodies to RG2 were also significantly positively correlated with IgG reactivity against human glomerular extract (Spearman, p<0.0001) (figure 2I), which is reported to identify the most nephritogenic anti-DNA antibodies.31 In contrast, there was neither an association with other types of lupus-associated autoantibodies, including anti-Ro52, nor with total IgG levels (online supplementary figure 1G, H). Relevant to clinical disease state, levels of IgG anti-RG2 antibodies also inversely correlated with C3 (Spearman, p<0.01) and C4 levels (Spearman, p<0.007) (figure 2J, K), biomarkers for in vivo activation of the complement cascade implicated in active immune-complex-mediated pathogenesis in LN.30 Indeed, serum IL-6 displayed a strong direct correlation with IgG anti-RG2 levels (Spearman, p=0.003) (figure 2L), while the relationship with anti-dsDNA was more modest (Spearman, p=0.02) (figure 2M). Furthermore, serum α2-interferon (IFN) also correlated with IgG anti-RG2 levels (Spearman, p=0.015), while there was no correlation with anti-dsDNA in the same samples (figure 2N, O). These findings provide circumstantial evidence of a connection between host immunity to RG2 antigen(s) and inflammatory cytokine drivers of lupus pathogenesis.
RG2 antigens cross-react with lupus anti-dsDNA antibodies
To evaluate for relationships between anti-self and anti-RG2 responses in patients with lupus, we assessed the capacity of native DNA and RG2 extract to inhibit lupus serum IgG binding (online supplementary figure 2A, B). As expected, in side-by-side assays, soluble RG2 extract was an efficient inhibitor of the binding of lupus serum IgG to immobilised treated RG2 extract (online supplementary figure 2C). Yet, native DNA only marginally inhibited IgG binding to RG2 extract (online supplementary figure 2D). In contrast, RG2 extracts displayed marked dose-dependent inhibition of binding of lupus serum IgG to native DNA, whereas RG1 extracts caused little or no inhibition of DNA binding (online supplementary figures 1I, 2B). Strikingly, RG2 displayed high-level (~90% maximal) inhibition of the binding to native mammalian DNA (online supplementary figure 2A) that was nearly as efficient in inhibitory activity as soluble native DNA itself (online supplementary figure 2B).
annrheumdis-2018-214856supp008.docx (5.1MB, docx)
To further confirm the relevance of RG2 epitopes with antinuclear antibody responses, we assessed the effect on IgG antibody reactivity with Crithidia luciliae, a protozoan with a kinetoplast organelle used to detect antibodies specific for chromatin.32 As expected, preincubation with native DNA inhibited this antibody interaction (online supplementary figure 2E-J), and RG2 strain was also inhibitory (online supplementary figure 2J), while a different RG strain (ie, RG1) was not (online supplementary table 5 and online supplementary figure 1I), suggesting that the relevant cross-reactive antigen(s) is strain-restricted (ie, associated with RG2 but not RG1). Collectively, we found that many patients with lupus have anti-dsDNA antibody responses that are cross-reactive with antigens in RG2, which is relevant to the notion that anti-DNA antibodies with great cross-reactivity more often have nephritogenic potential.10
RG2 contains lipoglycan(s) with antigenic properties recognised by patients with lupus
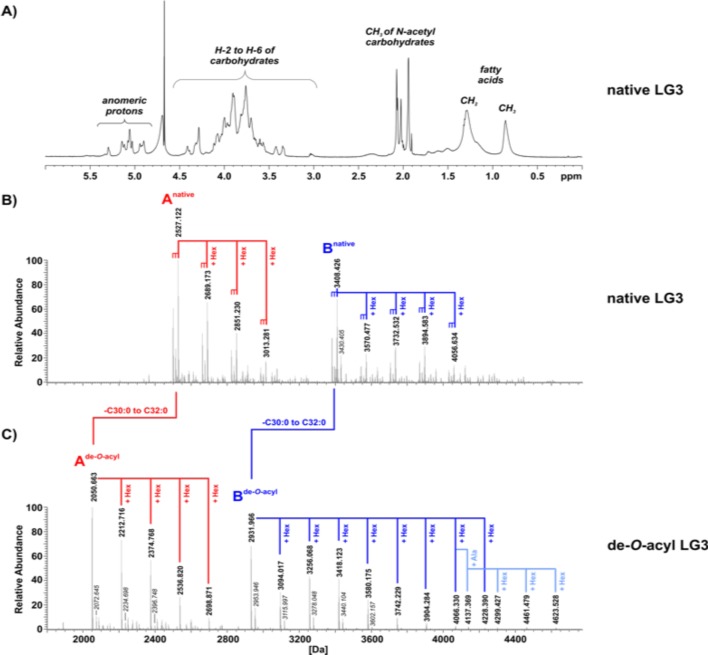
To isolate the lipoconjugates produced by RG2, we applied a validated Hydrophobic Interaction Chromatography (HIC)-based separation strategy to isolate specific cell wall moieties such as lipoteichoic acids (LTAs), lipoglycans (LGs or lipoproteins33 from the RG2 strain (online supplementary figure 1J). For further analyses, we combined fractions 23–27 (pool 2) and 28–31 (pool 3) (online supplementary figure 1J) and applied 1H NMR (figure 3A) as well as mass spectrometric (MS) analysis (figure 3B and online supplementary figures 1K, L), We thereby detected significant amounts of LGs, but the absence of 31P NMR signals (data not shown) indicated the absence of LTA molecules. We therefore termed these pools LG2 and LG3, respectively. Taken together, these analyses documented that LG2 and LG3 from the RG2 strain each represent a series of distinct LGs (online supplementary figure 1K, L).
Figure 3.
The LG3 pool contains two major series of lipoglycan molecules. (A) The 1H NMR spectrum of native LG3, recorded in D2O at 300 K, shows signals typical for lipoglycans, for example, for methyl groups of N-acetyl sugars, other carbohydrates and aliphatic chains of the lipid anchor. (B) The charge-deconvoluted MS spectrum of negative ion mode analysis of native LG3. (C) The charge-deconvoluted MS spectrum of negative ion mode analysis of de-O-acylated LG3. (Alternate version of this spectrum with additional assigned peaks is shown in online supplementary figure 1L.) (C) The charge-deconvoluted MS spectrum of negative ion mode analysis of de-O-acylated LG3. LG3 contains two major series of lipoglycans (termed series A [red] and B [dark blue]), starting from molecules with molecular weights of 2499.091/2513.107/2527.122 Da (series A) and 3380.395/3394.410/3408.426 Da (series B), respectively. These molecules are present in a series of further hexose-substituted (△m =+162.05 Da) molecular species. All peaks are present with variations in fatty acid composition (△m=14.02 Da is equivalent to CH2) (B). The complexity of the mass spectrum of de-O-acyl LG3 (C) is significantly decreased because of the removal of fatty acids. The overall signal pattern of the two series of lipoglycan species is well preserved. For series B, subspecies with a further alanine-substitution are apparent (light blue). Sodium adducts are represented as △m=21.98 Da, labelled in italic style. Abundances were normalised to the respective base peak of the depicted spectral region.
Further structural analyses were performed after hydrazine treatment that cleaves off all ester-bound residues, such as fatty acids.34 Compared with the MS spectrum of native LG3 (figure 3B and online supplementary Figure 1L), the de-O-acyl LG3 displayed significantly decreased complexity (figure 3C); in native LG3, the signals at 3380.395 Da, 3394.410 Da and 3408.426 Da (start of series Bnative [figure 3B]) shifted to the signal of 2931.966 Da in the spectrum of de-O-acyl LG3 (start of series Bde-O-acyl [figure 3C]), due to the removal of two fatty acids with combined chain length from C30:0 to C32:0. These changes reflected mass differences of 448.43 Da, 462.44 Da or 476.46 Da, respectively. For this series, molecules with varying number of hexoses were also detectable, as the signal pattern displays mass differences of 162.05 Da. A similar pattern was observed in the lower molecular range between 2000 and 2800 Da (figure 3C) for series Ade-O-acyl, for example, signals at 2050.663 Da, 2212.716 Da, 2374.768 Da and 2536.820 Da, representing the corresponding de-O-acylated molecules present in series Anative. Interestingly, LG2 mono-acyl variants of LGs of series A and B were also detected (online supplementary figure 1K), and LG2 had a third series (Cnative) of LG molecules that was also observed. Hence, although each of the three series most likely have differing core units, all LG variants display ‘hexose-capping’. The complete structural characterisation of these LGs from RG2 is beyond the scope of the current report. An analogous extraction of cell wall moieties from RG1 did not result in any yields of similar LGs, which suggests that RG strains differ in their capacity to produce cell wall LGs.
For investigations of the potential immune epitope features of the RG2 strain, we performed immunoblots with lupus IgG that detected high levels of oligomeric bands in the LG3 pool in the range of ~20–30 kDa protein markers, which resembled the reactivity pattern of protease-resistant antigens in the RG2 extract that were absent in other RG strains (figure 4A–D). In fact, preincubation of a lupus serum with LG3 at 2 µg/mL inhibited all immunoblot reactivity with the oligomeric antigenic bands in the LG2 and LG3 pools and with the nuclease-treated RG2 extract, but did not affect the IgG reactivity with the protease-sensitive bands (figure 4E, F). As predicted, lupus IgG immunoreactivity with these oligomeric bands was destroyed by periodate treatment (figure 4G), further supporting the notion that these immunodominant epitopes are on glyco-conjugate molecules. Side-by-side bead-based assays confirmed that the cell wall LG3 pool expresses the strain-restricted immunodominant epitopes in RG2 commonly recognised by lupus sera (figure 4H).
Figure 4.
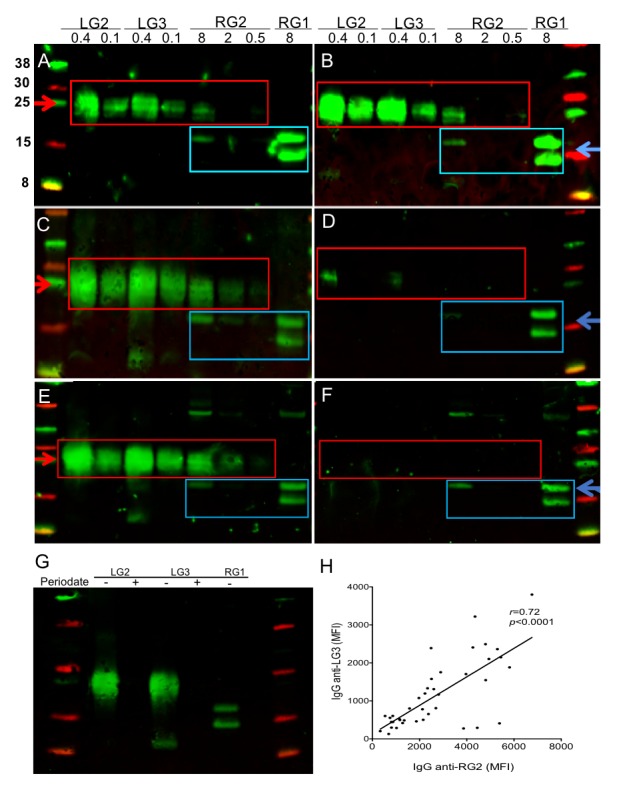
Lipoglycans from RG2 represent immunodominant antigens for patients with SLE. Immunoblots of purified LG2 and LG3 pools, and RG2 and RG1 extracts, were performed with sera from (A) SLE-07, (B) SLE-127, (C) SLE-080 that react with the same protease-resistant bands in LG2, LG3 and RG2 (red box), whereas (D) a healthy control reacts with distinct immunodominant antigens also recognised by patients with active SLE that are generally lower MW (blue box). (E) Immune recognition by serum antibodies in SLE-134 with oligomeric bands in LG2 and LG3, and in RG2, (F) are entirely inhibited by preincubation with LG2 at 2 µg/mL. (G) Periodate treatment before blotting destroys antigens in LG2 and LG3. Loading per lane is indicated above at µg/lane. Immunoblots were performed with serum at 1:200, with detection of IgG reactivity. (H) Serum levels of antibodies to LG3 pool and RG2 extract were highly correlated. Levels were determined by bead-based assay side-by-side in sera from 40 patients with SLE with diverse disease activity scores. SLE, systemic lupus erythematosus.
Furthermore, in vitro assays using a reporter cell line transfected with the human TLR2 demonstrated dose-dependent stimulation for the RG2 extract, and for the purified LG3, which was greater than for the LG2 sample, while the RG1 bacterial extract was devoid of detectable activity under these conditions (online supplementary figure 1M). To confirm the responsible mechanism, when studies were repeated in the presence of anti-TLR2 antibodies there was inhibition of NF-κB activation (online supplementary figure 1M). Hence, the LG pools, isolated from the RG2 strain, contain both strain-restricted carbohydrate-associated antigen(s) recognised by serum IgG in patients with SLE, and component(s) with immunostimulatory properties (online supplementary figure 1M), which might be lipoproteins that commonly copurify with LTA and/or LGs recovered using this HIC isolation protocol.33 34
Independent cohorts confirm the association of anti-RG2 responses with LN
In the NYU discovery cohort, patients with active nephritis at the time of blood sampling (based on activity criteria19 had significantly raised levels of IgG anti-RG2 strain-restricted antibodies (figure 5A). To confirm this association, we studied a second independent cohort of 27 patients (online supplementary table 1) (Temple University) in which 11/27 had elevated IgG anti-RG2 antibody reactivity, and of the 12 with active LN (based on level of proteinuria and/or concurrent biopsy data), 9 (75%) displayed high anti-RG2 antibody levels (figure 5B). In a third cohort of 17 patients with biopsy-documented active LN (online supplementary table 1) (The Ohio State University), 10 (59%) showed elevations of serum IgG anti-RG2 responses, but by contrast such responses were absent in patients with primary membranous glomerulonephritis and ANCA-associated vasculitis (Welch test, p=0.0025) (figure 5C). Furthermore, for a number of the patients in the NYU and Temple cohorts the LN diagnosis was confirmed by renal biopsy (online supplementary table 1). Inhibition studies of pooled sera from each of these three independent geographically separated cohorts confirmed that lupus-associated autoimmune IgG anti-native DNA responses were cross-reactive with epitope(s) in the RG2 strain (online supplementary figure 1N). Taken together, in three independent cohorts high IgG anti-RG2 antibody reactivity with cross-reactivity with anti-native DNA autoantibodies (online supplementary figure 1N) was commonly found in patients with LN from proliferative Class III and Class IV histopathology by ISN/RPS criteria35 (online supplementary table 1).
Figure 5.
Active lupus nephritis is associated with elevated IgG anti-RG2 antibodies. (A) Results from individuals in NYU cohort are shown, with comparisons to unaffected controls. (B) Results from individuals in the Temple University cohort with 16 patients with lupus without evidence of renal (ie, non-renal) are compared with 12 with active LN. (C) Results from 17 patients with biopsy-proven active LN from Ohio State University are compared with 6 patients with biopsy-proven idiopathic primary mesnagial glomerulonephritis (MGN) and 20 with IgA nephropathy (IgAN). The cut-off MFI value of 1900 for IgG anti-RG2 (dotted line) was determined by mean for 57 + 2 SD from unaffected 23 healthy controls with gut microbiome data, 15 other healthy individuals, 13 with psoriatic arthritis, and 12 with osteoarthritis). Renal disease was determined by clinical criteria and/or biopsy. Anti-RG2 values were determined by bead-based assay, with significance assigned by Mann-Whitney test.
SLE is a condition that overwhelmingly affects women, and due to concerns that gut microbiome can vary based on sex alone,36 our discovery cohort was restricted to female patients. Yet our confirmatory cohorts included several male patients, including individuals with active LN, who also commonly had elevated levels of serum IgG anti-RG2 antibodies (Figure 5, supplementary table 1 and data not shown). Furthermore, raised serum IgG antibody responses to RG2 were found in patients with active LN of diverse race/ethnicity: African-American, Asian, Caucasian, as well as Hispanic-White and Hispanic-Black (online supplementary table 1), which is consistent with the commonness of these immune responses in the three lupus cohorts examined.
Discussion
Our surveys of a cross-sectional discovery cohort documented that the severity of specific patterns of dysbiosis generally correlated with clinical disease activity. The gut microbiome of patients with higher SLEDAI scores had greater restrictions in taxonomic diversity. Indeed, our analyses provide the first evidence that lupus is associated with intestinal outgrowths of RG, an obligate anaerobic species, in the Blautia genus within the Lachnospiraceae family of the class Clostridia. 23 RG, which is a common (but generally low-level abundance) commensal in the faecal communities of healthy controls,37 was documented to be overrepresented, with >5-fold outgrowths in patients with lupus, with the greatest expansion in those with high disease activity and especially LN. Patients with lupus in these three geographically dispersed cohorts commonly displayed RG strain-restricted host immune responses that were cross-reactive with anti-DNA immune responses.
Patients with lupus also displayed higher levels of gut secretory IgA, a numerical trend towards higher abundance of faecal sIgA-coated RG bacteria and other abnormalities associated with gut barrier dysfunction. Importantly, there was a significant direct relationship with lupus disease activity and with LN, and the highest levels of serum IgG anti-RG antibody responses primarily directed against a RG strain-restricted cell wall-associated LG-containing antigen. Whereas these RG antigens may leak out of the intestine to induce IgG antibody responses, conversely IgG cannot enter the gut lumen and reduce the representation of taxa such as RG. Notably, RG2 LG-containing pools included moieties that were potent in vitro NF-κB immune activators via TLR2 (online supplementary figure 1M) that has previously been implicated in lupus pathogenesis.38 We speculate that the cross-reactivity of lupus autoreactive B cells to leaked stimulatory RG bacterial components, including TLR ligands(s), may contribute to the initiation and/or flares of lupus disease and especially LN that is a major source of early mortality and morbidity.39
In the human gut, the Lachnospiraceae family members fill a special niche, and degrade complex polysaccharides40 including those of the intestinal mucus layer, which may lead to a leaky gut.41 Yet RG strains vary greatly in their genomic composition, metabolic features and competitiveness. Indeed some isolates produce Lantibiotic polypeptides that suppress competing anaerobic species,42 and experimental colonisation can cause shifts in T cell balance toward IL-17 production.43 44
RG outgrowths have recently been reported in patients with ankylosing spondylitis, and abundance was reported to correlate with greater disease activity.45 In addition, patients with IBD have also been reported with RG blooms that comprised up to 69% of intestinal communities, and genomic analysis showed that these isolates were generally from a separate clade of RG than those in healthy individuals.46 In the IBD microbiome, strain-specific genes involved in oxidative stress responses have recently been postulated to confer an advantage for RG strains46 which could potentially be relevant to lupus pathogenesis as well.
The design of our observational studies has inherent limitations, as we do not currently know if RG expansions are truly causative or if lupus disease activity instead fosters preferential outgrowth of the candidate pathobiont. There is also a potential confounding effect of medications, as most patients were receiving treatment at the time of biosampling, and an enlarging list of medications is postulated to affect the balance of taxa within the gut microbiome community.47 48 However, we discount this potential effect as several of the recruited patients were in-flare while off medication and the same RG outgrowths and immune responses were nonetheless found (online supplementary table 1 and data not shown). The properties of the LGs in RG2, as well as other strains, will also require further investigation. In addition, Gram-positive opportunistic pathogens may have lipoproteins that are activators of innate immunity,49 and such lipoproteins can be copurified along with other lipoconjugates using the HIC-based methodology. Hence, future studies will also need to directly assess the specific immunostimulatory properties of the LGs themselves.
Intestinal communities are complex, and emerging data suggest many pathways may be intertwined with lupus pathogenesis. Expansions of the commensal pathobiont species, Segmented Filamentous Bacteria, in susceptible mouse strains can contribute to antinuclear autoantibody production.50 Furthermore, it has recently been reported that translocation and release of Enterococcus gallinarum from the small intestine may trigger the Aryl hydrocarbon receptor, resulting in autoantibody production and a type I interferon associated transcript signature in peripheral mononuclear cells.51 Indeed, we speculate that multiple pathobionts, singly or in combination, may contribute to lupus pathogenesis, and hence our finding of concurrent RG outgrowths merit further examination in longitudinal studies, including inception cohorts as well as more in-depth mechanistic studies.
Conclusion
Mounting evidence suggests that overt lupus disease is often preceded by years of a preclinical phase with prominent circulating autoantibody responses.52 Lupus disease has also been reported to be intertwined with greater inherent instability and lack of balance within the intestinal community53 which could underlie the restricted taxa diversity and disease-associated RG outgrowths that we found. Indeed, the abundance of RG outgrowths correlated with lupus disease activity, with the greatest expansions in those with active renal disease, and these immune responses were primarily directed against a strain-restricted cell wall lipoglycan. Notably, individual lupus communities also displayed concordant significant expansions of Veillonella species, along with reciprocal contractions of Bacteroides uniformis. Furthermore, our findings also contribute to evidence of a widening range of inflammatory and autoimmune conditions associated with a ‘leaky gut’,54 which may in part be responsible for the lupus-associated RG strain LG-directed antibody responses. We speculate that leakage of RG bacterial components contributes to nephritogenic autoantibody responses. To pursue these questions, it will be important to characterise the diversity and genetic features of RG strains and to understand when these strains colonise and expand in patients with lupus. In part, these investigations may lead to development of a biomarker assay that aids both earlier diagnosis and better prognostic determinations.
Acknowledgments
We acknowledge the assistance of the NYU Immune Monitoring Core and the NYU Flow Cytometry Core Facility, supported by NYU-HHC CTSI Grant UL1 TR000038, NYU Laura and Isaac Perlmutter Cancer Center support grant, P30CA016087 from the National Center for Advancing Translational Sciences (NCATS), the NYU Microscopy Laboratory and the NYU Genome Technology Core. In addition, we thank Simone Thomsen, Heiko Käßner and Brigitte Kunz (all at the RC Borstel) for their excellent technical assistance, Emma Allen-Vercoe for bacterial strains and technical advice, Karsten Zengler for assistance iwith bacterial cultures, and Dan Littman, Jeffrey Weiser and Martin Blaser for critical review of data and advice.
Footnotes
Handling editor: Josef S Smolen
Presented at: Parts of this manuscript were orally presented as a plenary presentation at the 2017 ACR/ARHP Annual Meeting and as such the abstract was published in an online supplementary of Arthritis & Rheumatology.
Contributors: DA, AVA and GJS designed research. DA, AO, DS and NG performed research. RC, BR and JPB recruited and characterised patients and critiqued the manuscript. DA, AVA and GJS analysed data and wrote the paper.
Funding: This work was supported in part by National Institutes of Health Grants; R01AI090118 (GJS), R01AI068063 (GJS), R01-AR42455 (GJS), P50 AR070591 (GJS, JPB), NIAID contract for B Cell Epitope Discovery and Mechanisms of Antibody Protection, HHSN272201400019C (GJS), N01-AR-4-2271, an American Recovery and Reinvestment Act supplement (GJS), NIH-NIAMS R01-AR061569-01A1 (RC), P50 AR070591-01A1/COMPEL (GJS and JPB), R01 LM012517 (AVA and GJS), the Lupus Research Institute and the Judith and Stewart Colton Autoimmunity Center (GJS). 16S rRNA gene sequence determinations and analysis were supported by the P Robert Majumder Charitable Trust (GJS).
Competing interests: NYU has filed intellectual property related to this report.
Patient consent for publication: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: Data in the text will be freely available. Microbiome community 16S rRNA data will be made available on reasonable request.
References
- 1. Tsokos GC. Systemic lupus erythematosus. N Engl J Med Overseas Ed 2011;365:2110–21. 10.1056/NEJMra1100359 [DOI] [PubMed] [Google Scholar]
- 2. Hochberg MC. Updating the American College of rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis & Rheumatism 1997;40 10.1002/art.1780400928 [DOI] [PubMed] [Google Scholar]
- 3. Borchers AT, Leibushor N, Naguwa SM, et al. Lupus nephritis: a critical review. Autoimmun Rev 2012;12:174–94. 10.1016/j.autrev.2012.08.018 [DOI] [PubMed] [Google Scholar]
- 4. Menzel AEO, Heidelberger MJ. Cell protein fractions of bovine and avian tubercle Bacillus strains and of the timothygrass Bacillus. J Biol Chem 1938;124:301–7. [Google Scholar]
- 5. Winkenwerder WL, Buell MV, Howard JE. The sensitizing properties of the nucleic acids and their derivatives. Science 1939;90:356 10.1126/science.90.2337.356 [DOI] [PubMed] [Google Scholar]
- 6. Sevag MGL, Smolens J. The isolation of the components of streptococcal nucleoproteins in serologically active form. J Biol Chem 1938;124:425–36. [Google Scholar]
- 7. Ceppellini R, Polli E, Celada F. A DNA-Reacting factor in serum of a patient with lupus erythematosus Diffusus. Exp Biol Med 1957;96:572–4. 10.3181/00379727-96-23544 [DOI] [PubMed] [Google Scholar]
- 8. Friou GJ. Identification of the nuclear component of the interaction of lupus erythematosus globulin and nuclei. J Immunol 1958;80:476–81. [PubMed] [Google Scholar]
- 9. Robbins WC, Holman HR, Deicher H, et al. Complement fixation with cell nuclei and DNA in lupus erythematosus. Exp Biol Med 1957;96:575–9. 10.3181/00379727-96-23545 [DOI] [PubMed] [Google Scholar]
- 10. Goilav B, Putterman C. The role of anti-DNA antibodies in the development of lupus nephritis: a complementary, or alternative, viewpoint? Semin Nephrol 2015;35:439–43. 10.1016/j.semnephrol.2015.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mannik M, Merrill CE, Stamps LD, et al. Multiple autoantibodies form the glomerular immune deposits in patients with systemic lupus erythematosus. J Rheumatol 2003;30:1495–504. [PubMed] [Google Scholar]
- 12. Pedersen HL, Horvei KD, Thiyagarajan D, et al. Murine and human lupus nephritis: pathogenic mechanisms and theoretical strategies for therapy. Semin Nephrol 2015;35:427–38. 10.1016/j.semnephrol.2015.08.004 [DOI] [PubMed] [Google Scholar]
- 13. Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature 2016;535:75–84. 10.1038/nature18848 [DOI] [PubMed] [Google Scholar]
- 14. Arnolds KL, Lozupone CA. Striking a balance with help from our little friends - how the gut microbiota contributes to immune homeostasis. Yale J Biol Med 2016;89:389–95. [PMC free article] [PubMed] [Google Scholar]
- 15. Rosser EC, Mauri C. A clinical update on the significance of the gut microbiota in systemic autoimmunity. J Autoimmun 2016;74:85–93. 10.1016/j.jaut.2016.06.009 [DOI] [PubMed] [Google Scholar]
- 16. Hevia A, Milani C, López P, et al. Intestinal dysbiosis associated with systemic lupus erythematosus. MBio 2014;5:e01548–14. 10.1128/mBio.01548-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. He Z, Shao T, Li H, et al. Alterations of the gut microbiome in Chinese patients with systemic lupus erythematosus. Gut Pathog 2016;8 10.1186/s13099-016-0146-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Luo XM, Edwards MR, Mu Q, et al. Gut microbiota in human systemic lupus erythematosus and a mouse model of lupus. Appl Environ Microbiol 2018;84:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Petri M, Kim MY, Kalunian KC, et al. Combined oral contraceptives in women with systemic lupus erythematosus. N Engl J Med 2005;353:2550–8. 10.1056/NEJMoa051135 [DOI] [PubMed] [Google Scholar]
- 20. Cole JR, Wang Q, Fish JA, et al. Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 2014;42:D633–D642. 10.1093/nar/gkt1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang Q, Garrity GM, Tiedje JM, et al. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007;73:5261–7. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schloissnig S, Arumugam M, Sunagawa S, et al. Genomic variation landscape of the human gut microbiome. Nature 2013;493:45–50. 10.1038/nature11711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu C, Finegold SM, Song Y, et al. Reclassification of Clostridium coccoides, Ruminococcus hansenii, Ruminococcus hydrogenotrophicus, Ruminococcus luti, Ruminococcus productus and Ruminococcus schinkii as Blautia coccoides gen. nov., comb. nov., Blautia hansenii comb. nov., Blautia hydrogenotrophica comb. nov., Blautia luti comb. nov., Blautia producta comb. nov., Blautia schinkii comb. nov. and description of Blautia wexlerae sp. nov., isolated from human faeces. Int J Syst Evol Microbiol 2008;58:1896–902. 10.1099/ijs.0.65208-0 [DOI] [PubMed] [Google Scholar]
- 24. Callahan BJ, McMurdie PJ, Rosen MJ, et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 2016;13:581–3. 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gonzalez ME, Schaffer JV, Orlow SJ, et al. Cutaneous microbiome effects of fluticasone propionate cream and adjunctive bleach baths in childhood atopic dermatitis. J Am Acad Dermatol 2016;75:481–93. 10.1016/j.jaad.2016.04.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gauffin Cano P, Santacruz A, Moya Á, et al. Bacteroides uniformis CECT 7771 ameliorates metabolic and immunological dysfunction in mice with high-fat-diet induced obesity. PLoS One 2012;7:e41079 10.1371/journal.pone.0041079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gisbert JP, Bermejo F, Pérez-Calle JL, et al. Fecal calprotectin and lactoferrin for the prediction of inflammatory bowel disease relapse. Inflamm Bowel Dis 2009;15:1190–8. 10.1002/ibd.20933 [DOI] [PubMed] [Google Scholar]
- 28. Rainone V, Schneider L, Saulle I, et al. Upregulation of inflammasome activity and increased gut permeability are associated with obesity in children and adolescents. Int J Obes 2016;40:1026–33. 10.1038/ijo.2016.26 [DOI] [PubMed] [Google Scholar]
- 29. Fotis L, Shaikh N, Baszis KW, et al. Serologic evidence of Gut-driven systemic inflammation in juvenile idiopathic arthritis. J Rheumatol 2017;44:1624–31. 10.3899/jrheum.161589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Davis P, Cumming RH, Verrier-Jones J. Relationship between anti-DNA antibodies complement consumption and circulating immune complexes in systemic lupus erythematosus. Clin Exp Immunol 1977;28:226–32. [PMC free article] [PubMed] [Google Scholar]
- 31. Lefkowith JB, Kiehl M, Rubenstein J, et al. Heterogeneity and clinical significance of glomerular-binding antibodies in systemic lupus erythematosus. J Clin Invest 1996;98:1373–80. 10.1172/JCI118924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tozzoli R, Bizzaro N, Tonutti E, et al. Guidelines for the laboratory use of autoantibody tests in the diagnosis and monitoring of autoimmune rheumatic diseases. Am J Clin Pathol 2002;117:316–24. 10.1309/Y5VF-C3DM-L8XV-U053 [DOI] [PubMed] [Google Scholar]
- 33. Hess N, Waldow F, Kohler TP, et al. Lipoteichoic acid deficiency permits normal growth but impairs virulence of Streptococcus pneumoniae. Nat Commun 2017;8;(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gisch N, Auger JP, Thomsen S, et al. Structural analysis and immunostimulatory potency of lipoteichoic acids isolated from three Streptococcus suis serotype 2 strains. J Biol Chem 2018;293:12011–25. 10.1074/jbc.RA118.002174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Weening JJ, D'Agati VD, Schwartz MM, et al. The classification of glomerulonephritis in systemic lupus erythematosus revisited. J Am Soc Nephrol 2004;15:241–50. 10.1097/01.ASN.0000108969.21691.5D [DOI] [PubMed] [Google Scholar]
- 36. Yurkovetskiy L, Burrows M, Khan AA, et al. Gender bias in autoimmunity is influenced by microbiota. Immunity 2013;39:400–12. 10.1016/j.immuni.2013.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nelson KE, Weinstock GM, Human Microbiome Jumpstart Reference Strains Consortium, et al. A catalog of reference genomes from the human microbiome. Science 2010;328:994–9. 10.1126/science.1183605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tsao JT, Hsieh SC, Chiang BL, et al. Altered IL-10 and TNF-α production in peripheral blood mononuclear cells of systemic lupus erythematosus patients after Toll-like receptor 2, 4, or 9 activation. Clin Exp Med 2012;12:153–8. 10.1007/s10238-011-0161-6 [DOI] [PubMed] [Google Scholar]
- 39. Almaani S, Meara A, Rovin BH. Update on Lupus nephritis. Clin J Am Soc Nephrol 2017;12:825–35. 10.2215/CJN.05780616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Flint HJ, Bayer EA, Rincon MT, et al. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Micro 2008;6:121–31. 10.1038/nrmicro1817 [DOI] [PubMed] [Google Scholar]
- 41. Crost EH, Tailford LE, Le Gall G, et al. Utilisation of mucin glycans by the human gut symbiont Ruminococcus gnavus is strain-dependent. PLoS One 2013;8:e76341 10.1371/journal.pone.0076341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dabard J, Bridonneau C, Phillipe C, et al. Ruminococcin A, a new lantibiotic produced by a Ruminococcus gnavus strain isolated from human feces. Appl Environ Microbiol 2001;67:4111–8. 10.1128/AEM.67.9.4111-4118.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Eun CS, Mishima Y, Wohlgemuth S, et al. Induction of bacterial antigen-specific colitis by a simplified human microbiota Consortium in gnotobiotic interleukin-10-/- mice. Infect Immun 2014;82:2239–46. 10.1128/IAI.01513-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hoffmann TW, Pham HP, Bridonneau C, et al. Microorganisms linked to inflammatory bowel disease-associated dysbiosis differentially impact host physiology in gnotobiotic mice. Isme J 2016;10:460–77. 10.1038/ismej.2015.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Breban M, Tap J, Leboime A, et al. Faecal microbiota study reveals specific dysbiosis in spondyloarthritis. Ann Rheum Dis 2017;76:1614–22. 10.1136/annrheumdis-2016-211064 [DOI] [PubMed] [Google Scholar]
- 46. Hall AB, Yassour M, Sauk J, et al. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med 2017;9 10.1186/s13073-017-0490-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gul'neva M, Shilkina NP. Intestinal microbial biocenosis in patients with systemic lupus erythematosus treated with prednisolone]. Klin Med 2009;87:42–5. [PubMed] [Google Scholar]
- 48. Maier L, Pruteanu M, Kuhn M, et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 2018;555:623–8. 10.1038/nature25979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kovacs-Simon A, Titball RW, Michell SL. Lipoproteins of bacterial pathogens. Infect Immun 2011;79:548–61. 10.1128/IAI.00682-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Van Praet JT, Donovan E, Vanassche I, et al. Commensal microbiota influence systemic autoimmune responses. Embo J 2015;34:466–74. 10.15252/embj.201489966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Manfredo Vieira S, Hiltensperger M, Kumar V, et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science 2018;359:1156–61. 10.1126/science.aar7201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Arbuckle MR, McClain MT, Rubertone MV, et al. Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N Engl J Med 2003;349:1526–33. 10.1056/NEJMoa021933 [DOI] [PubMed] [Google Scholar]
- 53. Apperloo-Renkema HZ, Bootsma H, Mulder BI, et al. Host-microflora interaction in systemic lupus erythematosus (SLE): colonization resistance of the indigenous bacteria of the intestinal tract. Epidemiol Infect 1994;112:367–73. 10.1017/S0950268800057770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fasano A. Leaky gut and autoimmune diseases. Clin Rev Allergy Immunol 2012;42:71–8. 10.1007/s12016-011-8291-x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
annrheumdis-2018-214856supp001.docx (53.4KB, docx)
annrheumdis-2018-214856supp002.xlsx (19.7KB, xlsx)
annrheumdis-2018-214856supp003.pdf (1.7MB, pdf)
annrheumdis-2018-214856supp004.docx (24.3KB, docx)
annrheumdis-2018-214856supp005.xlsx (13KB, xlsx)
annrheumdis-2018-214856supp007.pdf (19.4KB, pdf)
annrheumdis-2018-214856supp006.pdf (1.2MB, pdf)
annrheumdis-2018-214856supp008.docx (5.1MB, docx)